Abstract
Summary: CytoCom is an interactive plugin for Cytoscape that can be used to search, explore, analyse and visualize human disease comorbidity network. It represents disease–disease associations in terms of bipartite graphs and provides International Classification of Diseases, Ninth Revision (ICD9)-centric and disease name centric views of disease information. It allows users to find associations between diseases based on the two measures: Relative Risk (RR) and -correlation values. In the disease network, the size of each node is based on the prevalence of that disease. CytoCom is capable of clustering disease network based on the ICD9 disease category. It provides user-friendly access that facilitates exploration of human diseases, and finds additional associated diseases by double-clicking a node in the existing network. Additional comorbid diseases are then connected to the existing network. It is able to assist users for interpretation and exploration of the human diseases by a variety of built-in functions. Moreover, CytoCom permits multi-colouring of disease nodes according to standard disease classification for expedient visualization.
Availability and implementation: CytoCom is compatible with CytoScape 3.1.0 or later version. Please visit http://www.cl.cam.ac.uk/∼mam211/ for user tutorial and download.
Contact: Mohammad.Moni@cl.cam.ac.uk
1 Introduction
The term ‘comorbidity’ refers to the coexistence or presence of multiple diseases or disorders in relation to a primary disease or disorder in a patient (Moni and Liò, 2014). A comorbidity relationship between two diseases exists whenever they appear simultaneously in a patient more than chance alone (Hidalgo et al., 2009). It represents the co-occurrence of diseases or presence of different medical conditions one after another in the same patient (Moni and Liò, 2014; Hidalgo et al., 2009). Two diseases are connected if they are co-expressed in a significant number of patients in a population (Hidalgo et al., 2009). If two diseases have associated comorbidity, the occurrence of one of them in a patient may increase the likelihood of developing another disease.
The main challenge for research in the biomedical field is to understand the associations between human diseases. Clinical data contains many important clarifications regarding the occurrence of comorbidity in patients with two or more diseases. To measure a relationship based on disease co-occurrence, we need to quantify the strength of the comorbidity risk. We have adapted two comorbidity measures to quantify the strength of comorbidity associations between two diseases: (i) the Relative Risk - the probability that a number of patients will be diagnosed with both diseases relative to the probability that a number of patients will be similarly diagnosed according to random expectation based on disease prevalence, as the quantified measures of comorbidity tendency of two disease pairs; and (ii) -correlation - Pearsons correlation for binary variables, to measure the robustness of the comorbidity association.
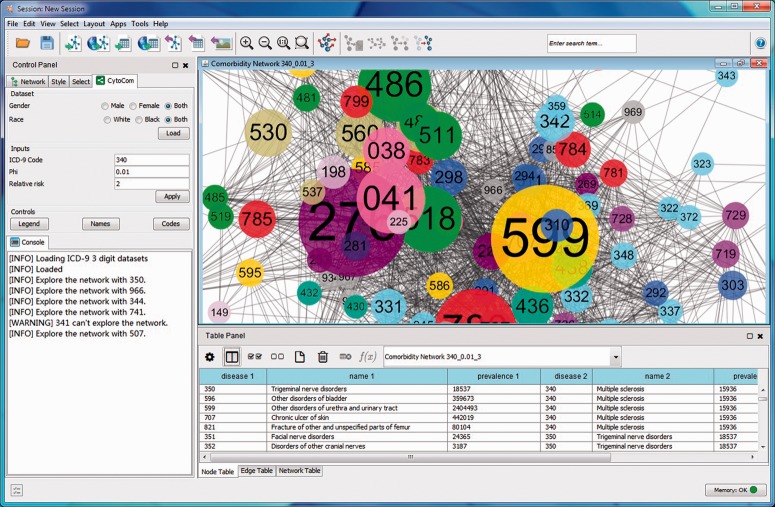
Cytoscape is a widely used Java-based, open-source platform for network visualization and analysis (Shannon et al., 2003). The Cytoscape framework is extendable through the implementation of apps. We have developed a Cytoscape app, named CytoCom, that enables a user to visualize the disease–disease network. To the best of our knowledge, CytoCom is the first app to provide a user with the facility to query, analyse and visualize disease–disease dynamic network using clinical information. CytoCom can be started from the control panel in Cytoscape. The main panel is shown in Figure 1.
Fig. 1.
Cytoscape screenshot of CytoCom. The centre of the screen shows an example of DCN, which is constructed based on clinical data. Each node represents a unique disease; the nodes are coloured according to their disease categories so that diseases of the same category have the same colour. Two nodes are considered to be linked if the diseases are significantly comorbid more than the randomly expected according to their specified RR and values
2 Overview
2.1 Network data sources
Patient medical records offer reliable data on diseases. To achieve a comprehensive study, we identify the comorbidity correlations of diseases based on the clinical information from the US Medicare claims database, http://www.icd9data.com, and collected from Hidalgo et al. (2009). Demographically, our dataset consists of the medical records of 13,039,018 patients in the ICD-9-CM format. International Classification of Diseases, Ninth Revision codes are 3–5 digits long and have hierarchical structures, with each 4-digit code being a subset of a 5-digit code, both of which also contain a further 3-digit code subset. In total, the ICD-9-CM includes 657 different categories at the 3-digit level. Each digit span for all diseases is then further classified based on gender and race.
2.2 Network construction
To understand the correlations between diseases, the disease–disease pair comorbidity correlations are visualized as a bipartite network graph. Two diseases are connected through an edge if they are associated. CytoCom constructs disease comorbidity network (DCN) in which nodes are diseases and edges indicate the comorbidity associations among them. CytoCom includes edges between disease pairs for which the co-occurrence is significantly greater than the random expectation based on the population prevalence of the diseases. For a pair of diseases i and j, we used two novel statistical measures to quantify the relationship between two diseases: Relative Risk (RRij) and -correlation () based on the Hidalgo et al. (2009). The two comorbidity measures are not completely independent each other. The RRij allows us to quantify the co-occurrence of disease pairs compared with the random expectation and allows us to measure the robustness of the comorbidity associations. Users can use integer or decimal numbers as input for both RRij and . When two diseases co-occur more frequently than expected by chance, we will get RRij > 1 and . The significance of the RRij is calculated using the Katz et al. method to estimate confidence intervals (Katz et al. 1978). To explore a particular disease, user needs to enter the seed disease code in the search box; CytoCom makes use of Cytoscape to construct the network and create a network view of disease association. The user can filter network using different input parameters based on gender, race and ICD-9 codes.
2.3 Network visualization
In the ICD-9 classification, diseases are assigned to different disease categories based on the first three digits of a given ICD-9 code. This classification is represented by a specific colouring of the nodes. The size of the nodes is visualized according to the prevalence of the diseases. More common diseases are represented as larger nodes. Additional associated diseases could be found by double clicking of a disease node of the existing network, which enables to access more association or comorbidity information of that disease. The additional diseases network is added to the existing network dynamically. Herein, the multi-colouring of disease nodes offers a convenient visualization of disease classifications in the networks. In addition, it offers a user-friendly means of conducting a visual exploration. CytoCom automatically creates a database table of mapping ICD-9 codes to disease names. It is also possible to export the network as an image or its data as a.txt or.CSV file (please see more details in user manual).
3 Conclusion
Exploring the associations between diseases could greatly enhance our understanding of pathogenesis and eventually lead to better diagnosis and treatment. CytoCom is an easy-to-use app to analyse and investigate the human DCN. This app will help users to advance their knowledge on disease mechanism and disease comorbidities in a quantitative and graphical way. We plan regular updates to the CytoCom app as well as the integration of further data sources.
References
- Hidalgo C.A., et al. (2009) A dynamic network approach for the study of human phenotypes. PLoS computational biology, 5, e1000353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz D., et al. (1978) Obtaining confidence intervals for the risk ratio in cohort studies. Biometrics, 34, 469–474. [Google Scholar]
- Moni M.A., Liò P. (2014) comoR: a software for disease comorbidity risk assessment. Journal of Clinical Bioinformatics, 4, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P., et al. (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]