Abstract
Mitochondrial diseases often exhibit tissue-specific pathologies, but this phenomenon is poorly understood. Here we present regulation of mitochondrial translation by the Mitochondrial Translation Optimization Factor 1, MTO1, as a novel player in this scenario. We demonstrate that MTO1 mediates tRNA modification and controls mitochondrial translation rate in a highly tissue-specific manner associated with tissue-specific OXPHOS defects. Activation of mitochondrial proteases, aberrant translation products, as well as defects in OXPHOS complex assembly observed in MTO1 deficient mice further imply that MTO1 impacts translation fidelity. In our mouse model, MTO1-related OXPHOS deficiency can be bypassed by feeding a ketogenic diet. This therapeutic intervention is independent of the MTO1-mediated tRNA modification and involves balancing of mitochondrial and cellular secondary stress responses. Our results thereby establish mammalian MTO1 as a novel factor in the tissue-specific regulation of OXPHOS and fine tuning of mitochondrial translation accuracy.
Introduction
Mitochondrial diseases are a group of multisystemic, progressive and fatal disorders that are often defined by defects in oxidative phosphorylation (OXPHOS), which affect the cellular ATP supply (1). A common feature of mitochondrial diseases is a strong tissue-specific phenotype. However, the molecular mechanisms that govern this tissue-specific regulation are only poorly understood (2–4). Recently, modulation of mitochondrial translation and its fidelity is increasingly being recognized as playing a significant role in the tissue-specific variation of OXPHOS in health and disease states (5,6).
A promising candidate that could be a novel regulator of this phenomenon is the Mitochondrial Translation Optimization Factor 1 (MTO1). MTO1 is an evolutionarily conserved protein. In mammals, MTO1 is predominantly expressed in high-energy demand tissue (7). In humans, various missense mutations causing early-onset mitochondrial disease associated with a strong heart-specific phenotype have been described (Table 1). The yeast (MTO1) and the Escherichia coli homolog (GidA) of the mammalian MTO1 are involved in the biosynthesis of the hypermodified 5-methylaminomethyl-2-thiouridine group of mnm5s2U34 in the wobble position of tRNALys, tRNAGlu and tRNAGln (10). In E. coli, this modification is important for maintaining tRNA structure and function by affecting its stability, aminoacylation and codon recognition (11–14). This tRNA modification is conferred by several proteins, within a complex pathway. However, many steps remain unclear (15). In mammals, the homologs of the thiolation-pathway Mss1 (E. coli MnmE), MTO1 (E. coli GidA) and TRMU (also termed Mtu1 or Mto2, E. coli TrmU) are supposed to be involved in taurine modification of tRNAs (τm5U), a modification that is unique to mammalian mitochondria (16). These taurine-modified tRNAs are then supposedly further modified by thiolation to yield the τm5S5-modified uracil position.
Table 1.
Clinical synopsis and biochemical features of MTO1 patients in present and published studies
| Mutation in MTO1 | Familiarity | Gender | Clinical features | Age/Cause of death | Metabolic finding | Biochemical MRC defects | References |
|---|---|---|---|---|---|---|---|
| P[Ile408Phe]; [Ile408Phe] |
Brother of four healthy siblings, three deceased siblings, consanguineous parents | M | Pyschomotor retardation, perinatal asphyxia, hypotonia, dysdiachokinesia, hypertrophic cardiomyopathy. | 15 years – |
Lactic acidemia, hyperalaninemia | Ms: CI ↓ Fbs: ↓ CI/CS: 0.02 (norm: 0.11–0.26) ↓ C13/CS: 0.08 (norm: 0.11–0.24) ↔ CII/CS: 0.34 (norm: 0.14–0.43) ↔ C23/CS: 0.37 (norm 0.10–0.29) ↔ CIII/CS: 1.61 (norm: 0.75–2.32) ↓ COX/CS: 0.66 (norm: 0.83–2.40) ↔ CV/CS: 0.75 (norm: 0.30–0.75) ↔ PDHC/CS: 0.031 (norm: 0.011–0.033) |
Present study |
| p. [Ala428Thr]; [Arg477His] |
No | F | Psychomotor delay, hypotonia, dystonia. Later, hypertrophic cardiomyopathy. | 14 years – |
Lactic acidemia, hyperalaninemia | Ms: ↓ CI and CIV Fbs: ↓ MRR |
(8) |
| p. [Thr411Ile]; [Thr411Ile] |
Brother of 3 siblings; consanguineous parents |
M | Poor feeding due to swallowing difficulties. Failure to thrive. Later, hypertrophic cardiomyopathy. Aspiration pneumonia. Hypotonia. | +12 months cardio-respiratory arrest |
Hypoglycemia, lactic acidemia | Ms: ↓ CI and CIV Fbs: ↓ MRR |
(8) |
| p. [Thr411Ile]; [Thr411Ile] |
Brother of 2 siblings; consanguineous parents |
M | Poor feeding due to swallowing difficulties. Failure to thrive. Early-onset hypertrophic cardiomyopathy. Hypotonia. | +3 months n.d. |
Lactic acidemia | n.d. | (8) |
| p. [Thr411Ile]; [Thr411Ile] |
Sister of 5 siblings; consanguineous parents |
F | Early-onset hypertrophic cardiomyopathy. Bronchiolitis-like illness. Encephalopathy and seizures. | 19 years – |
Lactic acidemia | Ms: ↓ CIV | (8) |
| p. [Thr411Ile]; [Thr411Ile] |
Sister of 4 siblings; consanguineous parents |
F | Upper respiratory illness. Hypertrophic cardiomyopathy and WPW. Psychomotor delay. | 12 years – |
Lactic acidemia, hyperalaninemia, ketonuria | Ms: ↓ CIV | (8) |
| p. [Ala428Thr]; [Arg620Lysfs*8] |
Brother of 7 siblings | M | Hypertrophic cardiomyopathy. | +19 days sudden bradycardia |
Lactic acidemia, hyperalaninemia | Ms: ↓ CI and CIV Fbs: ↓CIII and CIV; ↓ MRR |
(9) |
| p. [Ala428Thr]; [Arg620Lysfs*8] |
Sister of 6 siblings | F | Hypertrophic cardiomyopathy with tachycardia. Hypotonia. | +40 days sudden bradycardia | Lactic acidemia | Ms: ↓ CI and CIV Fbs: ↓ CI; ↓ MRR |
(9) |
| p. [Ala428Thr]; [Ala428Thr] |
No | M | Weakness, lack of ocular fixation. Hypertrophic cardiomyopathy with sinus bradycardia. Moderate bilateral optic atrophy. | 20 years – |
Lactic acidemia | Ms: ↓ CI and CIV | (9) |
Ms, muscle biopsy; Fbs, fibroblasts; MRC, mitochondrial respiratory chain; CI–CV, complexes I–V; C13, complex I + III activity; C23, complex II + III activity, CS, citrate synthase; PDHC, pyruvate dehydrogenase complex; WPW, Wolff–Parkinson–White syndrome.
Disturbed thiolation and taurine modification of mitochondrial tRNAs has been implicated in the pathomechanism of mitochondrial diseases caused by mutations in mitochondrial tRNAs such as MELAS and MERFF (17–21). However, the exact mechanism of the taurine and thiol modification of tRNAs in mammalian mitochondria and the function of each proposed component remains speculative, and it is, as yet, unclear whether these factors are truly dispensable for mammalian mitochondrial translation (22,23).
In the present study, we demonstrate that MTO1 is a major factor in the tissue-specific control of OXPHOS complexes by regulating tRNA modification and mitochondrial translation in a tissue-specific manner. We further highlight that MTO1 deficiency activates mitochondrial proteases and severely affects OXPHOS protein assembly and stability, suggesting an important role for MTO1 in ensuring translation fidelity. Crucially, for the patient population, we propose that a ketogenic diet (KD) may have therapeutic potential for MTO1-associated disorders by balancing secondary stress responses without affecting the tRNA modification defect, as these effects were seen in our mouse model. These findings not only highlight modulation of mitochondrial translation in response to cellular nutrient sensing, but also underline the importance of (mal)adaptive responses in the pathomechanism of mitochondrial disorders.
Results
A novel pathogenic mutation of MTO1 results in a combined CI + IV defect in patient fibroblasts associated with a defect in mitochondrial protein synthesis and tRNA modification
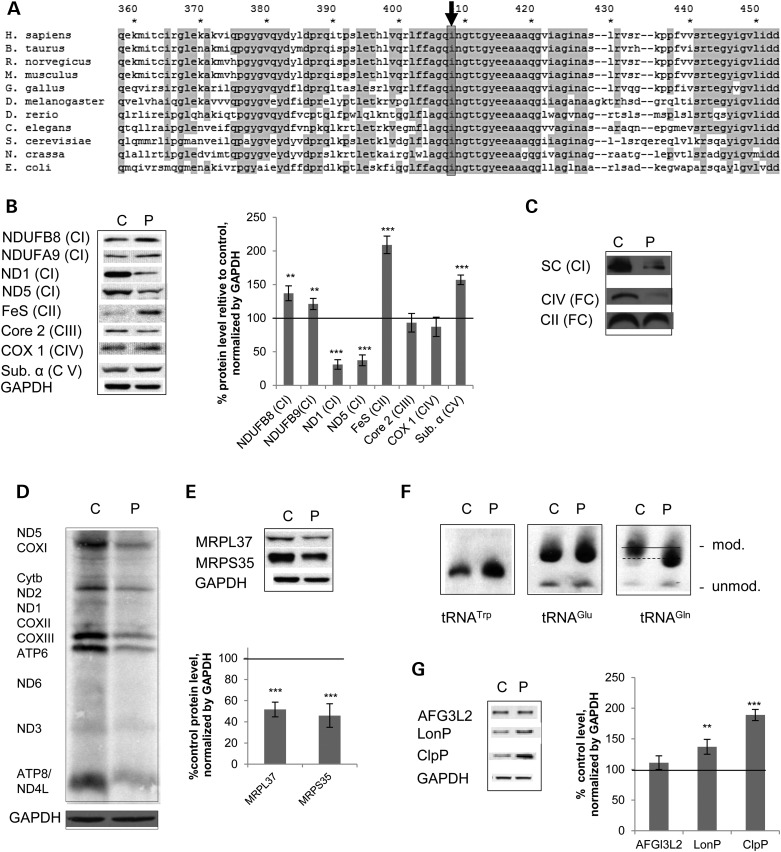
We identified a homozygous p.Ile408Phe mutation in MTO1 by next generation sequencing in a patient presenting with hypertrophic cardiomyopathy associated with lactic acidemia. Similar predominantly heart-related clinical symptoms were also observed in previously described patients with MTO1 mutations (Table 1). In our patient, Complex I was decreased in skeletal muscle biopsy. In fibroblasts, a selective defect was detected in Complex I and IV activity (Table 1). Human Ile408 is situated in a region that is highly conserved throughout the kingdoms (Fig. 1A). According to structural data of the MTO1 bacterial homolog GidA, human Ile408 is situated in the C-terminal end of β-sheet 22 adjacent to helix 9 in the designated FAD-binding domain (24). Mutations in this domain do not impact on protein stability but affect tRNA-modifying function and result in decreased thiourdinylation in bacteria (24).
Figure 1.
A novel pathological MTO1 mutation results in a Complex I + IV defect associated with a compromised mitochondrial translation, altered tRNA modification and activation of mitochondrial proteases in patient fibroblasts. (A) Alignment of MTO1 homologs from different species showing the area around the mutated residue Ile408 (highlighted). (B) Western blot analysis and quantification of whole cell extracts from healthy control (C) and patient fibroblasts (P). Subunits of OXPHOS Complexes I (NDUFB8, NDUFA9, ND1, ND5), II (FeS), III (Core2), IV (COX1) and V (ATPα) were probed. GAPDH was used as a loading control. (C) BN-PAGE analysis of digitonin-extracted fibroblasts. Presence of OXPHOS complexes in their free (FC) and supercomplex form (SC) as assessed by immunoblotting using antibodies directed against Complex I (SC) and the free complexes. (D) In vivo metabolic pulse-labeling experiment using [35S]-methionine in the presence of cyclohexamide to label mitochondrial-encoded proteins and thereby assess de novo mitochondrial translation in control and patient fibroblasts. (E) Western blot analysis and quantification of mitoribosomal proteins. Tested were components of the small (MRPS35) and the large mitoribosomal subunit (MRPL37). GAPDH was used as a loading control. (F) APM-northern blot of total RNA to visualize 2-thiouridiniylated tRNAGlu, tRNAGln and tRNATrp. The thio-modified and unmodified forms are indicated. The changed migration pattern of modified tRNAGln in the patient fibroblast is indicated by a dashed line. (G) Western blot analysis and quantification of mitochondrial proteases in control (C) and patient fibroblasts (P). VDAC was used as a loading control. *P < 0.05, **P < 0.01, ***P < 0.001.
To understand the pathological impact of the Ile408Phe mutation, we performed western blot analysis of whole cell lysates from control and patient fibroblasts. While the mitochondrial-encoded Complex I subunits ND1 and ND5 were significantly decreased to ∼25% of control levels, the protein level of the nuclear-encoded Complex I subunit NDUFB8 was significantly increased in patient cells. The α subunit of Complex V, as well as the FeS subunit of the Complex II, was also increased. No significant effect on the analyzed Complex IV (COXI) and Complex III (Core 2) subunits could be detected (Fig. 1B). Recently, the ratio between mitochondrial- and nuclear-encoded proteins, the so-called mito-nuclear protein ratio, has been associated with lifespan regulation (6). When we assessed this value in our patient fibroblasts, we observed a decreased mito-nuclear protein ratio not only for the mitochondrial-encoded subunits ND1 and ND5 showing decreased protein levels, but also for COXI (Supplementary Material, Fig. S1A).
We next performed Blue-Native (BN)-PAGE analysis to assess the impact of the MTO1 mutation on steady-state levels of fully assembled OXPHOS complexes and their supramolecular assemblies. These supercomplexes are particularly important to stabilize Complex I (25,26). In patient fibroblasts, supercomplexes containing Complex I as well as free Complex IV were decreased in line with decreased enzymatic activities (Fig. 1C; Table 1).
Human MTO1 was initially predicted to be involved in the optimization of mitochondrial translation. Expression of mutant MTO1 carrying pathogenic human mutations in yeast results in protein synthesis defects (8,9). However, the molecular basis has not been unraveled. To understand how the p.Ile408Phe MTO1 mutation affects human mitochondrial translation, we performed a metabolic pulse labeling of mtDNA encoded proteins. In patient fibroblasts, we found a general translation defect as indicated by an overall decrease in the labeling intensity (Fig. 1D). This result testifies for the first time that mutations in MTO1 affect protein synthesis not only in a yeast model system (8,9), but also in the original mammalian context. The signal in the patient samples was too weak to detect if any subunit was specifically affected. The mitochondrial translation defect was associated with up to 50% decreased levels of mitoribosomal proteins. Proteins of both the large (MRPL37) and small subunit (MRPS35) showed decreased steady-state levels in patients’ fibroblasts (Fig. 1E), suggesting that MTO1 might influence the mitoribosomal machinery itself.
Based on the homology with bacterial enzymes, MTO1 is hypothesized to be involved in the thiolation of selected mammalian mitochondrial tRNAs (16), but a demonstration is still lacking. To address this question, we analyzed the thiouridine modification of mitochondrial tRNAs by the retardation of electrophoretic mobility in a polyacrylamide gel containing [(N-acryloylamin) phenyl]mercuric chloride (APM) following northern blotting. In the resulting blot, the upper band represents the thiouridinylated tRNA (modified) and the lower band the unthioruridinylated species (unmodified). The gross thiouridinylation pattern for tRNAGlu did not change in our patient cells. However, the thiouridinylated form of tRNAGln exhibited an altered pattern compared with the control and migration more quickly indicating alterations in this tRNA modification (Fig. 1F). This finding indicates aberrant thiolation of tRNAGln in MTO1 mutant cells and demonstrates for the first time a role of mammalian MTO1 in modification of this mitochondrial tRNA. The altered thiolation pattern did not affect stability of tRNAGln (Supplementary Material, Fig. S1B). We did not observe any APM-modified form for tRNATrp, implying that there is no thiolation occurring for this tRNA within mitochondria (Fig. 1F).
Precise mitochondrial tRNA modification is necessary for correct codon–anti-codon recognition (20,27,28). This recognition is part of a complex network to maintain translation fidelity and accuracy (29–31). Translational infidelity causes amino acid misincorporation, resulting in protein misfolding and hence assembly defects or increased protein turnover. Amino acid misincorporation has been found in MELAS, a mitochondrial syndrome of encephalomyopathy, lactic acidosis with stroke-like episodes, which is associated with an OXPHOS defect (32). Intriguingly, aberrant tRNA modification is implicated in MELAS (16,33) and might be the underlying cause of the compromised translation accuracy. A readout of a compromised translation fidelity is the activation of mitochondrial proteases, which can serve as markers for the mitochondrial unfolded protein response (UPR) (34) and has been recently demonstrated to be sensitive to tRNA modification (35). We assessed protein levels of the mAAA-protease, Lon-protease and Clp protease by analyzing the steady-state levels of their subunits (Fig. 1G). The mAAA-protease has been implicated in the stability of OXPHOS proteins in a pathogenic context (36). ClpP is part of the defined mtUPR program (34), while LonP is a matrix-localized protease proposed to be involved in the quality control of soluble proteins in this mitochondrial compartment (37). While the protein level of the mAAA-subunit AFG3L2 remained unchanged in patient fibroblasts, the MTO1 mutation p.Ile408Arg induced a significant increase of LonP protein levels. Steady-state levels of ClpP approximately doubled implying a robust activation of mtUPR in the patient cell line (Fig. 1G). These findings are consistent with decreased translation accuracy in MTO1 mutant cells and hence activation of the mitochondrial quality control system as an adaptive response. A hyperactivation of the protease could also explain the decreased mito-nuclear protein ratio in MTO1 patient fibroblasts, which is a sign of disturbed mitochondrial protein homeostasis.
MTO1 deficiency in MEFs recapitulates the pathogenic CI + IV defect and is associated with aberrant supramolecular assemblies of OXPHOS proteins
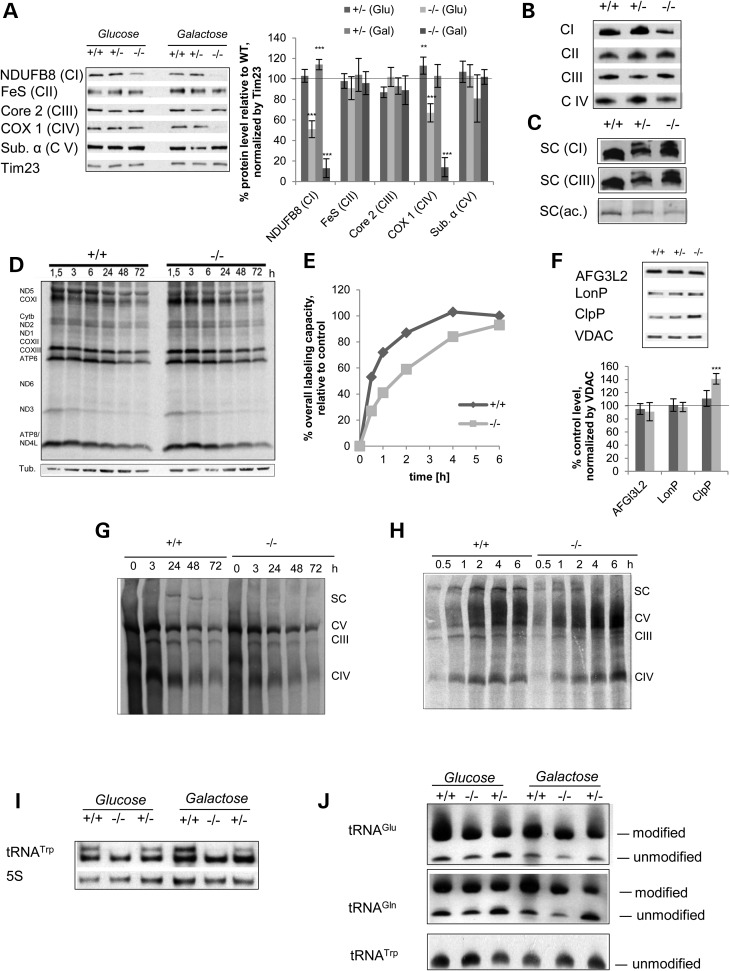
To further assess the molecular mechanism how MTO1 affects OXPHOS, we obtained mouse embryonic fibroblasts (MEFs) from a mouse model with of MTO1 deficiency (38). These mice develop cardiovascular symptoms including bradycardia and cardiomyopathy mimicking the clinical feature of patients with MTO1 mutations (38).
We grew MEFs obtained from mice which are wild-type (+/+), heterozygous (+/−) and homozygous (−/−) for the MTO1 cassette in high glucose media and in galactose-containing media. In the latter, metabolism switches from glycolytic to oxidative metabolism and hence relies on functional OXPHOS for cell survival (39,40), thereby exacerbating OXPHOS defects (41). To assess the effect of the MTO1 deficiency on OXPHOS, we performed western blot analysis of isolated mitochondria. The Complex I subunit NDUFB8 was reduced to ∼50% in MTO1-deficient mitochondria isolated from MEFs grown in high glucose media. This defect was worsened when cells had to rely on galactose: under these conditions, the NDUFB8 subunit was barely detectable (Fig. 2A). In glucose-based media, a significant decrease of the Complex IV subunit COX1 was also detected. This defect was again worsened by growth in galactose to only ∼10% of steady-state levels control COX1 protein (Fig. 2A). Further analysis revealed that other subunits of OXPHOS complexes were unaffected. As a consequence of the reduced COX1 protein, the mito-nuclear protein ratio was also decreased in MTO1 mutant MEFs with the most severe effect becoming evident when cells had to rely on galactose (Supplementary Material, Fig. S1C). Since the galactose-containing media revealed a severe OXPHOS defect, the cells did not tolerate longer periods of growth and expansion in this media. We were, therefore, forced to perform experiments that require a larger number of cells in high glucose conditions.
Figure 2.
MTO1 deficiency in MEFs induces a CI + CIV deficiency associated with a defect in mitochondrial protein synthesis and OXPHOS complex assembly. (A) Western blot analysis and quantification of mitochondria isolated from MEFs of WT mice (+/+) and mice that are heterozygous (+/−) or homozygous (−/−) for the MTO1 cassette. Subunits of OXPHOS Complexes I (NDUFB8), II (FeS), III (Core2), IV (COX1) and V (ATPα) were tested. The mitochondrial translocase Tim23 was used as a loading control. Shown are representative blots. (B) BN-PAGE analysis of fully assembled total OXPHOS complexes in mitochondria extracted with lauryl maltoside. Presence of complexes as probed by immunoblotting. (C) BN-PAGE analysis of respiratory supercomplexes (SC) in mitochondria extracted with digitonin. Presence of OXPHOS complexes in the SC as probed by immunoblotting. Complex I in-gel activity (SC (ac.)) was used to determine functionality of the different SC species in MTO1 mutant MEFs. (D) Metabolic pulse-chase labeling of newly synthesized mtDNA-encoded proteins resolved on a SDS-gel to determine the turnover of newly synthesized proteins. (E) Metabolic pulse titration of newly synthesized mtDNA-encoded proteins to determine changes in the rate of translation. Depicted is the quantification of total labeling intensity as a function of pulse time. See Supplementary Material, Figure S1E for gel image. (F) Western blot analysis and quantification of mitochondrial proteases in mitochondria isolated from MTO1 mutant and controls MEFs. (G) Metabolic pulse-chase labeling of newly synthesized mtDNA-encoded proteins resolved on a BN-PAGE to determine the turnover of supercomplexes (SC) and free OXPHOS Complexes III, IV and V. Loading was normalized according to protein content. (H) Metabolic pulse titration of newly synthesized mtDNA-encoded proteins resolved on BN-PAGE to visualize formation of OXPHOS complexes (CI, III, IV and V) and supercomplexes (SC). After the pulse, the cells were incubated for 24 h to allow assembly of the newly synthesized proteins into the subunits. Loading was normalized according to protein content. (I) Altered migration pattern of mitochondrial tRNATrp in a northern blot experiment. (J) APM-northern blot of total RNA to visualize 2-thiouridiniylated tRNAGlu and tRNAGln. The thio-modified and unmodified forms are indicated. **P < 0.01, ***P < 0.001.
To further analyze the observed OXPHOS defect and to assess steady-state levels of fully assembled complexes as well as their supramolecular assemblies, we performed BN-PAGE analysis. When assessing total complexes, we found that assembled Complex I and IV levels were reduced in homozygous MTO1 mutant MEFs while other OXPHOS complexes remained unaffected (Fig. 2B), consistent with our western blotting data. BN-PAGE analysis of supramolecular assemblies of OXPHOS Complexes I or III revealed an altered pattern of supercomplexes in MEFs containing one or two alleles of the cassette. Here, an additional band migrating at a higher molecular weight than the WT supercomplexes was observed (Fig. 2C). To gain insight into the functionality of these high-molecular CI + III positive species, we performed CI in gel activity assay (IGA). Both hetero- and homozygous MTO1 mutant MEFs generated an entity with the same migration height as WT. However, the staining intensity, was reduced in both mutant cells lines with a more striking effect in the homozygous MTO1 mutant. Interestingly, the higher molecular entities detected by immunoblotting were catalytically inactive by IGA, indicating that these higher CI + III positive supramolecular assemblies are non-functional with respect to CI activity (Fig. 2C) and may represent misfolded and/or misassembled complexes.
MTO1 deficiency compromises mitochondrial translation efficacy and activates mitochondrial proteases
To assess the cause of the selective Complex I + IV defect induced by MTO1 deficiency in MEFs and analyze the role of MTO1 in mammalian mitochondrial translation, we performed metabolic pulse-chase labeling experiments of newly synthesized mtDNA-encoded OXPHOS polypeptides to analyze mitochondrial protein synthesis capacity. We did not observe gross differences in the synthesis or turnover kinetics for the mtDNA-encoded subunits except for ND5 in MTO1-deficient cells that was markedly decreased to ∼50% of the WT levels of newly synthesized protein (Fig. 2D; Supplementary Material, Fig. S1D). All other subunits, including subunits COXI-III of Complex IV, remained unaffected by MTO1 deficiency. Stability of the newly synthesized subunits was not affected, as evidenced by an unchanged turnover pattern in our metabolic pulse-chase experiment (Fig. 2D). This finding suggests that post-translational mechanisms are likely to be involved in causing the CIV defect in MTO1-deficient MEFs.
The pulse-labeling assay of mitochondrial-encoded subunits is optimized on WT cells to assess the maximum protein synthesis capacity with pulse times that achieve maximal labeling intensity (42). To gain deeper insight into protein synthesis kinetics, we performed a time course of metabolic labeling to resolve time-dependent labeling intensity of mtDNA-encoded proteins (Fig. 2E; Supplementary Material, Fig. S1E). After ∼3 h of labeling, the maximal intensity is reached, but after only 0.5 h WT MEFs reached >50% and after 1 h ∼75% of the maximal labeling intensity. In comparison, MTO1-deficient MEFs show a translation defect in the form of a reduced translation rate. While a labeling intensity similar to maximal WT was observed at the 6 h time point, the intensity of MTO1 mutant MEFs was reduced relative to the WT control at all other time points. Further, the MTO1-deficient MEFs did not reach a plateau phase as did the WT within the analyzed timeframe. Instead labeling intensity continued to increase with prolonged labeling times, suggesting a reduced translation rate (Fig. 2E). This finding links MTO1 to the regulation of mitochondrial translation kinetics and concurs with a previous report of a reduced translation rate caused by impaired mitochondrial tRNA modification due to the MELAS mutation (43).
In patient fibroblasts, the MTO1 mutation causes a mitochondrial translation defect associated with an increase in mitochondrial proteases suggesting compromised translation fidelity. MTO1 deficiency in MEFs recapitulated the CI + IV defect observed in patients. This defect was associated with aberrant supramolecular assemblies of OXPHOS proteins, which lack enzymatic functionality (see above). To assess whether these misassembled complexes induce mitochondrial UPR, we tested the activation of mitochondrial proteases in this context. We found a selective increase of ClpP protease in MTO1-deficient MEFs (Fig. 2F) reminiscent of the observation in patient fibroblasts and consistent with the hypothesis that loss of MTO1 results in compromised translation accuracy and hence misfolded proteins.
Loss of MTO1 in MEFs results in an OXPHOS assembly and stability defect
Since MTO1 deficient MEFs show a defect in OXPHOS Complexes I and IV but only manifest a clear translational defect in the Complex I subunit ND5, we next addressed whether steps that follow protein synthesis might be involved in the observed OXPHOS defect. We therefore performed a pulse titration assay to assess the time-resolved formation of the different complexes as well as a pulse-chase experiment to analyze the turnover of the assembled complexes using BN-PAGE.
To address this question, we assessed the stability of newly assembled complexes by pulse-chase experiments that monitored the fate of the complexes for 72 h after the initial pulse. In WT cells, newly formed supercomplexes were stable and decay was detected only after 48 h. In MTO1-deficient cells, we were not able to detect supercomplexes at the assessed time points consistent with the previous experiment where we found an assembly defect of supercomplexes (Fig. 2G). With respect to CIV, the pulse-chase experiment revealed that also assembly of CIV is affected by MTO1 deficiency: despite equal labeling intensity, the appearance of the signal for assembled CIV in MTO1 mutant MEFs lacked behind compared with that of the control cells. Given that no specific defect in the de novo synthesis of COX subunits could be detected, we believe that the CIV defect in MTO1-deficient MEFs is caused by suboptimal synchronization of assembly and turnover of CIV. A WT pattern in a sucrose floatation assay implied that integration of the newly synthesized subunits into the membrane is not affected by loss of MTO1 (Supplementary Material, Fig. S1F).
The supercomplex defect in this pulse-chase experiment prompted us to investigate whether a longer labeling procedure and hence the possibility to produce more required subunits could enhance the assembly of the supercomplexes. We therefore performed a pulse titration experiment followed by BN-PAGE analysis. Here we observed a striking defect of supercomplex formation in MTO1-deficient MEFS. While supercomplex formation increases with prolonged pulse times and reaches a stationary phase at ∼2 h of pulse time in WT MEFs, the level of correctly formed supercomplexes is much lower in MTO1-deficient MEFs at 0.5, 1 and 2 h of time points. After 2 h, assembled supercomplexes are no longer visible implying increased instability of the newly synthesized and assembled complexes (Fig. 2H). This finding clearly indicates that a longer time frame for the production of labeled subunits does not increase the supercomplex levels in MTO1 mutant MEFs as is the case in WT cells. As also observed by immunoblotting, the steady-state level of supercomplexes in this pulse titration experiment detected a higher migrating entity in MTO1-deficient MEFs, which is also rapidly turned over (Fig. 2H). Taken together, these findings imply that loss of MTO1 causes aberrant assembly of supercomplexes which results in decreased levels and rapid turnover as deduced from the pulse-chase experiment. In addition, this pulse titration experiment gave insight into the cause of the CIV defect in MTO1-deficient MEFs: we observed that formation of CIV showed a slower kinetics in the assembly process. In MTO1-deficient cells, the maximal assembled level of CIV was reached at 6 h compared with 2 h in WT MEFs (Fig. 2H). It is possible that impaired translation fidelity in combination with the decreased translation rate in MTO1-deficient MEFs affects the assembly of supercomplexes and CIV. Moreover, potentially decreased translation accuracy and hence the presence of misfolded proteins, as suggested by the activation of mtUPR marker ClpP, might also contribute to both the assembly defect and the supercomplex instability.
MTO1 deficiency affects modification of tRNATrp independent of thiouridinylation and causes decreased mitochondrial tRNA levels
To investigate whether a disturbed RNA modification could contribute to the protein synthesis and OXPHOS assembly defect in MTO1-deficient MEFs, we assessed mitochondrial tRNA levels as well as the modification of tRNAGlu, tRNATrp and tRNAGln, which are hypothesized to be the targets of MTO1-mediated modification by northern blotting and APM-gel electrophoresis.
Loss of MTO1 affected the migration pattern of tRNATrp in northern blotting in both, glucose- and galactose-grown cells. In wild-type and heterozygous cells, tRNATrp migrated as two species, while MTO1 deficiency resulted in the loss of the more slowly migrating form (Fig. 2I). Although the loss of this species clearly correlates with the observed OXPHOS defect, the nature of this species as well as its significance is not clear at present. Potentially, this species could be the taurine modified form of tRNATrp that has been reported for this tRNA (44).
We then subjected mitochondrial tRNAs to APM-gel electrophoresis to investigate further the impact of MTO1 deficiency on tRNA thiouridinylation, which lies in the pathway MTO1 putatively acts in. In MTO1 mutant MEFs, thiouridinylation of tRNAGlu and tRNAGln was not abolished (Fig. 2J). We did not observe a thio-modified form when probing for tRNATrp, suggesting that the differences in migration pattern are not caused by this tRNA modification (Fig. 2J).
By standard northern blots, we observed that all tRNA levels analyzed except tRNAAla were decreased to 50–60% in MTO1-deficient MEFs. In response to galactose stress, the condition where the OXPHOS defect was more pronounced, tRNAAla steady levels also dropped to ∼50% of wild type (Supplementary Material, Fig. S1G), suggesting that loss of MTO1 results in decreased tRNA stability without apparent alteration in thiolation.
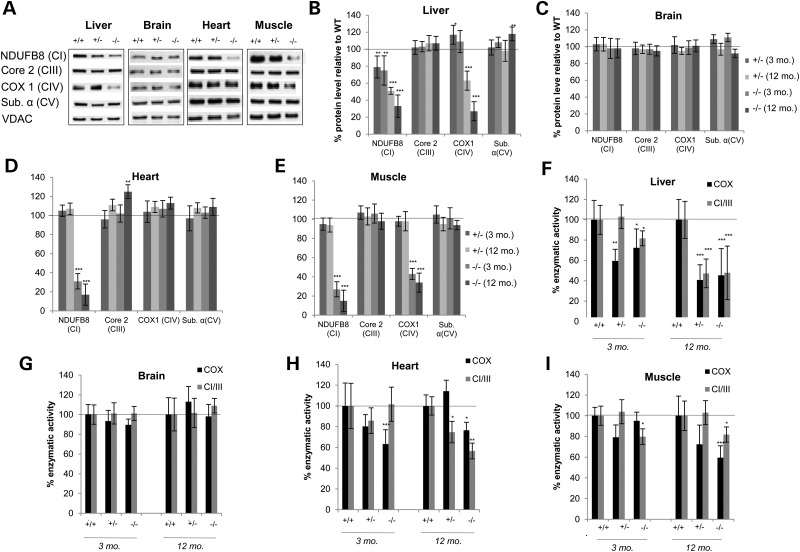
MTO1 deficiency results in a tissue-specific CI + IV defect in vivo
Mutations in MTO1 are associated with mitochondrial disorders and a strong heart-specific pathology. To shed light on the pathological consequence of loss of MTO1 in vivo, we examined a mouse model of MTO1 deficiency that recapitulates the cardiac disorder observed in patients (38). We focused on the high-energy demand tissues of liver, heart, brain and skeletal muscle, which are predominantly affected in many mitochondrial disorders. When examining OXPHOS subunits in mitochondria isolated from 3-month-old animals by western blotting, a marked decrease of the Complex I subunit NDUFB8 was detected in all tissues except brain, with heart and muscle being most severely affected (Fig. 3A–E). In addition, the Complex IV subunit COX1 was decreased in liver and muscle while no change was evident in brain and heart (Fig. 3A–E). Subunits of other OXPHOS complexes such as Core 2 (Complex III) and ATP α (Complex V) remained unaffected (Fig. 3A–E). This tissue-specific CI + CIV defect remained unchanged, and no additional defect in other complexes was observed during aging, underlining the CI + IV and tissue-specific effect as a consequence of the MTO1 deficiency. However, we did observe an age-related effect on the imbalance of the mito-nuclear protein ratio: here the ratio was decreased in MTO1-deficient liver to ∼50% of control and during aging decreased even further to ∼25% (Supplementary Material, Fig. S2A). In the heart, the CI defect resulted in a drastic increase in the mito-nuclear protein ratio, which increased further during aging. However, when other complexes were taken into account, no significant change was observed in this tissue. In contrast, in muscle, the CI + IV specific defect resulted in an increase in the mito-nuclear protein ratio based on Complex I subunits, while the ratio based on CIII and CII was decreased (Supplementary Material, Fig. S2B). The Complex I defect induced by loss of MTO1 was also evident when isolated mitochondria were subjected to BN-PAGE analysis followed by western blot (Supplementary Material, Fig. S2B). Complex I containing supercomplexes was strongly diminished in mitochondria isolated from liver, heart and muscle of MTO1 mutant mice.
Figure 3.
MTO1 deficiency induces a tissue-specific Complex I + IV defect (A) Western blot analysis of mitochondria from WT, heterozygous and homozygous MTO1 mice. The high-energy demand tissues of liver, brain, heart and muscle were analyzed. Subunits of OXPHOS Complexes I (NDUFB8), III (Core2), IV (COX1) and V (ATPα) were probed. Porin (VDAC) was used as a loading control. Depicted are representative blots. (B–E) Quantification of the protein steady-state levels at different ages relative to WT. (F–I) Enzymatic activity of CI + III and COX measured in isolated mitochondria from liver, brain, heart and muscle at 3 and 12 months of age. All activities were normalized relative to WT. *P < 0.05, **P < 0.01, ***P < 0.001.
The OXPHOS defect determined by western blot analysis was also evident when we measured enzymatic activities of Complex I and IV in isolated mitochondria (Fig. 3F–I). At 3 months of age, liver MTO1 mutant mitochondria exhibited a significant COX defect in both heterozygous and homozygous mutants. CI + III activity was also mildly, but significantly reduced in mitochondria isolated from homozygous MTO1 mutant animals. Upon aging, both COX and CI + III activity diminished dramatically in liver mitochondria of both hetero- and homozygous animals at 12 months of age, implying an age-related worsening of the OXPHOS defect (Fig. 3F). This age-related decline was also visible in western blot analysis, where a further decrease in levels of Complex I and IV subunits was detected (Fig. 3B). In MTO1-deficient heart mitochondria isolated from 3-month-old mice, we observed a decrease in enzymatic activity of COX (Fig. 3H), although no Complex IV defect was evident by western blot. The CI + III activity in MTO1 mutant heart mitochondria was not significantly affected in 3-month-old mice, but declined on aging (Fig. 3H). In western blots of MTO1-deficient heart mitochondria, we observed a trend towards a lowered steady-state level of Complex I subunit, but it did not reach significance (Fig. 3D). In skeletal muscle, we observed a mild but significant CI + III defect in MTO1-deficient mitochondria that worsened with age. COX activity in mutant MTO1 mitochondria was slightly decreased at 3 months of age, but this defect only reached statistical significance at 12 months of age (Fig. 3I). The same trend was observed by western blot analysis of skeletal muscle mitochondria, where the CIV and CI defect was mildly aggravated during aging, with a trend towards lowered protein levels of Complex I and IV subunits (Fig. 3E). Mitochondria isolated from brain did not exhibit an OXPHOS defect, neither on the enzymatic level nor in western blot analysis at any assessed age (Fig. 3C and G).
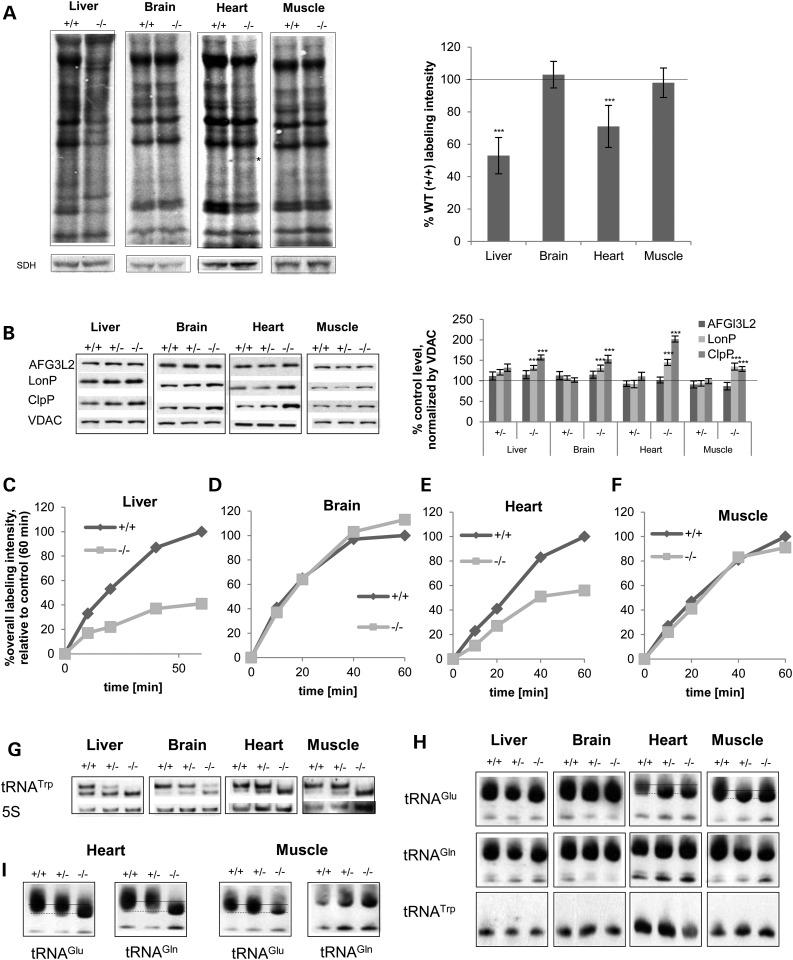
MTO1 deficiency results in a tissue-specific mitochondrial protein synthesis defect associated with a systemic activation of mitochondrial proteases
To gain further insight into the molecular cause of the OXPHOS defect and hence elucidate the molecular role of mammalian MTO1 in vivo, we assessed mitochondrial protein synthesis in different tissues, the process that MTO1 is hypothesized to interfere with and which we found to be affected in MTO1-deficient MEFs and patient fibroblasts harboring the MTO1 mutation. We therefore isolated mitochondria from different tissues and performed in organello metabolic labeling of newly synthesized mtDNA-encoded proteins. In liver, the loss of MTO1 results in a decreased overall labeling intensity and a change in the labeling pattern, implying a general mitochondrial protein synthesis defect and changes in translation of specific mt-mRNAs. In the heart, labeling intensity was also reduced and in addition a new species was detected. In brain and muscle, no gross alteration with respect to labeling intensity or pattern could be observed (Fig. 4A). Although the gross pattern was not disturbed, it remains possible that MTO1 deficiency causes translation infidelity and hence could result in the observed OXPHOS defect. Translation infidelity caused by amino acid misincorporation is observed in E. coli as a result of mutations in the bacterial homolog of MTO1 (15). In mammals, mutations in mitochondrial tRNAs such as in MELAS result in hypomodified tRNA (43,45,46) and amino acid misincorporation. These alterations result in OXPHOS complex instability, even so the translation pattern, determined by metabolic pulse labeling, remains unchanged (32). We also performed this assay, in mitochondria isolated from tissue of aged animals. In MTO1-deficient liver, we detected a decreased labeling intensity in comparison to WT tissues indicating that the translation defect observed in young animals persisted during aging. While neither brain nor heart showed any changes, we observed that the translation pattern in aged MTO1 mutant muscle mitochondria is altered indicating the ensuing of a translation defect during aging (Supplementary Material, Fig. S2C). The signal-to-noise ratio in these mitochondria was increased so that we were not able to confidently detect the aberrant translation products.
Figure 4.
MTO1 deficiency causes a tissue-specific defect in mitochondrial translation and tRNA modification associated with a systemic activation of mitochondrial proteases (A) In organello metabolic labeling of mtDNA-encoded proteins using mitochondria isolated from high-energy demand tissues in WT (+/+) mice and mice homozygous (−/−) for the MTO1 cassette. Shown are representative labeling images and the quantification of the overall labeling capacity. (B) Western blot analysis and quantification of mitochondrial proteases. VDAC was used as a loading control. (C–F) Kinetics of in organello labeling in mitochondria isolated from tissue of (+/+) and (−/−) mice. Different pulse times were used in an in organello pulse titration assay. The overall labeling capacity was quantified and depicted as a function of pulse time. Gels are depicted in Supplementary Material, Figure S2D. (G) Migration pattern of mitochondrial tRNATrp in a northern blot experiment in 3-month-old animals. (H) APM-northern blot of total RNA to visualize 2-thiouridiniylated tRNAGlu and tRNAGln of 3-month-old animals. The thio-modified and unmodified forms are indicated. The altered migration pattern of tRNAGlu in heart and muscle is indicated by a dashed line. (I) Same as (H) for 12-month-old animals to show age dependence of tRNA modification in muscular tissues. see Supplementary Material, Figure S3C for other tissues. ***P < 0.001.
As outlined earlier, an unaltered labeling pattern or kinetics does not exclude compromised labeling fidelity. As an indirect readout of translation accuracy, we assessed steady-state levels of mitochondrial proteases that are activated by unfolded and potentially mistranslated proteins. In MEFs and patient fibroblasts, these proteases were selectively upregulated and in line with this a decreased assembly and enhanced turnover of OXPHOS complexes and supercomplexes in MTO1-deficient MEFs was observed. These findings are consistent with the hypothesis that translation accuracy is compromised when MTO1 is deficient. In the assessed tissues, activation of mitochondrial proteases was recapitulated. Steady-state protein levels of LonP and ClpP were significantly increased with the strongest effect in heart tissue, where levels increased to 150 and 200% of the control level, respectively. Also in brain, which did not exhibit a detectable OXPHOS defect, we found a significant increase in these two proteases. Levels of AFG3L2 were not influenced by loss of MTO1 (Fig. 4B). This selective induction of LonP and ClpP in MTO1-deficient tissue further increased during aging (Supplementary Material, Fig. S3D). These findings indicate disturbed mitochondrial protein homeostasis when MTO1 is lost, which is consistent with compromised translation fidelity.
In MEFs, we had observed that MTO1 deficiency reduces the translation rate as deduced from a pulse titration experiment. To increase our understanding of the effect of MTO1 on translation kinetics in vivo, we performed a pulse titration in mitochondria isolated from different tissues to determine labeling capacity as a function of pulse time. In brain and muscle, there was essentially no difference in the timing of the labeling intensity and the translation kinetics of MTO1-deficient mitochondria (Fig. 4C and F; Supplementary Material, Fig. S2D). However, liver and heart show a reduction in the translation rate as well as in the final labeling intensity within the analyzed timeframe. In liver, the MTO1-deficient mitochondria lagged behind in the overall labeling capacity at all time points and reached only ∼40% of WT labeling intensity at 60 min pulsing time. The labeling intensity increased more slowly in MTO1-deficient mitochondria compared with WT controls indicating a slower translation rate (Fig. 4D; Supplementary Material, Fig. S2D). We also observed a translation defect in the heart. Here, the overall translation rate of labeling was reduced compared with WT reaching only ∼50% of the final labeling yield of WT heart mitochondria (Fig. 4E; Supplementary Material, Fig. S2D and E).
MTO1 regulates systematic tRNATrp modification and tissue-specific thiouridinylation of tRNAGlu and tRNAGln
To shed light on the role of MTO1 in mitochondrial RNA metabolism in tissues, we assessed mitochondrial tRNA levels along with their modification as well as mRNA levels in MTO1 mutant mice. As observed previously in MEFs, all MTO1 mutant tissues analyzed showed an altered migration pattern of tRNATrp in northern blot analysis: while WT tissues exhibited two species, RNA extracted from MTO1 mutant tissues showed a bias towards the lower migrating form (Fig. 4G). In wild-type tissues, the higher migrating form is the predominant entity. In all heterozygous and homozygous MTO1 mutant tissues, we observe a relative decrease of the higher migrating form and a relative increase of the lower migrating form, but the strongest effect was seen in tissue carrying the homozygous MTO1 mutation. Interestingly, this change in migration pattern is also observed in brain, where we did not detect an effect on OXPHOS complexes or their enzymatic activity. This alteration in the tRNATrp migration pattern did not change during aging (Supplementary Material, Fig. S3B). The higher migrating form may represent a putative taurine modification of mammalian mitochondrial tRNATrp. In liver, heart and muscle of MTO1 mutant mice, we could not detect any traces of this higher migrating form, indicating that loss of MTO1 abolishes this modification. However, in the brain of MTO1 mutant mice, low amounts of the higher migrating form were detectable. These findings are consistent with a hypothesized role of MTO1 in taurine modification of tRNATrp, which can be partially compensated in the brain, but not in other tissues. Remarkably, the level of this modification correlates well with the manifestation of a residual OXPHOS defect in the brain, the only assessed tissue with partial levels of modified tRNATrp.
Since MTO1 is also hypothesized to act in the thiouridinylation and carbomethylation pathway of tRNAs, we assessed the thiouridinylation-capacity of tRNATrp, tRNAGlu and tRNAGln in control and MTO1 mutant moue tissue by APM-electrophoresis. General thiouridinylation was not abolished by the deficiency of MTO1 as evidenced by the presence of the retarded, modified upper species in all analyzed tissues at 3 months of age (Fig. 4H). In heart and muscle, the modified band for tRNAGlu migrated faster in MTO1 mutant tissues compared with control samples implying an alteration in the thiouridinylation in these tissues. Upon aging, in addition to the alteration in the tRNAGlu thiouridinylation pattern, there was also a shift of tRNAGln to a lower migrating thiolated form in heart which recapitulates the change in tRNAGln migration seen in patient fibroblasts (Fig. 4I). No change in migration pattern of the modified tRNAGlu or tRNAGln was detected in liver or brain at any age analyzed (Fig. 4I; Supplementary Material, Fig. S3C). These findings support a role of MTO1 in correct thiouridinylation of selected tRNAs in muscle tissues and highlight the heart as being sensitive to disturbances in this system in accordance with the cardiac phenotype in patients. We could not detect thiolation of tRNATrp (Fig. 4H).
The alteration in the thiolation pattern in muscle tissues did not affect the stability of the respective tRNAs as revealed by northern blot analysis (Supplementary Material, Fig. S3A). Instead, we observed a tissue-specific response of tRNA levels to the MTO1 deficiency, which does not correlate with the changes in tRNA modification (Supplementary Material, Fig. S3A).
MTO1 interacts with the mitochondrial ribosome and loss of MTO1 affects mitoribosomal assembly in a tissue-specific manner
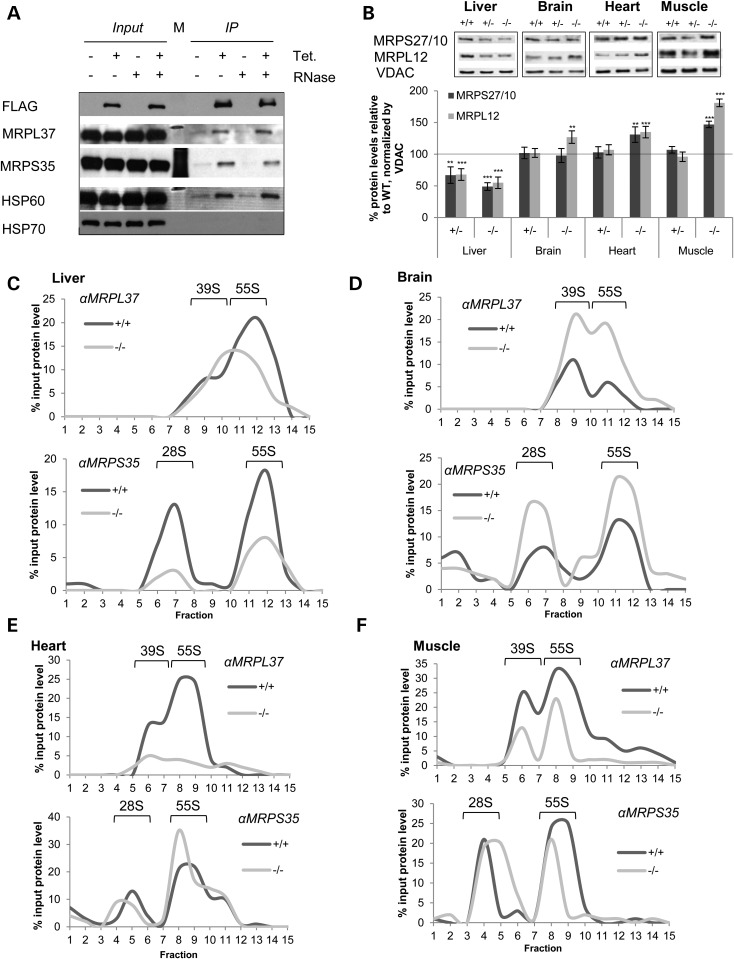
To further understand the effect of MTO1 on mitoribosomal proteins and hence mitochondrial translation, we performed immunoprecipitation using a HeLa cell line with inducible expression of FLAG-tagged MTO1. We performed the immunoprecipitation from isolated mitochondria in the absence or presence of RNase to determine the potential contribution of RNA to interactions. In induced cells, we see co-immunoprecipitation of MRPL37 and MRPS35 implying that MTO1 interacts with both large (LSU) and small (SSU) mitoribosomal subunits. This interaction was RNase insensitive, suggesting that MTO1 interacts directly with those proteins and not via RNA (Fig. 5A). This finding could be interpreted as (i) MTO1 interacts with the assembled monosome and aids in its assembly prior mRNA binding. (ii) Different pools of MTO1 interact with either the large or small subunit independent of each other. In addition, FLAG-tagged MTO1 appeared to interact with HSP60, which is thought to be involved in import and assembly of nuclear-encoded OXPHOS subunits but no interaction was seen with HSP70.
Figure 5.
MTO1 interacts with mitoribosomal subunits and affects mitoribosome assembly. (A) Immunoprecipitation of MTO1 from HeLa cells expressing flag-tagged MTO1 from a TET promoter. Immunoprecipitations were performed in the absence and presence of RNase. The eluate was probed by immunoblotting for the presence of the mitoribosomal proteins MRPL37 and MRPS35 as well as for HSP60 and HSP70. (B) Western blot analysis and quantification of mitochondria of WT, heterozygous and homozygous MTO1 mice to assess enzymes connected to the mitochondrial translation. Steady-state levels of mitoribosomal proteins MRPS27/10 and MRPL12 tested. Porin (VDAC) was used as a loading control. Depicted are representative blots. (C–F) Analysis of mitoribosomal assembly by sucrose gradient ultracentrifugation using mitochondrial extracts. Sedimentation of the 28S (small subunit), 39S (large subunit) and 55S (monosome) were determined by western blot analysis. Shown is the quantification of the western blots. The amount relative to the input is depicted. See also Supplementary Material, Figure S4. **P < 0.01, ***P < 0.001.
To further assess the influence of MTO1 on the mitoribosome, we next analyzed the effects of MTO1 on mitoribosomal subunits. In mutant MTO1 liver mitochondria, both small and large ribosomal subunits appeared decreased. In contrast, protein levels of MRPL12 were significantly elevated in brain, the tissue that lacked an OXPHOS defect. Mitoribosomal SSU and LSU proteins were also elevated in heart and even more so in skeletal muscle (Fig. 5B).
To determine whether the alterations in mitoribosomal components affected mitoribosome assembly, we separated control and MTO1 mutant mitochondria from various tissues by sucrose gradient centrifugation. Mutations in the yeast and bacterial homolog result in disturbed tRNA and mRNA levels (47,48), but we are not aware of any report on an effect on ribosome assembly. Western blotting on sucrose gradient fractions from MTO1 mutant brain mitochondria revealed increases in MRPS of SSU, LSU and 55S compared with the wild-type, implying upregulation of mitoribosomal assembly (Fig. 5D; Supplementary Material, Fig. S4B). In contrast, MTO1-deficient mouse liver mitochondria displayed decreased levels of MRPL37 associated with an altered migration pattern. Indeed LSU and assembled monosome could no longer be differentiated. MRPS35 levels in both free SSU and assembled monosome were also decreased (Fig. 5C; Supplementary Material, Fig. S4A). A similar MRPL37 phenotype was observed in MTO1-deficient heart, where a strong decrease in LSU and assembled ribosome was revealed, but with no apparent changes to the SSU. Surprisingly, the monosome fraction from MTO1 mutant mitochondria displayed a concurrent increase in MRPS25 signal together with a loss of MRPL37 signal in the same fractions. This finding indicates an alteration of mitoribosome assembly/composition (Fig. 5E; Supplementary Material, Fig. S4C). Skeletal muscle mitochondria of MTO1-deficient animals displayed decreased levels of MRPL37 in both the free LSU and assembled monosome; unchanged MRPS35 levels in the free SSU, but less MRPS35 in monosomes (Fig. 5F; Supplementary Material, Fig. S4D). Since the signals were normalized to the original input and hence the altered content of mitoribosomal proteins, the observed changes in mitoribosomal subunits and assembled monosome cannot be attributed to the altered steady-state protein levels in MTO1 mutant mitochondria. Decreased monosome assembly was also revealed by sucrose gradient fractionation of MTO1-deficient MEFs (Supplementary Material, Fig. S4E). Although the molecular basis of the tissue specificity remains elusive, these data highlight for the first time a role of MTO1 in mitoribosome assembly.
A ketogenic diet partially ameliorates the OXPHOS defect in MTO1-deficient mice
In bacteria, variations in nutritional conditions can exert an effect on tRNA modification (15), which is modulated by the MTO1-bacterial homolog GidA (49,50). This could suggest that tRNA modification and its impact on translation fidelity may be part of the cellular, and potentially in mammals of mitochondrial, nutrient sensing pathway. To assess whether the biochemical phenotype of MTO1 deficiency in mice can be modulated by metabolic measures, we subjected mice to a KD. This dietary intervention has been implicated in increased mitochondrial biogenesis (41,51) and hence could also impact mitochondrial translation as part of this process.
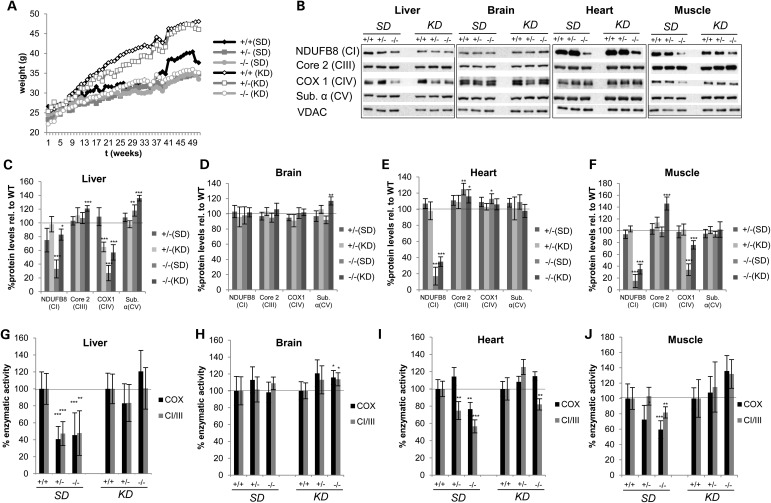
Dietary intervention started at 3 months of age and analysis at 12 months of age to assess the response to long-term KD feeding and compared with age-matched mice which have been fed the standard diet (SD). In some mouse models of mitochondrial dysfunction, a KD has a proven therapeutic effect (51), while in others the shift to increased mitochondrial biogenesis worsens or even induces an OXPHOS defect (41). This ketogenic regime inhibits the mTOR pathway akin to rapamycin (52), which has been recently shown to be a powerful therapeutic approach for mitochondrial disorders by an as yet unknown mechanism (53). The mTOR pathway is implicated in cytosolic ribosome biogenesis and a link between nutrient availability and translation fidelity is established (54). Feeding a KD protected MTO1-deficient mice from diet-induced weight gain (Fig. 6A). Since the therapeutic effects of a KD can be partially due to increasing mitochondrial mass, we assessed OXPHOS function in isolated mitochondria and normalized against mitochondrial markers to prevent masking of any potential positive effects.
Figure 6.
Long-term KD feeding partially ameliorates the OXPHOS defect in MTO1 mutant mice. (A) Weight curve of WT, heterozygous and homozygous MTO1 mice on SD and on KD (n = 10) (B–F) Western blot analysis and quantification of mitochondria of WT, heterozygous and homozygous MTO1 mice on SD and KD. All values were normalized relative to respective WT. Mice were 12 months old at the time of the analysis. (G–J) Enzymatic activity of CI + III and COX measured in isolated mitochondria from SD- and KD-fed mice. All activities were normalized relative to WT. Mice were 12 months old at the time of the analysis. *P < 0.05, **P < 0.01, ***P < 0.001.
The dietary intervention partially ameliorates the OXPHOS defect in mitochondria. In liver of KD mice, steady-state levels of NDUFB8, the Complex I subunit, recovered nearly reaching control levels (Fig. 6B and C). This increase was accompanied by a recovery of Complex I + III activity compared with age-matched MTO1 mutant mice on a SD (Fig. 6G). As described above, at this age (12 months), MTO1 mutant mice exhibit a clear CI + III and CIV defect in liver mitochondria. The CIV defect was also ameliorated in MTO1-deficient liver in response to a long-term KD as indicated by significantly increased COX1 protein steady-state levels and increased COX activity (Fig. 6B, C and G). In brain, no effect was observed on OXPHOS function or protein levels implying that KD does not induce a neurological OXPHOS defect in our MTO1 mutant mouse model (Fig. 6B, D and H) as had been observed in a MTERF2 mouse model (41). In heart, long-term KD rescued COX activity and partially rescued C + III activity in MTO1 mutant mice, both of which are reduced in SD-fed mutant mice (Fig. 6B, E and I). As seen before, protein levels of CIV subunit COX1 were not affected by MTO1 deficiency. KD modestly but significantly increased levels of NDUFB8 (Complex I subunit). In skeletal muscle, the OXPHOS defect was also partially ameliorated by the KD regime. Here, the reduced levels of NDUFB8 and COX1 (Complex IV) seen at 12 months of age on SD-fed mice were significantly increased in response to the KD. This partial rescue was also seen when measuring enzymatic activities (Fig. 6B, F and J).
A ketogenic diet in MTO1 mutant mice does not ameliorate the translation nor the tRNA modification defect, but balances the secondary stress responses induced by the loss of MTO1
To shed light on the molecular mechanism by which a long-term KD can ameliorate OXPHOS defects in MTO1-deficient mice, we assessed the key affected processes in animals on the SD. Analysis of de novo mitochondrial protein synthesis using mitochondria isolated from 12-month-old KD animals revealed no gross effect on the overall labeling capacity in MTO1-deficient mice relative to control mice on the same diet (Supplementary Material, Fig. S5A).
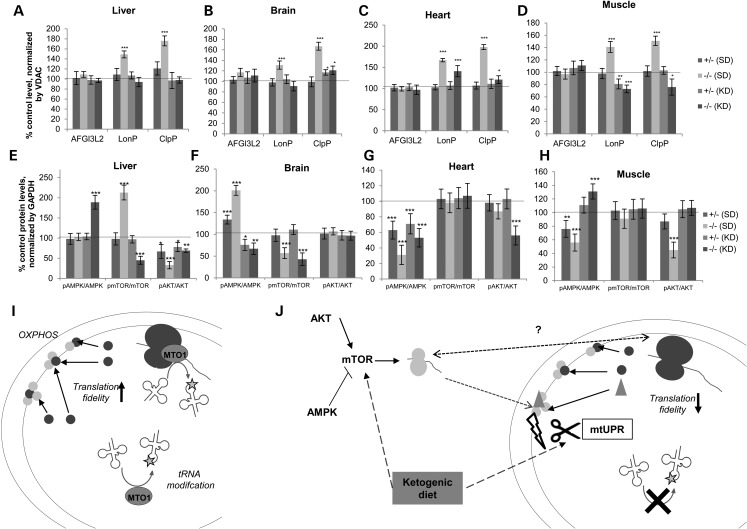
A long-term KD also did not alter the tRNATrp migration pattern in the analyzed tissues. KD and SD animals showed the same pattern with absence (heart, liver, muscle) or reduction (brain) of the more slowly, potentially modified form of tRNATrp (Supplementary Material, Fig. S5B). Nor was any modulatory effect of the KD on thiouridinylation detected. In all assessed tissues, thiouridinylation was not affected so that in KD-fed MTO1-deficient mice the altered migration pattern persisted for tRNAGlu in heart and muscle, and tRNAGln in heart. This suggests that the therapeutic effect of a KD is not mediated by correcting the tRNA modification defect (Supplementary Material, Fig. S5C and D). However, we did observe that the KD affected the steady-state levels of tRNA, thereby relieving the imbalance in liver, brain and muscle, which persisted in the heart (Supplementary Material, Fig. S5E–H). Since the KD did not correct the mutated MTO1-induced tRNA modification defect, we assessed the impact of the dietary intervention on secondary stress responses as potential disease-modifying mechanisms. We focused on the activation of mitochondrial proteases, which were hyperactivated in MTO1-deficient mice. Twelve-month-old MTO1 mutant KD mice showed an amelioration of the hyperactivation of LonP and ClpP in liver, brain and muscle. In the hearts of these animals, the hyperactivation of ClpP and reduced the levels of LonP were ameliorated. No effect on AFG3L2 as a marker for the mAAA-protease was observed (Fig. 6A–D). Activation of mitochondrial proteases can be a sign for misfolded proteins in the mitochondrial matrix compartment, resulting from translation infidelity. We did not observe any alteration of the gross pattern of de novo protein synthesis comparing mitochondria isolated from mice on a SD or a KD diet. However, an unchanged pattern in these studies does not preclude impaired translation accuracy as demonstrated in MELAS patient fibroblasts, where the labeling pattern is indistinguishable from controls, but in-depth analysis revealed impaired translation fidelity (32). The rebalancing of the mitochondrial proteases in MTO1-deficient mice on a KD would be consistent with improved translation accuracy. KD has been shown to reduce overall protein synthesis in muscle (55) in a mechanism that could involve mTOR inhibition (52). Reduction in protein synthesis rates via mTOR modulation could also contribute to less error-prone translation and hence higher translation fidelity (56). The same effect is conceivable as part of a mitochondrial nutrient sensing program.
To investigate the cellular responses that are modulated by the KD in MTO1-deficient mice, we assessed the major cellular metabolic regulators AKT, mTOR and AMPK. These proteins are highly sensitive to metabolic disturbances, serve as powerful nutrient sensors and are part of a cellular reprograming network to optimize nutrient utilization (57). In addition, their impact on cytosolic translation has been shown by us and others to contribute to the co-regulation of mitochondrial and cytosolic protein synthesis (58,59). MTO1 deficiency induces an imbalance in AKT, mTOR and AMPK signaling in all tissues independent of any OXPHOS defect (Fig. 7E–H). Feeding a KD reversed the mTOR hyperactivation in liver and partially recovered AKT phosphorylation. This rescue was associated with a hyperactivation of AMPK (Fig. 7E). In brain, KD feeding ameliorated the AMPK hyperactivation without a significant effect on the other pathways (Fig. 7F). In heart, KD mice had decreased AKT activation without any gross effect on the other markers (Fig. 7G), while in muscle, the KD regime reversed the activation defect in AMPK and AKT (Fig. 7H). Due to the complexity of these effects, it remains elusive which of them is adaptive or rather maladaptive and what the exact role is of the KD, with its own complex physiological consequences on this scenario.
Figure 7.
KD feeding balances mitochondrial protease and starvation signaling responses in MTO1-deficient mice. (A–D) Quantification of mitochondrial proteases in Mto1 mutant and control mice fed a SD and a KD. See Supplementary Material, Figure 6 for blots. (E–H) Quantification of markers of the starvation pathway AMPK, mTOR and AKT based on analysis of their unphosphorylated and their phosphorylated form. Values are normalized relative to the control. Mice are 12 months of age. (I) Model of the function of MTO1 in mammalian mitochondria: MTO1 is bound to the mitoribosome and involved in tRNA modification. Both functions could happen simultaneously or by different protein pools. MTO1 thereby controls the fidelity of mitochondrial translation and hence OXPHOS assembly and stability. (J) Model for the responses evoked due to MTO1 deficiency: loss of MTO1 results in a tissue-specific tRNA modification defect resulting in compromised translation fidelity which affects OXPHOS protein assembly. As a consequence, mitochondrial UPR in form of mitochondrial proteases is hyperactivated which impacts OXHOS protein stability and gives rise to the OXPHOS defect. The partially ameliorating effect of KD feeding could be caused by the impact of secondary effects of this mitochondrial defect on the cellular starvation signaling as well as on the mitochondrial UPR. It is also possible that the cytoplasmic and mitochondrial translation machinery communicate by an unknown mechanism. *P < 0.05, **P < 0.01, ***P < 0.001.
Discussion
Defects in the mitochondrial energy-generating system often result in tissue-specific phenotypes that might arise from highly tissue-specific regulation of the OXPHOS system in healthy but also pathological conditions. So far, this phenomenon is only poorly understood. Regulation of mitochondrial translation is increasingly recognized for contributing to this diversity (16,60,61). Our results underline the role of the Mitochondrial Translation Optimisation Factor 1, MTO1, in this scenario.
Genetic alterations in MTO1 have recently been associated with mitochondrial diseases and a growing number of patients are being reported, including a new one in this study (8,9). The reported MTO1 patients exhibit hypothrophic cardiomyopathy indicating a strong susceptibility of the heart for MTO1 deficiency independent of the exact mutation.
Mammalian MTO1 shares a high homology to the bacterial GidA and the yeast MTO1. GidA is involved in tRNA modification, which contributes to codon recognition, as well as stability and functionality of tRNAs (15,62). Yeast MTO1 is thought to be involved in the same process, and its deficiency in yeast results in reduced tRNA modification associated with a severe mitochondrial translation defect (47,48). Yeast has been also used to model the patient mutations, all of which confer a mitochondrial translation defect in yeast (8,9). However, no role of MTO1 in mammalian tRNA modification and translation could be established up to now. Here, we demonstrate for the first time that MTO1 is involved in tRNA modification and in the regulation of mitochondrial translation on a tissue-specific level (see Table 2 for summary) and thereby establish MTO1 as a novel player in the tissue-specific regulation of OXPHOS.
Table 2.
Summary of alterations in MTO1 mutant fibroblasts and tissues
| Patient fibroblasts | MEFs | Liver |
Brain |
Heart |
Muscle |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| SD | KD | SD | KD | SD | KD | SD | KD | |||
| OXPHOS | CI: ↓ CIV: ↓ |
CI: ↓ CIV: ↘ |
CI: ↓ CIV: ↓ |
CI: ↔ CIV: ↓ |
CI: ↔ CIV: ↔ |
CI: ↔ CIV: ↔ |
CI: ↓ CIV: ↔ |
CI: ↓ CIV: ↔ |
CI: ↓ CIV: ↓ |
CI: ↓ CIV: ↘ |
| Translation Maximal capacity Translation rate |
↓ | ND5 ↓ | ↓ Additional/missing bands |
↓ | ↔ | ↔ | ↓, additional bands | ↔ | ↔ | ↔ |
| tRNA steady-state levels thio-uridinylation tRNATrp migration |
↑ tRNAGln unchanged |
↓ ↔ altered |
↔/↑ ↔ altered |
↔ ↔ altered |
↔ ↔ altered |
↔ ↔ altered |
↔/↗ tRNAGlu, tRNAGln altered |
↑ tRNAGlu, tRNAGln altered |
↓ tRNAGlu altered |
↔ tRNAGlu altered |
| Mito. proteases | ↑ | ↑ | ↑ | ↔ | ↑ | ↔ | ↑ | ↔/↑ | ↑ | ↔ |
SD, standard diet; KD, ketogenic diet.
Here, we show for the first time that MTO1 mutations in patient fibroblasts result in a decreased translation capacity associated with an altered thiourdinylation pattern of mitochondrial tRNAs. The effect of MTO1 on mitochondrial translation and tRNA modification is recapitulated in MTO1 deficient MEFs and mice: here we found that loss of MTO1 reduced the mitochondrial translation rate. While we only detected an altered translation pattern in MTO1-deficient hearts, we observed a robust activation of mitochondrial proteases in all tissues independent of the OXPHOS defect. These findings indicate an increased abundance of mutated proteins in the mitochondrial compartment which is consistent with the hypothesis that MTO1 is a key factor for maintaining translation accuracy and that loss of MTO1 results in error-prone protein synthesis. In accordance with this hypothesis, we observed an assembly and stability defect of CI and CIV-containing entities which could be caused by misfolded or mutated proteins originating from error-prone mitochondrial translation.
In tissues and in MEFs, the activation of mitochondrial proteases correlated with an altered migration pattern of tRNATrp. Neither in tissues nor in cell models did we observe thiolation of tRNATrp, suggesting that the alteration is not induced by changed thiourdinylation. This tRNA has been shown to be taurine modified in mammalian mitochondria (44). It is plausible that loss of MTO1 causes an altered tRNATrp taurine modification resulting in the observed migration pattern. In addition, in heart and muscle, MTO1 deficiency results in aberrant thiolation of tRNAs without affecting the overall thiouridinylation capacity. In heart, this impairment increases with aging. These data demonstrate for the first time that MTO1 has indeed in a role in fine tuning the tRNA-thiolation pattern in mammalian mitochondria and that it is a crucial player in precise thiolation of tRNA in muscular tissue. This finding clearly demarcate MTO1 from MTO2, which affects in patient fibroblasts the thiouridinylation capacity without alteration of the thiolation pattern (22).
Patients carrying mutations in MTO1 have a cardiac-specific phenotype (Table 1). This heart-specific pathology is recapitulated by the MTO1 mouse model that we used in this study (38). Our findings show that the CI defect is most severe in the heart compared with that in other tissues that might be the driving force for this cardiac specific pathology. The heart is also most affected by alterations of tRNA modification, and the only assessed tissue that showed an age-related aggravation of this defect. Since precise tRNA modification is part of the translation fidelity control, this heart-specific alteration could result in a tissue-specific increased burden of translation inaccuracy. This hypothesis is in line with our finding of a heart-specific alteration in the mitochondrial translation pattern.
Our results also indicate a new and surprising role of MTO1 on mitoribosomal assembly. Interaction studies revealed that MTO1 interacts with mitoribosomal proteins in an RNA-independent manner. This finding is in line with the hypothesis that MTO1 interacts with the small and the large subunit and aids in the assembly or stability of the monosome. Consistently, loss of MTO1 results in an aberrant monosome assembly as well as an imbalance of mitoribosomal proteins. It is yet unclear whether the pool of MTO1 that interacts with the mitoribosome is different from the pool being involved in tRNA modification.
The function of MTO1-bacterial homolog GidA in tRNA modification depends on nutrient supply (49,50). We therefore used the mouse model to probe how metabolic intervention interacts with MTO1 deficiency to determine whether MTO1 and mitochondrial translation are involved in nutrient sensing as seen in the bacterial system. Long-term KD feeding partially ameliorated the OXPHOS defect in all affected tissue of MTO1 mutant mice. Intriguingly, this effect was independent of tRNA modification or the translation capacity. However, KD feeding had a balancing effect on secondary stress responses: the dietary intervention relieved the hyperactivation of mitochondrial proteases including the mtUPR marker ClpP. These responses are increasingly recognized for driving disease pathogenesis (35). MTO1 deficiency also resulted in strong and tissue-specific alteration of starvation signaling, independent of the OXPHOS defect. We and others could recently show that mitochondrial and cytosolic translation are co-regulated via mTOR/AMPK/AKT signaling and that this scenario is responsive to changes in nutrient supply (58,59). It is conceivable that the disturbance in mitochondrial translation induced by MTO1 deficiency is sensed by the cytoplasmic system and finally results in imbalanced co-regulation. In our mouse model, KD feeding reversed or diminished the hyperactivation of the tissue-specific starvation responses. While it is unclear how this effect contributes to the partial OXPHOS rescue, the dietary intervention contributes to a balanced co-regulation through affecting the mTOR/AMPK/AKT signaling cascade.
Based on our findings, we propose the following model: MTO1 is involved in controlling the accuracy of tRNA modification in mammalian mitochondria and thereby regulates translation efficiency and translation fidelity (Fig. 7I). Loss of MTO1 results in aberrant tRNA modification, which decreases translation rate and translation accuracy. As a consequence of the error-prone translation, the mitochondrial UPR becomes hyperactivated which can affect stability of OXPHOS proteins. The resulting mito-nuclear protein imbalance is also sensed by the starvation signaling which governs cytoplasmic translation and hence contributes to the co-regulation of both systems (Fig. 7J). Dietary intervention such as KD feeding alleviates this imbalance by resolving the hyperactivation of mitochondrial proteases and interfering with the starvation signaling which finally results in a partial OXPHOS rescue independent of the tRNA modification defect.
Overall, our data demonstrate for the first time a role of mammalian MTO1 in mitochondrial tRNA modification and translation efficiency and fidelity. These findings also highlight mitochondrial translation and MTO1 as important determinants in the tissue specificity of OXPHOS regulation. Importantly, we could demonstrate that KD feeding partially ameliorates the OXPHOS defects caused by MTO1 deficiency by bypassing tRNA modification and impaired translation fidelity through modulating downstream responses. These findings not only add a new dimension of developing therapies for disorders associated with impaired mitochondrial tRNA modification and compromised mitochondrial translation fidelity, but also imply MTO1 as part of a nutrient-sensing program within mitochondria and link translation fidelity to nutrient supply.
Materials and Methods
Cell culture
MEFs and patient fibroblasts were cultured at 37°C in humidified 5% CO2 and 95% air in DMEM medium supplemented with 10% fetal bovine serum, 1 mm pyruvate and 50 µg/ml uridine, 1× NEAA and l-glutamine. For galactose challenging, glucose was substituted with equimolar amounts of galactose, and dialyzed serum was used.
Animal husbandry
Mice were held in a 12/12 h light/dark cycle at room temperature, and tissue was extracted after PBS perfusion. The KD increased fat intake to 60% while maintaining the caloric intake of the control diet. Metabolic challenges were initiated at 3 months of age. This study was carried out in strict accordance with the recommendation of German Animal Protection laws. The protocol was approved by the Committee on the Ethics of Animal Experiments of the Office for Nature, Environment and Protection, North Rhine-Westphalia (Landesamt für Natur, Umwelt und Verbraucherschutz). All efforts were made to minimize suffering.
Measurement of mitochondrial enzymatic activities
Mitochondrial enzymatic complex activities were measured spectrophotometrically in isolated mitochondria using a Perkin Elmer Lambda 35 UV/VIS spectrophotometer (Perkin Elmer, Waltham, MA, USA) as reported before (63). All assays were performed at room temperature. Experiments were performed minimally in triplicate, followed by statistical analysis.
Western blot analysis
Extracts prepared from cells, tissue and mitochondria were separated by SDS-PAGE and transferred to polyvinylidene difluoride membranes (Bio-Rad, Munich, Germany). Membranes were blocked in 5% milk TBS and incubated with the primary antibody overnight followed by a secondary anti-mouse IgG conjugated to horseradish peroxidase (HRP). The chemiluminescent signal was detected using the Chemiluminiscence Detection Kit (Applichem, Darmstadt, Germany). Antibodies were obtained from Invitrogen (OXPHOS Cocktail, NUDFA8, NUDUFA9, COXI), Cell Signaling (pmTOR/mTOR, pAMPK/AMPK, pAKT/AKT) and Abcam (VDAC, MRPS35, MRPL37, UQCR2, ClpP, LonP). AFG3L2 was a gift from Prof. Thomas Langer.
from Abcam (Cambridge, UK), Invitrogen or Santa Cruz Biotechnology (Heidelberg, Germany). Experiments were performed in triplicate followed by densitometric and statistical analysis.
BN-PAGE
BN-PAGE was carried out as described to resolve fully assembled OXPHOS complexes in a lauryl maltoside extract as well as supramolecular assemblies of OXPHOS complexes in digitonin extracts (64,65). In-gel activity assays were performed following the protocols reported in Ref. (64). Metabolic chase of OXPHOS complexes and supercomplexes was carried out as described following extraction of OXPHOS proteins (42).
Metabolic pulse-chase labeling and flotation assay
The BN-pulse-chase experiment was carried out according to Ref. (66). Mitochondrial pulse-chase labeling was performed as described in Ref. (67). Flotation assay was carried out as published previously (68). In organello labeling was performed as previously described in Ref. (69). For the pulse titration, the incubation time with radiolabeled 35S-methionine/cysteine was varied as described in the text.
Northern blot analysis
Regular northern blot analysis was performed as previously described, on total RNA isolated from cells and tissue (70–72). [(N-acryloylamino)phenyl]mercuric chloride (APM)-northern blotting analysis was performed as described previously to assess the thiolation status of mitochondrial tRNAs (73). Experiments were performed in triplicates followed by densitometric and statistical analysis.
Mitoribosomal fractionation
Mitoribosomal fractionation of 28S and 39S ribosomal subunits as well as of 55S monosome was effected by ultracentrifugation through a linear density sucrose gradient as described previously in Ref. (74).
Immunoprecipitation
Mitochondria were isolated from cells and were resuspended in lysis buffer (50 mm Tris, pH 7.4, 150 mm NaCl, 1 mm MgCl2, 1% NP-40). Immunoprecipitation was performed with α-FLAG-Gel following manufacturer's recommendations (Sigma Aldrich, St Louis, MO, USA). Elution was effected with FLAG peptide
Statistics and quantification
ImageJ was used for densitometric analysis (75). Student’s T-test was used for statistical evaluation.
Supplementary Material
Supplementary Material is available at HMG online.
Conflict of Interest statement. None declared.
Funding
This work was supported by the Emmy-Noether-Program of the Deutsche Forschungsgemeinschaft (WE 4108/3-1) to T.W., the Cluster of Excellence: Cellular Stress Responses in Aging-Associated Diseases (CECAD) to A.H., J.S. and T.W., Collaborative Research Center SFB 635 to T.W., the Fritz-Thyssen-Foundation to T.W., the Care-for-Rare-Foundation to T.W., the NRW International Graduate School ‘From Embryo to Old Age: the Cell Biology and Genetics of Health and Disease’ (IGSDHD) to C.T., and I.D. and by the Bundesministerium für Bildung und Forschung (BMBF, mitoNET-German Network for Mitochondrial Diseases, 01GM1113A) to T.K. and (01GM1113C) to H.P. and T.W., as well as the European Community (Infrafrontier grant 01KX1012) to L.B. This work was supported by The Wellcome Trust (096919/Z/11/Z) to Z.C.L.
Supplementary Material
References
- 1.DiMauro S., Schon E.A. (2008) Mitochondrial disorders in the nervous system. Annu. Rev. Neurosci., 31, 91–123. [DOI] [PubMed] [Google Scholar]
- 2.Antonicka H., Sasarman F., Kennaway N.G., Shoubridge E.A. (2006) The molecular basis for tissue specificity of the oxidative phosphorylation deficiencies in patients with mutations in the mitochondrial translation factor EFG1. Hum. Mol. Genet., 15, 1835–1846. [DOI] [PubMed] [Google Scholar]
- 3.Lombes A., Aure K., Bellanne-Chantelot C., Gilleron M., Jardel C. (2014) Unsolved issues related to human mitochondrial diseases. Biochimie, 100C, 171–176. [DOI] [PubMed] [Google Scholar]
- 4.Konovalova S., Tyynismaa H. (2013) Mitochondrial aminoacyl-tRNA synthetases in human disease. Mol. Genet. Metab., 108, 206–211. [DOI] [PubMed] [Google Scholar]
- 5.Zhang Z., Falk M.J. (2014) Integrated transcriptome analysis across mitochondrial disease etiologies and tissues improves understanding of common cellular adaptations to respiratory chain dysfunction. Int. J. Biochem. Cell Biol., 50, 106–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Houtkooper R.H., Mouchiroud L., Ryu D., Moullan N., Katsyuba E., Knott G., Williams R.W., Auwerx J. (2013) Mitonuclear protein imbalance as a conserved longevity mechanism. Nature, 497, 451–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li R., Li X., Yan Q., Qin Mo J., Guan M.X. (2003) Identification and characterization of mouse MTO1 gene related to mitochondrial tRNA modification. Biochim. Biophys. Acta, 1629, 53–59. [DOI] [PubMed] [Google Scholar]
- 8.Baruffini E., Dallabona C., Invernizzi F., Yarham J.W., Melchionda L., Blakely E.L., Lamantea E., Donnini C., Santra S., Vijayaraghavan S., et al. (2013) MTO1 mutations are associated with hypertrophic cardiomyopathy and lactic acidosis and cause respiratory chain deficiency in humans and yeast. Hum. Mutat., 34, 1501–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ghezzi D., Baruffini E., Haack T.B., Invernizzi F., Melchionda L., Dallabona C., Strom T.M., Parini R., Burlina A.B., Meitinger T., et al. (2012) Mutations of the mitochondrial-tRNA modifier MTO1 cause hypertrophic cardiomyopathy and lactic acidosis. Am. J. Hum. Genet., 90, 1079–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hopper A.K., Phizicky E.M. (2003) tRNA transfers to the limelight. Genes Dev., 17, 162–180. [DOI] [PubMed] [Google Scholar]
- 11.Alexandrov A., Chernyakov I., Gu W., Hiley S.L., Hughes T.R., Grayhack E.J., Phizicky E.M. (2006) Rapid tRNA decay can result from lack of nonessential modifications. Mol. Cell, 21, 87–96. [DOI] [PubMed] [Google Scholar]
- 12.Hagervall T.G., Bjork G.R. (1984) Undermodification in the first position of the anticodon of supG-tRNA reduces translational efficiency. Mol. Gen. Genet., 196, 194–200. [DOI] [PubMed] [Google Scholar]
- 13.Sullivan M.A., Cannon J.F., Webb F.H., Bock R.M. (1985) Antisuppressor mutation in Escherichia coli defective in biosynthesis of 5-methylaminomethyl-2-thiouridine. J. Bacteriol., 161, 368–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kruger M.K., Sorensen M.A. (1998) Aminoacylation of hypomodified tRNAGlu in vivo. J. Mol. Biol., 284, 609–620. [DOI] [PubMed] [Google Scholar]
- 15.Bregeon D., Colot V., Radman M., Taddei F. (2001) Translational misreading: a tRNA modification counteracts a +2 ribosomal frameshift. Genes Dev., 15, 2295–2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hallberg B.M., Larsson N.G. (2014) Making proteins in the powerhouse. Cell Metab., 20, 226–240. [DOI] [PubMed] [Google Scholar]
- 17.Yasukawa T., Kirino Y., Ishii N., Holt I.J., Jacobs H.T., Makifuchi T., Fukuhara N., Ohta S., Suzuki T., Watanabe K. (2005) Wobble modification deficiency in mutant tRNAs in patients with mitochondrial diseases. FEBS Lett., 579, 2948–2952. [DOI] [PubMed] [Google Scholar]
- 18.Kirino Y., Goto Y., Campos Y., Arenas J., Suzuki T. (2005) Specific correlation between the wobble modification deficiency in mutant tRNAs and the clinical features of a human mitochondrial disease. Proc. Natl Acad. Sci. USA, 102, 7127–7132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yasukawa T., Suzuki T., Ohta S., Watanabe K. (2002) Wobble modification defect suppresses translational activity of tRNAs with MERRF and MELAS mutations. Mitochondrion, 2, 129–141. [DOI] [PubMed] [Google Scholar]
- 20.Suzuki T., Wada T., Saigo K., Watanabe K. (2002) Taurine as a constituent of mitochondrial tRNAs: new insights into the functions of taurine and human mitochondrial diseases. Embo. J., 21, 6581–6589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yasukawa T., Suzuki T., Ueda T., Ohta S., Watanabe K. (2000) Modification defect at anticodon wobble nucleotide of mitochondrial tRNAs(Leu)(UUR) with pathogenic mutations of mitochondrial myopathy, encephalopathy, lactic acidosis, and stroke-like episodes. J. Biol. Chem., 275, 4251–4257. [DOI] [PubMed] [Google Scholar]
- 22.Boczonadi V., Smith P.M., Pyle A., Gomez-Duran A., Schara U., Tulinius M., Chinnery P.F., Horvath R. (2013) Altered 2-thiouridylation impairs mitochondrial translation in reversible infantile respiratory chain deficiency. Hum. Mol. Genet., 22, 4602–4615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sasarman F., Antonicka H., Horvath R., Shoubridge E.A. (2011) The 2-thiouridylase function of the human MTU1 (TRMU) enzyme is dispensable for mitochondrial translation. Hum. Mol. Genet., 20, 4634–4643. [DOI] [PubMed] [Google Scholar]
- 24.Shi R., Villarroya M., Ruiz-Partida R., Li Y., Proteau A., Prado S., Moukadiri I., Benitez-Paez A., Lomas R., Wagner J., et al. (2009) Structure-function analysis of Escherichia coli MnmG (GidA), a highly conserved tRNA-modifying enzyme. J. Bacteriol., 191, 7614–7619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schagger H., de Coo R., Bauer M.F., Hofmann S., Godinot C., Brandt U. (2004) Significance of respirasomes for the assembly/stability of human respiratory chain complex I. J. Biol. Chem., 279, 36349–36353. [DOI] [PubMed] [Google Scholar]
- 26.Acin-Perez R., Bayona-Bafaluy M.P., Fernandez-Silva P., Moreno-Loshuertos R., Perez-Martos A., Bruno C., Moraes C.T., Enriquez J.A. (2004) Respiratory complex III is required to maintain complex I in mammalian mitochondria. Mol. Cell, 13, 805–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kurata S., Ohtsuki T., Wada T., Kirino Y., Takai K., Saigo K., Watanabe K., Suzuki T. (2003) Decoding property of C5 uridine modification at the wobble position of tRNA anticodon. Nucleic Acids Res. Suppl., 3, 245–246. [DOI] [PubMed] [Google Scholar]
- 28.Kurata K., Yanagisawa R., Ohira M., Kitagawa M., Nakagawara A., Kamijo T. (2008) Stress via p53 pathway causes apoptosis by mitochondrial Noxa upregulation in doxorubicin-treated neuroblastoma cells. Oncogene, 27, 741–754. [DOI] [PubMed] [Google Scholar]
- 29.Novoa E.M., Pavon-Eternod M., Pan T., Ribas de Pouplana L. (2012) A role for tRNA modifications in genome structure and codon usage. Cell, 149, 202–213. [DOI] [PubMed] [Google Scholar]
- 30.Rezgui V.A., Tyagi K., Ranjan N., Konevega A.L., Mittelstaet J., Rodnina M.V., Peter M., Pedrioli P.G. (2013) tRNA tKUUU, tQUUG, and tEUUC wobble position modifications fine-tune protein translation by promoting ribosome A-site binding. Proc. Natl Acad. Sci. USA, 110, 12289–12294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gingold H., Pilpel Y. (2011) Determinants of translation efficiency and accuracy. Mol. Syst. Biol., 7, 481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sasarman F., Antonicka H., Shoubridge E.A. (2008) The A3243G tRNALeu(UUR) MELAS mutation causes amino acid misincorporation and a combined respiratory chain assembly defect partially suppressed by overexpression of EFTu and EFG2. Hum. Mol. Genet., 17, 3697–3707. [DOI] [PubMed] [Google Scholar]
- 33.Kirino Y., Suzuki T. (2005) Human mitochondrial diseases associated with tRNA wobble modification deficiency. RNA Biol., 2, 41–44. [DOI] [PubMed] [Google Scholar]
- 34.Haynes C.M., Petrova K., Benedetti C., Yang Y., Ron D. (2007) ClpP mediates activation of a mitochondrial unfolded protein response in C. elegans. Dev. Cell, 13, 467–480. [DOI] [PubMed] [Google Scholar]
- 35.Dogan S.A., Pujol C., Maiti P., Kukat A., Wang S., Hermans S., Senft K., Wibom R., Rugarli E.I., Trifunovic A. (2014) Tissue-specific loss of DARS2 activates stress responses independently of respiratory chain deficiency in the heart. Cell Metab., 19, 458–469. [DOI] [PubMed] [Google Scholar]
- 36.Hornig-Do H.T., Tatsuta T., Buckermann A., Bust M., Kollberg G., Rotig A., Hellmich M., Nijtmans L., Wiesner R.J. (2012) Nonsense mutations in the COX1 subunit impair the stability of respiratory chain complexes rather than their assembly. Embo. J., 31, 1293–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Venkatesh S., Lee J., Singh K., Lee I., Suzuki C.K. (2012) Multitasking in the mitochondrion by the ATP-dependent Lon protease. Biochim. Biophys. Acta, 1823, 56–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Becker L., Kling E., Schiller E., Zeh R., Schrew A., Hölter S.M., Mossbrugger I., Calzada-Wach J., Strecker V., Wittig I., et al. (2014) MTO1-deficient mouse model mirrors the human phenotype showing complex I defect and cardiomyopathy. PLoS ONE, 9, e114918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marroquin L.D., Hynes J., Dykens J.A., Jamieson J.D., Will Y. (2007) Circumventing the Crabtree effect: replacing media glucose with galactose increases susceptibility of HepG2 cells to mitochondrial toxicants. Toxicol. Sci., 97, 539–547. [DOI] [PubMed] [Google Scholar]
- 40.Rossignol R., Gilkerson R., Aggeler R., Yamagata K., Remington S.J., Capaldi R.A. (2004) Energy substrate modulates mitochondrial structure and oxidative capacity in cancer cells. Cancer Res., 64, 985–993. [DOI] [PubMed] [Google Scholar]
- 41.Wenz T., Luca C., Torraco A., Moraes C.T. (2009) mTERF2 regulates oxidative phosphorylation by modulating mtDNA transcription. Cell Metab., 9, 499–511. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 42.Fernandez-Silva P., Acin-Perez R., Fernandez-Vizarra E., Perez-Martos A., Enriquez J.A. (2007) In vivo and in organello analyses of mitochondrial translation. Methods Cell Biol., 80, 571–588. [DOI] [PubMed] [Google Scholar]
- 43.Kirino Y., Yasukawa T., Ohta S., Akira S., Ishihara K., Watanabe K., Suzuki T. (2004) Codon-specific translational defect caused by a wobble modification deficiency in mutant tRNA from a human mitochondrial disease. Proc. Natl Acad. Sci. USA, 101, 15070–15075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Suzuki T. (2014) A complete landscape of post-transcriptional modifications in mammalian mitochondrial tRNAs. Nucleic Acids Res., 42, 7346–7357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yasukawa T., Suzuki T., Ishii N., Ohta S., Watanabe K. (2001) Wobble modification defect in tRNA disturbs codon-anticodon interaction in a mitochondrial disease. Embo. J., 20, 4794–4802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kirino Y., Yasukawa T., Marjavaara S.K., Jacobs H.T., Holt I.J., Watanabe K., Suzuki T. (2006) Acquisition of the wobble modification in mitochondrial tRNALeu(CUN) bearing the G12300A mutation suppresses the MELAS molecular defect. Hum. Mol. Genet., 15, 897–904. [DOI] [PubMed] [Google Scholar]
- 47.Colby G., Wu M., Tzagoloff A. (1998) MTO1 codes for a mitochondrial protein required for respiration in paromomycin-resistant mutants of Saccharomyces cerevisiae. J. Biol. Chem., 273, 27945–27952. [DOI] [PubMed] [Google Scholar]
- 48.Wang X., Yan Q., Guan M.X. (2009) Mutation in MTO1 involved in tRNA modification impairs mitochondrial RNA metabolism in the yeast Saccharomyces cerevisiae. Mitochondrion, 9, 180–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dittmar K.A., Sorensen M.A., Elf J., Ehrenberg M., Pan T. (2005) Selective charging of tRNA isoacceptors induced by amino-acid starvation. EMBO. Rep., 6, 151–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dong H., Nilsson L., Kurland C.G. (1996) Co-variation of tRNA abundance and codon usage in Escherichia coli at different growth rates. J. Mol. Biol., 260, 649–663. [DOI] [PubMed] [Google Scholar]
- 51.Ahola-Erkkila S., Carroll C.J., Peltola-Mjosund K., Tulkki V., Mattila I., Seppanen-Laakso T., Oresic M., Tyynismaa H., Suomalainen A. (2010) Ketogenic diet slows down mitochondrial myopathy progression in mice. Hum. Mol. Genet., 19, 1974–1984. [DOI] [PubMed] [Google Scholar]
- 52.McDaniel S.S., Rensing N.R., Thio L.L., Yamada K.A., Wong M. (2011) The ketogenic diet inhibits the mammalian target of rapamycin (mTOR) pathway. Epilepsia, 52, e7–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Johnson S.C., Yanos M.E., Kayser E.B., Quintana A., Sangesland M., Castanza A., Uhde L., Hui J., Wall V.Z., Gagnidze A., et al. (2013) mTOR inhibition alleviates mitochondrial disease in a mouse model of Leigh syndrome. Science, 342, 1524–1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Conn C.S., Qian S.B. (2013) Nutrient signaling in protein homeostasis: an increase in quantity at the expense of quality. Sci. Signal, 6, ra24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vazquez J.A., Adibi S.A. (1992) Protein sparing during treatment of obesity: ketogenic versus nonketogenic very low calorie diet. Metabolism, 41, 406–414. [DOI] [PubMed] [Google Scholar]
- 56.Huo Y., Iadevaia V., Proud C.G. (2011) Differing effects of rapamycin and mTOR kinase inhibitors on protein synthesis. Biochem. Soc. Trans, 39, 446–450. [DOI] [PubMed] [Google Scholar]
- 57.Yan L., Lamb R.F. (2012) Amino acid sensing and regulation of mTORC1. Semin. Cell Dev. Biol., 23, 621–625. [DOI] [PubMed] [Google Scholar]
- 58.DiDomenico A., Hofer A., Tundo F., Wenz T. (2014) Mitochondrial protein acetylation mediates nutrient sensing of mitochondrial protein synthesis and mitonuclear protein balance. IUBMB Life, doi:10.1002/iub.1328. [DOI] [PubMed] [Google Scholar]
- 59.Johnson M.A., Vidoni S., Durigon R., Pearce S.F., Rorbach J., He J., Brea-Calvo G., Minczuk M., Reyes A., Holt I.J., et al. (2014) Amino acid starvation has opposite effects on mitochondrial and cytosolic protein synthesis. PLoS ONE, 9, e93597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Boczonadi V., Horvath R. (2014) Mitochondria: impaired mitochondrial translation in human disease. Int. J. Biochem. Cell Biol., 48, 77–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rotig A. (2011) Human diseases with impaired mitochondrial protein synthesis. Biochim. Biophys. Acta, 1807, 1198–1205. [DOI] [PubMed] [Google Scholar]
- 62.Meyer S., Wittinghofer A., Versees W. (2009) G-domain dimerization orchestrates the tRNA wobble modification reaction in the MnmE/GidA complex. J. Mol. Biol., 392, 910–922. [DOI] [PubMed] [Google Scholar]
- 63.Barrientos A., Fontanesi F., Diaz F. (2009) Evaluation of the mitochondrial respiratory chain and oxidative phosphorylation system using polarography and spectrophotometric enzyme assays. Curr. Protoc. Hum. Genet., Chapter 19, Unit19 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Diaz F., Barrientos A., Fontanesi F. (2009) Evaluation of the mitochondrial respiratory chain and oxidative phosphorylation system using blue native gel electrophoresis. Curr. Protoc. Hum. Genet., Chapter 19, Unit19 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nijtmans L.G., Henderson N.S., Holt I.J. (2002) Blue Native electrophoresis to study mitochondrial and other protein complexes. Methods, 26, 327–334. [DOI] [PubMed] [Google Scholar]
- 66.McKenzie M., Lazarou M., Thorburn D.R., Ryan M.T. (2007) Analysis of mitochondrial subunit assembly into respiratory chain complexes using Blue Native polyacrylamide gel electrophoresis. Anal. Biochem., 364, 128–137. [DOI] [PubMed] [Google Scholar]
- 67.Chomyn A. (1996) In vivo labeling and analysis of human mitochondrial translation products. Methods Enzymol., 264, 197–211. [DOI] [PubMed] [Google Scholar]
- 68.Kim S.J., Kwon M.C., Ryu M.J., Chung H.K., Tadi S., Kim Y.K., Kim J.M., Lee S.H., Park J.H., Kweon G.R., et al. (2012) CRIF1 is essential for the synthesis and insertion of oxidative phosphorylation polypeptides in the mammalian mitochondrial membrane. Cell Metab., 16, 274–283. [DOI] [PubMed] [Google Scholar]
- 69.Formosa L.E., Hofer A., Tischner C., Wenz T., Ryan M.T. (2014) Methods in Molecular Biology. Springer, New York, NY. [DOI] [PubMed] [Google Scholar]
- 70.Hofer A., Noe N., Tischner C., Kladt N., Lellek V., Schauss A., Wenz T. (2014) Defining the action spectrum of potential PGC-1alpha activators on a mitochondrial and cellular level in vivo. Hum. Mol. Genet., 23, 2400–2415. [DOI] [PubMed] [Google Scholar]
- 71.Noe N., Dillon L., Lellek V., Diaz F., Hida A., Moraes C.T., Wenz T. (2013) Bezafibrate improves mitochondrial function in the CNS of a mouse model of mitochondrial encephalopathy. Mitochondrion, 13, 417–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wenz T., Wang X., Marini M., Moraes C.T. (2011) A metabolic shift induced by a PPAR panagonist markedly reduces the effects of pathogenic mitochondrial tRNA mutations. J. Cell Mol. Med., 15, 2317–2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Umeda N., Suzuki T., Yukawa M., Ohya Y., Shindo H., Watanabe K. (2005) Mitochondria-specific RNA-modifying enzymes responsible for the biosynthesis of the wobble base in mitochondrial tRNAs. Implications for the molecular pathogenesis of human mitochondrial diseases. J. Biol. Chem., 280, 1613–1624. [DOI] [PubMed] [Google Scholar]
- 74.Richter R., Rorbach J., Pajak A., Smith P.M., Wessels H.J., Huynen M.A., Smeitink J.A., Lightowlers R.N., Chrzanowska-Lightowlers Z.M. (2010) A functional peptidyl-tRNA hydrolase, ICT1, has been recruited into the human mitochondrial ribosome. Embo. J., 29, 1116–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Schneider C.A., Rasband W.S., Eliceiri K.W. (2012) NIH Image to ImageJ: 25 years of image analysis. Nat. Methods, 9, 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.