Abstract
Olfactomedin (OLF) domain-containing proteins play roles in fundamental cellular processes and have been implicated in disorders ranging from glaucoma, cancers and inflammatory bowel disorder, to attention deficit disorder and childhood obesity. We solved crystal structures of the OLF domain of myocilin (myoc-OLF), the best studied such domain to date. Mutations in myoc-OLF are causative in the autosomal dominant inherited form of the prevalent ocular disorder glaucoma. The structures reveal a new addition to the small family of five-bladed β-propellers. Propellers are most well known for their ability to act as hubs for protein–protein interactions, a function that seems most likely for myoc-OLF, but they can also act as enzymes. A calcium ion, sodium ion and glycerol molecule were identified within a central hydrophilic cavity that is accessible via movements of surface loop residues. By mapping familial glaucoma-associated lesions onto the myoc-OLF structure, three regions sensitive to aggregation have been identified, with direct applicability to differentiating between neutral and disease-causing non-synonymous mutations documented in the human population worldwide. Evolutionary analysis mapped onto the myoc-OLF structure reveals conserved and divergent regions for possible overlapping and distinctive functional protein–protein or protein–ligand interactions across the broader OLF domain family. While deciphering the specific normal biological functions, ligands and binding partners for OLF domains will likely continue to be a challenging long-term experimental pursuit, atomic detail structural knowledge of myoc-OLF is a valuable guide for understanding the implications of glaucoma-associated mutations and will help focus future studies of this biomedically important domain family.
Introduction
The olfactomedin (OLF) domain, first identified as part of an eponymous multidomain protein involved in olfactory chemoreception in bullfrogs >20 years ago (1,2), is now recognized as a large domain family (PFAM: PF02191) comprising seven phylogenetic branches (3) (Supplementary Material, Table S1). OLFs characterized to date are found largely as part of extracellular proteins (2,4–7) in multicellular organisms. They are implicated in an expansive and growing number of human disorders including a host of cancers (8), inflammatory bowel disorder (9), defense against infection (10), attention deficit hyperactivity disorder (11) and childhood obesity (12), among others.
Myocilin, the first to be associated with disease and the best studied OLF domain-containing protein, is one of the few veritable disease-modifying drug targets for glaucoma (13), the second leading cause of blindness worldwide (14). Non-synonymous lesions localized almost exclusively to its OLF domain (myoc-OLF) lead to an early-onset, autosomal-dominant inherited form of the most prevalent subtype, primary open angle glaucoma (15), and account for ∼3–10% of such patients (13). Nearly 100 myoc-OLF variants have been documented in individuals, through a combination of familial and population glaucoma case–control genetic studies (16). The leading proposed pathogenic mechanism involves a gain of toxic function, namely aggregation, in the region of the anterior eye called the trabecular meshwork (TM). In cell culture, disease-causing myocilin missense variants are all similarly prone to aggregation in the endoplasmic reticulum (ER) (17), leading to ER stress (18–23) and cell death (17,20). This cascade is presumed to lead to reduced TM-associated fluid outflow and accelerated onset of ocular hypertension, the major risk factor for vision loss and glaucoma. Although the ER chaperone machinery should be capable of efficient degradation of such mutant proteins, there appears to be an aberrant interaction between mutant myocilin and chaperones that subverts proper retrotranslocation for proteasomal degradation (19). Studies of purified recombinant myoc-OLF variants in vitro demonstrate that while wild-type myoc-OLF is a well-folded monomer, single glaucomatous point mutations, such as P370L, can be sufficient to impair folding to the extent that only insoluble aggregates are isolated (24). More generally, thermal stabilities of folded, disease-associated variants, measured by the melting temperature (Tm), are statistically correlated with the age the associated patients were diagnosed with glaucoma (24). Myoc-OLF aggregates exhibit hallmarks of amyloid (25,26) and appear either as long straight fibrils or an atypical large-diameter circular species. These morphologies are similar to aggregates formed by two well-conserved peptide stretches within myoc-OLF, P1 (G326AVVYSGSLYFQ) and P3 (V426ANAFIICGTLYTVSSY), respectively (25), suggesting that a propensity to misfold might be an inherent property of OLF domains. Finally, high levels of wild-type myocilin are associated with steroid-induced glaucoma, a secondary disease subtype (27). Although not directly linked to pathogenesis, myocilin expression is stimulated by glucocorticoids in TM cells (27), which, as seen with disease variants, may lead to aggregation and involve ER stress (28,29).
Like other disease-associated proteins that exhibit a toxic gain-of-function misfolding phenotype, studies of overexpressed wild-type or disease-associated missense variants of myocilin do not have an obvious connection to normal biological function(s), which remains poorly defined. Whereas mice expressing mutant myocilin display symptoms of glaucoma (30,31), myocilin knock out mice do not (32). Recent studies reveal that although there is no major systemic phenotype, myocilin null mice do exhibit structural changes in the retina (33), optic nerve (34), as well as in bone (35), and in the peripheral nervous system (36), suggesting a loss of function. Predominantly, but not exclusively, extracellular locations have been proposed for myocilin (8), and evidence suggests distinct functions in each region, including a regulatory role in signaling (37), cell proliferation and survival (33,38). Interacting partners identified to date have localized primarily to the myocilin N-terminal coiled-coil region (13). Individual studies have identified flotilin-1 (39), a membrane-bound protein involved in vesicular trafficking, and optimedin (40), an extracellular protein, as myoc-OLF-specific interacting partners. The implications of these interactions for myocilin function, however, remain unclear (39).
Here we report three crystal structures of myoc-OLF, revealing a new addition to the five-bladed β-propeller structural protein class. Propellers are known for their involvement in protein–protein interactions, and analysis of the structure reveals potential binding sites for protein–protein and/or ligand–protein interactions. Evolutionary considerations indicate that OLF domains share a common scaffold but likely interact with distinct surface binding partners. The myoc-OLF structure provides a three-dimensional framework from which to understand the molecular basis of glaucoma-associated misfolding, and differentiate between neutral and disease-causing non-synonymous mutations found in the human population. Our study offers new insight into and lays the groundwork for understanding pathogenesis and biology of this enigmatic yet important domain family.
Results
Overall architecture
The OLF crystal structure was solved by anomalous phasing using seleno-methionine (SeMet)-substituted myoc-OLF(E396D) (myoc-OLF(E396D)P21), a single nucleotide polymorphism (SNP) variant that grew superior quality crystals to those of wild type. Molecular replacement was then used to obtain the high resolution structure at 1.9 Å resolution of native myoc-OLF(E396D) (myoc-OLF(E396D)I222) and the 2.1 Å resolution structure of wild-type myoc-OLF (see Materials and Methods and Table 1). The final models include residues 244–502 (numbering scheme for full-length myocilin). The N-terminal 16 residues of myoc-OLF, which complete the C-terminal fragment of myocilin reported upon proteolytic cleavage in cell culture (41), are not visible in the structures, likely because they are part of a linker between the OLF domain and N-terminal regions of full-length myocilin.
Table 1.
Crystallographic statistics
| Myoc-OLF(E396D)P21 (SeMet, PDB code 4WXU) | Myoc-OLF(E396D)I222 (highest resolution, PDB code 4WXS) | Wild-type Myoc-OLF (PDB code 4WXQ) | |
|---|---|---|---|
| Data collection | |||
| Wavelength (Å) | 0.979370 | 1.000000 | 1.033160 |
| Space group | P1211 | I222 | I222 |
| Cell dimensions | |||
| a, b, c (Å) | 49.48, 50.56, 50.54 | 68.88, 86.65, 87.44 | 68.66, 85.82, 88.44 |
| α, β, γ (°) | 90, 97.27, 90 | 90, 90, 90 | 90, 90, 90 |
| Resolution (Å) | 19.6–2.09 (2.17–2.09) | 19.79–1.9 (1.97–1.9) | 19.78–2.15 (2.23–2.15) |
| Rmerge | 0.1174 (0.4071) | 0.0884 (0.4386) | 0.1025 (0.4144) |
| I/σI | 25.23 (5.15) | 15.52 (4.21) | 15.54 (5.47) |
| Completeness (%) | 97.22 (71.65) | 99.98 (100.00) | 99.93 (99.93) |
| Redundancy | 13.9 (5.5) | 6.2 (6.3) | 7.3 (7.4) |
| Refinement | |||
| Resolution (Å) | 19.6–2.09 | 19.79–1.9 | 19.78–2.15 |
| No. of reflections | 200 752 (5952) | 130 265 (12 954) | 106 349 (10 383) |
| Rwork/Rfree | 0.1782/0.2014 | 0.1724/0.2098 | 0.1934/0.2306 |
| No. of atoms | |||
| Protein | 2079 | 2072 | 2065 |
| Ligand/ion | 36 | 40 | 66 |
| Water | 244 | 304 | 138 |
| B-factors | |||
| Protein | 11.10 | 21.10 | 24.10 |
| Ligand/ion | 20.10 | 37.10 | 34.50 |
| Water | 28.50 | 37.30 | 31.00 |
| Root mean squared deviations | |||
| Bond lengths (Å) | 0.005 | 0.005 | 0.002 |
| Bond angles (°) | 0.83 | 0.78 | 0.62 |
| Ramachandran favored and additional allowed (%) | 100 | 100 | 100 |
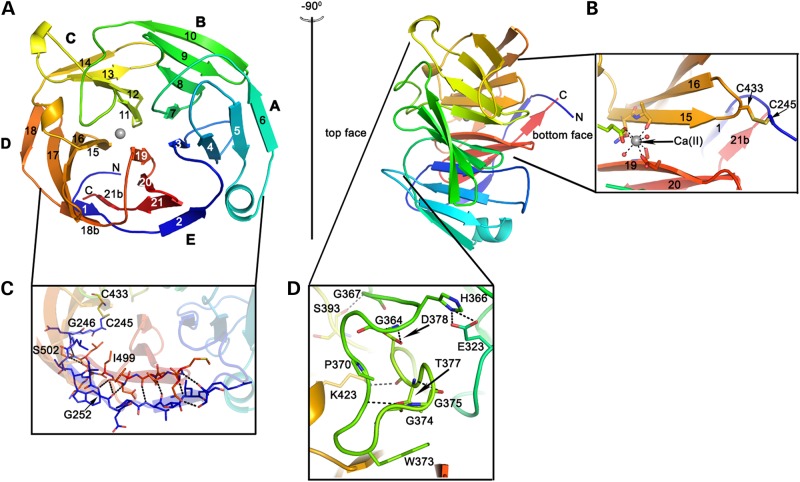
The OLF domain is an ∼40 Å diameter by ∼30 Å high β-propeller with five blades, each composed of four antiparallel β-strands (Fig. 1), arranged radially around a central water-filled cavity ∼11 Å in diameter (Supplementary Material, Fig. S1). Compared with other known propellers of varying blade number (42), the blades in OLF are notably asymmetric (see also comparison of five-bladed propellers below). Nearly half of the toroid-shaped molecule is occupied by Blades D and E. Discontinuous Strands D-18/D-18b and E-21/E-21b in the myoc-OLF(E396D) structures (Fig. 1a) appear as single continuous strands in the wild-type myoc-OLF structure (not shown). The electron density in this region is well defined in all structures, but for clarity, in our subsequent descriptions, we refer to the blades as labeled in Figure 1 for our highest resolution structure, that of myoc-OLF(E396D)I222. Other structural features include several helical turns and a short α-helix. The α-helix resides between myoc-OLF Strands A-5 and A-6, and packs against the E–A blade interface (Fig. 1a). Despite the lack of sequence similarity in this region (Supplementary Material, Fig. S2), the α-helix is predicted by secondary structure prediction algorithms (not shown) and is likely a general feature of the OLF domain.
Figure 1.
Structural features of the OLF domain. (A) Top and side view of representative myoc-OLF propeller. Blades A–E with corresponding numbered strands, listed according to myoc-OLF(E396D)I222 structure. (B) Internal calcium-binding site and disulfide bond at bottom face. (C) Molecular clasp region. (D) Loop B-10/C-11, comprising residues 360–379. Dashed lines represent relevant stabilizing interactions. Coloring: N (blue) to C (red).
Two features of OLF stabilize the propeller in a closed circular conformation. First, extensive interactions from the sequentially discontinuous two outer strands of Blade E, Strands E-1 to E-2 and E-21 (Fig. 1c), form the propeller feature colloquially termed ‘Velcro’ or ‘molecular clasp’ (43). Second, a single disulfide bond is found at the bottom face of the propeller (Fig. 1b), between Cys 245 at the N-terminus of the visible structural domain prior to the start of E-1, and Cys 433, located within Loop D-15/D-16 between the internal two strands of Blade D. A similar disulfide bond is seen in four-bladed propellers (43,44) but not five-bladed propeller structures. The disulfide bond is likely present in most OLF domains, as the C-terminal Cys is highly conserved, and in sequence alignments (Supplementary Material, Fig. S2), there is typically an N-terminal cysteine near the equivalent position of myoc-OLF Cys 245.
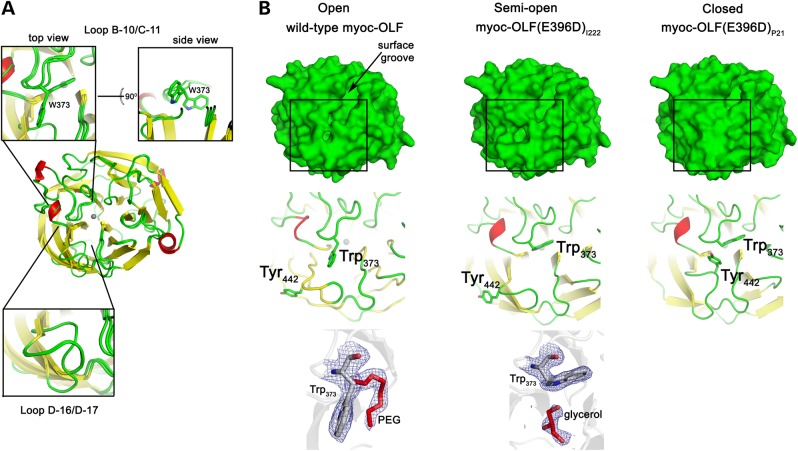
Nearly half of the residues are found within well-ordered loops above and below the propeller (Figs 1a and 2a), with minor differences among the solved structures (Fig. 2a). The longest loop is composed of residues 360–379 (Fig. 1d), connects Strands B-10 and C-11, and caps the top entrance to the central cavity (Supplementary Material, Fig. S1). Although no global changes are observed among our structures, Loop B-10/C-11 appears to serve as a gate for access to the central cavity via the positioning of the side chains of myoc-OLF Trp 373 and Tyr 442, which resides on Loop D-16/D-17. In total, three states were captured that are not attributable to crystal contacts (Fig. 2a and b). The side chain of myoc-OLF residue Trp 373 is visible in two conformations in our structures, one pointed into the central cavity and the other out towards the solvent. The ‘open’ conformation, seen in the wild-type myoc-OLF structure, has Trp 373 and Tyr 442 pointing outward towards the bulk solvent, with a polyethylene glycol (PEG) molecule bound just beneath the loop at the top of the cavity entrance. In the ‘semi open’ conformation, seen in myoc-OLF(E396D)I222, Trp 373 is nestled inward, but Tyr 442 on the opposite loop still points toward the solvent. This shift enlarges a small surface cavity where a bound glycerol was identified in a position distinct from the aforementioned PEG. Such PEG and glycerol molecules found fortuitously in our structures reflect sites likely to promote ligand or protein–protein interactions (45). Finally, the ‘closed’ state is represented by myoc-OLF(E396D)P21, whereby the position of the indole ring of Trp 373 is replaced with the phenol side chain of Tyr 442. This shifts the Tyr-containing loop into an inward conformation significantly different from that seen in the open conformations (Fig. 2a and b).
Figure 2.
OLF molecular surface. (A) Superposition of wild-type myoc-OLF and two myoc-OLF(E396D) structures. Yellow, β-strand; red, α-helix or turn; green, loop. Zoomed regions highlight where loop structures differ. (B) Top face surface representation and open–closed Loop B-10/C-11 snapshots observed crystallographically. Cartoon representations of boxed region appear below each surface representation as well as observed ligands in the corresponding structure (PEG or glycerol, red) with final 2Fo − Fc electron density contoured at 1σ.
Central cavity metal ion and other identified ligand-binding sites
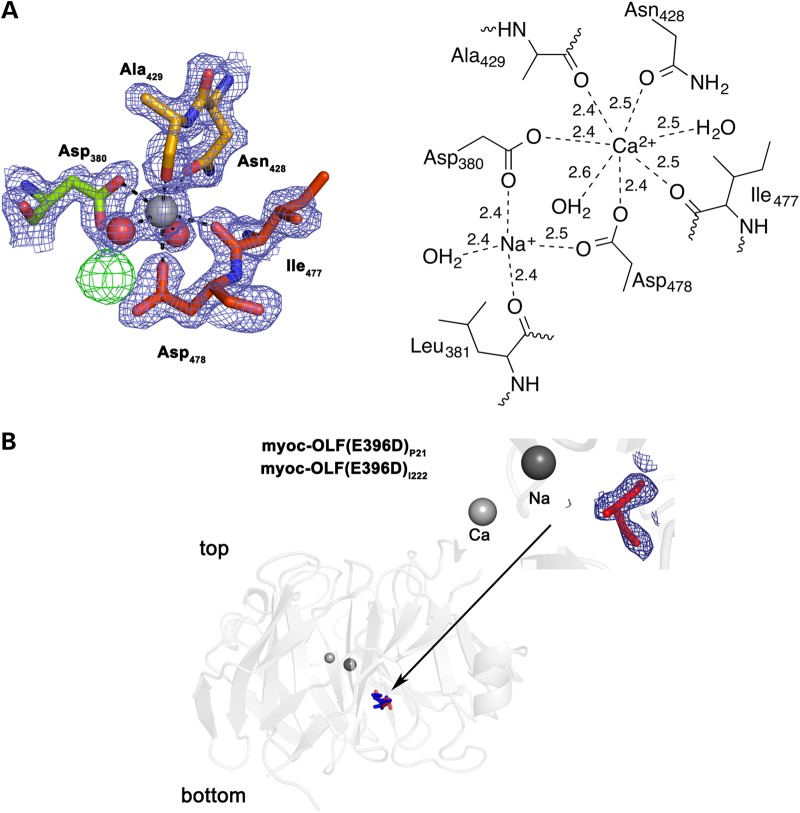
Within the central hydrophilic cavity of myoc-OLF, below the aforementioned gating loops, resides the calcium ion identified previously by metal analysis (46). The ion was modeled into a 25σ peak in the Fo− Fc difference electron density map present after initial model building. The heptacoordinate Ca2+ is ligated by the side chains of Asp 380, Asn 428 and Asp 478, all in a monodentate fashion, as well as the carbonyl backbones of Ala 429 and Ile 477, and two water molecules (Fig. 3a). The structure confirms Asp 380, the only previously identified Ca2+ ligand (46). The ligand environment in myoc-OLF is typical for Ca2+ (47), but not in a documented Ca2+ binding motif. Consistent with the previous finding that the apo disease variant D380A is a moderately stable protein (46), the internal position of the myoc-OLF Ca2+ ion appears as an ionic tether for Blades C, D and E (Fig. 1a) to confer stability. Adjacent to the Ca2+ site, at a distance of ∼3.4 Å deeper into the cavity toward the bottom face, a second major Fo − Fc difference electron density peak was identified (Fig. 3a). In our structures, based on final refined metal–ligand distances (48), this density is modeled as a Na+ ion coordinated by Asp 380 and Asp 478, as well as the carbonyl backbone of Leu 381 and a water molecule (Fig. 3a). Lastly, within the hydrophilic cavity in both closed and semi-open states of myoc-OLF(E396D), a glycerol molecule is bound, in the region between Strands A-3 and B-7 above the Ca2+ site (Fig. 3b). Glycerol was not introduced during crystallization of wild-type myoc-OLF and is thus not observed in that structure.
Figure 3.
Details of hydrophilic cavity. (A) Ca2+ site and coordination details. Final 2Fo −Fc electron density (blue) is contoured at 1.5σ and difference Fo −Fc density (green) depicting crystallographic Na+ site is contoured at 3σ (see text). Distances are listed in Å. (B) View of observed glycerol located in the internal hydrophilic cavity, with final 2Fo − Fc electron density for myoc-OLF(E396D)I222 contoured at 1σ.
Comparison to known five-bladed β-propellers
Myoc-OLF is distinct from the four different proteins with known five-bladed propeller folds (Supplementary Material, Fig. S3), with root mean squared deviations > 3 Å, consistent with sequence analyses that confirm OLF domains are not phylogenetically related to these propellers (<5–9% identity scattered throughout the sequence, not shown). Five-bladed propellers are primarily associated with glycosyl hydrolase or cyclotransferase activity; other five-bladed propellers include a human apyrase (49) and the highly symmetric founding member, tachylectin-2, which binds carbohydrates (42). The five tachylectin-2 binding sites, located between connecting segments of one sheet and the outer two strands of a second adjacent sheet (50), are absent in OLF (Supplementary Material, Fig. S3a). Superposition of myoc-OLF with arabinase (51) or glutaminyl cyclase (52) reveals that the only potential catalytic residue is the unlikely myoc-OLF Ca2+ ligand Asp 380 (Supplementary Material, Fig. S3b,c). While glutaminyl cyclase harbors a Ca2+ binding site, this ion is not part of the glutaminyl cyclase active site (52); arabinase has a His-ligated Cl− ion ∼3 Å away with no obvious relation to the myoc-OLF Ca2+ site. The surface substrate-binding cleft of arabinase is likewise not observed in OLF and clashes with Loop B-10/C-11 (Supplementary Material, Fig. S3b). Similar differences with apyrase are observed (Supplementary Material, Fig. S3d). In sum, the OLF domain expands the diversity of sequentially unrelated β-propellers (42) and is a new representative template for five-bladed propellers. However, as such, the ligand binding and active sites of these five-bladed propellers do not directly shed light on the specific function for the OLF domain (see Discussion).
Surface electrostatics
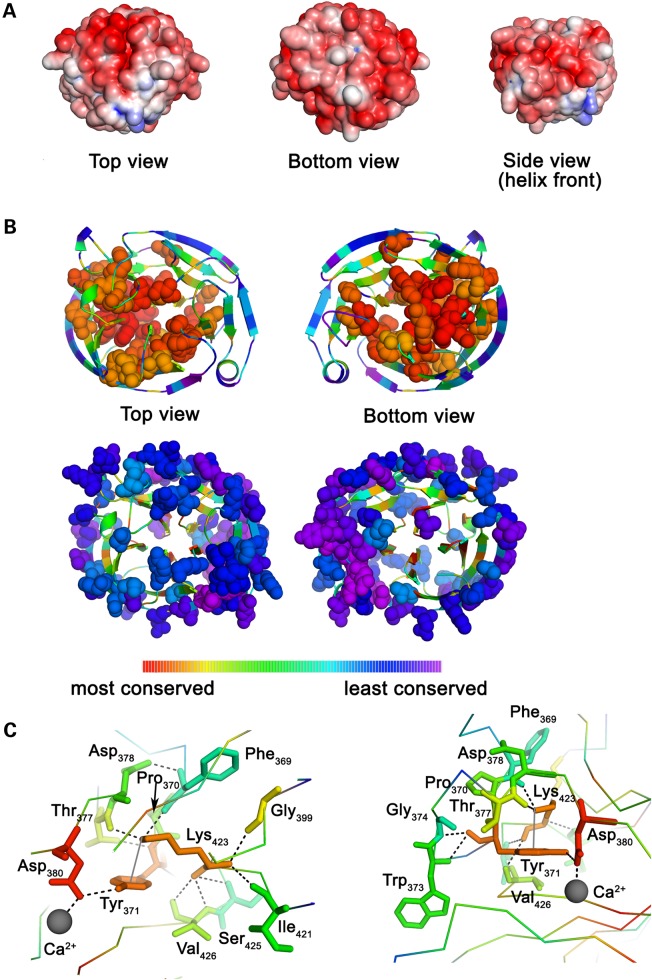
Inspection of the electrostatic surface potential (Fig. 4a) correlates with the predicted pI ∼5. The top surface of myoc-OLF contains several charged regions. There is a cluster of five positively charged surface Lys and Arg residues, in the loops connecting D-18/E-19 and E-20/E-21. A second positively charged surface on this face is in the loop that connects Strand C-14/D-15, which exposes two adjacent Lys/Arg residues at the helical turn. The main acidic patch on the top surface is formed by loops of Blade B, including those that connect to Blades A and C. At the bottom face is another acidic surface composed largely of loop residues from Blades C and D.
Figure 4.
Surface electrostatics and evolutionary analysis. (A) Electrostatic surfaces of myoc-OLF in three orientations. The surface potential is colored negative (red, −5 kT/e−) to positive (blue, +5 kT/e−). (B) Results of evolutionary trace mapped onto myoc-OLF structure highlighting regions of highest and lowest conservation. Range of conservation depicted as rainbow (53). (C) Details of highly conserved cation–π interaction in two orientations. Dashed lines indicate stabilizing interactions spanning 2.4–3.5Å. Color range as in (B).
Evolutionarily conserved and divergent features of the OLF domain
To gain better insight into the implications of the observed structural features of myoc-OLF on the OLF domain family as a whole, the evolutionary trace (54) of 1000 OLF domain sequences was mapped onto the OLF structure (Fig. 4b), revealing regions of conserved and divergent features of OLF domains. Spatially clustered, highly conserved residues define regions with a high likelihood of functional significance (54). Interestingly, the interior of OLF contains a high density of conserved residues, including Blade C and its loops, the interior strands of Blade D, the central cavity metal ion and ligand-binding sites, as well as near the site of a bound PEG at the bottom face (not shown). A major conserved feature revealed by this analysis is a cation–π interaction involving Tyr 371 within Loop B-10/C-11 and Lys 423 on Loop C-14/D-15 (Fig. 4c). Cation–π interactions are important for molecular recognition and other biochemical processes (55) and thus may prove to be a common functional design feature of OLFs. On the opposite end of the spectrum, the OLF surface clearly exhibits the highest degree of sequence divergence across the entire domain family (Fig. 4b; see Discussion).
Locations of familial glaucoma variants and identification of misfolding trigger zones
The myoc-OLF structure provides the first opportunity to understand the molecular basis of glaucoma-associated misfolding at the atomic level. Analysis of the locations of 21 myoc-OLF variants with Mendelian inheritance patterns in affected families or in selected groups of juvenile/early onset glaucoma patients, whose pathogenicity has been correlated quantitatively with thermal stability and aggregation propensity in cells and in vitro (17,24,56–58), and three non-disease-associated SNPs, reveals three distinct destabilizing regions—(i) core hydrophobic β-sheet belt including the molecular clasp, (ii) Loop B-10/C-11 and cation–π interaction, and (iii) Ca2+ environs (Table 2).
Table 2.
Glaucoma-associated lesions correlated with stability and myoc-OLF structurea
| Mutation | Tm (°C) | Structural location |
|---|---|---|
| I. Familial mutations | ||
| A. Hydrophobic β-sheet belt (40% ) | ||
| G246R | 42.5 ± 0.2 | 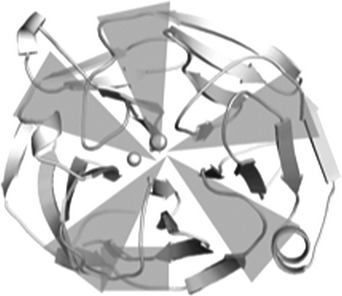 |
| G252R | 43.0 ± 0.2 | |
| R272G | 41.0 ± 0.3 | |
| W286R | N/Ab | |
| C433R | 40.4 ± 0.4 | |
| Y437H | 40.3 ± 0.4 | |
| I477Nb | 37.7 ± 0.8 | |
| I477Sb | 39.7 ± 0.2 | |
| N480Kb | 42.4 ± 0.2 | |
| I499F | 42.8 ± 0.1 | |
| S502P | 41.0 ± 0.3 | |
| B. Loop B-10/C-11 and cation–π (33%) | ||
| E323K | 44.0 ± 0.5 | 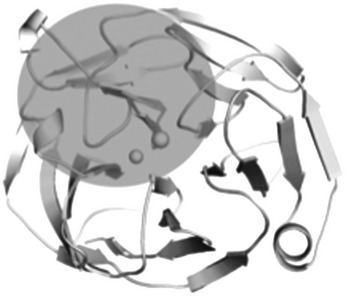 |
| G364V | 45.0 ± 0.4 | |
| G367R | 42.7 ± 0.1 | |
| P370L | N/Ac | |
| T377M | 44.3 ± 0.3 | |
| K423E | 34.2 ± 0.4 | |
| V426F | 41.5 ± 0.1 | |
| C. Ca2+ site environs (17%) | ||
| D380Ad | 46.6 ± 0.3 | 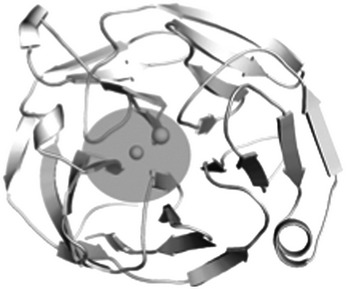 |
| A427T | 48.3 ± 0.3 | |
| P481L | 45.5 ± 0.4 | |
| II. Single nucleotide polymorphisms | ||
| E352Q | 54.8 ± 0.5c | Loop B-9/B-10 |
| E396D | 53.1 ± 0.1c | Loop C-12/C-13 |
| K398R | 53.8 ± 0.2c | Loop C-12/C-13 |
aPercentages in each category of (I) reflect those as listed in (16).
bI477N/S and N480K also belong to C. Tm values as reported in (46) except when denoted, cwhich are reported in (24). N/A indicates that no soluble recombinant protein could be isolated for Tm measurement. Tm value for wild-type myoc-OLF = 52.2°C (24).
dD380A also belongs to B.
The largest number of variants is found within the core β-sheet belt of the propeller (Table 2), particularly those variants with the lowest Tm, i.e. the most destabilized, such as W286R and I477N/S (24,56), where side chain alterations would disrupt hydrophobic packing and be least tolerated. Part of the core region, where substitutions overall appear better tolerated, is the molecular clasp (Fig. 1c). Such variants, namely moderate variants G246R, G252R, R272G, N480K, I499F and S502P, affect the tethering of the strands and/or alignment of the disulfide bond in this region. The C433R mutation abrogates disulfide bond formation. Interestingly, however, the effect of C433R on stability is not more drastic than that of Y437H (24), the variant used in myocilin glaucoma mouse models (21,30); both are highly conserved residues within OLF domains (Supplementary Material, Fig. S2). In the case of Tyr 437, a water-mediated side chain interaction stabilizes the region near the disulfide bond (Supplementary Material, Fig. S4).
Variants involving Loop B-10/C-11, and adjacent cation–π interaction (Table 2; Figs 1d and 4c), include those that are both moderately stable and severely destabilized. The substitution K423E, which would abolish the highly conserved cation–π interaction, is one of the most thermally unstable mutations measured to date (56). Similarly, although not assessed for stability in vitro due to the fact that it was reported just once, Y371D is associated with a teenage onset, severe glaucoma (59). Variants of moderate stability V426F and T377M affect the cation–π interaction: Val 426 holds Lys 423 in position via main chain interactions, and the side chain of Lys 423 interacts with the main chain carbonyl of Thr 377 (Fig. 4c). The side chain of Thr 377 also forms a hydrogen bonding interaction with the main chain of Tyr 371, which would be lost upon substitution to Met (Fig. 4c). P370L, a severe mutant (58,60), is a site that appears to organize nearby residues on Loop B-10/C-11. Curiously, Pro 370 itself does not appear to participate directly in regional stabilization (Fig. 1d and 4c), nor is it highly conserved among non-myocilin OLF domains (Supplementary Material, Fig. S2). Other moderately stable variants include charge inversion mutant E323K, as well as G364V and G367R, which may be better tolerated, because these residues are surface exposed and form no native side chain interactions.
Several additional substitutions cluster near the Ca2+ coordination sphere (Table 2; Fig. 1b and 3a). These include I477N/S, whose main chain interaction with Ca2+ may be compromised somewhat by the disruption of the β-sheet core but not enough to abolish Ca2+ binding, and the moderately stable apo variant D380A (46), which also serves to stabilize the side chain of cation–π residue Tyr 371 (Fig. 4c). Also in this region is A427T, identified within a small group of family members of variable glaucoma diagnosis (61), and a mild variant from a protein stability/aggregation standpoint (24). The substitution likely weakens Ca2+ coordination to Asn 428 and Ala 429; thus, our structure supports the initial pathogenic assignment.
Notably, neutral polymorphisms (http://www.ncbi.nlm.nih.gov/SNP), E352Q (rs61745146), K398R (rs56314834, (62,63)) and E396D (rs61730975), are located on surface loops, remote from the identified misfolding regions (Table 2). Similarly, A445V, identified in glaucoma patients (61–65), has a reported age of onset of 63 years, beyond the cutoff considered early onset (61,63). Like the SNPs (Table 2), Ala 445 is located on a remote surface and exhibits wild-type-like stability (24) (Table 3). Thus, our analysis would predict that the charge neutral A445V mutation would not be prone to misfolding; additional work will be required to strengthen the argument for glaucoma causality in associated individuals, for example, via one of the alternative mechanisms proposed for myocilin-associated glaucoma including altered endocytosis (66), mitochondrial membrane disruption (23) or apoptosis due to susceptibility toward reactive oxygen species (67).
Table 3.
Evaluated variants based on location in myoc-OLF structure
| Mutation | Tm (°C) | Location | Original assignmenta | Proposed new assignment |
|---|---|---|---|---|
| A445V | 54.2 ± 0.2 | Surface loop | Disease | SNP |
| V329M | 49.6 ± 0.4 | β-Sheet belt | SNP | Disease |
| S425P | 40.8 ± 0.3 | β-Sheet belt | SNP | Disease |
| R422C | 52.1 ± 0.1 | Surface loop | SNP | No change |
| Y473C | 54.2 ± 0.1 | Surface loop | SNP | No change |
| T293K | 52.9 ± 0.6 | Surface loop | Disease | SNP |
| T353I | 53.1 ± 0.3 | Surface loop | Ambiguous | SNP |
aAs listed in (16).
Assessment of population-based myoc-OLF variants
For variants identified from large population-based genetic studies, designation as pathogenic or neutral polymorphism is complicated by the heterogeneous nature of glaucoma and age onset of disease, and is largely dependent on whether the variant is identified in control subjects or relatives of an affected person (62). In addition, few of the many tens of variants identified through these studies have been biochemically characterized for stability and aggregation propensity. Similar to the treatment of A445V above, we applied the structural inferences from familial mutations and SNPs to intuit the likelihood of pathogenicity of an additional ∼60 disease-causing genetic lesions identified in such population studies but lacking confirmed protein behavior (16). Approximately 90% of the annotated disease or SNP variants in myoc-OLF fall within the categories described above. Six outliers were identified with possible alternative assignments, however. These proteins were recombinantly expressed and purified, and their Tms measured (Table 3) as a direct experimental determinant of structural deformation, and thus aggregation propensity and disease liability. First, V329M has been identified several times in glaucoma patients (63,68,69) but is labeled as a likely polymorphism, perhaps because Met is the residue found in the zebrafish myocilin homolog. Due to its location within a β-sheet, we predicted V329M to be a pathogenic variant. The V329M myoc-OLF variant is indeed somewhat thermally destabilized (Table 3), suggesting a mild pathogenic label, similar to the familial A427T variant described above, is likely more appropriate. S425P is also assigned as a neutral polymorphism variant (62), but we posited S425P would be prone to misfolding and aggregation, and thus instead be better characterized as pathogenic. Specifically, the Pro would compromise the position of Lys 423, which is involved in the stabilizing cation–π interaction (Fig. 4c). Indeed, the S425P myoc-OLF variant exhibits a low Tm (Table 3) and mediocre yield of folded protein previously associated with the least stable familial variants, even lower than that of Y437H (24). Variants harboring R422C and Y473C (62,63), both labeled as neutral polymorphisms, are further away from the aforementioned misfolding regions, but it was not clear whether a lone Cys would interfere with proper disulfide bond formation upon folding. Both the R422C and Y473C variants are thermally indistinguishable from wild type and are likely non-pathogenic, as originally suggested. Finally, labeled pathogenic mutation T293K (62–65) and T353I (63,68,70,71) whose pathogenicity is labeled as uncertain (16) are remote surface exposed residues unlikely to promote misfolding. In agreement with our structure-based prediction, these myoc-OLF variants are well folded, have wild-type-like stability and are more likely SNPs than causative via a misfolding phenotype in glaucoma.
Discussion
OLF domains are broadly involved in development and implicated in a host of human ailments, but molecular details of their biological activities and corresponding dysfunction in a variety of contexts remain active areas of investigation. Clarification of structure-(dys)function relationships continues to grow in importance as the biological reach of OLF domain-containing proteins expands in the context of the interactome (72–75). For myocilin and its relation to glaucoma, our results demonstrate how the myoc-OLF structure can be used to strengthen or dispute disease causality in identified mutants, and understand physicochemical features associated with a variant prone to pathogenic misfolding. Propellers do not fall into the traditional paradigm for globular proteins, namely propellers do not possess a hydrophobic core. Thus, they are both more difficult than others to accurately predict by sequence (76), and folding pathways are not well understood. We identified three structural trigger zones for misfolding—the hydrophobic inner blades, Loop B-10/C-11, and the cavity region with Ca2+—where substitutions correlate directly with destabilizing variants. Mutations within the categorized regions yield a folded, but non-native structure that enhances myoc-OLF fibrillization propensity (25). The previously identified amyloidogenic peptides P1 and P3 (Supplementary Material, Figs S2 and S5) are found within the innermost two strands of opposing Blades B (Strands B-7 and B-8) and D (Strands D-15 and D-16). Their internal position explains why fibrillization does not proceed except upon accessing a partially folded state, facilitated by non-native amino acid substitutions. In addition, we were able to shed new light onto a variant of ambiguous pathogenicity, suggest that selected mutations thought previously to be neutral polymorphisms are likely pathogenic, and identify probable neutral polymorphisms on the surface of myoc-OLF for further interrogation. As continued genetic testing of glaucoma patients throughout the world reveals new myocilin variants, it will now be possible to integrate such structural knowledge to help evaluate whether a patient is diagnosed with glaucoma because of or unrelated to the myoc-OLF amino acid substitution. Moreover, the structure affords the opportunity to identify myocilin-directed molecules to stabilize pathogenic mutants against aggregation using rational drug discovery as a new therapeutic direction. No such targeted treatment for any glaucoma subtype is currently available; indeed, a new glaucoma drug has not been approved in over a decade (77).
The discovery that the OLF domain possesses a propeller fold falls in line with the existing and complex functional picture. Propellers are generally known for their ability to orchestrate multiple signaling events across diverse pathways and have a broad portfolio of interacting partners [see, for example RACK1 (78)], both intracellularly and extracellularly (79). Evolutionary trace reveals convergent and divergent features of OLFs that could not be inferred simply from sequence alignments and homology considerations. Available evidence from our structures suggests that the central cavity of the OLF domain possesses a common ligand-binding site. Access to the central cavity appears gated in myoc-OLF at the top face where exogenous small molecules appear in our structures. In support of the solvent accessibility of the internal hydrophilic core of myoc-OLF, glycerol, only introduced during cryoprotection of myoc-OLF(E396D) crystals, is seen within the central cavity even when the structure reveals the closed Loop B-10/C-11 conformation. Entrance to the internal cavity could come from the observed movement of Trp 373 or via other loop motions on the top or bottom face that have not yet been detected. Biologically relevant metabolites that, like glycerol, could occupy the identified internal pocket may include those that regulate or signal extracellular matrix remodeling, development, or other changes in activity in neurons or aqueous humor. For example, lactate is thought to be an energy source for neurons (80,81), which, like urea and ascorbate, has been identified in aqueous humor (82). Ascorbate is also related to collagen (83) and elastin (84) synthesis, and may play a role in extraceulluar matrix remodeling by matrix metalloproteases (85). In support of these suggestions, lactate stabilizes myoc-OLF (86). Furthermore, though prior metal analysis of myoc-OLF clearly identified a single stable ion within a chelator-resistant site (46), we cannot rule out the possibility that the crystallographic Na+ site is a labile site for Ca2+. A secondary Ca2+ site could enable OLFs to be responsive to calcium release or other extracellular function (87), or, in appropriate orthologs, neurological triggers. Internal sensing could propagate relevant changes to other regions of the OLF domain to facilitate higher order protein–protein interactions or complex formation. Additional structures of OLF domains from different subfamilies will be valuable in expanding our appreciation of the structural and surface divergence of the OLF domain family.
Analysis of the structures further indicates the possibility of enzymatic function for the OLF domain is remote. Known five-bladed β-propeller enzymes have their active sites at the top face of the propeller near the central cavity. While there is noted overlap of the highly conserved myoc-OLF Asp 380 residue with that of a catalytic aspartate of arabinase, it is an unlikely nucleophile. Asp 380 is bound to the Ca2+ and Na+ ions as well as provides stabilization to and proper positioning of Tyr 371 of the cation–π interaction. Removal of Asp 380 as a Ca2+ ligand by its replacement with disease-associated Ala yields a destabilized apo protein (24,46) that readily aggregates in physiological buffers at 37°C (25). The remaining active site residues associated with hydrolysis in other five-bladed propellers are Thr residues in myoc-OLF, which are not commonly associated with catalysis (88). Major conformational changes in surface loops would be further required at the top face to create a binding pocket of sufficient size and depth to accommodate a small molecule substrate. Though it is possible to envision such motion, myoc-OLF variants that are expected to modulate the strength of the cation–π interaction are destabilized, suggesting that significant stabilization energy would need to be compensated upon such hypothetical substrate binding. The remaining loops other than B-10/C-11 are rather short and likewise may not be reconfigurable. Without candidate catalytic residues or a clear substrate-binding pocket, there is no obvious chemical reaction to propose and test.
In sum, the availability of OLF structures substantially advances our ability to interpret current data and design further experiments on function and dysfunction of the five-bladed β-propeller OLF domain. Knowledge gleaned through continued structure-(dys)function studies of myocilin-associated glaucoma can serve as a model to understand the molecular significance of identified mutations in other OLF-associated disorders (11,12,89) and adds to our comprehension of propeller protein folding more generally. Functionally, the OLF domain appears poised for interactions with other biological small molecules and/or binding partners. In the hydrophilic cavity, the binding entity is likely similar among OLFs. At the OLF surface, the lack of sequence conservation among subfamilies suggest dissimilar interactions and thus function. While the specific functions, ligands and binding partners for OLF domains will likely continue to be a challenging long-term experimental pursuit, atomic-detail structural knowledge will be a valuable guide to focus future studies of this family of biomedically important proteins.
Materials and Methods
Protein expression, purification and characterization
Wild-type myoc-OLF and myoc-OLF(E396D) were expressed and purified as described previously (24,56). Myoc-OLF variants T353I, R422C, S425P, Y473C, V329M and T293K were generated by site-directed mutagenesis (QuikChange Lightening kit, Stratagene), verified by DNA sequencing (MWG Operon), expressed, purified and Tms calculated as the midpoint of unfolding using differential scanning fluorimetry, as described previously (24,56). Primers are listed in Supplementary Material, Table S2. SeMet-substituted myoc-OLF(E396D) was produced by expression of the corresponding MBP-OLF(E396D) fusion protein (24) in the auxotrophic T7 Express Crystal E. coli cell line (New England Biolabs). Individual colonies were inoculated into 10 ml starter cultures, grown in Superior Broth (US Biological) overnight at 37°C, further diluted into 1 l of Superior Broth and shaken at 200 rpm at 37°C until an OD600 = 2.0 was reached. Cells were then pelleted by centrifugation and resuspended in pre-warmed M9 minimal media supplemented with 0.4% glucose, 1 mm MgSO4, 0.1 mm CaCl2 and 0.0002% ferric ammonium citrate, but lacking methionine or seleno-methionine. Cells were allowed to shake at 37°C for 2.5 h to deplete any remaining methionine. Fifty micrograms per milliliter of l-seleno-methionine (Sigma) were added to the cell growth media, and the temperature was dropped to 18°C. After 1 h, protein expression was induced with 0.5 mm isopropyl β-d-thiogalactopyranoside (IPTG), and cells were allowed to grow overnight. Cells were pelleted, lysed and purified as above. Overall, yield was ∼0.6 mg purified SeMet-substituted myoc-OLF(E396D)/L Superior Broth cell culture. Incorporation of SeMet was confirmed by using a Bruker S2 Picofox instrument.
Crystallization, data collection and structure determination
Purified SeMet-substituted myoc-OLF(E3396D)P21 was concentrated to 10 mg/ml, and crystals grown by the hanging drop method by equilibration against a solution containing 19% PEG 3350, 0.1m Bis–Tris pH 6 and 0.1m magnesium formate. Wild-type myoc-OLF (10 mg/ml, 50 mm HEPES, pH 7.5, supplemented with 0.36 mg/ml subtilisin A) grew crystals by equilibration against a reservoir solution containing 5% PEG 3000, 39% PEG 200, 100 mm MES, pH 6.0, and crystals of myoc-OLF(E396D)I222 (10 mg/ml in 10 mm HEPES, pH 7.5) were grown from 32% PEG 400, 5% PEG 3000 and 50 mm CAPSO, pH 9.5. Crystals of myoc-OLF(E396D) and myoc-OLF(E396D)P21 were cryo-cooled in a solution containing the respective reservoir solution supplemented with 5% glycerol and 20% PEG 200. The wild-type myoc-OLF crystals were cooled from mother liquor directly. Diffraction data were collected at the Advanced Photon Source, Argonne National Labs beamlines, The National Institute of General Medical Sciences and National Cancer Institute (GM/CA-CAT) 23-ID (wild-type myoc-OLF) and Southeast Regional Collaborative Access Team (SER-CAT) 22-ID (myoc-OLF(E396D)). Data were processed using XDS/XSCALE (90). The initial structure was solved by single wavelength Se anomalous phasing using AutoSol (91) (myoc-OLF(E396D)P21, mean anomalous difference in units of estimated standard deviation > 1 across all resolution bins in processed dataset, phasing figure of merit = 0.5) and the other two (wild-type myoc-OLF, high-resolution myoc-OLF(E396D)I222) by molecular replacement using Phaser (92), using a model generated from experimental phasing. The models were iteratively built and refined using Coot (93) and Phenix.refine (92). PEG 200 was modified from the PEG 400 entry from HicUP database (94). For wild-type myoc-OLF and myoc-OLF(E396D)I222, the pseudomerohedral twinning operator identified using Xtriage (95) was incorporated in the refinement algorithm. Structures have been deposited to the protein databank with PDB codes 4WXQ, 4WXS and 4WXU.
Structure analysis
Comparison to known structures in the PDB was enabled using DALI (96), crystal contact analysis using PDBePISA (97), metal coordination analysis conducted using FINDSITE (98) and MetalS(3) (99), and structural alignments using SSM (100). Sequence alignment for Supplementary Material, Figure S2 was prepared using PROMALS3D (101) and rendered in ESPript (102). Electrostatic surface was calculated using PDB2PQR (103) APBS (104) and figures generated in PyMOL (www.pymol.org), using the default secondary structure assignment settings. Evolutionary trace was conducted using default parameters except for increasing the number of OLF sequences to 1000, results of which were mapped on to the structure for rendering in PyMOL (53) (alignment in Supplementary Material, Table S3).
Supplementary Material
Funding
This work was supported by grants from the American Health Assistance Foundation, Glaucoma Research Foundation, National Institutes of Health (R01EY021205), and Pew Scholar in Biomedical Sciences program to R.L.L., Georgia Tech Molecular Biophysics Senior Award to R.K.D., US Department of Education Graduate Assistance in Areas of National Need (P200A060188) to S.D.O., and a Georgia Tech Presidential Undergraduate Research Award to D.M.F. GM/CA@APS has been funded in whole or in part with Federal funds from the National Cancer Institute (ACB-12002) and the National Institute of General Medical Sciences (AGM-12006). Institutions supporting SER-CAT may be found at www.ser-cat.org/members.html. The Advanced Photon Source is a US Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contracts No. DE-AC02-06CH11357 and W-31-109-Eng-38.
Supplementary Material
Acknowledgments
Conflict of Interest statement. None declared.
References
- 1.Snyder D.A., Rivers A.M., Yokoe H., Menco B.P., Anholt R.R. (1991) Olfactomedin: purification, characterization, and localization of a novel olfactory glycoprotein. Biochemistry, 30, 9143–9153. [DOI] [PubMed] [Google Scholar]
- 2.Yokoe H., Anholt R.R. (1993) Molecular cloning of olfactomedin, an extracellular matrix protein specific to olfactory neuroepithelium. Proc. Natl Acad. Sci. USA, 90, 4655–4659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zeng L.C., Han Z.G., Ma W.J. (2005) Elucidation of subfamily segregation and intramolecular coevolution of the olfactomedin-like proteins by comprehensive phylogenetic analysis and gene expression pattern assessment. FEBS Lett., 579, 5443–5453. [DOI] [PubMed] [Google Scholar]
- 4.Ando K., Nagano T., Nakamura A., Konno D., Yagi H., Sato M. (2005) Expression and characterization of disulfide bond use of oligomerized A2-Pancortins: extracellular matrix constituents in the developing brain. Neuroscience, 133, 947–957. [DOI] [PubMed] [Google Scholar]
- 5.Furutani Y., Manabe R., Tsutsui K., Yamada T., Sugimoto N., Fukuda S., Kawai J., Sugiura N., Kimata K., Hayashizaki Y., et al. (2005) Identification and characterization of photomedins: novel olfactomedin-domain-containing proteins with chondroitin sulphate-E-binding activity. Biochem. J., 389, 675–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hillier B.J., Vacquier V.D. (2003) Amassin, an olfactomedin protein, mediates the massive intercellular adhesion of sea urchin coelomocytes. J. Cell Biol., 160, 597–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zeng L.C., Liu F., Zhang X., Zhu Z.D., Wang Z.Q., Han Z.G., Ma W.J. (2004) hOLF44, a secreted glycoprotein with distinct expression pattern, belongs to an uncharacterized olfactomedin-like subfamily newly identified by phylogenetic analysis. FEBS Lett., 571, 74–80. [DOI] [PubMed] [Google Scholar]
- 8.Tomarev S.I., Nakaya N. (2009) Olfactomedin domain-containing proteins: possible mechanisms of action and functions in normal development and pathology. Mol. Neurobiol., 40, 122–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gersemann M., Becker S., Nuding S., Antoni L., Ott G., Fritz P., Oue N., Yasui W., Wehkamp J., Stange E.F. (2012) Olfactomedin-4 is a glycoprotein secreted into mucus in active IBD. J. Crohns Colitis, 6, 425–434. [DOI] [PubMed] [Google Scholar]
- 10.Liu W., Yan M., Liu Y., McLeish K.R., Coleman W.G., Jr., Rodgers G.P. (2012) Olfactomedin 4 inhibits cathepsin C-mediated protease activities, thereby modulating neutrophil killing of Staphylococcus aureus and Escherichia coli in mice. J. Immunol., 189, 2460–2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arcos-Burgos M., Jain M., Acosta M.T., Shively S., Stanescu H., Wallis D., Domene S., Velez J.I., Karkera J.D., Balog J., et al. (2010) A common variant of the latrophilin 3 gene, LPHN3, confers susceptibility to ADHD and predicts effectiveness of stimulant medication. Mol. Psychiatry, 15, 1053–1066. [DOI] [PubMed] [Google Scholar]
- 12.Bradfield J.P., Taal H.R., Timpson N.J., Scherag A., Lecoeur C., Warrington N.M., Hypponen E., Holst C., Valcarcel B., Thiering E., et al. (2012) A genome-wide association meta-analysis identifies new childhood obesity loci. Nat. Genet., 44, 526–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Resch Z.T., Fautsch M.P. (2009) Glaucoma-associated myocilin: a better understanding but much more to learn. Exp. Eye Res., 88, 704–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Quigley H.A. (1996) Number of people with glaucoma worldwide. Br. J. Ophthalmol., 80, 389–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stone E.M., Fingert J.H., Alward W.L., Nguyen T.D., Polansky J.R., Sunden S.L., Nishimura D., Clark A.F., Nystuen A., Nichols B.E., et al. (1997) Identification of a gene that causes primary open angle glaucoma. Science, 275, 668–670. [DOI] [PubMed] [Google Scholar]
- 16.Hewitt A.W., Mackey D.A., Craig J.E. (2008) Myocilin allele-specific glaucoma phenotype database. Hum. Mutat., 29, 207–211. [DOI] [PubMed] [Google Scholar]
- 17.Liu Y., Vollrath D. (2004) Reversal of mutant myocilin non-secretion and cell killing: implications for glaucoma. Hum. Mol. Genet., 13, 1193–1204. [DOI] [PubMed] [Google Scholar]
- 18.Joe M.K., Sohn S., Hur W., Moon Y., Choi Y.R., Kee C. (2003) Accumulation of mutant myocilins in ER leads to ER stress and potential cytotoxicity in human trabecular meshwork cells. Biochem. Biophys. Res. Commun., 312, 592–600. [DOI] [PubMed] [Google Scholar]
- 19.Suntharalingam A., Abisambra J.F., O'Leary J.C., 3rd, Koren J., 3rd, Zhang B., Joe M.K., Blair L.J., Hill S.E., Jinwal U.K., Cockman M., et al. (2012) Glucose-regulated protein 94 triage of mutant myocilin through endoplasmic reticulum-associated degradation subverts a more efficient autophagic clearance mechanism. J. Biol. Chem., 287, 40661–40669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yam G.H., Gaplovska-Kysela K., Zuber C., Roth J. (2007) Aggregated myocilin induces russell bodies and causes apoptosis: implications for the pathogenesis of myocilin-caused primary open-angle glaucoma. Am. J. Pathol., 170, 100–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zode G.S., Kuehn M.H., Nishimura D.Y., Searby C.C., Mohan K., Grozdanic S.D., Bugge K., Anderson M.G., Clark A.F., Stone E.M., et al. (2011) Reduction of ER stress via a chemical chaperone prevents disease phenotypes in a mouse model of primary open angle glaucoma. J. Clin. Invest., 121, 3542–3553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kanagavalli J., Pandaranayaka P.J., Krishnadas S.R., Krishnaswamy S., Sundaresan P. (2007) In vitro and in vivo study on the secretion of the Gly367Arg mutant myocilin protein. Mol. Vis., 13, 1161–1168. [PubMed] [Google Scholar]
- 23.Wang L., Zhuo Y., Liu B., Huang S., Hou F., Ge J. (2007) Pro370Leu mutant myocilin disturbs the endoplasm reticulum stress response and mitochondrial membrane potential in human trabecular meshwork cells. Mol. Vis., 13, 618–625. [PMC free article] [PubMed] [Google Scholar]
- 24.Burns J.N., Turnage K.C., Walker C.A., Lieberman R.L. (2011) The stability of myocilin olfactomedin domain variants provides new insight into glaucoma as a protein misfolding disorder. Biochemistry, 50, 5824–5833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hill S.E., Donegan R.K., Lieberman R.L. (2014) The glaucoma-associated olfactomedin domain of myocilin forms polymorphic fibrils that are constrained by partial unfolding and peptide sequence. J. Mol. Biol., 426, 921–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Orwig S.D., Perry C.W., Kim L.Y., Turnage K.C., Zhang R., Vollrath D., Schmidt-Krey I., Lieberman R.L. (2012) Amyloid fibril formation by the glaucoma-associated olfactomedin domain of myocilin. J. Mol. Biol., 421, 242–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Polansky J.R., Fauss D.J., Chen P., Chen H., Lutjen-Drecoll E., Johnson D., Kurtz R.M., Ma Z.D., Bloom E., Nguyen T.D. (1997) Cellular pharmacology and molecular biology of the trabecular meshwork inducible glucocorticoid response gene product. Ophthalmologica, 211, 126–139. [DOI] [PubMed] [Google Scholar]
- 28.Carbone M.A., Ayroles J.F., Yamamoto A., Morozova T.V., West S.A., Magwire M.M., Mackay T.F., Anholt R.R. (2009) Overexpression of myocilin in the Drosophila eye activates the unfolded protein response: implications for glaucoma. PLoS ONE, 4, e4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zode G.S., Sharma A.B., Lin X., Searby C.C., Bugge K., Kim G.H., Clark A.F., Sheffield V.C. (2014) Ocular-specific ER stress reduction rescues glaucoma in murine glucocorticoid-induced glaucoma. J. Clin. Invest., 124, 1956–1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou Y., Grinchuk O., Tomarev S.I. (2008) Transgenic mice expressing the Tyr437His mutant of human myocilin protein develop glaucoma. Invest. Ophthalmol. Vis. Sci., 49, 1932–1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zode G.S., Bugge K.E., Mohan K., Grozdanic S.D., Peters J.C., Koehn D.R., Anderson M.G., Kardon R.H., Stone E.M., Sheffield V.C. (2012) Topical ocular sodium 4-phenylbutyrate rescues glaucoma in a myocilin mouse model of primary open-angle glaucoma. Invest. Ophthalmol. Vis. Sci., 53, 1557–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim B.S., Savinova O.V., Reedy M.V., Martin J., Lun Y., Gan L., Smith R.S., Tomarev S.I., John S.W., Johnson R.L. (2001) Targeted disruption of the myocilin gene (Myoc) suggests that human glaucoma-causing mutations are gain of function. Mol. Cell Biol., 21, 7707–7713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koch M.A., Rosenhammer B., Koschade S.E., Braunger B.M., Volz C., Jagle H., Tamm E.R. (2014) Myocilin modulates programmed cell death during retinal development. Exp. Eye Res., 125, 41–52. [DOI] [PubMed] [Google Scholar]
- 34.Kwon H.S., Nakaya N., Abu-Asab M., Kim H.S., Tomarev S.I. (2014) Myocilin is involved in NgR1/Lingo-1-mediated oligodendrocyte differentiation and myelination of the optic nerve. J. Neurosci., 34, 5539–5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kwon H.S., Johnson T.V., Tomarev S.I. (2013) Myocilin stimulates osteogenic differentiation of mesenchymal stem cells through mitogen-activated protein kinase signaling. J. Biol. Chem., 288, 16882–16894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kwon H.S., Johnson T.V., Joe M.K., Abu-Asab M., Zhang J., Chan C.C., Tomarev S.I. (2013) Myocilin mediates myelination in the peripheral nervous system through ErbB2/3 signaling. J. Biol. Chem., 288, 26357–26371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kwon H.S., Lee H.S., Ji Y., Rubin J.S., Tomarev S.I. (2009) Myocilin is a modulator of Wnt signaling. Mol. Cell Biol., 29, 2139–2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Joe M.K., Kwon H.S., Cojocaru R., Tomarev S.I. (2014) Myocilin regulates cell proliferation and survival. J. Biol. Chem., 289, 10155–10167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Joe M.K., Sohn S., Choi Y.R., Park H., Kee C. (2005) Identification of flotillin-1 as a protein interacting with myocilin: implications for the pathogenesis of primary open-angle glaucoma. Biochem. Biophys. Res. Commun., 336, 1201–1206. [DOI] [PubMed] [Google Scholar]
- 40.Torrado M., Trivedi R., Zinovieva R., Karavanova I., Tomarev S.I. (2002) Optimedin: a novel olfactomedin-related protein that interacts with myocilin. Hum. Mol. Genet., 11, 1291–1301. [DOI] [PubMed] [Google Scholar]
- 41.Sanchez-Sanchez F., Martinez-Redondo F., Aroca-Aguilar J.D., Coca-Prados M., Escribano J. (2007) Characterization of the intracellular proteolytic cleavage of myocilin and identification of calpain II as a myocilin-processing protease. J. Biol. Chem., 282, 27810–27824. [DOI] [PubMed] [Google Scholar]
- 42.Chen C.K., Chan N.L., Wang A.H. (2011) The many blades of the beta-propeller proteins: conserved but versatile. Trends Biochem. Sci., 36, 553–561. [DOI] [PubMed] [Google Scholar]
- 43.Jawad Z., Paoli M. (2002) Novel sequences propel familiar folds. Structure, 10, 447–454. [DOI] [PubMed] [Google Scholar]
- 44.Paoli M., Anderson B.F., Baker H.M., Morgan W.T., Smith A., Baker E.N. (1999) Crystal structure of hemopexin reveals a novel high-affinity heme site formed between two beta-propeller domains. Nat. Struct. Biol., 6, 926–931. [DOI] [PubMed] [Google Scholar]
- 45.DeLano W.L. (2002) Unraveling hot spots in binding interfaces: progress and challenges. Curr. Opin. Struct. Biol., 12, 14–20. [DOI] [PubMed] [Google Scholar]
- 46.Donegan R.K., Hill S.E., Turnage K.C., Orwig S.D., Lieberman R.L. (2012) The glaucoma-associated olfactomedin domain of myocilin is a novel calcium binding protein. J. Biol. Chem., 287, 43370–43377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pidcock E., Moore G.R. (2001) Structural characteristics of protein binding sites for calcium and lanthanide ions. J. Biol. Inorg. Chem., 6, 479–489. [DOI] [PubMed] [Google Scholar]
- 48.Harding M.M. (2006) Small revisions to predicted distances around metal sites in proteins. Acta Crystallogr. D Biol. Crystallogr., 62, 678–682. [DOI] [PubMed] [Google Scholar]
- 49.Dai J., Liu J., Deng Y., Smith T.M., Lu M. (2004) Structure and protein design of a human platelet function inhibitor. Cell, 116, 649–659. [DOI] [PubMed] [Google Scholar]
- 50.Beisel H.G., Kawabata S., Iwanaga S., Huber R., Bode W. (1999) Tachylectin-2: crystal structure of a specific GlcNAc/GalNAc-binding lectin involved in the innate immunity host defense of the Japanese horseshoe crab Tachypleus tridentatus. EMBO J., 18, 2313–2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nurizzo D., Turkenburg J.P., Charnock S.J., Roberts S.M., Dodson E.J., McKie V.A., Taylor E.J., Gilbert H.J., Davies G.J. (2002) Cellvibrio japonicus alpha-L-arabinanase 43A has a novel five-blade beta-propeller fold. Nat. Struct. Biol., 9, 665–668. [DOI] [PubMed] [Google Scholar]
- 52.Huang W.L., Wang Y.R., Ko T.P., Chia C.Y., Huang K.F., Wang A.H. (2010) Crystal structure and functional analysis of the glutaminyl cyclase from Xanthomonas campestris. J. Mol. Biol., 401, 374–388. [DOI] [PubMed] [Google Scholar]
- 53.Lua R.C., Lichtarge O. (2010) PyETV: a PyMOL evolutionary trace viewer to analyze functional site predictions in protein complexes. Bioinformatics, 26, 2981–2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wilkins A., Erdin S., Lua R., Lichtarge O. (2012) Evolutionary trace for prediction and redesign of protein functional sites. Methods Mol. Biol., 819, 29–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ma J.C., Dougherty D.A. (1997) The Cation-Pi Interaction. Chem. Rev., 97, 1303–1324. [DOI] [PubMed] [Google Scholar]
- 56.Burns J.N., Orwig S.D., Harris J.L., Watkins J.D., Vollrath D., Lieberman R.L. (2010) Rescue of glaucoma-causing mutant myocilin thermal stability by chemical chaperones. ACS Chem. Biol., 5, 477–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gobeil S., Letartre L., Raymond V. (2006) Functional analysis of the glaucoma-causing TIGR/myocilin protein: integrity of amino-terminal coiled-coil regions and olfactomedin homology domain is essential for extracellular adhesion and secretion. Exp. Eye Res., 82, 1017–1029. [DOI] [PubMed] [Google Scholar]
- 58.Vollrath D., Liu Y. (2006) Temperature sensitive secretion of mutant myocilins. Exp. Eye Res., 82, 1030–1036. [DOI] [PubMed] [Google Scholar]
- 59.Avisar I., Lusky M., Robinson A., Shohat M., Dubois S., Raymond V., Gaton D.D. (2009) The novel Y371D myocilin mutation causes an aggressive form of juvenile open-angle glaucoma in a Caucasian family from the Middle-East. Mol. Vis., 15, 1945–1950. [PMC free article] [PubMed] [Google Scholar]
- 60.Gong G., Kosoko-Lasaki O., Haynatzki G.R., Wilson M.R. (2004) Genetic dissection of myocilin glaucoma. Hum. Mol. Genet., 13 (Spec No 1), R91–R102. [DOI] [PubMed] [Google Scholar]
- 61.Faucher M., Anctil J.L., Rodrigue M.A., Duchesne A., Bergeron D., Blondeau P., Cote G., Dubois S., Bergeron J., Arseneault R., et al. (2002) Founder TIGR/myocilin mutations for glaucoma in the Quebec population. Hum. Mol. Genet., 11, 2077–2090. [DOI] [PubMed] [Google Scholar]
- 62.Alward W.L., Fingert J.H., Coote M.A., Johnson A.T., Lerner S.F., Junqua D., Durcan F.J., McCartney P.J., Mackey D.A., Sheffield V.C., et al. (1998) Clinical features associated with mutations in the chromosome 1 open-angle glaucoma gene (GLC1A). N. Engl. J. Med., 338, 1022–1027. [DOI] [PubMed] [Google Scholar]
- 63.Fingert J.H., Heon E., Liebmann J.M., Yamamoto T., Craig J.E., Rait J., Kawase K., Hoh S.T., Buys Y.M., Dickinson J., et al. (1999) Analysis of myocilin mutations in 1703 glaucoma patients from five different populations. Hum. Mol. Genet., 8, 899–905. [DOI] [PubMed] [Google Scholar]
- 64.Vincent A.L., Billingsley G., Buys Y., Levin A.V., Priston M., Trope G., Williams-Lyn D., Héon E. (2002) Digenic Inheritance of early-onset glaucoma: CYP1B1, a potential modifier gene. Am. J. Hum. Genet., 70, 448–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Weisschuh N., Neumann D., Wolf C., Wissinger B., Gramer E. (2005) Prevalence of myocilin and optineurin sequence variants in German normal tension glaucoma patients. Mol. Vis., 11, 284–287. [PubMed] [Google Scholar]
- 66.McKay B.S., Congrove N.R., Johnson A.A., Dismuke W.M., Bowen T.J., Stamer W.D. (2013) A role for myocilin in receptor-mediated endocytosis. PLoS ONE, 8, e82301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Joe M.K., Tomarev S.I. (2010) Expression of myocilin mutants sensitizes cells to oxidative stress-induced apoptosis: implication for glaucoma pathogenesis. Am. J. Pathol., 176, 2880–2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liu W., Liu Y., Challa P., Herndon L.W., Wiggs J.L., Girkin C.A., Allingham R.R., Hauser M.A. (2012) Low prevalence of myocilin mutations in an African American population with primary open-angle glaucoma. Mol. Vis., 18, 2241. [PMC free article] [PubMed] [Google Scholar]
- 69.Shimizu S., Lichter P.R., Johnson A.T., Zhou Z., Higashi M., Gottfredsdottir M., Othman M., Moroi S.E., Rozsa F.W., Schertzer R.M., et al. (2000) Age-dependent prevalence of mutations at the GLC1A locus in primary open-angle glaucoma. Am. J. Ophthalmol., 130, 165–177. [DOI] [PubMed] [Google Scholar]
- 70.Pang C.P., Leung Y.F., Fan B., Baum L., Tong W.C., Lee W.S., Chua J.K., Fan D.S., Liu Y., Lam D.S. (2002) TIGR/MYOC gene sequence alterations in individuals with and without primary open-angle glaucoma. Invest. Ophthalmol. Vis. Sci., 43, 3231–3235. [PubMed] [Google Scholar]
- 71.Yoon S.-J.K., Kim H.-S., Moon J.-I., Lim J.M., Joo C.-K. (1999) Mutations of the TIGR/MYOC gene in primary open-angle glaucoma in Korea. Am. J. Hum. Genet., 64, 1775–1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Stelzl U., Worm U., Lalowski M., Haenig C., Brembeck F.H., Goehler H., Stroedicke M., Zenkner M., Schoenherr A., Koeppen S., et al. (2005) A human protein-protein interaction network: a resource for annotating the proteome. Cell, 122, 957–968. [DOI] [PubMed] [Google Scholar]
- 73.Giot L., Bader J.S., Brouwer C., Chaudhuri A., Kuang B., Li Y., Hao Y.L., Ooi C.E., Godwin B., Vitols E., et al. (2003) A protein interaction map of Drosophila melanogaster. Science, 302, 1727–1736. [DOI] [PubMed] [Google Scholar]
- 74.Dyer M.D., Neff C., Dufford M., Rivera C.G., Shattuck D., Bassaganya-Riera J., Murali T.M., Sobral B.W. (2010) The human-bacterial pathogen protein interaction networks of Bacillus anthracis, Francisella tularensis, and Yersinia pestis. PLoS ONE, 5, e12089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Camargo L.M., Collura V., Rain J.C., Mizuguchi K., Hermjakob H., Kerrien S., Bonnert T.P., Whiting P.J., Brandon N.J. (2007) Disrupted in Schizophrenia 1 Interactome: evidence for the close connectivity of risk genes and a potential synaptic basis for schizophrenia. Mol. Psychiatry, 12, 74–86. [DOI] [PubMed] [Google Scholar]
- 76.Fulop V., Jones D.T. (1999) Beta propellers: structural rigidity and functional diversity. Curr. Opin. Struct. Biol., 9, 715–721. [DOI] [PubMed] [Google Scholar]
- 77.Zhang K., Zhang L., Weinreb R.N. (2012) Ophthalmic drug discovery: novel targets and mechanisms for retinal diseases and glaucoma. Nat. Rev. Drug Discov., 11, 541–559. [DOI] [PubMed] [Google Scholar]
- 78.Adams D.R., Ron D., Kiely P.A. (2011) RACK1, a multifaceted scaffolding protein: structure and function. Cell Commun. Signal, 9, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pons T., Gmez R., Chinea G., Valencia A. (2003) Beta-propellers: associated functions and their role in human diseases. Curr. Med. Chem., 10, 505–524. [DOI] [PubMed] [Google Scholar]
- 80.Genc S., Kurnaz I.A., Ozilgen M. (2011) Astrocyte-neuron lactate shuttle may boost more ATP supply to the neuron under hypoxic conditions--in silico study supported by in vitro expression data. BMC Syst. Biol., 5, 162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tarczyluk M.A., Nagel D.A., O'Neil J.D., Parri H.R., Tse E.H., Coleman M.D., Hill E.J. (2013) Functional astrocyte-neuron lactate shuttle in a human stem cell-derived neuronal network. J. Cereb. Blood Flow Metab., 33, 1386–1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang S.Y., Hasty C.E., Watson P.A., Wicksted J.P., Stith R.D., March W.F. (1993) Analysis of metabolites in aqueous solutions by using laser Raman spectroscopy. Appl. Opt., 32, 925–929. [DOI] [PubMed] [Google Scholar]
- 83.Chan D., Lamande S.R., Cole W.G., Bateman J.F. (1990) Regulation of procollagen synthesis and processing during ascorbate-induced extracellular matrix accumulation in vitro. Biochem. J., 269, 175–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Davidson J.M., LuValle P.A., Zoia O., Quaglino D., Jr., Giro M. (1997) Ascorbate differentially regulates elastin and collagen biosynthesis in vascular smooth muscle cells and skin fibroblasts by pretranslational mechanisms. J. Biol. Chem., 272, 345–352. [DOI] [PubMed] [Google Scholar]
- 85.Philips N., Keller T., Holmes C. (2007) Reciprocal effects of ascorbate on cancer cell growth and the expression of matrix metalloproteinases and transforming growth factor-beta. Cancer Lett., 256, 49–55. [DOI] [PubMed] [Google Scholar]
- 86.Orwig S.D., Lieberman R.L. (2011) Biophysical characterization of the olfactomedin domain of myocilin, an extracellular matrix protein implicated in inherited forms of glaucoma. PLoS ONE, 6, e16347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Maurer P., Hohenester E. (1997) Structural and functional aspects of calcium binding in extracellular matrix proteins. Matrix Biol., 15, 569–580; discussion 581. [DOI] [PubMed] [Google Scholar]
- 88.Dodson G., Wlodawer A. (1998) Catalytic triads and their relatives. Trends Biochem. Sci., 23, 347–352. [DOI] [PubMed] [Google Scholar]
- 89.Domene S., Stanescu H., Wallis D., Tinloy B., Pineda D.E., Kleta R., Arcos-Burgos M., Roessler E., Muenke M. (2011) Screening of human LPHN3 for variants with a potential impact on ADHD susceptibility. Am. J. Med. Genet. B, 156B, 11–18. [DOI] [PubMed] [Google Scholar]
- 90.Kabsch W. (2010) XDS. Acta Crystallogr. D Biol. Crystallogr., 66, 125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Terwilliger T.C., Adams P.D., Read R.J., McCoy A.J., Moriarty N.W., Grosse-Kunstleve R.W., Afonine P.V., Zwart P.H., Hung L.W. (2009) Decision-making in structure solution using Bayesian estimates of map quality: the PHENIX AutoSol wizard. Acta Crystallogr. D Biol. Crystallogr., 65, 582–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J. (2007) Phaser crystallographic software. J. Appl. Crystallogr., 40, 658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Emsley P., Lohkamp B., Scott W.G., Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr., 66, 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kleywegt G.J. (2007) Crystallographic refinement of ligand complexes. Acta Crystallogr. D Biol. Crystallogr., 63, 94–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W., et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr., 66, 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Holm L., Rosenstrom P. (2010) Dali server: conservation mapping in 3D. Nucleic Acids Res., 38, W545–e4549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Krissinel E., Henrick K. (2007) Inference of macromolecular assemblies from crystalline state. J. Mol. Biol., 372, 774–797. [DOI] [PubMed] [Google Scholar]
- 98.Brylinski M., Skolnick J. (2011) FINDSITE-metal: integrating evolutionary information and machine learning for structure-based metal-binding site prediction at the proteome level. Proteins, 79, 735–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Valasatava Y., Rosato A., Cavallaro G., Andreini C. (2014) MetalS(3), a database-mining tool for the identification of structurally similar metal sites. J. Biol. Inorg. Chem., 19, 937–945. [DOI] [PubMed] [Google Scholar]
- 100.Krissinel E., Henrick K. (2004) Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D Biol. Crystallogr., 60, 2256–2268. [DOI] [PubMed] [Google Scholar]
- 101.Pei J., Kim B.H., Grishin N.V. (2008) PROMALS3D: a tool for multiple protein sequence and structure alignments. Nucleic Acids Res., 36, 2295–2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Gouet P., Robert X., Courcelle E. (2003) ESPript/ENDscript: extracting and rendering sequence and 3D information from atomic structures of proteins. Nucleic Acids Res., 31, 3320–3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Dolinsky T.J., Czodrowski P., Li H., Nielsen J.E., Jensen J.H., Klebe G., Baker N.A. (2007) PDB2PQR: expanding and upgrading automated preparation of biomolecular structures for molecular simulations. Nucleic Acids Res., 35, W522–W525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Baker N.A., Sept D., Joseph S., Holst M.J., McCammon J.A. (2001) Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl Acad. Sci. USA, 98, 10037–10041. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.