Abstract
Glucosylceramides (GlcCers) are important lipid components of the membrane systems of eukaryotes. Recent studies have suggested the roles for GlcCers in regulating fungal growth and pathogenesis. In this study, we report the identification and functional characterization of PdGcs1, a gene encoding GlcCer synthase (GCS) essential for the biosynthesis of GlcCers, in Penicillium digitatum genome. We demonstrated that the deletion of PdGcs1 in P. digitatum resulted in the complete loss of production of GlcCer (d18:1/18:0 h) and GlcCer (d18:2/18:0 h), a decrease in vegetation growth and sporulation, and a delay in spore germination. The virulence of the PdGcs1 deletion mutant on citrus fruits was also impaired, as evidenced by the delayed occurrence of water soaking lesion and the formation of smaller size of lesion. These results suggest that PdGcs1 is a bona fide GCS that plays an important role in regulating cell growth, differentiation, and virulence of P. digitatum by controlling the biosynthesis of GlcCers.
Keywords: Penicillium digitatum, GlcCers, GlcCers synthase, Virulence
1. Introduction
Glucosylceramides (GlcCers) are the compositions of membrane lipids in animals, plants and fungi. A typical fungal GlcCer structure consisting of 4,8-diunsaturated, C9-methylated sphingadienine amidic linked to N-2′-hydroxyoct/hexadecanoic acid and glucose or galactose as the monosaccharide residue was reported in a number of fungi including Pichia pastoris and Aspergillus fumigatus [1]. The important step in GlcCer biosynthesis is catalyzed by uridine diphosphate-glucose: ceramide glucosyltransferase (GCS, EC 2.4.1.80) encoded by GlcCer synthase gene (Gcs1), which transferred a glucose group to ceramides [2].
Although the structure of fungal GlcCers and their biosynthesis pathway have been studied in detail in several model fungi [3], much remains unknown about their biological function. Recently GlcCers were found to play important roles in spore germination, hyphal development, fungal growth and differentiation by regulating the physical properties of membrane [4–7]. A Fusarium graminearum mutant lacking GlcCers exhibited remarkable changes in the morphology of the conidia and defects in the polar growth of fungal hyphae [5]. More interestingly, GlcCers were reported to be required for virulence in human fungal pathogens Cryptococcus neoformans and Candida albicans. Compared with the wild type strains, Gcs1-deleted mutants of C. neoformans and C. albicans lost their most virulence [8–9]. The involvement of GlcCers in virulence was also observed in plant pathogen F. graminearum [5].
Because GlcCers are involved in the growth and virulence of fungi, targeting the biosynthesis pathways of GlcCers may kill fungal pathogens. Plant defensin RsAFP2 isolated from Raphanus sativus and MsDef1/4 from Medicago spp. interacted with GlcCers and lead to the growth arrest of C. albicans and F. graminearum [5,10]. Although GlcCers were described as cell membrane constituents of mammal, plant and fungi [11], the structure of fungal GlcCers were remarkable distinct from their counterparts in animal cells [12], suggesting that GlcCers and GCS are the ideal targets for containing pathogenic fungi.
Penicillium digitatum (Pers.:Fr) Sacc., the causative agent of green mold decay on postharvest citrus, is one of the most destructive pathogens in citrus industry. It usually caused a significant decay loss of postharvest citrus during post-harvest storing, packaging, transportation and marketing [13]. The genome sequences of three strains of P. digitatum have been published recently [14–15], The biological functions of several transcription factors and protein kinases were explored recently through gene manipulation technology [16–19]. However, the roles for GlcCers in the biological responses of P. digitatum have not been investigated. In this study, we report the identification and functional analyses of an ortholog of Gcs1 in P. digitatum.
2. Materials and methods
2.1. Fungal strains
P. digitatum strain PdKH8 was used as a wild type strain for creating the mutant strains.
2.2. Sequence analysis of PdGcs1
The PdGcs1 gene was originally identified through a BLASTp search of the P. digitatum genome sequence [14–15] using the amino acid sequence of C. albicans Gcs1p (XP_722809) and Magnaporthe grisea Gcs1p (AAK73019) as queries. The coding sequence of the putative PdGcs1 was amplified from the genomic DNA and cDNA, respectively, with primers Gcs1-F/Gcs1-R (Table S1).
2.3. Construction of PdGcs1 deletion mutants
The PdGcs1 deletion mutants were generated using the gene replacement strategy. Two flanking sequences of PdGcs1 amplified from P. digitatum genomic DNA were inserted into the two sides of the hph (hygromycin resistance) gene of pTFCM vector. Briefly, a 1714 bp upstream flanking fragment of PdGcs1 was amplified with Gcs1-up-F and Gcs1-up-R (Table S1) and cloned into the KpnI and SacI sites of pTFCM vector to construct PTFCM-PdGcs1-up. Subsequently, a 957 bp downstream fragment of the PdGcs1 was amplified using primers Gcs1-down-F and Gcs1-down-R (Table S1) and inserted into the SpeI and XhoI sites of pTFCM-PdGcs1-up vector to obtain the final PdGcs1 replacement vector pTFCM-ΔPdGcs1. The resultant replacement vector was then transformed into Agrobacterium tumefaciens AGL-1 by electroporation using ECM630 (BTX, California, USA). Transformants were obtained by A. tumefaciens-mediated transformation (ATMT) as described previously and selected on PDA medium supplemented with 70 μg/mL hygromycin B [20]. Initial identification of gene replacement mutants was performed by PCR using primers Gcs1–check-F1/Gcs1–check-R1 and Gcs1–check-F2/Gcs1–check-R2 (Table S1). Southern hybridization analysis was used to further confirm the PdGcs1 replacement. Genomic DNA isolated from the strains of PdGcs1 deletion mutant or wild type strain of P. digitatum was cut with the EcoRV and XhoI restriction enzyme. A 972 bp DNA fragment upstream of the flanking sequence of PdGcs1 gene was amplified from genomic DNA of the wild type strain with the primers Gcs1-SB-F and Gcs1-SB-R (Table S1) and used as a probe for Southern blot. Southern blot was carried out using the DIG high prime DNA labeling and detection starter kit I with NBT/BCIP (Roche, Mannheim, Germany) following the protocol of the manufacturer.
2.4. Genetic complementation of PdGcs1 deletion mutant
The PdGcs1 complement mutant, termed as CPPdGcs1, was generated by introducing the full-length PdGcs1 sequence into a PdGcs1 deletion mutant. Briefly, an approximate 4566 bp full-length PdGcs1 gene including the PdGcs1 coding region, its 1.6-kb upstream and 0.9-kb downstream sequence was amplified from genomic DNA of wild type strain PdKH8 with Gcs1-com-F and Gcs1-com-R (Table S1). This fragment was then cloned into pCA-neo [18] to generate the complementation vector pCA-neo-PdGcs1. Transformation of PdGcs1 deletion mutant with this complementation vector pCA-neo-PdGcs1 was conducted as described above, except that neomycin was used as a selection agent. The identification of PdGcs1 complement mutant by PCR and Southern blot was performed as method mentioned previously.
2.5. HPLC-MS/MS analysis of GlcCers
GlcCers were extracted as described [21]. Conidial suspension (100 μL 1.0 × 106 conidia/mL) of the wild type, PdGcs1 deletion mutant or its corresponding complement mutant strains of P. digitatum was added into 100 mL PDB media and cultured at 25 °C for 4 days with 180 rpm shaking. Total sphingolipids were extracted from 5 g of the mycelial powder with ultrasonic extraction in 50 mL iso-propanol:water:ethyl acetate (30:10:60, by vol) for 10 min. The organic phase containing the extracted GlcCers was dried under nitrogen and then dissolved in 500 μL MeOH with 25 mmol/l ammonium formate.
HPLC-MS/MS analyses were performed on an Agilent 6460 triple quadruple mass spectrometer coupled with Agilent 1200 infinity LC modules (Agilent, California, USA). Ten-microliter sample was separated on ZORBAX Eclipse XDB-C8 column (150 mm × 4.6 mm, 5 μm particle size, Waters) with a flow rate of 0.3 mL/min at 30 °C. Two mobile phase solvents were used in this method. Solvent A was 25 mmol/L ammonium formate. Solvent B was acetonitrile. GlcCers were eluted under the following gradient elution conditions: 20–95% B at 0–15 min, 95–100% B at 15–30 min, and 100% B at 30–40 min. The source parameters were as follows: gas temperature, 350 °C; gas flow rate, 10 L/min; nebulizer pressure, 50 psi; capillary voltage, 3500 V; The fragmentor voltage was 100 V and collision energy was 20 V. Soybean GlcCer standard (Avanti Polar Lipids, Alabama, USA) was used as a standard.
2.6. Assay of vegetative growth, sporulation and conidium germination
The conidial suspension (5 μL 1.0 × 106 conidia/mL) of PdGcs1 deletion mutant, CPPdGcs1 mutant or the wild type strain of P. digitatum was prepared and spotted on the center of PDA, CYA MEA and OA plates. These plates were incubated at 25 °C. Radial growth was determined at 5 dpi (days post inoculation) by calculating the mean of two perpendicular colony diameters in each plate. Conidiation were assayed at the same time by washing the agar surface with 5 mL sterile distilled water. The conidial suspension was diluted with sterile distilled water and counted in a haemocytometer. The experiments were performed twice with three replicates.
The conidial suspension (100 μL 1.0 × 104 conidia/mL) of PdGcs1 deletion mutant, CPPdGcs1 mutant and the wild-type strains were spread evenly onto the surface of PDA medium and incubated at 25 °C. The germination of spores were observed under microscope Eclipse 80i (Nikon, Japan) at 8, 12, 16 and 24 h after inoculation. Conidia were considered to be germinated when the germ tube extended to at least twice the length of the conidia itself. Germination percent was assessed for at least 100 conidia/sample and determined as germination rate (%) = [germinated conidia/(germinated conidia + ungerminated conidia)] × 100%. The experiment was performed twice with three replicates.
2.7. Gene expression analysis
The dynamic expression of PdGcs1 during the infection of citrus fruits was assayed using qRT-PCR. The peel of mature and un-wounded Ponkan (Citrus reticulata Blanco) was punctured (1–2 mm deep) with a bunch of 5 needles followed by inoculation with conidial suspension (10 μL 1.0 × 106 conidia/mL) of P. digitatum wild type strain PdKH8. At 12, 24, 48, 72 and 96 h post inoculation, total RNA was extracted from inoculated tissues and reversely transcribed into cDNA. The expression levels of PdGcs1 were determined by using specific primers PdGcs1-qF and PdGcs1-qR (Table S1). The qRT-PCR was conducted on Applied Biosystems 7300 Real Time PCR system (ABI, Foster City, USA) using the kit of SYBR® Premix Ex Taq™ II (TaKaRa, Dalian, China) following the instruction. The β-actin gene of P. digitatum (accession number: AB030227) was used as an internal control. The relative changes for PdGcs1 expression during infection were analyzed according to the 2−ΔΔCt method [22]. The experiment was conducted twice with three independent biological replicates. Data were calculated using the analysis of t-test in SPSS version 13.0 for Windows (SPSS, Chicago, USA).
2.8. Virulence assay
The health and un-wounded Mature Ponkan fruits were selected to investigate the role of PdGcs1 on P. digitatum virulence. The fruits were injured as described previously, conidial suspensions (10 μL 1 × 106 conidia/mL) of PdGcs1 deletion mutant, CPPdGcs1 mutant and the wild type strains were added on the wounds, respectively. Equivalent volume of sterile water was used as the negative control. The inoculated fruits were placed in a plastic tray which was covered with plastic film and incubated at room temperature. The lesion diameters were recorded at 5 dpi. At least twenty fruits were used for each strain and the experiment was repeated twice.
3. Results
3.1. Sequence analysis of PdGcs1
Through BLASTp search, EKV12123 (GenBank number) of P. digitatum was found to be homologous to Gcs1p (XP722809) of C. albicans and Gcs1p (AAK73019) of M. grisea with 32% and 48% amino acid identity, respectively. Sequencing analyses indicated that the sequence of the putative PdGcs1 amplified from P. digitatum strain PdKH8 was identical to that of EKV12123 published for a Spain strain Pd1 of P. digitatum [15]. PdGcs1p had two nucleotide recognition domains NRD2L and NRD2S, and also had D1, D2, D3 and Q/RXXRW motif, which are conserved for glucosyltransferase family [1].
3.2. Creation and identification of PdGcs1 deletion and complementation mutants
PdGcs1 deletion mutants were created by using a homologous recombination strategy (Fig. 1A). Two transformants were identified as PdGcs1 deletion mutants by PCR (Fig. 1B). With the internal primer pair Gcs1-check-F1/Gcs1-check-R1, a 662-bp fragment could be amplified from the ectopic transformants and the wild type strain PdKH8, but not PdGcs1 deletion mutants. On the other hand, with the outer primer pair Gcs1-check-F2/Gcs1-check-R2, a fragment of 2566-bp could be amplified from the PdGcs1 deletion mutants, but not from the wild type strain PdKH8 or ectopic trans-formants (Fig. 1B). One replacement mutant, termed as ΔPdGcs1, was chosen for further studies. Southern blot using the probe specific to the 5′ region of PdGcs1 revealed a 4691 bp fragment in PdGcs1 deletion mutant and a 2180 bp fragment in the wild type strain of P. digitatum (Fig. 1C). This Southern hybridization pattern validated that PdGcs1 deletion mutant was a null mutant resulting from a single homologous recombination event at the PdGcs1 locus.
Fig. 1.
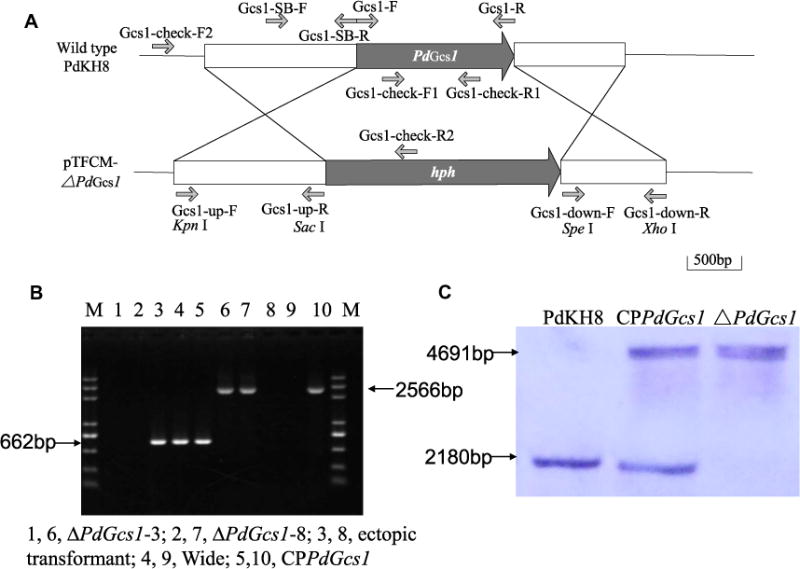
Construction and identification of P. digitatum PdGcs1 deletion mutant. (A) Schematic diagram showed the construction of PdGcs1 deleting plasmid. (B) Identification of putative PdGcs1 deletion mutant and CPPdGcs1 mutant by PCR using primer pairs Gcs1–check-F1/Gcs1–check-R1 (lane 1–5) and Gcs1–check-F2/Gcs1–check-R2 (lane 6–10). (C) The fragment sizes from the PdGcs1 deletion mutant and the wild type strain of P. digitatum were 4691 and 2180 bp in length, respectively.
3.3. PdGcs1 encoded GCS and catalyzed the synthesis of GlcCers
Qualitative analysis of GlcCers purified and concentrated from equal mass of mycelia were resolved using HPLC–MS/MS (Fig. 2). Soybean GlcCer standard served as a positive control (data not shown). Precursor scans revealed that P. digitatum wild type strain produced two kinds of GlcCers. According to the structure of fungal GlcCers, we conjectured the peak at m/z 742.4 and 744 corresponding to [M+H]+ charge for GlcCer (d18:2/18:0 h) and GlcCer (d18:1/18:0 h), respectively. Their HPLC retention times were at 21–24 min, being consistent with that of the soybean GlcCer. Expectedly, these GlcCers were undetectable in the sphingolipid extraction of PdGcs1 deletion mutant, but were reconstituted in CPPdGcs1 mutant strain, suggesting that PdGcs1 is a bona fide GCS that catalyzes the synthesis of GlcCers in P. digitatum.
Fig. 2.
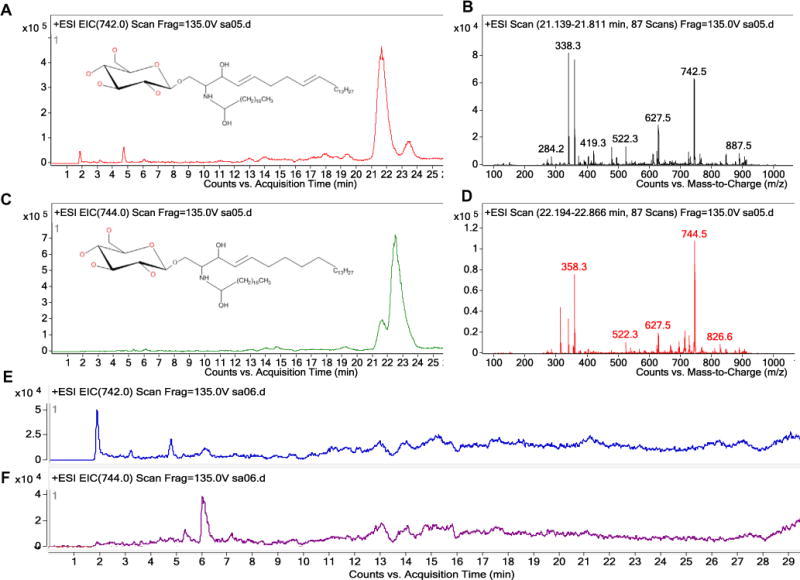
Detection of GlcCer (d18:2/18:0 h) and GlcCer (d18:1/18:0h) in the PdGcs1 deletion mutant and its wild-type control. Chromatogram of GlcCer (d18:2/18:0h) (A) and GlcCer (d18:1/18:0h) (C) from wild type strain PdKH8. (B) and (D) were the mass spectrums of GlcCer (d18:2/18:0h) (m/z 742) and GlcCer (d18:1/18:0h) (m/z 744), respectively. Neither GlcCer (d18:2/18:0 h) nor GlcCer (d18:1/18:0 h) were found in PdGcs1 deletion mutant (E and F).
3.4. PdGcs1 was required for radial growth, sporulation and conidial germination
The radial growth of PdGcs1 deletion mutant on different media was compared with that of P. digitatum wild type strain PdKH8. The radial growth of PdGcs1 deletion mutant was weaker than that of the parental strain on these media (Fig. 3A). Compared to parental strain PdKH8, the colony diameter of the PdGcs1 deletion mutant on PDA, CYA, OA and MEA medium was reduced to 66%, 77%, 73% and 68%, respectively. While the sporulation of PdGcs1 deletion mutant was also severely impaired with approximate 32% reduction on PDA (Fig. 3B).
Fig. 3.
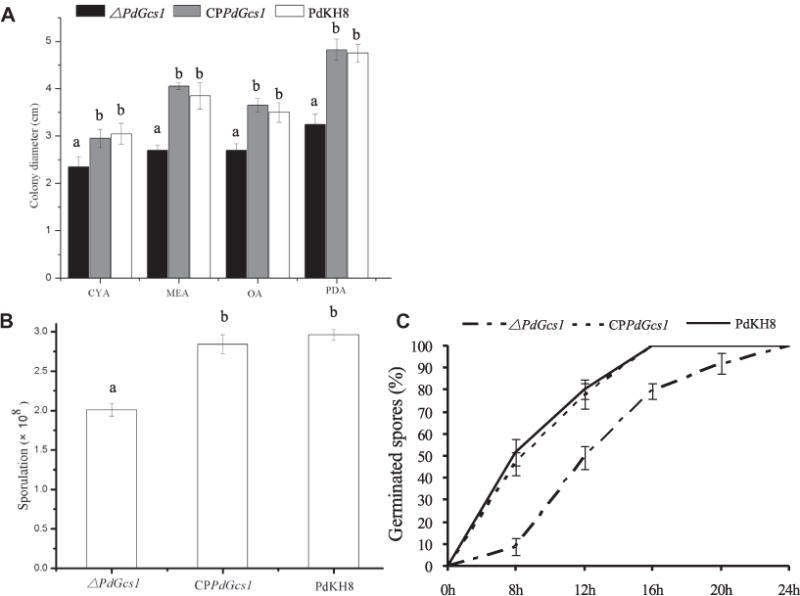
The comparison of the mycelial growth (A), conidiation (B) and germination (C) of PdGcs1 deletion mutant and wild type strain PdKH8.
The germination tendency of conidia for the wide type strain PdKH8, PdGcs1 deletion mutant, or CPPdGcs1 mutant strain was measured (Fig. 3C). For the wide type strain PdKH8, 51.6 ± 6.2% of conidia germinated at 8 h after incubation on PDA. However, only 8.9 ± 3.9% of conidia of PdGcs1 deletion mutant germinated at the same time. After incubation for 12 h, the germination percent of conidia of the wide type strain PdKH8 increased to 80.2 ± 4.6%, but only 49.3 ± 5.3% for PdGcs1 deletion mutant. Almost all conidia germinated in the wild type strain at 16 h after incubation. While only 79.4 ± 3.6% of conidia of PdGcs1 deletion mutant germinated. The conidia of PdGcs1 deletion mutant were all germinated until 24 h after incubation.
As shown in Fig. 3, the reduction of growth, sporulation, and conidial germination in PdGcs1 deletion mutant was recovered when PdGcs1 was reintroduced into the PdGcs1 deletion mutant.
3.5. Expression of PdGcs1 during infection
During the infection of P. digitatum on citrus fruit, the dynamic expression pattern of PdGcs1 was analyzed. As shown in Fig. 4A, transcript accumulation of PdGcs1 increased and reached a peak at 36 h after inoculation. From 36 to 72 h, there was a sustained phase, and then the expression of PdGcs1 was decreased gradually. At 96 and 120 h after infection, the expression levels were decreased to the level as the beginning (0 h).
Fig. 4.
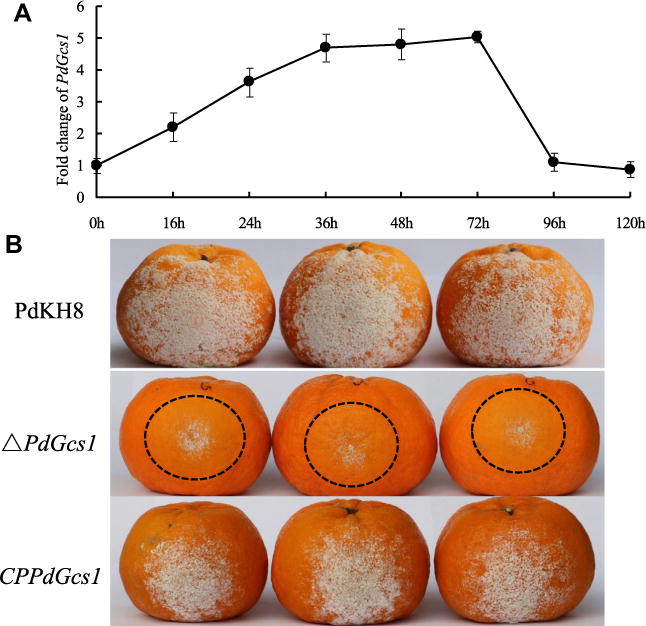
The dynamic expression pattern of PdGcs1 during the infection of P. digitatum on citrus fruit (A) and the virulence assays of PdGcs1 deletion mutant and wild type strain PdKH8 (B). PdGcs1 mRNA levels were determined by using specific primers PdGcs1-qF and PdGcs1-qR using qRT-PCR. The β-actin gene of P. digitatum was used as an internal control.
3.6. Disruption of PdGcs1 affected the virulence
The effect of PdGcs1 on the virulence of P. digitatum was assayed by inoculating Ponkan fruits with the conidial suspension of the PdGcs1 deletion mutant and the wild type strain PdKH8 (Fig. 4B). Our results showed that the water-soaked lesion occurred on the inoculated spots of the wild type strain PdKH8 at 1 day post infection (dpi), while, there was no symptom on citrus fruits inoculated with PdGcs1 deletion mutant. At 2 dpi, the maceration lesions were observed on the citrus fruits inoculated with the PdGcs1 deletion mutant. The maceration lesions of the wild-type P. digitatum-inoculated fruits were about two times larger than those of PdGcs1 deletion mutant-inoculated fruits during the infection process. At 5 dpi, the average diameter of maceration lesions caused by the wild type strain PdKH8 was 11.7 ± 0.6 cm. However, the diameter of lesions caused by PdGcs1 deletion mutant decreased significantly, and the average diameter was only 5.1 ± 0.3 cm. Statistic analyses showed that there was a significant difference between the average diameter of maceration lesions caused by PdGcs1 deletion mutant and the wild type strain PdKH8 (P < 5%). The reduction in the virulence of PdGcs1 deletion mutant was reversed when PdGcs1 was introduced into the PdGcs1 deletion mutant. These results suggest that the disruption of PdGcs1 reduces the virulence of P. digitatum.
4. Discussion
GlcCers are important membrane lipids in most eukaryotic organisms. However, much remains unknown about their roles in filamentous fungi. In this study, for the first time, we report the identification and functional characterization of the GCS gene, termed PdGcs1, in P. digitatum. We demonstrate that the protein product of PdGcs1 is a bona fide GCS that plays a key role in regulating hyphal growth, sporulation, conidial germination and virulence in P. digitatum likely by synthesizing GlcCers.
The first gene encoding GlcCer synthase was cloned from human [23]. Recently, Gcs1 was found in a number of fungi including A. fumigatus and M. grisea [6,24]. The deletions of the putative glycosyltransferase genes in C. albicans, P. pastoris, C. neoformans and F. graminearum resulted in the complete loss of GlcCers [1,5,9]. In this study, we found that the deletion of PdGcs1 also resulted in the complete loss of GlcCer (d18:1/18:0 h) and GlcCer (d18:2/18:0 h). This result confirmed that PdGcs1 indeed encodes GCS that catalyzes the synthesis of GlcCers.
GlcCers have been shown to regulate fungal growth. Compared with the wild type strain, the PdGcs1 deletion mutant appear to have pleiotropic phenotypes, including slow vegetative growth, sporulation decline and conidial germination delay in this study. These results are consistent with those reported in A. fumigatus, A. nidulans, F. graminearum and. C. albicans. Without GlcCers, the F. graminearum Gcs1 deletion mutant exhibited marked changes in the morphology of the conidia and defects in the polar growth of fungal hyphae [5,25]. Due to a defect in its cell membrane structure, the C. albicans Gcs1 deletion mutant had a decreased hyphal growth rate compared with the wild type strain [26]. Previous study confirmed that GlcCers are the composition of fungal cell membranes, and GlcCers control membrane fluidity and stability by modulating membrane lipid topography [7]. So the efficiency of nutrient absorption and transportation decreased in the Glc-Cers-lacking cell. Another mechanism involved is a second messenger ceramide, a substrate of GCS. Ceramide was reported as the central core in sphingolipid metabolism, high concentration of ceramide caused many biological processes such as cell cycle arrest and apoptosis [27]. Based on these observations, we postulated that PdGcs1 deficiency changes the fluidity and stability of cell membrane due to the absence of GlcCers and/or causes the growth defects by increasing the level of ceramide.
In this study, GCS and its product GlcCers were found to be required for the full virulence of P. digitatum. Other studies also suggested a role for GlcCers in the regulation of fungal pathogenesis. Disruption of the GlcCers biosynthetic pathway altered the virulence of C. albicans and C. neoformans [8–9]. Previous study showed that GlcCers organized the lipid raft of the plasma membranes and altered the bilayer structure [7]. In C. neoformans, the lipid raft had a function to cluster the virulence determinant phospholipase B1 and the antioxidant virulence factor Cu/Zn superoxide. The lipid raft also participated the release of these virulence molecules to the host cell [28]. The virulence determinants of P. digitatum are retained to be explored, and if there is a similar mechanism present in lipid raft of P. digitatum, it is retained to be experimentally elucidated.
In conclusion, this study shows that GlcCers play important roles in the growth and virulence of P. digitatum mainly through a structure change of the lipid raft. Since a significant difference between human GlcCer and fungal GlcCer, the researches about GlcCers and Gcs1 may provide a new approach to control pathogenic fungi.
Supplementary Material
Acknowledgments
This work was supported by China Agriculture Research System (CARS-27), the Special Funding for Agro-scientific Research in the Public Interest (201303023), Zhejiang nonprofit technology research programs (20012C22100) and National Institutes of Health (NIH) Grant R01CA163825 (to C.M.). We are very grateful to Ms. Xiaodan Wu for the assistance in HPLC–MS/MS analysis of GlcCers.
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.bbrc.2014.10.142.
References
- 1.Leipelt M. Glucosylceramide synthases, a gene family responsible for the biosynthesis of glucosphingolipids in animals, plants, and fungi. J Biol Chem. 2001;276:33621–33629. doi: 10.1074/jbc.M104952200. [DOI] [PubMed] [Google Scholar]
- 2.Ichikawa S, Sakiyama H, Suzuki G, Hidari K, Hirabayashi Y. Expression cloning of a cDNA for human ceramide glucosyltransferase that catalyzes the first glycosylation step of glycosphingolipid synthesis. Proc Natl Acad Sci USA. 1996;93:4638–4643. doi: 10.1073/pnas.93.10.4638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ternes P, Wobbe T, Schwarz M, Albrecht S, Feussner K, Riezman I, Cregg JM, Heinz E, Riezman H, Feussner I, Warnecke D. Two pathways of sphingolipid biosynthesis are separated in the yeast Pichia pastoris. J Biol Chem. 2011;286:11401–11414. doi: 10.1074/jbc.M110.193094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oura T, Kajiwara S. Disruption of the sphingolipid 8-desaturase gene causes a delay in morphological changes in Candida albicans. Microbiology. 2008;154:3795–3803. doi: 10.1099/mic.0.2008/018788-0. [DOI] [PubMed] [Google Scholar]
- 5.Ramamoorthy V, Cahoon EB, Li J, Thokala M, Minto RE, Shah DM. Glucosylceramide synthase is essential for alfalfa defensin-mediated growth inhibition but not for pathogenicity of Fusarium graminearum. Mol Microbiol. 2007;66:771–786. doi: 10.1111/j.1365-2958.2007.05955.x. [DOI] [PubMed] [Google Scholar]
- 6.Levery SB, Momany M, Lindsey R, Toledo MS, Shayman JA, Fuller M, Brooks K, Doong RL, Straus AH, Takahashi HK. Disruption of the glucosylceramide biosynthetic pathway in Aspergillus nidulans and Aspergillus fumigatus by inhibitors of UDP-Glc: ceramide glucosyltransferase strongly affects spore germination, cell cycle, and hyphal growth. FEBS Lett. 2002;525:59–64. doi: 10.1016/s0014-5793(02)03067-3. [DOI] [PubMed] [Google Scholar]
- 7.Del Poeta M, Nimrichter L, Rodrigues ML, Luberto C. Synthesis and biological properties of fungal glucosylceramide. PLoS Pathog. 2014;10:e1003832. doi: 10.1371/journal.ppat.1003832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Noble SM, French S, Kohn LA, Chen V, Johnson AD. Systematic screens of a Candida albicans homozygous deletion library decouple morphogenetic switching and pathogenicity. Nat Genet. 2010;42:590–598. doi: 10.1038/ng.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rittershaus PC, Kechichian TB, Allegood JC, Merrill AH, Hennig M, Luberto C, Del Poeta M. Glucosylceramide synthase is an essential regulator of pathogenicity of Cryptococcus neoformans. J Clin Invest. 2006;116:1651–1659. doi: 10.1172/JCI27890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thevissen K. Defensins from insects and plants interact with fungal glucosylceramides. J Biol Chem. 2003;279:3900–3905. doi: 10.1074/jbc.M311165200. [DOI] [PubMed] [Google Scholar]
- 11.Nimrichter L, Cerqueira MD, Leitao EA, Miranda K, Nakayasu ES, Almeida SR, Almeida IC, Alviano CS, Barreto-Bergter E, Rodrigues ML. Structure, cellular distribution, antigenicity, and biological functions of fonsecaea pedrosoi ceramide monohexosides. Infect Immun. 2005;73:7860–7868. doi: 10.1128/IAI.73.12.7860-7868.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nimrichter L, Rodrigues ML. Fungal glucosylceramides: from structural components to biologically active targets of new antimicrobials. Front Microbiol. 2011;2:1–7. doi: 10.3389/fmicb.2011.00212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Macarisin D, Cohen L, Eick A, Rafael G, Belausov E, Wisniewski M, Droby S. Penicillium digitatum suppresses production of hydrogen peroxide in host tissue during infection of citrus fruit. Phytopathology. 2007;97:1491–1500. doi: 10.1094/PHYTO-97-11-1491. [DOI] [PubMed] [Google Scholar]
- 14.Sun X, Ruan R, Lin L, Zhu C, Zhang T, Wang M, Li H, Yu D. Genomewide investigation into DNA elements and ABC transporters involved in imazalil resistance in Penicillium digitatum. FEMS Microbiol Lett. 2013;348:11–18. doi: 10.1111/1574-6968.12235. [DOI] [PubMed] [Google Scholar]
- 15.Marcet-Houben M, Ballester AR, de la Fuente B, Harries E, Marcos JF, González-Candelas L, Gabaldón T. Genome sequence of the necrotrophic fungus Penicillium digitatum, the main postharvest pathogen of citrus. BMC Genomics. 2012;13:646. doi: 10.1186/1471-2164-13-646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang T, Sun X, Xu Q, Zhu C, Li Q, Li H. PdSNF1, a sucrose non-fermenting protein kinase gene, is required for Penicillium digitatum conidiation and virulence. Appl Microbiol Biotechnol. 2013;97:5433–5445. doi: 10.1007/s00253-012-4593-z. [DOI] [PubMed] [Google Scholar]
- 17.Zhang T, Sun X, Xu Q, Candelas LG, Li H. The pH signaling transcription factor PacC is required for full virulence in Penicillium digitatum. Appl Microbiol Biotechnol. 2013:1–12. doi: 10.1007/s00253-013-5129-x. [DOI] [PubMed] [Google Scholar]
- 18.Zhang T, Xu Q, Sun X, Li H. The calcineurin-responsive transcription factor Crz1 is required for conidation, full virulence and DMI resistance in Penicillium digitatum. Microbiol Res. 2012;168:211–222. doi: 10.1016/j.micres.2012.11.006. [DOI] [PubMed] [Google Scholar]
- 19.Wang M, Chen C, Zhu C, Sun X, Ruan R, Li H. Os2 MAP kinase-mediated osmostress tolerance in Penicillium digitatum is associated with its positive regulation on glycerol synthesis and negative regulation on ergosterol synthesis. Microbiol Res. 2014;169:511–521. doi: 10.1016/j.micres.2013.12.004. [DOI] [PubMed] [Google Scholar]
- 20.Wang JY, Li HY. Agrobacterium tumefaciens-mediated genetic transformation of the phytopathogenic fungus Penicillium digitatum. J Zhejiang Univ Sci B. 2008;9:823–828. doi: 10.1631/jzus.B0860006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bielawski J, Szulc ZM, Hannun YA, Bielawska A. Simultaneous quantitative analysis of bioactive sphingolipids by high-performance liquid chromatography-tandem mass spectrometry. Methods. 2006;39:82–91. doi: 10.1016/j.ymeth.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 22.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using realtime quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 23.Marks DL, Wu K, Paul P, Kamisaka Y, Watanabe R, Pagano RE. Oligomerization and topology of the Golgi membrane protein glucosylceramide synthase. J Biol Chem. 1999;274:451–456. doi: 10.1074/jbc.274.1.451. [DOI] [PubMed] [Google Scholar]
- 24.Koga J, Yamauchi T, Shimura M, Ogawa N, Oshima K, Umemura K, Kikuchi M, Ogasawara N. Cerebrosides A and C, sphingolipid elicitors of hypersensitive cell death and phytoalexin accumulation in rice plants. J Biol Chem. 1998;273:31985–31991. doi: 10.1074/jbc.273.48.31985. [DOI] [PubMed] [Google Scholar]
- 25.Yu JH, Rittenour WR, Chen M, Cahoon EB, Harris SD. Control of glucosylceramide production and morphogenesis by the Bar1 ceramide synthase in Fusarium graminearum. PLoS ONE. 2011;6:e19385. doi: 10.1371/journal.pone.0019385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oura T, Kajiwara S. Candida albicans sphingolipid C9-methyltransferase is involved in hyphal elongation. Microbiology. 2010;156:1234–1243. doi: 10.1099/mic.0.033985-0. [DOI] [PubMed] [Google Scholar]
- 27.Arana L, Gangoiti P, Ouro A, Trueba M, Gómez-Muñoz A. Review ceramide and ceramide 1-phosphate in health and disease. Lipids Health Dis. 2010;9:2–12. doi: 10.1186/1476-511X-9-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Siafakas AR, Wright LC, Sorrell TC, Djordjevic JT. Lipid rafts in Cryptococcus neoformans concentrate the virulence determinants phospholipase B1 and Cu/Zn superoxide dismutase. Eukaryot Cell. 2006;5:488–498. doi: 10.1128/EC.5.3.488-498.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


