Abstract
We have previously isolated nucleic acid ligands (aptamers) that bind the surface envelope glycoprotein, gp120, of HIV-1, and neutralize infection of diverse sub-types of virus. Our earlier studies have identified the overall structure of one of these aptamers, B40, and have indicated that it binds to gp120 in a manner that competes with that of the HIV-1 co-receptor, CCR5, and select “CD4i” antibodies with epitopes overlapping this region. Here, we sought to map the B40 binding site on gp120 more precisely by analysing its interaction with a panel of alanine substitution mutants of gp120. Furthermore, we tested our hypothesis concerning the structure of the 40 nucleotide functional core of the aptamer by the solid-phase synthesis of truncated and chemically modified derivatives. The results confirm our structural predictions and demonstrate that aptamer B40 neutralizes a diverse range of HIV-1 isolates as a result of binding to relatively conserved residues on gp120 at the heart of the CCR5-binding site. These structural insights may provide the basis for the development of potential antiviral agents with high specificity and robustness.
Introduction
HIV-1 entry is a multi-step process; the trimeric surface glycoprotein of HIV-1, gp120, binds to CD4 on the host cell surface, and undergoes a conformational change enabling an interaction with the alternative coreceptors, CCR5 or CXCR4. Thereafter, the viral and cellular membranes fuse and the viral RNA enters the cell. Agents that inhibit these processes might allow further investigation of the molecular interactions involved, and also have potential as viral therapeutics.
Accordingly, we have isolated nucleic acid ligands (aptamers) that bind gp120, and thereby neutralize infection of virus (Khati et al., 2003). The massively combinatorial library from which the aptamers were selected comprised single stranded nucleic acids with 2′-F pyrimidine and 2′-OH purine nucleotides, 117 nucleotides in length. Aptamers were selected using an in vitro evolution method (flow cell SELEX) to bind to gp120 from a CCR5-utilizing strain of HIV-1 (BaL) (Khati et al., 2003). Of over 60 monoclonal aptamers that bound to gp120, more than 20 were able to neutralize HIV-1 infection of PBMC cultures. Most interestingly, these aptamers were able to neutralize clinical isolates of HIV-1 representing all subtypes, not just the subtype B strain BaL against which they were selected (Khati et al., 2003).
The RNA structure and mechanism of action of one such anti-gp120 aptamer, B40, have been investigated in detail. Aptamer B40 RNA forms two alternative conformations in solution, and we have hypothesized that it is the three-way junction structure rather than the linear structure that binds to gp120 (Dey et al., 2005a). We had previously noted partial competition between the binding of B40 and monoclonal antibodies defining the CD4i (CD4-induced) epitope (Khati et al., 2003). Consistent with this, the aptamer competed with a synthetic peptide corresponding to the N-terminus of CCR5 for binding to gp120 (Dey et al., 2005b).
Here we sought to map the B40 binding site on gp120 (its “aptatope”) more precisely by analysing its interaction with a panel of alanine substitution mutants of gp120. A similar method has previously been employed successfully to determine the epitopes of a number of anti-gp120 antibodies (Darbha et al., 2004; Pantophlet et al., 2003; Scanlan et al., 2002). Furthermore, we tested our hypothesis concerning the structure of the 40 nucleotide (nt) functional core of the aptamer by the solid-phase synthesis of truncated and chemically modified derivatives. Our results confirm the structural predictions and demonstrate that aptamer B40 neutralizes HIV-1 infectivity as a result of binding to conserved residues of gp120 at the heart of the binding site for the N-terminus of CCR5.
Results
Confirmation of the branched structure of functional aptamer B40
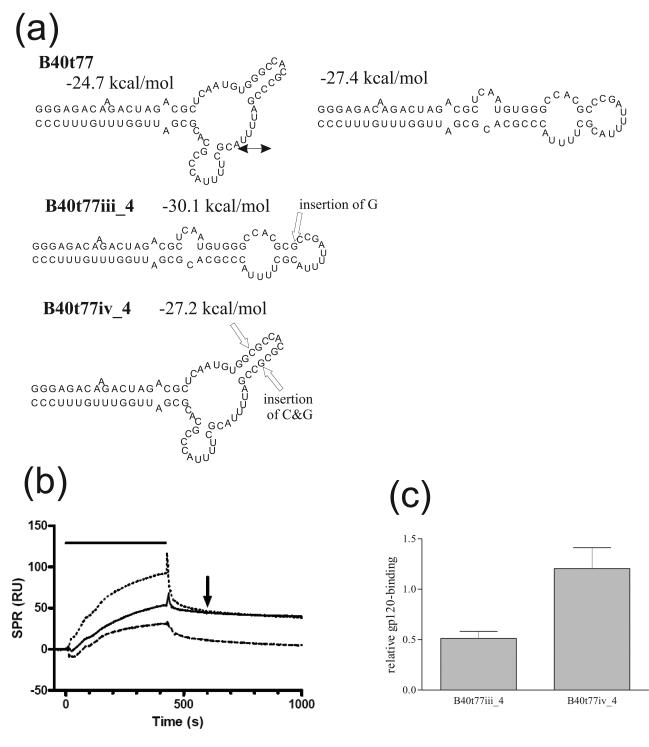
Thermodynamic predictions suggest that both full-length aptamer B40 and a 77 nt shortened derivative, B40t77, can exist in two conformations – linear and branched – that are difficult to distinguish using low-resolution analysis with structure-sensitive nucleases. It was important to define the functionally relevant conformers, in order to guide the subsequent development of chemically synthesized derivatives. Accordingly, we devised point mutants of B40t77 designed to bias the respective proportions of each conformer in the aptamer population. Ideally a pair of mutations lying outside the previously defined gp120-binding site of the aptamer would be chosen (Dey et al., 2005a), of which one favoured the folding of the molecule into one conformer, and the second of which, a compensatory mutation, would favour the alternative conformer. A large number of mutational pairs were evaluated in silico, and of these, three pairs appeared to meet the design requirements. When evaluated for gp120 binding in vitro, two of these pairs proved to be uninformative, as the first mutation, which inactivated binding, was not reverted phenotypically by the structure-correcting second mutation (see Supplementary Figure 1). In such cases, it is possible that one or other of the mutations directly disrupted the aptamer-gp120 contact site, and so the effect on structure per se could not be evaluated. The third pair of aptamers, B40t77iii_4 and B40t77iv_4, did give decisive results (Figure 1). Aptamer B40t77iii_4 contains an inserted G after residue C35 that is predicted to stabilize the linear conformation, and this aptamer bound poorly to gp120. Insertion of an additional C residue after G28 in aptamer B40t77iii_4, to generate aptamer B40t77iv_4 was predicted to stabilize the branched conformation, and gp120-binding was restored to wild-type levels. The restoration of gp120-binding by a second mutation that compensates for the structural bias introduced by the first is strong evidence that the branched conformer is the active form of the aptamer. Not only have the mutations in B40t77iii_4 and B40t77iv_4 increased the stability of the intended conformer, as shown by more negative free energies compared to the same structure in B40t77, but also they have decreased the stability of the alternative conformers, which are energetically unfavourable. Nevertheless, we concede that within a population of RNA molecules additional conformations may still exist, and this could explain why only modest changes in binding were seen with these aptamer derivatives compared to the parental aptamer B40t77.
Figure 1. Confirmation of the branched structure of functional aptamer B40t77.
(a) Structures of aptamer B40t77, which is predicted to fold into both the linear or branched conformations, and its mutant derivatives designed to adopt solely the linear (B40t77iii_4) or branched (B40t77iv_4) conformation. The positions of nucleotide insertions are shown by open arrows, together with the predicted free energy of each structure and double-headed arrows indicating conformational interconversion. Structures and free energies were predicted using mfold. Unless otherwise stated, in this figure and elsewhere, all purine nucleotides (G, A) have 2′-OH, and all pyrimidine nucleotides (U, C) are 2′-F modified. (b) Representative BIAcore sensorgram overlay showing binding of aptamers B40t77 (solid line), B40t77iii_4 (dashed line) and B40t77iv_4 (dotted line) to gp120. In this and subsequent figures binding responses were normalized by subtracting the response from a blank flow cell, the duration of the injection is indicated by the horizontal bar and the 600 s time-point is indicated by an arrow. (c) Aptamer binding responses to gp120 obtained in (b). Here and subsequently responses observed 600 seconds after the start of injection are expressed relative to the binding of parental aptamer B40t77 following subtraction of the response in the blank flow cell, and normalized for the molecular mass of each aptamer. Error bars represent the standard deviation of three replicates.
Chemical synthesis and stabilization of truncated aptamer B40 derivatives
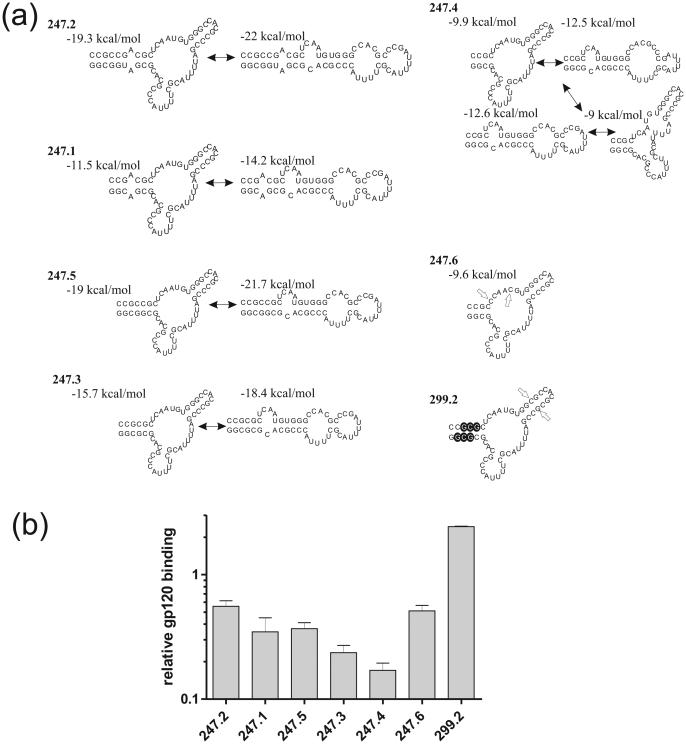
As earlier foot printing results had indicated that the terminal stem of aptamer B40 was not in contact with gp120, but simply required to stabilize its structure, we undertook the design and synthesis of an extensive series of derivatives of B40 in which we aimed to minimize the size of this stem while maintaining its stabilizing effect (Figure 2a). The shortest derivative that we succeeded in producing by in vitro transcription using T7 RNA polymerase, B40t77, possessed a stem 1 comprising 17 base pairs, interrupted by a single A bulge and an A:A mismatch (Figure 1). We used solid phase synthesis to produce derivatives in which the extent of base-paired stem was progressively reduced, and in which the interruptions to the stem were removed (Figure 2a). The data show that the terminal stem can be shortened substantially while retaining good binding activity. However, once the stability of the branched form drops relative to alternate conformers by overly shortening the stem (indicated by an increased number of structures predicted by mfold to have free energies within 5% of the optimal structure), for example with aptamer 247.4, then binding activity also drops (Figure 2b). By introducing additional mutations into the non-contact region of the internal 3-way junction loop of the poorly binding derivative 247.4 (4 base pair stem 1), to generate aptamer 247.6, we hypothesized that the multiple alternative conformers that were predicted to exist in populations of the former would be suppressed. This is supported by the mfold prediction which only showed one structure for 247.6 with a free energy within 5% of the optimal structure (Figure 2b). The increase of gp120 binding resulting from these mutations (Figure 2b) confirmed the functional importance of the branched form.
Figure 2. Binding of truncated aptamers to gp120.
(a) Structures of synthetic aptamers based on B40t77 with truncations in the terminal stem. Predicted structures and their free energies are shown as predicted by mfold. Double-headed arrows indicate conformational interconversions. Note that 247.4 is predicted to form four principal structures, whereas 247.6 and 299.2 are predicted to form just one each. In aptamer 247.6, open arrows mark the positions of residues mutated with respect to aptamer 247.4. In aptamer 299.2, open arrows mark the G:C insertion and the 2′-O-dimethylallyl residues are indicated by white letters on black-filled circles. (b) Aptamer gp120-binding responses obtained by SPR at 600 seconds post-injection (see sensorgrams in Supplementary Figure 2). All aptamers showed a statistically significant change in binding compared to B40t77 using a T-test (P<0.005).
In order to test whether alternative approaches to stabilizing the short stem 1 of synthetic aptamer derivatives of B40 might result in full restoration of binding activity, we took advantage of the properties of 2′-O-dimethylallylribonucleotides (DMA; (Iribarren et al., 1990; Lamond and Sproat, 1993)) This hydrophobic substitution has the useful characteristic that it destabilizes duplexes when incorporated into just one strand, but stabilizes them when incorporated on opposite strands, presumably through hydrophobic interactions across the minor groove of the A-form helix. We found that incorporation of a total of six DMA nucleotides in stem 1, together with the insertion of the additional base pair in stem 2 that stabilizes the helix (aptamer 299.2) increased binding to above the levels seen with B40t77 (Figure 2b).
Role of gp120 variable loops in aptatope exposure
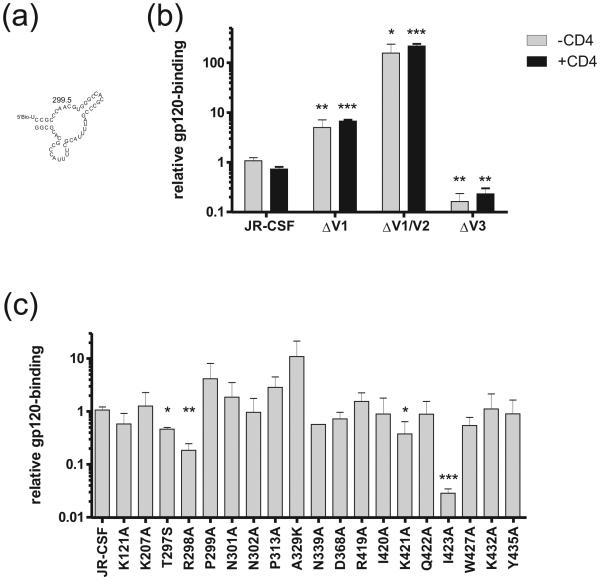
We next sought to determine the aptamer-binding site (aptatope) on gp120 using a novel aptamer-gp120 binding assay similar to an ELISA. This method used aptamer 299.5 (Figure 3a), a 5′-biotinylated version of aptamer 247.6, chosen for its small size and gp120-binding activity (see above). This aptamer was immobilized on a streptavidin-coated plate and serial dilutions of gp120 mutants that were pre-incubated with or without CD4 were added. Bound gp120 was detected with antibody 2G12, which binds to the carbohydrate face of gp120, a secondary HRP-conjugated anti-IgG, and a luminescent substrate. Several optimizations were performed in order to maximize the sensitivity of the assay, and we were finally able to test the aptamer binding to gp120 over a concentration range of four orders of magnitude. Examples of binding curves are shown in Supplementary Figure 3.
Figure 3. Binding of aptamer to mutants of gp120.
Truncation mutants (b) and point mutants (c) of gp120 were tested for binding to aptamer 299.5 (a) in an aptamer-gp120 binding assay. In (b) the gp120 was pre-incubated in the presence or absence of CD4 prior to aptamer binding. EC50 values generated from binding curves are expressed relative to binding to wild type gp120. Mutants whose binding is significantly different from that of the wild-type as determined by a T-test are indicated by asterisks, where *, ** and *** indicate p values of less than 0.05, 0.01 and 0.001 respectively.
Since earlier work had indicated that the aptamer binding site overlaps with the CCR5 binding site on gp120 (Dey et al., 2005b), gp120 truncation and point mutants were selected that had previously been shown to affect binding of CCR5, a sulfated N-terminal CCR5 peptide, or CD4i antibodies 17b, 48d and X5 to gp120 (Cormier and Dragic, 2002; Cormier et al., 2001; Darbha et al., 2004; Rizzuto and Sodroski, 2000; Rizzuto et al., 1998; Thali et al., 1993). Our first approach was to analyse aptamer binding to gp120 molecules with truncations of three of the variable loops.
Figure 3b shows the binding of aptamer 299.5 to gp120 truncation mutants lacking the first (ΔV1), first and second (ΔV1/V2) or third (ΔV3) variable loops, in the presence or absence of CD4 (Figure 3b). The EC50 of aptamer binding was significantly lower to the ΔV1 and ΔV1/V2 mutants compared to the wild-type gp120, indicating an increase in relative binding, whereas the EC50 of aptamer binding to the ΔV3 mutant was substantially increased, corresponding to a large decrease in relative binding. This pattern differs in important respects from that seen in binding experiments using CD4i antibodies and CCR5. For example, although the binding of antibody X5 is also increased by truncation of V1 and V2 simultaneously, no effect is seen following truncation of V1 alone (Labrijn et al., 2003; Moulard et al., 2002). Moreover, truncation of V3 often does not affect the binding of CD4i antibodies. Binding of the aptamer to these truncation mutants and wild-type gp120 was unaffected by the addition of CD4 (Figure 3b), which again is in contrast to that of CCR5 and CD4i antibodies whose binding to gp120 is enhanced by the addition of CD4 (Moulard et al., 2002; Wyatt et al., 1995). These findings show that although the variable loops of gp120 affect the accessibility of the B40 aptatope, their relation to it differs from their relation the CD4i epitope and the CCR5 binding site. Because removal of all three loops simultaneously does not impair aptamer binding (Dey et al., 2005b), we conclude that the aptatope lies largely within the gp120 core and that the V1 and V2 loops most likely occlude it, at least partially, in the native structure of the envelope spike. As the effect of deleting V3 on aptamer binding is different in the presence than in the absence of V1/V2, we could not firmly conclude the putative role of V3 in aptamer binding using truncated loops. Accordingly, we turned to a panel of single-point gp120 mutants in order to define the aptamer binding site more precisely.
Definition of B40 aptatope using gp120 single-point mutants
Figure 3c shows aptamer 299.5 binding to point mutants of gp120. Four of the 19 mutants tested, T297S, R298A, K421A and I423A, showed a significant increase in EC50, corresponding to a decrease in binding relative to wild type. We speculate that the positively charged residues R298 and K421 interact with negatively charged backbone phosphates of the aptamer. In the absence of a co-crystal structure of gp120 with the aptamer, it is difficult to speculate on the manner in which the side chains of residues T297 and I423 are involved in the interaction with the aptamer molecule. We do note that the substantial reduction in binding of the aptamer to mutant I423A indicates that the Ile is critical for the interaction, suggesting direct interaction with the aptamer. Irrespective of the precise mode of interaction, the identification of all four residues as constituting the aptatope provide important clues as to its cross-neutralizing activity (Khati et al., 2003) and a comparison of how these residue substitutions affect CD4i and CCR5 binding allows us to model the aptatope relative to the binding sites of these other ligands. For example, residue K421 is highly conserved amongst all primate immunodeficiency viruses (Kwong et al., 1998); mutation of K421 to alanine does not affect the binding of antibodies X5 or 17b, but does decrease binding of 48d and CCR5 to gp120. Residue R298 binds to the sulphated tyrosines in the N-terminus of CCR5 and is a critical residue for the gp120-CCR5 interaction; mutation of R298 results in a loss of CCR5 binding (Cormier et al., 2001; de Parseval et al., 2005; Rizzuto and Sodroski, 2000) and also the binding of antibody X5 (Darbha et al., 2004), but not 17b or 48d (Thali et al., 1993). Residue I423 is important for the binding of antibodies X5and 17b to gp120 (Darbha et al., 2004; Kwong et al., 1998), but is only somewhat important for CCR5 binding (Rizzuto et al., 1998); whether this residue is important for 48d binding has not been reported (Thali et al., 1993). These observations are consistent with previous mapping studies that show contiguous residues R419, I420, K421 and Q422 as highly important for CCR5 interaction with gp120 (Cormier et al., 2001; Rizzuto and Sodroski, 2000; Rizzuto et al., 1998). Residue T297 in gp120 was not tested in previous studies that used gp120 mutants to map the CCR5, X5, 17b or 48d binding site (Cormier and Dragic, 2002; Cormier et al., 2001; Darbha et al., 2004; Rizzuto and Sodroski, 2000; Rizzuto et al., 1998; Thali et al., 1993). However, the recent structure of gp120 in complex with the CD4i antibody 412d and docking models of the CCR5 N terminus with gp120-CD4 indicate that residue T297 is unlikely to be important for coreceptor binding (Huang et al., 2007).
Neutralization of HIV-1 infectivity by minimal aptamers
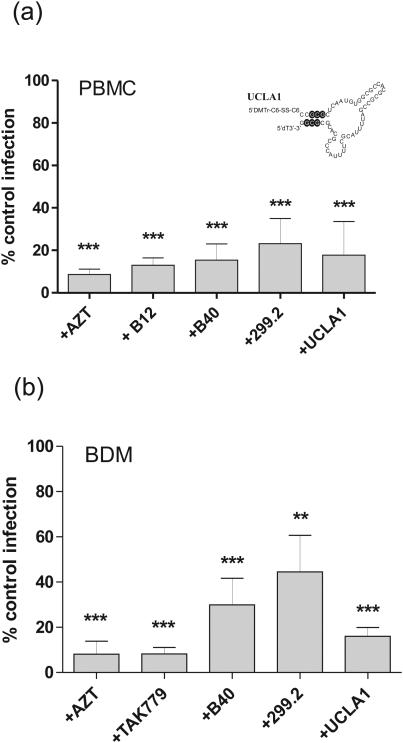
Although some short synthetic aptamers show an increase in gp120 binding activity, when compared to the full length B40 (see above), it was necessary to test whether this is reflected in their ability to limit HIV-1 infectivity. Accordingly, we compared the neutralization activity of the parental aptamer, B40, with that of minimal aptamer 299.2 and a further derivative, UCLA1, in which there is, in addition, an inverted thymidine at the 3’-end (to block degradation by 3’-exonucleases) and a dimethoxyltrityloxy-(CH2)6-SS-(CH2)6-phospho linker at the 5’-end (Figure 4). These modifications were introduced in order to increase the stability of the RNA to degradation by nucleases. The neutralization activity of each aptamer was tested against HIV-1BaL in both peripheral blood mononuclear cells (PBMCs) and blood monocyte derived macrophages (BDMs) at a final concentration of 100nM. We monitored HIV-1 neutralization after a single cycle of infection by using quantitative PCR, which quantifies the number of copies of proviral DNA in each infected cell, in the presence and absence of a potentially neutralizing agent. The neutralizing activity of the aptamers was compared to the neutralizing monoclonal antibody b12 (Burton et al., 1991) and the CCR5 antagonist, TAK 779 (Baba et al., 1999).
Figure 4. Neutralization of HIV-1 by Aptamer.
The ability of aptamers B40 (Khati et al., 2003), 299.2 (Figure 2a) and UCLA1 (inset) to neutralize HIV-1 infection was tested in (a) PBMCs and (b) BDMs isolated from three different donors. Each infection was carried out in biological duplicates. Thirty-six hours post challenge, cellular DNA was extracted and assayed by real time qPCR for HIV-1 late reverse transcription products and the human β-actin gene. These data were expressed as proviral DNA copies per cell. Data from multiple donors, whose cells are found to vary in their susceptibility to HIV-1, were combined by normalizing against the respective positive control samples. Significant neutralization, as determined by the t-test, is indicated as follows: ** (P<0.01) or *** (P<0.001).
Our results indicate that the parental aptamer, B40, reduces virus infectivity by an average of 85% in PBMCs, the minimized aptamer 299.2 by 77%, and the closely-related derivative UCLA1 by 82%. Interestingly, neutralization was generally less potent in BDMs, with a similar but more noticeable order of potency between B40, 299.2 and UCLA1 (70%, 56% and 84%, respectively). All neutralizations were statistically significant in relation to the aptamer-free control, but the potency differences between aptamers did not reach statistical significance (Figure 4).
Discussion
In this study we have demonstrated that the binding site of aptamer 299.5 on gp120, and by extension we presume, that of the parental aptamer B40, involves four key gp120 residues: K421, which is conserved in all primate immunodeficiency viruses (Kwong et al., 1998); R298, which is conserved in >99% of HIV-1 isolates (Kwong et al., 1998); and T297 and I423, which are both moderately conserved among HIV-1 isolates (Kwong et al., 1998). These data, in conjunction with previous evidence (Dey et al., 2005b), indicate that the B40 aptatope on gp120 overlaps most closely with that of the CCR5 N-terminus, thus, explaining the ability of this aptamer to block the infectivity of a broad range of viral isolates. In order to obtain these data, we generated a minimized, synthetic aptamer to serve as a ligand for binding assays, and in doing so were able to confirm the secondary structure of the active form.
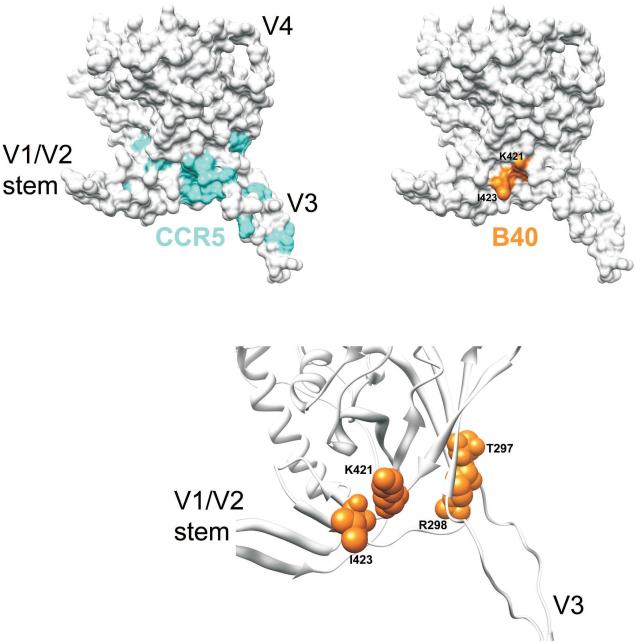
The B40 aptatope, as defined here by alanine-scan mapping, is illustrated in Figure 5 in the context of the structure of a V3-containing gp120 core (Huang et al., 2005). The aptatope overlaps with the location of residues near the base of the V3 loop that have been shown previously to be important for CCR5 binding (Cormier et al., 2001; Rizzuto and Sodroski, 2000). Importantly, these results are in high agreement with a docking model of the CCR5 N terminus with a recent CD4-complexed gp120 core structure {Huang, 2007 #33}. This model suggests that the site of CCR5 N terminus interaction is located in a crevice formed at the base of the V3 loop with the gp120 bridging sheet, which would partially overlap with the B40 aptatope and indicates that the anti-viral activity of the B40 aptamer is due to partial steric blockage of the CCR5 N terminus-gp120 interaction. It is perhaps worth noting here that while the B40 aptatope overlaps with the interaction sites of CCR5 and CD4i antibodies such as 17b or X5, it appears to involve fewer residues. We reason that this is because the aptamer (17 kDa) is likely to have a smaller footprint on gp120 than larger molecules such as CCR5 or an antibody. Moreover, in contrast to these antibody epitopes and the CCR5-binding site, our data suggest that the aptamer binding site may be already fully formed on unliganded gp120. This interpretation is consistent with the known structure of unliganded SIV-1 surface glycoprotein (Chen et al., 2005) but in the absence of equivalent data on HIV-1 gp120 (see review by (Pantophlet and Burton, 2006)) our conclusion must remain provisional.
Figure 5. Aptamer binding site.
Location of gp120 residues important for B40 aptamer binding mapped onto a V3-containing gp120 core structure derived from the primary isolate JR-FL (Huang et al., 2005). Top panel: Surface rendering of gp120JR-FL core+V3 depicting residues identified by mutagenesis as important for CCR5 binding (cyan) (Cormier et al., 2001; Rizzuto and Sodroski 2000; Rizzuto et al., 1998) and residues identified in this study by alanine mutagenesis that significantly reduce aptamer binding (orange). The locations of the V1/V2 stem, V3 and V4 are denoted for orientation purposes. Bottom panel: Ribbon model of gp120JR-FL+V3 depicting a close-up of the residues important for B40 binding (orange; space-filling models).
The mapping of the aptamer-binding site to a core part of the coreceptor-binding site provides a satisfying explanation for the previously described neutralization activity of the parental aptamer (Khati et al., 2003). It was important to determine whether the short, synthetic derivative aptamers, which had been used to accomplish this mapping, retained the ability to inhibit virus infection. We tested this possibility using a newly optimized neutralization assay involving real-time PCR-based quantitation of proviral DNA produced during the first cellular infection cycle. We compared the activity of the full length (117 nt) aptamer B40, with two synthetic derivatives, 299.2 (54 nt) and UCLA1 (55 nt plus a bulky 5′ blocking group) that retain full gp120-binding activity. While aptamer 299.2 exhibited lower neutralization potency relative to B40, this was restored by the addition of a bulky 5′-protecting group and an inverted nucleotide cap at the 3′ in UCLA1.
The difficulties encountered in recent clinical trials of generic microbicides have prompted a reinvestigation of the use of more targeted antiviral agents in microbicide strategies (Cohen, 2008; Padma, 2008). It is possible that aptamers such as B40 and its derivatives have potential in this regard, as they are exquisitely targeted to the virus, while sharing the chemical composition of already proven nucleic acid-based therapeutic agents such as Macugen (for a review, see (James, 2007)). Furthermore, aptamers may prove useful as molecular tools to study the HIV-1 entry process, particularly conformational changes in the envelope spike.
Materials and Methods
Synthesis of aptamer variants and in silico structural analysis
The sequences of the aptamers are given in Figures 1-4. Unless otherwise indicated, all purine nucleotides are standard ribonucleotides and all pyrimidine nucleotides are 2′-F modified. Aptamers B40t77iii_4 and B40t77iv_4 were synthesized by in vitro transcription as previously described (Dey et al., 2005a). Synthetic aptamers were produced by Integrated DNA Technologies, BVBA, Leuven by solid-phase β-cyanoethylphosphoramidite chemistry on a 1 μmole scale, except for UCLA1 which was synthesized on a 52 μmole scale. 5’-O-Dimethoxytrityl-2’-O-dimethylallyl-N4-(4-tert-butylphenoxyacetyl)cytidine-3’-O-(β-cyanoethylphosphoramidite and 5’-O-dimethoxytrityl-2’-O-dimethylallyl-N2-(4-tert-butylphenoxyacetyl)-3’-O-(β-cyanoethylphosphoramidite) were custom synthesized by SAFC Proligo (Hamburg). 5’-O-Dimethoxytrityl-N(tac)-2’-O-TBDMS-3’-O-(β-cyanoethylphosphoramidites) of A and G and 5’-O-dimethoxytrityl-2’-fluoro-2’-deoxy-N4-acetylcytidine-3’-O-(β-cyanoethylphosphoramidite) and 5’-O-dimethoxytrityl-2’-fluoro-2’-deoxyuridine-3’-O-(β-cyanoethylphosphoramidite) were obtained from SAFC Proligo. Polystyrene based solid-phase RNA supports were from GE Healthcare. The thiol-modifier C6 S-S CE phosphoramidite used at the 5’-terminus of UCLA1 was obtained from Glen Research. The synthesis of UCLA1 was performed on an inverted dT polystyrene support loaded at 40 μmoles/g, which was obtained from GE Healthcare.
The aptamers were all synthesized “trityl-on” using 5-benzylmercapto-1H-tetrazole as coupling agent following previously described methods (Sproat, 2005) with some minor modifications. The deprotection of the support was performed with anhydrous 7 M methanolic ammonia for 12 h at room temperature followed by 3 h at 65 °C. When cool the supernatant was removed by filtration, the support was washed 3 times with sterile ethanol/water (1:1 v/v) and the combined supernatants were dried in vacuo at a temperature below 30 °C. Subsequently the standard “trityl-on” desilylation protocol was employed. The precipitated fully deprotected, still “trityl-on” aptamers were purified by reversed phase HPLC and then detritylated, except for UCLA1. UCLA1 was purified on a Source 15 RPC FineLINE 35 column eluted with a gradient of acetonitrile in aqueous 0.1 M ammonium bicarbonate. After desalting and removal of solvent in vacuo all aptamers were repurified and simultaneously converted to their sodium salts by anion-exchange HPLC on Source 15Q, eluting with a linear gradient of aqueous sodium perchlorate. The desired product peaks were desalted by gel filtration and lyophilized. QC was performed by analytical anion-exchange HPLC on a DNAPac PA200, 4 × 250 mm column (Dionex) and by reversed phase HPLC on an XTerra RP8 5 μm, 4.6 × 250 mm column (Waters) and the desired aptamer mass was confirmed by electrospray mass spectroscopy.
The mfold algorithm (Zuker, 2003), implemented on the Rensselaer bioinformatics web server (http://mfold.bioinfo.rpi.edu/) was used to predict optimal and sub-optimal secondary structures and their free energy values for the aptamer sequences. We have previously found the standard parameters adequate to predict the empirically verified structures of aptamer B40 (Dey et al., 2005a).
SPR binding assays
Aptamer binding to gp120 by BIAcore surface plasmon resonance (SPR) was performed essentially as described (Dey et al., 2005a; Dey et al., 2005b). Approximately 5000 units of gp120 were coupled to the surface of flow cell 1 of a CM5 chip using amine coupling chemistry, and flow cell 2 was left blank as a control. Aptamers were refolded in CHBS buffer at 1 μM and were injected over flow cells 1 and 2 for 7 minutes at 5 μL/min, then allowed to dissociate for 10 minutes. The chip was regenerated by injection of 5 μL 10 mM NaOH then flow of buffer for 10 minutes, and this did not affect subsequent aptamer binding (data not shown). Each aptamer was injected three times and the order of injection was randomized. Analysis was performed using BIAevaluation software and the binding response at 600 s, following subtraction of the curve from the control flow cell, was noted. This time point was chosen to coincide with maximal aptamer binding just after the injection was completed. Since SPR response correlates with molecule size, the aptamer binding responses were normalized for the aptamer molecular mass and are expressed as binding relative to B40t77, where B40t77 response was normalized to 1 for each set of experiments. As earlier attempts to obtain detailed kinetic data of aptamer-gp120 binding by SPR were unsuccessful, we pursued the semi-quantitative approach described above.
Generation of wild-type and mutant gp120s
Expression of HIV-1BaL gp120 from a baculovirus vector in Sf9 insect cells was performed as described (Dey et al., 2005a; Dey et al., 2005b). Additional recombinant HIV-1BaL gp120 was obtained from the AIDS Research and Reference Reagent Program, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, MD (NIH ARRRP) (#4961).
HIV-1 particles competent for a single round of infection and pseudotyped with JR-CSF Env containing truncation or point mutations were generated as described previously (Pantophlet et al., 2003). 293T cells were transiently transfected with pNL4.3.Luc.R-.E- and pSVIII-Env using FuGENE 6 (Roche), according to manufacturer’s instructions. After 48 hours, viral particles were lysed with the addition of 1 % Empigen to supernatants. The concentration of gp120 in pseudovirus samples was estimated by quantitative Western blot detected with monoclonal antibody 2G12 (NIH ARRRP) at 1 μg/mL and peroxidase-conjugated anti-human IgG secondary antibody (Sigma) and developed by enhanced chemiluminescence (GE Healthcare). The film was used to generate an electronic image and sample concentrations were estimated according to band density in comparison to standards, using ImageQuant™ software version 5.2 (GE Healthcare).
Aptamer-gp120 binding assay
A novel binding assay was developed in order to investigate the interaction of aptamer 299.5 (a 5′-biotinylated, synthetic derivative of B40, see Figure 3a) with various gp120 mutants. All steps were performed at room temperature except where indicated, reaction volumes were 100 μ L, and 3-5 washes with 0.05 % Tween-20 in PBS were performed between the additions of each reagent. Aptamer 299.5 was refolded as previously described (Dey et al., 2005a) in CHBS buffer to a final concentration of 100 nM and was added to Reacti-bind HBC streptavidin-coated 96-well plates (Pierce) for 30 minutes. A blocking solution of 0.8 μM biotin and 1% BSA in PBS was added for 30 min. Serially-diluted pseudovirus-derived gp120 was pre-incubated in the presence or absence of 100 nM soluble hCD4 for 1 hour at 37 °C, and then added to the reaction plate for at least two hours. Gp120 was detected with antibody 2G12 at 1 μg/mL in CHBS for 1 hour followed by HRP-conjugated anti-human IgG (Sigma) diluted 1/1000 in CHBS for 1 hour. Aptamer-gp120 interaction was detected by the addition of SuperSignal luminescent substrate (Pierce) and was recorded using a luminometer. Luminescence was plotted against the log of gp120 concentration and binding was modeled on a sigmoidal dose-response curve using GraphPad Prism (GraphPad Software, Inc. CA). From this model EC50 values were obtained for each mutant, and these values were transformed using (EC50mutant/EC50WT) to calculate binding relative to wild-type. Each Env mutant was analyzed in at least two independent assays.
Cultivation of human PBMCs and BDMs.
Peripheral blood mononuclear cells (PBMCs) and blood monocyte-derived macrophages (BDMs) were isolated by Ficoll-Hypaque (Pharmacia-Amersham) density gradient centrifugation from heparinized buffy coats of normal, HIV-negative donors. The diluted, autologous plasma was saved, heat-inactivated, and clarified to provide autologous serum (AS) supplement for the culture. The PBMCs were washed six times in PBS (Sigma) at 4 °C and were essentially free of platelets and granulocytes. PBMC cultures were cultivated without mitogen-activation or exogenous interleukin-2 (IL-2). The cells were maintained in X-VIVO 10 (BioWhittaker) containing 2 % AS. BDMs were isolated by CD14+ selection using anti-CD14 magnetic beads (Miltenyi Biotec Ltd) according to the manufacturer’s protocol. CD14+ cells were then plated in 12-well plates at a density of 5x105cells/well. These cells were cultured for 7 days in RPMI 1640 media supplemented with 10 % FCS and 50 ng/ml of recombinant human M-CSF (R&D Systems) growth factor until they were fully attached and differentiated into macrophages.
Virus stock
HIV-1BaL strain used in this study was obtained through the NIH ARRRP AIDS Research and Reference Reagent Program, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, MD. Virus was grown in human PBMC cultures, harvested and centrifuged at 3,000 rpm for 5 min to remove any cellular debris. The collected supernatant was then filtered through a 0.45 μm filter, and treated with DNase I (Sigma-Aldrich) to remove any traces of cellular DNA. Aliquots were stored at −130 °C.
Neutralization assays
RNA aptamers (1 μM in CHBS) were refolded as previously described (Dey et al., 2005a) and kept on ice. Virus (HIV-1BaL; 105 infectious units/mL) was incubated with aptamer (100 nM) for 45 min at room temperature in a total volume of 500 μL. Positive neutralization control was the neutralizing anti-gp120 monoclonal antibody, b12 (Burton et al., 1991) (reagent #2640 from the NIH ARRRP; 10 μg/mL). For comparison, the CCR5 antagonist, TAK 779 (Baba et al., 1999) (reagent #4983 from the NIH ARRRP; 300 nM) was pre-incubated with target cells for 1 hour before virus challenge.
PBMCs and BDMs were infected in 12-well plates with 500 μL of DNase I-treated HIV-1BaL in the presence or absence of inhibitor. Virus was spinoculated onto the cells by centrifugation at 2000 × g for 90 min at 4 °C. Cells were then incubated for a further 30 min at 37 °C, before removal of the inoculum and replacement with fresh media. The infections were left to proceed for 36 h, after which the cells were harvested by scraping. DNA was extracted using DNeasy Blood and Tissue Kit (Qiagen) according to the manufacturer’s instructions. Real-time PCR using primers MH531 and MH532, and the fluorescent probe, LRT-P (Butler, Hansen, and Bushman, 2001), was used to quantify late stage HIV-1 reverse transcripts. The conditions for qPCR comprised forty cycles, each comprising 1.5 min annealing and extension at 58 °C. In parallel, real-time PCR quantitation of the host gene, β-actin was performed using the control kit, RT-CKYD-ACTB (Eurogentec) in order to normalize for cell numbers. Quantitative PCR master mix and methods were from Eurogentec, and equipment from MJ Research (Chromo4™ PTC-200).
Supplementary Material
Supplementary figure 1. Uninformative structure-testing mutants.
(a) Structures of aptamer pairs B40t77i_2 and B40t77ii_2 (i) and B40t77viii_5 and B40t77vii_5 (ii) and their free energies as predicted using mfold. Deletions and substitutions from the parental apatmer, B40t77, are indicated. (b) Representative BIAcore sensorgram overlays showing binding of aptamers (i) B40t77 (solid line), B40t77i_2 (dashed line), B40t77ii_2 (dotted line) and (ii) B40t77 (solid line) B40t77viii_5 (dashed line) and B40t77vii_5 (dotted line) to gp120. (c) Aptamer gp120-binding responses obtained by SPR at 600 seconds post-injection.
Supplementary figure 2. SPR analysis of binding of synthetic aptamer derivatives to gp120.
Representative BIAcore sensorgram overlays showing binding of synthetic aptamers (i) 247.2, (ii) 247.1, (iii) 247.5, (iv) 247.3, (v) 247.4, (vi) 247.6 and (vii) 299.2 (dotted line) compared to B40t77 (solid line) to gp120.
Supplementary figure 3. Representative raw data from the aptamer-gp120 binding assay.
Representative data showing binding of aptamer 299.5 to wild-type gp120 (●) and mutants T297S (■) and P299A (▲) by aptamer-gp120 binding assay. Non-linear regression curves were fitted to a sigmoidal-dose response with variable slope using parameters bottom = 0; top = 100 and are depicted with a solid, dashed and dotted lines for wild-type, T297S and P299A gp120s respectively.
Acknowledgements
This work was funded by the Medical Research Council (grant # G9826944 to WJ), the American Foundation for AIDS Research (grant # 106603-36-RGGN to WJ) and NIH grants 5U01AI066734-03 (to IMcG, WJ and BS) and AI33292 (to DRB).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Baba M, Nishimura O, Kanzaki N, Okamoto M, Sawada H, Iizawa Y, Shiraishi M, Aramaki Y, Okonogi K, Ogawa Y, Meguro K, Fujino M. A small-molecule, nonpeptide CCR5 antagonist with highly potent and selective anti-HIV-1 activity. Proc Natl Acad Sci U S A. 1999;96(10):5698–703. doi: 10.1073/pnas.96.10.5698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton DR, Barbas CF, 3rd, Persson MA, Koenig S, Chanock RM, Lerner RA. A large array of human monoclonal antibodies to type 1 human immunodeficiency virus from combinatorial libraries of asymptomatic seropositive individuals. Proc Natl Acad Sci U S A. 1991;88(22):10134–7. doi: 10.1073/pnas.88.22.10134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler SL, Hansen MS, Bushman FD. A quantitative assay for HIV DNA integration in vivo. Nat Med. 2001;7(5):631–4. doi: 10.1038/87979. [DOI] [PubMed] [Google Scholar]
- Chen B, Vogan EM, Gong H, Skehel JJ, Wiley DC, Harrison SC. Structure of an unliganded simian immunodeficiency virus gp120 core. Nature. 2005;433(7028):834–41. doi: 10.1038/nature03327. [DOI] [PubMed] [Google Scholar]
- Cohen J. AIDS research. Microbicide fails to protect against HIV. Science. 2008;319(5866):1026–7. doi: 10.1126/science.319.5866.1026b. [DOI] [PubMed] [Google Scholar]
- Cormier EG, Dragic T. The crown and stem of the V3 loop play distinct roles in human immunodeficiency virus type 1 envelope glycoprotein interactions with the CCR5 coreceptor. J Virol. 2002;76(17):8953–7. doi: 10.1128/JVI.76.17.8953-8957.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cormier EG, Tran DN, Yukhayeva L, Olson WC, Dragic T. Mapping the determinants of the CCR5 amino-terminal sulfopeptide interaction with soluble human immunodeficiency virus type 1 gp120-CD4 complexes. J Virol. 2001;75(12):5541–9. doi: 10.1128/JVI.75.12.5541-5549.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darbha R, Phogat S, Labrijn AF, Shu Y, Gu Y, Andrykovitch M, Zhang MY, Pantophlet R, Martin L, Vita C, Burton DR, Dimitrov DS, Ji X. Crystal structure of the broadly cross-reactive HIV-1-neutralizing Fab X5 and fine mapping of its epitope. Biochemistry. 2004;43(6):1410–7. doi: 10.1021/bi035323x. [DOI] [PubMed] [Google Scholar]
- de Parseval A, Bobardt MD, Chatterji A, Chatterji U, Elder JH, David G, Zolla-Pazner S, Farzan M, Lee TH, Gallay PA. A highly conserved arginine in gp120 governs HIV-1 binding to both syndecans and CCR5 via sulfated motifs. J Biol Chem. 2005;280(47):39493–504. doi: 10.1074/jbc.M504233200. [DOI] [PubMed] [Google Scholar]
- Dey AK, Griffiths C, Lea SM, James W. Structural characterization of an anti-gp120 RNA aptamer that neutralizes R5 strains of HIV-1. Rna. 2005a;11(6):873–84. doi: 10.1261/rna.7205405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey AK, Khati M, Tang M, Wyatt R, Lea SM, James W. An Aptamer That Neutralizes R5 Strains of Human Immunodeficiency Virus Type 1 Blocks gp120-CCR5 Interaction. J. Virol. 2005b;79(21):13806–13810. doi: 10.1128/JVI.79.21.13806-13810.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CC, Lam SN, Acharya P, Tang M, Xiang SH, Hussan SS, Stanfield RL, Robinson J, Sodroski J, Wilson IA, Wyatt R, Bewley CA, Kwong PD. Structures of the CCR5 N terminus and of a tyrosine-sulfated antibody with HIV-1 gp120 and CD4. Science. 2007;317(5846):1930–4. doi: 10.1126/science.1145373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CC, Tang M, Zhang MY, Majeed S, Montabana E, Stanfield RL, Dimitrov DS, Korber B, Sodroski J, Wilson IA, Wyatt R, Kwong PD. Structure of a V3-containing HIV-1 gp120 core. Science. 2005;310(5750):1025–8. doi: 10.1126/science.1118398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iribarren AM, Sproat BS, Neuner P, Sulston I, Ryder U, Lamond AI. 2'-O-alkyl oligoribonucleotides as antisense probes. Proc Natl Acad Sci U S A. 1990;87(19):7747–51. doi: 10.1073/pnas.87.19.7747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James W. Aptamers in the virologists’ tool kit. J Gen Virol. 2007;2007(88):351–364. doi: 10.1099/vir.0.82442-0. [DOI] [PubMed] [Google Scholar]
- Khati M, Schuman M, Ibrahim J, Sattentau Q, Gordon S, James W. Neutralization of infectivity of diverse R5 clinical isolates of human immunodeficiency virus type 1 by gp120-binding 2'F-RNA aptamers. J Virol. 2003;77(23):12692–8. doi: 10.1128/JVI.77.23.12692-12698.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong PD, Wyatt R, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393(6686):648–59. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labrijn AF, Poignard P, Raja A, Zwick MB, Delgado K, Franti M, Binley J, Vivona V, Grundner C, Huang CC, Venturi M, Petropoulos CJ, Wrin T, Dimitrov DS, Robinson J, Kwong PD, Wyatt RT, Sodroski J, Burton DR. Access of antibody molecules to the conserved coreceptor binding site on glycoprotein gp120 is sterically restricted on primary human immunodeficiency virus type 1. J Virol. 2003;77(19):10557–65. doi: 10.1128/JVI.77.19.10557-10565.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamond AI, Sproat BS. Antisense oligonucleotides made of 2'-O-alkylRNA: their properties and applications in RNA biochemistry. FEBS Lett. 1993;325(1-2):123–7. doi: 10.1016/0014-5793(93)81427-2. [DOI] [PubMed] [Google Scholar]
- Moulard M, Phogat SK, Shu Y, Labrijn AF, Xiao X, Binley JM, Zhang MY, Sidorov IA, Broder CC, Robinson J, Parren PW, Burton DR, Dimitrov DS. Broadly cross-reactive HIV-1-neutralizing human monoclonal Fab selected for binding to gp120-CD4-CCR5 complexes. Proc Natl Acad Sci U S A. 2002;99(10):6913–8. doi: 10.1073/pnas.102562599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padma TV. After microbicide failures, hope that antiviral approach will gel. Nat Med. 2008;14(4):354. doi: 10.1038/nm0408-354a. [DOI] [PubMed] [Google Scholar]
- Pantophlet R, Burton DR. GP120: target for neutralizing HIV-1 antibodies. Annu Rev Immunol. 2006;24:739–69. doi: 10.1146/annurev.immunol.24.021605.090557. [DOI] [PubMed] [Google Scholar]
- Pantophlet R, Ollmann Saphire E, Poignard P, Parren PW, Wilson IA, Burton DR. Fine mapping of the interaction of neutralizing and nonneutralizing monoclonal antibodies with the CD4 binding site of human immunodeficiency virus type 1 gp120. J Virol. 2003;77(1):642–58. doi: 10.1128/JVI.77.1.642-658.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzuto C, Sodroski J. Fine definition of a conserved CCR5-binding region on the human immunodeficiency virus type 1 glycoprotein 120. AIDS Res Hum Retroviruses. 2000;16(8):741–9. doi: 10.1089/088922200308747. [DOI] [PubMed] [Google Scholar]
- Rizzuto CD, Wyatt R, Hernandez-Ramos N, Sun Y, Kwong PD, Hendrickson WA, Sodroski J. A conserved HIV gp120 glycoprotein structure involved in chemokine receptor binding. Science. 1998;280(5371):1949–53. doi: 10.1126/science.280.5371.1949. [DOI] [PubMed] [Google Scholar]
- Scanlan CN, Pantophlet R, Wormald MR, Ollmann Saphire E, Stanfield R, Wilson IA, Katinger H, Dwek RA, Rudd PM, Burton DR. The broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2G12 recognizes a cluster of alpha1-->2 mannose residues on the outer face of gp120. J Virol. 2002;76(14):7306–21. doi: 10.1128/JVI.76.14.7306-7321.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sproat BS. RNA synthesis using 2'-O-(tert-butyldimethylsilyl) protection. Methods Mol Biol. 2005;288:17–32. doi: 10.1385/1-59259-823-4:017. [DOI] [PubMed] [Google Scholar]
- Thali M, Moore JP, Furman C, Charles M, Ho DD, Robinson J, Sodroski J. Characterization of conserved human immunodeficiency virus type 1 gp120 neutralization epitopes exposed upon gp120-CD4 binding. J Virol. 1993;67(7):3978–88. doi: 10.1128/jvi.67.7.3978-3988.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt R, Moore J, Accola M, Desjardin E, Robinson J, Sodroski J. Involvement of the V1/V2 variable loop structure in the exposure of human immunodeficiency virus type 1 gp120 epitopes induced by receptor binding. J Virol. 1995;69(9):5723–33. doi: 10.1128/jvi.69.9.5723-5733.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31(13):3406–15. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figure 1. Uninformative structure-testing mutants.
(a) Structures of aptamer pairs B40t77i_2 and B40t77ii_2 (i) and B40t77viii_5 and B40t77vii_5 (ii) and their free energies as predicted using mfold. Deletions and substitutions from the parental apatmer, B40t77, are indicated. (b) Representative BIAcore sensorgram overlays showing binding of aptamers (i) B40t77 (solid line), B40t77i_2 (dashed line), B40t77ii_2 (dotted line) and (ii) B40t77 (solid line) B40t77viii_5 (dashed line) and B40t77vii_5 (dotted line) to gp120. (c) Aptamer gp120-binding responses obtained by SPR at 600 seconds post-injection.
Supplementary figure 2. SPR analysis of binding of synthetic aptamer derivatives to gp120.
Representative BIAcore sensorgram overlays showing binding of synthetic aptamers (i) 247.2, (ii) 247.1, (iii) 247.5, (iv) 247.3, (v) 247.4, (vi) 247.6 and (vii) 299.2 (dotted line) compared to B40t77 (solid line) to gp120.
Supplementary figure 3. Representative raw data from the aptamer-gp120 binding assay.
Representative data showing binding of aptamer 299.5 to wild-type gp120 (●) and mutants T297S (■) and P299A (▲) by aptamer-gp120 binding assay. Non-linear regression curves were fitted to a sigmoidal-dose response with variable slope using parameters bottom = 0; top = 100 and are depicted with a solid, dashed and dotted lines for wild-type, T297S and P299A gp120s respectively.