Abstract
Extended-spectrum beta-lactamase (ESBL)-producing Enterobacteriaceae have been isolated from humans and animals across the world. However, data on prevalence of ESBL-producing Enterobacteriaceae from rural water reservoirs is limited. This study aimed to isolate and characterize ESBL-producing Enterobacteriaceae in rural water reservoirs in Guantao, China. ESBL-producing Enterobacteriaceae were found in 5 (16.7%) of 30 sampled rural water reservoirs. Sixty-six individual isolates expressing an ESBL phenotype were obtained in the present study. Species identification showed that 42 representatives of Escherichia coli, 17 Klebsiella pneumoniae, 4 Raoultella planticola, and 3 Enterobacter cloacae. Twenty isolates contained a single bla gene, including CTX-M (17 strains), TEM (2 strains), and SHV (1 strain). Forty-six isolates contained more than one type of beta-lactamase genes. ESBL-producing Enterobacteriaceae isolated in this study were all multidrug resistant. These findings indicated that the serious contamination of ESBL-producing Enterobacteriaceae in rural water reservoirs existed in Guantao, China.
Keywords: ESBL, Enterobacteriaceae, rural water reservoirs, multidrug resistance, bla genes
Introduction
The rational use of antibiotics helps control infectious diseases of humans and animals. Abuse and overuse of antibiotics in clinical practice has selected drug resistant bacteria and “superbugs” (Thompson et al., 2007; Yong et al., 2009; Pruden et al., 2012). Extended-spectrum beta-lactamases (ESBLs), resulting from amino acid substitutions in TEM-1, TEM-2, and SHV-1 enzymes were described in the 1980s and 1990s (Bush and Jacoby, 2010). ESBLs can hydrolyze penicillins, oxyimino-cephalosporins (e.g., cefotaxime, ceftazidime, ceftriaxone, cefuroxime, cefepime) and aztreonam but not cephamycins (e.g., cefoxitin, cefotetan) or carbapenems (Bush and Jacoby, 2010; El Salabi et al., 2013). ESBLs are predominantly found among Enterobacteriaceae, which are inhabitants of intestinal flora and important pathogens in nosocomial and community settings (Laurent et al., 2008; Azap et al., 2010; Song et al., 2011; Kang et al., 2013).
Extended-spectrum beta-lactamase-producing Enterobacteriaceae can spread between humans via contaminated food or water (Oteo et al., 2010; Piednoir et al., 2011) and acquire resistance to antibiotics by plasmids, transposons or other mobile vectors that carry resistance elements (Oteo et al., 2010; Peirano et al., 2012). Water environments are considered as important reservoirs for resistance genes (Gao et al., 2009), and maybe play an important role in transfer of drug-resistant genes between bacteria (Malakoff, 2002; Kummerer, 2004). More importantly, once ESBL-producing Enterobacteriaceae enter the intestine of humans and animals via drinking water, these bacteria could lead to the spread of resistance genes and to serious infections.
To date, numerous studies on ESBL-producing Enterobacteriaceae isolated from water environments have focused on wastewaters of hospitals and animal farms, and waters from rivers and lakes (Cabello et al., 2013; Varela and Manaia, 2013; Yang et al., 2013; Zurfluh et al., 2013; Haque et al., 2014). However, data on ESBL-producing Enterobacteriaceae isolated from drinking water in rural areas is very limited. In China, the main drinking sources for rural residents in many villages are water reservoirs. Therefore, the present study was conducted to describe the isolation and characterization of ESBL-producing Enterobacteriaceae in rural water reservoirs in Guantao, China.
Materials and Methods
Sampling Sites and Water Sample Collection
Between July and September of 2013, water sampling was conducted in Guantao, China (Figures 1 and 2). Five samples each were collected at six locations for a total of 30 samples. The water samples were collected from 50 cm below the water surface using sterile bottles (100 ml/bottle, one bottle/each reservoir). The collected water samples were stored on ice and immediately transported to our lab for further analyses within 3 h.
FIGURE 1.

The rural water reservoir commonly found in Guantao villages.
FIGURE 2.
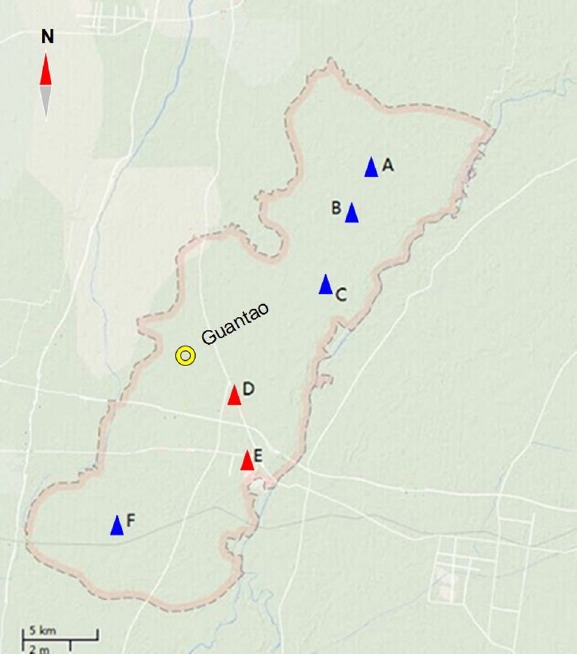
Sampling sites. Blue triangles (A–C,F) represent the communities where no ESBL-producing Enterobacteriaceae were detected in water samples. Red triangles (D, E) represent the communities where ESBL-producing Enterobacteriaceae were found in water samples. Yellow circle represents urban areas of Guantao County.
Microbiological Analysis
Hundred milliliters of water was filtrated through a sterile 0.45 μm membrane (Millipore, Billerica, MA, USA), and then the filters were incubated in 20 ml of enterobacteria enrichment (EE) Broth (Becton Dickinson, Heidelberg, Germany) at 37°C for 24 h. One loopful of enrichment cultures was spread onto chromogenic Brilliance ESBL agar (Oxoid, Hampshire, UK) and incubated at 37°C for 24 h. The colonies with different color and morphology were picked and sub-cultured on sheep blood agar for 24 h at 37°C (Huang et al., 2010). Conventional biochemical methods and API ID 32 E (bioMérieux, Marcy l’Etoile, France) were used to identify the isolates. If isolates showed doubtful results, they were subjected to genetic identification based on sequencing of rpoB gene fragments (Mollet et al., 1997).
Antimicrobial Susceptibility Testing and ESBL Confirmation
According to the protocols of the Clinical and Laboratory Standards Institute (CLSI, 2011), the disk diffusion method was used to test susceptibility of the isolates against 17 antimicrobial agents. The tested antibiotics were: ampicillin (AMP), cefaclor (CEC), cefazolin (CFZ), cefepime (FEP), cefotaxime (CTX), ceftazidime (CAZ), ceftriaxone (CRO), cefuroxime (CXM), aztreonam (AZT), ciprofloxacin (CIP), gentamicin (GEN), imipenem (IPM), ofloxacin (OFX), piperacillin (PIP), amikacin (AMK), chloramphenicol (CHL) and tetracycline (TET). According to the manufacturer’s protocols, Etest-ESBL strips (bioMérieux, Marcy l’Etoile, France) were used to confirm ESBL production. Isolates showing resistance to three or more antibiotic classes were defined as multidrug resistant (MDR). E. coli ATCC 25922 and K. pneumoniae ATCC 700603 were used as quality control strains.
Polymerase Chain Reaction (PCR) to Detect bla Genes
The DNA of the isolates confirmed for producing ESBLs was extracted separately using a DNA extraction kit (Biospin plasmid extraction, Bioflux, Japan). According to previously published work, PCR was used to detect blaTEM, blaCTX-M, and blaSHV genes using specific primers (Chen et al., 2010).
Results
Detection of ESBL-Producing Enterobacteriaceae
Extended-spectrum beta-lactamase-producing Enterobacteriaceae were detected in five water reservoirs of two rural communities (D: 2, E: 3). The five water reservoirs were all located close to chicken farms (approximately 12–15 m). No ESBL-producing Enterobacteriaceae were found in the other reservoirs, which were far away from rural villages and animal farms (approximately 1.0–1.5 km).
Sixty-six different isolates exhibiting an ESBL phenotype were obtained (D: 28, E: 38). The results of species identification showed that 42 E. coli, 17 K. pneumoniae, 4 Raoultella planticola, and 3 Enterobacter cloacae (Table 1).
TABLE 1.
Species composition of ESBL-producing Enterobacteriaceae from water samples.
| Species composition | Number | Percent (%) | Sources D/E |
|---|---|---|---|
| E. coli | 42 | 63.6 | 18/24 |
| K. pneumoniae | 17 | 25.8 | 7/10 |
| Raoultella planticola | 4 | 6.1 | 2/2 |
| Enterobacter cloacae | 3 | 4.5 | 1/2 |
| Total | 66 | 100.0 | 28/38 |
Antibiotic Susceptibility of ESBL-producing Enterobacteriaceae
Extended-spectrum beta-lactamase-producing Enterobacteriaceae isolates displayed similar drug-resistant trends. Nearly all ESBL-producing Enterobacteriaceae were resistant to the first- and second-generation cephalosporins (cefazolin, cefaclor, cefuroxime). These isolates were also resistant to the third-generation cephalosporins: cefotaxime (91.5%), ceftriaxone (67.9%), and ceftazidime (31.1%). Moreover, 48.1% of the isolates were resistant to cefepime (the fourth-generation cephalosporin), 51.9% to aztreonam (a monocyclic β-lactam antibiotic), and 89.6% to ampicillin.
Extended-spectrum beta-lactamase-producing Enterobacteriaceae were resistant to non-β-lactam antibiotics: resistant to ciprofloxacin (78.3%), gentamicin (60.4%), ofloxacin (76.4%), piperacillin (68.9%), chloramphenicol (55.7%), and tetracycline (65.1%). But the majority of ESBL-producing Enterobacteriaceae isolates were susceptible to amikacin (95.3%) and imipenem (97.2%) (Figure 3).
FIGURE 3.
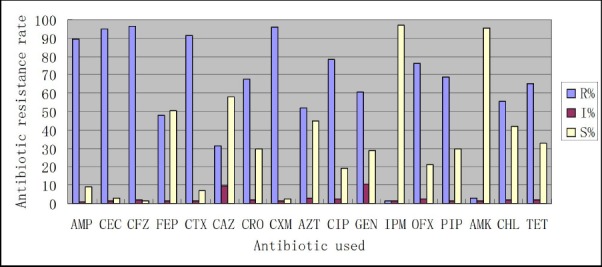
Antibiotic resistance rates of ESBL-producing Enterobacteriaceae. R: Resistance; I: intermediate; S: susceptible; AMP, ampicillin; CEC, cefaclor; CFZ, cefazolin; FEP, cefepime; CTX, cefotaxime; CAZ, ceftazidime; CRO, ceftriaxone; CXM, cefuroxime; AZT, aztreonam; CIP, ciprofloxacin; GEN, gentamicin; IPM, imipenem; OFX, ofloxacin; PIP, piperacillin; AMK, amikacin; CHL, chloramphenicol; TET, tetracycline.
Characterization of bla Genes in ESBL-producing Enterobacteriaceae
All 66 ESBL-producing Enterobacteriaceae carried bla genes. Among 66 ESBL-carriers, 20 strains carried only one bla gene (20/66, 30.3%), including 17 carrying blaCTX-M (17/66, 25.8%), 2 isolates carrying blaTEM (2/66, 3.0%), and 1 carrying blaSHV (1/66, 1.5%). The other 46 isolates carried at least two bla genes (46/66, 69.7%), including 38 carrying blaTEM+CTX-M (38/66, 57.6%), 6 carrying blaSHV+CTX-M (6/66, 9.1%), and the other 2 carrying three bla genes (2/66, 3.0%) (Table 2).
TABLE 2.
bla gene types of ESBL-producing Enterobacteriaceae from water samples.
| bla gene types | Number | Percent (%) | Sources (D/E) |
|---|---|---|---|
| TEM | 2 | 3.0 | 1/1 |
| CTX-M | 17 | 25.8 | 6/11 |
| SHV | 1 | 1.5 | 0/1 |
| TEM+CTX-M | 38 | 57.6 | 18/20 |
| SHV+CTX-M | 6 | 9.1 | 2/4 |
| TEM+SHV+CTX-M | 2 | 3.0 | 1/1 |
| Total | 66 | 100.0 | 28/38 |
Discussion
In this study, ESBL-producing Enterobacteriaceae were detected in five out of 30 rural reservoirs (5/30, 16.7%), but not found in the other water reservoirs far away from villages and animal farms. This suggests that ESBL-producing Enterobacteriaceae are being shed into reservoirs from animal farms and anthropogenic activities.
Extended-spectrum beta-lactamase-producing Enterobacteriaceae isolates in this study were MDR. These isolates showed high resistance against the third-generation cephalosporins: cefotaxime (91.5%), and ceftriaxone (67.9%). Importantly, 48.1% of these isolates were resistant to cefepime, the fourth-generation cephalosporin. In addition, antibiotics resistance rates of these isolates to non-β-lactam antibiotics were also worrisome. But 95.3% and 97.2% of these isolates were respectively susceptible to amikacin and imipenem, which may be related with relatively low use of these medicines in this region.
In mainland China, previous investigations about ESBL-producing Enterobacteriaceae in water bodies and food-producing animals showed that blaCTX-M gene was the dominant ESBL producer (Jin and Ling, 2006; Chen et al., 2010; Rao et al., 2014). Our data also identified blaCTX-M gene as an important ESBL producer. Additionally, 46 out of 66 ESBL-producing Enterobacteriaceae isolates carried at least two bla genes and blaCTX-M+TEM has become the dominant phenotype of ESBL, which was different from the previous result in clinical isolates (Zhang et al., 2014).
There were some limitations in this study: water sampling was carried out only in 30 rural water reservoirs, so the results may not be representative of the whole area; sequence analyses of bla genes encoding TEM, CTX-M, and SHV were not further conducted to identify variants of these types of enzymes; the risk factors associated with rural reservoir water carriage of ESBL-producing Enterobacteriaceae were not further analyzed.
In summary, these findings indicated that the contamination of ESBL-producing Enterobacteriaceae in rural water environments existed in Guantao, China, and the pollution may be closely related to local animal farms and anthropogenic activities.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Azap O. K., Arslan H., Serefhanoğlu K., Colakoğlu S., Erdoğan H., Timurkaynak F., et al. (2010). Risk factors for extended-spectrum β-lactamase positivity in uropathogenic Escherichia coli isolated from community-acquired urinary tract infections. Clin. Microbiol. Infect. 16, 147–151. 10.1111/j.1469-0691.2009.02941.x [DOI] [PubMed] [Google Scholar]
- Bush K., Jacoby G. A. (2010). Updated functional classification of β-lactamases. Antimicrob. Agents Chemother. 54, 969–976. 10.1128/AAC.01009-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabello F. C., Godfrey H. P., Tomova A., Ivanova L., Dölz H., Millanao A., et al. (2013). Antimicrobial use in aquaculture re-examined: its relevance to antimicrobial resistance and to animal and human health. Environ. Microbiol. 15, 1917–1942. 10.1111/1462-2920.12134 [DOI] [PubMed] [Google Scholar]
- Chen H., Shu W. Q., Chang X. S., Chen J. A., Guo Y. B., Tan Y. (2010). The profile of antibiotics resistance and integrons of extended-spectrum β-lactamase producing thermotolerant coliforms isolated from the Yangtze River basin in Chongqing. Environ. Pollut. 158, 2459–2464. 10.1016/j.envpol.2010.03.023 [DOI] [PubMed] [Google Scholar]
- Clinical and Laboratory Standards Institute (CLSI). (2011). “Performance Standards for Antimicrobial Susceptibility Testing: Twenty-first Informational Supplement. Document M100-S21s. Wayne, PA: Clinical and Laboratory Standards Institute. [Google Scholar]
- El Salabi A., Walsh T. R., Chouchani C. (2013). Extended spectrum β-lactamases, carbapenemases and mobile genetic elements responsible for antibiotics resistance in Gram-negative bacteria. Crit. Rev. Microbiol. 39, 113–122. 10.3109/1040841X.2012.691870 [DOI] [PubMed] [Google Scholar]
- Gao P. P., Luo Y., Zhou Q. X., Mao D. Q. (2009). Research advancement of antibiotics resistance genes in aquaculture environment. Asian J. Ecotoxicol. 4, 770–779. [Google Scholar]
- Haque A., Yoshizumi A., Saga T., Ishii Y., Tateda K. (2014). ESBL-producing Enterobacteriaceae in environmental water in Dhaka, Bangladesh. J. Infect. Chemother. 20, 735–737. 10.1016/j.jiac.2014.07.003 [DOI] [PubMed] [Google Scholar]
- Huang T. D., Bogaerts P., Berhin C., Guisset A., Glupczynski Y. (2010). Evaluation of Brilliance ESBL agar, a novel chromogenic medium for detection of extended-spectrum-β-lactamase-producing Enterobacteriaceae. J. Clin. Microbiol. 48, 2091–2096. 10.1128/JCM.02342-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y., Ling J. M. (2006). CTX-M-producing Salmonella spp. in Hong Kong: an emerging problem. J. Med. Microbiol. 55, 1245–1250. 10.1099/jmm.0.46637-0 [DOI] [PubMed] [Google Scholar]
- Kang C. I., Wi Y. M., Ko K. S., Chung D. R., Peck K. R., Lee N. Y., et al. (2013). Outcomes and risk factors for mortality in community-onset bacteremia caused by extended-spectrum β-lactamase-producing Escherichia coli, with a special emphasis on antimicrobial therapy. Scand. J. Infect. Dis. 45, 519–525. 10.3109/00365548.2013.775479 [DOI] [PubMed] [Google Scholar]
- Kummerer K. (2004). Resistance in the environment. J. Antimicrob. Chemother. 54, 311–320. 10.1093/jac/dkh325 [DOI] [PubMed] [Google Scholar]
- Laurent C., Rodriguez-Villalobos H., Rost F., Strale H., Vincent J. L., Deplano A., et al. (2008). Intensive care unit outbreak of extended-spectrum β-lactamase–producing Klebsiella pneumoniae controlled by cohorting patients and reinforcing infection control Measures. Infect. Control Hosp. Epidemiol. 29, 517–524. 10.1086/588004 [DOI] [PubMed] [Google Scholar]
- Malakoff D. (2002). Microbiologists on the trail of polluting bacteria. Science 29, 2352–2353. 10.1126/science.295.5564.2352 [DOI] [PubMed] [Google Scholar]
- Mollet C., Drancourt M., Raoult D. (1997). rpoB sequence analysis as a novel basis for bacterial identification. Mol. Microbiol. 26, 1005–10011. 10.1046/j.1365-2958.1997.6382009.x [DOI] [PubMed] [Google Scholar]
- Oteo J., Perez-Vazquez M., Campos J. (2010). Extended-spectrum β-lactamase-producing Escherichia coli: changing epidemiology and clinical impact. Curr. Opin. Infect. Dis. 23, 320–326. 10.1097/QCO.0b013e3283398dc1 [DOI] [PubMed] [Google Scholar]
- Peirano G., van der Bij A. K., Gregson D. B., Pitout J. D. (2012). Molecular epidemiology over an 11-year period (2000–2010) of extended-spectrum β-lactamase-producing Escherichia coli causing bacteremia in a centralized Canadian region. J. Clin. Microbiol. 50, 294–299. 10.1128/JCM.06025-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piednoir E., Thibon P., Borderan G. C., Godde F., Borgey F., Le Coutour X., et al. (2011). Long-term clinical and economic benefits associated with the management of a nosocomial outbreak resulting from extended-spectrum β-lactamase-producing Klebsiella pneumoniae. Crit. Care Med. 39, 2672–2677. 10.1097/CCM.0b013e31822827e0 [DOI] [PubMed] [Google Scholar]
- Pruden A., Arabi M., Storteboom H. N. (2012). Correlation between upstream human activities and riverine antibiotic resistance genes. Environ. Sci. Technol. 46, 11541–11549. 10.1021/es302657r [DOI] [PubMed] [Google Scholar]
- Rao L., Lv L., Zeng Z., Chen S., He D., Chen X., et al. (2014). Increasing prevalence of extended-spectrum cephalosporin-resistant Escherichia coli in food animals and the diversity of CTX-M genotypes during 2003–2012. Vet. Microbiol. 172, 534–541. 10.1016/j.vetmic.2014.06.013 [DOI] [PubMed] [Google Scholar]
- Song J. H., Thamlikitkul V., Hsueh P. R. (2011). Clinical and economic burden of community-acquired pneumonia amongst adults in the Asia-Pacific region. Int. J. Antimicrob. Agents 38, 108–117. 10.1016/j.ijantimicag.2011.02.017 [DOI] [PubMed] [Google Scholar]
- Thompson S. A., Maani E. V., Lindell A. H., King C. J., McArthur J. V. (2007). Novel tetracycline resistance determinant isolated from an environmental strain of Serratia marcescens. Appl. Environ. Microbiol. 73, 2199–2206. 10.1128/AEM.02511-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varela A. R., Manaia C. M. (2013). Human health implications of clinically relevant bacteria in wastewater habitats. Environ. Sci. Pollut. Res. Int. 20, 3550–3569. 10.1007/s11356-013-1594-0 [DOI] [PubMed] [Google Scholar]
- Yang F. X., Mao D. Q., Luo Y., Wang Q., Mu Q. H. (2013). Horizontal transfer of antibiotic resistance genes in the environment. Chin. J. Appl. Ecol. 24, 2993–3002. [PubMed] [Google Scholar]
- Yong D., Toleman M. A., Giske C. G., Cho H. S., Sundman K., Lee K., et al. (2009). Characterization of a new metallo-beta-lactamase gene, b/a(NDM–1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob. Agents Chemother. 53, 5046–5054. 10.1128/AAC.00774-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Zheng B., Zhao L., Wei Z., Ji J., Li L., et al. (2014). Nationwide high prevalence of CTX-M and an increase of CTX-M-55 in Escherichia coli isolated from patients with community-onset infections in Chinese county hospitals. BMC Infect. Dis. 14:659–669. 10.1186/s12879-014-0659-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurfluh K., Hächler H., Nüesch-Inderbinen M., Stephan R. (2013). Characteristics of extended-spectrum β-lactamase (ESBL)- and carbapenemase-producing Enterobacteriaceae isolated from rivers and lakes in Switzerland. Appl. Environ. Microbiol. 79, 3021–3026. 10.1128/AEM.00054-13 [DOI] [PMC free article] [PubMed] [Google Scholar]


