Abstract
Anaplastic thyroid carcinoma (ATC) is a frequently lethal malignancy that is often unresponsive to available therapeutic strategies. The tumorigenesis of ATC and its relationship to the widely prevalent well-differentiated thyroid carcinomas are unclear. We have analyzed 22 cases of ATC as well as 4 established ATC cell lines using whole-exome sequencing. A total of 2674 somatic mutations (121/sample) were detected. Ontology analysis revealed that the majority of variants aggregated in the MAPK, ErbB and RAS signaling pathways. Mutations in genes related to malignancy not previously associated with thyroid tumorigenesis were observed, including mTOR, NF1, NF2, MLH1, MLH3, MSH5, MSH6, ERBB2, EIF1AX and USH2A; some of which were recurrent and were investigated in 24 additional ATC cases and 8 ATC cell lines. Somatic mutations in established thyroid cancer genes were detected in 14 of 22 (64%) tumors and included recurrent mutations in BRAF, TP53 and RAS-family genes (6 cases each), as well as PIK3CA (2 cases) and single cases of CDKN1B, CDKN2C, CTNNB1 and RET mutations. BRAF V600E and RAS mutations were mutually exclusive; all ATC cell lines exhibited a combination of mutations in either BRAF and TP53 or NRAS and TP53. A hypermutator phenotype in two cases with >8 times higher mutational burden than the remaining mean was identified; both cases harbored unique somatic mutations in MLH mismatch-repair genes. This first comprehensive exome-wide analysis of the mutational landscape of ATC identifies novel genes potentially associated with ATC tumorigenesis, some of which may be targets for future therapeutic intervention.
Introduction
Anaplastic thyroid carcinoma (ATC) is a rare endocrine malignancy with therapeutic options of limited effectiveness. In contrast to the much more common malignancies of the thyroid follicular epithelium, such as papillary or follicular thyroid cancers (collectively known as ‘well-differentiated thyroid cancers’—WDTC) which boast excellent cure rates and long-term survival, prognosis in ATC patients is exceedingly poor, nearly uniformly fatal, and has changed little in the past half century. In the USA, ATC is responsible for 1–2% of all thyroid cancers, but accounts for over 50% of deaths attributable to thyroid malignancy (1,2). Over 80% of patients present with loco-regional invasion and approximately half have distant metastases at the time of diagnosis (3–5). Considering the explosive clinical course of ATC, the recommended therapy is generally aggressive multimodal treatment for most patients (4). Despite maximal intervention, the median survival time is <6 months in most series, and cancer-specific mortality at 1 year exceeds 80% (4,6,7). The dismal prognosis and high prevalence of distant disease at the time of diagnosis highlights the urgent need for novel, targeted systemic treatments for ATC.
Approximately 20–25% of patients with ATC have a history of a previous WDTC and an additional 20–30% have a coexisting WDTC found following surgery for ATC (3,4,8,9). Although ATC can also arise de novo or in the setting of chronic goiter in occasional patients, these data suggest that a significant fraction of ATC cases are the result of progression from WDTC. Furthermore, genetic analysis of ATC has characterized frequent somatic mutations in several genes such as BRAF and RAS that are commonly mutated in WDTC as well, implying a common initial tumorigenic pathway (10–12). Mutations unique to ATC, in genes such as TP53, CTNNB1 and others, are thought to be involved in dedifferentiation and progression to ATC. However, due to the rarity of ATC and correspondingly small sample size of most studies evaluating its genetic landscape, conclusively determining the incidence and impact of these mutations is difficult. For example, CTNNB1 mutations have been variably observed at a rate of 0–61.3% of ATC cases (13–15). Similarly, widespread copy number and structural genomic instability have been repeatedly demonstrated in ATC with copy number gain present in over 80% of tumors, but wide spectrum of variation reported coupled with small sample sizes limit the generalizability of these studies (2,16). While current knowledge of the molecular pathogenesis of ATC has led to several clinical trials of existing targeted pharmaceuticals, the results have thus far not shown dramatic improvements in outcomes and the precise molecular mechanisms of ATC dedifferentiation and tumorigenesis remain unknown (17–19), although individual successes have been described (20,21).
Whole-exome capture coupled with next-generation sequencing technology is a proven method for identifying functionally relevant genetic variants underlying both Mendelian and complex disease states, such as neoplasia (22–24). The ability of whole-exome sequencing (WES) to resolve single-nucleotide variants is particularly suited for characterizing previously unknown drivers of tumorigenesis and survey the landscape of somatic mutations present in a malignancy (25,26). Furthermore, WES has been successfully utilized in the clinical environment for both genetic diagnosis and tumor genotyping (23,27,28); a possibly useful application in ATC, where conventional cancer therapies are largely ineffective and targeted therapies have not been widely applied but have been shown to be beneficial in individual cases. Thus, we hypothesized that WES would be an ideal platform to (i) better characterize the mutational landscape of ATC, (ii) identify novel potential driver mutations for further study in ATC tumorigenesis and (iii) detect possible candidates for targeted therapy in patients with ATC. Accordingly, we applied WES to a cohort of 22 unique cases of ATC and four established ATC cell lines in an effort to further these goals.
Results
Exome sequencing cohort
Demographic and pathologic information describing the exome sequencing cohort are shown in Table 1. Thirteen patients (59%) were women, nine (41%) were men and the mean age at surgery was 73 years (median 74 years). Tumor tissue from 19 of the patients (86%) were derived from primary tumor tissue, whereas three patients (14%) had previously undergone total thyroidectomy for papillary thyroid cancer and ATC specimens were obtained during subsequent procedures for recurrent disease or palliative purposes. Thirteen patients (59%) presented with distant metastases at diagnosis. Eight patients had been treated with neoadjuvant external beam irradiation. Ten patients had demonstrable evidence of concurrent or previous WDTC in either the ipsi- or contralateral thyroid lobe, nine with papillary thyroid cancer and one with follicular thyroid cancer.
Table 1.
Clinical characteristics of ATC tumor samples subjected to WES
| Case | Source | Age | Presentation | Primary tumor size | Nodal stage | Metastases at diagnosis | Alive? | Survival (months) | Additional thyroid disease? |
|---|---|---|---|---|---|---|---|---|---|
| T1 | FFPE | 53 | Neck mass, cord paralysis | 5.0 cm | N0 | None | Yes | 50.9 | No |
| T2 | FFPE | 70 | Stridor, dyspnea | Debulking only | N1b | Bone | No | 2.8 | No |
| T3 | FFPE | 72 | Surveillance ultrasound | N/A | N1b | Lung, chest wall | No | 24.1 | WDTC |
| T4 | FFPE | 70 | Dysphagia | 8.0 cm | N1b | Lung | N/A | N/A | Goiter |
| T5 | FFPE | 61 | Neck mass | 5.8 cm | N1b | Lung, brain, retroperitoneum | No | 0.9 | No |
| T6 | FFPE | 45 | Neck mass | 8.0 cm | N1b | None | No | 11.3 | No |
| T7 | FFPE | 89 | Dysphagia | 4.5 cm | Nx | Lung, chest wall | No | 1 | WDTC |
| T8 | FFPE | 78 | Neck mass | 6.0 cm | N1a | Lung | Yes | 40.3 | Graves' disease |
| T9 | FFPE | 59 | Neck mass | 7.5 cm | N1a | None | No | 4.3 | No |
| T10 | FFPE | 59 | Dysphagia | N/A | N1b | Lung, chest wall | No | 24.9 | WDTC |
| T11 | FFPE | 65 | Neck mass | 8.0 cm | Nx | Lung | No | 39.9 | Goiter, WDTC |
| T12 | FFPE | 93 | Airway collapse | 8.0 cm | Nx | Lung | No | 0.8 | WDTC |
| T13 | FFPE | 75 | Neck mass | 3.7 cm | N1b | Lung | N/A | N/A | No |
| T14 | FFPE | 63 | Hemoptysis | N/A | N1b | Lung, adrenal | N/A | N/A | WDTC |
| T15 | FFPE | 79 | Stridor, dyspnea | Debulking only | Nx | Lung, liver | N/A | N/A | No |
| T16 | Fresh Frozen | 77 | Neck mass | 7.0 cm | Nx | Lung | No | 3 | WDTC |
| T17 | Fresh Frozen | 83 | Neck mass | 7.0 cm | Nx | None | No | 11 | Goiter |
| T18 | Fresh Frozen | 82 | Neck mass | 4.0 cm | Nx | N/A | No | 1 | WDTC |
| T19 | Fresh Frozen | 84 | Neck mass | 5.0 cm | N1b | None | No | 3 | No |
| T20 | Fresh Frozen | 72 | Neck mass, dyspnea | 10.0 cm | Nx | None | No | 1 | Goiter |
| T21 | Fresh Frozen | 89 | Neck mass | 8.0 cm | N1b | None | No | 0.5 | WDTC |
| T22 | Fresh Frozen | 85 | Neck mass | 4.0 cm | Nx | None | No | 4 | WDTC |
FFPE, fresh-frozen paraffin-embedded; N/A, not available (lost to follow-up); WDTC, well-differentiated thyroid cancer.
Whole-exome sequencing
In the 22 matched tumor/normal pairs that underwent WES, a total of 2674 somatic variants were identified, 1972 of which were non-synonymous (74%). In total, 68% constituted coding missense mutations, 5% non-sense mutations and 1% were exon–intron boundary mutations. The number of somatic variants per tumor ranged from 4 to 1805, with a mean number of total and non-synonymous variants of 121.5 and 89.6 per tumor, respectively. Two samples demonstrated a significantly higher mutation burden compared with all other samples (T2—253 mutations, T11—1805 mutations). When samples T2 and T11 were excluded, the mean number of total and non-synonymous variants was 30.8 and 23.7 per tumor, respectively. An overview of the WES results by sample is shown in Figure 1, and a detailed list of the sequencing data for all 22 cohort tumor-normal pairs is included in Supplementary Material, Tables S1 and S2. The percentage of reads on target was 66% and 65% for tumor and normal tissue, respectively, and the percentage of bases covered >20 times was 96% (tumor) and 92% (normal tissue). Tumor samples were deliberately sequenced to a greater depth than normal tissue (mean 264 and 138 reads/sample, respectively) in order to minimize the impact of the perceived lower quality of DNA libraries derived from fresh-frozen, paraffin-embedded (FFPE) samples (T1–T15) and also to maximize detection of heterozygous mutations in tumor samples admixed with adjacent normal tissue. Thirty mutations identified by exome sequencing throughout the cohort were selected for confirmatory Sanger sequencing. The mutations selected for confirmation were chosen based on perceived relevance in tumorigenesis, availability of sample for confirmatory studies and to ensure mutations from multiple FFPE-derived samples were confirmed. As expected based on confirmatory sequencing from prior whole exome studies, 29 of 30 (96.7%) mutations examined were confirmed when subjected to Sanger sequencing (Supplementary Material, Table S3).
Figure 1.
Overall mutational profile of the 22 ATCs studied by exome sequencing. All panels are aligned with vertical tracks representing 22 individuals with ATC (left) and 4 established ATC cell lines (right). The underlying heatmap shows the distribution of somatic coding mutations in thyroid malignancy-related genes, novel ATC gene variants [established driver genes as suggested by Vogelstein et al. (24) or recurrently mutated COSMIC genes found in ≥3 samples], and finally mutated Mut homolog genes in which a correlation to the amount of mutational rate was observed.
Potential novel driver gene mutations
While a majority of cases harbored somatic mutations in known thyroid malignancy-related genes (discussed below), 16 different cancer-related genes denoted as ‘driver genes' by Vogelstein et al. (29) not typically identified as drivers of thyroid malignancy were found to be present among the studied tumors. Several of these mutations were present in multiple samples and are strongly correlated with non-thyroidal malignancies, such as NF1, mTOR (two cases each), ERBB2, DAXX, MLL2, and NOTCH2 (one case each—Fig. 1). Cases T2 and T11, which exhibited a ‘hypermutator phenotype’ with a somatic mutation frequency well above the average were found to have missense and non-sense somatic mutations in evolutionary conserved positions of the mismatch repair (MMR) genes MLH1 (T2) and MLH3 (T11) (Table 2). Case T11, which also exhibited a mutation in mutS homolog gene MSH5, had 1805 somatic mutations compared with 253 in Case T2. These mutations were unique to cases T2 and T11 and the mutational burden of these two cases combined comprises 77% of the total somatic burden across the entire cohort.
Table 2.
Summary of individual mutations and associated protein coding change in non-synonymous mut homolog genes, USH2A, and EIF1AX among all ATC tissue samples assayed
| Case | Gene | Location | Mutation (AA) | Position (AA/total AA) | P-value | Nucleotide position | Nucleotide change | PolyPhen2 score |
|---|---|---|---|---|---|---|---|---|
| T2 | MLH1 | Chr3 | I19M | 19/756 | 1.16E−31 | 37 010 099 | C>G | 1 |
| T3 | MLH1 | Chr3 | I68M | 68/756 | 2.63E−21 | 37 013 201 | C>G | 1 |
| T4 | MLH1 | Chr3 | Q60X | 60/756 | 8.23E−18 | 37 013 175 | C>T | Truncating |
| T11 | MLH3 | Chr14 | L264V | 264/1429 | 5.10E−08 | 74 585 322 | G>C | 0.968 |
| T12 | MSH5 | Chr6 | A199V | 199/822 | 4.43E−06 | 31 819 953 | C>T | 0.999 |
| T10 | MSH6 | Chr2 | D736H | 736/1360 | 9.08E−06 | 47 880 832 | G>C | 1 |
| T5 | USH2A | Chr1 | I2189V | 2189/5202 | 2.18E−22 | 214 238 944 | T>C | 0 |
| T11 | USH2A | Chr1 | D798V | 798/5202 | 2.79E−05 | 214 486 966 | T>A | 0.921 |
| T21 | USH2A | Chr1 | E4571K | 4571/5202 | 6.33E−13 | 213 914 165 | C>T | 0.549 |
| T22 | USH2A | Chr1 | L1727F | 1727/5202 | 2.08E−34 | 214 323 540 | G>A | 0.961 |
| T9 | EIF1AX | ChrX | Ex-in boundary | 1 bp upstream of exon 6 | 1.14E−88 | 20 058 647 | C>G | Splice site |
| T13 | EIF1AX | ChrX | G9R | 9/144 | 1.40E−15 | 20 066 653 | C>G | 0.996 |
| T21 | EIF1AX | ChrX | P2R | 2/144 | 2.57E−12 | 20 069 675 | G>C | 0.028 |
AA, amino acid.
Novel recurrently mutated COSMIC genes
COSMIC genes found to be recurrently mutated that have not previously been described in ATC development included four cases of non-synonymous USH2A missense mutations (4/22; 18%). Additionally, three cases with mutations in EIF1AX and HECTD1 were found (3/22; 14%). These are summarized in Figure 1, Table 2 (USH2A, EIF1AX), and Supplementary Material, Table S2 (HECTD1).
ATC-related gene mutations
Multiple genes previously found to be involved in thyroid neoplasia demonstrated somatic mutations in the study group. These findings are summarized in Table 3 and include several well-described oncogenes and tumor suppressors. The most common recurrently mutated genes were TP53 and BRAF with six cases each, followed by NRAS (3 cases), KRAS (2 cases), PIK3CA (2 cases) as well as HRAS, CDKN1B, CDKN2C, CTNNB1, HRAS and RET (one case each, Fig. 1, Table 3). Previously recognized ATC-related recurrent mutations present in the cohort included BRAF V600E (six cases), NRAS Q61R (two cases) and PIC3CA H1047R/L (one case each). The mean number of single-nucleotide variants in tumors harboring BRAF V600E mutations versus RAS mutations was 47.8 (±31.5) versus 22.2 (±6.7). For tumors harboring neither BRAF nor RAS mutations, the mean number of single-nucleotide variants (SNVs) was 24.5 (±18.3) (comparison of three groups; P-value 0.09). Previously reported frequencies of commonly described ATC gene variants are compared with the frequencies described here in Supplementary Material, Table S4.
Table 3.
Overview of somatic mutations across all analyzed ATC tissue samples annotated in the COSMIC (Catalog of Somatic Mutations in Cancer) database (n = 22)
| Gene | Location (cytoband) | No. of cases | Mutation(s) observed | Mutation type | No. with LOH | % tumors mutated in COSMICa |
|---|---|---|---|---|---|---|
| All genes with recurrent mutations | ||||||
| BRAF | 7q34 | 6 | V600E | Missense | None | 19.61% |
| NRAS | 1p13.2 | 3 | Q61R/K | Missense | None | 5.73% |
| PIK3CA | 3q26.32 | 2 | H1047R/L | Missense | None | 11.85% |
| Recurrently mutated genes (n ≥ 3) | ||||||
| TP53 | 17p13.1 | 6 | Various | Various | LOH (2/6) | 29.00% |
| BRAF | 7q34 | 6 | V600E | Missense | None | 19.61% |
| MUC16 | 19p13.2 | 4 | Various | Various | None | 9.46% |
| USH2A | 1q41 | 4 | Various | Various | None | 9.15% |
| GPR112 | Xq26.3 | 4 | Various | Various | None | 4.11% |
| NRAS | 1p13.2 | 3 | Q61R/K | Missense | None | 5.73% |
| PCDH15 | 10q21.1 | 3 | Various | Various | None | 4.80% |
| LRP1 | 12q13.3 | 3 | Various | Missense | None | 3.80% |
| KIAA1109 | 4q27 | 3 | Various | Missense | LOH (1/3) | 3.76% |
| EIF1AX | Xp22.12 | 3 | Various | Various | None | 0.25% |
| HECTD1 | 14q12 | 3 | Various | Missense | None | 1.60% |
| Genes with damaging mutations + LOH | ||||||
| NF2 | 22q12.2 | 1 | E103X | Non-sense | LOH | 7.57% |
| MPDZ | 9p23 | 1 | Q124X | Non-sense | LOH | 1.82% |
| OR8K3 | 11q12.1 | 1 | R292X | Non-sense | LOH | 1.04% |
| SMARCAL1 | 2q35 | 1 | Q34X | Non-sense | LOH | 0.99% |
| C3orf77 | 3p21.31 | 1 | W1446X | Non-sense | LOH | 0.55% |
| EIF5A2 | 3q26.2 | 1 | S44X | Non-sense | LOH | 0.22% |
LOH, loss of heterozygosity.
aMutational frequency as reported in COSMIC in February 2014.
Five cases did not exhibit somatic mutations in either the ATC-related or novel ATC gene variant gene groups (Fig. 1), and the mutational status of these cases are summarized in Supplementary Material, Table S5. The mean number of mutations present among these five cases was 12.4 (range 4–22). A subset of the cases (T6, T8) did exhibit damaging mutations with concurrent loss of heterozygosity (LOH) in COSMIC genes with largely unknown functions. The recent discoveries of RASAL1 as a thyroid-specific tumor suppressor gene with a mutational frequency of 17% in ATC (30) and the role of ALK rearrangements in thyroid tumorigenesis (31,32) prompted investigation of whether RASAL1 and ALK mutations were present in the cohort. No somatic mutations in ALK were found; however, a missense mutation in RASAL1 (R774C) was identified in case T11 (Fig. 1).
Gene ontology and pathway analyses
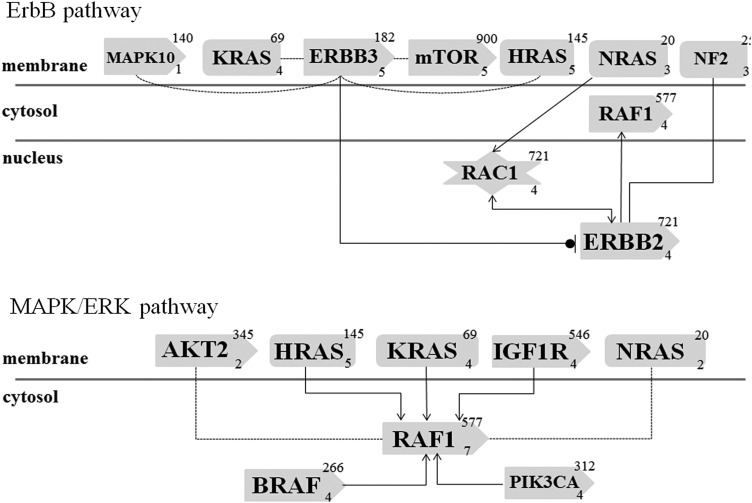
Using the DAVID (Database for Annotation, Visualization and Integrated Discovery) Bioinformatics Resources 6.7 database for identification of enriched biological processes in the pool of somatically altered genes, wide and generic ontology terms were identified. These included metabolic processes, intracellular signaling, phosphorylation and apoptosis. Further analyses using KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway annotations identified several pathways with significant representation in the ATC cohort, including general pathways in cancer, MAPK kinase and ErbB pathways as enriched among the mutated genes. The involvement of the MAPK kinase and ErbB pathways were further characterized and validated using an independent ontology analysis (Genomatix Pathway System), yielding P-values of 2.01e−6 and 6.88e−5, respectively. This analysis identified somatic mutations among the tumor cohort in both the MAPK/ERK pathway (BRAF, HRAS, KRAS, NRAS, RAF1, AKT2 and PIK3CA) and the ErbB pathway (MAPK10, ERBB2, ERBB3, RAF1, RAC1 and NF2) (Fig. 2). Additional analysis using DAPPLE (Disease Association Protein-Protein Link Evaluator) version 2.0 to examine the significance of protein–protein interactions among the gene products of those genes identified as mutated by exome sequencing was performed. Cases T2 and T11 were excluded from this analysis as well as the gene ontology analysis as they harbored a much greater mutational burden due to mutations in the mismatch repair process (described above). A pattern was observed in which proteins involved in the ErbB/RAS/MAPK signaling pathways separated from apoptosis-related pathways (Supplementary Material, Fig. S1).
Figure 2.
Simplistic overview of the top two mutation-enriched transduction signaling pathways in ATC as according to the Genomatix Pathway System. The ErbB (top) and MAPK/Erk (bottom) signaling pathways are illustrated with each mutated gene depicted. Gene products are drawn as rounded rectangles, snip-sided rectangles denote proteins with kinase properties and star-shaped symbols denote co-factors. The top right number denotes the number of sources for a chemical association with established molecules (for therapeutic purposes), the lower right number refers to the number of established interactions within the pathway in addition to the drawn association. Filled lines with arrows denote an activating effect; filled lines with stop line and circle denote inhibitory effect. Dotted lines indicate evidence at the level of co-citation, in contrast to filled lines which indicate evidence at expert-curation level.
Exome sequencing of ATC cell lines
The results from the exome sequencing of four established ATC cell lines are detailed in Figure 1 and Supplementary Material, Table S6. The number of mutations ranged from 358 (BHT101) to 565 (ACT1), averaging 457 mutations per cell line. All four cell lines exhibited mutations in TP53. Three cell lines were found to have the BRAF V600E mutation (8305C, 8505C, BHT101). The remaining cell line (ACT1) had mutations in NRAS and EIF1AX. Furthermore, ACT1 and BHT101 both exhibited USH2A mutations.
LOH and copy number variation analyses
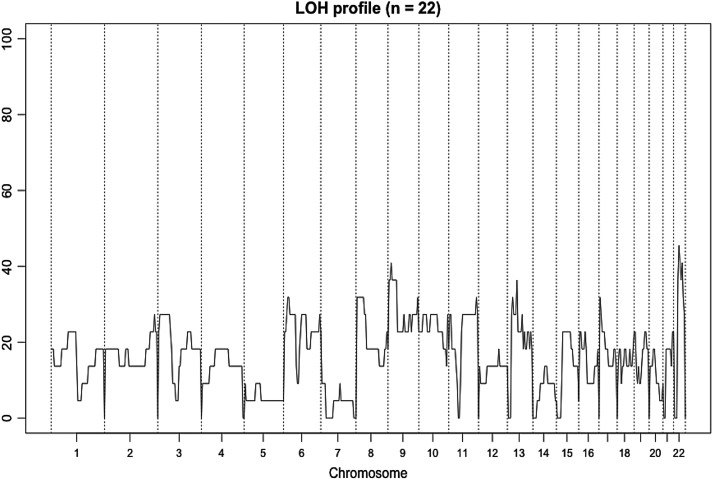
A schematic overview of the LOH profile of the 22 ATCs studied is shown in Figure 3. In general, LOH events were most frequently identified in chromosomes 9p, 13p and 22q, averaging 40% of the cases. Eight of the ATCs had discernible copy number variations (CNVs), and there were no significant focal or arm-level CNVs from the GISTIC (Genomic Identification of Significant Targets in Cancer) analysis.
Figure 3.
LOH profile for the exome cohort. The graph depicts the overall LOH profile of the 22 ATC cases. Chromosome numbers are annotated on the x-axis and frequency of LOH on the y-axis. LOH events were most frequently identified in chromosomes 9p, 13p and 22q, averaging 40% of the cases.
Correlations to clinical characteristics
The mean somatic mutation burden for the group that received neoadjuvant external radiotherapy (n = 8) was 51.6 mutations, compared with the 161.5 mutations seen in the group that did not receive neoadjuvant treatment (n = 14; P = NS). However, when excluding the hypermutator tumors T2 and T11 (did, and did not, receive neoadjuvant radiotherapy, respectively), almost comparable levels were seen (35.1 versus 22.9 mutations respectively; P = NS). Furthermore, no significant correlations were seen between number of somatic mutations and length of survival (Spearman's correlation r = 0.169, P = 0.592), lymph node status (Kruskal–Wallis P = 0.909, ANOVA P = 0.800) or metastases at presentation (t-test P = 0.250). There was no statistically significant difference in the number of mutations observed in the FFPE- versus fresh-frozen-derived tissues subjected to exome sequencing. All described correlations remained insignificant when cases with a hypermutator phenotype were excluded.
Validation cohort analysis
To further examine the novel mutations discussed above in mTOR and ERBB2 that could have significant clinical impact, a validation cohort of 24 primary ATC cases and 8 ATC cell lines were subjected to targeted Sanger sequencing of the relevant mutation loci. In addition, EIF1AX, recently shown to be a potential driver in papillary thyroid carcinoma (33) and was one of the most frequent recurrently mutated novel genes among the exome sequencing cohort in the current study (three primary tumors and one cell line), was also selected for further examination, as was the well-recognized BRAF V600E mutation.
A mutation in EIF1AX (Chr X:20058647; G>A) was found in ATC cell line c643, which corresponds to the identical chromosomal position of a heterozygous somatic mutation found in exome sample T9 (Chr X:20058647; G>C, Supplementary Material, Tables S2 and S7). This mutation, a single base pair upstream from the splice site of exon 6, is predicted as damaging by computational analysis (34). Furthermore, a recurrent SNP in the 5′ UTR of EIF1AX (rs201653081, C>T) four base pairs upstream of exon 1 (Chr X:20141644) were observed in two validation cohort samples; greater than the expected minor allele frequency of 0.24%. No additional mutations at the examined mTOR or ERBB2 loci were found in the validation cohort (R164Q/M2327I and D387N, respectively). Five BRAF V600E mutations were found; four were within the ATC cell lines and served as positive controls for the Sanger sequencing as they have been previously described (35).
Discussion
The dismal prognosis of ATC is directly related to the ineffectiveness of mainstream treatment options and the incomplete understanding of the mechanisms of ATC tumorigenesis. This study sought to address these challenges using an exome-wide sequencing approach in 22 cases of ATC to survey the frequency of established genetic events in tumorigenesis, identify potential novel driver genetic events and characterize future candidates for targeted therapy. In summary, the results confirm that the genetic background of ATC is highly heterogeneous, but several novel genetic events not previously described in ATC were demonstrated. Some of these, such as mutations mTOR, NF1, or ERBB2, are particularly intriguing as these genes have been targeted previously by pharmaceuticals currently in clinical or experimental use. Additionally, two tumors were found to have defects in a mismatch repair genes and feature a hypermutator phenotype, a previously poorly characterized finding in ATC. Finally, the mutational landscape of the cohort was described with known thyroid malignancy genes present at varying frequencies in most, but not all, ATC cases.
The prototypic progression to ATC can be considered as a multi-step de-differentiation process in many cases, arising from a pre-existing WDTC such as papillary thyroid cancer or follicular thyroid cancer. This is best illustrated by the frequent observation of mutations in the BRAF and RAS oncogenes in ATC. The well-described BRAF V600E variant is the most common mutation in papillary thyroid cancer, while NRAS/KRAS/HRAS variants are commonly found in follicular thyroid cancer. In the current study, the observed frequencies for the BRAF V600E and RAS mutations (27% for both) mirrored the prevalence reported in previous manuscripts [Supplementary Material, Table S4 (12,15,36–38)]. As BRAF and RAS mutations are thought to be the drivers for development of WDTC prior to ATC development, the prevalence of these two mutations in a particular ATC cohort would be expected to be static. The cumulative prevalence of ∼50–60% for these two mutations in this and other ATC cohorts may represent the fraction of ATCs that genuinely arise from prior WDTCs. All BRAF mutations in the current study were found to be the V600E variant and were mutually exclusive with RAS mutations, presumably reflecting the underlying WDTC variant (papillary or follicular thyroid cancer, respectively). For instance, in six cases of ATC found to harbor the BRAF V600E mutation described here, four had either been treated for pre-existing PTC or were found to have concurrent PTC elsewhere in the thyroid following resection.
The mechanism by which de-differentiation results in progression from WDTC to ATC have been a topic of frequent study with accumulation of mutations in recognized malignancy-associated genes such as TP53 and the phosphatidylinositol 3-kinase/Akt (PI3-K/Akt) and Ras/Raf/mitogen-activated protein kinase (Ras/Raf/MAPK) pathways being regularly mentioned (12,15,39). Indeed, half of the cases of ATC with BRAF V600E mutations in this cohort harbored coexisting mutations in TP53; mice with loss of TP53 function with BRAF V600E mutations are known to develop ATC (40). Among those BRAF V600E cases that did not have TP53 mutations, two-thirds demonstrated activating mutations in PIK3CA (H1047R/L in T15/T11). In a mouse model, PIK3CA H1047L is unable to drive thyroid tumorigenesis independently; however, in mice with both PIK3CA H1047L and BRAF V600E mutations, ATC will occur (41). Notably, in these ATC cases featuring BRAF V600E in coexistence with PIK3CA or TP53 mutations, few, if any, mutations in other known ATC-related or COSMIC genes are observed.
Among cases harboring NRAS/HRAS/KRAS mutations, the de-differentiation process is less clear. Only a single TP53 mutation (T9) and no PIK3CA mutations were observed. RAS mutations in thyroid cancer appear to be independently activating of the PI3K-Akt pathway (38), yet many such cases of WDTC do not progress to ATC. An intriguing candidate not previously reported in thyroid malignancy identified by exome sequencing is EIF1AX. This gene encodes an essential eukaryotic translation initiation factor and was observed to be recurrently mutated in half of ATC cases harboring RAS mutations. It was not observed in any cases without a coexisting RAS mutation, nor was it observed in any cases with BRAF V600E mutations. Two of the three mutations were missense and the third was at a splice site; all were predicted to be damaging (Table 2). Furthermore, cell line ACT-1, which features wild-type BRAF but mutant NRAS, also demonstrated the EIF1AX mutation in the same residue mutated in sample T21. Moreover, in the validation cohort assessed here, cell line c643 (known to be BRAF wild-type) demonstrated the same splice site mutation as sample T9 from the whole exome cohort mutations in EIF1AX have recently been identified (via WES) in a significant fraction of uveal melanomas (42). These mutations are postulated to be late events in melanoma progression and have prognostic significance (43,44). Moreover, the recently released analysis of the genetic landscape of papillary thyroid cancer by the Cancer Genome Atlas Research Network has also identified EIF1AX as a possible novel driver of thyroid tumorigenesis (33). While the contribution of EIF1AX to tumor progression remains unclear, it has been shown that increased EIF1AX activity triggers cell proliferation in vitro (45). As a recurrent event coexisting with RAS-mutated ATC cases, EIF1AX is an attractive target for further investigation.
ATC also appears to arise de novo, without previous overt signs of differentiated cancer (39). Potential driver mutations in ErbB pathway genes (ERBB2 (D387N), NF2 (E103X) and mTOR (R164Q) were identified in two cases. These three genes have driver properties based on experimental findings in unrelated tumor types, and both ERBB2 (HER-2) and mTOR are overexpressed or mutated in various malignancies (46,47). The specific ERBB2 and mTOR mutations described in T5 have not previously been annotated as COSMIC variants. The ERBB2 D387N mutation is located in the extracellular domain; this region has unknown biological significance but is a commonly mutated domain in urothelial cancer (48). The truncating NF2 mutation in Case T17 was found to overlap a region of LOH, implying that the bi-allelic inactivation of NF2 in this case could represent a crucial event in progression of this tumor. Such alterations in the ErbB pathway alterations may potentially have an independent driver role in the development of ATC. Several additional recurrently mutated genes annotated in the COSMIC database previously not described in ATC were also identified. In addition to EIF1AX (discussed above), the most frequent were USH2A (four cases) and HECTD1 (three cases). Of these cases, USH2A is of particular interest. The gene encodes uscherin, which is ubiquitously expressed and involved in extracellular matrix binding via basement membrane-based interaction with collagen IV and fibronectin. USH2A is a COSMIC-annotated gene with various frequencies of somatic mutations observed in malignant lesions, and the gene was recently postulated to be one of the top 10 mutated genes across various human tumor types (49). Germline mutations in USH2A are associated with Usher syndrome type 2A (OMIM: 276901), a disorder characterized by hearing deficiencies and retinitis pigmentosa (50). Potential alterations in ECM or cellular adhesion functions merit close attention in highly invasive and metastatic tumors such as ATC.
Two ATC cases exhibited a ‘hypermutator’ phenotype that has been observed in other cancers but has not been described in ATC (51,52). The probable etiology of this phenotype is the presence of highly damaging MutL homolog mutations in both cases (MLH1 in T2 and MLH3 in T11; T11 also having an MSH5 mutation). Damaging mutations in this family of genes results in hypermutability in both human cancers and experimental yeast models (53,54). A single case of ATC has been reported in an individual with hereditary nonpolyposis colon cancer (HNPCC—Lynch syndrome) (55). Both cases here demonstrated TP53 mutations as well, but as both were BRAF and RAS wild-type, the MutL homolog mutations may be crucial contributors to tumor initiation or progression. A third case demonstrated a mutation in the MutS homolog MSH6 (T10); this tumor did exhibit BRAF V600E.
Five ATC cases (23%) did not demonstrate somatic mutations in either known thyroid cancer-related genes or genes with known driver properties. The mechanism of ATC tumorigenesis in these samples is limited to speculation, but might be attributable to somatic mutations in COSMIC genes of unknown function (Supplementary Material, Table S5), alternatively due to epigenetic or genetic alterations such as gene fusions; this is noteworthy as several rearrangements are recognized to drive WDTC tumorigenesis, such as RET, ALK or NTRK3 fusions in PTC and PAX8/PPARγ in FTC (33,39). Interestingly, the lack of potential driver mutations in 22.7% (5/22) cases in the exome sequencing cohort roughly mirrors the 15.3% (74/484) of cases found to harbor chromosomal rearrangements and translocations in a recent analysis of the genetic landscape of PTC (33). Regarding the most common previously reported gene variants characterized in ATC, significant heterogeneity exists in the reported frequency of mutation. However, no significant difference in the rate of TP53, BRAF or RAS-family mutations were observed when historical reports were compared with the frequencies observed in the current study (12,15,36–38,56) (Supplementary Material, Table S4). Garcia-Rostan et al. (57) initially reported mutations in exon 3 of CTNNB1 occurred in 65.5% of ATCs; a statistically significant difference in prevalence compared with the single CTNNB1 mutation is observed here (also in exon 3). However, subsequent reports have found that the rate of CTNNB1 mutation seems to approximate the prevalence observed here (0–4.5% of ATC) (14,15). Other uncommon genetic events reported in thyroid malignancy were observed only rarely or not at all (such as mutations in RASAL1, PTEN, or Akt), likely due to the small sample size mandated by the rarity of ATC and the infrequency of these events (16,30,58).
Ontology analysis of all somatic mutations demonstrated during WES revealed clusters in the MAPK and RAS/ErbB pathways (Fig. 2). Prior studies had surmised the importance of these pathways in ATC tumorigenesis. However, a significant number of somatic mutations in genes associated with these pathways but previously not shown to be altered in ATCs were confirmed here. An overall enrichment of mutations in apoptosis-related genes was also appreciated. This persisted even when limiting analysis to COSMIC-annotated gene variants; presumably, these variants are involved in dedifferentiation as large-cohort landscape analyses of WDTC have not demonstrated similar findings. DAPPLE analysis confirmed the above findings; also identifying clusters in ErbB/RAS/MAPK pathway and apoptosis-related proteins (Supplementary Material, Fig. S1). This may correlate with the global up-regulation of TGF-β genes described by Pita et al. in ATC (15); as RAS/MAPK signaling is known to integrate with the TGF-β pathway to regulate transcriptional activity. This synergy appears to have a significant effect on gene expression in ATC and should be a topic for ongoing study (59,60).
Examination of the mutational landscape of the study cohort revealed several trends. Three broad, mutually exclusive groups of ATC can be characterized based on their defining mutation: (1) tumors harboring BRAF V600E, (2) tumors with mutations within Ras-family genes, (3) tumors without either BRAF or Ras mutations (non-BRAF/non-Ras). Presumably, the majority of tumors in Groups (1) and (2) arose from pre-existing WDTC; a subset of tumors in Group (3) may also have arisen from WDTC via driver mechanisms not completely detectable by exome sequencing (i.e. gene rearrangements) or by novel mechanisms suggested here (such as microsatellite instability). Within Groups (1) and (2), dedifferentiation occurs via a small number well-recognized mechanisms including additional mutations in genes such as TP53, PIK3CA and others; EIF1AX may be a novel driver within this group identified here by exome sequencing in ATC. Within the non-BRAF/non-Ras group of tumors, potential drivers of dedifferentiation including NF1, ERBB2 and MTOR and hypermutator MLH mutations.
Although the genetic and epigenetic events that result in thyroid malignancy continue to be defined with increasing clarity, some patients with certain subtypes of thyroid cancer, such as ATC, persistently have dismal outcomes. Of the recognized or novel driver genes identified here, many have pharmaceuticals targeting their action already in clinical or research usage. While several intriguing findings meriting further investigation are described here, perhaps the most noteworthy conclusion from this study is the potential role for next-generation sequencing of all ATC cases upon diagnosis. Such an effort may identify candidates for empiric targeted therapy in order to mitigate the highly lethal burden of this disease.
Materials and Methods
Sample acquisition
The exome sequencing cohort included matched tumor and normal samples from 22 ATC cases. Regardless of tissue source, ATC samples are histologically heterogeneous as different areas of the tumor contain a mix of viable and necrotic tumor cells, normal parenchyma and stromal tissue. In order to maximize sample accrual, we utilized formalin-fixed, paraffin-embedded (FFPE) archival tumor samples for tissue acquisition for 15 out of 22 cases (T1–15; 68%). Recently, FFPE-derived tissue has been demonstrated to be viable for effective WES (61,62). These patients underwent surgical resection at Yale-New Haven Hospital (New Haven, CT, USA). Briefly, all surgical pathology reports containing ‘anaplastic’ and ‘thyroid’ search terms from 1988 to 2012 were manually reviewed and tissue blocks were obtained for appropriate cases. Blocks with adequate remaining tissue were sectioned and re-reviewed by an experienced pathologist to confirm the presence of ATC and matched normal tissue for WES. The histopathological diagnosis was established according to WHO classification. Three 1 mm tissue cores were obtained and post-core sectioning was performed and reviewed to ensure tissue fidelity. Paraffin was then enzymatically removed and genomic DNA was extracted and sheared via sonication. Seven additional cases of ATC (T16–22, 32%) were obtained from fresh-frozen tissue samples with matched normal tissue from fresh-frozen thyroid (n = 5) or leukocyte DNA (n = 2) from the Karolinska University Hospital (Stockholm, Sweden). Genomic DNA was extracted using the DNeasy Blood and Tissue DNA isolation kit (Qiagen, Hilden, Germany) in accordance with the manufacturer's instruction. All fresh-frozen specimens were sectioned in parallel to the DNA extraction and subsequently re-analyzed by light microscopy to verify appropriate representation of tumor or normal tissue. The procurement of tissue and subsequent genomic analyses were approved by the Yale University Institutional Review Board, New Haven, CT, USA, and the local ethical review board at Karolinska Institutet, Stockholm, Sweden.
The validation cohort consisted of samples of fresh-frozen tissues from a total of 16 histologically confirmed ATCs collected at the Karolinska University Hospital, Stockholm, Sweden. Tissue samples were snap-frozen following resection and genomic DNA was extracted using the DNeasy Blood and Tissue DNA isolation kit (Qiagen, Hilden, Germany) in accordance with the manufacturer's instruction. Fourteen of sixteen tissue samples were examined and confirmed by an experienced endocrine pathologist to demonstrate adequate representation of ATC tumor cells (Supplementary Material, Table S7).
ATC cell lines
Four established ATC cell lines (8305C, 8505C, ACT-1 and BHT101) were also exome-sequenced in addition to the 22 matched pairs of ATC and normal tissues described above using identical methodology for DNA isolation. 8305C and 8505C were purchased from Sigma, MO, USA. BHT101 was purchased from DSMZ Germany, while ACT-1 was generously provided by R.E. Schweppe, University of Colorado School of Medicine, Aurora, CO, USA (originator—Naoyoshi Onoda, Osaka City University, Japan). Cell lines used for the exome analysis (8505C, 8305C, BHT101 and ACT-1) were authenticated to be distinct and of human ATC origin by short tandem repeat and single-nucleotide polymorphism array analysis (63). Additional ATC cell lines used for the validation cohort (Supplementary Material, Table S7) were kindly provided by Dr Nils-Erik Heldin (Uppsala University, Sweden). These lines were reported previously to be of human origin and authenticated by genotyping of short tandem repeats (35,64).
Exome capture, massively parallel sequencing and analysis
Genomic DNA samples generating an adequate high-quality library were subjected to exome capture and sequencing as previously described (23). Briefly, adaptors of known sequence were ligated to genomic DNA fragments which were amplified by ligation-mediated PCR and were then subjected to capture utilizing the NimbleGen 2.1M human exome array. Exome-specific DNA was eluted and then underwent 74 base paired-end sequencing on the Illumina HiSeq 2000 instrument according to the manufacturer's instructions. SAMtools was utilized for base calling and removal of PCR duplicates. Reads were then mapped to the human reference genome GRCh37/hg18 using the ELAND program. Significance of somatic variant calls was assessed by comparing reference and non-reference reads utilizing Fisher's exact test and were manually curated on a sample-by-sample basis to determine high-quality reads. Known variants in annotated databases were excluded and novel variants were evaluated for impact on transcriptional and/or translational processing as well as sequence conservation.
CNV prediction and LOH analysis
LOH loci were identified by comparing B allele frequencies (BAF) of a known array of SNPs in matched tumor and normal samples. Regions with apparent BAF shift were then manually curated to determine LOH. Comparative analysis of coverage depth between tumor and normal samples in 500 kb capture intervals was utilized to identify regions of somatic CNV. Significance was assessed by randomly distributing CNVs in >108 permutations and assessing the likelihood of observing the distribution by chance alone. Again, a false-discovery cut-off of <0.25 was considered significant. A GISTIC-like peel algorithm was utilized to assess the significance of individual CNV peaks.
Sequence validation
Thirty somatic mutations of interest were selected for validation via Sanger sequencing. Flanking primers were designed using an NCBI PrimerBlast (http://blast.ncbi.nlm.nih.gov) and genomic DNA was amplified via PCR and verified to be of correct size by agarose gel electrophoresis (Supplementary Material, Table S3). Putative mutations were sequenced using forward and reverse primers and confirmed by two independent reads. All cases in the validation cohort were analyzed for the demonstrated mTOR R164Q/M2327I, ErbB2 D387N, BRAF V600E mutations defined during exome sequencing and the seven coding exons of EIF1AX were sequenced in a similar fashion. Chromatograms were analyzed using CodonCode Aligner software (CodonCode Corporation; Centerville, MA, USA).
Ontology analysis
We applied the DAVID Bioinformatics Resources 6.7 database (http://david.abcc.ncifcrf.gov) to aid in the identification of significantly altered biological processes and pathways among the 22 ATC cases. By using a functional annotation tool and a user-customized gene background, gene-annotation enrichment analyses as well as KEGG pathway mapping were performed. Significantly enriched GO terms and pathways were selected based on Benjamini-corrected P-values, to allow a balance between discovery of statistically significant genes and restriction of false-positive occurrences. Pathway analyses were also carried out using the Genomatix Pathway System (www.genomatix.de), in which information is extracted from public databases to present >400 canonical pathways as well as extended networks based on literature data rather than GO terms. We furthermore employed the Disease Association Protein-Protein Link Evaluator (DAPPLE—www.broadinstitute.org/mpg/dapple/dapple.php) to evaluate important physical associations between proteins encoded by mutated genes in the ATC cohort. The analysis was made allowing 10 000 permutations and a common interactor binding degree cut-off of 2. Associations are reported with the Bonferroni-corrected P-values representing the statistical significance of a number of network connectivity parameters reported in the literature.
Statistical analyses
Statistical calculations were carried out using the IBM SPSS Statistics 19 software (IBM, Armonk, NY, USA). Tests for normality of continuous variables were performed using the Shapiro–Wilk test, and Spearman's correlation, t-test, Kruskal–Wallis and ANOVA tests were used to test for significance between various clinical parameters and overall mutational burden. P-values of <0.05 were considered as statistically significant.
Supplementary Material
Funding
This work was supported by the Damon Runyon Cancer Research Foundation (T.C.); the Stockholm County Council (C.C.J.); the Swedish Cancer Society (C.C.J., J.Z.); the Swedish Research Council (C.L., A.S.); the Agency for Science, Technology and Research, Singapore (G.G.); and the Howard Hughes Medical Institute (R.P.L.).
Supplementary Material
Acknowledgements
The authors would like to thank Aruna Madan, MD, Lisa Ånfalk and John Overton, PhD for their invaluable assistance in sample collection, submission and assistance. Dr Robert Udelsman, MD, MBA, FACS, FACE, is thanked for his unwavering support of this project.
Conflict of Interest statement. The funding sources had no role in the design, conduct, or reporting of this study and the authors have no conflict of interest to report.
References
- 1.Ragazzi M., Ciarrocchi A., Sancisi V., Gandolfi G., Bisagni A., Piana S. (2014) Update on anaplastic thyroid carcinoma: morphological, molecular, and genetic features of the most aggressive thyroid cancer. Int. J. Endocrinol., 2014, 790834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smallridge R.C., Marlow L.A., Copland J.A. (2009) Anaplastic thyroid cancer: molecular pathogenesis and emerging therapies. Endocr. Relat. Cancer, 16, 17–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McIver B., Hay I.D., Giuffrida D.F., Dvorak C.E., Grant C.S., Thompson G.B., van Heerden J.A., Goellner J.R. (2001) Anaplastic thyroid carcinoma: a 50-year experience at a single institution. Surgery, 130, 1028–1034. [DOI] [PubMed] [Google Scholar]
- 4.Smallridge R.C., Ain K.B., Asa S.L., Bible K.C., Brierley J.D., Burman K.D., Kebebew E., Lee N.Y., Nikiforov Y.E., Rosenthal M.S., et al. (2012) American Thyroid Association guidelines for management of patients with anaplastic thyroid cancer. Thyroid, 22, 1104–1139. [DOI] [PubMed] [Google Scholar]
- 5.Sugitani I., Miyauchi A., Sugino K., Okamoto T., Yoshida A., Suzuki S. (2012) Prognostic factors and treatment outcomes for anaplastic thyroid carcinoma: ATC Research Consortium of Japan cohort study of 677 patients. World J. Surg., 36, 1247–1254. [DOI] [PubMed] [Google Scholar]
- 6.Kebebew E., Greenspan F.S., Clark O.H., Woeber K.A., McMillan A. (2005) Anaplastic thyroid carcinoma. Treatment outcome and prognostic factors. Cancer, 103, 1330–1335. [DOI] [PubMed] [Google Scholar]
- 7.Neff R.L., Farrar W.B., Kloos R.T., Burman K.D. (2008) Anaplastic thyroid cancer. Endocrinol. Metab. Clin. North Am., 37, 525–538 xi. [DOI] [PubMed] [Google Scholar]
- 8.Lam K.Y., Lo C.Y., Chan K.W., Wan K.Y. (2000) Insular and anaplastic carcinoma of the thyroid: a 45-year comparative study at a single institution and a review of the significance of p53 and p21. Ann. Surg., 231, 329–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Venkatesh Y.S., Ordonez N.G., Schultz P.N., Hickey R.C., Goepfert H., Samaan N.A. (1990) Anaplastic carcinoma of the thyroid. A clinicopathologic study of 121 cases. Cancer, 66, 321–330. [DOI] [PubMed] [Google Scholar]
- 10.Guerra A., Di Crescenzo V., Garzi A., Cinelli M., Carlomagno C., Tonacchera M., Zeppa P., Vitale M. (2013) Genetic mutations in the treatment of anaplastic thyroid cancer: a systematic review. BMC Surg., 13(Suppl 2), S44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nikiforov Y.E. (2004) Genetic alterations involved in the transition from well-differentiated to poorly differentiated and anaplastic thyroid carcinomas. Endocr. Pathol., 15, 319–327. [DOI] [PubMed] [Google Scholar]
- 12.Santarpia L., El-Naggar A.K., Cote G.J., Myers J.N., Sherman S.I. (2008) Phosphatidylinositol 3-kinase/akt and ras/raf-mitogen-activated protein kinase pathway mutations in anaplastic thyroid cancer. J. Clin. Endocrinol. Metab., 93, 278–284. [DOI] [PubMed] [Google Scholar]
- 13.Garcia-Rostan G., Tallini G., Herrero A., D'Aquila T.G., Carcangiu M.L., Rimm D.L. (1999) Frequent mutation and nuclear localization of beta-catenin in anaplastic thyroid carcinoma. Cancer Res., 59, 1811–1815. [PubMed] [Google Scholar]
- 14.Kurihara T., Ikeda S., Ishizaki Y., Fujimori M., Tokumoto N., Hirata Y., Ozaki S., Okajima M., Sugino K., Asahara T. (2004) Immunohistochemical and sequencing analyses of the Wnt signaling components in Japanese anaplastic thyroid cancers. Thyroid, 14, 1020–1029. [DOI] [PubMed] [Google Scholar]
- 15.Pita J.M., Figueiredo I.F., Moura M.M., Leite V., Cavaco B.M. (2014) Cell cycle deregulation and TP53 and RAS mutations are major events in poorly differentiated and undifferentiated thyroid carcinomas. J. Clin. Endocrinol. Metab., 99, E497–E507. [DOI] [PubMed] [Google Scholar]
- 16.Liu Z., Hou P., Ji M., Guan H., Studeman K., Jensen K., Vasko V., El-Naggar A.K., Xing M. (2008) Highly prevalent genetic alterations in receptor tyrosine kinases and phosphatidylinositol 3-kinase/akt and mitogen-activated protein kinase pathways in anaplastic and follicular thyroid cancers. J. Clin. Endocrinol. Metab., 93, 3106–3116. [DOI] [PubMed] [Google Scholar]
- 17.Sosa J.A., Balkissoon J., Lu S.P., Langecker P., Elisei R., Jarzab B., Bal C.S., Marur S., Gramza A., Ondrey F. (2012) Thyroidectomy followed by fosbretabulin (CA4P) combination regimen appears to suggest improvement in patient survival in anaplastic thyroid cancer. Surgery, 152, 1078–1087. [DOI] [PubMed] [Google Scholar]
- 18.Nixon I.J., Shaha A.R., Tuttle M.R. (2013) Targeted therapy in thyroid cancer. Curr. Opin. Otolaryngol. Head Neck Surg., 21, 130–134. [DOI] [PubMed] [Google Scholar]
- 19.Jin N., Jiang T., Rosen D.M., Nelkin B.D., Ball D.W. (2009) Dual inhibition of mitogen-activated protein kinase kinase and mammalian target of rapamycin in differentiated and anaplastic thyroid cancer. J. Clin. Endocrinol. Metab., 94, 4107–4112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wagle N., Grabiner B.C., Van Allen E.M., Amin-Mansour A., Taylor-Weiner A., Rosenberg M., Gray N., Barletta J.A., Guo Y., Swanson S.J., et al. (2014) Response and acquired resistance to everolimus in anaplastic thyroid cancer. N. Engl. J. Med., 371, 1426–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rosove M.H., Peddi P.F., Glaspy J.A. (2013) BRAF V600E inhibition in anaplastic thyroid cancer. N. Engl. J. Med., 368, 684–685. [DOI] [PubMed] [Google Scholar]
- 22.Bilguvar K., Ozturk A.K., Louvi A., Kwan K.Y., Choi M., Tatli B., Yalnizoglu D., Tuysuz B., Caglayan A.O., Gokben S., et al. (2010) Whole-exome sequencing identifies recessive WDR62 mutations in severe brain malformations. Nature, 467, 207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Choi M., Scholl U.I., Ji W., Liu T., Tikhonova I.R., Zumbo P., Nayir A., Bakkaloglu A., Ozen S., Sanjad S., et al. (2009) Genetic diagnosis by whole exome capture and massively parallel DNA sequencing. Proc. Natl Acad. Sci. USA, 106, 19096–19101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cromer M.K., Starker L.F., Choi M., Udelsman R., Nelson-Williams C., Lifton R.P., Carling T. (2012) Identification of somatic mutations in parathyroid tumors using whole-exome sequencing. J. Clin. Endocrinol. Metab., 97, E1774–E1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao S., Choi M., Overton J.D., Bellone S., Roque D.M., Cocco E., Guzzo F., English D.P., Varughese J., Gasparrini S., et al. (2013) Landscape of somatic single-nucleotide and copy-number mutations in uterine serous carcinoma. Proc. Natl Acad. Sci. USA, 110, 2916–2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krauthammer M., Kong Y., Ha B.H., Evans P., Bacchiocchi A., McCusker J.P., Cheng E., Davis M.J., Goh G., Choi M., et al. (2012) Exome sequencing identifies recurrent somatic RAC1 mutations in melanoma. Nat. Genet., 44, 1006–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haimovich A.D. (2011) Methods, challenges, and promise of next-generation sequencing in cancer biology. Yale J. Biol. Med., 84, 439–446. [PMC free article] [PubMed] [Google Scholar]
- 28.Vater I., Montesinos-Rongen M., Schlesner M., Haake A., Purschke F., Sprute R., Mettenmeyer N., Nazzal I., Nagel I., Gutwein J., et al. (2014) The mutational pattern of primary lymphoma of the central nervous system determined by whole-exome sequencing. Leukemia, doi:10.1038/leu.2014.264. [DOI] [PubMed] [Google Scholar]
- 29.Vogelstein B., Papadopoulos N., Velculescu V.E., Zhou S., Diaz L.A., Jr., Kinzler K.W. (2013) Cancer genome landscapes. Science, 339, 1546–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu D., Yang C., Bojdani E., Murugan A.K., Xing M. (2013) Identification of RASAL1 as a major tumor suppressor gene in thyroid cancer. J. Natl Cancer Inst., 105, 1617–1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kelly L.M., Barila G., Liu P., Evdokimova V.N., Trivedi S., Panebianco F., Gandhi M., Carty S.E., Hodak S.P., Luo J., et al. (2014) Identification of the transforming STRN-ALK fusion as a potential therapeutic target in the aggressive forms of thyroid cancer. Proc. Natl Acad. Sci. USA, 111, 4233–4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perot G., Soubeyran I., Ribeiro A., Bonhomme B., Savagner F., Boutet-Bouzamondo N., Hostein I., Bonichon F., Godbert Y., Chibon F. (2014) Identification of a recurrent STRN/ALK fusion in thyroid carcinomas. PLoS One, 9, e87170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Agrawal N., Akbani R., Aksoy B.A., Ally A., Arachchi H., Asa S.L., Auman J.T., Balasundaram M., Balu S., Baylin S.B., et al. (2014) Integrated genomic characterization of papillary thyroid carcinoma. Cell, 159, 676–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schwarz J.M., Cooper D.N., Schuelke M., Seelow D. (2014) MutationTaster2: mutation prediction for the deep-sequencing age. Nat. Methods, 11, 361–362. [DOI] [PubMed] [Google Scholar]
- 35.Lee J.J., Foukakis T., Hashemi J., Grimelius L., Heldin N.E., Wallin G., Rudduck C., Lui W.O., Hoog A., Larsson C. (2007) Molecular cytogenetic profiles of novel and established human anaplastic thyroid carcinoma models. Thyroid, 17, 289–301. [DOI] [PubMed] [Google Scholar]
- 36.Garcia-Rostan G., Zhao H., Camp R.L., Pollan M., Herrero A., Pardo J., Wu R., Carcangiu M.L., Costa J., Tallini G. (2003) ras mutations are associated with aggressive tumor phenotypes and poor prognosis in thyroid cancer. J. Clin. Oncol., 21, 3226–3235. [DOI] [PubMed] [Google Scholar]
- 37.Milosevic Z., Pesic M., Stankovic T., Dinic J., Milovanovic Z., Stojsic J., Dzodic R., Tanic N., Bankovic J. (2014) Targeting RAS-MAPK-ERK and PI3K-AKT-mTOR signal transduction pathways to chemosensitize anaplastic thyroid carcinoma. Transl. Res., 164, 411–423. [DOI] [PubMed] [Google Scholar]
- 38.Ricarte-Filho J.C., Ryder M., Chitale D.A., Rivera M., Heguy A., Ladanyi M., Janakiraman M., Solit D., Knauf J.A., Tuttle R.M., et al. (2009) Mutational profile of advanced primary and metastatic radioactive iodine-refractory thyroid cancers reveals distinct pathogenetic roles for BRAF, PIK3CA, and AKT1. Cancer Res., 69, 4885–4893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xing M. (2013) Molecular pathogenesis and mechanisms of thyroid cancer. Nat. Rev. Cancer, 13, 184–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McFadden D.G., Vernon A., Santiago P.M., Martinez-McFaline R., Bhutkar A., Crowley D.M., McMahon M., Sadow P.M., Jacks T. (2014) p53 constrains progression to anaplastic thyroid carcinoma in a Braf-mutant mouse model of papillary thyroid cancer. Proc. Natl Acad. Sci. USA, 111, E1600–E1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Charles R.P., Silva J., Iezza G., Phillips W.A., McMahon M. (2014) Activating BRAF and PIK3CA mutations cooperate to promote anaplastic thyroid carcinogenesis. Mol. Cancer Res., 12, 979–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martin M., Masshofer L., Temming P., Rahmann S., Metz C., Bornfeld N., van de Nes J., Klein-Hitpass L., Hinnebusch A.G., Horsthemke B., et al. (2013) Exome sequencing identifies recurrent somatic mutations in EIF1AX and SF3B1 in uveal melanoma with disomy 3. Nat. Genet., 45, 933–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ewens K.G., Kanetsky P.A., Richards-Yutz J., Purrazzella J., Shields C.L., Ganguly T., Ganguly A. (2014) Chromosome 3 status combined with BAP1 and EIF1AX mutation profiles are associated with metastasis in uveal melanoma. Invest. Ophthalmol. Vis. Sci., 55, 5160–5167. [DOI] [PubMed] [Google Scholar]
- 44.Field M.G., Harbour J.W. (2014) Recent developments in prognostic and predictive testing in uveal melanoma. Curr. Opin. Ophthalmol., 25, 234–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yu C., Luo C., Qu B., Khudhair N., Gu X., Zang Y., Wang C., Zhang N., Li Q., Gao X. (2014) Molecular network including eIF1AX, RPS7, and 14-3-3gamma regulates protein translation and cell proliferation in bovine mammary epithelial cells. Arch. Biochem. Biophys., 564, 142–155. [DOI] [PubMed] [Google Scholar]
- 46.Hardt M., Chantaravisoot N., Tamanoi F. (2011) Activating mutations of TOR (target of rapamycin). Genes Cells, 16, 141–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roskoski R., Jr. (2014) The ErbB/HER family of protein-tyrosine kinases and cancer. Pharmacol. Res., 79, 34–74. [DOI] [PubMed] [Google Scholar]
- 48.Ross J.S., Wang K., Gay L.M., Al-Rohil R.N., Nazeer T., Sheehan C.E., Jennings T.A., Otto G.A., Donahue A., He J., et al. (2014) A high frequency of activating extracellular domain ERBB2 (HER2) mutation in micropapillary urothelial carcinoma. Clin. Cancer Res., 20, 68–75. [DOI] [PubMed] [Google Scholar]
- 49.Kim N., Hong Y., Kwon D., Yoon S. (2013) Somatic mutaome profile in human cancer tissues. Genomics Inform., 11, 239–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Eudy J.D., Weston M.D., Yao S., Hoover D.M., Rehm H.L., Ma-Edmonds M., Yan D., Ahmad I., Cheng J.J., Ayuso C., et al. (1998) Mutation of a gene encoding a protein with extracellular matrix motifs in Usher syndrome type IIa. Science, 280, 1753–1757. [DOI] [PubMed] [Google Scholar]
- 51.Yoshida R., Miyashita K., Inoue M., Shimamoto A., Yan Z., Egashira A., Oki E., Kakeji Y., Oda S., Maehara Y. (2011) Concurrent genetic alterations in DNA polymerase proofreading and mismatch repair in human colorectal cancer. Eur. J. Hum. Genet., 19, 320–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Loeb L.A. (2011) Human cancers express mutator phenotypes: origin, consequences and targeting. Nat. Rev. Cancer, 11, 450–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Birkbak N.J., Kochupurakkal B., Izarzugaza J.M., Eklund A.C., Li Y., Liu J., Szallasi Z., Matulonis U.A., Richardson A.L., Iglehart J.D., et al. (2013) Tumor mutation burden forecasts outcome in ovarian cancer with BRCA1 or BRCA2 mutations. PLoS One, 8, e80023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shcherbakova P.V., Kunkel T.A. (1999) Mutator phenotypes conferred by MLH1 overexpression and by heterozygosity for mlh1 mutations. Mol. Cell. Biol., 19, 3177–3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stulp R.P., Herkert J.C., Karrenbeld A., Mol B., Vos Y.J., Sijmons R.H. (2008) Thyroid cancer in a patient with a germline MSH2 mutation. Case report and review of the Lynch syndrome expanding tumour spectrum. Hered. Cancer Clin. Pract., 6, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fagin J.A., Matsuo K., Karmakar A., Chen D.L., Tang S.H., Koeffler H.P. (1993) High prevalence of mutations of the p53 gene in poorly differentiated human thyroid carcinomas. J. Clin. Invest., 91, 179–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Garcia-Rostan G., Camp R.L., Herrero A., Carcangiu M.L., Rimm D.L., Tallini G. (2001) Beta-catenin dysregulation in thyroid neoplasms: down-regulation, aberrant nuclear expression, and CTNNB1 exon 3 mutations are markers for aggressive tumor phenotypes and poor prognosis. Am. J. Pathol., 158, 987–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Paes J.E., Ringel M.D. (2008) Dysregulation of the phosphatidylinositol 3-kinase pathway in thyroid neoplasia. Endocrinol. Metab. Clin. North Am., 37, 375–387 viii–ix. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cordenonsi M., Montagner M., Adorno M., Zacchigna L., Martello G., Mamidi A., Soligo S., Dupont S., Piccolo S. (2007) Integration of TGF-beta and Ras/MAPK signaling through p53 phosphorylation. Science, 315, 840–843. [DOI] [PubMed] [Google Scholar]
- 60.Guo X., Wang X.F. (2009) Signaling cross-talk between TGF-beta/BMP and other pathways. Cell Res., 19, 71–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Holley T., Lenkiewicz E., Evers L., Tembe W., Ruiz C., Gsponer J.R., Rentsch C.A., Bubendorf L., Stapleton M., Amorese D., et al. (2012) Deep clonal profiling of formalin fixed paraffin embedded clinical samples. PLoS One, 7, e50586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Menon R., Deng M., Boehm D., Braun M., Fend F., Boehm D., Biskup S., Perner S. (2012) Exome enrichment and SOLiD sequencing of formalin fixed paraffin embedded (FFPE) prostate cancer tissue. Int. J. Mol. Sci., 13, 8933–8942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schweppe R.E., Klopper J.P., Korch C., Pugazhenthi U., Benezra M., Knauf J.A., Fagin J.A., Marlow L.A., Copland J.A., Smallridge R.C., et al. (2008) Deoxyribonucleic acid profiling analysis of 40 human thyroid cancer cell lines reveals cross-contamination resulting in cell line redundancy and misidentification. J. Clin. Endocrinol. Metab., 93, 4331–4341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wennerberg E., Pfefferle A., Ekblad L., Yoshimoto Y., Kremer V., Kaminskyy V.O., Juhlin C.C., Hoog A., Bodin I., Svjatoha V., et al. (2014) Human anaplastic thyroid carcinoma cells are sensitive to NK cell-mediated lysis via ULBP2/5/6 and chemoattract NK cells. Clin. Cancer Res., 20, 5733–5744. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.