Abstract
Proper functioning of cilia, hair-like structures responsible for sensation and locomotion, requires nephrocystin-5 (NPHP5) and a multi-subunit complex called the Bardet–Biedl syndrome (BBS)ome, but their precise relationship is not understood. The BBSome is involved in the trafficking of membrane cargos to cilia. While it is known that a loss of any single subunit prevents ciliary trafficking of the BBSome and its cargos, the mechanisms underlying ciliary entry of this complex are not well characterized. Here, we report that a transition zone protein NPHP5 contains two separate BBS-binding sites and interacts with the BBSome to mediate its integrity. Depletion of NPHP5, or expression of NPHP5 mutant missing one binding site, specifically leads to dissociation of BBS2 and BBS5 from the BBSome and loss of ciliary BBS2 and BBS5 without compromising the ability of the other subunits to traffic into cilia. Depletion of Cep290, another transition zone protein that directly binds to NPHP5, causes additional dissociation of BBS8 and loss of ciliary BBS8. Furthermore, delivery of BBSome cargos, smoothened, VPAC2 and Rab8a, to the ciliary compartment is completely disabled in the absence of single BBS subunits, but is selectively impaired in the absence of NPHP5 or Cep290. These findings define a new role of NPHP5 and Cep290 in controlling integrity and ciliary trafficking of the BBSome, which in turn impinge on the delivery of ciliary cargo.
Introduction
In animal cells, centrioles are composed of nine sets of microtubule triplets and constitute the core of centrosomes, essential organelles that modulate various cellular processes including cell division, cell cycle progression, aging, cell morphology, polarity and motility (1,2). A pair of centrioles, termed the mother and daughter centrioles, recruit an amorphous mass of protein called the pericentriolar matrix (PCM), which is responsible for microtubule nucleation and anchoring (3,4). In quiescent cells, mother centrioles, but not daughter centrioles, transform into basal bodies and become competent to template cilia, hair-like protuberances that possess sensory and/or motility functions (5–7). Regardless of functionality, every cilium is made up of an axoneme, the microtubular backbone, surrounded by a ciliary membrane that is continuous with the plasma membrane. Cilia malfunction is increasingly recognized as a major cause of ciliary diseases or ciliopathies, a heterogeneous group of genetic disorders affecting many parts of the body, including the kidney, eye, liver and brain (8,9). Clinically distinct disorders often display overlapping phenotypes, but the molecular basis of this overlap is not fully understood and remains an open question.
Bardet–Biedl Syndrome (BBS) is a ciliopathy characterized by retinal degeneration, renal failure, obesity, diabetes, male infertility, polydactyly and cognitive impairment (10,11). To date, 19 genes had been identified as disease loci, and the majority encode products that are essential for the formation and proper functioning of a multi-subunit complex called the BBSome. The BBSome is comprised of eight distinct BBS subunits (BBS1, BBS2, BBS4, BBS5, BBS7, BBS8, BBS9 and BBIP10/BBS18) and its assembly occurs in several stages (12,13). In brief, three chaperonin-like subunits, BBS6, BBS10 and BBS12 first bind to and stabilize BBS7, leading to the generation of an assembly intermediate known as the BBSome core, which consists of BBS2, BBS7 and BBS9 (14,15). Subsequent incorporation of peripheral subunits BBS1, BBS5, BBS8, and finally BBS4, to the core completes its transformation to the holo-complex (15). BBS4 is also known to interact with BBIP10, although it is not certain when and how the latter is integrated into the BBSome (13). BBSome subunits possess domains known to mediate protein–protein interactions. BBS1, BBS2, BBS7 and BBS9 contain β-propeller domains. BBS4 and BBS8 contain solenoid or tetratricopeptide repeat domains, while BBIP10 possesses two alpha helices. In contrast, BBS5 contains pleckstrin homology domains, binds to phosphoinositides and is believed to be the only BBSome subunit in direct contact with the ciliary membrane (12). Recently, BBS3/ARL6, an Arf-like GTPase, is shown to be a major effector of the BBSome. BBS3 recruits the BBSome to the membrane, wherein it assembles a coat that selectively sorts membrane cargos to cilia (16). In the nematode Caenorhabditis elegans, the BBSome regulates the assembly of intraflagellar transport (IFT) particles (17), a multi-subunit complex responsible for transporting the BBSome and its associated cargos into and out of the ciliary compartment (18). Unlike the BBSome which is generally not required for cilia assembly (13,19), the IFT complex controls the bidirectional motility along the axoneme that is essential for the formation, maintenance and function of cilia.
Despite our knowledge of the BBSome, the precise mechanisms by which its ciliary trafficking is regulated remain enigmatic. Previous studies have demonstrated that all BBSome subunits are essential for BBSome assembly, and only a fully assembled holo-complex can gain entry to the ciliary compartment (20–22). Ciliary entry requires the passage of the BBSome through a special region between the basal body and the axoneme called the transition zone, which acts as a permeability barrier to control the entry and exit of ciliary proteins (23). The transition zone contains several multi-subunit complexes including Cep290/NPHP5 (24–26), NPHP1/NPHP4/NPHP8 (24,27,28), MKS (27,29,30) and nucleoporin (31); however, the precise manner in which these complexes function remains shrouded in mystery. Here, we established a previously unknown connection between the BBSome and NPHP5, a transition zone protein whose deficiency is associated with ciliopathies. We demonstrated that NPHP5 and its binding partner Cep290, another transition zone protein, modulate not only BBSome integrity, but also trafficking of the holo-complex and its associated cargos into the cilium.
Results
NPHP5 interacts with the BBSome through two distinct binding sites
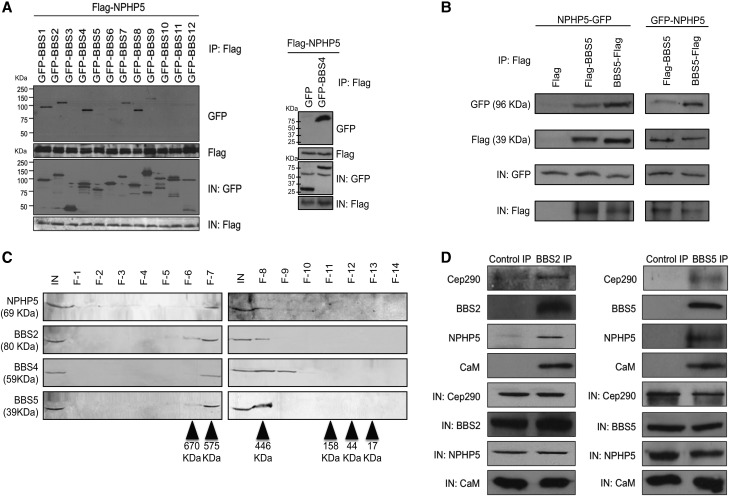
NPHP5 is a ciliopathy protein localized to the distal ends of centrioles, including the ciliary base (24,25). Pathogenic mutations in the NPHP5 gene render the resulting protein non-functional (25) and cause two ciliary diseases, Leber congenital amaurosis (LCA; retinal degeneration) and Senior-Løken syndrome (SLS; retinal degeneration and renal failure) (32–35). Because LCA and SLS share overlapping clinical manifestations with BBS, we hypothesize that NPHP5 and BBS proteins could interact to regulate cilia homeostasis. Thus, the ability of NPHP5 to associate with the first 12 BBS subunits was examined. We immunoprecipitated recombinant, N-terminal tagged NPHP5 (Flag-NPHP5) from HEK293 cell extracts and found that it interacts with all GFP-tagged BBSome subunits BBS1, BBS2, BBS4, BBS7 and BBS8 except BBS5 (Fig. 1A). No or weak co-immunoprecipitation was observed between NPHP5 and non-BBSome subunits, including an Arf-like GTPase BBS3, chaperonin-like BBS proteins BBS6, BBS10 and BBS12 and an E3 ubiquitin ligase BBS11 (36) (Fig. 1A). Similar results were obtained when a C-terminal tagged NPHP5 (NPHP5-Flag) was used instead of Flag-NPHP5 (Supplementary Material, Fig. S1a) or when immunoprecipitations were performed in a less stringent lysis buffer designed to emulate physiological conditions (150 mm salt, pH 7.4) (Supplementary Material, Fig. S1b). Because every BBSome subunit apart from BBS5 associates with NPHP5, we wondered whether the position and/or size of the tag on BBS5 or NPHP5 interfere(s) with binding. We co-expressed Flag-BBS5 or BBS5-Flag and GFP-NPHP5 or NPHP5-GFP, performed anti-Flag immunoprecipitations and confirmed that recombinant NPHP5 and BBS5 interact in all possible combinations (Fig. 1B). Notably, NPHP5 may bind to the N-terminal region of BBS5, because recombinant NPHP5 interacted strongly with BBS5-Flag but poorly with Flag-BBS5 (Fig. 1B), and did not interact at all with GFP-BBS5 (Fig. 1a and supplementary material Fig. S1a–b). Moreover, no interaction was observed between GFP-NPHP5 and non-BBSome subunits BBS3-Flag, BBS6-Flag or BBS10-Flag (Supplementary Material, Fig. S1c), suggesting that NPHP5 specifically associates with the holo-complex. To provide further proof that NPHP5 interacts with the BBSome, endogenous NPHP5, BBS2, BBS4, BBS5 and BBS8 co-fractionated in a discrete protein complex at ∼500 kDa (Figs 1C and 6C), which is in close agreement with the reported molecular weight of the BBSome (12). In addition, antibodies against BBS2 or BBS5 co-precipitated NPHP5, along with known NPHP5-interacting proteins, Cep290 and calmodulin (CaM) (Fig. 1D) (25,26,33,37). An in situ proximity ligation assay (PLA) designed to detect interaction at a distance below 40 nm (38) also revealed an association of NPHP5 with BBSome subunits (Table 1 and Fig. 4). Given that some BBSome subunits are efficiently co-immunoprecipitated with, and physically closer to, NPHP5 than others (Figs. 1 and 4, Supplementary Material, Fig. S1 and Table 1), these data together suggest that NPHP5 interacts with the holo-complex through certain subunits. Moreover, NPHP5 most likely forms a complex with the BBSome, Cep290 and CaM.
Figure 1.
NPHP5 interacts with the BBSome. (A) Flag-NPHP5 and the indicated GFP proteins were co-expressed in HEK293 cells, and lysates were immunoprecipitated with an anti-Flag antibody. The resulting immunoprecipitates were western blotted with anti-Flag or anti-GFP antibodies. IN, input. (B) The indicated N-terminal or C-terminal tagged recombinant NPHP5 or BBS5 proteins were co-expressed in HEK293 cells, and lysates were immunoprecipitated with an anti-Flag antibody. The resulting immunoprecipitates were western blotted with anti-Flag or anti-GFP antibodies. IN, input. (C) HEK293 cell extract was chromatographed on a Superose-6 gel filtration column, and the resulting fractions were western blotted with indicated antibodies. Estimated molecular weights are indicated. IN, input; F, fraction. (D) Western blotting of endogenous Cep290, NPHP5, BBS2, BBS5 and CaM after immunoprecipitation of HEK293 cell extracts with anti-Flag (control), anti-BBS2 or anti-BBS5 antibodies. IN, input.
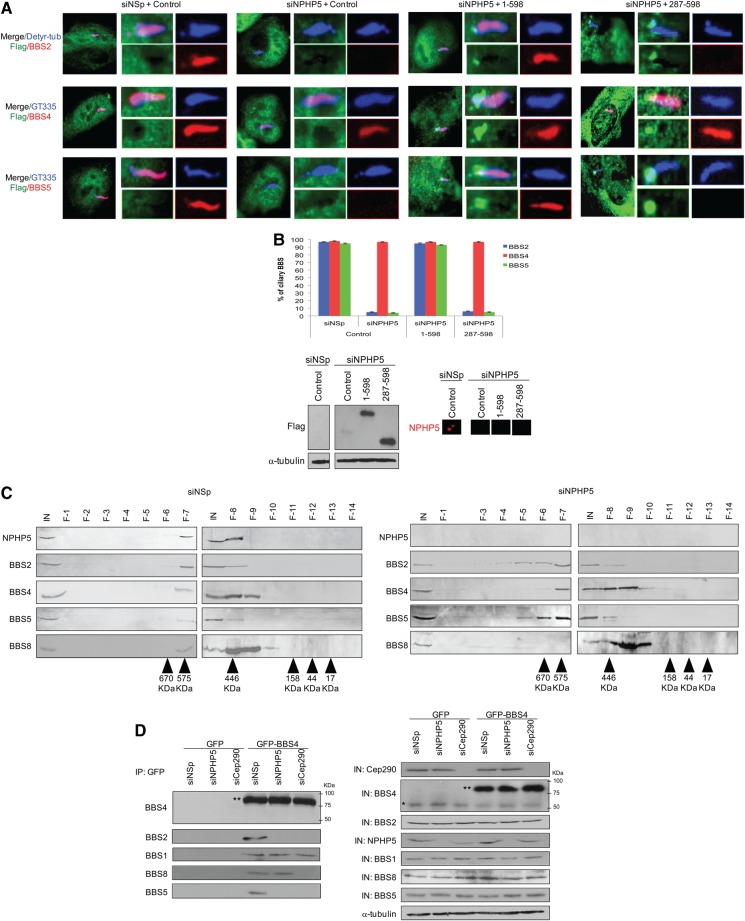
Figure 6.
NPHP5 and Cep290 regulate BBSome integrity. (A) ARPE-19 cells transfected with control siRNA (siNSp) or siRNA targeting NPHP5 3′UTR (siNPHP5) and plasmid expressing an irrelevant Flag-tagged protein (control), full-length Flag-NPHP5 (1-598) or a C-terminal fragment of Flag-tagged NPHP5 (287-598) lacking a BBSome-binding site, induced to quiescence, were stained with antibodies against Flag (green), detyrosinated-tubulin or GT335 (blue), and BBS2, BBS4 or BBS5 (red). (B) (Top) The percentages of quiescent cells with ciliary BBS2, BBS4 or BBS5 staining were determined using either detyrosinated tubulin or GT335 as a ciliary marker. At least 100 transfected cells were counted per condition, and error bars represent average of three independent experiments. (Bottom left) Western blotting of Flag, with α-tubulin used as loading control. (Bottom right) Cells were processed for immunofluorescence and stained with anti-NPHP5 (red) antibody. (C) Extract from control (siNSp) or NPHP5 siRNA-depleted (siNPHP5) HEK293 cells was chromatographed on a Superose-6 gel filtration column, and the resulting fractions were western blotted with indicated antibodies. Estimated molecular weights are indicated. IN, input; F, fraction. (D) GFP or GFP-BBS4 were expressed in HEK293 cells treated with control (siNSp), NPHP5 (siNPHP5) or Cep290 (siCep290) siRNAs and immunoprecipitated from lysates. Endogenous (*) and recombinant BBS4 (**) along with endogenous BBS1, BBS2, BBS5 and BBS8 were detected after western blotting the resulting immunoprecipitates. Western blotting of NPHP5 and Cep290 were performed to monitor knockdown efficiency. IN, input. α-tubulin was used as loading control.
Table 1.
PLA signals generated from in situ PLAs using the indicated combinations of antibodies were quantitated and normalized
| % Cells | Proliferating (PLA signal intensity) |
Quiescent (PLA signal intensity) |
||||||
|---|---|---|---|---|---|---|---|---|
| No | Weak | Medium | Strong | No | Weak | Medium | Strong | |
| NPHP5 + BBS1 | 2 | 6 | 60 | 32 | 46 | 50 | 4 | 0 |
| NPHP5 + BBS2 | 3 | 15 | 75 | 7 | 12 | 76 | 12 | 0 |
| NPHP5 + BBS4 | 14 | 78 | 8 | 0 | 0 | 6 | 16 | 78 |
| NPHP5 + BBS5 | 2 | 10 | 62 | 26 | 4 | 42 | 54 | 0 |
| NPHP5 + BBS7 | 42 | 50 | 8 | 0 | 6 | 18 | 74 | 2 |
| NPHP5 + BBS8 | 10 | 84 | 6 | 0 | 2 | 38 | 58 | 2 |
| NPHP5 + BBS9 | 8 | 42 | 48 | 2 | 0 | 10 | 66 | 24 |
| NPHP5 + BBS10 | 16 | 82 | 2 | 0 | 6 | 20 | 72 | 2 |
| Cep290 + BBS1 | 46 | 52 | 2 | 0 | 48 | 50 | 2 | 0 |
| Cep290 + BBS2 | 12 | 76 | 12 | 0 | 4 | 46 | 48 | 2 |
| Cep290 + BBS4 | 46 | 50 | 4 | 0 | 0 | 6 | 14 | 80 |
| Cep290 + BBS5 | 48 | 50 | 2 | 0 | 47 | 50 | 3 | 0 |
| Cep290 + BBS7 | 8 | 44 | 46 | 2 | 47 | 53 | 0 | 0 |
| Cep290 + BBS8 | 48 | 50 | 2 | 0 | 46 | 50 | 4 | 0 |
| Cep290 + BBS9 | 4 | 10 | 54 | 32 | 10 | 46 | 42 | 2 |
| Cep290 + BBS10 | 48 | 50 | 2 | 0 | 9 | 42 | 48 | 1 |
| Cep290 + NPHP5 | 0 | 6 | 24 | 70 | 0 | 2 | 16 | 82 |
The percentages of proliferating (non-ciliated) or quiescent (ciliated) RPE-1 cells possessing no, weak, medium and strong signal for each antibody combination were determined.
Figure 4.
NPHP5/Cep290 interaction with the BBSome in ciliated and non-ciliated cells. (A) In situ PLAs using antibodies against NPHP5 and Cep290 were performed to visualize protein–protein interaction (PLA signal in red) in proliferating (non-ciliated) and quiescent (ciliated) RPE-1 cells. Cells were co-stained with γ-tubulin (green) to visualize the centrosome. No PLA signal was detected when one or both antibodies were missing, or when an NPHP5 antibody was combined with an irrelevant antibody, glutamylated tubulin (GT335). No Ab, no antibody. (B and C) In situ PLAs using the indicated combinations of antibodies were performed to visualize protein–protein interaction (PLA signal in red) in proliferating (non-ciliated) and quiescent (ciliated) RPE-1 cells. Cells were co-stained with γ-tubulin (green) to visualize the centrosome.
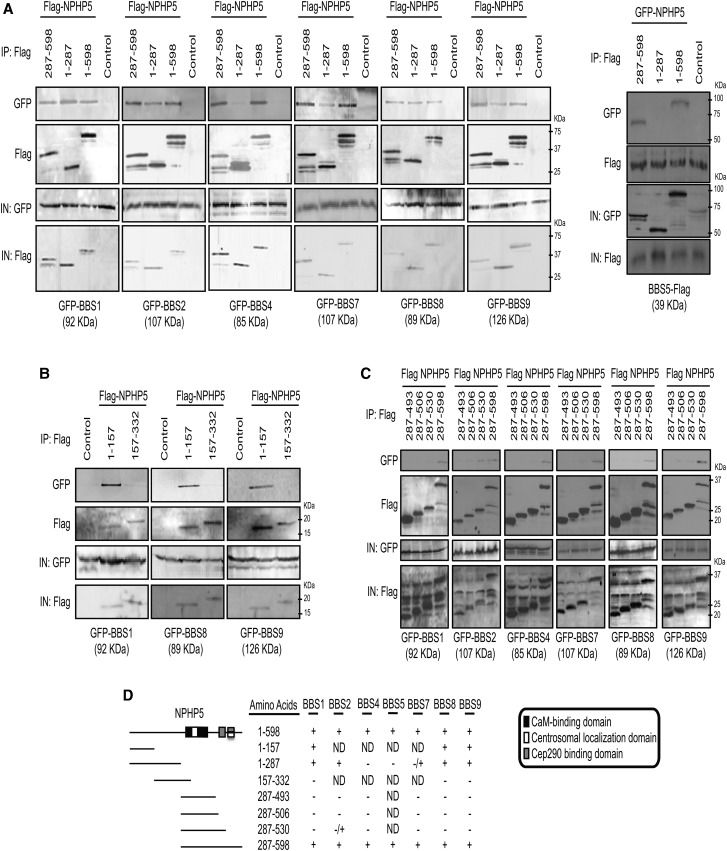
Next, we sought to map the BBSome-binding domain(s) of NPHP5 by expressing a series of epitope-tagged NPHP5 truncation mutants and BBSome subunits and examining their ability to co-immunoprecipitate in cell extracts. We found that there are two distinct BBSome-binding sites, one at the N-terminal region and the other at the C-terminal region (Fig. 2A). Further mapping studies revealed that the first 157 residues (1–157) and the last 68 residues (530–598) of NPHP5 are critical for binding (Fig. 2B–D).
Figure 2.
NPHP5 possesses two distinct BBSome-binding sites. (A) Left panel: Flag (control), Flag-tagged full-length NPHP5 (1–598) or the indicated fragments of Flag-tagged NPHP5 were co-expressed with the indicated GFP-BBS proteins in HEK293 cells, and lysates were immunoprecipitated with an anti-Flag antibody. Flag-NPHP5 and GFP-BBS were detected after western blotting the resulting immunoprecipitates. IN, input. Right panel: GFP (control), GFP-tagged full-length NPHP5 (1–598) or the indicated fragments of GFP-tagged NPHP5 were co-expressed with BBS5-Flag in HEK293 cells, and lysates were immunoprecipitated with an anti-Flag antibody. BBS5-Flag and GFP-NPHP5 were detected after western blotting the resulting immunoprecipitates. IN, input. (B and C) Flag (control) or the indicated fragments of Flag-tagged NPHP5 were co-expressed with the indicated GFP-BBS proteins in HEK293 cells, and lysates were immunoprecipitated with an anti-Flag antibody. Flag-NPHP5 and GFP-BBS were detected after western blotting the resulting immunoprecipitates. IN, input. (D) Summary of the results of in vivo binding experiments. Known domains of NPHP5 (centrosomal localization, CaM-binding and Cep290-binding domains) are indicated as in (25). Each fragment used for the study takes into account the position of these domains so that a given domain is not prematurely truncated. +, interaction; –, no interaction; ND, not determined.
During the course of our mapping studies, an unexpected and reproducible phenomenon was noticed with regard to the specificity of the two BBSome-binding sites of NPHP5. We found that BBS1 interacted more strongly with a N-terminal fragment of NPHP5 (1–287), encompassing the N-terminal BBSome-binding site, than full-length (1–598) or a C-terminal fragment of NPHP5 (287–598) (Fig. 2A). Likewise, two other N-terminal fragments of NPHP5 (1–157 and 1–332) also exhibited a stronger interaction with BBS1 compared with full-length NPHP5 (data not shown). Conversely, BBS9 bound more robustly to 287–598 than 1–598 or 1–287 (Fig. 2A). BBS2, BBS4, BBS5 and BBS7 mostly interacted with the C-terminal BBSome-binding site, because they bound equally well to 1–598 and 287–598 but poorly to 1–287 (Fig. 2A). BBS8, meanwhile, bound to 1–598, 1–287 and 287–598 with similar affinity (Fig. 2A). Our data suggest that the N-terminal site, when present alone and/or acting independently, may preferentially bind BBS1, whereas the C-terminal site is favorably occupied by BBS9 and several other subunits.
NPHP5 interacts with the BBSome independently of its associated partner, Cep290
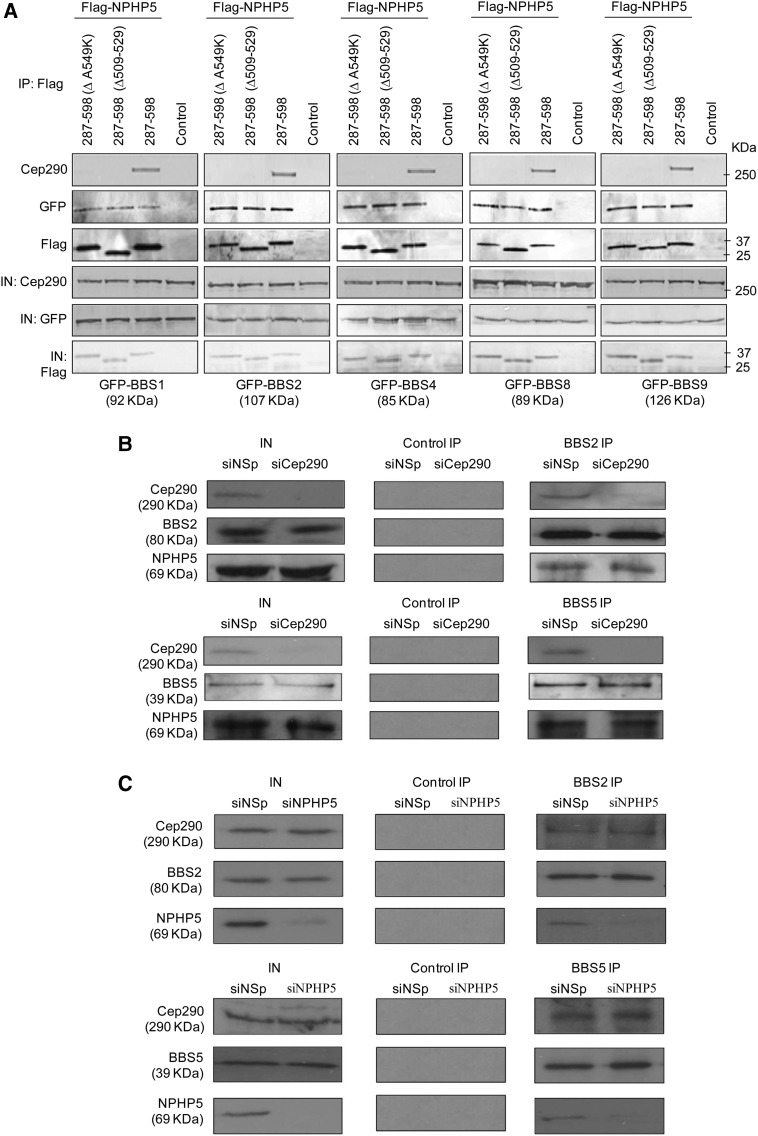
We had previously demonstrated that residues 509–529 and 549 of NPHP5 are critical for binding to Cep290 (25). Given that Cep290 interacts with the BBSome (39) and that the Cep290- and the C-terminal BBSome-binding sites are mapped to the C-terminal region of NPHP5, we determined if these two sites overlap, and whether NPHP5 and Cep290 could associate with the BBSome independently of each other. First, we showed that a C-terminal fragment of NPHP5 (Flag-NPHP5(287–598)) containing only one BBSome-binding site interacted with Cep290 and the BBSome (Fig. 3A). In contrast, the same fragment carrying a point mutation (Flag-NPHP5 (287–598A549K)) or a deletion (Flag-NPHP5 (287–598Δ509–529)) known to disrupt Cep290 binding still associated with the BBSome (Fig. 3A). Second, antibodies against BBS2 and BBS5 co-precipitated endogenous NPHP5 in extracts specifically depleted of Cep290 (Fig. 3B). Likewise, the same antibodies co-precipitated Cep290 in extracts depleted of NPHP5 (Fig. 3C). Taken together, our findings suggest that NPHP5 and Cep290 can independently bind to the BBSome.
Figure 3.
NPHP5 and Cep290 bind to the BBSome independently of each other. (A) Flag (control) or the indicated fragments and mutants of Flag-tagged NPHP5 were co-expressed with the indicated GFP-BBS proteins in HEK293 cells, and lysates were immunoprecipitated with an anti-Flag antibody. Flag-NPHP5, GFP-BBS and endogenous Cep290 were detected after western blotting the resulting immunoprecipitates. IN, input. (B and C) Western blotting of endogenous Cep290, NPHP5, BBS2 and BBS5 after immunoprecipitation of HEK293 cell extracts with anti-Flag (control), anti-BBS2 or anti-BBS5 antibodies. Extracts were transfected with control (siNSp), NPHP5 (siNPHP5) or Cep290 (siCep290) siRNAs prior to immunoprecipitation. IN, input.
NPHP5 and Cep290 interact with the BBSome in non-ciliated and ciliated cells
A number of studies showed that BBS proteins are localized to centrosomes and cilia (12,13,16,17,22,40–46). Because NPHP5 and the BBSome interact, we therefore examined the localization of these proteins in greater detail by performing immunofluorescence experiments to co-stain NPHP5 with BBSome subunits in proliferating (non-ciliated) and quiescent (ciliated) human retinal pigmented epithelial (RPE-1) cells. We found that NPHP5 staining partially overlapped with every subunit at the centrosome/ciliary base (Supplementary Material, Fig. S2a–b). Of note, BBSome subunits generally exhibited weak staining at the centrosome in proliferating cells but accumulated at the cilium in quiescent cells (Supplementary Material, Fig. S2a–b). Thus, it is conceivable that when cilia have yet to form, the BBSome complex may be present in minute amounts and is not fully assembled. Because ciliary accumulation of the BBSome coincides with ciliogenesis and because the BBSome has to pass through the transition zone at the ciliary base prior to entry into cilia, we asked whether NPHP5 at the centrosome/ciliary base could interact with the BBSome in proliferating and quiescent cells. In situ PLA was used to identify endogenous interaction between NPHP5 and BBSome subunits and to access how close these two entities are in space. To validate assay specificity, we first visualized the interaction between NPHP5 and Cep290, which is known to be direct (24,25). Both proteins are also shown to localize to the distal ends of centrioles/ciliary base during interphase and quiescence (25,47,48). We observed strong PLA signals in proliferating and quiescent cells when anti-NPHP5 and anti-Cep290 antibodies were used (Fig. 4A and Table 1). These signals substantially overlapped with a centrosomal marker, γ-tubulin, suggesting that NPHP5 and Cep290 interact at the centrosome/ciliary base under all conditions (Fig. 4A). Negligible signal was detected when one or more antibodies were omitted, or when anti-NPHP5 antibody was mixed with an irrelevant antibody (Fig. 4A). In situ PLAs performed using proximity probes against NPHP5 and different BBSome subunits revealed that NPHP5 interacts with, and/or is in close proximity to, every BBSome subunit (Fig. 4B and Table 1). PLA signal intensity varies between different NPHP5/subunit combinations and between proliferative and quiescence states for a given combination. Notably, the NPHP5/BBS4 combination in quiescent cells yielded the strongest signal (Fig. 4B and Table 1). PLA signals of varying intensity were likewise detected when antibodies against Cep290 and BBSome subunits were combined. Overall, these signals tended to be weaker than those of NPHP5/BBSome subunits (Fig. 4C and Table 1), suggesting that Cep290 may be physically further from the BBSome than NPHP5. Interestingly, a strong PLA signal was also reported for the Cep290/BBS4 combination in quiescent cells (Fig. 4C and Table 1). Because NPHP5 interacts with Cep290 and the BBSome, contains two BBSome-binding sites with different subunit specificities, and is in close proximity to different subunits depending on proliferation status, our data suggest that NPHP5 and Cep290 could control some aspects of BBSome function.
NPHP5 and Cep290 control BBSome integrity and ciliary trafficking
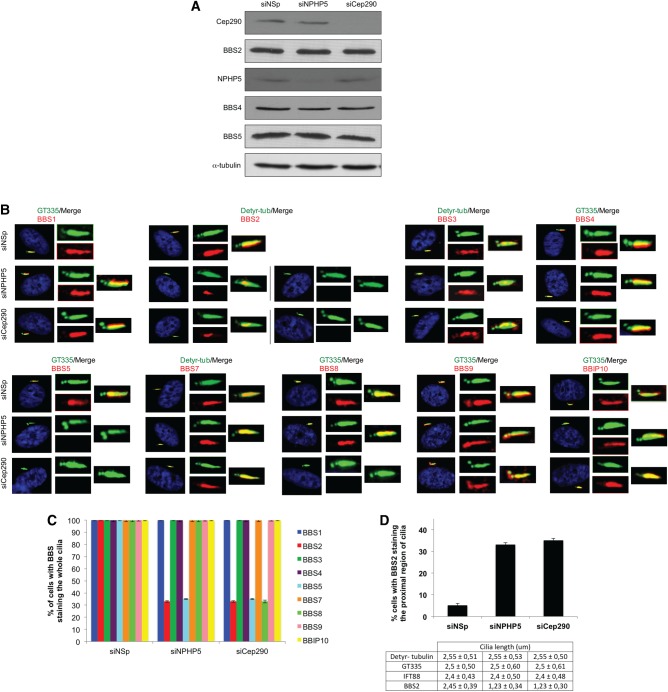
In an effort to reveal the biological relevance of the interaction between NPHP5 and the BBSome, we performed reciprocal depletion of NPHP5 and two BBS subunits, BBS2 and BBS5, using small-interfering RNAs (siRNAs) in HEK293, RPE-1 and ARPE-19 cells. Depletion of BBS2 or BBS5 had no effect on the levels and localization of NPHP5 (Supplementary Material, Fig. S3a–b), and likewise, ablation of NPHP5 did not alter the levels of BBS2, BBS4 or BBS5 (Fig. 5A). We and others have previously shown that although siRNA depletion of NPHP5 greatly compromises ciliogenesis (24,25), cilia can still form in cells where the silencing is not as efficient. Remarkably, we found that a loss of NPHP5 specifically affects ciliary localization of BBS2 and BBS5 without affecting other BBSome subunits (BBS1, BBS4, BBS7, BBS8, BBS9 and BBIP10) and BBS3 in cells that have retained their cilia (Fig. 5B–C). In particular, BBS5 was no longer detected at the cilium, whereas BBS2 was either completely absent from, or confined to a proximal region of, the cilium (Fig. 5B–D), reminiscent of the so-called inversin compartment or EvC zone (49–51). Of note, proximal ciliary confinement of BBS2 was not due to shortened cilia, because cilia length (as judged by staining of three different ciliary markers, detyrosinated tubulin, glutamylated tubulin and IFT88) was unaffected in NPHP5-depleted cells (Fig. 5B and D). Furthermore, depletion of NPHP5 did not result in enhanced staining or accumulation of other BBSome subunits (BBS1, BBS4, BBS7, BBS8, BBS9 and BBIP10) at the cilium (Fig. 5B), suggesting that this protein, unlike BBS17 or AZI1 (22,52), is not a negative regulator of BBSome ciliary trafficking. Next, we conducted rescue experiments in which we expressed full-length NPHP5 (1-598) or a NPHP5 mutant lacking the N-terminal BBSome-binding site (287–598) in quiescent cells depleted of endogenous NPHP5, using a siRNA oligo that targets the 3′UTR of NPHP5 mRNA. We found that although 1–598 and 287–598 both localize to centrosomes, the two phenotypes associated with NPHP5 depletion, namely, loss of ciliary BBS2/BBS5 and proximal ciliary staining of BBS2, were largely rescued by 1–598 but not 287–598 expression (Fig. 6A–B). These results suggest that NPHP5-binding to the BBSome is crucial for BBSome integrity and ciliary trafficking of certain subunits.
Figure 5.
Depletion of NPHP5 or Cep290 impairs ciliary localization of a subset of BBSome subunits. (A) Western blotting of Cep290, BBS2, BBS4, BBS5 and NPHP5 in HEK293 cells treated with control (siNSp), NPHP5 (siNPHP5) or Cep290 (siCep290) siRNAs. α-tubulin was used as loading control. (B) RPE-1 cells transfected with control (siNSp), NPHP5 (siNPHP5) or Cep290 (siCep290) siRNAs and induced to quiescence were stained with the indicated combinations of antibodies. Detyr-tub, detyrosinated tubulin; GT335, polyglutamylated tubulin. (C) The percentages of quiescent RPE-1 cells showing BBS staining along the entire length of cilia were determined using detyrosinated tubulin or GT335 as a ciliary marker. (D) (Top) The percentages of quiescent RPE-1 cells showing BBS2 staining along the proximal region of cilia were determined. (Bottom) Cilia length was measured with different markers (BBS2, detyrosinated tubulin, polyglutamylated tubulin and IFT88). In (C and D), at least 100 cells were counted and/or measured per siRNA condition, and error bars represent average of three independent experiments.
The mislocalization of BBS2 and BBS5 provoked by NPHP5 depletion strongly suggests that these two subunits are separated from the rest of the BBSome. To examine this possibility, we noticed that in size exclusion chromatography, BBS2, BBS4, BBS5 and BBS8 peak at around fractions 7–9 (∼500 kDa) in control cells (Fig. 6C) (12,22). In NPHP5-depleted cells, however, a significant amount of BBS2 and BBS5 was found in early fractions (fractions 5 and 6) (Fig. 6C), indicating that these two subunits co-fractionate in a high molecular weight complex distinct from the BBSome. The overall size of the remaining BBSome is expected to decrease after losing two critical subunits, and indeed, we reproducibly detected a shift of BBS4 and BBS8 to lower molecular weight fractions (fractions 9–11) in NPHP5-depleted cells, but not in control cells (Fig. 6C). Importantly, we also demonstrated that while endogenous BBS1, BBS2, BBS5 and BBS8 co-immunoprecipitated with GFP-BBS4 in control extracts, only BBS1 and BBS8 were co-immunoprecipitated in the absence of NPHP5 (Fig. 6D). Taken together, our data suggest the existence of a BBSome sub-complex, devoid of BBS2 and BBS5, which retains the capacity to traffic into cilia.
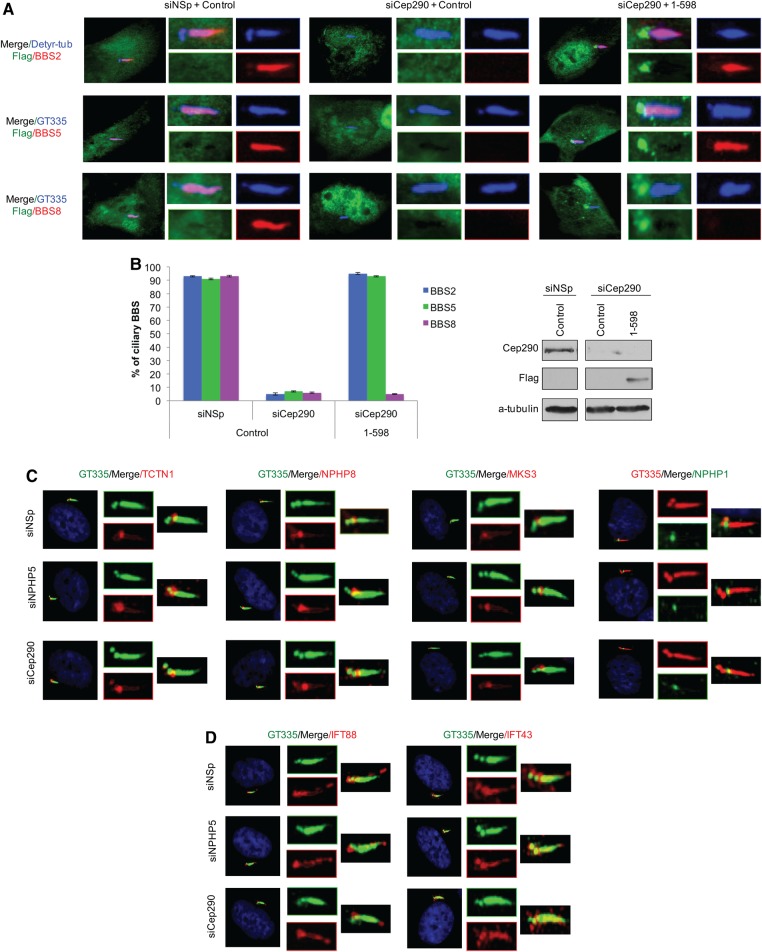
Because Cep290 docks NPHP5 at the centrosome/ciliary base and is required for ciliogenesis (24,25,53), we surmise that ablation of Cep290 should phenocopy NPHP5 loss of function. Depletion of Cep290 indeed prevented ciliary localization of BBS2 and BBS5, in addition to restricting BBS2 to the proximal region of cilia in a subpopulation of ciliated cells (Fig. 5B–D). Interestingly, ciliary localization of BBS8 was also abolished, a result consistent with previous work (54), whereas the remaining BBSome subunits (BBS1, BBS4, BBS7, BBS9 and BBIP10) and BBS3 were efficiently targeted to the cilium (Fig. 5B–C). Furthermore, GFP-BBS4 associated with BBS1 but not with BBS2, BBS5 or BBS8 in Cep290-depleted cells (Fig. 6D), suggesting that BBS2, BBS5 and BBS8 disintegrate and are no longer in a complex with BBS1/BBS4. To ascertain that the BBS8 phenotype is direct consequence of Cep290 loss and that suppression of Cep290 indirectly affects BBS2/BBS5, we performed rescue experiments in which we expressed full-length NPHP5 (1–598) in quiescent cells depleted of endogenous Cep290. We found that exclusion of BBS2/BBS5 from cilia and proximal ciliary confinement of BBS2 were largely rescued, whereas loss of ciliary BBS8 was not (Fig. 7A–B). Finally, we showed that the phenotype induced by NPHP5 or Cep290 loss is unique, because ciliary entry of the BBSome or any single subunit, including BBS2, BBS4, BBS5, BBIP10, BBS8 (22) and BBS9 (22), was precluded in BBS2- or BBS5-depleted cells (Supplementary Material, Fig. S3c). Considered together, our data suggest that NPHP5 maintains association of BBS2 and BBS5 with the BBSome, while Cep290 keeps BBS8 glued to the complex. In the absence of NPHP5 or Cep290, the BBSome is missing some subunits, and yet its trafficking to cilia is not compromised. Because only a holo-BBSome enters cilia (20–22), we reason that NPHP5 and Cep290 may perform an additional gatekeeping function to restrain entry of a malformed BBSome into cilia.
Figure 7.
Cep290 regulates BBSome integrity and depletion of NPHP5 or Cep290 does not impair the localization of transition zone proteins. (A) ARPE-19 cells transfected with control (siNSp) or Cep290 (siCep290) siRNA and plasmid expressing an irrelevant Flag-tagged protein (control) or full-length Flag-NPHP5 (1-598), induced to quiescence, were stained with antibodies against Flag (green), detyrosinated-tubulin or GT335 (blue), and BBS2, BBS5 or BBS8 (red). (B) (Left) The percentages of quiescent cells with ciliary BBS2, BBS5 or BBS8 staining were determined using either detyrosinated tubulin or GT335 as a ciliary marker. At least 100 transfected cells were counted per condition, and error bars represent average of three independent experiments. (Right) Western blotting of Flag and Cep290. α-tubulin was used as loading control. (C and D) RPE-1 cells transfected with control (NSp), NPHP5 (siNPHP5) or Cep290 (siCep290) siRNAs and induced to quiescence were stained with the indicated antibodies.
To assess the potential role of NPHP5 and Cep290 as a gatekeeper, we sought to determine if their inactivation triggers gross malformation of the transition zone at the ciliary base. We found that two components of the NPHP1/NPHP4/NPHP8 complex, NPHP1 and NPHP8, and two components of the MKS complex, TCTN1 and MKS3, were efficiently recruited to the ciliary base in NPHP5- or Cep290-depleted cells (Fig. 7C). In addition, no aberrant accumulation of two IFT subunits, IFT43 and IFT88 at the ciliary tip, indicative of impaired retrograde transport, was observed (Fig. 7D). Likewise, there was no apparent loss of ciliary IFT43 or IFT88 (Fig. 7D), suggesting that anterograde transport is not compromised. Moreover, depletion of NPHP5 or Cep290 did not affect ciliary trafficking or accumulation of most BBSome subunits (Fig. 5B–C). These data suggest that the transition zone is not grossly malformed, and ciliary content is not substantially altered. Therefore, it appears that these two proteins specifically regulate ciliary trafficking of the BBSome.
Loss of NPHP5 or Cep290 partially impairs ciliary targeting of BBSome cargoes
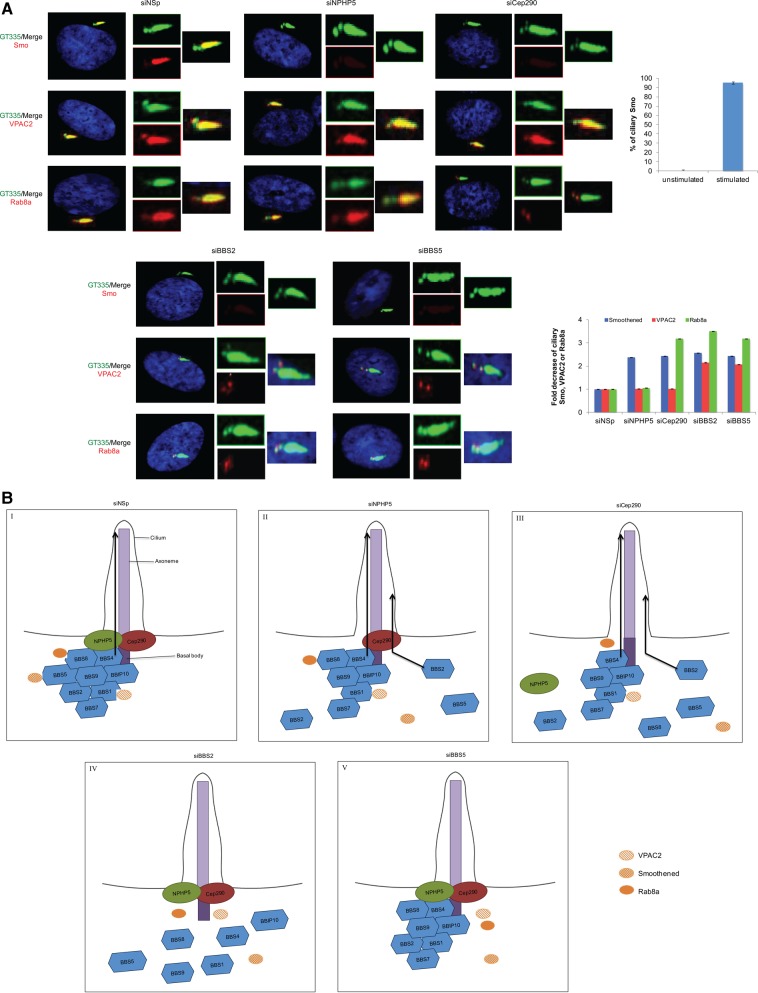
Next, we asked whether a malformed BBSome induced by a loss of NPHP5 or Cep290 could impinge on ciliary trafficking of its cargos. We hypothesize that such BBSome sub-complex is proficient in delivering some, but not all cargos to cilia. In mammals, the BBSome is known to traffic a subset of cargos, including G protein-coupled receptors smoothened (Smo), melanin-concentrating hormone receptor 1 (MCHR1), somatostatin receptor 3 (SSTR3), vasoactive intestinal peptide receptor 2 (VPAC2) and dopamine receptor 1 (D1R) into and/or out of cilia (16,22,55–59). In addition, this complex cooperates with the GTPase Rab8a to regulate vesicular trafficking and ciliary membrane biogenesis (12). Although previously reported in other cell lines and tissues (58,59), we were unable to detect endogenous MCHR1 or SSTR3 at the cilium or abnormal accumulation of ciliary D1R caused by depletion of BBS proteins in quiescent RPE-1 cells. Nevertheless, endogenous VPAC2 and Rab8a were localized to the centrosome and cilium, and although Smo was not detectable in unstimulated cells, it exhibited strong ciliary accumulation in response to a Smo agonist in quiescent RPE-1 cells (Fig. 8A). We first confirmed earlier reports (22,57) by showing that ciliary localization of Smo in response to stimulation, along with ciliary localization of VPAC2, was greatly diminished in cells depleted of BBS proteins (Fig. 8A). Ablation of BBS proteins also abolished ciliary targeting of Rab8a (Fig. 8A), consistent with the notion that this GTPase is a downstream effector of the BBSome. In contrast, centrosomal targeting of VPAC2 or Rab8a remained unaffected, suggesting that the BBSome may play a more prominent role in cargo delivery to cilia than cargo loading (Fig. 8A). In NPHP5 or Cep290-depleted cells, ciliary Smo was also dramatically reduced but was never confined to the proximal region of cilia (Fig. 8A), reminiscent of BBS5 mislocalization. Because BBS5 and Smo are known to interact (22), our data strongly suggest that a loss of ciliary BBS5 impairs Smo trafficking to cilia. Remarkably, we found that ciliary trafficking of VPAC2 remained unaffected upon suppression of NPHP5 or Cep290, whereas depletion of Cep290, but not NPHP5, resulted in exclusion of Rab8a from the ciliary membrane (Fig. 8A). Centrosomal targeting of Rab8a, on the other hand, remains unaffected. Thus, despite losing BBS2 and BBS5, the BBSome sub-complex owing to NPHP5 loss can still undergo ciliary trafficking and deliver Rab8a and VPAC2 to cilia. In the absence of Cep290, the BBSome sub-complex lacks BBS2, BBS5 and BBS8, and our data suggest that an additional loss of BBS8 leads to defective trafficking of Rab8a to cilia.
Figure 8.
Depletion of NPHP5 or Cep290 partially disrupts ciliary trafficking of BBSome cargos. (A) (Left) RPE-1 cells transfected with control (NSp), NPHP5 (siNPHP5), Cep290 (siCep290), BBS2 (siBBS2) or BBS5 (siBBS5) siRNAs, induced to quiescence, were stained with antibodies against Smo, VPAC2 or Rab8a (red) and GT335 (green). In the case of Smo, cells were treated with a Smo agonist prior to immunofluorescence. (Top right) The percentage of quiescent cells with ciliary Smo, untreated (unstimulated) or treated with a Smo agonist (stimulated), was determined using GT335 as a ciliary marker. (Bottom right) The fold decrease in the percentage of cells with ciliary Smo, VPAC2 or Rab8a was determined using GT335 as a ciliary marker. At least 100 cells were scored per condition, and error bars represent average of three independent experiments. (B) Model illustrating the roles of NPHP5 and Cep290 in BBSome homeostasis. (I) NPHP5 and Cep290 at the ciliary base serve two purposes: they regulate BBSome integrity and form a diffusion barrier that allows the selective passage of the holo-complex into the cilium. Although NPHP5 and Cep290 interact with multiple subunits, their interaction with BBS4/BBS8 is shown for the sake of simplicity. (II) In the absence of NPHP5, BBS2 and BBS5 dissociate from the BBSome, and BBS5 is completely mislocalized from the cilium. A fraction of BBS2 exhibits similar mislocalization pattern, whereas another fraction is confined to the proximal region of the cilium through an undefined mechanism. A faulty barrier permits the BBSome sub-complex to traffic into the cilium. Ciliary trafficking of Smo is impaired, since this cargo normally interacts with BBS5. (III) A loss of Cep290 induces NPHP5 mislocalization and promotes further dissociation of BBS8 from the BBSome; yet this sub-complex is also allowed to undergo ciliary trafficking. Rab8a ciliary trafficking is additionally impaired, but its centrosomal localization is unaffected. (IV and V) In contrast, a loss of any single BBS subunit, irrespective of its effects on BBSome assembly, is excluded from the cilium due to an intact barrier created by NPHP5/Cep290. Inactivation of BBS2 is known to destabilize BBS7 and prevents early BBSome assembly, whereas inactivation of BBS5 has minor impact on assembly (22,59). As a consequence, Smo, Rab8a and VPAC2 are unable to enter the ciliary compartment, and Rab8a and VPAC2 remain at the centrosome. We speculate that Rab8a is transported into the cilium by tethering to BBS8, whereas ciliary trafficking of VPAC2 likely requires interaction with BBS1, BBS4, BBS7, BBS9 and/or BBIP10. The interaction between VPAC2 and BBS1 is shown for simplicity.
Discussion
In recent years, cilia have become a topic of intense research interest because of their role in different signaling pathways and human disease. The BBSome is a multi-subunit complex required for proper cilia function, and defects in any one subunit can have detrimental consequences on complex formation and/or function, giving rise to disease (10,12,22). Therefore, current efforts focus on understanding how this complex is assembled and trafficked into cilia, and how it delivers cargos into the ciliary compartment. In order for the BBSome to gain access to cilia, it has to transit through a control barrier called the transition zone located at the ciliary base (23). The transition zone houses several protein complexes, including the Cep290/NPHP5 complex (24–26), whose function is incompletely understood. Here, we demonstrated for the first time that NPHP5 interacts with, and is located in close proximity to, every subunit of the BBSome, suggesting that NPHP5 associates with the holo-complex. We further showed that NPHP5 exists in a complex with Cep290 and the BBSome. Thus, these data add another layer of complexity to protein interaction network at the ciliary base, because NPHP5 is known to directly interact with Cep290 (24,25), while Cep290 interacts with BBS4 (39).
How exactly does NPHP5/Cep290 modulate the function of the BBSome, or vice versa? Our data strongly suggest that NPHP5 and Cep290 specifically regulate BBSome integrity. In the absence of NPHP5 or Cep290, the BBSome sub-complex is missing at least two subunits, BBS2 and BBS5. Because BBS2 is an early and essential player in BBSome assembly (15), it is difficult to envisage how this sub-complex, or for that matter the BBSome core, is formed without BBS2. Depletion of NPHP5 does not impinge on the levels or localization of BBS2 at the centrosome/ciliary base (Fig. 5A and data not shown), suggesting that BBSome assembly, which probably takes place in this location, is not affected. Although we cannot rule out the possibility that the BBSome is assembled at another sub-cellular compartment, such process would still require BBS2. We propose that the BBSome is either partially formed or not formed in non-ciliated cells, and its assembly could only take place at full throttle at the centrosome/ciliary base during cilia formation. At this critical juncture, protein interactions between different BBSome subunits are highly dynamic and are presumed to be broken and formed readily, with NPHP5/Cep290 serving as the glue to keep certain subunits together. It is currently unknown whether the holo-complex is first formed and a subset of subunits subsequently falls apart, or whether the sub-complex represents a late assembly intermediate. Further studies are needed to distinguish these two possibilities.
In addition to BBSome integrity, a number of our observations suggest that NPHP5 and Cep290 could act as a gatekeeper to specifically control entry of the BBSome to cilia. First, the transition zone does not appear to be not grossly malformed in the absence of NPHP5 or Cep290, and further structural analysis will be needed to unequivocally prove that the transition zone maintains its normal architecture. Second, depletion of NPHP5 or Cep290 does not affect anterograde and retrograde IFT. Third, with the exception of a subset of BBSome subunits, ciliary content of several other ciliary proteins tested remains normal. Forth, ciliary trafficking of BBSome cargos due to NPHP5 or Cep290 loss is partially impaired. In this regard, Cep290 has been postulated to form a diffusion barrier between the cilium and the cytoplasm, based on studies that it associates with a cohort of proteins at the transition zone, localizes to Y-shaped linkers that connect the axoneme to the ciliary membrane and directly binds to membranes and microtubules (30,60–62). The gatekeeping role of NPHP5/Cep290 is also consistent with the idea that only the holo-BBSome can traffic into cilia under normal conditions (Fig. 8B). When NPHP5/Cep290 is missing, a compromised gate mistakenly allows the malformed BBSome and a subset of associated cargos to enter cilia (Fig. 8B). In contrast, the malformed sub-complex provoked by depletion of any single BBS subunit is always incompetent to undergo ciliary trafficking, because ciliary entry is restricted by NPHP5/Cep290 at the transition zone (Fig. 8B).
In addition to being present at the transition zone (24,30, 31,48,60–62), Cep290 is known to localize to centriolar satellites (46,47) where it may facilitate the relocalization of BBS4 from the satellites to the cilium (54). This relocalization event is thought to be crucial for ciliary recruitment of BBS8 and possibly other BBSome subunits (54). Indeed, a previous report has shown that depletion of Cep290 diminishes ciliary localization of BBS4 and BBS8 in RPE-1 cells that are still able to form cilia (54). We also reported a loss of ciliary BBS8 here; however, we did not observe an impairment of ciliary BBS4 upon Cep290 knockdown. We speculate that this discrepancy may be due to efficiency of RNA silencing and/or detection of endogenous (our present study) versus recombinant BBS4 (54). Further studies will be needed to examine the relative contribution of the two pools of Cep290 (transition zone and satellites) to BBSome ciliary trafficking. It is important to note that unlike Cep290, NPHP5 is not recruited to centriolar satellites (data not shown).
The BBSome is thought to deliver specific membrane cargos to cilia (13,19). However, the majority of BBSome cargos have not been identified, and how different cargos are tethered to the holo-complex for transport is not known. We propose that different BBSome subunits tether a unique set of cargos to the complex for delivery to the ciliary compartment (Fig. 8B). In support of this, Smo is a BBSome cargo that interacts with BBS5 (22), and we have shown here that a loss of ciliary BBS5 and Smo can be triggered by ablation of NPHP5, Cep290 or BBS proteins. Likewise, the BBSome sub-complex induced by NPHP5 (or Cep290) depletion is able to promote ciliary trafficking of VPAC2 despite missing BBS2/BBS5 (and BBS8), raising the possibility that the remaining subunit(s) BBS1, BBS4, BBS7, BBS9 and/or BBIP10 tether(s) VPAC2 to the BBSome. Moreover, because a loss of ciliary BBS8 correlates with impaired trafficking of Rab8a to cilia, Rab8a may preferentially bind to BBS8. Further analysis will be needed to identify additional cargoes that are uniquely associated with each individual subunit.
Previously, it was shown that depletion of NPHP5 or Cep290 has no effect on hedgehog (Hh) signaling in osteoblasts (24). This result appears to contradict our findings that ciliary Smo, a key component of the Hh pathway, is reduced in NPHP5- or Cep290-depleted retinal epithelial cells. We speculate that NPHP5/Cep290 could function differently in different cell/tissue types. Because mutations in NPHP5/Cep290 often give rise to renal and retinal failure, these two proteins may specifically be involved in Hh signaling in these cell/tissue types. Indeed, abnormal Hh signaling has recently been reported in kidneys from Cep290 mutant mice (63), and it would be interesting to determine if such abnormality also contributes to retinal failure.
Materials and Methods
Cell culture and plasmids
Human ARPE-19, hTERT-RPE-1 and HEK293 cells were grown in dulbecco's modified eagle's medium supplemented with 10% FBS at 37°C in a humidified 5% CO2 atmosphere. To generate Flag-tagged NPHP5 fusion proteins, human NPHP5 cDNA fragments encoding residues 1–287, 157–332, 287–598(A549K), 287–598(Δ509–529) were amplified by PCR using Phusion High-Fidelity DNA Polymerase (New England Biolabs) and sub-cloned into mammalian expression vector pCBF-Flag. Human NPHP5 cDNA was also sub-cloned into mammalian vector pFlag-C-CMV to generate NPHP5-Flag. Other Flag-NPHP5 constructs were previously described (25). To generate GFP-tagged NPHP5 protein, human NPHP5 cDNAs were sub-cloned into mammalian vector pEGFP-C1 or pEGFP-N1. To generate GFP-BBS proteins, human BBS cDNAs (BBS1 to BBS12) were sub-cloned into mammalian vector pEGFP-C1. Human BBS cDNAs were also sub-cloned into mammalian vector pFlag-CMV5 or pFlag-C-CMV to generate Flag-BBS or BBS-Flag. All constructs were verified by DNA sequencing.
Antibodies
Antibodies used in this study included anti-NPHP5 (25), anti-Rab8a (64), anti-Cep290 (Bethyl Laboratories), anti-centrin, anti-detyrosinated tubulin, anti-BBS4, anti-BBIP10 (Millipore), anti-BBS1, anti-BBS2, anti-BBS3, anti-BBS4, anti-BBS7, anti-BBS9, anti-Cep290, anti-γ-tubulin-FITC, anti-VPAC2, anti-NPHP1 (Santa Cruz), anti-α-tubulin, anti-acetylated tubulin, anti-BBS8, anti-Flag, anti-GFP, anti-γ-tubulin, anti-NPHP8, anti-IFT43 (Sigma-Aldrich), anti-glutamylated tubulin GT335 (Invitrogen), anti-IFT88, anti-BBS5, anti-TCTN1, anti-MKS3 (ProteinTech), anti-Smoothened, anti-CaM and anti-NPHP5 (Abcam). Specificity of antibodies against BBS1, BBS2, BBS4, BBS5 and BBS7 is presented in Supplementary Material, Figs. S3 and S4.
Immunoprecipitation, immunoblotting and immunofluorescence microscopy
Cells were lysed with ELB buffer [50 mm HEPES/pH 7.4, 250 mm NaCl (or 150 mm NaCl as indicated), 5 mm ethylenediaminetetraacetic acid/pH 8, 0.1% NP-40, 1 mm dithiothreitol, 0.5 mm PMSF, 2 μg/ml leupeptin, 2 μg aprotinin, 10 mm NaF, 50 mm β-glycerophosphate and 10% glycerol] at 4°C for 30 min and extracted proteins were recovered in the supernatant after centrifugation at 16,000g. For immunoprecipitation, 2 mg of the resulting supernatant was incubated with an appropriate antibody at 4°C for 1 h and collected using protein A- or G-Sepharose. The resin was washed with lysis buffer, and bound proteins were analyzed by SDS-PAGE and immunoblotting with primary antibodies and horseradish peroxidase (HRP)-conjugated secondary antibodies (VWR). 100 μg of lysate was typically loaded into the input (IN) lane. For experiments involving recombinant protein expression, Flag-tagged and/or GFP-tagged constructs were (co-)transfected into HEK293 cells, and cells were harvested 48–72 h after transfection. Anti-Flag M2 beads (Sigma-Aldrich) or anti-GFP coupled to protein G-Sepharose were used for immunoprecipitations. For indirect immunofluorescence, cells were grown on glass coverslips, fixed with cold methanol or 4% paraformaldehyde and permeabilized with 1% Triton X-100/phosphate-buffered saline (PBS). Slides were blocked with 3% Bovine serum albumin (BSA) in 0.1% Triton X-100/PBS prior to incubation with primary antibodies. Secondary antibodies used were Cy3-, Cy5- or Alexa488- conjugated donkey anti-mouse, anti-rat or anti-rabbit IgG (Jackson Immunolabs and Molecular Probes). Cells were then stained with DAPI, and slides were mounted, observed and photographed using a Leitz DMRB (Leica) microscope (100×, NA 1.3) equipped with a Retiga EXi cooled camera. For ciliary staining, cells were pre-fixed in 0.4% paraformaldehyde for 5 min at 37°C, extracted with 0.5% Triton X-100 in PHEM buffer (60 mm PIPES, 25 mm HEPES, 10 mm EGTA, 2 mm MgCl2, pH 6.9) for 2 min at 37°C, and washed with PBS before proceeding with the normal protocol. To detect ciliary Smo, cells were treated with 1 µm purmorphamine (Abcam), a Smo agonist, for 24 h at 37°C prior to indirect immunofluorescence. Cilia length was determined using the Matlab software.
In situ PLA
Duolink in situ PLA kit (Sigma) was used following manufacturer's instructions. Proliferating or quiescent RPE-1 cells grown on glass coverslips were fixed and permeabilized according to the normal immunoflourescence protocol. After pre-incubation with a blocking reagent for 1 h, samples were incubated with two primary antibodies raised in different host species (rabbit, mouse or goat) for 1 h at room temperature. Following this, samples were washed in Duolink Wash Buffer A twice at room temperature. PLA anti-mouse/goat MINUS probe and anti-rabbit/mouse PLUS probe were then applied to samples in a preheated humidity chamber for 1 h at 37°C. Samples were washed in Wash Buffer A twice for 5 min, incubated with the Duolink Ligation Stock for 30 min at 37°C, washed in Wash Buffer A twice for 2 min, incubated with the Duolink Amplification Stock for 100 min at 37°C, washed in Wash Buffer B twice for 10 min, incubated with anti-γ-tubulin-FITC for 45 min and washed in Wash Buffer B once for 5 min. Finally, slides were mounted with the Duolink Mounting Medium with DAPI. ImageJ (v1.43m; Rasband, W.S., ImageJ, US National Institutes of Health, Bethesda, MD, USA) was used to quantify the PLA signal of each image. The PLA signal was assigned an arbitrary range of 0–1.00 (no signal, 0–0.25; weak signal, 0.26–0.50; medium signal, 0.51–0.75; strong signal, 0.76–1.00) and normalized according to the PLA signal of NPHP5/Cep290 pair, which had a value of 1.00.
RNA interference
Transfection of siRNAs was performed using siImporter (Millipore) per manufacturer's instructions. siRNAs for Cep290, NPHP5 and the non-specific control were previously described (25). Smartpool siRNAs against human BBS1, BBS2, BBS4, BBS5 and BBS7 were obtained from Thermo Fisher Scientific.
Induction of primary cilia
RPE-1 or ARPE-19 cells were brought to quiescence by serum starvation for 48–72 h and examined for cilia markers such as detyrosinated tubulin or glutamylated tubulin. Approximately 80–90% of cells formed cilia under this condition as opposed to ∼10% when cells were proliferating.
Size exclusion chromatography
Two milligrams of cell extract was chromatographed (ÄKTA FPLC; GE Healthcare) over a Superose-6 10/300 GL column (GE Healthcare). Then, 1 ml fractions were collected, TCA precipitated and analyzed by SDS-PAGE. The column was calibrated with Gel Filtration Standard (Bio-Rad) containing a mixture of molecular weight markers from 17 to 670 kDa.
Supplementary Material
Funding
This work was supported by the Canadian Institutes of Health Research (MOP-115033 to W.Y.T.) and the IRCM (Emmanuel Triassi scholarship to M.B.).
Supplementary Material
Acknowledgements
We thank all members of the Tsang Laboratory for constructive advice. W.Y.T. was a Canadian Institutes of Health Research New Investigator and a Fonds de recherche Santé Junior 1 Research Scholar.
Conflict of Interest statement: The authors have no conflicts of interest to declare.
References
- 1.Bornens M. (2012) The centrosome in cells and organisms. Science, 335, 422–426. [DOI] [PubMed] [Google Scholar]
- 2.Hossain D., Tsang W.Y. (2013) Centrosome dysfunction and senescence: coincidence or causality? Aging Sci., 1, 113. [Google Scholar]
- 3.Nigg E.A., Stearns T. (2011) The centrosome cycle: centriole biogenesis, duplication and inherent asymmetries. Nat. Cell Biol., 13, 1154–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Debec A., Sullivan W., Bettencourt-Dias M. (2010) Centrioles: active players or passengers during mitosis? Cell Mol. Life Sci., 67, 2173–2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim S., Dynlacht B.D. (2013) Assembling a primary cilium. Curr. Opin. Cell Biol., 25, 506–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Avasthi P., Marshall W.F. (2012) Stages of ciliogenesis and regulation of ciliary length. Differentiation, 83, S30–S42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tsang W.Y., Dynlacht B.D. (2013) CP110 and its network of partners coordinately regulate cilia assembly. Cilia, 2, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nigg E.A., Raff J.W. (2009) Centrioles, centrosomes, and cilia in health and disease. Cell, 139, 663–678. [DOI] [PubMed] [Google Scholar]
- 9.Hildebrandt F., Benzing T., Katsanis N. (2011) Ciliopathies. N. Engl. J. Med., 364, 1533–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Daniels A.B., Sandberg M.A., Chen J., Weigel-DiFranco C., Fielding Hejtmancic J., Berson E.L. (2012) Genotype–phenotype correlations in Bardet–Biedl syndrome. Arch. Ophthalmol., 130, 901–907. [DOI] [PubMed] [Google Scholar]
- 11.Zaghloul N.A., Katsanis N. (2009) Mechanistic insights into Bardet–Biedl syndrome, a model ciliopathy. J. Clin. Invest., 119, 428–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nachury M.V., Loktev A.V., Zhang Q., Westlake C.J., Peranen J., Merdes A., Slusarski D.C., Scheller R.H., Bazan J.F., Sheffield V.C., et al. (2007) A core complex of BBS proteins cooperates with the GTPase Rab8 to promote ciliary membrane biogenesis. Cell, 129, 1201–1213. [DOI] [PubMed] [Google Scholar]
- 13.Loktev A.V., Zhang Q., Beck J.S., Searby C.C., Scheetz T.E., Bazan J.F., Slusarski D.C., Sheffield V.C., Jackson P.K., Nachury M.V. (2008) A BBSome subunit links ciliogenesis, microtubule stability, and acetylation. Dev. Cell, 15, 854–865. [DOI] [PubMed] [Google Scholar]
- 14.Seo S., Baye L.M., Schulz N.P., Beck J.S., Zhang Q., Slusarski D.C., Sheffield V.C. (2010) BBS6, BBS10, and BBS12 form a complex with CCT/TRiC family chaperonins and mediate BBSome assembly. Proc. Natl. Acad. Sci. USA, 107, 1488–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Q., Yu D., Seo S., Stone E.M., Sheffield V.C. (2012) Intrinsic protein–protein interaction-mediated and chaperonin-assisted sequential assembly of stable Bardet–Biedl syndrome protein complex, the BBSome. J. Biol. Chem., 287, 20625–20635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jin H., White S.R., Shida T., Schulz S., Aguiar M., Gygi S.P., Bazan J.F., Nachury M.V. (2010) The conserved Bardet–Biedl syndrome proteins assemble a coat that traffics membrane proteins to cilia. Cell, 141, 1208–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wei Q., Zhang Y., Li Y., Zhang Q., Ling K., Hu J. (2012) The BBSome controls IFT assembly and turnaround in cilia. Nat. Cell. Biol., 14, 950–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kozminski K.G., Johnson K.A., Forscher P., Rosenbaum J.L. (1993) A motility in the eukaryotic flagellum unrelated to flagellar beating. Proc. Natl. Acad. Sci. USA, 90, 5519–5523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mykytyn K., Mullins R.F., Andrews M., Chiang A.P., Swiderski R.E., Yang B., Braun T., Casavant T., Stone E.M., Sheffield V.C. (2004) Bardet–Biedl syndrome type 4 (BBS4)-null mice implicate Bbs4 in flagella formation but not global cilia assembly. Proc. Natl. Acad. Sci. USA, 101, 8664–8669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blacque O.E., Li C., Inglis P.N., Esmail M.A., Ou G., Mah A.K., Baillie D.L., Scholey J.M., Leroux M.R. (2006) The WD repeat-containing protein IFTA-1 is required for retrograde intraflagellar transport. Mol. Biol. Cell, 17, 5053–5062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lechtreck K.F., Johnson E.C., Sakai T., Cochran D., Ballif B.A., Rush J., Pazour G.J., Ikebe M., Witman G.B. (2009) The Chlamydomonas reinhardtii BBSome is an IFT cargo required for export of specific signaling proteins from flagella. J. Cell. Biol., 187, 1117–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seo S., Zhang Q., Bugge K., Breslow D.K., Searby C.C., Nachury M.V., Sheffield V.C. (2011) A novel protein LZTFL1 regulates ciliary trafficking of the BBSome and Smoothened. PLoS Genet., 7, e1002358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reiter J.F., Blacque O.E., Leroux M.R. (2012) The base of the cilium: roles for transition fibres and the transition zone in ciliary formation, maintenance and compartmentalization. EMBO Rep., 13, 608–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sang L., Miller J.J., Corbit K.C., Giles R.H., Brauer M.J., Otto E.A., Baye L.M., Wen X., Scales S.J., Kwong M., et al. (2011) Mapping the NPHP-JBTS-MKS protein network reveals ciliopathy disease genes and pathways. Cell, 145, 513–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barbelanne M., Song J., Ahmadzai M., Tsang W.Y. (2013) Pathogenic NPHP5 mutations impair protein interaction with Cep290, a prerequisite for ciliogenesis. Hum. Mol. Genet., 22, 2482–2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schafer T., Putz M., Lienkamp S., Ganner A., Bergbreiter A., Ramachandran H., Gieloff V., Gerner M., Mattonet C., Czarnecki P.G., et al. (2008) Genetic and physical interaction between the NPHP5 and NPHP6 gene products. Hum. Mol. Genet., 17, 3655–3662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Williams C.L., Li C., Kida K., Inglis P.N., Mohan S., Semenec L., Bialas N.J., Stupay R.M., Chen N., Blacque O.E., et al. (2011) MKS and NPHP modules cooperate to establish basal body/transition zone membrane associations and ciliary gate function during ciliogenesis. J. Cell. Biol., 192, 1023–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Awata J., Takada S., Standley C., Lechtreck K.F., Bellve K.D., Pazour G.J., Fogarty K.E., Witman G.B. (2014) Nephrocystin-4 controls ciliary trafficking of membrane and large soluble proteins at the transition zone. J. Cell. Sci, 127, 4714–4727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chih B., Liu P., Chinn Y., Chalouni C., Komuves L.G., Hass P.E., Sandoval W., Peterson A.S. (2012) A ciliopathy complex at the transition zone protects the cilia as a privileged membrane domain. Nat. Cell. Biol., 14, 61–72. [DOI] [PubMed] [Google Scholar]
- 30.Garcia-Gonzalo F.R., Corbit K.C., Sirerol-Piquer M.S., Ramaswami G., Otto E.A., Noriega T.R., Seol A.D., Robinson J.F., Bennett C.L., Josifova D.J., et al. (2011) A transition zone complex regulates mammalian ciliogenesis and ciliary membrane composition. Nat. Genet., 43, 776–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kee H.L., Dishinger J.F., Blasius T.L., Liu C.J., Margolis B., Verhey K.J. (2012) A size-exclusion permeability barrier and nucleoporins characterize a ciliary pore complex that regulates transport into cilia. Nat. Cell. Biol., 14, 431–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Otto E.A., Helou J., Allen S.J., O'Toole J.F., Wise E.L., Ashraf S., Attanasio M., Zhou W., Wolf M.T., Hildebrandt F. (2008) Mutation analysis in nephronophthisis using a combined approach of homozygosity mapping, CEL I endonuclease cleavage, and direct sequencing. Hum. Mutat., 29, 418–426. [DOI] [PubMed] [Google Scholar]
- 33.Otto E.A., Loeys B., Khanna H., Hellemans J., Sudbrak R., Fan S., Muerb U., O'Toole J.F., Helou J., Attanasio M., et al. (2005) Nephrocystin-5, a ciliary IQ domain protein, is mutated in Senior-Loken syndrome and interacts with RPGR and calmodulin. Nat. Genet., 37, 282–288. [DOI] [PubMed] [Google Scholar]
- 34.Stone E.M., Cideciyan A.V., Aleman T.S., Scheetz T.E., Sumaroka A., Ehlinger M.A., Schwartz S.B., Fishman G.A., Traboulsi E.I., Lam B.L., et al. (2011) Variations in NPHP5 in patients with nonsyndromic leber congenital amaurosis and Senior-Loken syndrome. Arch. Ophthalmol., 129, 81–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Estrada-Cuzcano A., Koenekoop R.K., Coppieters F., Kohl S., Lopez I., Collin R.W., De Baere E.B., Roeleveld D., Marek J., Bernd A., et al. (2011) IQCB1 mutations in patients with leber congenital amaurosis. Invest. Ophthalmol. Vis. Sci., 52, 834–839. [DOI] [PubMed] [Google Scholar]
- 36.Chiang A.P., Beck J.S., Yen H.J., Tayeh M.K., Scheetz T.E., Swiderski R.E., Nishimura D.Y., Braun T.A., Kim K.Y., Huang J., et al. (2006) Homozygosity mapping with SNP arrays identifies TRIM32, an E3 ubiquitin ligase, as a Bardet–Biedl syndrome gene (BBS11). Proc. Natl. Acad. Sci. USA, 103, 6287–6292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Luo X., He Q., Huang Y., Sheikh M.S. (2005) Cloning and characterization of a p53 and DNA damage down-regulated gene PIQ that codes for a novel calmodulin-binding IQ motif protein and is up-regulated in gastrointestinal cancers. Cancer Res., 65, 10725–10733. [DOI] [PubMed] [Google Scholar]
- 38.Soderberg O., Gullberg M., Jarvius M., Ridderstrale K., Leuchowius K.J., Jarvius J., Wester K., Hydbring P., Bahram F., Larsson L.G., et al. (2006) Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat. Methods, 3, 995–1000. [DOI] [PubMed] [Google Scholar]
- 39.Zhang Y., Seo S., Bhattarai S., Bugge K., Searby C.C., Zhang Q., Drack A.V., Stone E.M., Sheffield V.C. (2014) BBS mutations modify phenotypic expression of CEP290-related ciliopathies. Hum. Mol. Genet., 23, 40–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wiens C.J., Tong Y., Esmail M.A., Oh E., Gerdes J.M., Wang J., Tempel W., Rattner J.B., Katsanis N., Park H.W., et al. (2010) Bardet–Biedl syndrome-associated small GTPase ARL6 (BBS3) functions at or near the ciliary gate and modulates Wnt signaling. J. Biol. Chem., 285, 16218–16230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dawe H.R., Smith U.M., Cullinane A.R., Gerrelli D., Cox P., Badano J.L., Blair-Reid S., Sriram N., Katsanis N., Attie-Bitach T., et al. (2007) The Meckel–Gruber syndrome proteins MKS1 and meckelin interact and are required for primary cilium formation. Hum. Mol. Genet., 16, 173–186. [DOI] [PubMed] [Google Scholar]
- 42.Shah A.S., Farmen S.L., Moninger T.O., Businga T.R., Andrews M.P., Bugge K., Searby C.C., Nishimura D., Brogden K.A., Kline J.N., et al. (2008) Loss of Bardet–Biedl syndrome proteins alters the morphology and function of motile cilia in airway epithelia. Proc. Natl. Acad. Sci. USA, 105, 3380–3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ansley S.J., Badano J.L., Blacque O.E., Hill J., Hoskins B.E., Leitch C.C., Kim J.C., Ross A.J., Eichers E.R., Teslovich T.M., et al. (2003) Basal body dysfunction is a likely cause of pleiotropic Bardet–Biedl syndrome. Nature, 425, 628–633. [DOI] [PubMed] [Google Scholar]
- 44.Kim J.C., Badano J.L., Sibold S., Esmail M.A., Hill J., Hoskins B.E., Leitch C.C., Venner K., Ansley S.J., Ross A.J., et al. (2004) The Bardet–Biedl protein BBS4 targets cargo to the pericentriolar region and is required for microtubule anchoring and cell cycle progression. Nat. Genet., 36, 462–470. [DOI] [PubMed] [Google Scholar]
- 45.Kim J.C., Ou Y.Y., Badano J.L., Esmail M.A., Leitch C.C., Fiedrich E., Beales P.L., Archibald J.M., Katsanis N., Rattner J.B., et al. (2005) MKKS/BBS6, a divergent chaperonin-like protein linked to the obesity disorder Bardet–Biedl syndrome, is a novel centrosomal component required for cytokinesis. J. Cell. Sci., 118, 1007–1020. [DOI] [PubMed] [Google Scholar]
- 46.Kim J., Krishnaswami S.R., Gleeson J.G. (2008) CEP290 interacts with the centriolar satellite component PCM-1 and is required for Rab8 localization to the primary cilium. Hum. Mol. Genet., 17, 3796–3805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chang B., Khanna H., Hawes N., Jimeno D., He S., Lillo C., Parapuram S.K., Cheng H., Scott A., Hurd R.E., et al. (2006) In-frame deletion in a novel centrosomal/ciliary protein CEP290/NPHP6 perturbs its interaction with RPGR and results in early-onset retinal degeneration in the rd16 mouse. Hum. Mol. Genet., 15, 1847–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tsang W.Y., Bossard C., Khanna H., Peranen J., Swaroop A., Malhotra V., Dynlacht B.D. (2008) CP110 suppresses primary cilia formation through its interaction with CEP290, a protein deficient in human ciliary disease. Dev. Cell, 15, 187–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shiba D., Yamaoka Y., Hagiwara H., Takamatsu T., Hamada H., Yokoyama T. (2009) Localization of Inv in a distinctive intraciliary compartment requires the C-terminal ninein-homolog-containing region. J. Cell. Sci., 122, 44–54. [DOI] [PubMed] [Google Scholar]
- 50.Shiba D., Manning D.K., Koga H., Beier D.R., Yokoyama T. (2010) Inv acts as a molecular anchor for Nphp3 and Nek8 in the proximal segment of primary cilia. Cytoskeleton (Hoboken), 67, 112–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dorn K.V., Hughes C.E., Rohatgi R. (2012) A smoothened–Evc2 complex transduces the Hedgehog signal at primary cilia. Dev. Cell, 23, 823–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chamling X., Seo S., Searby C.C., Kim G., Slusarski D.C., Sheffield V.C. (2014) The centriolar satellite protein AZI1 interacts with BBS4 and regulates ciliary trafficking of the BBSome. PLoS Genet., 10, e1004083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Graser S., Stierhof Y.D., Lavoie S.B., Gassner O.S., Lamla S., Le Clech M., Nigg E.A. (2007) Cep164, a novel centriole appendage protein required for primary cilium formation. J. Cell Biol., 179, 321–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stowe T.R., Wilkinson C.J., Iqbal A., Stearns T. (2012) The centriolar satellite proteins Cep72 and Cep290 interact and are required for recruitment of BBS proteins to the cilium. Mol. Biol. Cell, 23, 3322–3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang Q., Seo S., Bugge K., Stone E.M., Sheffield V.C. (2012) BBS proteins interact genetically with the IFT pathway to influence SHH-related phenotypes. Hum. Mol. Genet., 21, 1945–1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Berbari N.F., Lewis J.S., Bishop G.A., Askwith C.C., Mykytyn K. (2008) Bardet–Biedl syndrome proteins are required for the localization of G protein-coupled receptors to primary cilia. Proc. Natl. Acad. Sci. USA, 105, 4242–4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Soetedjo L., Glover D.A., Jin H. (2013) Targeting of vasoactive intestinal peptide receptor 2, VPAC2, a secretin family G-protein coupled receptor, to primary cilia. Biol. Open., 2, 686–694. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 58.Domire J.S., Green J.A., Lee K.G., Johnson A.D., Askwith C.C., Mykytyn K. (2011) Dopamine receptor 1 localizes to neuronal cilia in a dynamic process that requires the Bardet–Biedl syndrome proteins. Cell. Mol. Life Sci., 68, 2951–2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang Q., Nishimura D., Vogel T., Shao J., Swiderski R., Yin T., Searby C., Carter C.S., Kim G., Bugge K., et al. (2013) BBS7 is required for BBSome formation and its absence in mice results in Bardet–Biedl syndrome phenotypes and selective abnormalities in membrane protein trafficking. J. Cell Sci., 126, 2372–2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Craige B., Tsao C.C., Diener D.R., Hou Y., Lechtreck K.F., Rosenbaum J.L., Witman G.B. (2010) CEP290 tethers flagellar transition zone microtubules to the membrane and regulates flagellar protein content. J. Cell Biol., 190, 927–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Drivas T.G., Holzbaur E.L., Bennett J. (2013) Disruption of CEP290 microtubule/membrane-binding domains causes retinal degeneration. J. Clin. Invest., 123, 4525–4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gorden N.T., Arts H.H., Parisi M.A., Coene K.L., Letteboer S.J., van Beersum S.E., Mans D.A., Hikida A., Eckert M., Knutzen D., et al. (2008) CC2D2A is mutated in Joubert syndrome and interacts with the ciliopathy-associated basal body protein CEP290. Am. J. Hum. Genet., 83, 559–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hynes A.M., Giles R.H., Srivastava S., Eley L., Whitehead J., Danilenko M., Raman S., Slaats G.G., Colville J.G., Ajzenberg H., et al. (2014) Murine Joubert syndrome reveals Hedgehog signaling defects as a potential therapeutic target for nephronophthisis. Proc. Natl. Acad. Sci. USA, 111, 9893–9898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Peranen J., Auvinen P., Virta H., Wepf R., Simons K. (1996) Rab8 promotes polarized membrane transport through reorganization of actin and microtubules in fibroblasts. J. Cell Biol., 135, 153–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.