ABSTRACT
Caveolae are an abundant feature of the plasma membrane in many cells. Until recently, they were generally considered to be membrane invaginations whose formation primarily driven by integral membrane proteins called caveolins. However, the past decade has seen the emergence of the cavin family of peripheral membrane proteins as essential coat components and regulators of caveola biogenesis. In this Commentary, we summarise recent data on the role of cavins in caveola formation, highlighting structural studies that provide new insights into cavin coat assembly. In mammals, there are four cavin family members that associate through homo- and hetero-oligomerisation to form distinct subcomplexes on caveolae, which can be released into the cell in response to stimuli. Studies from several labs have provided a better understanding of cavin stoichiometry and the molecular basis for their oligomerisation, as well as identifying interactions with membrane phospholipids that may be important for caveola function. We propose a model in which coincident, low-affinity electrostatically controlled protein–protein and protein–lipid interactions allow the formation of caveolae, generating a meta-stable structure that can respond to plasma membrane stress by release of cavins.
KEY WORDS: Caveolae, Caveolin, Cavin, Coiled coil, Electron microscopy, X-ray crystallography
Introduction
Caveolae (or ‘little caves’) are bulb-shaped invaginations of 50–60 nm that are a main feature of the plasma membrane in many cell types, including adipocytes, endothelial and muscle cells (Fig. 1A). Caveolae generally display an uneven cellular distribution, and caveola-associated proteins have distinct tissue-specific expression profiles suggestive of highly specialised functions. A large number of studies have now revealed a remarkably diverse array of functions for caveolae – including lipid turnover and organisation of the plasma membrane – providing a platform for regulation of signalling molecules and enzymes, intracellular trafficking, as well as supplying a membrane reservoir for mechanosensing and mechanoprotection at the cell surface (reviewed in Parton and del Pozo, 2013).
Fig. 1.
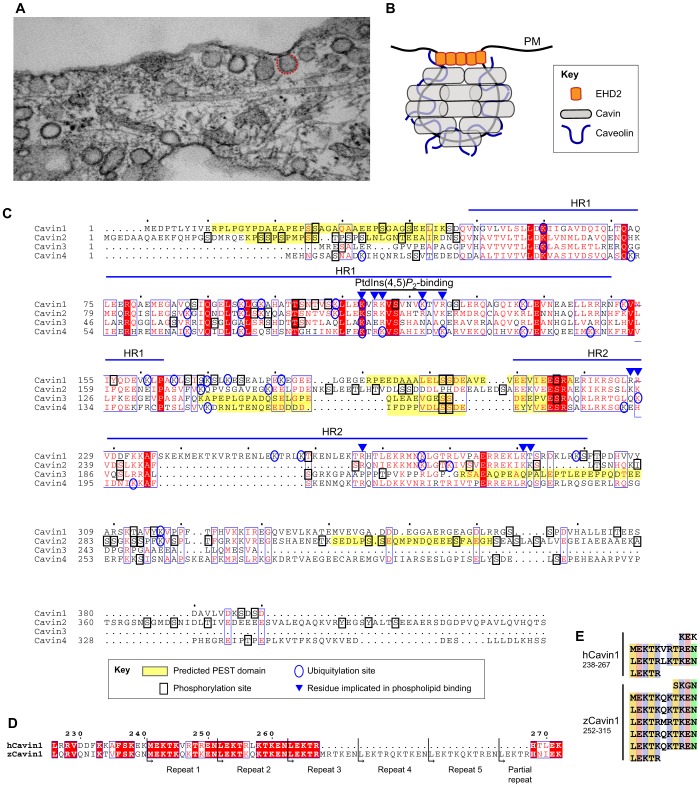
Caveolae and the cavin family of proteins. (A) Electron micrograph of caveolar bulbs (see caveolae marked by red dotted line) at the cell surface of a fibroblast cell. (B) Simplified model of the caveolar coat. Cavins are peripheral membrane proteins that coat the caveolar surface, with caveolins embedded in the interior membrane layer. EHD2 is known to associate with the caveolar neck and can modulate the dynamics of caveolae. (C) Secondary-structure-based sequence alignment of human cavins. Indicated on the alignment are predicted PEST domains (predicted by sPESTfind), and identified phosphorylation and ubiquitylation sites from the Phosphosite database (Hornbeck et al., 2012), as well as previously identified residues implicated in phospholipid interactions (Hansen et al., 2009; Kovtun et al., 2014). Amino acid residues in red are strictly conserved in the four cavins; amino acid residues surrounded by a blue box are highly conserved; amino acid residues highlighted red are conserved in residue type. (D) Putative 11-residue (hendecad/undecad) repeats unique to the HR2 domain of Cavin1, which – we propose – might form a coiled-coil structure. Portions of the Cavin1 sequences are shown highlighting the two repeats in human (h) Cavin1, and the five in zebrafish (z) Cavin1. (E) Alignment of the repeat elements shown in Fig. 1D; colour-coding indicates conserved hydrophobic (yellow), acidic (orange), basic (blue) and polar (green) residues.
Although caveolae were originally observed by electron microscopy (EM) in the 1950s (Palade, 1953; Yamada, 1955), the first protein component caveolin-1 (CAV1) was only identified 40 years later (Kurzchalia et al., 1992; Rothberg et al., 1992). Following this, two other homologues of CAV1 were found, CAV2 (Scherer et al., 1996) and the muscle specific CAV3 (Way and Parton, 1995). Around the same time, specialised EM techniques revealed that caveolae possess a distinctive striated or spiral coating (Peters et al., 1985; Rothberg et al., 1992), which was initially ascribed to the only known protein components, the membrane-embedded caveolins. However, in the past decade, a second abundant and essential structural component of caveolae, the cavin family of proteins, has been defined, demanding a revision of our understanding of caveolar assembly and architecture (Aboulaich et al., 2004; Bastiani et al., 2009; Hansen et al., 2009; Hill et al., 2008; Liu et al., 2008; Vinten et al., 2005; Vinten et al., 2001). There are four cavins in mammals, Cavin1 (PTRF), which displays a relatively broad expression profile and, like CAV1, is essential for the formation of all caveolae, Cavin2 (SDPR), Cavin3 (PRKCDBP), and the muscle-specific isoform Cavin4 (MURC). Recent studies of the cavin proteins by using both EM and X-ray crystallography have provided new insights into cavin structure and assembly, which strongly suggest the striated coats of caveolae are, in fact, polymerised cavin proteins (Gambin et al., 2014; Kovtun et al., 2014; Ludwig et al., 2013) (Fig. 1B).
In addition to caveolins and cavins, several other proteins with inherent membrane-binding and -remodelling properties have also been found to be associated with caveolae, and are concentrated, in particular, at the narrow neck region; the dynamin GTPase, the dynamin-like ATPase EHD2, and the BAR-domain-containing protein PACSIN2 (alternatively known as Syndapin2). However, although these proteins contribute to a network of interactions that influence caveola morphology, dynamics and endocytosis, none are actually essential for caveola formation (Hansen et al., 2011; Henley et al., 1998; Morén et al., 2012; Senju et al., 2011). Moreover, recent analyses of crosslinked CAV1-containing complexes detected only cavins and caveolins as the major caveolar coat components (Ludwig et al., 2013). This suggests a highly cooperative mechanism of caveola formation that is primarily mediated by just two families of protein, both of which show no clear structural similarity to any other known coat proteins. Understanding the molecular mechanisms that govern the assembly and disassembly of the cavin coat is of major interest in terms of defining their role in membrane remodelling, caveolar biogenesis and cell signalling, and their role in diseases including cancer, and lipid and muscle dystrophies (reviewed in Parton and del Pozo, 2013). Recent studies have provided new data that are beginning to shed light on these processes, including biochemical, single-molecule and ultrastructural analysis of the caveolar coat, the first X-ray crystallographic structure of the universal cavin association domain and the visualisation of isolated cavin oligomers by EM (Gambin et al., 2014; Hansen et al., 2013; Kovtun et al., 2014; Ludwig et al., 2013). Although there are comprehensive reviews that provide timely updates on the biology of caveolae (Bastiani and Parton, 2010; Chidlow and Sessa, 2010; Parton and del Pozo, 2013; Shvets et al., 2014), in this Commentary, we examine these recent structural data and propose a refined model of caveola formation that is based on the emerging molecular insights into the unique properties of the cavin protein family.
The cavin family of coat proteins
Remarkably, despite the fact that all four mammalian cavins are now recognised components of caveolae, each member of the family has originally been described in studies implicating them in unrelated cellular processes (see Box 1). The cavins constitute a unique protein family at sequence and structural levels. All cavins share sequence homology and conserved α-helical secondary structure elements within two clearly delineated regions called helical region (HR) 1 and HR2 (Gustincich et al., 1999; Kovtun et al., 2014) (Fig. 1C). HR1 and HR2 are separated by three disordered regions (DR) we refer to as DR1, DR2 and DR3; these are not well conserved at the sequence level, but do share a highly acidic sequence profile and predicted disordered secondary structure. The genomic conservation of cavin orthologues is limited to vertebrates, and is likely to coincide with the appearance of morphologically observable caveolae (Hansen and Nichols, 2010). In contrast, caveolins can also be found in non-vertebrate species, specifically in worms and some insects, suggesting roles in these organisms that are unrelated to the formation of caveolae (Kirkham et al., 2008).
Box 1. Discovering and defining the cavin protein family.
Cavin1 (also known as Cav-p60, BBP), was first identified as a factor involved in transcription termination, receiving the official name polymerase I and transcript release factor (PTRF) (Jansa et al., 1998). Cavin2 (also known as PS-p68, SDR) was initially discovered for its abundance and high affinity for PtdSer (PS) in platelets (Burgener et al., 1990). It was later described as a protein induced upon serum deprivation and was given the official name serum-deprivation-response protein (SDPR) (Gustincich and Schneider, 1993). Cavin3 was first discovered for its function as an adaptor for protein kinase Cδ homologous to SDPR (Izumi et al., 1997) and named SDPR-related gene product that binds to C-kinase (SRBC) but is now officially known as protein kinase C delta-binding protein (PRKCDBP). Cavin4 was initially identified in muscle, and received the official name muscle-related coiled-coil protein (MURC, Ogata et al., 2008).
The identification of cavins as structural components of caveolae began with the discovery of a 60-kDa protein that is abundant in caveolae of several cell types (Vinten et al., 2001), which later was identified as PTRF (Vinten et al., 2005). As PTRF was found to be a main constituent of caveolae, the protein was given the moniker ‘Cavin1’. These data coincided with proteomics-based identification of caveola-associated PTRF and SRBC (Aboulaich et al., 2004) that, taken together with earlier observations (Izumi et al., 1997; Mineo et al., 1998), suggested grouping of these proteins into a single family. Subsequently Cavin1/PTRF was shown to be required for caveola formation and function (Hill et al., 2008; Liu et al., 2008; Liu and Pilch, 2008), and Cavin2/SDPR, Cavin3/SRBC and Cavin4/MURC were also found to associate with caveolae (Bastiani et al., 2009; Hansen et al., 2009; McMahon et al., 2009). The four proteins were, therefore, united into a single protein family on the grounds of sequence similarity and common function as peripheral membrane proteins that associate with and stabilise caveolae, and it was suggested that a common naming system was adopted for the proteins from Cavin1 to Cavin4 (Bastiani et al., 2009; McMahon et al., 2009).
Whereas their functional significance is still not clear, all cavins have been shown to undergo extensive post-translational modification (Fig. 1C). PEST motifs (sequences enriched in proline, glutamic acid, serine and threonine) residing in the DRs of cavins are a characteristic feature of this family and sites of proteolytic sensitivity. The abundance of these sites suggests that protein degradation is an important regulator of cavin homeostasis and function. Interestingly, a significant amount of Cavin1 is cleaved in situ (Vinten et al., 2005), presumably at the PEST regions (Aboulaich et al., 2004). This limited proteolysis might modulate localisation of cavin because removal of the C-terminal DR3 region leads to association with microtubules (Liu and Pilch, 2008) and flanking DRs are involved in cavin oligomerisation (Kovtun et al., 2014). SUMOylation and ubiquitylation sites have also been identified in cavins (Williams and Palmer, 2014), and the observation of accelerated proteasome-dependent degradation of non-caveolar Cavin2 suggests a potential role for ubiquitylation in cavin turnover (Breen et al., 2012). Phosphorylation is an abundant modification of all cavins, with dozens of experimentally identified sites distributed primarily within the DRs (reviewed in Hansen and Nichols, 2010; Kovtun et al., 2014). Cavins have been found to be rapidly phosphorylated following stimulation of adipocytes (Aboulaich et al., 2011; Humphrey et al., 2013; Krüger et al., 2008), suggesting that their regulation is intimately coupled to cellular signalling pathways, and the mobilisation of caveolae is reportedly connected to kinase activity (Pelkmans et al., 2002; Pelkmans and Zerial, 2005). None of these phosphorylation events have yet been linked to caveolae assembly, except for one study that suggests phosphorylation of Cavin1 within DR3 at Ser365 and/or Ser366 can enhance caveolar association (Bai et al., 2011). Likewise, insulin-induced Ser/Thr phosphorylation of Cavin1 is associated with its loss from the plasma membrane (Aboulaich et al., 2011; Aboulaich et al., 2006). However, the precise functional significance of this extensive phosphorylation remains an important unanswered question.
Cavin subcomplexes – homo- and hetero-oligomerisation
One of the most strikingly consistent observations is that cavins possess the ability to associate into higher order homo- and hetero-oligomers. Cavin–cavin interactions are independent of membrane or caveolins (Gambin et al., 2014; Hansen et al., 2013; Hayer et al., 2010; Kovtun et al., 2014; Ludwig et al., 2013). In detergent-solubilised cellular lysates, the main cavin pool is incorporated into large complexes with apparent sedimentation coefficients varying between 40S and 60S (Hayer et al., 2010; Ludwig et al., 2013). This variability of the weight distribution appears to be sensitive to the expression ratios between the different cavins, suggesting that the oligomerisation and stability of the cavin coat is tuned by the relative abundance of the cavin isoforms. Various tissues and cell types demonstrate strikingly different cavin expression patterns (Bastiani et al., 2009; Hansen et al., 2013). Cavin1 can be considered a ‘gatekeeper cavin’ because it is ubiquitously expressed and required for the formation of all caveolae. High levels of Cavin2 are seen in lung and fat tissue; they correlate with a smaller average size of the cavin complex as a result of the accumulation of low-molecular mass cavin oligomers. In addition, Cavin2 knockout resulted in the loss of endothelial caveolae in lung but not heart, where Cavin2 is expressed at low levels (Hansen et al., 2013). These data suggest the importance of cavin isoforms in tuning caveola properties for tissue-specific functions and highlights the importance of understanding the different modes of cavin oligomerisation.
Observations of various cavins expressed in in vitro systems or cell models using single-molecule fluorescence correlation approaches revealed rapid, probably co-translational, oligomerisation that results in the formation of protein complexes containing on average 50 cavin monomers (Gambin et al., 2014), which potentially represents the 60S-size cavin complex reported previously (Hayer et al., 2010; Ludwig et al., 2013). Despite the similar homo-oligomerisation properties of different cavins, only Cavin1 homo-oligomers have the ability to engage with caveolin at the plasma membrane to trigger caveolar formation (Bastiani et al., 2009; Hansen et al., 2013; Hill et al., 2008). This is in agreement with studies of Cavin1-knockout animals, which show a complete loss of caveolae in all tissues (Hill et al., 2008; Liu et al., 2008), and Cavin1-deficient patients (Hayashi et al., 2009). All other cavins require Cavin1 in order to be incorporated into the caveolar coat (Bastiani et al., 2009). Analysis of cavins following co-expression revealed highly promiscuous patterns of homo- and hetero-association, with the major exception that Cavin2 and Cavin3 are unable to associate with each other (Gambin et al., 2014; Kovtun et al., 2014; Ludwig et al., 2013). Moreover, Cavin2 and Cavin3 compete for binding to Cavin1, and segregate into mutually exclusive caveolar domains (Gambin et al., 2014; Ludwig et al., 2013). It is unclear what is the underlying basis of this particular property of Cavin1 because it is the only family member required for formation of all caveolae, as well as being able to form caveolae in the absence of other cavins. One unique feature of Cavin1 not previously noticed is the presence of an inserted sequence in the centre of the HR2 domain (Fig. 1C) (O.K. and B.M.C., unpublished observation). Closer examination reveals that the sequence consists of two tandem-repeat elements of eleven residues. This becomes even clearer when human Cavin1 is compared to its orthologue from zebrafish (Fig. 1D). Zebrafish Cavin1 possesses an even longer insertion, with five highly conserved tandem repeats. Alignment of the repeats suggests they potentially comprise a hendecad (also called undecad), i.e. 11-residue, repeat, previously found to fold into a right-handed coiled-coil structure (Burkhard et al., 2001; Stetefeld et al., 2000) (Fig. 1E). It will, therefore, be of interest to determine whether the Cavin1 HR2 domain does, indeed, possess this structural element and, if so, whether it is able to confer unique functional properties on Cavin1 relative to other family members.
Recent studies permitted a quantitative protein composition analysis of the caveolar coat. In isolated complexes, Cavin1 was found to associate with either Cavin2 or Cavin3 at a ratio of ∼2–3 to 1 (Gambin et al., 2014; Ludwig et al., 2013). The Cavin1 to CAV1 ratio was estimated to be ∼1 to 4 and was independent of other cavins. In addition, single-molecule coincidence analysis of various cavin oligomers with CAV1 suggested that the caveolar coat comprises 150–200 caveolin subunits and approximately 50 cavins (Gambin et al., 2014), which overall correlates well with previous reports (Ludwig et al., 2013; Pelkmans and Zerial, 2005).
The caveolar coat enters the molecular era – structure of the cavin oligomerisation domain
As outlined above, the cavins comprise an apparently unique protein family in terms of their architecture and structure. Early sequence analyses identified a potential N-terminal leucine zipper motif, suggestive of a supercoil structure (Aboulaich et al., 2004; Bastiani et al., 2009; Gustincich et al., 1999; Izumi et al., 1997). Overall secondary structure predictions indicate that cavins are predominantly α-helical proteins, with a high content of disordered/extended sequence and negligible β-sheet content (Kovtun et al., 2014). There is an extraordinarily conserved alternating electrostatic pattern to the cavin proteins, whereby basic, positively charged HR sequences are linked together by acidic, negatively charged DRs (Fig. 1C). This is seen in all cavin isoforms, and in all species.
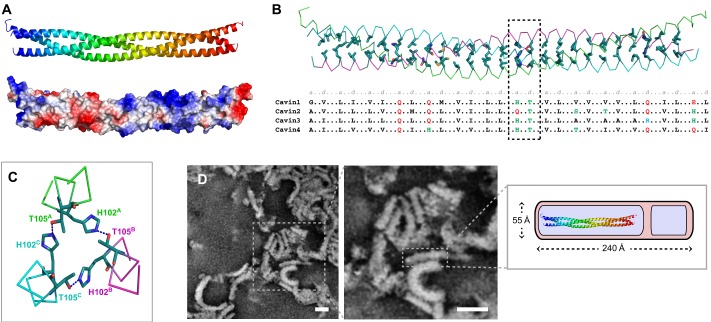
Isolation of cavins as recombinant proteins is generally hampered by their tendency to form larger oligomers and undergo degradation (Hansen et al., 2009), but it has recently been found that an ∼100-residue cavin fragment, the N-terminal HR1, can be isolated in a homogeneous form and readily crystallised (Kovtun et al., 2014). Most importantly, this fragment recapitulates the specific homo- and heteromeric interactions observed for the full-length proteins, indicating that this domain drives these underlying functional cavin–cavin interactions (Kovtun et al., 2014; Mohan et al., 2015). X-ray crystallographic structures of the HR1 domain from both mouse Cavin1 and zebrafish Cavin4a reveal a conserved and highly extended parallel trimeric coiled-coil (Fig. 2A). This structure incorporates the shorter predicted leucine-zipper motifs and has important implications for the assembly of the cavin coat. First, the trimeric nature of the cavin HR1 domain immediately provides an explanation for the apparent trimeric nature of Cavin1 following crosslinking (Ludwig et al., 2013) (Mohan et al., 2015). The consequence of this is that the cavin coat must be composed of trimeric building blocks, although how these trimers assemble into membrane-associated polymers remains an important question (discussed below). Second, the hetero-oligomers formed by Cavin1–Cavin2 and Cavin1–Cavin3 are most likely to be composed of two copies of Cavin1, whereby the third helix of the coiled-coil is replaced with either Cavin2 or Cavin3 in a mutually exclusive arrangement. This appears to be the most reasonable explanation satisfying both the structural constraints and the previous biochemical studies indicating the presence of two to three copies of Cavin1 per molecule of Cavin2 or Cavin3 in caveolar complexes (Gambin et al., 2014; Ludwig et al., 2013), although it remains to be confirmed by high-resolution structural studies of mixed HR1 assemblies. Third, the structure shows that a large fraction of each cavin sequence folds into a highly rigid and elongated rod assembly (∼16 nm in length and ∼3 nm in width) that, as discussed below, is relevant to the overall architecture of the cavin complexes.
Fig. 2.
Structure of the cavin proteins by crystallography and EM. (A) Crystal structure of the mouse Cavin1 HR (Kovtun et al., 2014) (PDB ID 4QKV). Top panel shows the homotrimeric coiled-coil in ribbon representation coloured blue to red from N- to C-terminus. Bottom panel shows the surface representation of the same structure coloured for electrostatic potential (red = −0.5 V, blue = +0.5 V). Note the region of strong positive charge (blue) within the C-terminal half of the HR1 domain. (B) The heptad repeat of the cavin proteins. The top panel shows the structure of the mouse Cavin1 HR1 domain in ‘worm-like’ representation with the central residues of the heptad repeat shown as side chains. Below this, the corresponding residues of human Cavin1, Cavin2, Cavin3 and Cavin4 are shown. The dashed box highlights the central His–Thr pairing (His102 and Thr105 in Cavin1). (C) Cross section of the Cavin1 structure showing the hydrogen-bond pairing between the central His–Thr side chains, creating a central hydrophilic layer of the coiled coil. (D) Negatively stained electron micrograph of recombinant bacterially expressed Cavin1 full-length protein after crosslinking and lipid removal. Arrays of rod-like elements are seen that are generally uniform in dimension but display both linear and curved topologies. The inset shows a schematic representation of a single rod element with dimensions indicated. The HR1 coiled coil is shown to scale for comparison.
Two other notable features of the HR1 coiled-coil were also observed. First, cavins share a conserved hydrogen-bonded pair of His and Thr side chains lying at the centre of the structure (Fig. 2B,C). Coiled-coils are characterised by a hydrophobic pattern repeat of seven amino acids, labelled abcdefg, termed the heptad repeat, which defines the central residues at the interface between protomers. However, the a and d positions of this repeat can vary with the inclusion of side-chains capable of hydrogen-bond pairing, and this is often what defines the specificity of coiled-coil formation. In Fig. 2B, the core residues of the Cavin1 HR1 are shown, and conservation of these residues in other cavins is indicated below. In cavins, there are several conserved Gln residues that require pairing to an equivalent Gln side chain from adjoining protomers, thus excluding unrelated coiled-coil sequences from interacting. The sequences of these heptads will also contribute to the differing specificity of cavin–cavin interactions, but it is not clear yet which residues are crucial. The His–Thr pair at the centre of the coiled-coil is highly unusual in coiled-coil structures (Fig. 2C), and its conservation suggests an important function. Cavin2 is an exception in that it possesses Gln instead of His, and it will be interesting to determine the structural impact of this on cavin–cavin interactions. Interestingly, proteomics studies indicate that the Thr residue can be phosphorylated in several cavins, despite the fact that it is buried within the structure (Humphrey et al., 2013; Kovtun et al., 2014) (Fig. 1C). Clearly, phosphorylation of this residue will disrupt the hydrogen-bonded pairing, and we speculate it to have consequences for the stable structure of the cavins and, potentially, represent a mechanism for regulating their assembly. Remarkably, this region also contains multiple ubiquitylation sites (Fig. 1C), suggesting an additional level of regulation of cavin assembly and turnover. Second, the cavin HR1 domain has a highly conserved basic surface at its C-terminus (Fig. 2A), composed of clusters of Lys and Arg residues. As discussed below, these residues play an important role in membrane association by the cavin polymers.
Higher order structures of the cavin proteins
Whereas the HR1 determines cavin–cavin specificity, it is not sufficient to promote the formation of larger complexes required for caveolar formation. Other domains within the cavins are clearly functionally important and, through poorly understood mechanisms, can support the assembly of higher order cavin oligomers (Kovtun et al., 2014). The second putative helical domain of the cavins is HR2, whose sequence can vary in length but is also highly basic in nature. It was found that, in contrast to HR1, HR2 (of Cavin4 at least) is unfolded in isolation. However, when combined with HR1 it adopts an α-helical conformation of unknown topology, presumably driven by self-association established through HR1 trimerisation. This increase in secondary structure also coincides with an additional oligomerisation step, yielding a large protein complex with a molecular mass in the sub-MDa range. Interestingly, inclusion of the N- and C-terminal DR1 and DR3 that flank the α-helical core promotes the formation of an even larger assembly that is relatively heterogeneous in size, i.e. ∼1–2 MDa.
When isolated from bacteria, recombinant cavins are found to co-fractionate with a substantial quantity of lipids (Kovtun et al., 2014). The purified homo-oligomeric protein complexes display relatively isomorphous spherical structures of ∼20–40 nm in diameter. Following gentle crosslinking and detergent extraction, however, the purified cavins are revealed to possess a distinct rod-like structure, ∼23 nm long and 5 nm wide (Fig. 2D) (Kovtun et al., 2014). The size and morphology of these cavin rods is strikingly similar to the striated elements that coat caveolae, as observed in vivo (Ludwig et al., 2013; Rothberg et al., 1992). Furthermore, recent ultra-structural analyses by using EM that focused specifically on the cavins strongly suggest that they adopt peripheral membrane striations on the surface of the caveolar bulbs (Gambin et al., 2014; Ludwig et al., 2013). Altogether, it now seems almost certain that the striated densities that are accepted as a defining feature of caveolae are, in fact, formed by the peripheral membrane-associated cavins, with perhaps much less contribution from the membrane-embedded caveolins as previously assumed.
Cavin interactions with membrane lipids
Caveolae are generally considered to comprise a distinct species of lipid raft, with a characteristic invaginated topology, and a membrane leaflet enriched in various lipid species (Ortegren et al., 2004). The assembly of caveolae is dependent on their specific lipid composition and, in particular, requires the presence of cholesterol (Breen et al., 2012; Rothberg et al., 1992). In addition, the most abundant negatively charged phospholipids of the plasma membrane, PtdSer and the phosphoinositide phosphatidylinositol (4,5)-bisphosphate [PtdIns(4,5)P2] have been suggested to be concentrated in caveolae, forming physiologically distinct lipid pools (Fairn et al., 2011; Fujita et al., 2009; Pike and Casey, 1996). Furthermore, depletion of caveolae by knockdown of CAV1 results in re-organisation of PtdSer domains within the plasma membrane (Ariotti et al., 2014). Cytoplasmically exposed headgroups of these phospholipids are often involved in membrane association and allosteric regulation of membrane-associated proteins (Li et al., 2014; McLaughlin et al., 2002), with the prime example being the coordinated assembly of clathrin-coated endocytic vesicles (Antonescu et al., 2011; Kelly et al., 2014).
Phospholipids
The question, therefore, arises as to how the distinct protein and lipid species of caveolae cooperate to control caveola assembly and function. Cavin2 was originally isolated as a major PtdSer-binding protein of platelet cells (Burgener et al., 1990). Further reports identified similar Ca2+-independent PtdSer-binding sites in all members of the family (Gustincich et al., 1999; Hill et al., 2008; Izumi et al., 1997; Mineo et al., 1998). A PtdSer-binding site was previously postulated to reside in a conserved region at the end of HR1 (Izumi et al., 1997; McMahon et al., 2009), but recent data on lipid affinities of systematically truncated cavins have revealed two independent PtdSer-binding regions that are composed of the conserved polybasic HR2 domain and, to a lesser extent, HR1 (Kovtun et al., 2014) (Figs 1C, 3). The same work has also established a connection between caveolar PtdInsP2 and cavins, revealing a high-affinity phosphoinositide-binding site within HR1. The model, thus, proposed is that cavins possess a general affinity for negatively charged phospholipid membranes, but HR1 promotes a somewhat specific recognition of phosphoinositide headgroups. It has been shown that concomitant mutation of the conserved Arg and Lys residues in either HR1 or HR2, substantially affects the recruitment of Cavin1 or Cavin2 to caveolar membranes (Hansen et al., 2009; Kovtun et al., 2014). However, mutating these residues individually does not result in an absolute block in membrane localisation, and it is thought that redundancy in membrane binding between HR1 and HR2 may explain this intermediate phenotype (Kovtun et al., 2014). Given the fact that between 15 and 20 cavin trimers are thought to be present in each caveola vesicle (Gambin et al., 2014), and that each trimer will possess at least two independent phospholipid-binding sites, membrane association and polymerisation of cavins are likely to, in turn, enhance the localised enrichment of PtdSer and PtdIns(4,5)P2 observed at caveolae. This suggests a cooperative feedback mechanism between association of cavins with negatively charged phospholipids and increased localised concentrations of those lipids, which may then be important in regulating other caveola-associated factors that are involved in signalling and transport (Izumi et al., 1997; Simone et al., 2013). Overall, however, despite the clear activity of cavins in phospholipid binding, the actual role of these lipids in caveolar function remains quite poorly studied.
Fig. 3.
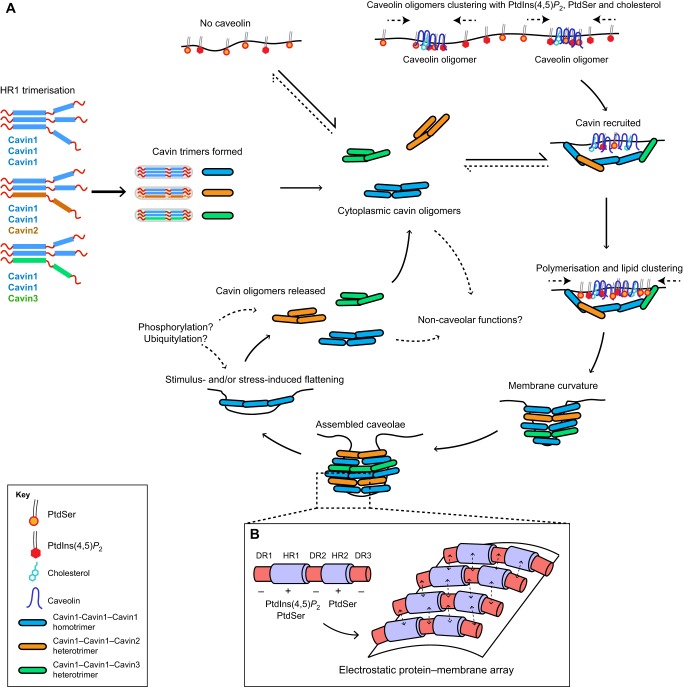
A model for cavin function in caveolar formation. (A) The cavin life cycle involves initial trimerisation through the HR1 domain and subsequent assembly into cytoplasmic cavin oligomers. These can bind negatively charged membranes but only become stably associated with the plasma membrane in the presence of caveolins. Caveolins are believed to cluster with cholesterol, PtdIns(4,5)P2 and PtdSer (shown at the top); subsequent recruitment of cavins will further increase the local concentration of the negatively charged lipids in particular. This can then nucleate membrane curvature and formation of the distinctive caveola bud. Certain stimuli, such as membrane stretching or cholesterol depletion, can lead to flattening of caveolae and the release of cavins back into the cytoplasmic pool. Cavins are extensively modified by posttranslational modifications, but the negative or positive consequences for caveolar formation are still unknown. (B) An expanded view of the membrane surface, showing a speculative model for cavin–cavin interactions through an array of electrostatic interactions involving both membrane lipids and domains of opposite charge in adjoining protomers.
Cholesterol
Cholesterol is another abundant component of caveolar membrane domains (Ortegren et al., 2004; Yao et al., 2009) and is essential for caveola formation (Breen et al., 2012; Hailstones et al., 1998; Rothberg et al., 1992). Pharmacological depletion of cholesterol results in caveola flattening and dissociation of cavin into the cytoplasm (Breen et al., 2012; Rothberg et al., 1992). Although there is a rich history of literature implicating caveolae in the transport and homeostasis of cellular cholesterol (reviewed in Martin and Parton, 2005), the reason why cholesterol is required for caveola formation and cavin localisation is not clear. A direct cavin–cholesterol interaction is unlikely to be the explanantion because cavins do not bind significantly to cholesterol-containing membranes in vitro (Kovtun et al., 2014) and do not contain obvious hydrophobic stretches that are required for insertion into the acyl layer of the lipid membrane to associate with cholesterol –as seen in bona fide cholesterol-interacting proteins (Epand et al., 2010). We suggest that caveola flattening and cavin disassembly following cholesterol depletion are likely to reflect a more general perturbation of the physical organisation of the lipid bilayer within the caveolar domain, potentially resulting from perturbed caveolin–cholesterol interactions (Hulce et al., 2013; Murata et al., 1995).
The role of cavins in membrane remodelling
When heterologously expressed in bacteria, membrane-embedded caveolins have an inherent ability to generate spherical particles with a morphology that is similar to mammalian caveolae (Walser et al., 2012). CAV1 overexpression induces tubule formation in cell models and this activity can be suppressed by Cavin1 (Verma et al., 2010). Nevertheless, caveolins cannot by themselves generate caveolae in mammalian cells, probably because of differences in the biophysical properties of the membrane and an increased energetic cost of membrane bending. A recent review by Stachowiak and colleagues (Stachowiak et al., 2013) provides an excellent description of the principles that lead to increased energetic cost upon bending of more complex biological membranes. Instead, the cavins and, in particular Cavin1, are required to stabilise the invagination of caveolar membrane domains in vivo. In fact, like caveolins, cavins alone are capable of remodelling membranes to generate extended tubular membrane structures, both when overexpressed in mammalian cells (Hansen et al., 2009), and when purified cavins and synthetic PtdSer–PtdIns(4,5)P2-containing liposomes are used in vitro (Kovtun et al., 2014). This places the cavins firmly within the broader family of membrane curvature-inducing molecules, such as BAR-domain- and ENTH domain-containing peripheral membrane proteins. However, despite the individual abilities of both caveolins and cavins to shape membrane structure, the formation of morphological caveolae in vivo requires that both caveolin and cavin proteins act in concert. This suggests that key factors, either proteins, lipids and/or biophysical properties of the plasma membrane in mammalian cells, present a barrier to caveola generation, and surmounting this barrier requires the coordinated assembly of the cavin-caveolin coat.
A model for the role of cavins in caveolar assembly
Many types of vesicular coat, such as the COPI, COPII and clathrin machineries, have been relatively well characterised in terms of their protein and membrane requirements, structural organisation and mechanism of assembly (Faini et al., 2013; McMahon and Mills, 2004). In general, proteins including small GTPases promote recruitment and nucleation, adaptors interact with membrane lipids and transmembrane ‘cargos’, specialised proteins promote membrane remodelling and curvature, and polymeric scaffolding proteins are coupled to the adaptors to coordinate membrane invagination. In contrast, the process of caveola formation appears to involve quite different and still poorly understood mechanisms. In this final section, we attempt to summarise and combine the known biochemical, spatial, temporal and structural features of cavins presented above to propose a testable model for their role in caveolae formation (Fig. 3).
Caveolin oligomerisation and insertion in the membrane occurs in early exocytic compartments of the cell, and results in caveolin oligomers being directed to the plasma membrane independently of caveola formation and interaction with cavins (Hayer et al., 2010). Caveolin export to the plasmalemma also relies on lipids, in particular cholesterol (Pol et al., 2005). The peripheral cavin coat complex is believed to assemble in the cytoplasm prior to association with caveolin on the plasma membrane, and observations in vitro and in cells suggest that cavin oligomerisation is autonomous and results in the formation of multimers that contain on average ∼50 monomers (Gambin et al., 2014), similar to the number of cavin molecules present in caveolae (Gambin et al., 2014; Ludwig et al., 2013). The exact mechanism of cavin oligomerisation and recruitment is not yet fully clear but, as outlined above, some key principles are now emerging. First, the cavin proteins form homo- and heterotrimers through the association of their coiled-coil structures in the N-terminal HR1 domain (Kovtun et al., 2014). Second, there is a specific stoichiometry of the cavin–cavin association, with heterotrimers consisting of Cavin1 and Cavin2, and Cavin1 and Cavin3 containing two copies of Cavin1 and one copy of either Cavin2 or Cavin3 (Cavin1–Cavin1–Cavin2 or Cavin1–Cavin1–Cavin3, respectively). Third, the formation of Cavin2–Cavin3 complexes is strictly prohibited, thereby dictating their segregation into biochemically and morphologically separate subcomplexes on the surface of caveolae (Gambin et al., 2014; Kovtun et al., 2014; Ludwig et al., 2013). Last, cavin trimerisation induces the formation of α-helical secondary structures within the HR2 domain and this coincides with oligomerisation of the cavins, whereby the final step of cavin oligomersaion requires the disordered N- and C-terminal regions DR1 and DR3. Overall, this suggests a high degree of cooperativity is involved in the assembly of functional cavin complexes. As a consequence, cavin ratios in the caveolar coat are likely to be defined by relative expression levels of each cavin, thus highlighting the importance of upstream control processes – such as transcriptional regulation, processing, transport and protein degradation – to regulate cavins and, ultimately, exert control over caveola abundance and function.
Based on biochemical analyses of isolated recombinant cavins, they are most likely to exist in an equilibrium between a cytoplasmic and membrane-associated state, dependent on the presence of plasma-membrane-enriched phospholipids, such as phosphatidylserine (PtdSer) and PtdIns(4,5)P2 (Kovtun et al., 2014). Both cavins and caveolins possess intrinsic abilities to reshape membrane structure (Hansen et al., 2009; Kovtun et al., 2014; Walser et al., 2012) but, in what remains perhaps the most poorly understood aspect of caveolar biogenesis, cavins must cooperate with the membrane-embedded caveolins to form the typical invaginated, spherically shaped buds. There is some evidence for direct cavin–caveolin association that may play a role in this process (Mohan et al., 2015), although we have been unable to detect a stoichiometric association between purified cavin and caveolin proteins (our unpublished data), which argues against a direct protein–protein interaction as the primary driver of cavin recruitment to caveolae.
We propose that the formation of a cooperative network of protein and lipid interactions is the most important factor in the dual cavin–caveolin requirement for caveola formation. Caveolae have long been suggested to be important in binding to various membrane lipids and regulating their organisation in microdomains, including cholesterol, PtdSer and PtdIns(4,5)P2 (Ariotti et al., 2014; Fairn et al., 2011; Fujita et al., 2009; Murata et al., 1995; Parton and del Pozo, 2013; Pike and Casey, 1996; Yang et al., 2014). The affinity of these lipids for caveolae is reciprocated in the sense that depletion of either cholesterol or PtdIns(4,5)P2 can, in turn, lead to loss of caveolae or dissociation of caveolar proteins (Breen et al., 2012; Rothberg et al., 1992; Simone et al., 2013). Liposome-based experiments suggest that the caveolin ‘scaffolding domain’, a 20-residue sequence of basic and hydrophobic amino acids, possesses an intrinsic ability to locally increase cholesterol, PtdSer and PtdIns(4,5)P2 to a concentration several fold higher than in the surrounding membrane (Wanaski et al., 2003). Thus, caveolin oligomers on the plasma membrane are likely to be enriched with PtdIns(4,5)P2 and PtdSer, and we hypothesise that this enrichment locally attracts membrane-sensing cavin proteins. This interaction will then initiate a positive feedback loop causing additional association of caveolin with the raft, therefore, further increasing PtdIns(4,5)P2 and PtdSer locally through binding of caveolin to membrane-binding stretches in cavins (i.e. HR1 and HR2). We imagine this to be a type of ‘electrostatic trapping’ of the cavin complex on the membrane. The resulting elevated concentration of phospholipids may also be required for localisation of other protein components, such as EHD2 (Simone et al., 2013), or enzymatic activities (Izumi et al., 1997). Phospholipid redistribution caused by caveola formation was also reported to modulate lipid-raft-associated Ras signalling (Ariotti et al., 2014). To accommodate the cavin complex at the membrane, the cooperative action of cavin and caveolin bends the membrane and stabilises the characteristic morphology of caveolae. The importance of cholesterol binding to caveolins to form caveolae is also clear (Breen et al., 2012; Murata et al., 1995) and, as mentioned above, we believe this is likely to reflect an important role in modulating the localised biophysical properties of the lipid bilayer to allow appropriate membrane bending by the cavin–caveolin coat to occur.
An intriguing question now is to understand how the rod-shaped cavin building blocks assemble at the membrane interface to create the characteristic spiral polymers. Although it remains highly speculative, we believe that at least part of the answer to this question lies in two general properties of the cavin proteins; their DRs allowing overall cavin flexibility and their highly conserved alternating electrostatic profile. Whereas basic surfaces within HR1 and HR2 can contact negatively charged phospholipid bilayers (see below), we envisage that the acidic DRs may, in turn, be able to recognise HRs of neighbouring cavins, in essence creating an electrostatically assembled lattice as illustrated in Fig. 3B. A probable consequence of such an arrangement is that membrane-associated cavin polymers would be highly elastic, whereas contacts between subunits may be quite dynamic locally and allow relatively flexible coats to form around the curved membrane. This would be in contrast with the highly rigid and tightly organised arrays that are formed by the polymeric clathrin coats. That such a flexibility, indeed, exists is at least partly supported by the fact that the spiral cavin coats can adopt both densely packed structures around the invaginated caveolae, as well as more loosely arranged structures – such as flattened caveolae observed following cholesterol depletion (Rothberg et al., 1992).
Many questions remain regarding how caveola formation is regulated. As cavins undergo extensive post-translation modification, they are likely to play an important role in the life cycle of caveolae and their response to stimuli, yet the roles of these modifications are essentially unknown. Furthermore, in addition to the assembly of caveolae, understanding the mechanisms that control the dynamic disassembly of caveolae, and the subsequent redistribution and turnover of cavin proteins in response to stimuli – such as cholesterol depletion (Breen et al., 2012; Liu and Pilch, 2008; Murata et al., 1995), signal transduction (Aboulaich et al., 2006), and membrane stress (Gambin et al., 2014; Sinha et al., 2011) – remains relatively unexplored. As a final comment, it is known that changes in microtubule and actin assembly can alter caveolar transport and stability (Hernandez et al., 2013; Mundy et al., 2002; Verma et al., 2010; Wickström et al., 2010) (reviewed in Parton and del Pozo, 2013). Furthermore, caveolae are morphologically associated with cytoskeletal structures (Richter et al., 2008); however, it remains to be addressed whether interactions with cytoskeletal proteins have a direct role in caveolar biogenesis.
Concluding remarks
Caveolae are crucial regulators of many cellular processes, possess a highly characteristic morphology and are defined by a relatively restricted set of core protein ingredients. And yet, despite their importance, the dynamic nature of caveolae and the inherently flexible structures of the core proteins have made it difficult to study the molecular principles that govern their formation. In stark contrast to clathrin- or COP-coated vesicles, where the structures of most of the primary constituents are now well defined, the underlying ultrastructure of caveolar vesicles is very poorly understood. The work highlighted here has begun to shed some light on the role of cavins in caveolar organisation. Yet, many issues still remain, such as the architecture of full-length cavins, understanding how cavins self-assemble at the membrane interface to form the distinctive spiral coats and, of course, how caveolins and cavins come together to generate invaginated membrane domains. Although obtaining new crystal structures of cavins and their complexes are an obvious priority, we suggest that combining these structures with in vitro reconstitution of the caveolar components and imaging approaches, for instance the rapidly improving EM techniques currently under development, may also provide answers to these fundamental questions of caveolae biology.
Acknowledgments
We thank Richard Lundmark (Umeå University, Sweden) for providing data on cavin–caveolin interactions prior to publication, now included in Mohan et al., 2015.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Funding
This work was supported by grants from the National Health and Medical Research Council of Australia (NHMRC) to R.G.P. [grant numbers APP569542 and APP1037320] and the Australian Research Council (ARC) to B.M.C. [grant number DP120101298] and partly by the ARC Centre of Excellence in Convergent Bio-Nano Science and Technology [project number CE140100036]. R.G.P. is supported by an NHMRC Senior Principal Research Fellowship [APP1058565] and BMC by an NHMRC Career Development Fellowship [APP1061574].
References
- Aboulaich N., Vainonen J. P., Strålfors P., Vener A. V. (2004). Vectorial proteomics reveal targeting, phosphorylation and specific fragmentation of polymerase I and transcript release factor (PTRF) at the surface of caveolae in human adipocytes. Biochem. J. 383, 237–248 10.1042/BJ20040647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aboulaich N., Ortegren U., Vener A. V., Strålfors P. (2006). Association and insulin regulated translocation of hormone-sensitive lipase with PTRF. Biochem. Biophys. Res. Commun. 350, 657–661 10.1016/j.bbrc.2006.09.094 [DOI] [PubMed] [Google Scholar]
- Aboulaich N., Chui P. C., Asara J. M., Flier J. S., Maratos-Flier E. (2011). Polymerase I and transcript release factor regulates lipolysis via a phosphorylation-dependent mechanism. Diabetes 60, 757–765 10.2337/db10-0744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonescu C. N., Aguet F., Danuser G., Schmid S. L. (2011). Phosphatidylinositol-(4,5)-bisphosphate regulates clathrin-coated pit initiation, stabilization, and size. Mol. Biol. Cell 22, 2588–2600 10.1091/mbc.E11-04-0362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariotti N., Fernández-Rojo M. A., Zhou Y., Hill M. M., Rodkey T. L., Inder K. L., Tanner L. B., Wenk M. R., Hancock J. F., Parton R. G. (2014). Caveolae regulate the nanoscale organization of the plasma membrane to remotely control Ras signaling. J. Cell Biol. 204, 777–792 10.1083/jcb.201307055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai L., Deng X., Li J., Wang M., Li Q., An W., A D., Cong Y. S. (2011). Regulation of cellular senescence by the essential caveolar component PTRF/Cavin-1. Cell Res. 21, 1088–1101 10.1038/cr.2011.56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastiani M., Parton R. G. (2010). Caveolae at a glance. J. Cell Sci. 123, 3831–3836 10.1242/jcs.070102 [DOI] [PubMed] [Google Scholar]
- Bastiani M., Liu L., Hill M. M., Jedrychowski M. P., Nixon S. J., Lo H. P., Abankwa D., Luetterforst R., Fernandez-Rojo M., Breen M. R. et al. (2009). MURC/Cavin-4 and cavin family members form tissue-specific caveolar complexes. J. Cell Biol. 185, 1259–1273 10.1083/jcb.200903053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breen M. R., Camps M., Carvalho-Simoes F., Zorzano A., Pilch P. F. (2012). Cholesterol depletion in adipocytes causes caveolae collapse concomitant with proteosomal degradation of cavin-2 in a switch-like fashion. PLoS ONE 7, e34516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgener R., Wolf M., Ganz T., Baggiolini M. (1990). Purification and characterization of a major phosphatidylserine-binding phosphoprotein from human platelets. Biochem. J. 269, 729–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhard P., Stetefeld J., Strelkov S. V. (2001). Coiled coils: a highly versatile protein folding motif. Trends Cell Biol. 11, 82–88 10.1016/S0962-8924(00)01898-5 [DOI] [PubMed] [Google Scholar]
- Chidlow J. H., Jr, Sessa W. C. (2010). Caveolae, caveolins, and cavins: complex control of cellular signalling and inflammation. Cardiovasc. Res. 86, 219–225 10.1093/cvr/cvq075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epand R. M., Thomas A., Brasseur R., Epand R. F. (2010). Cholesterol interaction with proteins that partition into membrane domains: an overview. Subcell. Biochem. 51, 253–278 10.1007/978-90-481-8622-8_9 [DOI] [PubMed] [Google Scholar]
- Faini M., Beck R., Wieland F. T., Briggs J. A. (2013). Vesicle coats: structure, function, and general principles of assembly. Trends Cell Biol. 23, 279–288 10.1016/j.tcb.2013.01.005 [DOI] [PubMed] [Google Scholar]
- Fairn G. D., Schieber N. L., Ariotti N., Murphy S., Kuerschner L., Webb R. I., Grinstein S., Parton R. G. (2011). High-resolution mapping reveals topologically distinct cellular pools of phosphatidylserine. J. Cell Biol. 194, 257–275 10.1083/jcb.201012028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita A., Cheng J., Tauchi-Sato K., Takenawa T., Fujimoto T. (2009). A distinct pool of phosphatidylinositol 4,5-bisphosphate in caveolae revealed by a nanoscale labeling technique. Proc. Natl. Acad. Sci. USA 106, 9256–9261 10.1073/pnas.0900216106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambin Y., Ariotti N., McMahon K. A., Bastiani M., Sierecki E., Kovtun O., Polinkovsky M. E., Magenau A., Jung W., Okano S. et al. (2014). Single-molecule analysis reveals self assembly and nanoscale segregation of two distinct cavin subcomplexes on caveolae. eLife 3, e01434 10.7554/eLife.01434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustincich S., Schneider C. (1993). Serum deprivation response gene is induced by serum starvation but not by contact inhibition. Cell Growth Differ. 4, 753–760. [PubMed] [Google Scholar]
- Gustincich S., Vatta P., Goruppi S., Wolf M., Saccone S., Della Valle G., Baggiolini M., Schneider C. (1999). The human serum deprivation response gene (SDPR) maps to 2q32-q33 and codes for a phosphatidylserine-binding protein. Genomics 57, 120–129 10.1006/geno.1998.5733 [DOI] [PubMed] [Google Scholar]
- Hailstones D., Sleer L. S., Parton R. G., Stanley K. K. (1998). Regulation of caveolin and caveolae by cholesterol in MDCK cells. J. Lipid Res. 39, 369–379. [PubMed] [Google Scholar]
- Hansen C. G., Nichols B. J. (2010). Exploring the caves: cavins, caveolins and caveolae. Trends Cell Biol. 20, 177–186 10.1016/j.tcb.2010.01.005 [DOI] [PubMed] [Google Scholar]
- Hansen C. G., Bright N. A., Howard G., Nichols B. J. (2009). SDPR induces membrane curvature and functions in the formation of caveolae. Nat. Cell Biol. 11, 807–814 10.1038/ncb1887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen C. G., Howard G., Nichols B. J. (2011). Pacsin 2 is recruited to caveolae and functions in caveolar biogenesis. J. Cell Sci. 124, 2777–2785 10.1242/jcs.084319 [DOI] [PubMed] [Google Scholar]
- Hansen C. G., Shvets E., Howard G., Riento K., Nichols B. J. (2013). Deletion of cavin genes reveals tissue-specific mechanisms for morphogenesis of endothelial caveolae. Nat. Commun. 4, 1831 10.1038/ncomms2808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi Y. K., Matsuda C., Ogawa M., Goto K., Tominaga K., Mitsuhashi S., Park Y. E., Nonaka I., Hino-Fukuyo N., Haginoya K. et al. (2009). Human PTRF mutations cause secondary deficiency of caveolins resulting in muscular dystrophy with generalized lipodystrophy. J. Clin. Invest. 119, 2623–2633 10.1172/JCI38660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayer A., Stoeber M., Bissig C., Helenius A. (2010). Biogenesis of caveolae: stepwise assembly of large caveolin and cavin complexes. Traffic 11, 361–382 10.1111/j.1600-0854.2009.01023.x [DOI] [PubMed] [Google Scholar]
- Henley J. R., Krueger E. W., Oswald B. J., McNiven M. A. (1998). Dynamin-mediated internalization of caveolae. J. Cell Biol. 141, 85–99 10.1083/jcb.141.1.85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez V. J., Weng J., Ly P., Pompey S., Dong H., Mishra L., Schwarz M., Anderson R. G., Michaely P. (2013). Cavin-3 dictates the balance between ERK and Akt signaling. eLife 2, e00905 10.7554/eLife.00905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill M. M., Bastiani M., Luetterforst R., Kirkham M., Kirkham A., Nixon S. J., Walser P., Abankwa D., Oorschot V. M., Martin S. et al. (2008). PTRF-Cavin, a conserved cytoplasmic protein required for caveola formation and function. Cell 132, 113–124 10.1016/j.cell.2007.11.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornbeck P. V., Kornhauser J. M., Tkachev S., Zhang B., Skrzypek E., Murray B., Latham V., Sullivan M. (2012). PhosphoSitePlus: a comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucleic Acids Res. 40, D261–D270 10.1093/nar/gkr1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulce J. J., Cognetta A. B., Niphakis M. J., Tully S. E., Cravatt B. F. (2013). Proteome-wide mapping of cholesterol-interacting proteins in mammalian cells. Nat. Methods 10, 259–264 10.1038/nmeth.2368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey S. J., Yang G., Yang P., Fazakerley D. J., Stöckli J., Yang J. Y., James D. E. (2013). Dynamic adipocyte phosphoproteome reveals that Akt directly regulates mTORC2. Cell Metab. 17, 1009–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi Y., Hirai S., Tamai Y., Fujise-Matsuoka A., Nishimura Y., Ohno S. (1997). A protein kinase Cdelta-binding protein SRBC whose expression is induced by serum starvation. J. Biol. Chem. 272, 7381–7389 10.1074/jbc.272.11.7381 [DOI] [PubMed] [Google Scholar]
- Jansa P., Mason S. W., Hoffmann-Rohrer U., Grummt I. (1998). Cloning and functional characterization of PTRF, a novel protein which induces dissociation of paused ternary transcription complexes. EMBO J. 17, 2855–2864 10.1093/emboj/17.10.2855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly B. T., Graham S. C., Liska N., Dannhauser P. N., Höning S., Ungewickell E. J., Owen D. J. (2014). Clathrin adaptors. AP2 controls clathrin polymerization with a membrane-activated switch. Science 345, 459–463 10.1126/science.1254836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkham M., Nixon S. J., Howes M. T., Abi-Rached L., Wakeham D. E., Hanzal-Bayer M., Ferguson C., Hill M. M., Fernandez-Rojo M., Brown D. A. et al. (2008). Evolutionary analysis and molecular dissection of caveola biogenesis. J. Cell Sci. 121, 2075–2086 10.1242/jcs.024588 [DOI] [PubMed] [Google Scholar]
- Kovtun O., Tillu V. A., Jung W., Leneva N., Ariotti N., Chaudhary N., Mandyam R. A., Ferguson C., Morgan G. P., Johnston W. A. et al. (2014). Structural insights into the organization of the cavin membrane coat complex. Dev. Cell 31, 405–419. [DOI] [PubMed] [Google Scholar]
- Krüger M., Kratchmarova I., Blagoev B., Tseng Y. H., Kahn C. R., Mann M. (2008). Dissection of the insulin signaling pathway via quantitative phosphoproteomics. Proc. Natl. Acad. Sci. USA 105, 2451–2456 10.1073/pnas.0711713105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurzchalia T. V., Dupree P., Parton R. G., Kellner R., Virta H., Lehnert M., Simons K. (1992). VIP21, a 21-kD membrane protein is an integral component of trans-Golgi-network-derived transport vesicles. J. Cell Biol. 118, 1003–1014 10.1083/jcb.118.5.1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Shi X., Guo X., Li H., Xu C. (2014). Ionic protein-lipid interaction at the plasma membrane: what can the charge do? Trends Biochem. Sci. 39, 130–140 10.1016/j.tibs.2014.01.002 [DOI] [PubMed] [Google Scholar]
- Liu L., Pilch P. F. (2008). A critical role of cavin (polymerase I and transcript release factor) in caveolae formation and organization. J. Biol. Chem. 283, 4314–4322 10.1074/jbc.M707890200 [DOI] [PubMed] [Google Scholar]
- Liu L., Brown D., McKee M., Lebrasseur N. K., Yang D., Albrecht K. H., Ravid K., Pilch P. F. (2008). Deletion of Cavin/PTRF causes global loss of caveolae, dyslipidemia, and glucose intolerance. Cell Metab. 8, 310–317 10.1016/j.cmet.2008.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig A., Howard G., Mendoza-Topaz C., Deerinck T., Mackey M., Sandin S., Ellisman M. H., Nichols B. J. (2013). Molecular composition and ultrastructure of the caveolar coat complex. PLoS Biol. 11, e1001640 10.1371/journal.pbio.1001640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin S., Parton R. G. (2005). Caveolin, cholesterol, and lipid bodies. Semin. Cell Dev. Biol. 16, 163–174 10.1016/j.semcdb.2005.01.007 [DOI] [PubMed] [Google Scholar]
- McLaughlin S., Wang J., Gambhir A., Murray D. (2002). PIP(2) and proteins: interactions, organization, and information flow. Annu. Rev. Biophys. Biomol. Struct. 31, 151–175 10.1146/annurev.biophys.31.082901.134259 [DOI] [PubMed] [Google Scholar]
- McMahon H. T., Mills I. G. (2004). COP and clathrin-coated vesicle budding: different pathways, common approaches. Curr. Opin. Cell Biol. 16, 379–391 10.1016/j.ceb.2004.06.009 [DOI] [PubMed] [Google Scholar]
- McMahon K. A., Zajicek H., Li W. P., Peyton M. J., Minna J. D., Hernandez V. J., Luby-Phelps K., Anderson R. G. (2009). SRBC/cavin-3 is a caveolin adapter protein that regulates caveolae function. EMBO J. 28, 1001–1015 10.1038/emboj.2009.46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mineo C., Ying Y. S., Chapline C., Jaken S., Anderson R. G. (1998). Targeting of protein kinase Calpha to caveolae. J. Cell Biol. 141, 601–610 10.1083/jcb.141.3.601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohan J., Morén B., Larsson E., Holst M., Lundmark R. (2015). Cavin3 interacts with cavin1 and caveolin1 to increase surface dynamics of caveolae. J. Cell Sci 10.1242/jcs.161463 [DOI] [PubMed] [Google Scholar]
- Morén B., Shah C., Howes M. T., Schieber N. L., McMahon H. T., Parton R. G., Daumke O., Lundmark R. (2012). EHD2 regulates caveolar dynamics via ATP-driven targeting and oligomerization. Mol. Biol. Cell 23, 1316–1329 10.1091/mbc.E11-09-0787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mundy D. I., Machleidt T., Ying Y. S., Anderson R. G., Bloom G. S. (2002). Dual control of caveolar membrane traffic by microtubules and the actin cytoskeleton. J. Cell Sci. 115, 4327–4339 10.1242/jcs.00117 [DOI] [PubMed] [Google Scholar]
- Murata M., Peränen J., Schreiner R., Wieland F., Kurzchalia T. V., Simons K. (1995). VIP21/caveolin is a cholesterol-binding protein. Proc. Natl. Acad. Sci. USA 92, 10339–10343 10.1073/pnas.92.22.10339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata T., Ueyama T., Isodono K., Tagawa M., Takehara N., Kawashima T., Harada K., Takahashi T., Shioi T., Matsubara H. et al. (2008). MURC, a muscle-restricted coiled-coil protein that modulates the Rho/ROCK pathway, induces cardiac dysfunction and conduction disturbance. Mol. Cell. Biol. 28, 3424–3436 10.1128/MCB.02186-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortegren U., Karlsson M., Blazic N., Blomqvist M., Nystrom F. H., Gustavsson J., Fredman P., Strålfors P. (2004). Lipids and glycosphingolipids in caveolae and surrounding plasma membrane of primary rat adipocytes. Eur. J. Biochem. 271, 2028–2036 10.1111/j.1432-1033.2004.04117.x [DOI] [PubMed] [Google Scholar]
- Palade G. E. (1953). Fine structure of blood capillaries. J. Appl. Phys. 24, 1424. [Google Scholar]
- Parton R. G., del Pozo M. A. (2013). Caveolae as plasma membrane sensors, protectors and organizers. Nat. Rev. Mol. Cell Biol. 14, 98–112 10.1038/nrm3512 [DOI] [PubMed] [Google Scholar]
- Pelkmans L., Zerial M. (2005). Kinase-regulated quantal assemblies and kiss-and-run recycling of caveolae. Nature 436, 128–133 10.1038/nature03866 [DOI] [PubMed] [Google Scholar]
- Pelkmans L., Püntener D., Helenius A. (2002). Local actin polymerization and dynamin recruitment in SV40-induced internalization of caveolae. Science 296, 535–539 10.1126/science.1069784 [DOI] [PubMed] [Google Scholar]
- Peters K. R., Carley W. W., Palade G. E. (1985). Endothelial plasmalemmal vesicles have a characteristic striped bipolar surface structure. J. Cell Biol. 101, 2233–2238 10.1083/jcb.101.6.2233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pike L. J., Casey L. (1996). Localization and turnover of phosphatidylinositol 4,5-bisphosphate in caveolin-enriched membrane domains. J. Biol. Chem. 271, 26453–26456 10.1074/jbc.271.43.26453 [DOI] [PubMed] [Google Scholar]
- Pol A., Martin S., Fernández M. A., Ingelmo-Torres M., Ferguson C., Enrich C., Parton R. G. (2005). Cholesterol and fatty acids regulate dynamic caveolin trafficking through the Golgi complex and between the cell surface and lipid bodies. Mol. Biol. Cell 16, 2091–2105 10.1091/mbc.E04-08-0737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter T., Floetenmeyer M., Ferguson C., Galea J., Goh J., Lindsay M. R., Morgan G. P., Marsh B. J., Parton R. G. (2008). High-resolution 3D quantitative analysis of caveolar ultrastructure and caveola-cytoskeleton interactions. Traffic 9, 893–909 10.1111/j.1600-0854.2008.00733.x [DOI] [PubMed] [Google Scholar]
- Rothberg K. G., Heuser J. E., Donzell W. C., Ying Y. S., Glenney J. R., Anderson R. G. (1992). Caveolin, a protein component of caveolae membrane coats. Cell 68, 673–682 10.1016/0092-8674(92)90143-Z [DOI] [PubMed] [Google Scholar]
- Scherer P. E., Okamoto T., Chun M., Nishimoto I., Lodish H. F., Lisanti M. P. (1996). Identification, sequence, and expression of caveolin-2 defines a caveolin gene family. Proc. Natl. Acad. Sci. USA 93, 131–135 10.1073/pnas.93.1.131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senju Y., Itoh Y., Takano K., Hamada S., Suetsugu S. (2011). Essential role of PACSIN2/syndapin-II in caveolae membrane sculpting. J. Cell Sci. 124, 2032–2040 10.1242/jcs.086264 [DOI] [PubMed] [Google Scholar]
- Shvets E., Ludwig A., Nichols B. J. (2014). News from the caves: update on the structure and function of caveolae. Curr. Opin. Cell Biol. 29, 99–106 10.1016/j.ceb.2014.04.011 [DOI] [PubMed] [Google Scholar]
- Simone L. C., Caplan S., Naslavsky N. (2013). Role of phosphatidylinositol 4,5-bisphosphate in regulating EHD2 plasma membrane localization. PLoS ONE 8, e74519 10.1371/journal.pone.0074519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha B., Köster D., Ruez R., Gonnord P., Bastiani M., Abankwa D., Stan R. V., Butler-Browne G., Vedie B., Johannes L. et al. (2011). Cells respond to mechanical stress by rapid disassembly of caveolae. Cell 144, 402–413 10.1016/j.cell.2010.12.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stachowiak J. C., Brodsky F. M., Miller E. A. (2013). A cost-benefit analysis of the physical mechanisms of membrane curvature. Nat. Cell Biol. 15, 1019–1027 10.1038/ncb2832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stetefeld J., Jenny M., Schulthess T., Landwehr R., Engel J., Kammerer R. A. (2000). Crystal structure of a naturally occurring parallel right-handed coiled coil tetramer. Nat. Struct. Biol. 7, 772–776 10.1038/79006 [DOI] [PubMed] [Google Scholar]
- Verma P., Ostermeyer-Fay A. G., Brown D. A. (2010). Caveolin-1 induces formation of membrane tubules that sense actomyosin tension and are inhibited by polymerase I and transcript release factor/cavin-1. Mol. Biol. Cell 21, 2226–2240 10.1091/mbc.E09-05-0417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinten J., Voldstedlund M., Clausen H., Christiansen K., Carlsen J., Tranum-Jensen J. (2001). A 60-kDa protein abundant in adipocyte caveolae. Cell Tissue Res. 305, 99–106 10.1007/s004410100389 [DOI] [PubMed] [Google Scholar]
- Vinten J., Johnsen A. H., Roepstorff P., Harpøth J., Tranum-Jensen J. (2005). Identification of a major protein on the cytosolic face of caveolae. Biochim. Biophys. Acta 1717, 34–40 10.1016/j.bbamem.2005.09.013 [DOI] [PubMed] [Google Scholar]
- Walser P. J., Ariotti N., Howes M., Ferguson C., Webb R., Schwudke D., Leneva N., Cho K. J., Cooper L., Rae J. et al. (2012). Constitutive formation of caveolae in a bacterium. Cell 150, 752–763. [DOI] [PubMed] [Google Scholar]
- Wanaski S. P., Ng B. K., Glaser M. (2003). Caveolin scaffolding region and the membrane binding region of SRC form lateral membrane domains. Biochemistry 42, 42–56 10.1021/bi012097n [DOI] [PubMed] [Google Scholar]
- Way M., Parton R. G. (1995). M-caveolin, a muscle-specific caveolin-related protein. FEBS Lett. 376, 108–112 10.1016/0014-5793(95)01256-7 [DOI] [PubMed] [Google Scholar]
- Wickström S. A., Lange A., Hess M. W., Polleux J., Spatz J. P., Krüger M., Pfaller K., Lambacher A., Bloch W., Mann M. et al. (2010). Integrin-linked kinase controls microtubule dynamics required for plasma membrane targeting of caveolae. Dev. Cell 19, 574–588 10.1016/j.devcel.2010.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams J. J., Palmer T. M. (2014). Cavin-1: caveolae-dependent signalling and cardiovascular disease. Biochem. Soc. Trans. 42, 284–288 10.1042/BST20130270 [DOI] [PubMed] [Google Scholar]
- Yamada E. (1955). The fine structure of the gall bladder epithelium of the mouse. J. Biophys. Biochem. Cytol. 1, 445–458 10.1083/jcb.1.5.445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang G., Xu H., Li Z., Li F. (2014). Interactions of caveolin-1 scaffolding and intramembrane regions containing a CRAC motif with cholesterol in lipid bilayers. Biochim. Biophys. Acta 1838, 2588–2599 10.1016/j.bbamem.2014.06.018 [DOI] [PubMed] [Google Scholar]
- Yao Y., Hong S., Zhou H., Yuan T., Zeng R., Liao K. (2009). The differential protein and lipid compositions of noncaveolar lipid microdomains and caveolae. Cell Res. 19, 497–506 10.1038/cr.2009.27 [DOI] [PubMed] [Google Scholar]