Abstract
Background
The duodenal ulcer promoting gene (dupA) and dupA cluster in Helicobacter pylori have been described as a risk factor for duodenal ulcer development in some populations. Polymorphic gene dupA can be divided into two groups, intact dupA1 (long or short type based on the presence or absence of 615-bp extra sequences at the 5′ region) having complete reading frame and other truncated dupA2 having frame-shift mutation. This study was aimed to elucidate the role of dupA of H. pylori and their clusters in the disease manifestation of Indian population.
Methods
A total of 170 H. pylori strains were screened for the presence of dupA, dupA alleles and dupA cluster by PCR and sequencing. Pro-inflammatory cytokine (IL-8) with different dupA variant H. pylori stimulated gastric epithelial cells (AGS cells) was measured by ELISA.
Results
A total of 50 strains (29.4%) were positive for dupA among the tested 170 strains. The prevalence of dupA1 in duodenal ulcer (DU) and non-ulcer dyspepsia (NUD) populations was found to be 25.5% (25/98) and 11.1% (8/72), respectively and 16.4% (28/170) of the tested strains had dupA1, cagA and vacAs1m1 positive. The distribution of long and short type dupA1 has not been significantly associated with the disease outcome. The dupA cluster analysis showed that 10.2% (10/98) and 8.3% (6/72) strains were positive among DU and NUD, respectively. IL-8 production was significantly higher in dupA1+, cagA+, vacA+ (902.5 ± 79.01 pg/mL) than dupA2+, cagA+, vacA+ (536.0 ± 100.4 pg/mL, P = 0.008) and dupA−, cagA+, vacA+ (549.7 ± 104.1 pg/mL, P = 0.009). Phylogenetic analysis of dupA indicated that the Indian H. pylori strains clustered with East Asian strains but distinct from Western strains. This is the first known genetic element of Indian H. pylori that is genetically closer to the East Asian strains but differed from the Western strains.
Conclusions
The intact dupA1 was significantly associated with DU than NUD (P = 0.029) but the dupA1 cluster has no role in the disease manifestation at India (P = 0.79). Thus, dupA1 can be considered as a biomarker for DU patients in India.
Keywords: Helicobacter pylori, Duodenal ulcer, dupA, dupA cluster, Non-ulcer dyspepsia, Disease association
Introduction
Infection caused by H. pylori is a growing concern as this pathogen is involved in chronic gastritis, peptic ulcer and multi-step carcinogenic processes of gastric cancer. Gastric cancer is the fourth most common cancer worldwide and the second cause of cancer related deaths [1-3]. Epidemiologically, more than 50% of the world population has been infected by this bacterium with persistent inflammation in their stomachs, which lasts for decades unless treated with antibiotics. However, only 15-20% of infected patients develop gastric or duodenal ulcer (DU) and less than 1% develop gastric adenocarcinoma [4]. Several studies demonstrate that about 50-80% Indian populations have been infected with the H. pylori [5,6]. It was shown that mere presence of H. pylori in the stomach is not associated with any gastric disease. Besides bacterial genetics, host genetic factors, hygiene, microbiome, medication, food habits along with life-style of the individuals are said to enhance the infections caused by H. pylori. In this connection, H. pylori bear an arsenal of specific virulence factors. Among them, the cytotoxin-associated gene pathogenicity island (cag-PAI) and vacuolating associated cytotoxin gene A (vacA) are associated with virulence in Western countries. However, the association of cag-PAI and vacA of H. pylori are not established in the Indian population [7]. A novel virulence factor duodenal ulcer promoting gene (dupA), located in the plasticity region of the H. pylori genome, homologues to virB4 gene, which encodes a component protein of the type IV secretion system (T4SS) has been associated with increased risk of DU and protection against gastric cancer (GC) in East Asian and Western countries [8]. However, the role of dupA as a virulence marker is still debated [9-17]. It has been reported that dupA gene is highly polymorphic as frameshift mutation found along the length of the gene that leads to truncated protein and the rate of frameshift mutation varies geographically around the world [12,18-20]. Thus, mere detection of dupA gene by PCR is not adequate to characterize this variable gene. Accordingly, the dupA gene has been classified into two groups; i.e., (i) intact dupA without frameshift mutation called dupA1 and (ii) dupA with frameshift mutation that leads to stop codon called dupA2.
The full genome sequence analysis of strains SNT49, Shi470 and G27 along with the study conducted on the Okinawa population in Japan revealed that the intact dupA has two genotypes: short type of 1.8 kb and long type of 2.5 kb due to an additional length of 615 bp in the 5′ region [21]. The latter has been primarily associated with gastric ulcer and gastric cancer in the Okinawa population [21]. A recent study reported that dupA and adjacent 6 vir genes homologues (virB8, virB9, virB10, virB11, virD2 and virD4) in the plasticity region predicting to form a third T4SS (tfs3a) [22] termed as “dupA cluster” (dupA along with six surrounding gene) has been associated with DU in the United States population [23]. However, the functional role of this dupA cluster in endorsing the DU formation in the Iraqi population is not very clear [24]. It is well known that the Indian H. pylori strains are genetically distinct than East Asian and Western strains [7].
H. pylori infection in vivo induces the mucosal production of various cytokines e.g. interleukin-8 (IL-8), IL-1b, IL-6 and tumor necrosis factor alpha (TNF-α). IL-8, a potent neutrophil-activating chemokine produced by various cell types, including macrophages, epithelial cells, endothelial cells and T cells. Elevated levels of IL-8 have been reported in a number of inflammatory conditions, including inflammatory bowel disease, cystic fibrosis, psoriasis, rheumatoid arthritis, septic shock, and acute meningococcal infections [25]. IL-8 a chemokine, central to the pathogenesis of H. pylori-induced tissue injury [26] and previous reports from other research groups showed that dupA-positive strains are associated with high-level of IL-8 production [8,19]. Moreover, our recent study showed that intact cag-PAI containing H. pylori strains were found more frequently in Kolkata than in southern India indicating regional variations in H. pylori gene pools [27]. Considering the genetic diversity of H. pylori and associated infection, we have undertaken this study to elucidate the role of dupA alleles and their cluster in disease manifestation in Indian population.
Methods
H. pylori strains
A total of 170 H. pylori strains archived in the National Institute of Cholera and Enteric Diseases (NICED) Kolkata were used in this study. These strains were isolated from 98 DU patients and 72 NUD subjects of both sexes (aged between 20 and 65 years) with upper gastrointestinal disorder who underwent endoscopy or gastric surgery at the Hospital of the Institute of Post Graduate Medical Education and Research, Kolkata, and St. John’s Medical College Hospital, Bangalore, India during the year 2004–2010. The NICED Ethical committee had approved the study. The patient information or record was kept blind during the experimental procedures and the disease status was decoded during the data analysis. The H. pylori strains were stored in brain heart infusion (BHI) broth (Difco Laboratories) with 15% glycerol at −70°C until further use. These strains were revived using BHI agar (Difco) supplemented with 7% sheep blood, 0.4% IsoVitaleX, amphotricin B (8 μg/ml) (Sigma Chemicals Co., St. Louis, MO), trimethoprim (5 μg/ml), vancomycin (6 μg/ml) and nalidixic acid (8 μg/ml) (all from Sigma). Plates were incubated at 37°C in a double gas incubator (Heraeus Instrument, Germany), which maintains an atmosphere of 5% O2, 10% CO2 and 85% N2. The H. pylori culture was reconfirmed by positive reactions in urease, catalase and oxidase tests along with the urease PCR.
PCR Amplification
H. pylori genomic DNA isolation and PCR of dupA, cagA and vacA were done using primers and protocols described previously [17]. The six vir genes virB8, virB9, virB10, virB11, virD4 and virD2 surrounding the dupA and forming the dupA cluster were amplified with primers described elsewhere [23]. Gene specific primers were used for the amplification of dupA genotypes under different PCR condition listed in Table 1. Long type and short type dupA1 was amplified with primer pair (SNT49F/dupA2499R) and (dupAF/dupA2499R), respectively. Primer SNT49F was designed from the start codon of dupA of full genome sequenced Indian strain SNT49, located 615 bp upstream to the earlier proposed dupA of C142 strain (AB 196363). Primer dupA2499R was located on the stop codon (end) of dupA of SNT49 strain and 5′ end of the jhp0919 region of J99 strain. Primer dupAF was designed from the start codon of jhp0917 of strain J99. The PCR conditions were standardized to a final volume of 20 μl containing template DNA (2–20 ng), 2 μl of 10x Buffer (Genei, India), 2.5 mM dNTPs (Genei, India) and 10 pmol of corresponding primers in the presence of 1U of Taq DNA Polymerase (Genei, India) [Table 1].
Table 1.
List of primer used for Amplification and sequencing of dupA gene
| Primer | Sequence (5′-3′) | PCR cycling condition | Amplicon size (bp) |
|---|---|---|---|
| SNT49F | ATGTTTCTTGGTTTAGAGGG | 94°C 30 sec, 55°C 30 sec and 72°C 2.5 mins | 2499 |
| dupA2499R | TCACACATATTGAACATTCTCG | ||
| dupAF | ATGAGTTCTGTATTAACAGACTTTG | 94°C 30 sec, 50°C 30 sec and 72°C 2 mins | 1884 |
| dupA2499R | TTAAATACTCTTCCTTATAAGTTTC | ||
| SNT49F | ATGTTTCTTGGTTTAGAGGG | 94°C 30 sec, 55°C 30 sec and 72°C 1 mins | 685 |
| SNT49R685 | CAGCGTATAAATTCAATAGATC | ||
| dupAF | ATGAGTTCTGTATTAACAGACTTTG | 94°C 30 sec, 50°C 30 sec and 72°C 1.5 mins | 1172 |
| dupA20R | CCTAAATTTTTGGCAATTTCTAATAAG | ||
| dupA16F | ACAATACTGCTAATACAGATG | 94°C 30 sec, 55°C 30 sec and 72°C 1 min | 947 |
| dupA2499R | TCACACATATTGAACATTCTCG | ||
| SNT49_470F | ATGATTTTAAATTATGTAGAGACC | 94°C 30 sec, 55°C 30 sec and 72°C 40 secs | 623 |
| SNT49R350 | GCATTAACAATTTTTTTAGCG | ||
| 918 F | CCTATATCGCTAACGCGCTC | 94°C 30 sec, 55°C 30 sec and 72°C 40 secs | 791 |
| jhp0919R | CTTTTTGTGATTTCATGAAACTC |
DNA sequencing and phylogenetic analysis
Different fragments of dupA were amplified with different set of primers listed in Table 1. Primer walking method was used for the sequencing of full length of dupA gene. We used primer pair SNT49_470F and SNT49R350 for confirmation of start codon of long type dupA gene and 918 F and jhp0919R for the confirmation of stop codon of dupA gene. Primer jhp0919R was located from 326 bp to 350 of ORF jhp0919 of strain J99. The amplified products were purified using the QIAquick PCR Purification Kit (QIAGEN). The purified PCR product was quantified on gel. The intensity of the band compared with λ hind III digest. The PCR purified products were sequenced with a Big Dye Terminator v3.1 cycle sequencing kit on an ABI PRISM 3100 genetic Analyzer (Applied Biosystem, USA). The sequences obtained in this study were deposited in GenBank under accession numbers KC894688-KC894692. We performed BLAST to get the identical sequence available on the NCBI. All the sequences obtained were multiple aligned with the known dupA sequence from different geographic areas using Clustal W of MEGA6 software (version 6.0.5, AZ, USA). This Maximum Likelihood tree was generated using Tamura 3 parameter model.
Cell culture and H. pylori infection
To measure in vitro IL-8 secretion from gastric epithelial cells, AGS (human gastric adenocarcinoma cell line) were cultured in RPMI 1640 (HiMedia, Mumbai, India) medium supplemented with 10% fetal bovine serum (Invitrogen, UK) for 3 days at 37°C, under 5% CO2. The cells were trypsinized (Gibco BRL), microscopically enumerated, and distributed in a 24-well microtitre plate at a final concentration of 5 X 105 cells/ml (1 ml/well) and incubated for 24 h at 37°C prior to infection. H. pylori (multiplicity of infection [MOI] of 100) was added to cultured cells for 8 hours and IL-8 levels in the supernatant was measured in duplicates using a commercially available ELISA kit (Amersham Biosciences Biotrak™ System) following the manufacturer’s instructions.
Statistical analysis
A univariate analysis was performed to determine the risk of dupA alleles and dupA cluster in relation to clinical outcome. For univariate analysis, χ2 test was used. A Probability levels (P) value of ≤ 0.05 was considered statistically significant.
Results
Distribution of dupA alleles
A total of 170 H. pylori strains were isolated from the following two groups: (i) 98 DU patients and (ii) 72 NUD. PCR and sequencing were performed to screen the presence of dupA gene with published primers [17] and found that 29.4% (50/170) strains were positive for dupA. Frameshift mutations were screened in these dupA positive strains by sequencing with primers described in Table 1. It was found that 19.4% (33/170) strains had intact gene without frameshift mutation and hence were considered as dupA1. In 10% (17/170) of the strains, frameshift mutation has been detected leading to a premature stop codon and hence the dupA2 in these strains were truncated. The dupA1 alleles namely, long type and short type dupA1 with 2499 bp and 1884 bp amplicons [Figure 1] were detected in 12.3% (21/170) and 7% (12/170) strains, respectively [Table 2]. This data showed that the distribution of long type dupA1 was more than the short type dupA1 in the Indian population.
Figure 1.
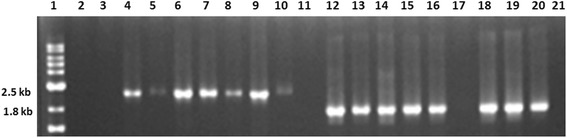
PCR results of dupA 1 ( long type and short type) amplified with snt49F/2499R and dupAF/2499R sets of primers in representative H. pylori strains. Lane 1 denoted 1 kb marker (NEB). Lane 2–11 showed the long type dupA1 of 2499 bp and lane 12–21 yielded the short type dupA1 of 1884 bp, lane 2, 3 and 17 did not produce any amplicon. Lanes 11 and 21 were taken as negative controls.
Table 2.
Prevalence of dupA (dupA 1and dupA 2 ), dupA1 alleles and dupA1 cluster in Indian population
| Total | DU | NUD | |
|---|---|---|---|
| Number | 170 | 98 | 72 |
| dupA | 50/170 (29.4%) | 38/98 (38.7%) | 12/72 (16.6%) |
| dupA1 | 33/170 (19.4%) | 25/98 (25.5%) | 8/72 (11.1%) |
| long type dupA1 | 21/170 (12.3%) | 15/98 (15.3%) | 6/72 (8.3%) |
| short type dupA1 | 12/170 (7%) | 10/98 (10.2%) | 2/72(2.7%) |
| dupA1 with cluster | 16/170 (9.4%) | 10/98 (10.2%) | 6/72 (8.3%) |
| dupA2 | 17/170 (10%) | 13/98 (13.2%) | 4/72 (5.5%) |
| cagA | 142/170 (83.5%) | 86/98 (87.7) | 56/72 (77.7%) |
| vacAs1m1 | 118/170 (69.4) | 71/98 (72.4%) | 47/72 (65.2%) |
| dupA1, cagA, vacAs1m1 | 28/170 (16.4%) | 22/98 (22.4%) | 6/72 (8.3%) |
Prevalence of vir genes homologues and cagA, vacA
All the 6 vir genes in 170 strains were tested by primer described by Jung et al. [23] and their distribution showed 44.1% of virB8 (75/170), 30% of virB9 (51/170), 24.1% of virB10 (41/170), 27% of virB11 (46/170), 64.1% of virD2 (109/170) and 42.9% of virD4 (73/170) [Table 3]. Twenty strains (11.7%) had dupA and all 6 vir genes homologues indicating positive for dupA cluster and 35 strains had no vir gene homologues. A total of 115 strains were positive for various combinations of vir gene homologues while lacking some vir genes. Further analysis showed that dupA1 cluster (intact dupA gene with 6 vir homologues) and dupA2 cluster (truncated dupA with 6 vir homologues) was found in 9.4% (16/170) and 2% (4/170) strains, respectively. The cagA gene was present in 83.5% (142/170) of the tested strains and 91% (30/33) of the dupA1 positive strains from this region. The vacA s1m1 was present in 69.4% (118/170) of the total tested strains and 85% (28/33) of the dupA1 positive strains [Table 2]. The other two alleles of vacA, s1m2 and s2m2, were present in 17.6% (30/170) and 12.9% (22/140) of the strains, respectively (data not shown).
Table 3.
Distribution of all six vir gene in Indian population
| Gene | Total (n = 170) | DU (n-98) | NUD (n = 72) |
|---|---|---|---|
| virB8 | 75/170 (44.1%) | 50/98 (51%) | 25/72 (34.7%) |
| virB9 | 51/170 (30%) | 32/98 (32.6%) | 19/72 (26.3%) |
| virB10 | 41/170 (24.1%) | 27/98 (27.5%) | 14/72 (19.4%) |
| virB11 | 46/170 (27%) | 28/98 (28.5%) | 18/72 (25%) |
| virD2 | 109/170 (64.1%) | 65/98 (66.3%) | 44/72 (61.1%) |
| virD4 | 73/170 (42.9%) | 42/98 (42.8%) | 31/72 (43%) |
| All six vir genes with dupA (dupA1 and dupA2) | 20/170 (11.7%) | 12/98 (12.2%) | 8/72 (11.1%) |
Association of dupA genotypes and dupA cluster with disease outcome
In this study, 38.7% (38/98) DU and 16.6% (12/72) NUD strains were positive for dupA gene by PCR and sequencing of intergenic region of jhp0917- jhp0918 ORF. Sequence analysis indicated that 66% (33/50) of the dupA positive strains had intact dupA gene without any frameshift mutation (dupA1) and 34% (17/50) strains had insertion or deletion of adenine at different positions leading to a premature stop codon (dupA2). Interestingly, the intact dupA1 was found significantly higher in DU patients (25/98, 25.5%) than in NUD patients (8/72, 11.1%) (P =0.02, odds ratio = 2.73, 95% confidence interval = 1.15-6.50). Additional analysis on the type of dupA1 alleles of H. pylori present in the Indian population showed the presence of both long and short types dupA1 were more in DU patients (15/98, 10/98) than NUD subjects (6/72, 2/72). However, the difference did not reach up to the significant level (P = 0.23, odds ratio = 1.98, 95% confidence interval = 0.731-5.40, P = 0.073, odds ratio = 3.9, 95% confidence interval = 0.84-18.74). The 6 vir genes cluster was detected in 10.2% (10/98) and 8.3% (6/72) of the strains from DU and NUD subjects, respectively with intact dupA. This result indicated that there was no significant association of intact dupA1 cluster with DU (P =0.79, odds ratio = 1.25, 95% confidence interval = 0.43-3.61) [Table 2].
Intact dupA and IL-8 production in gastric cancer cells
In general, there was no difference in IL-8 production between dupA-positive but truncated (dupA2) and dupA-negative strains when tested in AGS cells co-cultured with H. pylori. However, IL-8 production was significantly higher (902.5 ± 79.01 pg/mL) in the strains with an intact dupA1 compared with a truncated dupA2 (536.0 ± 100.4 pg/mL, P = 0.008) or with dupA-negative strains (549.7 ± 104.1 pg/mL, P = 0.009) [Figure 2]. All the strains taken for IL-8 assay were positive for cagA and vacA which helped us to focus specifically the role of different dupA groups.
Figure 2.
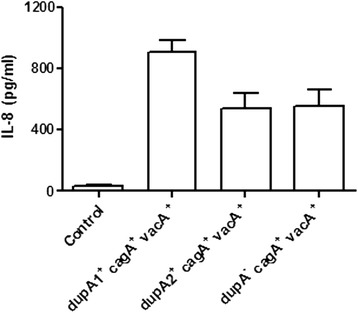
In vitro IL-8 production from AGS cells co-cultured with 6 ( dupA 1 + , cagA + , vacA + ) strains, 6 ( dupA 2 + , cagA + , vacA + ) and 6 ( dupA − , cagA + , vacA + ) strains. IL-8 production was significantly higher in dupA1+ (cagA +, vacA +) compared to dupA2+ (cagA +, vacA +) (536.0 ± 100.4 pg/mL, P = 0.008) and dupA-negative strains (cagA +, vacA + ) (549.7 ± 104.1 pg/mL, P = 0.009). MOI of H. pylori strain was 100 for 8 hours. Experiments were repeated 3 times for the 18 strains. IL-8 in the supernatant was assayed by an ELISA in duplicate. Data are expressed as mean ± standard error.
Phylogenetic analysis of dupA gene
Sequence heterogeneity of dupA gene was analysed with 22 randomly selected strains from India and other countries (assembled using GenBank data). Phylogenetic analysis of dupA sequence revealed the existence of 2 distinct clusters [Figure 3]. The first cluster designated as “Group I” included 5 strains from Japan, 5 from China and 6 from different parts of India. The second group included 6 strains (5 from Brazil and one each from Colombia) and was designated as “Group II”, which was called as the European cluster. Most strikingly, phylogenetic analysis of dupA sequence showed that the Indian strains clustered more closely with the East Asian strains.
Figure 3.
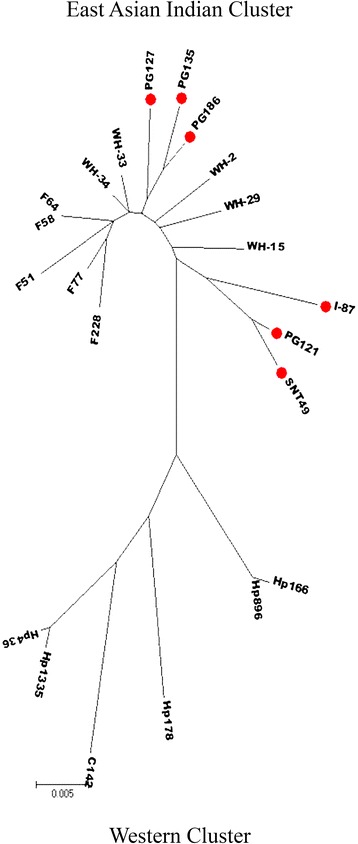
Phylogenetic tree constructed based on 1.8 kb segment of dupA gene of H. pylori (determined in this study and reported by others). This Maximum Likelihood tree was generated using Tamura 3 parameter model in MEGA6 software (version 6.0.5, AZ, USA). Representative strains from India are marked in Red dots (●, red circle). Sequences of non Indian strains were used here from public database. Indian and East Asian strains formed one cluster called group I and Brazilian and Colombian strains formed another cluster called group II. The length of the vertical bar indicates the number of nucleotide substitution per site. Bootstrap values of ≥ 70 are indicated at the nodes. H. pylori strain designations indicate the geographic origins, as follows: F, Japan; WH, China; PG or I or SNT, India; HP, Brazil; C, Colombia. GenBank: accession no. for the strains used in the study are given in the parentheses: F228 [GenBank: AB617836.1], F77 [GenBank: AB617834.1], F58 [GenBank: AB617835.1], F64 [GenBank: AB617833.1], WH-34 [GenBank: KC707844.1], WH-33 [GenBank: KC707843.1], F51 [GenBank: AB617832.1], PG127 [GenBank: Submission in process], PG135-1a [GenBank: JN379048.1], PG186 [GenBank: KC894690.1], WH-2 [GenBank: KC707837.1], WH-29 [GenBank: KC707842.1], WH-15 ([GenBank: KC707839.1], I-87 [GenBank: KC894692.1], I-121 [GenBank: KC894689.1], SNT49 GenBank: CP002983.1], Hp166.03 [GenBank: HM770857.1], Hp896.95 [GenBank: HM770862.1], Hp178.02 [GenBank: HQ228198.1], C142 [GenBank: AB196363.1], Hp1335.95 [GenBank: HQ228195.1], Hp436.95 [GenBank: HQ228197.1].
Discussion
A novel virulence factor encoded by the duodenal ulcer promoting (dupA) gene (homologous to virB4 and a component of the T4SS) has been found to be associated with DU and increased expression of IL-8 [8]. However, the role of dupA as a virulence marker is still a debated issue [9-11,17,19,28,29]. The frameshift mutations in the dupA gene with a premature stop codon may have a considerable influence on the protein expression or function of DupA. [12,18-21]. In our study, we have found the rate of frameshift mutations along the length of dupA gene was [34% (17/50)] high in the Indian population. Similar trend has also been observed in other populations [19,20,24]. On the other hand, 66% (33/50) of the dupA positive strains had no stop codon and were considered as intact dupA known as dupA1. In consistent with the findings of the other studies, the intact dupA (dupA1) without frameshift mutation was significantly associated with DU in the Indian population [20,24]. This result reflects that the detection of dupA by PCR is not adequate to identify an intact dupA because frameshift mutation is common along the length of gene. Hence, dupA PCR along with sequencing is mandatory for the detection of intact dupA gene. Moura et al. [20] reported that intact dupA was independently associated with DU and can be used as a disease marker in the Brazilian population. Findings from another study [22] showed that intact dupA was more frequent in DU than gastritis but the data did not reach up to the significant level. Nevertheless the intact dupA was negatively associated with gastric carcinoma as previously observed by Lu et al. [8] and Zhang et al. [13]. Takahashi et al. [21] reported that there was an additional 615-bp in the 5′ region of dupA gene in some Okinawa strains, which classified the dupA gene into two alleles: long type (2.5 kb) and short type (1.8 kb) dupA gene [21]. The long type dupA was significantly associated with gastroduodenal diseases as compared to short type dupA [21]. However, this trend has not been detected in the H. pylori strains from the Indian population.
Jung et al. [23] reported that six additional vir genes homologues (virB8, virB9, virB10, virB11, virD2 and virD4) present around the dupA gene forming the dupA cluster was associated with duodenal ulcer and might play a pathogenic role like other T4SS cluster, similar to cag PAI [23]. The present study on additional six genes was carried out to understand the pathogenic associations of dupA cluster in Indian strains as they are geographically distinct from Western strains. Results showed that the prevalence of intact dupA1 with six vir genes was not associated with disease outcome in Indian population. The vir genes were randomly distributed among DU and NUD patients. In our population, intact dupA1 (long type or short type) with vir genes cluster was not important in promoting DU formation, which is not comparable to the one reported by Jung et al. [23] where the intact dupA cluster was associated with DU. This trend can be linked to our earlier finding in which we have showed that the Indian H. pylori strains are genetically distinct from Western and East Asian strains and the intact cag-PAI from Kolkata has not associated with disease outcome [7,24]. One study showed that none of the strains from Iraq had all six vir gene homologues, but the gene dupA1 was found to be significantly associated with DU [28]. Considering the importance of dupA1 cluster in relation to the disease manifestation, there is a need for comprehensive studies around the world. Additionally, intact dupA without frameshift mutation should be detected with DupA protein using immunoblot techniques. The intact long type as well as short type dupA1 gene might produce a functional DupA protein. The primary sequence analysis of dupA gene showed that the dupA protein was involved in cell division and peptidoglycan synthesis or modification and was implicated in intercellular chromosomal DNA transfer encodes homologues of VirB4 ATPase, as jhp0917 region (position 3–201 of dupA) contains CagE_TrbE_VirB domain and FtsK/SpoIIIE family. The region from the 3′ region jhp0917 to jhp0918 region (position 203–610) is homologue to TraG/TraD family. The FtsK/SpoIIIE domain contains a putative ATP-binding P-loop motif [30,31]. The phylogenetic analysis of dupA gene of Indian H. pylori strains showed a different pattern as compared to the distribution of other potential virulence genes such as cagA and vacA. cagA sequence of Indian H. pylori strains intermingled with the Western strains but distinct from the east Asian strains. On the other hand, the vacA mid region sequence of Indian strains formed a separate cluster from both the Western and the east Asian strains [7]. However, the dupA gene from the Indian H. pylori strains showed phylogenetic similarity with the East Asian strains and distinct from the Western strains. This is the first known genetic element of Indian H. pylori which intermingled with the East Asian strains but differed with the European strains. The exact reason for this dupA cluster difference is not known. This dupA gene is located in the hypervariable plasticity region and hence there is a possibility that dupA gene in particular or whole plasticity region of Indian H. pylori might have been acquired from East Asian strains. However, there is a need for independent studies to elucidate the dynamics of dupA in different populations around the world. Additionally, it was found that the IL-8 production was significantly associated with DU in intact dupA1 rather than truncated dupA2 or dupA negative strains. This finding is in accordance with the observation of Hussein et al. [24] in the Iraqi population, which showed that IL-8 production was significantly higher in intact dupA1 strain than truncated dupA2 or dupA negative strains. Our study showed that H. pylori strains containing cagA and vacA can induce IL-8 in cell line assay. We have also demonstrated that H. pylori strains containing cagA, vacA and dupA1 can induce IL-8 significantly higher than the strains containing only cagA and vacA. This result indicates that dupA1 is an important virulent marker of H. pylori in Indian population. Six dupA2 and 6 dupA negative strains included in this study were positive for cagA and vacA s1m1 genotypes. The presence of cagA and vacA in these dupA2 and dupA negative strains are capable to induce the IL-8 secretion up to a certain level. Moreover, the prevalence of the six vir genes of dupA cluster were almost similar in both DU and NUD groups among Indian population. This data suggests that dupA1 cluster has no role in disease manifestation in the Indian population but dupA1 might be considered as an important virulent factor of H. pylori for causing duodenal ulcer in Indian population.
Another recent study demonstrated that patients infected with dupA-positive H. pylori strains had significantly elevated gastric acid output than the dupA-negative strains [32]. In addition, Abadi et al. [16] detected higher acid resistance of dupA-positive strains making them to adopt well under high acidic condition in the stomach milieu. This increased gastric acid output is thought to be typical for an antrum-predominant H. pylori infection with an increased risk for DU and it also reduces the risk for the genesis of GUs and GC. Together, these results may explain the associations between the dupA gene and an increased risk for DU formation. Several studies conducted in different countries have shown the variable nature of dupA with respect to the clinical outcomes. Our findings emphasize the need of examining the frame-shift-mutations or dupA1 gene polymorphisms in parallel with the disease outcome. In conclusion, the presence of intact dupA seems to be important in DU development rather than complete dupA cluster (dupA with six vir genes). Thus, dupA1 may possibly act as a potential biomarker for disease manifestation in the Indian population.
Acknowledgement
JA acknowledges Indian Council of Medical Research for providing a Senior Research Fellowship. This work was supported by the intramural project of NICED, ICMR, Government of India.
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
Conceived and design the experiments AKM. Performed the experiments JA, PG, MG, AS and RD. Analyzed the data JA, AS and AKM. Contributed reagents/materials/analysis tools AKM. Wrote the paper AKM and JA. All authors read and approved the final manuscript.
Contributor Information
Jawed Alam, Email: jawedalam81@yahoo.com.
Prachetash Ghosh, Email: prache.bio@gmail.com.
Mou Ganguly, Email: moumukhopadhyay6@gmail.com.
Avijit Sarkar, Email: vijit2me@gmail.com.
Ronita De, Email: dronita@gmail.com.
Asish K Mukhopadhyay, Email: asish_mukhopadhyay@yahoo.com.
References
- 1.Fukase K, Kato M, Kikuchi S, Inoue K, Uemura N, Okamoto S, et al. Effect of eradication of Helicobacter pylori on incidence of metachronous gastric carcinoma after endoscopic resection of early gastric cancer: an open-label, randomised controlled trial. Lancet. 2008;372:392–7. doi: 10.1016/S0140-6736(08)61159-9. [DOI] [PubMed] [Google Scholar]
- 2.Suerbaum S, Michetti P. Helicobacter pylori infection. N Engl J Med. 2002;347:1175–86. doi: 10.1056/NEJMra020542. [DOI] [PubMed] [Google Scholar]
- 3.Uemura N, Okamoto S, Yamamoto S, Matsumura N, Yamaguchi S, Yamakido M, et al. Helicobacter pylori infection and the development of gastric cancer. N Engl J Med. 2001;345:784–9. doi: 10.1056/NEJMoa001999. [DOI] [PubMed] [Google Scholar]
- 4.Brown LM. Helicobacter pylori: epidemiology and routes of transmission. Epidemiol Rev. 2000;22:283–97. doi: 10.1093/oxfordjournals.epirev.a018040. [DOI] [PubMed] [Google Scholar]
- 5.Misra V, Pandey R, Misra SP, Dwivedi M. Helicobacter pylori and gastric cancer: Indian enigma. World J Gastroenterol. 2014;20:1503–9. doi: 10.3748/wjg.v20.i6.1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Patra R, Chattopadhyay S, De R, Ghosh P, Ganguly M, Chowdhury A, et al. Multiple infection and microdiversity among Helicobacter pylori isolates in a single host in India. PLoS One. 2012;7:e43370. doi: 10.1371/journal.pone.0043370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mukhopadhyay AK, Kersulyte D, Jeong JY, Datta S, Ito Y, Chowdhury A, et al. Distinctiveness of genotypes of Helicobacter pylori in Calcutta, India. J Bacteriol. 2000;182:3219–27. doi: 10.1128/JB.182.11.3219-3227.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu H, Hsu PI, Graham DY, Yamaoka Y. Duodenal ulcer promoting gene of Helicobacter pylori. Gastroenterology. 2005;28:833–48. doi: 10.1053/j.gastro.2005.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arachchi HS, Kalra V, Lal B, Bhatia V, Baba CS, Chakravarthy S, et al. Prevalence of duodenal ulcer-promoting gene (dupA) of Helicobacter pylori in patients with duodenal ulcer in North Indian population. Helicobacter. 2007;12:591–7. doi: 10.1111/j.1523-5378.2007.00557.x. [DOI] [PubMed] [Google Scholar]
- 10.Argent RH, Burette A, Miendje Deyi VY, Atherton JC. The presence of dupA in Helicobacter pylori is not significantly associated with duodenal ulceration in Belgium, South Africa, China, or North America. Clin Infect Dis. 2007;45:1204–6. doi: 10.1086/522177. [DOI] [PubMed] [Google Scholar]
- 11.Douraghi M, Mohammadi M, Oghalaie A, Abdirad A, Mohagheghi MA, Hosseini ME, et al. dupA as a risk determinant in Helicobacter pylori infection. J Med Microbiol. 2008;57:554–62. doi: 10.1099/jmm.0.47776-0. [DOI] [PubMed] [Google Scholar]
- 12.Gomes LI, Rocha GA, Rocha AM, Soares TF, Oliveira CA, Bittencourt PF, et al. Lack of association between Helicobacter pylori infection with dupA-positive strains and gastroduodenal diseases in Brazilianpatients. Int J Med Microbiol. 2008;298:223–30. doi: 10.1016/j.ijmm.2007.05.006. [DOI] [PubMed] [Google Scholar]
- 13.Zhang Z, Zheng Q, Chen X, Xiao S, Liu W, Lu H. The Helicobacter pylori duodenal ulcer promoting gene, dupA in China. BMC Gastroenterol. 2008;8:49. doi: 10.1186/1471-230X-8-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hussein NR, Mohammadi M, Talebkhan Y, Doraghi M, Letley DP, Muhammad MK, et al. Differences in virulence markers between Helicobacter pylori strains from Iraq and those from Iran: potential importance of regional differences in H. pylori-associated disease. J Clin Microbiol. 2008;46:1774–9. doi: 10.1128/JCM.01737-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nguyen LT, Uchida T, Tsukamoto Y, Kuroda A, Okimoto T, Kodama M, et al. Helicobacter pylori dupA gene is not associated with clinical outcomes in the Japanese population. Clin Microbiol Infect. 2010;16:1264–9. doi: 10.1111/j.1469-0691.2009.03081.x. [DOI] [PubMed] [Google Scholar]
- 16.Abadi AT, Taghvaei T, Wolfram L, Kusters JG. Infection with Helicobacter pylori strains lacking dupA is associated with an increased risk of gastric ulcer and gastric cancer development. J Med Microbiol. 2012;61:23–30. doi: 10.1099/jmm.0.027052-0. [DOI] [PubMed] [Google Scholar]
- 17.Alam J, Maiti S, Ghosh P, De R, Chowdhury A, Das S, et al. Significant association of the dupA gene of Helicobacter pylori with duodenal ulcer development in a South-east Indian population. J Med Microbiol. 2012;61:1295–302. doi: 10.1099/jmm.0.038398-0. [DOI] [PubMed] [Google Scholar]
- 18.Hussein NR. The association of dupA and Helicobacter pylori-related gastroduodenal diseases. Eur J Clin Microbiol Infect Dis. 2010;29:817–21. doi: 10.1007/s10096-010-0933-z. [DOI] [PubMed] [Google Scholar]
- 19.Queiroz DM, Rocha GA, Rocha AM, Moura SB, Saraiva IE, Gomes LI, et al. dupA polymorphisms and risk of Helicobacter pylori-associated diseases. Int J Med Microbiol. 2011;301:225–8. doi: 10.1016/j.ijmm.2010.08.019. [DOI] [PubMed] [Google Scholar]
- 20.Moura SB, Costa RF, Anacleto C, Rocha GA, Rocha AM, Queiroz DM. Single nucleotide polymorphisms of Helicobacter pylori dupA that lead to premature stop codons. Helicobacter. 2012;17:176–80. doi: 10.1111/j.1523-5378.2011.00933.x. [DOI] [PubMed] [Google Scholar]
- 21.Takahashi A, Shiota S, Matsunari O, Watada M, Suzuki R, Nakachi S, et al. Intact long-type dupA as a marker for gastroduodenal diseases in Okinawan subpopulation, Japan. Helicobacter. 2013;18:66–72. doi: 10.1111/j.1523-5378.2012.00994.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kersulyte D, Lee W, Subramaniam D, Anant S, Herrera P, Cabrera L, et al. Helicobacter Pylori’s plasticity zones are novel transposable elements. PLoS One. 2009;4:e6859. doi: 10.1371/journal.pone.0006859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jung SW, Sugimoto M, Shiota S, Graham DY, Yamaoka Y. The intact dupA cluster is a more reliable Helicobacter pylori virulence marker than dupA alone. Infect Immun. 2012;80:381–7. doi: 10.1128/IAI.05472-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hussein NR, Abdullah SM, Salih AM, Assafi MA. dupA1 is associated with duodenal ulcer and high interleukin-8 secretion from the gastric mucosa. Infect Immun. 2012;80:2971–2. doi: 10.1128/IAI.00076-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Crabtree JE, Lindley IJ. Mucosal interleukin-8 and Helicobacter pylori-associated gastroduodenal disease. Eur J Gastroenterol Hepatol. 1994;6:33–8. [PubMed] [Google Scholar]
- 26.Sharma SA, Tummuru MK, Miller GG, Blaser MJ. Interleukin 8 response of gastric epithelial cell lines to Helicobacter pylori stimulation in vitro. Infect Immun. 1995;63:1681–7. doi: 10.1128/iai.63.5.1681-1687.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Patra R, Chattopadhyay S, De R, Datta S, Chowdhury A, Ramamurthy T, et al. Intact cag pathogenicity island of Helicobacter pylori without disease association in Kolkata, India. Int J Med Microbiol. 2011;301:293–302. doi: 10.1016/j.ijmm.2010.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schmidt HM, Andres S, Kaakoush NO, Engstrand L, Eriksson L, Goh KL, et al. The prevalence of the duodenal ulcer promoting gene (dupA) in Helicobacter pylori isolates varies by ethnic group and is not universally associated with disease development: a case–control study. Gut Pathog. 2009;1:5. doi: 10.1186/1757-4749-1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hussein NR, Argent RH, Marx CK, Patel SR, Robinson K, Atherton JC. Helicobacter pylori dupA is polymorphic, and its active form induces pro-inflammatory cytokine secretion by mononuclear cells. J Infect Dis. 2010;202:261–9. doi: 10.1086/653587. [DOI] [PubMed] [Google Scholar]
- 30.Begg KJ, Dewar SJ, Donachie WD. A new Escherichia coli cell division gene, ftsK. J Bacteriol. 1995;177:6211–22. doi: 10.1128/jb.177.21.6211-6222.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu LJ, Errington J. Bacillus subtilis SpoIIIE protein required for DNA segregation during asymmetric cell division. Science. 1994;264:572–5. doi: 10.1126/science.8160014. [DOI] [PubMed] [Google Scholar]
- 32.Imagawa S, Ito M, Yoshihara M, Eguchi H, Tanaka S, Chayama K. Helicobacter pylori dupA and gastric acid secretion are negatively associated with gastric cancer development. J Med Microbiol. 2010;59:1484–9. doi: 10.1099/jmm.0.021816-0. [DOI] [PubMed] [Google Scholar]


