Abstract
Reprogrammed glucose metabolism as a result of increased glycolysis and glucose uptake is a hallmark of cancer. Here we show that cancer cells can suppress glucose uptake by non-tumour cells in the pre-metastatic niche, by secreting vesicles that carry high levels of the miR-122 microRNA. High miR-122 levels in the circulation have been associated with metastasis in breast cancer patients and we show that cancer-cell-secreted miR-122 facilitates metastasis by increasing nutrient availability in the pre-metastatic niche. Mechanistically cancer-cell-derived miR-122 suppresses glucose uptake by niche cells in vitro and in vivo by downregulating the glycolytic enzyme pyruvate kinase (PKM). In vivo inhibition of miR-122 restores glucose uptake in distant organs, including brain and lungs, and decreases the incidence of metastasis. These results demonstrate that by modifying glucose utilization by recipient pre-metastatic niche cells, cancer-derived extracellular miR-122 is able to reprogram systemic energy metabolism to facilitate disease progression.
Reprogrammed energy metabolism to fuel rapid cell growth and proliferation is an emerging hallmark of cancer1. Most cancer cells use aerobic glycolysis with reduced mitochondrial oxidative phosphorylation for glucose metabolism even when oxygen is sufficient. This phenomenon, known as the “Warburg effect”, favours the uptake and incorporation of nutrients needed to produce a new cell2. To compensate for the consequent reduction in ATP production, cancer cells often adopt mechanisms to increase glucose uptake and utilization. One mechanism involves the regulation of glucose transporters, among which GLUT1 is responsible for basal levels of glucose uptake in all cells3. GLUT1 can be regulated by the PI3K/Akt/mTOR pathway which is frequently activated in cancer4, 5. Additionally, hypoxia can stimulate glucose uptake and metabolism through HIF-1 by inducing GLUT3 and glycolytic genes ALDA, PGK1 and PKM 6, 7. It was recently reported that phosphorylation or sumoylation of PKM2 leads to translocation to the nucleus, where it acts as a transcriptional co-activator to induce GLUT1, PDK1 and HK18–13. Here we focus on a mechanism mediated by a cancer-secreted miRNA that reallocates glucose to favour uptake by cancer cells.
MiRNA negatively regulates gene expression by binding to the 3′ untranslated region (3′UTR) of mRNA, leading to degradation or translation blockade14. Deregulation of miRNA is tightly linked to cancer15, and circulating miRNA has emerged as potential biomarkers for cancer diagnosis and prognosis16–19. MiRNA can be secreted into the extracellular environment through membrane-enclosed vesicles (such as exosomes) or in complexes with protein or lipid-based carriers20, 21. Accumulating evidence demonstrates that miRNA as well as proteins can be transferred to neighbouring or distant cells in these secretory forms to modulate cell function22–27. Extracellular miRNA is therefore emerging as a new group of messengers and effectors in intercellular communication.
Several miRNAs have been implicated in metabolism and metabolic disorders28. Among them, miR-122 regulates cholesterol efflux, liver triglyceride content, and the rate of β-oxidation29. Potential miR-122 targets have been analyzed by luciferase reporter-based 3′UTR screening, identifying PKM as one of the targets30, which suggests miR-122 may play a role in glucose metabolism. Our recent study in breast cancer (BC) patients identified higher levels of circulating miR-122 as a marker for predicting metastatic progression in early-stage BC18. This urged us to investigate the function of extracellular miR-122 in cancer progression and metastasis. Here we demonstrate that cancer-secreted miR-122 can be transferred to normal cells in the pre-metastatic niches, thereby suppressing glucose utilization in these cells to accommodate the massive energy needs of cancer cells during metastatic growth.
RESULTS
MiR-122 is highly secreted by cancer cells
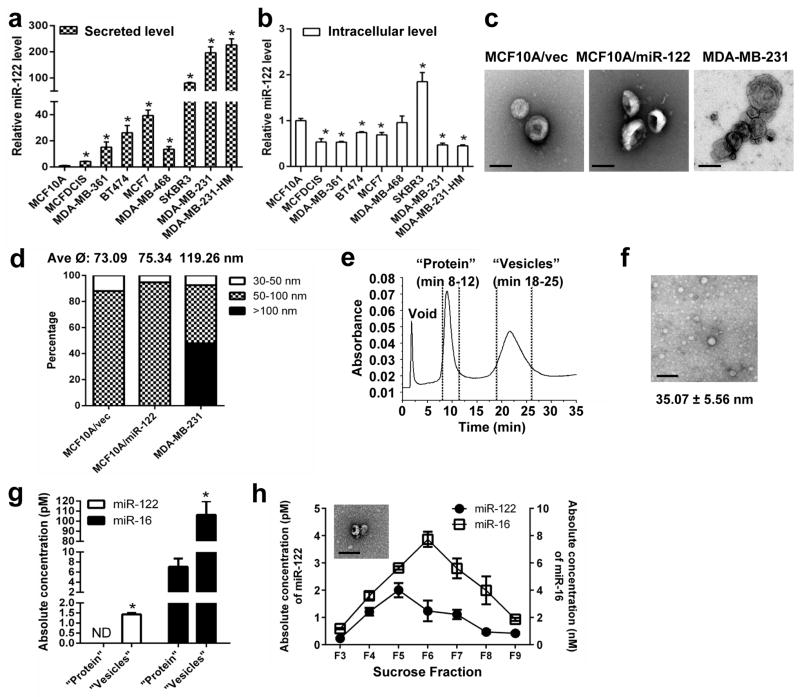
We first examined the conditioned media of various breast cell lines for miR-122 secretion. We focused on the 110,000 ×g medium pellet that is known to contain extracellular vesicles (EVs) including exosomes and that carried the majority of extracellular miR-122 compared to the supernatant fraction (Supplementary Fig. 1a). All BC lines secreted significantly elevated miR-122 compared to non-cancerous MCF10A (Fig. 1a). This was not accompanied by an elevated intracellular level, as most cancer lines exhibited reduced intracellular miR-122 (Fig. 1b). While MCF10A-derived vesicles all exhibited a diameter of 30–100 nm representing exosomes, vesicles from the BC line MDA-MB-231 were more heterogeneous and contained >50% exosomes with the rest being microvesicles larger than 100 nm (Fig. 1c–d), consistent with a previous study31. Further characterization of the medium pellet by asymmetrical flow field flow fractionation (AF4)32 revealed two peaks representing proteins (eluted at 8–11 min) and vesicles (eluted at 18–25 min) measuring 30–60 nm (averaged ≈5 nm for MDA-MB-231), but lack of high-density lipoproteins (HDL; eluted at 11–16 min) (Fig. 1e–f, Supplementary Fig. 1b, 32). For MDA-MB-231, miR-122 was exclusively detected in the vesicle but not the protein fraction, whereas forced overexpression of miR-122 in MCF10A increased miR-122 secretion predominantly in vesicles with a slight induction also detected in protein-associated form (Fig. 1g, Supplementary Fig. 1c). Secretion of miR-122 by MCF10A/vec was below the detection limit in fractionated samples. By gradient centrifugation of the medium pellet we further determined that for both MDA-MB-231 and MCF10A-derived lines, miR-122 and miR-16 peaked in fractions 5–6 that contained vesicles measuring 30–100 nm (Fig. 1h, Supplementary Fig. 1d–e). Overall, our results indicate that cancer cells specifically secrete high levels of miR-122 into EVs including exosomes, and suggest that the potential effect of cancer-derived miR-122 may be ectopically observed in the recipient cells upon EV-mediated transfer rather than in the cancer cells producing it.
Figure 1.
MiR-122 is highly secreted by cancer cells. RNA were extracted from the 110,000 ×g medium pellet (a) and PBS-washed cells (b) and analysed for miR-122 by RT-qPCR. Data was normalized to levels of total proteins (secreted; a) or U6 (cellular; b), and compared to the non-tumour line MCF10A (n = 6 extracts). (c) Representative EM images of vesicles in the 110,000 ×g medium pellet. Bar equals 100 nm. (d) Size distribution of vesicles identified in the 110,000 ×g medium pellets (n = 25 vesicles for MCF10A/vec; n = 38 for MCF10A/miR-122; n = 94 for MDA-MB-231). (e) Fractogram (UV absorption at 280 nm) for the AF4 eluates characterizing the MDA-MB-231 110,000 ×g medium pellet. (f) A representative EM image of MDA-MB-231-derived vesicles in the fraction eluted at 18–25 min. The measured diameter of vesicles was shown as mean ± SD (n = 41). Bar equals 100 nm. (g) RT-qPCR-determined levels of miR-122 and miR-16 in MDA-MB-231-derived protein and vesicle fractions separated by AF4 (n = 6 extracts). Absolute miRNA levels are calculated based on standard curves. ND: not detected. (h) After sucrose gradient centrifugation of MDA-MB-231-derived 110,000 ×g medium pellet, absolute miRNA levels in each gradient fraction was determined by RT-qPCR and calculated based on standard curves (n = 6 extracts). A representative EM image of MDA-MB-231-derived vesicles in sucrose fraction 5 (F5) is shown. Bar equals 100 nm. * p < 0.05 for all panels derived from Kruskal-Wallis test. Data are represented as mean ± SD in all panels except (c–e).
MiR-122 suppresses glucose metabolism by downregulating PKM
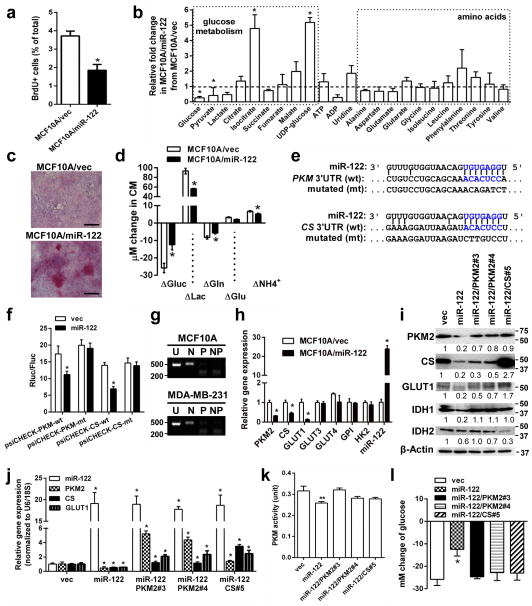
To study the function of miR-122, we first used MCF10A cells engineered to stably overexpress miR-122 (MCF10A/miR-122) or the control vector (MCF10A/vec). MCF10A/miR-122 had significantly reduced proliferation measured by BrdU incorporation (Fig. 2a). Metabolome analysis of the cells revealed significantly decreased intracellular glucose and pyruvate in MCF10A/miR-122 (Fig. 2b, Supplementary Fig. 2), along with increased UDP-glucose (Fig. 2b) and glycogen staining (Fig. 2c) that is likely due to the excessive glucose spared from glycolysis going towards storage. In contrast, levels of amino acids were not significantly altered by miR-122. ATP level in MCF10A/miR-122 was not significantly changed, which may reflect an adaptation of other metabolic pathways to meet the energy demand. Analyses of metabolites in the culture media of MCF10A/miR-122 indicated ~50% decreased glucose uptake and ~40% reduced lactate production (Fig. 2d). Modestly diminished glutamine metabolism was also observed in these cells, possibly reflecting cell adaptation for the altered overall metabolic rate.
Figure 2.
MiR-122 suppresses glucose metabolism by downregulating PKM. (a) BrdU uptake in indicated cells were analysed by flow cytometry (n = 6 biological replicates). (b) Quantification of intracellular metabolites by NMR spectroscopy (n = 3 biological replicates). (c) Glycogen staining (red) in MCF10A/vec and MCF10A/miR-122 cells. Bar equals 100 μm. (d) Change of metabolites in the media after 72 h of culture (n = 6 biological replicates). (e) The predicted miR-122 binding site in the 3′UTR of the human PKM and CS gene. The corresponding sequence in the mutated (mt) version is also shown. (f) The psiCHECK reporters containing 3′UTR of human PKM and CS gene with wild-type (wt) or mutated (mt) miR-122 binding site were used to transfect MCF10A cells stably expressing miR-122 or the empty vector (as control). Luciferase activity was analysed at 48 h post-transfection (n = 6 extracts). (g) Determination of PKM isoforms expressed in MCF10A and MDA-MB-231. RNA was subjected to RT-PCR followed by digestion with NcoI (N), PstI (P), or both enzymes (NP), plus an uncut control (U). Products were separated on an agarose gel with Sybr safe. The presence of a PstI digestion site indicates the splicing isoform M2 whereas the NcoI site indicates isoform M1. Size of markers (in bp) are indicated. (h) RT-qPCR analysis showing the relative expression of indicated genes in MCF10A/miR-122 and MCF10A/vec cells (n = 6 extracts). (i) Western blot analysis in MCF10A/miR-122 and MCF10A/vec cells with restored expression of PKM2 and CS. Size of markers (in kDa) are indicated. (j) RT-qPCR analysis in selected colonies with restored expression of PKM2 and CS (n = 6 extracts). (k) PKM activity (unit) in 5 μg proteins in indicated cells (n = 6 extracts). (l) Change of glucose in the media after 72 h of culture of selected clones normalized to cell number (n = 3 biological replicates). * p < 0.05, ** p < 0.01 for all panels derived from Kruskal-Wallis test. Data are represented as mean ± SD in all panels except (c, e, g, i). Uncropped images of blots and gels are shown in Supplementary Fig. 5.
The metabolomic changes observed in MCF10A/miR-122 suggested a role of miR-122 in glucose metabolism. TargetScan and microRNA.org algorithms predicted a single, species-conserved miR-122 binding site in the 3′UTRs of pyruvate kinase (PKM) and citrate synthase (CS) genes (Fig. 2e). Therefore, we PCR-cloned the 3′UTRs and their seed-sequence-mutated version downstream to the ORF of a Renilla luciferase reporter gene and assessed the ability of miR-122 to downregulate luciferase expression. For both PKM and CS, the wild-type but not mutated 3′UTR responded to miR-122 by directing ~50% reduction of reporter gene expression (Fig. 2f). Among a panel of genes controlling glucose metabolism, MCF10A/miR-122 exhibited significantly reduced PKM2 (isoform determined in Fig. 2g), CS, and GLUT1 (Fig. 2h–j). Consistent with PKM2 downregulation, miR-122 also caused a significant reduction of PKM enzymatic activity (Fig. 2k).
To further determine if the miR-122-induced decrease in glucose consumption was mediated by PKM2 and/or CS downregulation, we restored the expression of these genes in MCF10A/miR-122 by overexpressing the PKM2 or CS cDNA that lacked the 3′UTR. Both clones with fully or partially restored PKM activity (Fig. 2k) showed restored GLUT1 expression and glucose uptake from the media that were comparable to MCF10A/vec (Fig. 2i–j, l). This is consistent with the previously reported ability of PKM2 to induce c-Myc and GLUT1 expression as a nuclear co-activator of β-catenin9. We noticed that restoration of CS by exogenous expression was always accompanied by elevated expression of endogenous PKM2 (Fig. 2i–j), possibly reflecting a natural feedback mechanism to accommodate the increased need for pyruvate by enhanced CS activity. Although this hindered us from dissecting the role of CS downregulation in mediating miR-122’s function without the concomitant regulation of PKM2, our results indicate that restoration of PKM2 alone is sufficient to abolish the effect of miR-122 on glucose uptake; therefore we chose to focus on this miR-122/PKM-mediated effect for the rest of this study.
Cancer-secreted miR-122 downregulates glucose consumption in niche cells
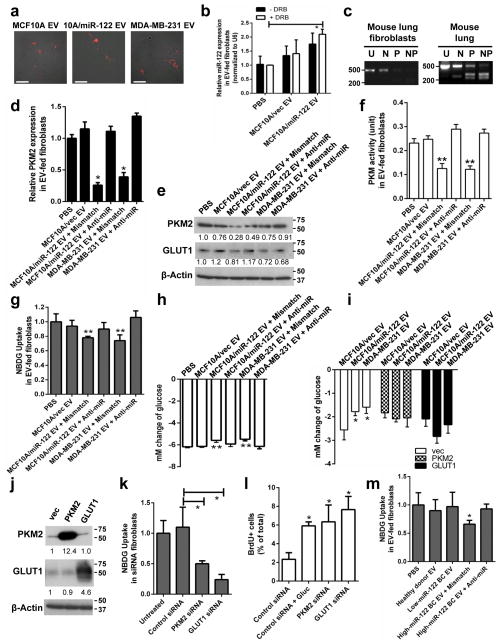
To study the ectopic effect of cancer-secreted miR-122, we focused on lung fibroblasts, brain astrocytes, and neurons that are abundantly present in the pre-metastatic sites of BC. Primary lung fibroblasts exhibited efficient uptake of exosome-containing EVs regardless of the producer cells as indicated by the internalization of DiI-labelled EVs (Fig. 3a). In these cells, EVs that are high in miR-122 caused significantly increased intracellular miR-122, which was not affected by an RNA polymerase II inhibitor (Fig. 3b), indicating that this increase of miR-122 reflects the EV-mediated miRNA transfer but not an induction of miR-122’s endogenous expression. Also observed in high-miR-122 EV-treated fibroblasts were the decreased expression of PKM2 and GLUT1 (Fig. 3c–e), along with decreased PKM activity (Fig. 3f). In addition, high-miR-122 EVs significantly reduced recipient cells’ uptake of 2-NBDG, a fluorescent analogue of glucose which has been used to assess glucose transport in various cell types33–36 (Fig. 3g), as well as their glucose consumption from the medium (Fig. 3h). These effects were significantly suppressed by treating recipient cells with anti-miR-122 (Fig. 3d–h).
Figure 3.
Cancer-secreted miR-122 downregulates glucose uptake in lung fibroblasts. (a) Uptake of DiI-labelled exosome-containing EVs (prepared from the 110,000 ×g medium pellet) at 48 h. Bar equals 60 μm. (b) Levels of miRNAs in fibroblasts pre-treated with 10 μM DRB for 2 h followed by treatment with EVs for 16 h. miR-122 levels were normalized to U6 (n = 6 extracts). (c) Determination of PKM isoforms in fibroblasts and mouse tissues by RT-PCR as indicated in Fig. 2g. Size of markers (in bp) are indicated. (d–h) Fibroblasts were treated with two doses of EVs from indicated producer cells given 48 h apart and transfected with anti-miR-122 oligos or mismatch control oligos, before analysis at 96 h by (d) RT-qPCR (n = 6 extracts), (e) Western blot analysis (with marker size indicated in kDa), (f) PKM activity assay using 5 μg of proteins (n = 5 biological replicates), (g) 2-NBDG uptake (n = 6 biological replicates), and (h) change of glucose in the CM (n = 9 biological replicates). (i) Change of glucose in the CM of fibroblasts transfected with expression plasmids containing the ORF but not 3′UTR of PKM2 or GLUT1, or the empty vector, and treated with EVs from indicated producer cells (n = 5 biological replicates). (j) Western blot of fibroblasts transfected with expression plasmids for PKM2 or GLUT1. Size of markers (in kDa) are indicated. (k) 2-NBDG uptake in siRNA-transfected fibroblasts (n = 5 biological replicates). (l) CM was collected from siRNA-transfected fibroblasts cultured for 72 h and the glucose concentration measured. The CM was then fed to MDA-MB-231-HM cells before proliferation was assessed by BrdU-incorporation at 72 h (n = 6 biological replicates). (m) Circulating EVs were extracted from pooled healthy donor sera and from BC patients’ sera with low or high vesicular miR-122, used to treat fibroblasts and 2-NBDG uptake was measured (n = 8 biological replicates). * p < 0.05, ** p < 0.01 for all panels derived from Kruskal-Wallis test. Data are represented as mean ± SD in all panels except (a, c, e, j). Uncropped images of blots and gels are shown in Supplementary Fig. 5.
Restored expression of PKM2 or GLUT1 by transfecting fibroblasts with the corresponding cDNA construct lacking 3′UTR abolished the effect of high-miR-122 EVs on fibroblast glucose consumption (Fig. 3i–j). In contrast, knockdown of either PKM2 or GLUT1 significantly reduced 2-NBDG uptake (Fig. 3k). We next assessed the effect of fibroblast glucose metabolism on proliferation of cancer cells sharing the same media. Media from fibroblasts treated with siRNA against PKM2 or GLUT1 and therefore contained higher levels of residual glucose significantly enhanced proliferation of MDA-MB-231-HM cells, whereas media collected from control fibroblasts needed to be supplemented with glucose to achieve this effect (Fig. 3l). We further examined the effect of circulating EVs isolated from the sera of healthy donors or BC patients. High-miR-122 EVs derived from a BC patient significantly reduced 2-NBDG uptake in fibroblasts compared to healthy donor- or patient-derived low-miR-122 EVs, and this effect was abolished by treating the fibroblasts with anti-miR-122 (Fig. 3m).
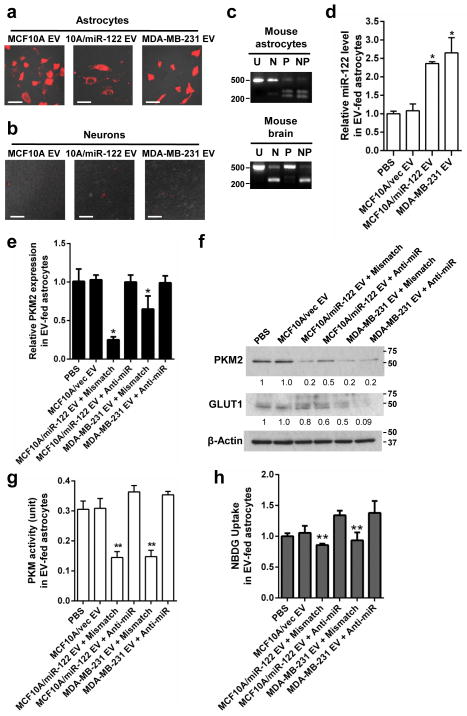
Glucose is the primary energy substrate in the mammalian brain with astrocytes and neurons being the major consumers among other cell types36–38. While primary astrocytes exhibited efficient uptake of EVs (Fig. 4a), neurons differentiated from embryonic neural stem cells exhibited poor EV uptake in vitro (Fig. 4b) and were therefore excluded from the current study. Similar to lung fibroblasts, astrocytes treated with high-miR-122 EVs had significantly increased miR-122 in recipient cells, resulting in significantly decreased expression of PKM2 and GLUT1 (Fig. 4c–f), PKM activity (Fig. 4g), and 2-NBDG uptake (Fig. 4h) in a miR-122-dependent manner. Overall, these in vitro data indicate that vesicular transfer of miR-122 reduces glucose uptake by niche cells through downregulation of PKM2 and GLUT1, leading to enhanced cancer cell proliferation partially mediated by increased glucose availability to cancer cells.
Figure 4.
Cancer-secreted miR-122 downregulates glucose uptake in astrocytes. (a–b) EV uptake by astrocytes and neurons. Indicated cells were incubated with DiI-labelled EVs (red) for 48 h before fluorescent and phase contrast images were captured. Bar equals 60 μm. (c) Determination of PKM isoforms in primary mouse cells and mouse tissues by RT-PCR as indicated in Fig. 2g. Size of markers (in bp) are indicated. (d–h) Primary mouse astrocytes were treated with two doses of EVs from indicated producer cells given 48 h apart, before subjected to (d–e) RT-qPCR at 72 h for miR-122 (n = 6 extracts) (d) and PKM2 (n = 6 extracts) (e), (f) Western blot analysis for PKM2 and GLUT1 at 96 h (with marker size indicated in kDa), (g) PKM activity assay using 10 μg of proteins (n = 5 biological replicates), and (h) 2-NBDG uptake assay (n = 5 biological replicates). * p < 0.05, ** p < 0.01 for all panels derived from Kruskal-Wallis test. Data are represented as mean ± SD in (d–e & g–h). Uncropped images of blots and gels are shown in Supplementary Fig. 5.
Cancer-secreted miR-122 reprograms glucose consumption in niche tissues and promotes metastasis
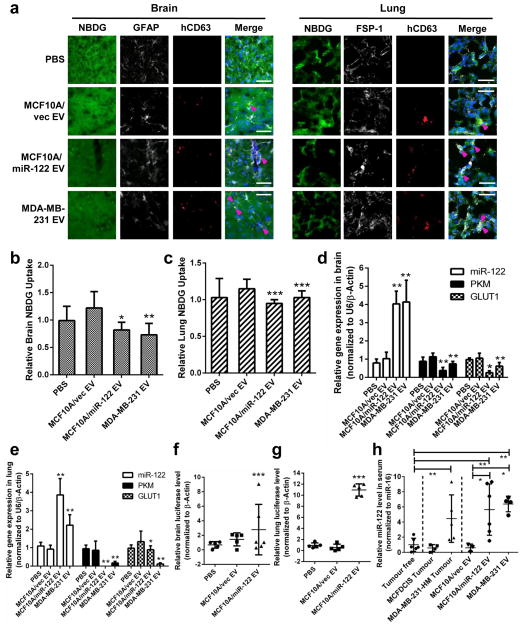
To verify that cancer-secreted miR-122 modulates glucose metabolism in the pre-metastatic niches in vivo, we intravenously injected exosome-containing EVs with low or high levels of miR-122 into mice and measured glucose uptake in brain and lungs. Co-immunofluorescence of cell type specific markers for astrocytes and fibroblasts and a human-specific exosomal marker CD63 demonstrated that human-derived EVs can be received by these niche cell types in vivo (Fig. 5a). Both brain and lungs showed reduced 2-NBDG uptake as a consequence of receiving vesicular miR-122 which resulted in reduced expression of PKM and GLUT1 (Fig. 5b–e).
Figure 5.
Vesicular transfer of miR-122 alters glucose uptake in niche tissues. Indicated EVs were intravenously injected into the tail vein of NSG mice biweekly for 3.5 weeks. (a) Co-immunofluorescence of exosome marker CD63 (detected by a human-specific antibody; red) with astrocyte marker GFAP (white) or fibroblast marker FSP-1 (white) in brain and lung tissues of mice injected with 2-NBDG (green). Nuclei were counterstained with DAPI (blue). White bar represents 20 μm. (b) Quantification of 2-NBDG uptake in brain (n = 20 fields from 4 mice). (c) Quantification of 2-NBDG uptake in lung (n = 20 fields from 4 mice). (d–e) RT-qPCR in brain (d) and lungs (e) (n = 9 extracts from 3 mice). (f–g) Luciferase qPCR for the detection of metastases in the brain (f) and lungs (g) of mice pre-treated with EVs followed by an intracardiac injection with luciferase-labelled MDA-MB-231-HM tumour cells (n = 15 extracts from 5 mice). (h) MiR-122 levels determined by RT-qPCR in the serum of non-tumour bearing mice (group 1), mice bearing MCFDCIS or MDA-MB-231-HM tumours (groups 2–3), and non-tumour-bearing mice treated with EVs from MCF10A/vec, MCF10A/miR-122, or MDA-MB-231 cells (groups 4–6) (n = 15 extracts from 5 mice). Data was normalized to the levels of miR-16. Data are represented as mean ± SD for all panels except (a). * p < 0.05, ** p < 0.01, *** p < 0.001 for all panels derived from Kruskal-Wallis test.
In another experiment, mice were pretreated with EVs before an intracardiac injection of luciferase-labelled MDA-MB-231-HM cells. Three weeks later, metastases in the brain and lungs were quantified by luciferase qPCR and confirmed by histology. Among mice receiving high-miR-122 EVs, all exhibited significant metastatic colonization in lungs and brain, whereas no metastases were observed in mice treated with MCF10A/vec EVs or PBS (Fig. 5f–g). Serum miR-122 levels were comparable between mice received high-miR-122 EVs and those bearing MDA-MB-231-HM tumours that naturally secrete miR-122 (Fig. 5h). Therefore, vesicular miR-122 results in reprogramming of niche tissue glucose utilization as a possible mechanism to promote circulating tumour cell colonization.
MiR-122 overexpression reduces primary tumour growth while enhancing metastasis
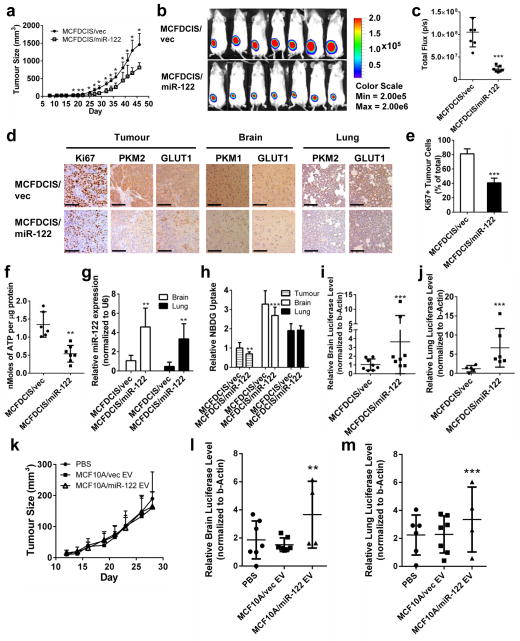
To determine if miR-122 level in primary tumours regulates tumour growth and if the primary tumour-secreted miR-122 adapts the pre-metastatic niches to promote metastasis, we stably overexpressed miR-122 in an MCF10A-derived tumourigenic line MCFDCIS, which forms comedo ductal carcinoma in situ-like lesions that spontaneously progress to invasive tumours39. MiR-122 overexpression reduced cell proliferation, glucose uptake, and the expression of PKM2, CS, and GLUT1 in vitro (Supplementary Fig. 3a–d). Orthotopic xenograft tumours of MCFDCIS/miR-122 were significantly smaller than MCFDCIS/vec tumours (Fig. 6a–c), containing decreased number of Ki67+ tumour cells, reduced expression of PKM2 and GLUT1, and decreased ATP level (Fig. 6d–f).
Figure 6.
In vivo effect of miR-122 on primary tumour growth and metastasis. (a) Tumour growth curve in mice carrying MCFDCIS/vec and MCFDCIS/miR-122 orthotopic xenografts (n = 7 mice). (b) BLI imaging at week 3. (c) Luciferase quantification of (b) (n = 7 mice). (d) IHC for Ki67, PKM1/2, and GLUT1 in tumour, brain, and lung sections. Scale bar is 100 μm. (e) Quantification of Ki67+ tumour cells from 3 fields per tumour (n = 6 mice per group). (f) Intratumoural levels of ATP were assessed in tumour lysates by ENLIGHTEN ATP assay (n = 6 mice per group). (g) miR-122 levels in the brain and lungs determined by RT-qPCR (n = 18 extracts from 6 mice). (h) 2-NBDG uptake quantification in the tumour, brain, and lungs (n = 12 fields from 4 mice per group). (i–j) Luciferase qPCR in the brain (i, n = 24 extracts from 8 mice per group) and lungs (j, n = 18 extracts from 6 mice per group) of MCFDCIS tumour-bearing mice. (k) Tumour growth curve in mice carrying orthotopic xenografts of MDA-MB-231 with stable knockdown of miR-122 (MDA-MB-231/122KD) and also receiving EV treatments as indicated (n = 7 mice per group). No significant difference (p > 0.05) between groups based on Kruskal-Wallis test. (l–m) Luciferase qPCR for the detection of metastases in the brain (l) and lungs (m) of mice bearing MDA-MB-231/122KD tumours and treated with indicated EVs (n = 12 extracts from 4 mice per group). * p < 0.05, ** p < 0.01, *** p < 0.001 for all panels derived from Kruskal-Wallis test. Data are represented as mean ± SD in all panels except (b & d).
Mice bearing MCFDCIS/miR-122 tumours had increased miR-122 and decreased PKM1/2 and GLUT1 in the brain and lungs as well as significantly reduced 2-NBDG uptake in the brain which was not observed in the lungs at the time of tissue collection (Fig. 6d, g–h). These mice also had significantly enhanced metastases to the brain and lungs (Fig. 6i–j), suggesting that miR-122 reduces primary tumour cell proliferation by restricting glucose uptake while simultaneously reprogramming the pre-metastatic niches to promote tumour cell colonization and metastatic formation.
To further focus on the niche-adapting effect of extracellular miR-122 by blocking its’ function inside of cancer cells, we generated luciferase-labelled MDA-MB-231 cells stably expressing an anti-miR-122, which exhibited significantly reduced intracellular and secreted miR-122 and increased PKM2 (Supplementary Fig. 3e). Orthotopic xenograft tumours of these cells were established in mice that also received EV treatments. Although no difference in primary tumour growth was observed among all groups, mice receiving high-miR-122 EVs developed more metastases in brain and lungs (Fig. 6k–m).
Systemic miR-122 intervention alleviates cancer-induced glucose reallocation in vivo and reduces metastasis
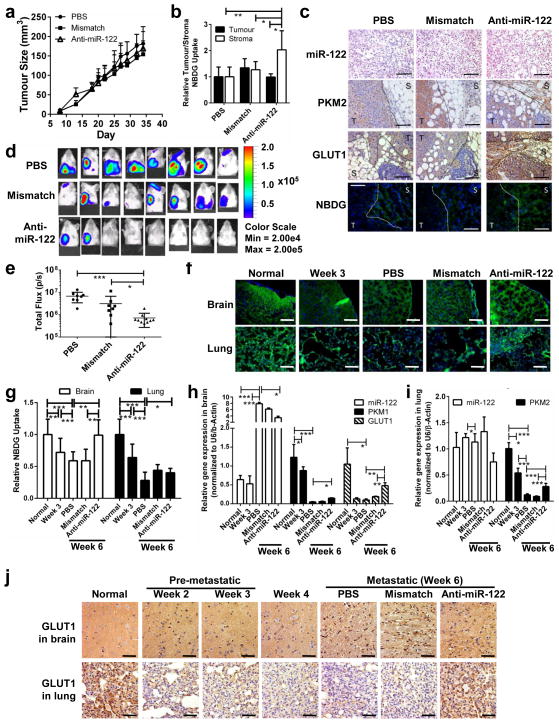
To study the in vivo effect of systemic miR-122 intervention, xenograft tumours of MDA-MB-231-HM that naturally secreted high-miR-122 EVs were established and mice were randomized into three treatment groups that received PBS, mismatch control oligos, or anti-miR-122 oligos. Although there was no difference in primary tumour size among the three groups (Fig. 7a, Supplementary Fig. 4), increased 2-NBDG uptake and enhanced staining of PKM2 and GLUT1 were observed in tumour-adjacent stromal cells in anti-miR-122-treated mice (Fig. 7b–c).
Figure 7.
MiR-122 intervention alleviates cancer-induced glucose reallocation in vivo and reduces metastasis. Luciferase-labelled MDA-MB-231-HM cells were injected into the No. 4 mammary fat pad of NSG mice. Mice were divided into 3 groups (n = 8 mice per group) for treatment with PBS, anti-miR-122, or mismatch control oligos. (a) Tumour growth curve (n = 8 mice). No significant difference (p > 0.05) between groups based on Kruskal-Wallis test. (b) 2-NBDG uptake in the tumour and tumour-adjacent stroma (n = 20 fields from 4 mice per group). (c) Primary tumour sections were analysed by IHC (for PKM2 and GLUT1) and ISH (for miR-122). For 2-NBDG (green) uptake, sections were counterstained with DAPI (blue) to show nuclei. Dotted line delineates tumour (T) from stroma (S). White bar represents 100 μm. (d) BLI at week 5 indicating extensive brain and lung metastases in PBS and mismatch groups and reduced incidence of metastasis in anti-miR-122 group. (e) Quantification of BLI at week 5 (n = 8 mice per group). (f) Representative images of 2-NBDG uptake fluorescence. White bar represents 60 μm. (g) Quantification of 2-NBDG uptake in the brain and lungs of tumour-free (normal) NSG mice and tumour bearing mice that were untreated when sacrificed at week 3 after tumour cell implantation or treated as indicated and sacrificed at week 6 (n = 20 fields from 4 mice per group). (h–i) RT-qPCR in brain (h) and lungs (i) of tumour-free and tumour bearing mice (n = 12 extracts from 4 mice per group). (j) GLUT1 IHC in brain and lungs. Scale bar is 60 μm. Data are represented as mean ± SD in all panels except (c–d, f, & j). * p < 0.05, ** p < 0.01, *** p < 0.001 for all panels derived from Kruskal-Wallis test.
At week 5 we started to observe a lower incidence of metastasis to the brain and lungs in mice receiving anti-miR-122 treatment (Fig. 7d–e). At the pre-metastatic stage when there were no detectable metastases by luciferase PCR, both organs showed reduced 2-NBDG uptake compared to non-tumour-bearing mice (Fig. 7f–g), suggesting that factors secreted by the primary tumour can regulate glucose utilization in a distant organ in preparation for metastasis. This effect became more pronounced as cancer progressed, since a further reduction in 2-NBDG uptake was observed in the metastasis-free areas of brain and lungs at week 6 when metastases had developed in these organs (Fig. 7f–g; PBS group). Notably, treatment with anti-miR-122 but not the mismatch control oligos significantly alleviated tumour-derived suppression of glucose uptake in the brain, although the restoration was not significant in the lungs (Fig. 7f–g). In the brain, miR-122 levels increased with tumour progression with a significant reduction in mice receiving anti-miR-122 oligos. Supporting our in vitro data, we noticed a concomitant decrease in the levels of PKM1 (isoform determined in Fig. 4c) and GLUT1 during tumour progression, which was alleviated by anti-miR-122 treatment (Fig. 7h, j). In the lung, anti-miR-122 oligos alleviated cancer-induced suppression of PKM2 and GLUT1 (Fig. 7i–j). Taken together, our in vitro and in vivo data indicates that cancer cells can induce glucose reallocation in the pre-metastatic microenvironments by suppressing glucose utilization in niche cells and allowing for more glucose to be available to cancer cells, thereby facilitating metastatic cancer growth. This effect is at least partially mediated by cancer-secreted miR-122.
DISCUSSION
Our study demonstrates that miR-122 is highly secreted by BC cells and can promote metastasis by adapting the metabolic environment in a pre-metastatic niche, providing an understanding of our previous observation that miR-122 levels in the circulation are associated with metastatic progression in BC patients18. In addition to EVs, other protein and lipoprotein carriers of circulating miRNAs have been identified40, 41. Although we cannot exclude the potential role of these additional forms of extracellular miR-122 in our herein identified mechanism, characterization of the 110,000 ×g medium pellet used in our study by AF4 and gradient centrifugation indicates that EVs of 30–100 nm are a major component of this material and capable of transferring miR-122 from cancer to normal niche cells to promote metastasis. An interesting phenomenon of stromal–epithelial metabolic coupling, termed the reverse Warburg effect for stromal glycolysis and cancer cell oxidative phosphorylation, has been recently recognized42. In BC, cancer-associated stromal cells rely on glycolysis to provide energy metabolites to cancer cells via monocarboxylate transporters during disease progression43. Endothelial cells also rely on glycolytic metabolism to support vessel sprouting for angiogenesis43. Although extracellular miR-122 does not seem to contribute to increased glycolysis in cells in the primary tumour microenvironment, based on the concomitant decrease in lactate production upon miR-122-mediated reduction of glucose uptake (Fig. 2d, Supplementary Fig. 3b) and lack of a change in most TCA cycle metabolites (Fig. 2b), other miRNAs in cancer-secreted EVs may contribute to this effect which would be an interesting future direction. In exploring what caused the increase of isocitrate in MCF10A/miR-122 (Fig. 2b), we found that isocitrate dehydrogenase (IDH) 1 and 2 were both downregulated by miR-122 (Fig. 2i). While a search of the 3′UTR of IDH1/2 did not reveal any miR-122 binding site, restoration of PKM2 was able to restore IDH1/2 levels in MCF10A/miR-122, suggesting that like GLUT1, IDH1/2 might be directly or indirectly regulated by PKM2. We also examined the NMR spectrum for α-ketoglutarate which could not be reliably quantified due to the low concentrations. However, the spectrum did suggest a reduction of α-ketoglutarate in MCF10A/miR-122 (Supplementary Fig. 2e), which is consistent with decreased IDH1/2 and increased isocitrate in these cells.
Adaptation of a pre-metastatic niche, initially defined by Kaplan and Lyden et al.44, 45, prior to the arrival of tumour cells has been recognized as an important means for cancer to facilitate their sustained growth and metastasis46, 47. Exosomes from highly metastatic melanomas “educate” bone marrow progenitor cells towards a pro-metastatic phenotype and induce vascular leakiness at pre-metastatic sites to facilitate metastasis26. Melanoma-derived exosomes also prepare sentinel lymph node for metastasis by inducing cell recruitment, extracellular matrix remodelling, and vascular growth factors48. In renal cell carcinoma, microvesicles derived from CD105+ tumour-initiating cells trigger angiogenesis which serves to enhance lung metastasis49. Additional studies have highlighted the importance of other tumour-secreted factors resulting in establishment of a pre-metastatic niche50–54.
Here we added a unique aspect of nutrient utilization to this paradigm of cancer-host crosstalk. Enhanced glucose uptake is common in cancer as a result of the high energy demand in cancer cells and the low ATP-generating efficiency due to the Warburg effect. GLUT1 and glycolytic enzymes have been shown to be upregulated in BC55, 56 as potential mechanisms for increasing glucose uptake. Cancer cells also develop strategies to increase their availability to glucose, such as angiogenesis to gain nutrients from the blood. Here we provide evidence that cancer cells also systemically suppress the nutrient utilization by other cell types to favour themselves. This miR-122-mediated mechanism may be more important at an early stage prior to cancer-induced angiogenesis, when the availability of nutrients in the tumour microenvironment becomes limited to sustain tumour growth, and when disseminated tumour cells arrive to a distant tissue to prepare for rapid expansion among the surrounding normal niche cells which are native competitors for nutrients. Indeed, we observed that BC cells at the primary site were able to affect glucose uptake by brain and lungs at a pre-metastatic stage (Fig. 7f–j). Importantly, miR-122 intervention using antisense oligos significantly reduced BC metastasis to brain and lungs (Fig. 7d–e). Thus, our previous18 and current studies indicate that cancer-derived circulating miR-122 has the potential to be both a predictive marker and therapeutic target for metastatic BC. As miR-122 antagonists are in clinical trials for patients with hepatitis C infection and exhibit good tolerance with a low propensity for drug interactions57–59, miR-122-targeted therapy in cancer patients seems highly feasible, while the non-invasive blood test for circulating miR-122 would enable accurate selection of patients who may benefit from this treatment.
METHODS
Cells, plasmids, and viruses
Human cancer cell lines and the non-cancerous cell line MCF10A were obtained from American Type Culture Collection (Manassas, VA) and cultured in the recommended media. Mouse astrocytes were purchased from Lonza and cultured following the manufacturer’s instructions. MCF10DCIS.com (MCFDCIS) cells were purchased from Asterand (Detroit, MI). The MDA-MB-231-HM cells were generated in our lab through explant culture of a spontaneous meningeal metastasis of MDA-MB-231 cells from an immunocompromised mouse and have been used in our previous study27. Mouse lung fibroblasts were isolated from minced lung tissue grown in DMEM supplemented with 10% fetal bovine serum (FBS) and 10 μg/ml bFGF (Life Technologies; Carlsbad, CA). Purity of the fibroblasts was confirmed by the expression of FSP-1 in >95% of the cells. Neurons were generated from mouse E14.5 embryo. Briefly, the embryonic brain cortical neurons were differentiated from primary culture of embryonic neural stem cells by culturing in medium containing N2 supplement (Life Technologies), 0.5% FBS, 20 ng/ml FGF and 20 ng/ml EGF for 5 days on plates coated with Matrigel (BD Biosciences; San Jose, CA). All cells used herein were tested to be free of mycoplasma contamination. PCR primers 5′-GCACGTCCTCGAGTAGGCCAGCAACGCTTGTAG and 5′-TTATAATGCGGCCGCAGGTGGAGGGTGGAGTGTTTGCTGC (for wild-type miR-122 site) or 5′-TTATAATGCGGCCGCAGGTGGAGGGAGATCTGTTTGCTGC (for mutated miR-122 site) were used to clone the 3′UTR of human PKM. Primers 5′-GCACGTCCTCGAGTGGAGACTGGGTGAAAGTGA and 5′-TTATAATGCGGCCGCACAGCAGGAGTGTATCTTAATCC (for wild-type miR-122 site) or 5′-TTATAATGCGGCCGCACAGCAGGACAAGATCTTAATCC (for mutated miR-122 site) were used to clone the 3′UTR of human CS. The PCR fragments were digested with XhoI and NotI and then inserted into the same sites of psiCHECK-2 vector (Promega; Madison, WI) downstream of the Renilla luciferase gene. For miR-122 overexpression, the hsa-mir-122 gene was cloned by PCR using primers 5′-GCAGCTGAATTCGAGCTGACAAGGTTCCCCTA and 5′-TAGTACGTCGACAAAGCAAACGATGCCAAGAC, and ligated into the EcoRI/SalI sites of pBABE-Puro or pBABE-GFP retroviral vector. PKM2 and CS overexpressing plasmids were constructed by PCR cloning the full-length ORF of PKM2 or CS using primers 5′-GAAGTTGGATCCAGATCAGGACCTCAGCA and 5′-GAAGTTGAATTCGGCTCTGGGGTCCATCAC for PKM2 or 5′-GAAGTTGGATCCTTACCTCCCCACCAGATCC and 5′-GAAGTTGAATTCACTTTCACCCAGTCTCCA for CS from MCF10A cells, and inserting the ORF into the BamHI/EcoRI sites of pBABE-Puro. The GLUT1 overexpressing plasmid pcDNA3.2/v5-DEST hGlut160 was obtained from Addgene (Cambridge, MA). A lentiviral construct expressing an anti-miR-122 was purchased from GeneCopoeia (Rockville, MD) to generated MDA-MB-231 cells with stable knockdown of miR-122 (MDA-MB-231/122KD). All constructs were verified by sequencing. Target gene knockdown was accomplished using the GeneSolution system which contains 4 FlexiTube siRNAs against Pkm2 (Cat# GS18746) or Slc2a1 (Cat# GS20525, Qiagen; Valencia, CA). Cell transfection, reporter assays, production of viruses, as well as infection and selection of transduced cells were carried out as previously described61. The RNA polymerase II inhibitor 5,6-dichloro-1-β-D-ribofuranoside (DRB) was purchased from Sigma-Aldrich (St. Louis, MO).
Extracellular vesicle (EV) purification and characterization
Conditioned media (CM) was collected from cells grown in media containing vesicle-depleted FBS or horse serum (prepared by overnight ultracentrifugation at 156,000 ×g at 4°C) for 48 h. Dead cells and contaminating cell debris were removed by centrifugation at 500 ×g for 15 min and then at 12,500 ×g for 20 min at 4°C. Media was then subjected to ultracentrifugation at 110,000 ×g for 70 min at 4°C, and the pellet was washed with PBS and subjected to an additional round of ultracentrifugation under the same conditions to produce the so-called 110,000 ×g pellet that is known to enrich for extracellular vesicles including exosomes. The supernatant collected after 110,000 ×g spins was concentrated using Corning Spin-X UF Concentrator with a 30,000 MWCO (Sigma-Aldrich) and subjected to RNA extraction. When indicated, the 110,000 ×g pellet was incubated in 1 μM 1,1′-dioctadecyl-3,3,3′3′-tetramethylindocarbocyanine perchlorate (DiI) for 20 min in PBS, followed by an additional round of PBS wash. PBS was then used to resuspend the EV-enriched pellets for cell treatment or animal injection. For cell treatment, 2 μg of EVs (equivalent to those collected from ~5 × 106 producer cells) based on protein measurement using Bradford protein assay (Bio-Rad; Hercules, CA) were added to 2 × 105 recipient cells. For patient-derived EVs, 1.35 mL of human sera was diluted in PBS to 9.5 mL and filtered though a 0.22-μm-pore filter before ultracentrifugation at 110,000 ×g for 2 h at 4°C, followed by an additional round of PBS wash. Human sera from breast cancer patients were obtained from voluntarily consenting patients at the City of Hope Medical Center (Duarte, CA) under institutional review board (IRB)-approved protocols. The low-miR-122 serum was from a 62-year-old female patient with ER+PR+HER2+ invasive ductal carcinoma, and the high-miR-122 serum from a 53-year-old female patient with ER+PR−HER2+ invasive ductal carcinoma. Pooled normal serum (Cat# IPLA-SER) was purchased from Innovative Research (Novi, MI).
To further characterize materials in the 110,000 ×g pellet for a more precisely defined miR-122 distribution profile, we first adopted a recently established method using asymmetrical flow field flow fractionation (AF4) following the previously reported procedure32. In brief, the 110,000 ×g pellet was resuspended in PBS and injected into an AF2000 system (Postnova Analytics; Salt Lake City, UT), with an injection loop of 50 μL and a channel thickness of 350 μm. The accumulation wall for the AF4 was a regenerated cellulose ultrafiltration membrane (Postnova), with a molecular weight cut-off of 10 kDa. The flow program for the vesicle separation consisted of a 5-min constant segment with the outlet and cross flows of 0.3 and 3 mL/min, respectively, followed by a 15-min rampdown of the cross flow to zero flow. Using PBS as running buffer, the eluate exiting AF4 passed through a SPD-20A absorbance detector detecting at 280 nm (Shimadzu; Kyoto, Japan), followed by minute collections on a fraction collector (Bio-Rad; Model 2110). Based on the established AF4 profile32 and our sample distribution (Fig. 1e), we combined eluates collected from 8 to 11 min as the “protein fraction” and those from 18 to 25 min as the “vesicle fraction”. Although vesicles and low-density lipoproteins (LDL) are both eluted from 18 to 25 min, LDL is undetectable in the 110,000 ×g medium pellet. Each fraction was then divided into two aliquots; one was subjected to RNA extraction using TRIZOL LS reagent (Life Technologies) followed by miRNA assessment by RT-qPCR (see the corresponding section below), and the other was analyzed by electron microscopy (EM) to verify the absence (in “protein fraction”) or presence (in “vesicle fraction”) of vesicles and characterize their size. For EM, samples were fixed with 2% paraformaldehyde, loaded on 200-mesh Formvar-coated grids, and then contrasted and embedded as described in 62. The grids were observed under an FEI Tecnai12 transmission electron microscope equipped with a CCD camera.
We also adopted another method to fractionate the 110,000 ×g pellet using buoyant sucrose gradient centrifugation following a reported protocol31. The pellet was resuspended in 100 μL PBS, diluted to 1 mL in 2.5 M sucrose, and loaded into the bottom of a sucrose density gradient ranging from 2.0 to 0.25 M (in PBS) with a 1-mL 2.5 M sucrose cushion loaded below the sample. After ultracentrifugation at 100,000 ×g for 6 h at 4°C, each 1-mL gradient fractions were collected and subjected to RNA extraction followed by miRNA assessment and to EM as described above. Microvesicle measurements were quantified using ImagePro Premier software (Media Cybernetics; Rockville, MD).
BrdU incorporation assay
Cells were plated at 5 × 105 per well on a 6-well plate in triplicate. BrdU labelling reagent (Life Technologies) was diluted 1:100 in growth media. Cells were labelled for 1 h at 37°C prior to fixation with 1% formaldehyde and stained using anti-BrdU-APC diluted 1:100 (BU20A, Cat# 17-5071, eBiosciences; San Diego, CA) before assessment by flow cytometry. Data was analysed using FlowJo software.
2-NBDG uptake assay
After 96 h post-EV treatment, recipient cells were labelled with 100 μM 2-[N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)amino]-2-deoxy-D-glucose (2-NBDG) diluted in glucose-free media and incubated for 40 min at 37°C. Fluorescence quantification was performed using a SpectraMax M3 (Molecular Devices; Sunnyvale, CA) fluorometer at em 465/ex 540 nm. Values were normalized to protein content.
RNA extraction and reverse transcription (RT) real-time qPCR
These procedures were performed as described previously63, 64. Primers used in RT-qPCR are indicated in Supplementary Table 1. The miR-122, miR-16 (as internal control for vesicular miR-122), and U6 primers (as internal control for intracellular miR-122) were purchased from Qiagen. Determination of PKM isoforms was performed as described65. Products of RT-PCR were digested with NcoI, PstI, or both enzymes, plus an uncut control, and separated on an agarose gel with Sybr safe. The presence of a PstI digestion site indicates the splicing isoform M2 whereas the NcoI site indicates isoform M1.
Western blot analysis and PKM activity assay
PKM2 antibody diluted 1:500 (C-11, Cat# sc-365684) was purchased from Santa Cruz Biotechnology (Santa Cruz, CA), CS antibody diluted 1:1000 (Cat# 16131-1-AP) from Proteintech Group (Chicago, IL), and GLUT1 antibody diluted 1:1000 (Cat# ab652) from AbCam (Cambridge, MA). IDH1 diluted 1:500 (D2H1, Cat# 8137) and IDH2 diluted 1:1000 (D7H6Q, Cat# 12652) antibodies were purchased from Cell Signaling (Boston, MA). β-actin antibody diluted 1:4000 (AC-15, Cat# A1978) was purchased from Sigma-Aldrich. PKM activity was assessed using a protocol modified from Edwards and Watts66 by diluting each protein sample in 228 μl Solution A (110 mmol/L Imidazole-HCl, 165 mmol/L KCl, 5.5 mmol/L MgCl2, 0.19 mmol/L NADH, 5.5 mmol/L ADP, 5.5 mmol/L DTT, pH 7.4) supplemented with 2.5 unit lactate dehydrogenase and 62.5 nmol phosphoenolpyruvate (Sigma-Aldrich). Decreases in absorbance of NADH at 340 nm were followed every minute for 10 min after initiation of the reaction. One unit of PKM activity was defined as the amount that will consume 1 μmol of NADH (molar absorptivity 6.22 cm2/μmol) per minute under the assay condition.
Medium metabolite analysis
MCF10A-derived cells seeded at equal number were cultured in growth media containing 3 g/L glucose but no pyruvate for 72 h before CM was collected, cleared by centrifugation, and subjected to metabolite measurement using a BioProfile 100 Plus (Nova Biomedical; Waltham, MA). Media collected from cell-free plates after 72 h incubation was used as the baseline control to calculate the consumption or production of each metabolite, which was further normalized to the cell number in each plate determined at the time of CM collection. MCFDCIS-derived cells were cultured for 48 h before CM was collected for analysis.
Cell metabolome analysis by NMR spectroscopy
Sample preparation, NMR spectroscopy, and data analyses were performed as described67. Hydrophilic metabolites dried from the methanol-water fractions were resuspended in 500 μL 100% D2O containing 3.2 μM of 4,4-dimethyl-4-silapentane-1-sulfonic acid (DSS), which serves as an internal chemical shift reference and a concentration standard. 1D NMR spectra were acquired at 25°C on a Bruker Avance spectrometer equipped with a cryoprobe operating at 600.19 MHz 1H frequency. Pre-saturation was used to suppress water signal, and the spectra was collected with spectral width of 10 kHz, 32 k data points, 3 s relaxation delay and 1024 transients. 1H NMR spectra of the samples were processed using the Chenomx NMR Suite Processor (version 7.5, Chenomx Inc., Edmonton, Canada), and the metabolites were identified and quantified using the Chenomx NMR Suite Profiler. Standard deviation was calculated from triplicate samples.
Animal models: EV conditioning and xenografts
All animal procedures were approved by the Institutional Animal Care and Use Committee at City of Hope and in compliance with ethical regulations. Female NOD/SCID/IL2Rγ-null (NSG) mice of 6–8 week old were used in this study. Exosome-containing EVs were isolated from MCF10A/vec, MCF10A/miR-122, and MDA-MB-231 by the above procedure (for preparation of the 110,000 ×g medium pellet), resuspended in PBS, and followed by centrifugation at 16,000 ×g for 10 min at 4°C. The supernatant was then transferred to a new tube for mouse injection. When indicated, EVs were injected into the tail vein of NSG mice biweekly for 3.5 weeks (~6 μg/injection). For cancer cell chase, 1 × 104 luciferase-labelled MDA-MB-231-HM cells were injected intracardiac after pre-conditioning with 6 biweekly EV injections; after the cancer cell injection, EV injections continued biweekly for 3 weeks. Orthotopic mammary xenografts were established in NSG mice by injecting 1 × 105 luciferase-labelled MCFDCIS cells or 2 × 105 luciferase-labelled MDA-MB-231/122KD cells or MDA-MB-231-HM cells combined with Matrigel (BD Biosciences; San Jose, CA) in a 1:1 ratio into the No. 4 mammary fat pad. Weekly bioluminescence imaging (BLI) was carried out using a Xenogen system (Caliper Life Sciences; Alameda, CA). Tumour volume (mm3) was assessed by calliper measurements using the formula (width2 × length)/2. For the miR-122 intervention study, mice were divided into 3 groups for treatment with PBS, anti-miR-122 oligos (sequence: 5′-CcAttGTcaCaCtCC; 2′-deoxy-2′-fluoro-RNA in capitals, DNA in lower case) or mismatch control oligos (5′-CcAttCTcaCaCtGC) with a phosphorothioate backbone synthesized at the City of Hope Core of Synthetic and Biopolymer Chemistry. Starting from day 3 after cancer cell transplantation, oligos (25 mg/kg) were intraperitoneally (i.p.) injected daily for 5 days and then twice weekly until the end of experiment.
For 2-NBDG uptake, 15 mg/kg of 2-NBDG was injected through the tail vein 45 min before a transcardiac perfusion with 1% PFA in PBS was carried out to remove the excess dye. Tissues were embedded in Tissue-Tek O.C.T. Compound (Sakura; Torrance, CA) to make frozen blocks for sectioning and 2-NBDG analysis by fluorescence microscopy. The 2-NBDG signals were quantified using ImagePro 6.3/Premier software. ATP levels in the tumour samples were prepared by TCA extraction as previous described68 and assessed by Enlighten ATP assay (Promega) following the manufacturer’s instructions.
Co-immunofluorescence (IF)
O.C.T. sections were fixed with 4% PFA in PBS, blocked and permeabilised with PBS containing 10% goat serum and 0.05% saponin, prior to incubation with rabbit anti-mouse GFAP diluted 1:500 (AbCam; Cat# ab7260) for brain or rabbit anti-mouse FSP-1 diluted 1:100 (AbCam; Cat# ab27957) for lungs together with mouse anti-human CD63 diluted 1:50 (MEM-259, Cat# NB100-77913; Novus Biologicals; Littleton, CO). Primary antibodies were then visualised with goat anti-rabbit Alexa 647 IgG diluted 1:300 (Cat# A-21244) and goat anti-mouse Alexa 594 IgG diluted 1:300 (Cat# A-11032, Life Technologies; Grand Island, NY). Images were obtained by fluorescence microscopy then pseudo-coloured and merged using ImagePro Premier software.
Immunohistochemistry & in situ hybridization
IHC was performed as previously described69 using a 1:400 antibody dilution for PKM2 (C-11), 1:600 dilution for PKM1 (Cat# NBP2-14833, Novus Biologicals; Littleton, CO), and 1:250 dilution for GLUT1 (AbCam; Cat# ab652). ISH was performed as described70 using LNA™ microRNA ISH miR-122 optimization kit (Cat# 90003, Exiqon; Woburn, MA) followed by incubation of sheep anti-digoxigenin-AP (Cat# 11093274910, Roche Diagnostics; Mannheim, Germany), and developed with NBT:BCIP (Cat# SK-5400, Vector Laboratories; Burlingame, CA) at 30°C overnight. Nuclear fast red was used to counterstain nuclei (Cat# H-3403, Vector Laboratories).
Glycogen staining
Glycogen staining was performed using a Periodic Acid-Schiff (PAS) kit (Sigma-Aldrich) following the manufacturer’s protocol.
Statistical analyses
All quantitative data are presented as mean ± standard deviation. For all quantitative data, statistical analyses were performed using Kruskal-Wallis tests. Values of p < 0.05 were considered significant. Sample size was generally chosen based on preliminary data indicating the variance within each group and the differences between groups. For animal studies, sample size was predetermined to allow an 80% power to detect a difference of 50%. All samples/animals that have received the proper procedures with confidence were included for the analyses. Animals were randomized before treatments in Figs. 5–7. For animal studies, the investigators were blinded to allocation during outcome assessment. For every figure, statistical tests are justified as appropriate, and the data meet the assumptions of the tests.
Repeatability of experiments
For the experiments in which no quantification is shown, images representative of at least 3 independent experiments are shown.
Supplementary Material
Acknowledgments
This work was supported by the United States Army Research and Material Command grant W81-14-1-0029 (MYF), National Institutes of Health (NIH)/National Cancer Institute (NCI) grants R01CA166020 (SEW) and R01CA163586 (SEW), California Breast Cancer Research Program grant 20IB-0118 (SEW), Breast Cancer Research Foundation-AACR grant 12-60-26-WANG (SEW), and the City of Hope Women’s Cancer Program. Research reported in this publication included work performed in Core facilities supported by the NIH/NCI under grant number P30CA33572. We thank Drs. Arthur Riggs, Eugene Roberts, Linda Malkas, Susan Kane, Shiuan Chen, Joanne Mortimer, and Peter Sarnow for valuable comments, as well as the core facilities at City of Hope for highly professional services.
Footnotes
AUTHOR CONTRIBUTIONS
S.E.W. conceived ideas, and M.Y.F., Y.C., and X.R. contributed to project planning. M.Y.F. and S.E.W. designed and performed the experiments. W. Zhou, L.L., A.C., S.T.F.O., S.L., A.R.C., and J.R.T. assisted with EV preparation and mouse experiments. G. Somlo and M.P. assisted with patient serum samples. Z.L. assisted with EM. A.T. assisted with mouse lung fibroblast culture. A.Y.A., M.C., and Y.C. assisted with NMR analysis. J.A. and W. Zhong assisted with AF4 analysis. G. Sun and Y.S. assisted with neuron culture. M.A.R. and M.K. assisted with medium metabolite analysis. X.W. assisted with bioinformatics analysis of miR-122 targets. P.S. assisted with anti-miR-122 and mismatch oligo synthesis. S.E.W. and M.Y.F. wrote the manuscript.
The authors declare no conflict of interest.
References
- 1.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhao FQ, Keating AF. Functional properties and genomics of glucose transporters. Curr Genomics. 2007;8:113–128. doi: 10.2174/138920207780368187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008;7:11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 5.Barthel A, et al. Regulation of GLUT1 gene transcription by the serine/threonine kinase Akt1. J Biol Chem. 1999;274:20281–20286. doi: 10.1074/jbc.274.29.20281. [DOI] [PubMed] [Google Scholar]
- 6.O’Rourke JF, Pugh CW, Bartlett SM, Ratcliffe PJ. Identification of hypoxically inducible mRNAs in HeLa cells using differential-display PCR. Role of hypoxia-inducible factor-1. Eur J Biochem. 1996;241:403–410. doi: 10.1111/j.1432-1033.1996.00403.x. [DOI] [PubMed] [Google Scholar]
- 7.Semenza GL, Roth PH, Fang HM, Wang GL. Transcriptional regulation of genes encoding glycolytic enzymes by hypoxia-inducible factor 1. J Biol Chem. 1994;269:23757–23763. [PubMed] [Google Scholar]
- 8.Spoden GA, et al. The SUMO-E3 ligase PIAS3 targets pyruvate kinase M2. J Cell Biochem. 2009;107:293–302. doi: 10.1002/jcb.22125. [DOI] [PubMed] [Google Scholar]
- 9.Yang W, et al. ERK1/2-dependent phosphorylation and nuclear translocation of PKM2 promotes the Warburg effect. Nat Cell Biol. 2012;14:1295–1304. doi: 10.1038/ncb2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gao X, Wang H, Yang JJ, Liu X, Liu ZR. Pyruvate kinase M2 regulates gene transcription by acting as a protein kinase. Mol Cell. 2012;45:598–609. doi: 10.1016/j.molcel.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang W, et al. Nuclear PKM2 regulates beta-catenin transactivation upon EGFR activation. Nature. 2011;480:118–122. doi: 10.1038/nature10598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee J, Kim HK, Han YM, Kim J. Pyruvate kinase isozyme type M2 (PKM2) interacts and cooperates with Oct-4 in regulating transcription. Int J Biochem Cell Biol. 2008;40:1043–1054. doi: 10.1016/j.biocel.2007.11.009. [DOI] [PubMed] [Google Scholar]
- 13.Luo W, et al. Pyruvate kinase M2 is a PHD3-stimulated coactivator for hypoxia-inducible factor 1. Cell. 2011;145:732–744. doi: 10.1016/j.cell.2011.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 16.Mitchell PS, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Taylor DD, Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol Oncol. 2008;110:13–21. doi: 10.1016/j.ygyno.2008.04.033. [DOI] [PubMed] [Google Scholar]
- 18.Wu X, et al. De novo sequencing of circulating miRNAs identifies novel markers predicting clinical outcome of locally advanced breast cancer. J Transl Med. 2012;10:42. doi: 10.1186/1479-5876-10-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhu W, Qin W, Atasoy U, Sauter ER. Circulating microRNAs in breast cancer and healthy subjects. BMC Res Notes. 2009;2:89. doi: 10.1186/1756-0500-2-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zen K, Zhang CY. Circulating microRNAs: a novel class of biomarkers to diagnose and monitor human cancers. Med Res Rev. 2012;32:326–348. doi: 10.1002/med.20215. [DOI] [PubMed] [Google Scholar]
- 21.Redis RS, Calin S, Yang Y, You MJ, Calin GA. Cell-to-cell miRNA transfer: from body homeostasis to therapy. Pharmacol Ther. 2012;136:169–174. doi: 10.1016/j.pharmthera.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Valadi H, et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 23.Wang K, Zhang S, Weber J, Baxter D, Galas DJ. Export of microRNAs and microRNA-protective protein by mammalian cells. Nucleic Acids Res. 2010;38:7248–7259. doi: 10.1093/nar/gkq601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vickers KC, Remaley AT. Lipid-based carriers of microRNAs and intercellular communication. Curr Opin Lipidol. 2012;23:91–97. doi: 10.1097/MOL.0b013e328350a425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Skog J, et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10:1470–1476. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peinado H, et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat Med. 2012;18:883–891. doi: 10.1038/nm.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou W, et al. Cancer-Secreted miR-105 Destroys Vascular Endothelial Barriers to Promote Metastasis. Cancer Cell. 2014;25:501–515. doi: 10.1016/j.ccr.2014.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rottiers V, Naar AM. MicroRNAs in metabolism and metabolic disorders. Nat Rev Mol Cell Biol. 2012;13:239–250. doi: 10.1038/nrm3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moore KJ, Rayner KJ, Suarez Y, Fernandez-Hernando C. The role of microRNAs in cholesterol efflux and hepatic lipid metabolism. Annu Rev Nutr. 2011;31:49–63. doi: 10.1146/annurev-nutr-081810-160756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boutz DR, et al. Two-tiered approach identifies a network of cancer and liver disease-related genes regulated by miR-122. J Biol Chem. 2011;286:18066–18078. doi: 10.1074/jbc.M110.196451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Palma J, et al. MicroRNAs are exported from malignant cells in customized particles. Nucleic Acids Res. 2012;40:9125–9138. doi: 10.1093/nar/gks656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ashby J, et al. Distribution Profiling of Circulating MicroRNAs in Serum. Anal Chem. 2014;86:9343–9349. doi: 10.1021/ac5028929. [DOI] [PubMed] [Google Scholar]
- 33.Lloyd PG, Hardin CD, Sturek M. Examining glucose transport in single vascular smooth muscle cells with a fluorescent glucose analog. Physiol Res. 1999;48:401–410. [PubMed] [Google Scholar]
- 34.Loaiza A, Porras OH, Barros LF. Glutamate triggers rapid glucose transport stimulation in astrocytes as evidenced by real-time confocal microscopy. J Neurosci. 2003;23:7337–7342. doi: 10.1523/JNEUROSCI.23-19-07337.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yamada K, et al. Measurement of glucose uptake and intracellular calcium concentration in single, living pancreatic beta-cells. J Biol Chem. 2000;275:22278–22283. doi: 10.1074/jbc.M908048199. [DOI] [PubMed] [Google Scholar]
- 36.Itoh Y, Abe T, Takaoka R, Tanahashi N. Fluorometric determination of glucose utilization in neurons in vitro and in vivo. J Cereb Blood Flow Metab. 2004;24:993–1003. doi: 10.1097/01.WCB.0000127661.07591.DE. [DOI] [PubMed] [Google Scholar]
- 37.Chuquet J, Quilichini P, Nimchinsky EA, Buzsaki G. Predominant enhancement of glucose uptake in astrocytes versus neurons during activation of the somatosensory cortex. J Neurosci. 2010;30:15298–15303. doi: 10.1523/JNEUROSCI.0762-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bak LK, Schousboe A, Sonnewald U, Waagepetersen HS. Glucose is necessary to maintain neurotransmitter homeostasis during synaptic activity in cultured glutamatergic neurons. J Cereb Blood Flow Metab. 2006;26:1285–1297. doi: 10.1038/sj.jcbfm.9600281. [DOI] [PubMed] [Google Scholar]
- 39.Hu M, et al. Regulation of in situ to invasive breast carcinoma transition. Cancer Cell. 2008;13:394–406. doi: 10.1016/j.ccr.2008.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vickers KC, Palmisano BT, Shoucri BM, Shamburek RD, Remaley AT. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat Cell Biol. 2011;13:423–433. doi: 10.1038/ncb2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Arroyo JD, et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci U S A. 2011;108:5003–5008. doi: 10.1073/pnas.1019055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pavlides S, et al. The reverse Warburg effect: aerobic glycolysis in cancer associated fibroblasts and the tumor stroma. Cell Cycle. 2009;8:3984–4001. doi: 10.4161/cc.8.23.10238. [DOI] [PubMed] [Google Scholar]
- 43.Martinez-Outschoorn UE, Lisanti MP, Sotgia F. Catabolic cancer-associated fibroblasts transfer energy and biomass to anabolic cancer cells, fueling tumor growth. Semin Cancer Biol. 2014;25:47–60. doi: 10.1016/j.semcancer.2014.01.005. [DOI] [PubMed] [Google Scholar]
- 44.Kaplan RN, Psaila B, Lyden D. Bone marrow cells in the ‘pre-metastatic niche’: within bone and beyond. Cancer Metastasis Rev. 2006;25:521–529. doi: 10.1007/s10555-006-9036-9. [DOI] [PubMed] [Google Scholar]
- 45.Kaplan RN, et al. VEGFR1-positive haematopoietic bone marrow progenitors initiate the pre-metastatic niche. Nature. 2005;438:820–827. doi: 10.1038/nature04186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Peinado H, Lavotshkin S, Lyden D. The secreted factors responsible for pre-metastatic niche formation: old sayings and new thoughts. Seminars in cancer biology. 2011;21:139–146. doi: 10.1016/j.semcancer.2011.01.002. [DOI] [PubMed] [Google Scholar]
- 47.Sethi N, Kang Y. Unravelling the complexity of metastasis - molecular understanding and targeted therapies. Nat Rev Cancer. 2011;11:735–748. doi: 10.1038/nrc3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hood JL, San RS, Wickline SA. Exosomes released by melanoma cells prepare sentinel lymph nodes for tumor metastasis. Cancer Res. 2011;71:3792–3801. doi: 10.1158/0008-5472.CAN-10-4455. [DOI] [PubMed] [Google Scholar]
- 49.Grange C, et al. Microvesicles released from human renal cancer stem cells stimulate angiogenesis and formation of lung premetastatic niche. Cancer Res. 2011;71:5346–5356. doi: 10.1158/0008-5472.CAN-11-0241. [DOI] [PubMed] [Google Scholar]
- 50.Erler JT, et al. Hypoxia-induced lysyl oxidase is a critical mediator of bone marrow cell recruitment to form the premetastatic niche. Cancer Cell. 2009;15:35–44. doi: 10.1016/j.ccr.2008.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hiratsuka S, et al. Primary tumours modulate innate immune signalling to create pre-metastatic vascular hyperpermeability foci. Nature communications. 2013;4:1853. doi: 10.1038/ncomms2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hiratsuka S, et al. Endothelial focal adhesion kinase mediates cancer cell homing to discrete regions of the lungs via E-selectin up-regulation. Proc Natl Acad Sci U S A. 2011;108:3725–3730. doi: 10.1073/pnas.1100446108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hiratsuka S, Watanabe A, Aburatani H, Maru Y. Tumour-mediated upregulation of chemoattractants and recruitment of myeloid cells predetermines lung metastasis. Nat Cell Biol. 2006;8:1369–1375. doi: 10.1038/ncb1507. [DOI] [PubMed] [Google Scholar]
- 54.Hiratsuka S, et al. The S100A8-serum amyloid A3-TLR4 paracrine cascade establishes a pre-metastatic phase. Nat Cell Biol. 2008;10:1349–1355. doi: 10.1038/ncb1794. [DOI] [PubMed] [Google Scholar]
- 55.Bos R, et al. Biologic correlates of (18)fluorodeoxyglucose uptake in human breast cancer measured by positron emission tomography. J Clin Oncol. 2002;20:379–387. doi: 10.1200/JCO.2002.20.2.379. [DOI] [PubMed] [Google Scholar]
- 56.Kang SS, et al. Clinical significance of glucose transporter 1 (GLUT1) expression in human breast carcinoma. Jpn J Cancer Res. 2002;93:1123–1128. doi: 10.1111/j.1349-7006.2002.tb01214.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Elmen J, et al. Antagonism of microRNA-122 in mice by systemically administered LNA-antimiR leads to up-regulation of a large set of predicted target mRNAs in the liver. Nucleic Acids Res. 2008;36:1153–1162. doi: 10.1093/nar/gkm1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Elmen J, et al. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452:896–899. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- 59.Rai N. News. Santaris Pharma A/S; Hoersholm, Denmark; San Diego, CA: 2011. [Google Scholar]
- 60.Takanaga H, Frommer WB. Facilitative plasma membrane transporters function during ER transit. FASEB J. 2010;24:2849–2858. doi: 10.1096/fj.09-146472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang Y, et al. Transforming growth factor-beta regulates the sphere-initiating stem cell-like feature in breast cancer through miRNA-181 and ATM. Oncogene. 2011;30:1470–1480. doi: 10.1038/onc.2010.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Thery C, Amigorena S, Raposo G, Clayton A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr Protoc Cell Biol. 2006;Chapter 3(Unit 3):22. doi: 10.1002/0471143030.cb0322s30. [DOI] [PubMed] [Google Scholar]
- 63.Tsuyada A, et al. CCL2 mediates cross-talk between cancer cells and stromal fibroblasts that regulates breast cancer stem cells. Cancer Res. 2012;72:2768–2779. doi: 10.1158/0008-5472.CAN-11-3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yu Y, et al. Context-dependent bidirectional regulation of the MutS homolog 2 by transforming growth factor beta contributes to chemoresistance in breast cancer cells. Mol Cancer Res. 2010;8:1633–1642. doi: 10.1158/1541-7786.MCR-10-0362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Clower CV, et al. The alternative splicing repressors hnRNP A1/A2 and PTB influence pyruvate kinase isoform expression and cell metabolism. Proc Natl Acad Sci U S A. 2010;107:1894–1899. doi: 10.1073/pnas.0914845107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Edwards RJ, Watts DC. Specific spectrophotometric assay for the M isoenzymes of pyruvate kinase in plasma samples containing mixtures of the muscle (M) and liver (L) isoenzymes. Clin Chem. 1981;27:906–909. [PubMed] [Google Scholar]
- 67.Cano KE, Li YJ, Chen Y. NMR metabolomic profiling reveals new roles of SUMOylation in DNA damage response. J Proteome Res. 2010;9:5382–5388. doi: 10.1021/pr100614a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chida J, Yamane K, Takei T, Kido H. An efficient extraction method for quantitation of adenosine triphosphate in mammalian tissues and cells. Analytica chimica acta. 2012;727:8–12. doi: 10.1016/j.aca.2012.03.022. [DOI] [PubMed] [Google Scholar]
- 69.Fong MY, et al. Withaferin A Synergizes the Therapeutic Effect of Doxorubicin through ROS-Mediated Autophagy in Ovarian Cancer. PloS one. 2012;7:e42265. doi: 10.1371/journal.pone.0042265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jorgensen S, Baker A, Moller S, Nielsen BS. Robust one-day in situ hybridization protocol for detection of microRNAs in paraffin samples using LNA probes. Methods. 2010;52:375–381. doi: 10.1016/j.ymeth.2010.07.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.