Abstract
The Rbfox family of RNA-binding proteins is highly conserved with established roles in alternative splicing (AS) regulation. High-throughput studies aimed at understanding transcriptome remodeling have revealed skeletal muscle as displaying one of the largest number of AS events. This finding is consistent with requirements for tissue-specific protein isoforms needed to sustain muscle-specific functions. Rbfox1 is abundant in vertebrate brain, heart and skeletal muscle. Genome-wide genetic approaches have linked the Rbfox1 gene to autism, and a brain-specific knockout mouse revealed a critical role for this splicing regulator in neuronal function. Moreover, a Caenorhabditis elegans Rbfox1 homolog regulates muscle-specific splicing. To determine the role of Rbfox1 in muscle function, we developed a conditional knockout mouse model to specifically delete Rbfox1 in adult tissue. We show that Rbfox1 is required for muscle function but a >70% loss of Rbfox1 in satellite cells does not disrupt muscle regeneration. Deep sequencing identified aberrant splicing of multiple genes including those encoding myofibrillar and cytoskeletal proteins, and proteins that regulate calcium handling. Ultrastructure analysis of Rbfox1−/− muscle by electron microscopy revealed abundant tubular aggregates. Immunostaining showed mislocalization of the sarcoplasmic reticulum proteins Serca1 and Ryr1 in a pattern indicative of colocalization with the tubular aggregates. Consistent with mislocalization of Serca1 and Ryr1, calcium handling was drastically altered in Rbfox1−/− muscle. Moreover, muscle function was significantly impaired in Rbfox1−/− muscle as indicated by decreased force generation. These results demonstrate that Rbfox1 regulates a network of AS events required to maintain multiple aspects of muscle physiology.
Introduction
Analysis of metazoan transcriptomes by deep sequencing has revealed that a high level of mRNA diversity is generated through alternative splicing (AS) (1,2). More than 90% of human genes undergo AS producing multiple protein isoforms, which can differ in localization, expression level and biological function (1,3). AS is tightly regulated, particularly, during development and cell differentiation (4–7). AS regulation is achieved through an interplay between trans-acting splicing factors and cis-acting intronic and exonic regulatory elements within pre-mRNAs. Highly specialized organs, such as brain, testis, heart and skeletal muscle, display the highest levels of AS events (4,7–10). Tissue-specific splicing regulation is often achieved through expression of tissue-restricted splicing factors modulating inclusion or skipping of target alternative exons (10–12).
The Rbfox family of RNA-binding proteins contains three genes: Rbfox1, Rbfox2 and Rbfox3. All three family members regulate AS of target exons closely associated with the binding motif (U/A)GCAU/CG (13–15). While Rbfox3 expression is restricted to neurons, Rbfox2 is widely expressed in whole embryo, ovary, stem cells, brain, skeletal muscle and heart, and Rbfox1 is selectively expressed in brain, heart and skeletal muscle (16). In the brain, loss of function of Rbfox1 was shown to alter AS of genes involved in neuronal function (17). Rbfox1 deletion in CNS-derived stem and progenitor cells revealed altered splicing patterns of genes with a role in synaptic function, thus correlating the brain phenotype to splicing changes. In humans, point mutations, chromosomal translocation or deletions affecting RBFOX1 have been found in patients with severe neurological disorders such as epilepsy, mental retardation (18), schizophrenia (19) and autism (20,21). Notably, deletion of 1.3 kb in the RBFOX1 locus has been reported in an individual affected by autism, who also showed muscle weakness (22), implying potential involvement of this splicing regulator in muscle function. A role for Rbfox1 in skeletal muscle is supported by the finding that the Rbfox-binding motif is enriched and conserved within introns surrounding alternative exons that are regulated during muscle differentiation (23). Furthermore, concomitant depletion of Rbfox1 and Rbfox2 was shown to affect normal muscle development in zebrafish and worms by regulating muscle-specific splicing events required for proper muscle function (24,25). However, a comprehensive study on the role of Rbfox proteins in mammalian skeletal muscle has not been performed, leaving an open question about their involvement in muscle function.
AS contributes to a tissue-specific repertoire of transcripts critical to the regulation of skeletal muscle function (26) and aberrant splicing is a major feature of several muscle diseases, such as myotonic dystrophy (27). Although several muscle-specific splicing factors have been described and their role has been investigated in vitro (28), the impact of AS regulation on muscle function remains unknown in part because of the absence of studies using in vivo models of skeletal muscle-specific loss of function. Given the high expression level of Rbfox1 in skeletal muscle and evidence from individuals with altered RBFOX1 expression and non-mammalian experimental systems, we hypothesized that Rbfox1 plays a crucial role in regulating muscle physiology. To investigate this role, we generated conditional Rbfox1 loss of function specifically in skeletal muscle. Deep sequencing of RNA from Rbfox1−/− muscle identified a number of genes involved in muscle function whose splicing was altered compared with Rbfox1loxP/loxP litter mate controls. In particular, we found aberrant splicing in genes involved in maintenance of myofibrillar and cytoskeletal organization. Accordingly, Rbfox1−/− muscle showed abnormal myofibrillar structure and sarcolemma fragility consequent to exercise. Furthermore, Rbfox1 loss of function causes formation of tubular aggregates as shown by electron microscopy (EM). Tubular aggregates are proposed to be derived from the sarcoplasmic reticulum (SR) and immunofluorescent studies showed mislocalization of the calcium channel ryanodine receptor 1 (Ryr1) and the sarco(endo)plasmic reticulum calcium ATPase 1 (Serca1) in cytoplasmic aggregates, which resemble the tubular aggregates observed by EM. Physiological analysis of Rbfox1−/− showed loss of muscle performance as a consequence of alterations in myoplasmic calcium handling. Stimulated muscle fibers from Rbfox1−/− mice exhibited a delay in agonist-induced calcium release and alterations of both the amplitude and timing of calcium release. These defects lead to decreased force generation as assayed by force-frequency experiments.
Our study demonstrates that Rbfox1 plays a pivotal role in skeletal muscle by directing an AS program necessary for adult skeletal muscle function. These findings indicate that selective alteration of muscle-specific AS signatures can exert a strong impact on muscle functionality in vivo.
Results
Rbfox1 expression in physiological and pathological conditions
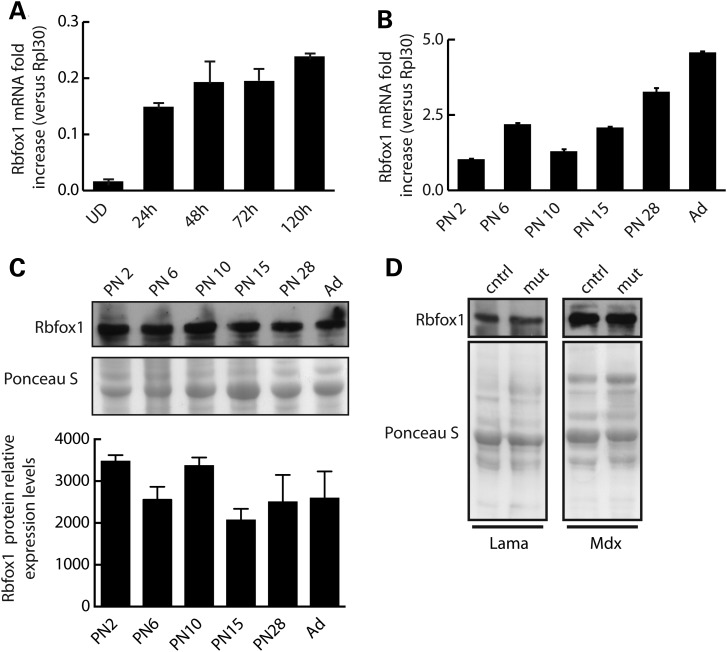
To determine the pattern of Rbfox1 expression during skeletal muscle development, we first analyzed Rbfox1 mRNA levels during a differentiation time course of primary myoblasts into myotubes (Fig. 1A). Using quantitative RT-PCR, we found that Rbfox1 mRNA is barely detectable in proliferating myoblasts and gradually increases after induction of differentiation, reaching a peak by 120 h of myotube formation (Fig. 1A). The expression of Rbfox1 protein similarly increased during differentiation of primary myoblast cultures (Supplementary Material, Fig. S1). We also determined Rbfox1 mRNA expression throughout post-natal (PN) muscle development and identified a substantial increase in mRNA levels from newborn to adult (Fig. 1B). Interestingly, we found that Rbfox1 protein expression does not parallel mRNA expression. Western blot analysis revealed only modest changes in the level of Rbfox1 protein during PN muscle development (Fig. 1C). We also asked whether Rbfox1 expression was altered in conditions of muscle pathology. Previously published results revealed altered expression of Rbfox1 associated with two diseases affecting muscle: myotonic dystrophy (29) and facioscapulohumeral muscular dystrophy (30). We assayed Rbfox1 protein levels by western blot in two additional mouse models of muscular dystrophy: congenital merosin-deficient muscular dystrophy (129P1/ReJ-Lama2dy/J) and Duchenne muscular dystrophy (mdx) (Fig. 1D). We found that Rbfox1 protein expression was not altered in either of the muscular dystrophy models we analyzed (Fig. 1D), indicating that there is not a general response to muscle pathology. These results demonstrate that Rbfox1 mRNA and protein is induced during skeletal muscle differentiation in vitro. The discordance between mRNA and protein expression levels during PN development indicates that protein levels are regulated in part by posttranscriptional mechanisms.
Figure 1.
Rbfox1 is expressed throughout skeletal muscle development. (A and B) Total RNA was extracted from primary myoblast cultures at the indicated times relative to addition of differentiation media (A) or tibialis anterior muscle isolated at the indicated ages (B). Quantitative RT-PCR was performed in triplicate using an Rbfox1 TaqMan probe. Data were normalized to Rpl30 and expressed as mean fold increase ±s.e.m. (C) Western blot analysis of Rbfox1 protein levels assayed using total protein extracted from the samples used in (B). Quantitative densitometry of Rbfox1 protein expression relative to Ponceau staining. Error bars represent the standard error for n = 2 sets of experiments. (D) Gastrocnemius muscle from congenital merosin-deficient muscular dystrophy (129P1/ReJ-Lama2dy/J) at 1 to 2 months of age, and Duchenne muscular dystrophy (mdx) mouse models were harvested, total protein extracted and used for western blot analysis. Age-matched wild-type mice on either the 129 background (for 129P1/ReJ-Lama2dy/J mice) or C57BL/6 (for mdx mice) were used as controls.
Rbfox1 depletion in satellite cells does not disrupt skeletal muscle regeneration
Following injury, exercise or under pathological conditions, skeletal muscle stem cells, called satellite cells, exit quiescence, proliferate, migrate to the site of injury and fuse to pre-existing fibers to repair damage (31). We observed that Rbfox1 mRNA is barely detectable in proliferating primary myoblasts (satellite cells), and its expression increases with differentiation (Fig. 1A). This finding suggested a potential role for Rbfox1 during late steps of muscle regeneration, i.e. fusion of newly generated myoblasts to pre-existing fibers. To determine whether loss of Rbfox1 affects regeneration, we generated mice for tamoxifen-inducible knockout of Rbfox1 specifically in satellite cells. Mice carrying Rbfox1 floxed alleles (Rbfox1loxP/loxP) were crossed with mice carrying Cre recombinase knocked in to the Pax7 gene (Pax7Cre/Cre) (32) to obtain double homozygous Rbfox1loxP/loxP; Pax7Cre/Cre mice (Supplementary Material, Fig. S2A). Recombination was induced by daily intraperitoneal administration of 5 mg of tamoxifen for 5 days (Supplementary Material, Fig. S2B). Rbfox1loxP/loxP litter mates were injected with the same amount of tamoxifen and used as controls. To assess the efficiency of recombination, primary myoblasts were isolated from Rbfox1loxP/loxP; Pax7Cre/Cre (referred to as Rbfox1−/−) and Rbfox1loxP/loxP litter mates (control mice) 1 week after the last tamoxifen injection. Since Rbfox1 mRNA is difficult to detect in myoblasts (Fig. 1A), primary myoblast cultures were differentiated in vitro for 5 days to induce Rbfox1 mRNA expression. Quantitative RT-PCR using RNA from differentiated myotubes showed a 71% reduction of Rbfox1 mRNA levels (Supplementary Material, Fig. S2C), comparable with published reports using this Cre line (32). To determine the role of Rbfox1 in muscle regeneration, the tibialis anterior muscle of knockout mice and control litter mates was injured by cardiotoxin injection (33). Hematoxylin and eosin staining of cross-sections from Rbfox1−/− mice and controls was used to monitor regeneration 4, 14 and 30 days after injury. We did not observed differences in the timing or efficiency of muscle regeneration between controls and Rbfox1−/− mice (Supplementary Material, Fig. S2D). Our results indicate that >70% deficiency of Rbfox1 in satellite cells does not substantially affect skeletal muscle regeneration. However, we cannot rule out that elimination of Rbfox1 expression from a higher fraction of satellite cells would reveal a role for Rbfox1 in muscle regeneration or that the paralog Rbfox2 provides functional redundancy.
Loss of Rbfox1 in adult skeletal muscle affects myofiber size
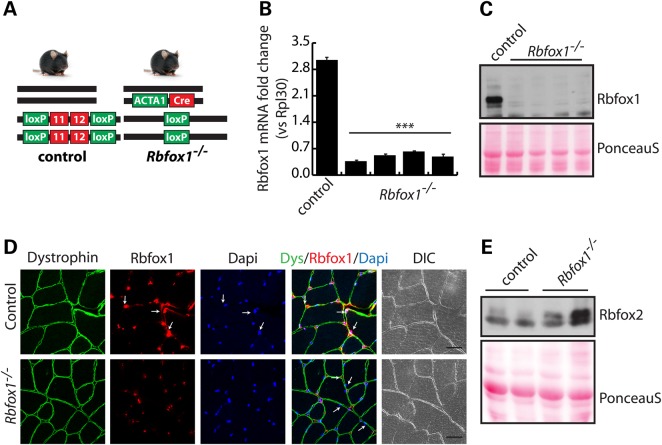
To evaluate the role of Rbfox1 in adult skeletal muscle, we generated inducible and muscle-specific Rbfox1 knockout mice by breeding Rbfox1loxP/loxP mice with ACTA1-rtTAcre/+ mice (34). In ACTA1-rtTAcre/+ mice, Cre expression is driven by the tetracycline-responsive regulatory element and reverse tetracycline-controlled transactivator (rtTA) is driven by the promoter of the human alpha skeletal muscle actin 1 (ACTA1) gene (Fig. 2A). Deletion of the Rbfox1 gene was achieved by feeding Rbfox1loxP/loxP; ACTA1-rtTAcre/+ (referred to as Rbfox1−/−) mice chow containing 2 g/kg doxycycline for 1 week starting at PN Day 21, after which mice were put on regular diet. Quantitative RT-PCR and western blot analyses confirmed >95% loss of of Rbfox1 in adult Rbfox1−/− skeletal muscle (Fig. 2B and C). Immunofluorescence staining for Rbfox1 performed on cross-sections from tibialis anterior muscle showed a substantial loss of signal in knockout mice (Fig. 2D).
Figure 2.
Myofiber-specific Rbfox1 knockout mice. (A) The genotype of control and Rbfox1−/− mice is schematically represented. In the control mouse (left), the floxed Rbfox1 exons 11 and 12 are represented as red boxes, flanked by two loxP sites (green boxes). Wild-type alleles are represented as solid black bars. In Rbfox1−/− mice (right), doxycycline-induced expression of the Cre recombinase under the control of the human skeletal muscle alpha actin 1 (ACTA1) gene promoter (green box), drives recombination between the loxP sites and deletion of Rbfox1 exons 11 and 12. (B) Total RNA was extracted from tibialis anterior muscle of Rbfox1loxP/loxP control or Rbfox1−/− mice. Quantitative RT-PCR analysis of Rbfox1 knockout efficiency was performed in triplicate using a TaqMan probe for Rbfox1. *** indicates P ≤ 0.001 as determined by two-tailed Student's t-test. (C) Western blot analysis of the same tissues as in (B) confirmed efficient Rbfox1 knockout at the protein level. Ponceau staining shows equal loading between samples. (D) Immunofluorence staining for Rbfox1 in tibialis anterior muscle shows nuclear localization in control tissues and a substantial loss of signal in myofibers from Rbfox1−/− mice. Non-muscle nuclei will retain Rbfox1 staining. Dystrophin staining was used to visualize myofiber membranes. Arrows indicate either Rbfox1-positive (top panels) or -negative (bottom panels) nuclei within myofibers. Scale bar: 20 μm. (E) Western blot analysis of Rbfox2 protein expression in control and Rbfox1−/− tibialis anterior muscles.
We also observed a slight increase of Rbfox2 protein expression in Rbfox1−/− muscles (Fig. 2E). These data correlate with increased expression (1.7-fold) of Rbfox2 mRNA as identified by RNA-Seq analysis (Supplementary Material, Table S2). In contrast, Rbfox3 was not detected by RNA-Seq (Supplementary Material, Table S2).
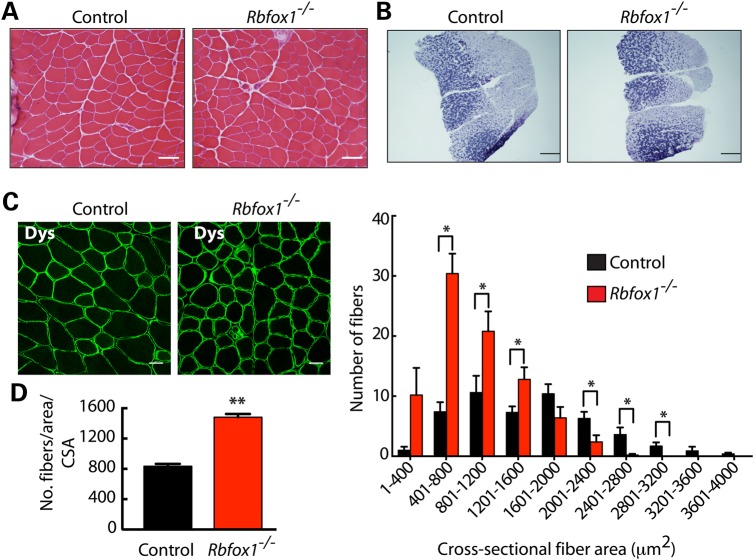
Rbfox1−/− mice were indistinguishable from Rbfox1loxP/loxP litter mate controls with regard to weight gain through development. Histological analysis did not reveal obvious myopathy (Fig. 3A). Succinate dehydrogenase staining revealed normal distribution of glycolytic and oxidative fibers, suggesting that Rbfox1 loss does not affect fiber type (Fig. 3B). This result was supported by analysis of tibialis anterior muscle by immunofluorescence staining for slow or fast myosin heavy chain (MHC), which showed the expected enrichment of fast-twitch fibers in both control and Rbfox1−/− mice (Supplementary Material, Fig. S3A, left and right panels, respectively).
Figure 3.
Rbfox1−/− muscle has reduced myofiber size. (A) Hematoxylin and eosin staining of control and Rbfox1−/− tibialis anterior muscle shows no overt myopathic features. Scale bar: 25 μm. (B) Succinate dehydrogenase staining on gastrocnemius muscle from control and Rbfox1−/− mice reveals normal distribution of glycolytic and oxidative fibers. Scale bar: 500 μm. (C) Left panel. Dystrophin staining of tibialis anterior muscle from control and Rbfox1−/− mice was used to visualize the myofiber membrane. Scale bar: 20 μm. Right panel. Crosssectional area of myofibers in control and Rbfox1−/− muscles was measured by ImageJ based on dystrophin staining of tibialis anterior muscle sections. **indicates P≤ 0.01 as determined by two-tailed Student's t-test. (D) Number of fibers per area was counted in control and Rbfox1−/− tibialis anterior muscles using ImageJ based on dystrophin staining. * indicates P≤ 0.05 as determined by two-tailed Student's t-test.
Detailed analysis demonstrated that myofibers in Rbfox1−/− mice appeared smaller than those of control animals (Fig. 3C, left). Indeed, quantification of cross-sectional area (CSA) of myofibers revealed reduced fiber size in Rbfox1−/− mice compared with control litter mates (Fig. 3C, right). Moreover, knockout mice had a significant increase of myofiber number per area (Fig. 3D) relative to the CSA of the muscle analyzed. To determine whether the increased number of smaller fibers was a consequence of muscle regeneration, we analyzed the expression of regeneration markers. Immunostaining for desmin, which marks fetal as well as regenerative fibers (35,36), revealed no differences in expression between control and Rbfox1−/− mice (Supplementary Material, Fig. S3B, top panels). Similarly, neonatal MHC was not expressed in Rbfox1−/− skeletal muscle (Supplementary Material, Fig. S3B, bottom panels). Taken together, these results demonstrated that loss of Rbfox1 produced smaller myofibers without increasing muscle regeneration. We monitored the expression of two atrophy-related genes, the ubiquitin ligases Fbxo32 (atrogin-1) and Trim63 (MuRF1) (Supplementary Material, Fig. S3C). Quantitative RT-PCR showed no difference in Trim63 mRNA and slightly reduced Fbxo32 that did not reach significant levels between Rbfox1−/− and control mice, indicating that atrophy markers are not significantly affected in Rbfox1−/− muscle.
Rbfox1 regulates splicing of genes necessary for calcium signaling and cytoskeleton organization
The data presented above indicated that decreased fiber size caused by loss of Rbfox1 was independent from regeneration and atrophy-related pathways. The Rbfox1 gene encodes a well-known splicing regulator. Disruption of the Rbfox1 splicing program in the brain was found to impair neuronal function, altering synaptic transmission (17). To determine whether transcriptome changes triggered by loss of Rbfox1 could account for the phenotype observed in muscle, we analyzed AS and gene expression differences in Rbfox1 knockout and control muscles using mRNA deep sequencing (RNA-Seq). RNA isolated from the tibialis anterior of two Rbfox1−/− and two control mice were used to prepare complementary DNA libraries after polyA selection for 100 bp paired-end reads using the Illumina HiSeq2000. We obtained >130 million read pairs per sample, 90% of which mapped to the mouse genome (Supplementary Material, Table S1).
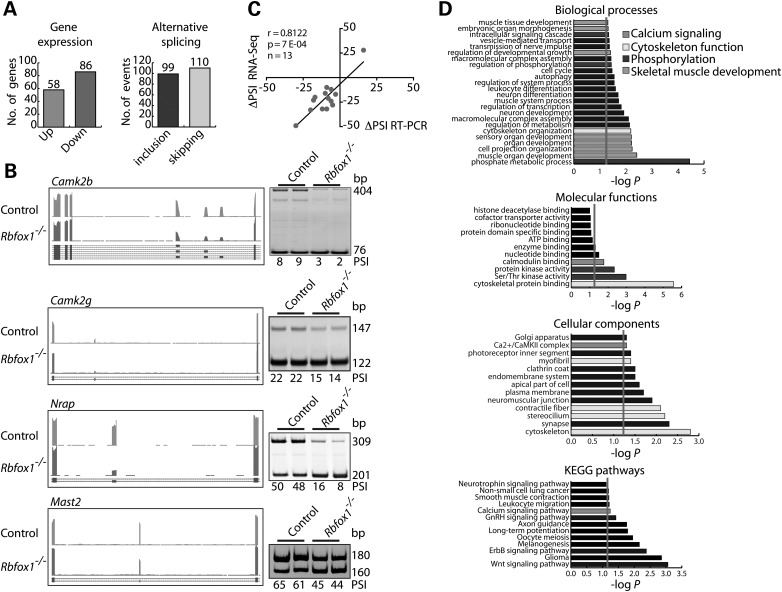
Analysis of the RNA-Seq data identified 144 genes that were differentially expressed (≥1.5-fold, P≤ 0.0010) between control and Rbfox1−/− muscle, 58 of which were up- and 96 downregulated (Fig. 4A, left panel, Supplementary Material, Table S2). Analysis of transcript variants between control and Rbfox1−/− identified 209 AS events with ≥10% change in percent spliced in (PSI) (3). Among the identified exons, 99 showed increased inclusion and 110 increased skipping relative to control mice (Fig. 4A, right panel; Supplementary Material, Table S3). The level of expression of these genes was not affected, suggesting no overlap between AS and gene expression changes in Rbfox1−/−.
Figure 4.
Rbfox1−/− muscles show splicing changes in genes implicated in multiple aspects of muscle function. (A) Differentially expressed genes (left panel) and altered alternative RNA splicing events (right panel) between control and Rbfox1−/− tibialis anterior muscles. (B) RNA-Seq data (UCSC browser, mm10) and RT-PCR validation assay (n= 2 biological replicates) for AS events (ΔPSI ≥ 10%) regulated by Rbfox1. (C) Pearson correlation between RNA-Seq and RT-PCR ΔPSI value (ΔPSI ≥ 20%). (D) GO and Kyoto Encyclopedia of Genes and Genome Pathways analyses on AS genes identified by RNA-Seq between control and Rbfox1−/− muscles.
Gene Ontology (GO) analysis of genes that was differentially expressed between control and Rbfox1−/− muscle did not reveal enrichment of categories with relevance for the observed phenotype. For this reason and given the known role of Rbfox1 in AS regulation, we focused on AS transitions as the primary contributors to the skeletal muscle phenotype. To determine the extent to which the RNA-seq data for AS could be validated, we compared the change in PSI (ΔPSI) (3) between Rbfox1−/− and control based on RNA-Seq and RT-PCR. We assayed 14 splicing events by RT-PCR, nine with ΔPSI ≥ 20% and five with ΔPSI ≤ 20% based on RNA-Seq. The results showed a high Pearson correlation (r= 0.86) between ΔPSI values from RT-PCR and RNA-Seq data (Fig. 4B and C; Supplementary Material, Fig. S4). Binding motif analysis (37) of the validated events revealed the presence of the Rbfox1 consensus binding site (U/A)GCAU/CG) (13–15) in the region upstream or downstream of the alternative exon, suggesting they are likely to be directly regulated by Rbfox1 (Table 1). We note, however, that the Rbfox consensus binding site was found within a minority of the genes that responded to Rbfox1 loss of function suggesting a large impact of secondary effects.
Table 1.
Location of the Rbfox1 binding site in the proximal upstream and downstream region of the alternative exon
| AS event | Alternative exon genomic coordinates | RNA-Seq ΔPS | Positiona | Genomic coordinateb | k-mer | Z-scorec | P-valuec | Occurrence |
|---|---|---|---|---|---|---|---|---|
| Ablim1 | chr19:57059085–57059204 | −32 | 246 | chr19:57059459 | gcaug | 1.991 | 2.32E−02 | Upstream |
| Camk2b | chr11:5976765-5976893 | −24 | 455 | chr11:5976939 | gcaug | 2.028 | 2.13E−02 | Upstream |
| Camk2g | chr14:20747819-20747851 | −21 | 639 | chr14:20747713 | gcaug | 2.648 | 4.05E−03 | Downstream |
| Camta1 | chr4:151071426-151071456 | −33 | 602 | chr4:151071355 | gcaug | 2.843 | 2.23E−03 | Downstream |
| Kcnma1 | chr14:23336040-23336120 | −20 | 849 | chr14:23335772 | gcauc | 1.991 | 2.32E−02 | Downstream |
| Ldb3 | chr14:34569713-34569727 | −12 | 531 | chr14:34569697 | gcauc | 2.463 | 6.89E−03 | Downstream |
| Mast2 | chr4:116333413-116333433 | −23 | 565 | chr4:116333369 | gcaug | 2.102 | 1.78E−02 | Downstream |
| Mbnl1 | chr3:60621423-60621458 | 28 | 513 | chr3:60621435 | gcaug | 2.500 | 6.21E−03 | Upstream |
| Mybpc1 | chr10:88582007-88582048 | −17.0 | 274 | chr10:88582275 | gcaug | 2.648 | 4.05E−03 | Upstream |
| Ndrg3 | chr2:156929932-156929970 | −32.0 | 606 | chr2:156929865 | gcaug | 2.565 | 5.16E−03 | Upstream |
| Nrap | chr19:56374364-56374468 | −49.0 | 721 | chr19:56374248 | gcaug | 3.028 | 1.23E−03 | Downstream |
| Rock2 | chr12:16973440-16973610 | −10.0 | 806 | chr12:16973745 | gcaug | 2.574 | 5.03E−03 | Upstream |
| Sorbs2 | chr8:45782802-45782960 | −28.0 | 1136 | chr8:45783437 | gcaug | 3.259 | 5.59E−04 | Upstream |
aPosition: the starting position of the binding site in the input sequence (relative to the sequence itself and not to the genomic position).
bGenomic coordinate: the genomic coordinate from which the binding site starts.
cZ-score and P-value: The Z-score (standard score) measures the deviation of the WR score from the mean. The mean WR score was calculated using specific background datasets. The P-value represents the probability of obtaining a specific Z-score considering a normal distribution (one tailed).
GO analysis of regulated AS events between control and Rbfox1−/− muscle showed significant enrichment (P< 5E–03) in categories related to cytoskeleton organization and myofibrillar structure (Table 2). Transcripts encoding proteins regulating actin filament dynamics such as the protein phosphatase Ssh1 and the actin binding proteins Ablim1 and Ablim2 were altered in Rbfox1−/− muscles; key regulators of the actin cytoskeleton organization and muscle contraction Rock2 and MyBP-C are mispliced in Rbfox1−/− muscle (Table 2). Furthermore, we also found splicing changes in genes encoding sarcomeric proteins, including the nebulin-related anchoring protein N-rap, known to link the terminal actin filaments of myofibrils to the sarcolemma; the myofibrillar protein obscurin, important for the organization of sarcomere contractil proteins; the LIM domain-binding protein-3 Ldb3, which has been proposed to support Z-line structure and muscle contraction (38). Interestingly, LDB3 exon 4 shows increased inclusion in Rbfox1−/− which has been reported in myotonic dystrophy skeletal muscle with a potential role in muscle pathogenesis (38). In addition to genes with cytoskeleton function, a number of genes involved in calcium signaling (Camta1, Camk2γ, Myo7a, Camk2δ and Camk2β) were also found aberrantly spliced in mice lacking Rbfox1. These data clearly indicate that Rbfox1 regulates AS of genes involved in multiple aspects of adult muscle function. These results strongly suggest that the phenotype of the muscle-specific Rbfox1 knockout results from combinatorial effects of Rbfox1-coordinated regulation of several genes.
Table 2.
Transcript involved in cytoskeleton organization and myofibril structure altered in Rbfox1−/− muscle
| Gene | Protein | Function |
|---|---|---|
| Ablim1 | Actin-binding LIM protein 1 | Cytoskeletal LIM protein that binds to actin filaments, mediates interactions between actin filaments and cytoplasmic targets |
| Obscn | Obscurin | Belongs to the family of giant sarcomeric signaling proteins, may have a role in the organization of myofibrils during assembly and mediates interactions between the sarcoplasmic reticulum and myofibrils |
| Ssh1 | Slingshot homolog 1 | Regulates actin filament dynamics |
| Rock2 | Rho-associated protein kinase | Key regulator of actin cytoskeleton and cell polarity, involved in regulation of smooth muscle contraction and actin cytoskeleton organization |
| Mybpc1 | Myosin-binding protein C | Binds to myosin playing a critical role in maintaining thick filaments structure and regulating contraction |
| Myo7A | Myosin VII A | Member of the myosin gene family. Myosins are mechanochemical proteins |
| Ldb3 | LIM domain binding 3 | Mutations in this gene have been associated with myofibrillar myopathy and dilated cardiomyopathy |
| Mast2 | Microtubule-associated Ser/Thr-protein kinase 2 | Appears to link the dystrophin/utrophin network with microtubule filaments |
| Nrap | Nebulin-related protein | May be involved in anchoring the terminal actin filaments in the myofibril to the membrane and in transmitting tension from the myofibrils to the extracellular matrix |
Loss of Rbfox1 increases sarcolemma fragility alters the cytoskeleton organization and causes formation of tubular aggregates
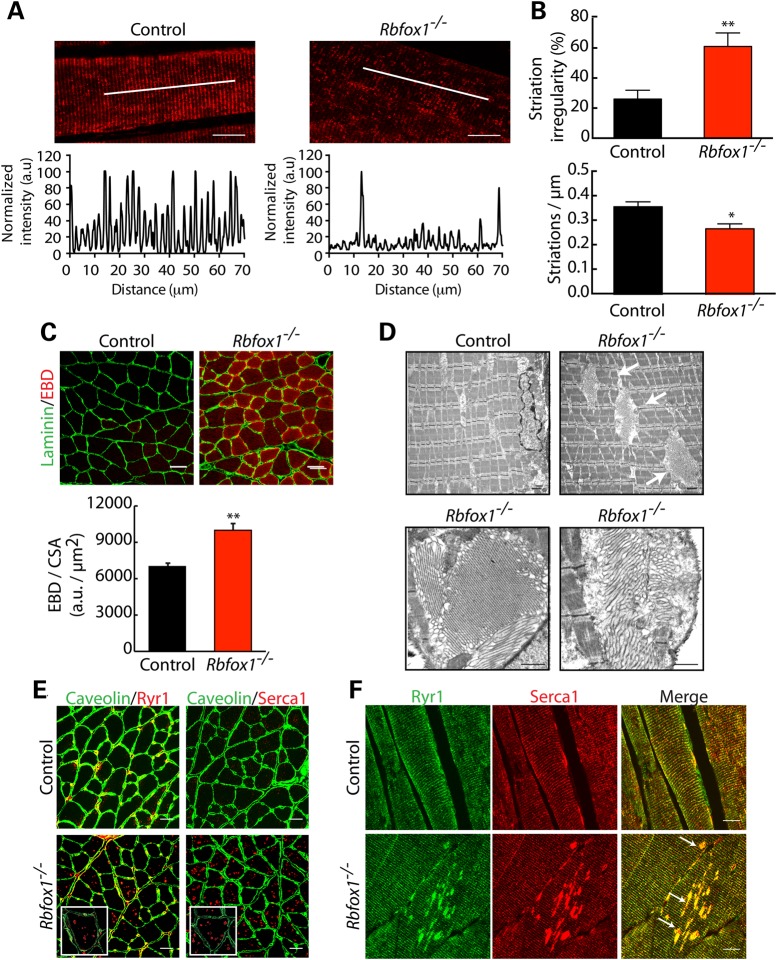
Within the most enriched categories found in the GO analysis of RNA-Seq, AS data were cytoskeleton organization (Fig. 4D and Table 2). To identify the role of Rbfox1 in the maintenance of cytoskeleton structure, we performed immunostaining for sarcomeric α-actin in transverse sections from control and Rbfox1−/− mice. The organization of the I-band was clearly altered, with loss of the normal striation pattern (Fig. 5A, top panels). To quantify the irregularity of the striation pattern, we quantified the fluorescence intensity along a line drawn within the fiber (Fig. 5A, bottom panels), which showed lost of regularity in Rbfox1−/− mice (Fig. 5B). We also tested whether sarcolemma integrity was compromised by systemic delivery of Evans blue dye (EBD). An EBD solution (1%) was delivered by intraperitoneal injection of 5-month-old control and Rbfox1−/− mice as described (39). Mice were sacrificed 24 h after EBD injection and dye uptake by muscle fibers was analyzed by microscopic evaluation. No differences were observed between control and Rbfox1−/− mice (data not shown), indicating that the absence of Rbfox1−/− does not affect sarcolemma integrity in conditions of normal activity. We then evaluated membrane damage under stress conditions. Ninety minutes after EBD injection, mice were run on a downhill treadmill for 30 min. Analysis of gastrocnemius muscle showed a higher fraction of EBD-positive fibers in Rbfox1−/− compare with control animals (Fig. 5C), indicating that loss of Rbfox1 affects sarcolemma integrity under stress conditions. This result, together with the altered cytoskeleton organization observed by actin immunostaining, confirms that loss of function of Rbfox1−/− in skeletal muscle causes muscle cytoskeletal aberrations.
Figure 5.
Rbfox1 loss of function alters skeletal muscle ultrastructure and leads to sarcolemma fragility after exercise. (A) Confocal imaging of alpha sarcomeric actin on longitudinal sections from control and Rbfox1−/− tibialis anterior muscles. Scale bar: 20 μm. Bottom panels: plot of fluorescence intensity. (B) Bottom panel: actin striation per micrometer (n= 11 fibers per genotype); Top panel: striation irregularity (%) (n= 11 fibers per genotype). ** indicates P≤ 0.01 as determined by two-tailed Student's t-test. (C) EBD was injected intraperitoneally; 90 min later mice were run downhill for 30 min and sacrificed 24 h later. EBD uptake was evaluated on gastrocnemius muscles from control and Rbfox1−/− mice by confocal microscopy. MATLAB script was used to calculate fluorescence intensity within fibers relative to fiber area. Rbfox1−/− muscles show a higher number of EBD positive fibers compare with controls. The results were expressed as the mean ± s.e.m. and the P-values were estimated using two-tailed Student's t-test (**P≤ 0.01). Laminin staining was used to visualize the myofiber membrane. (D) Electron microscopy of control (top left panel) and Rbfox1−/− (top right panel) gastrocnemius muscles showing tubular aggregates (arrows) in Rbfox1−/− muscle. Bottom panels show tubular aggregates at higher magnification in Rbfox1−/− muscle. Scale bar: 500 nm. (E and F) Immunostaining for Ryr1 and Serca1 in control and Rbfox1−/− gastrocnemius muscles reveal mislocalization of both calcium handling proteins in Rbfox1−/− muscle. (E) Immunofluorescence on control and Rbfox1−/− gastrocnemius muscle; higher magnification shown in the insets. Scale bar: 20 μm. (F) Immunostaining on longitudinal sections for Ryr1 and Serca1. Arrows in the merged image show colocalization of Ryr1 and Serca1 in the aggregates. Scale bar: 10 μm.
We perfomed EM on gastrocnemius muscle of control and knockout animals to identify effects on myofiber ultrastructure (Fig. 5D). Tubular aggregates were a prominent feature in Rbfox1−/− mice that were absent in control mice (Fig. 5D). Tubular aggregates are arrangements of expanded SR tubules, and have been described in patients with specific myopathies (40). The mechanism(s) leading to their formation is unknown; however, SR reshaping is thought to be triggered by the loss of a connection of the SR with myofibrils and T tubules (41–43). In mature muscle, the SR is stabilized by the contractile apparatus which participates in maintenance of longitudinal SR architecture. Tubular aggregates have been found to contain several SR components, including Serca1 and Ryr1 (40,44,45). To determine whether Serca1 and Ryr1 localization is altered in Rbfox1−/− mice, we performed immunofluorescence analysis on both cross- and transverse-sections of gastrocnemius muscle from control and Rbfox1−/− animals (Fig. 5E and F). In Rbfox1−/− mice, both Serca1 and Ryr1 are present in aggregates within the myofibers as well as within T-tubules (Fig. 5F). Interestingly, we observed appearance of these aggregates as early as after 1 month from Rbfox1 deletion (Supplementary Material, Fig. S5A), suggesting that lack of Rbfox1 triggers Serca1 and Ryr1 mislocalization. These results indicate that Rbfox1 is required for maintenance of skeletal muscle SR structure and correct localization of Serca1 and Ryr1.
Rbfox1 loss of function results in altered calcium release and reduced force generation
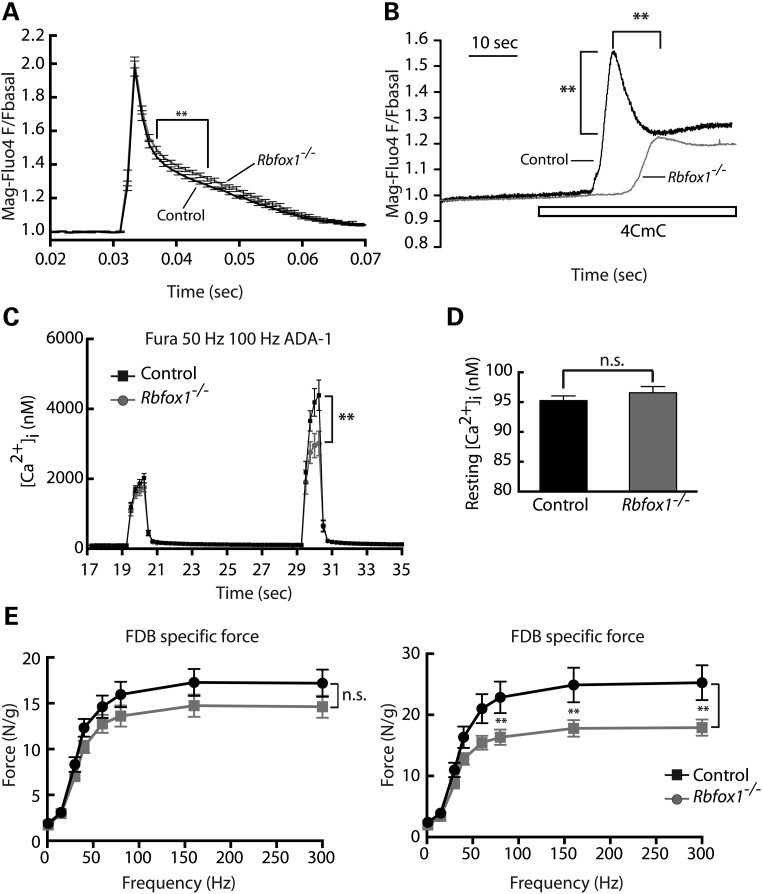
In skeletal muscle, calcium release and reuptake from and into the SR is orchestrated by Ryr1 and Serca1, respectively, during excitation–contraction (E–C) coupling. After electrical stimulation of the muscle membrane, the voltage-gated L-type calcium channels (named dihydropyridine receptors, DHPRs) are activated and in turn activate the Ryr1, which opens and releases a burst of calcium from the SR, leading to muscle contraction. Muscle relaxation results from pumping of calcium from the cytoplasm into the SR by Serca1. The appearance of tubular aggregates in Rbfox1−/− (Fig. 5D) with consequent re-localization of the SR proteins, Serca1 and Ryr1 (Fig. 5E and F), suggested that Rbfox1 loss of function could significantly affect SR calcium release and uptake, thereby impacting muscle contraction. To determine whether myoplasmic calcium handling was altered in Rbfox1−/− mice, flexor digitorum brevis (FDB)-derived myofibers were electrically stimulated by a single pulse, and both the amplitude of calcium release and the post-stimulation time required for calcium to return to baseline were measured. In myofibers from Rbfox1−/− mice compared with controls, the amplitude of the calcium transient with one twitch was not altered (Fig. 6A). However, the return of myoplasmic calcium levels to baseline was mildly but significantly slowed in the FDB fibers from Rbfox1−/− mice compared with controls (Fig. 6A). These results suggest that the activity of Serca1 is decreased in Rbfox1−/ muscle. To assess releasable SR calcium stores, we used the ryanodine receptor activator 4-Chloro-m-cresol (4CmC). We observed a significant delay in the 4CmC-induced release of calcium via Ryr1 opening in Rbfox1−/− compared with controls (Fig. 6B; Supplementary Material, Fig. S5B). SR calcium stores also appeared to be significantly depleted in the Rbfox1−/− fibers compared with controls (Fig. 6B; Supplementary Material, Fig. S5B), as would be expected for decreased Serca1 activity and disrupted SR structure. We also measured calcium concentration after electrical stimulation of FDB fibers with 50 and 100 Hz (Fig. 6C; Supplementary Material, Fig. S5C). First, we measured cytoplasmic calcium concentration before stimulus was applied. Although no differences were observed in the amount of calcium present in the cytoplasm of control and knockout mice (Fig. 6D), we found significant reduction in the amount of calcium released at 100 Hz in Rbfox1−/− (Fig. 6C; Supplementary Material, Fig. S5C).
Figure 6.
Loss of Rbfox1 alters calcium handling and decreases muscle force generation. (A) Normalized average twitch evoked calcium release in Mag-Fluo4 loaded fibers from FDB myofibers isolated from control and Rbfox1−/−mice (n= 15 fibers for each genotype). (B) Myofibers were stimulated with 1 mm 4-CMC to determine the relative amount of calcium stored in the SR (n= 13 fibers for each genotype). The graph shows representative traces of calcium response in Mag-fluo4-loaded FDB myofibers. (C) Evaluation of free calcium in Fura-2-loaded FDB myofibers electrically stimulated with trains at 50 or 100 Hz. ** indicates P≤ 0.01 as determined by two-tailed Student's t-test. (D) Average of the resting cytosolic calcium level. (E) Force–frequency relationship obtained from ex vivo FBD muscle (n= 6 mice for each genotype) isolated from 2-month-old (left panel) and 5-month-old (right panel) control and Rbfox1−/− mice (1 month and 4 months after induction of Rbfox1 knockout, respectively). Each graph combines data obtained from analysis performed on male and female mice.
The data presented above indicate that loss of Rbfox1 in skeletal muscle causes significant abnormalities in calcium handling. To determine whether these alterations in Rbfox1−/− mice affect muscle performance, we obtained force frequency measurements using ex vivo FDB muscle from mice 1 month and 4 months after Rbfox1 deletion, at frequencies from 15 to 300 Hz. Tetanic stimulation of Rbfox1−/− muscle 1 month following knockout revealed a decrease in specific force compared with control that did not reach statistical significance (Fig. 6E, left panel). However, 4 months after Rbfox1 deletion, we observed a strong and significant reduction in specific force (Fig. 6E, right panel). The trend was observed in both males and females (Supplementary Material, Fig. S5D). These results suggest that loss of muscle performance as a consequence of loss of Rbfox1 is progressive, consistent with the progressive appearance of mislocalized Serca and Ryr1. Moreover, in agreement with previous results (Fig. 6C), we observed force reduction using frequency from 70 to 300 Hz, when calcium release is most affected. Taken together, these results indicate that loss of Rbfox1 affects E–C coupling and calcium handling leading to impairment of force generation.
Discussion
Our results indicate that Rbfox1 drives an AS program in adult skeletal muscle that is required to maintain muscle structural and functional homeostasis. We provide evidence that Rbfox1 regulates AS of multiple transcripts, which are involved either in maintenance of muscle architecture or in regulating muscle contraction. Indeed, RNA-Seq analysis of Rbfox1−/− muscle revealed alterations of the splicing patterns of genes controlling myofibril structure and cytoskeleton organization. Depletion of Rbfox1 resulted in loss of regularity in the sarcomere architecture and consequent sarcolemma fragility under stress. Rbfox1−/− muscle showed formation of tubular aggregates and mislocalization of the calcium channel Ryr1 and the calcium pump Serca1 (Fig. 5D and F). We demonstrated that Rbfox1 loss of function results in misregulated calcium handling and reduced muscle force generation. When Rbfox1 was depleted >70% in satellite cells, we did not observe an effect on muscle regeneration. While one conclusion is that Rbfox1 is not required for muscle regeneration, it is also possible that residual Rbfox1-positive satellite cells are sufficient for normal regeneration. A third possibility is that expression of the paralog Rbfox2 compensates for loss of Rbfox1 activity. We conclude that >70% loss of Rbfox1 in satellite cells is not sufficient to cause aberrant muscle regeneration.
The results we obtained by inactivating the Rbfox1 gene specifically in skeletal muscle myofibers reveal a critical role for this splicing regulator in muscle function. Although Rbfox1−/− muscles do not present major myopathic features (Fig. 3A), such as centralized nuclei and/or necrotic fibers, histological analysis showed reduced fiber size (Fig. 3C and D). The lack of centralized nuclei together with unchanged expression of regenerative markers (Supplementary Material, Fig. S3B and C) indicate that reduced fiber size is not produced by regeneration. Interestingly, RNA-Seq analysis revealed that the Tmem8c gene (encoding Myomaker) is upregulated 24-fold in Rbfox1−/− muscle compared with control. Tmem8c encodes a transmembrane protein required for muscle formation (46). Tmem8c is highly expressed during embryogenesis and its expression drops dramatically at birth, after muscles have formed. The Tmem8c upregulation we observed in Rbfox1−/− muscles may represent a feature of fiber immaturity which can be linked to the smaller size of the fibers observed in Rbfox1−/− muscle.
Loss of Rbfox1 in skeletal muscle causes myofibril disorganization and alterations of the normal striated myofibrillar pattern in Rbfox1−/− muscle (Fig. 5A and B). Consistent with disrupted myofibrillar structure, RNA-Seq revealed altered splicing of genes encoding myofibrillar proteins (i.e. Nrap, Obscurin, Ablim1 and Ldb3) in Rbfox1−/− muscle. Moreover, a role for Rbfox1 in myofibril organization is conserved within vertebrates, as double knockdown of the zebrafish homologs of mammalian Rbfox1 and Rbfox2 genes, named rbfox1l and rbfox2, respectively, showed myofibril defects (24). However, in the mouse, deletion of Rbfox1 alone is sufficient to trigger the phenotype, suggesting a pivotal role played by this splicing factor for myofibril organization in mammals. More importantly, disorganization of the myofibrillar compartment resulted in sarcolemma fragility under stress, as shown by increased uptake of EBD by Rbfox1−/− muscles after exercise (Fig. 5C). These results demonstrate the importance of Rbfox1 in maintaining muscle architecture.
In support of a role for Rbfox1 in skeletal muscle structure, EM analysis of Rbfox1−/− muscles showed formation of tubular aggregates (Fig. 5D). Tubular aggregates are regular arrays of membrane tubules of SR origin (47,48), the formation of which is the major feature of a rare inherited disorder, named tubular aggregate myopathy. The molecular mechanism(s) leading to development of tubular aggregates are yet to be identified. SR reshaping during tubular aggregate formation is thought to occur through loss of connection between the SR and the contractile apparatus (47). We hypothesize that the altered myofibrillar organization observed in Rbfox1−/− muscles contributes to the loss of a stable SR, leading to SR reshaping and tubular aggregate formation.
Recently, mutations in the store operated calcium entry ORAI1 and in the endoplasmic reticulum/SR calcium sensor STIM1 have been identified in patients affected by tubular aggregate myopathies (TAMs) (49,50). Gain-of-function mutations in the STIM1 gene disrupt its calcium-sensor domain, which is believed to be the main mechanism underlying STIM1-driven TAMs (49). Missense mutations in the ORAI1 gene cause constitutive opening of the calcium release-activated calcium channel. As a consequence, cytosolic calcium influx increases, leading to altered calcium homeostasis (50). These reports imply calcium mishandling as one of the causes leading to TAs formation.
Interestingly, we observed altered calcium homeostasis in mice lacking Rbfox1, possibly as a consequence of relocalization of Serca1 and Ryr1 in Rbfox1−/− muscles. Immunostaining performed on cross-sections show mislocalization of both Serca1 and Ryr1 in myoplasmic aggregates (Fig. 5E) while staining of longitudinal sections revealed aggregates which resemble the tubular aggregates observed by EM (Fig. 5F). We hypothesize that the relocalization of Serca1 and Ryr1 observed in Rbfox1−/− has functional consequences on muscle performance. Indeed, we demonstrated that Rbfox1 loss of function leads to calcium mishandling, as shown by a delay in calcium release and reuptake (Fig. 6A, B; Supplementary Material, Fig. S5B), and causes decreased force generation in Rbfox1−/− muscles (Fig. 6E). Ryr1 activity is regulated through phosphorylation by several protein kinases, including the calcium-activated serine/threonine kinase, calmodulin kinase 2 (Camk2) (51). Interestingly, our RNA-Seq data revealed aberrant splicing in mRNAs from three Camk2 γενεσ, Camk2γ, Camk2δ and Camk2β. This work gives new insight into the critical role played by the tissue-specific splicing factor Rbfox1 as an AS regulator affecting multiple genes required to sustain the physiological requirements of the skeletal muscle tissue.
Materials and Methods
Mice
Rbfox1loxP/loxP (B6.129S2-Rbfox1tm1.1Dblk/J) and ACTA1-rtTAcre/+ (B6;C3-Tg(ACTA1-rtTA,tetO-cre)102Monk/J) were purchased from the Jackson Laboratory. Male Rbfox1loxP/loxP mice were bred to female ACTA1-rtTAcre/+ to generate Rbfox1loxP/loxP; ACTA1-rtTAcre/+. Cre-mediated recombination was achieved by feeding 21-day-old Rbfox1loxP/loxP; ACTA1-rtTAcre/+ (Rbfox1−/−) and Rbfox1loxP/loxP (control) with 2 g/kg doxycycline containing chow for 7 days. All experiments were performed on 5-month-old mice (4 month after achievement of Rbfox1 knockout), except where indicated.
129P1/ReJ-Lama2dy/J (stock #641) mice were obtained from Jackson Laboratory (Bar Harbor, ME, USA). Mdx mice on the C57BL/6 background were obtained from Dr Marta Fiorotto (Baylor College of Medicine, Houston, TX, USA).
Rbfox1loxP/loxP and Pax7Cre/Cre were previously described (17,32). To obtain Rbfox1loxP/loxP; Pax7Cre/Cre(Rbfox1−/−) mice, Rbfox1loxP/loxP male mice were crossed to Pax7Cre/Cre female mice. Seven-week-old Rbfox1loxP/loxP; Pax7Cre/+ and Rbfox1loxP/loxP (control) were intraperitoneal (i.p.) injected with 5 mg of tamoxifen per day for 5 days to induce Cre-mediated recombination.
Histological analyses
Skeletal muscles were isolated and flash frozen in liquid nitrogen-cooled isopentane using tragacanth gum (Sigma-Aldrich) as support, or fixed in 10% buffered formalin phosphate and processed for routine paraffin histology. Hematoxylin and eosin and succinate dehydrogenase staining were performed using standard procedures.
Treadmill exercise and EBD uptake experiments
Control and Rbfox1−/− 5-month-old (n= 4 control and n= 6 Rbfox1−/−) mice were injected via i.p. with 1% EBD solution (1 mg/0.1 ml per 10 g of body weight) (Sigma-Aldrich). For the resting experiments, mice were sacrificed 24 h after EBD injection. The gastrocnemius muscle was snap frozen in liquid nitrogen-cooled isopentane. Dye uptake was evaluated on cross sectioned (12 μm) muscles using confocal Nikon A1-Rs inverted laser scanning microscope with a ×20 objective. To evaluate EBD uptake under stress conditions, 90 min after dye injection mice were treadmill exercised (30 min, 16° downhill angle, 10 m/min) (Columbus Instruments Animal Treadmills, Exer-6M open treadmill). Twenty-four hours after the running session, EBD uptake was evaluated as described above.
Isolation of FDB muscle myofibers
Fiber isolation has been performed as previously described (52). Briefly, the FDB muscle was removed and placed in Dulbecco's modified Eagle's medium (DMEM) containing 3 mg/ml collagenase and 10% (v/v) fetal bovine serum. After a 2 h incubation at 37°C, muscles were transferred to 1 ml of DMEM and triturated 10 times through a 1 ml pipette tip to separate individual fibers. Next, 150 μl of DMEM containing separated FDB fibers was placed onto a 25 mm glass coverslip coated with laminin in PBS. Fibers were incubated overnight at 37°C in DMEM containing antibiotic-antimycotic (Gibco, Carlsbad, CA, USA).
Evaluation of calcium release with single twitch
The kinetic behavior of a single twitch generated with electrical stimulation was performed in single isolated FDB fibers [as in (52)] loaded with 5 µm of Mag-Fluo4 for 30 min at room temperature (RT) in the presence of 20 µm of BTS (4-methyl-N-(phenylmethyl)benzenesulfonamide). Preloaded FDBs were placed on the stage of a confocal microscope with an adapted perfusion system (Tyrode with 20 μm BTS at 0.5 ml/min) and using the ×20 objective (EC Plan-Neofluar) mounted in the confocal microscope (Zeiss LSM 510 meta) preconfigured for line scan mode, lines were acquired every 1.15 ms (3.66 µs/pixel time) with a 488 nm excitation laser and the LP 505 emission filter. FDBs were stimulated with five square electrical pulses (200 µs duration) at 1 Hz using two platinum wires placed at each side of the fiber, the generated transients were normalized to basal fluorescence (Fx/F0) and then averaged to generate a single transient for each fiber (counted as n =1).
Force-frequency measurement
For force-frequency experiments, 2- and 5-month-old control and Rbfox1−/− mice were used. Intact FDB muscles were removed and immediately immersed in incubation medium comprised of Kreb's ringer solution oxygenated with a 95/5% mixture of O2/CO2. Muscles were tied with sutures at the musculotendinous junction and suspended between a force transducer and stationary anchor within a test chamber filled with warm (35°C) oxygenated incubation medium. After a 20 min rest to allow mounted muscles to equilibrate, muscle optimal length (l0) was determined via single twitch force generation measurements. Next, force frequency measurements were obtained at l0 using frequencies from 15 to 300 Hz under 200 ms trains of 0.2 ms pulses. Muscle stimulation occurred within the test chamber using platinum electrodes attached to a Grass S48 stimulator and recorded within Chart5 (version 5.2) software. Data were normalized to the weight of each muscle used and are reported as force (N)/cm3 ± s.e.m.
Frequency-calcium response measurement
FDB fibers were preloaded with 10 µm Fura-2AM for 1 h, in the presence of 20 µm BTS. Loaded Fura-2 fibers were excited at four cycles per second (with 340/380 nm excitation filters) using a lamda-DG4 system as light source. The emitted fluorescence was collected through a Nikon S Fluor objective (×20, 0.75 NA), coupled to an inverted microscope Nikon Eclipse TE-200, and digitized with a Rolera MGi-Plus CCD camera using 510 × 252 pixels with six binning. Data were collected using Metafluor software (Ver.6.2r6). Fibers were electrically stimulated at 50 or 100 Hz for 1 s using the same stimulation system described above for twitch stimulation, each stimulus was separated for a 10 s resting period. Response was estimated as area under curve during the stimulation period.
Calcium store evaluation
Mag-Fluo4-loaded FDBs fibers were excited at 20 Hz with a 500 nm filter, using the same system described for Fura-2 experiments. Normal Tyrode (121 mm NaCl, 5 mm KCl, 1.8 mm CaCl2, 500 μm MgCl2, 400 μm NaH2PO4, 100 μm EDTA, 5.5 mm glucose and 24 mm NaHCO3) was replaced with calcium free Tyrode containing 1 mm of 4-CMC, and perfused at rate of 3 ml/min. To estimate response amplitude, the peak and time of the peak were collected and fluorescent data normalized to basal fluorescence 10 s before the peak.
RNA extraction
RNA was isolated from primary cultures using TRIzol reagent (Invitrogen). RNeasy fibrous tissue mini-kit (Qiagen) was used for RNA extraction from skeletal muscle tissue. RNA was resuspended in RNase-free water (Ambion) and DNase treated.
RT-PCR and real-time PCR analyses
RT-PCR to validate AS events identified by RNA-Seq was performed using GoTaq DNA Polymerase (Promega). Primers were designed annealing to the flanking constitutive exon regions (Supplementary Material, Table S2). PCR products were separated on a 5% acrylamide gel and the PSI (3) was calculated after ethidium bromide staining using Kodak El Logic 2200 imaging system. Real-time PCR was performed using TaqMan probes (Applied Biosystems) for Rbfox1, Trim63, MuRF1 and Rpl30 transcripts, using 7500 Fast real-time PCR (Applied Biosystems).
Western blot analysis
Total protein samples were separated on 10% Tris-glycine SDS–PAGE gels and transferred to PVDF membranes (Immobilon) for western blot analysis. The membranes were incubated with monoclonal anti-Rbfox1 (1 : 250; generated in-house) over night at 4°C. Membranes were then washed three times in PBST (0.1% Tween 20) and incubated for 1 h at RT with HRP-conjugated goat anti-mouse (1 : 5000; Jackson). After washing in PBST (0.1% Tween 20), immunoreactivity was detected by using HRP-chemiluminescence system (Thermo Scientific).
Immunofluorescence
Frozen sections were fixed in 4% PFA for 10 min at RT and washed in distilled water. Autofluorescence was quenched by incubating slides with 1 mg/ml NaBH4 in PBS for 15 min. Sections were then incubated with 0.1 m glycine in PBST (0.05% Tween 20) for 1 h and rinsed with water then PBS. Sections were permeabilized with 0.2% Triton in PBST for 5 min. Mouse IgG blocking was performed by incubating sections with M.O.M. IgG-blocking reagent (Vector Lab) over night at 4°C. Sections were then incubated with 10% donkey serum, 5% BSA for 1 h. Primary antibodies were diluted in M.O.M. diluent reagent and applied to sections over night at 4°C, washed three times with PBST and incubated with secondary antibodies diluted in M.O.M. diluent reagent for 1 h at RT. After three washes in PBST, slides were mounted using Ultramount Aqueous Permanent mounting medium (Dako). Confocal microscopy was performed using a Nikon A1-Rs inverted laser scanning microscope. Primary antibodies used are the following: monoclonal anti-Rbfox1 (1 : 50, in house made), monoclonal anti-MHC neonatal (1 : 500, Developmental Studies Hybridoma Bank, DSHB), monoclonal anti-MHC slow (1 : 500, Sigma-Aldrich), monoclonal anti-MHC fast (1 : 500, Sigma-Aldrich), monoclonal anti-Desmin (1 : 250, Abcam), rabbit anti-Laminin (1 : 200, Sigma-Aldrich), monoclonal anti-Dystrophin (1 : 200, Abcam), monoclonal anti-Ryr1 (1 : 200, Pierce), rabbit anti-Ryr1 (1 : 200, Millipore), monoclonal anti-Serca1 (1 : 250, Sigma-Aldrich) and monoclonal anti-alpha sarcomeric actin (Sigma-Aldrich). Secondary antibodies were: Alexa-Fluo-488 and Alexa-Fluo-555 (1 : 500, Invitrogen).
Electron microscopy
Freshly dissected gastrocnemius muscles were fixed in EM grade glutaraldehyde. Tissue samples were cut into 1 mm3 cubes, rinsed and exposed to 1% osmium tetroxide, dehydrated and embedded in an araldite-epon mixture. Semi-thin tissue sections were prepared (0.6 mm) and stained with uranyl acetate and lead citrate. The prepared samples were examined with a JEOL 1210 transmission electron microscope (JEOL Corporation).
RNA-Seq
RNA from tibialis anterior muscle from two controls and two Rbfox1−/− mice was isolated using TRIzol (Invitrogen). Hi-Seq library preparation and Illumina sequencing of 100 bp paired-end reads were performed by the Genomic and RNA Profiling Core (GARP) at Baylor College of Medicine (BCM) on a Hi-Seq2000. RNA was poly-A selected, barcoded and run in three lanes. To increase the map-ability, we trimmed the low-quality nucleotides (nts) from both ends of the reads (10 nts from the 5′ end and 1 nt from the 3′). The resulting 90 nt pair-ended reads were mapped to the mouse genome (UCSC mm10) using Tophat (53) with NCBI RefSeq genes as the reference and up to two possible mismatches. To reduce possible PCR biases, we removed the read duplicates using Picard (http://picard.sourceforge.net). HTseq (http://www-huber.embl.de/users/anders/HTSeq) was used to determine the number of reads falling in the known genes. DESeq (54) was used to analyze the gene-based transcript counts to detect differentially expressed genes between the control and treatment groups. The false discovery rate of the differentially expressed genes was estimated using Benjamini and Hochberg method. MISO (55) was used to analyze AS events, which include skipped exons, alternative 3′/5′ splice sites (A3SS/A5SS), mutually exclusive exons and retained introns. Alternative splicing was quantified by PSI (percentage spliced in) value, which denotes the fraction of splicing events that are included in the mature mRNAs. The splicing events with |PSI| between two conditions and Bayes factor 3 were considered significant.
Binding motif analysis
Rbfox1 consensus sequence was searched within 500 nt upstream and downstream the alternative exon by using the RBPmap computational tool (http://rbpmap.technion.ac.il/index.html). Rbfox1 binding motif sequences were given: GCAUG, GCACG. High stringency was applied as filter.
Quantification of fiber area and distribution
A script was generated using MATLAB. A median filter was applied and pixels with an intensity >20 were considered. Fiber boundaries were defined applying a threshold (triangle) and a binary closing using five iterations and alternating connectivity. Fibers were defined as the regions that were not under the mask. Only whole fibers were considered for analysis (partial fibers because at the edge of the image were discarded). Fibers were labeled and automatically counted. The number of fibers obtained was normalized against the CSA of the entire muscle analyzed. Muscle CSA was calculated using ImageJ (National Institutes of Health) as the average of all the muscles analyzed for both control (seven) and Rbfox1−/− mice (four). CSA of each fiber was measured automatically using MATLAB script. Fibers were grouped based on their area (1–400, 401–800, 801–1200, 1201–1600, 1601–2000, 2001–2400, 2401–2800, 2801–3200, 3201–3600 and 3601–4000 μm2). Five images were used from four Rbfox1−/− mice and seven images from two control animals. The results were expressed as the mean ± s.e.m. and the P-values were estimated using two-tailed Student's t-test (*P≤ 0.05).
Measurement of EBD fluorescence per fiber area
A script was generated using MATLAB. Background for EBD channel was calculated and subtracted from each image measuring the fluorescence intensity in an empty region. The laminin signal was used delimit each fiber and the CSA for each fiber was calculated as described in the previous section. EBD intensity was measured as ratio between fluorescence intensity and CSA of each fiber. The results were expressed as the mean ± s.e.m. and the P-values were estimated using two-tailed Student's t-test (*P≤ 0.05; n = 90 fibers from two control animals; n = 129 fibers from three Rbfox1−/− mice).
Analysis of actin striation and organization
Analysis of actin striation regularity was performed as we previously reported (7). Briefly, background of an empty region was measured using ImageJ software and was subtracted from each image. Three lines (100 μm each) were drawn over each fiber and plots were generated along each line. Each plot was normalized to its maximum. The cutoff was determined as 20 units above the baseline. Striations per length unit (μm) were obtained by counting the number of picks (striations) above the cutoff. To analyze striation regularity, the number of picks (striations) was counted above the cutoff between 0–20, 10–30, 20–40 and 30–50 μm. A regular region was defined as containing 7–10 striations/μm on average. The results were expressed as the mean ± s.e.m. and the P-values were estimated using two-tailed Student's t-test (* P≤ 0.05; n = 11–10 fibers per genotype).
Isolation and in vitro differentiation of primary myoblasts
To obtain primary myoblasts from 3-month-old Rbfox1loxP/loxP; Pax7Cre/Cre(Rbfox1−/−) and Rbfox1loxP/loxP (control), four mice for each genotype were used. Primary myoblast isolation was performed as previously described (56). Briefly, gastrocnemius and tibialis anterior muscles were isolated from both hindlimbs of each mouse. Muscles were pooled, minced and digested with 0.1% pronase (Roche) for 1 h at 37°C, followed by trituration. The cell suspension was filtered through a 40 μm strainer (Cornig). Cells were pelleted by centrifugation and pre-plated for 2 h in DMEM, 10% FBS with 2 mm l-glutamine and 0.01% penicillin and streptomycin (growth medium), allowing exclusion of fibroblast contamination based on primary myoblast and fibroblast adherence differences (57). After 2 h, primary myoblast cells-containing supernatant was collected and re-plated into new culture dishes overnight (10 h). The next day primarily myoblasts were plated onto matrigel-coated dishes for expansion. For differentiation, 90% confluent cultures were placed in differentiation medium (growth medium with 2% horse serum replacing FBS). Cells were harvested 96 h after differentiation medium was added and processed for RNA analysis.
Cardiotoxin injury
Cardiotoxin (CTX) from Naja mossambica mossambica (Sigma-Aldrich) was dissolved in sterile saline solution to a final concentration of 10 μm. Mice were anesthetized using 2.5% Avertin and legs shaved and cleaned with alcohol. Tibialis anterior muscles were injured by injecting 50 μl of CTX using a 26-gauge needle. Mice were kept over a warming pad until recovery.
Supplementary Material
Funding
This project is funded by the NIH R01HL045565, R01AR060733 and R01AR045653 (T.A.C.) and the Muscular Dystrophy Association (T.A.C.); R01AR053349, R01AR041802 and NIH T32 HL007676 to S.H. This project was supported by three cores at BCM: Genomic and RNA Profiling Core (Lisa D. White), Integrated Microscopy Core (HD007495, DK56338 and CA125123) (Michael Mancini), and Baculovirus/Monoclonal Antibody Facility (P30 CA125123).
Supplementary Material
Acknowledgements
We thank Dr Charles Keller for providing the Pax7Cre/Cre mice and members of the Cooper lab for reading the manuscript and giving suggestions. The authors acknowledge the Texas Advanced Computing Center (TACC) at The University of Texas at Austin and the Rice University for providing HPC resources for the RNA-seq data analysis. The Rice University HPC resources were supported in part by NIH award NCRR S10RR02950 and an IBM Shared University Research (SUR) Award in partnership with CISCO, Qlogic and Adaptive Computing.
Conflict of Interest statement. None declared.
References
- 1.Pan Q., Shai O., Lee L.J., Frey B.J., Blencowe B.J. (2008) Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet., 40, 1413–1415. [DOI] [PubMed] [Google Scholar]
- 2.Nilsen T.W., Graveley B.R. (2010) Expansion of the eukaryotic proteome by alternative splicing. Nature, 463, 457–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang E.T., Sandberg R., Luo S., Khrebtukova I., Zhang L., Mayr C., Kingsmore S.F., Schroth G.P., Burge C.B. (2008) Alternative isoform regulation in human tissue transcriptomes. Nature, 456, 470–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kalsotra A., Xiao X., Ward A.J., Castle J.C., Johnson J.M., Burge C.B., Cooper T.A. (2008) A postnatal switch of CELF and MBNL proteins reprograms alternative splicing in the developing heart. Proc. Natl Acad. Sci. USA, 105, 20333–20338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chawla G., Lin C.H., Han A., Shiue L., Ares M., Jr., Black D.L. (2009) Sam68 regulates a set of alternatively spliced exons during neurogenesis. Mol. Cell. Biol., 29, 201–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Licatalosi D.D., Mele A., Fak J.J., Ule J., Kayikci M., Chi S.W., Clark T.A., Schweitzer A.C., Blume J.E., Wang X., et al. (2008) HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature, 456, 464–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Giudice J., Xia Z., Wang E.T., Scavuzzo M.A., Ward A.J., Kalsotra A., Wang W., Wehrens X.H., Burge C.B., Li W., et al. (2014) Alternative splicing regulates vesicular trafficking genes in cardiomyocytes during postnatal heart development. Nat. Commun., 5, 3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ule J., Darnell R.B. (2007) Functional and mechanistic insights from genome-wide studies of splicing regulation in the brain. Adv. Exp. Med. Biol., 623, 148–160. [DOI] [PubMed] [Google Scholar]
- 9.Schmid R., Grellscheid S.N., Ehrmann I., Dalgliesh C., Danilenko M., Paronetto M.P., Pedrotti S., Grellscheid D., Dixon R.J., Sette C., et al. (2013) The splicing landscape is globally reprogrammed during male meiosis. Nucleic Acids Res., 41, 10170–10184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pedrotti S., Busa R., Compagnucci C., Sette C. (2012) The RNA recognition motif protein RBM11 is a novel tissue-specific splicing regulator. Nucleic Acids Res., 40, 1021–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de la Grange P., Gratadou L., Delord M., Dutertre M., Auboeuf D. (2010) Splicing factor and exon profiling across human tissues. Nucleic Acids Res., 38, 2825–2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grosso A.R., Gomes A.Q., Barbosa-Morais N.L., Caldeira S., Thorne N.P., Grech G., von Lindern M., Carmo-Fonseca M. (2008) Tissue-specific splicing factor gene expression signatures. Nucleic Acids Res., 36, 4823–4832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lambert N., Robertson A., Jangi M., McGeary S., Sharp P.A., Burge C.B. (2014) RNA Bind-n-Seq: quantitative assessment of the sequence and structural binding specificity of RNA binding proteins. Mol. Cell., 54, 887–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lovci M.T., Ghanem D., Marr H., Arnold J., Gee S., Parra M., Liang T.Y., Stark T.J., Gehman L.T., Hoon S., et al. (2013) Rbfox proteins regulate alternative mRNA splicing through evolutionarily conserved RNA bridges. Nat. Struct. Mol. Biol., 20, 1434–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weyn-Vanhentenryck S.M., Mele A., Yan Q., Sun S., Farny N., Zhang Z., Xue C., Herre M., Silver P.A., Zhang M.Q., et al. (2014) HITS-CLIP and integrative modeling define the Rbfox splicing-regulatory network linked to brain development and autism. Cell. Rep., 6, 1139–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang C., Zhang Z., Castle J., Sun S., Johnson J., Krainer A.R., Zhang M.Q. (2008) Defining the regulatory network of the tissue-specific splicing factors Fox-1 and Fox-2. Genes Dev., 22, 2550–2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gehman L.T., Stoilov P., Maguire J., Damianov A., Lin C.H., Shiue L., Ares M., Jr, Mody I., Black D.L. (2011) The splicing regulator Rbfox1 (A2bp1) controls neuronal excitation in the mammalian brain. Nat. Genet., 43, 706–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bhalla K., Phillips H.A., Crawford J., McKenzie O.L., Mulley J.C., Eyre H., Gardner A.E., Kremmidiotis G., Callen D.F. (2004) The de novo chromosome 16 translocations of two patients with abnormal phenotypes (mental retardation and epilepsy) disrupt the A2bp1 gene. J. Hum. Genet., 49, 308–311. [DOI] [PubMed] [Google Scholar]
- 19.Xu B., Roos J.L., Levy S., van Rensburg E.J., Gogos J.A., Karayiorgou M. (2008) Strong association of de novo copy number mutations with sporadic schizophrenia. Nat. Genet., 40, 880–885. [DOI] [PubMed] [Google Scholar]
- 20.Martin C.L., Duvall J.A., Ilkin Y., Simon J.S., Arreaza M.G., Wilkes K., Alvarez-Retuerto A., Whichello A., Powell C.M., Rao K., et al. (2007) Cytogenetic and molecular characterization of A2BP1/FOX1 as a candidate gene for autism. Am. J. Med. Genet. B Neuropsychiatr. Genet., 144B, 869–876. [DOI] [PubMed] [Google Scholar]
- 21.Sebat J., Lakshmi B., Malhotra D., Troge J., Lese-Martin C., Walsh T., Yamrom B., Yoon S., Krasnitz A., Kendall J., et al. (2007) Strong association of de novo copy number mutations with autism. Science, 316, 445–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Davis L.K., Maltman N., Mosconi M.W., Macmillan C., Schmitt L., Moore K., Francis S.M., Jacob S., Sweeney J.A., Cook E.H. (2012) Rare inherited A2BP1 deletion in a proband with autism and developmental hemiparesis. Am. J. Med. Genet. A, 158A, 1654–1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bland C.S., Wang E.T., Vu A., David M.P., Castle J.C., Johnson J.M., Burge C.B., Cooper T.A. (2010) Global regulation of alternative splicing during myogenic differentiation. Nucleic Acids Res., 38, 7651–7664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gallagher T.L., Arribere J.A., Geurts P.A., Exner C.R., McDonald K.L., Dill K.K., Marr H.L., Adkar S.S., Garnett A.T., Amacher S.L., et al. (2011) Rbfox-regulated alternative splicing is critical for zebrafish cardiac and skeletal muscle functions. Dev. Biol., 359, 251–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuroyanagi H., Ohno G., Mitani S., Hagiwara M. (2007) The Fox-1 family and SUP-12 coordinately regulate tissue-specific alternative splicing in vivo. Mol. Cell. Biol., 27, 8612–8621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Das D., Clark T.A., Schweitzer A., Yamamoto M., Marr H., Arribere J., Minovitsky S., Poliakov A., Dubchak I., Blume J.E., et al. (2007) A correlation with exon expression approach to identify cis-regulatory elements for tissue-specific alternative splicing. Nucleic Acids Res., 35, 4845–4857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cooper T.A., Wan L., Dreyfuss G. (2009) RNA and disease. Cell, 136, 777–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kalsotra A., Cooper T.A. (2011) Functional consequences of developmentally regulated alternative splicing. Nat. Rev. Genet., 12, 715–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klinck R., Fourrier A., Thibault P., Toutant J., Durand M., Lapointe E., Caillet-Boudin M.L., Sergeant N., Gourdon G., Meola G., et al. (2014) RBFOX1 cooperates with MBNL1 to control splicing in muscle, including events altered in myotonic dystrophy type 1. PLoS ONE, 9, e107324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pistoni M., Shiue L., Cline M.S., Bortolanza S., Neguembor M.V., Xynos A., Ares M., Jr, Gabellini D. (2013) Rbfox1 downregulation and altered calpain 3 splicing by FRG1 in a mouse model of facioscapulohumeral muscular dystrophy (Fshd). PLoS Genet., 9, e1003186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Le Grand F., Rudnicki M.A. (2007) Skeletal muscle satellite cells and adult myogenesis. Curr. Opin. Cell. Biol., 19, 628–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nishijo K., Hosoyama T., Bjornson C.R., Schaffer B.S., Prajapati S.I., Bahadur A.N., Hansen M.S., Blandford M.C., McCleish A.T., Rubin B.P., et al. (2009) Biomarker system for studying muscle, stem cells, and cancer in vivo. FASEB J., 23, 2681–2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Couteaux R., Mira J.C., d'Albis A. (1988) Regeneration of muscles after cardiotoxin injury. I. cytological aspects. Biol. Cell., 62, 171–182. [PubMed] [Google Scholar]
- 34.Rao P., Monks D.A. (2009) A tetracycline-inducible and skeletal muscle-specific Cre recombinase transgenic mouse. Dev. Neurobiol., 69, 401–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Helliwell T.R. (1988) Lectin binding and desmin staining during bupivicaine-induced necrosis and regeneration in rat skeletal muscle. J. Pathol., 155, 317–326. [DOI] [PubMed] [Google Scholar]
- 36.Goebel H.H. (1995) Desmin-related neuromuscular disorders. Muscle Nerve, 18, 1306–1320. [DOI] [PubMed] [Google Scholar]
- 37.Paz I., Kosti I., Ares M., Jr., Cline M., Mandel-Gutfreund Y. (2014) RBPmap: a web server for mapping binding sites of RNA-binding proteins. Nucleic Acids Res, 42, W361–W367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yamashita Y., Matsuura T., Shinmi J., Amakusa Y., Masuda A., Ito M., Kinoshita M., Furuya H., Abe K., Ibi T., et al. (2012) Four parameters increase the sensitivity and specificity of the exon array analysis and disclose 25 novel aberrantly spliced exons in myotonic dystrophy. J. Hum. Genet., 57, 368–374. [DOI] [PubMed] [Google Scholar]
- 39.Hamer P.W., McGeachie J.M., Davies M.J., Grounds M.D. (2002) Evans Blue Dye as an in vivo marker of myofibre damage: optimising parameters for detecting initial myofibre membrane permeability. J. Anat., 200, 69–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boncompagni S., Protasi F., Franzini-Armstrong C. (2012) Sequential stages in the age-dependent gradual formation and accumulation of tubular aggregates in fast twitch muscle fibers: SERCA and calsequestrin involvement. Age (Dordr), 34, 27–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bagnato P., Barone V., Giacomello E., Rossi D., Sorrentino V. (2003) Binding of an ankyrin-1 isoform to obscurin suggests a molecular link between the sarcoplasmic reticulum and myofibrils in striated muscles. J. Cell. Biol., 160, 245–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gokhin D.S., Fowler V.M. (2011) Cytoplasmic gamma-actin and tropomodulin isoforms link to the sarcoplasmic reticulum in skeletal muscle fibers. J. Cell. Biol., 194, 105–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kontrogianni-Konstantopoulos A., Jones E.M., Van Rossum D.B., Bloch R.J. (2003) Obscurin is a ligand for small ankyrin 1 in skeletal muscle. Mol. Biol. Cell, 14, 1138–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chevessier F., Bauche-Godard S., Leroy J.P., Koenig J., Paturneau-Jouas M., Eymard B., Hantai D., Verdiere-Sahuque M. (2005) The origin of tubular aggregates in human myopathies. J. Pathol., 207, 313–323. [DOI] [PubMed] [Google Scholar]
- 45.Chevessier F., Marty I., Paturneau-Jouas M., Hantai D., Verdiere-Sahuque M. (2004) Tubular aggregates are from whole sarcoplasmic reticulum origin: alterations in calcium binding protein expression in mouse skeletal muscle during aging. Neuromuscul. Disord., 14, 208–216. [DOI] [PubMed] [Google Scholar]
- 46.Millay D.P., O'Rourke J.R., Sutherland L.B., Bezprozvannaya S., Shelton J.M., Bassel-Duby R., Olson E.N. (2013) Myomaker is a membrane activator of myoblast fusion and muscle formation. Nature, 499, 301–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schiaffino S. (2012) Tubular aggregates in skeletal muscle: just a special type of protein aggregates? Neuromuscul. Disord., 22, 199–207. [DOI] [PubMed] [Google Scholar]
- 48.Engel W.K., Bishop D.W., Cunningham G.G. (1970) Tubular aggregates in type Ii muscle fibers: ultrastructural and histochemical correlation. J. Ultrastruct. Res., 31, 507–525. [DOI] [PubMed] [Google Scholar]
- 49.Bohm J., Chevessier F., Maues De Paula A., Koch C., Attarian S., Feger C., Hantai D., Laforet P., Ghorab K., Vallat J.M., et al. (2013) Constitutive activation of the calcium sensor STIM1 causes tubular-aggregate myopathy. Am. J. Hum. Genet., 92, 271–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Endo Y., Noguchi S., Hara Y., Hayashi Y.K., Motomura K., Miyatake S., Murakami N., Tanaka S., Yamashita S., Kizu R., et al. (2014) Dominant mutations in ORAI1 cause tubular aggregate myopathy with hypocalcemia via constitutive activation of store-operated Ca2+ channels. Hum. Mol. Genet., 23, 637–648. [DOI] [PubMed] [Google Scholar]
- 51.Tavi P., Allen D.G., Niemela P., Vuolteenaho O., Weckstrom M., Westerblad H. (2003) Calmodulin kinase modulates Ca2+ release in mouse skeletal muscle. J. Physiol., 551, 5–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Knoblauch M., Dagnino-Acosta A., Hamilton S.L. (2013) Mice with RyR1 mutation (Y524 s) undergo hypermetabolic response to simvastatin. Skelet. Muscle, 3, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Trapnell C., Pachter L., Salzberg S.L. (2009) Tophat: discovering splice junctions with RNA-Seq. Bioinformatics, 25, 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Anders S., Huber W. (2010) Differential expression analysis for sequence count data. Genome Biol., 11, R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Katz Y., Wang E.T., Airoldi E.M., Burge C.B. (2010) Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat. Methods, 7, 1009–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Danoviz M.E., Yablonka-Reuveni Z. (2012) Skeletal muscle satellite cells: background and methods for isolation and analysis in a primary culture system. Methods Mol. Biol., 798, 21–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sabourin L.A., Girgis-Gabardo A., Seale P., Asakura A., Rudnicki M.A. (1999) Reduced differentiation potential of primary Myod-/- myogenic cells derived from adult skeletal muscle. J. Cell. Biol., 144, 631–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.