Abstract
Dynamic packaging of DNA into strings of nucleosomes is a major mechanism whereby eukaryotic cells regulate gene expression. Intricate control of nucleosomal structure and assembly governs access of RNA polymerase II to DNA and consequent RNA synthesis. As part of this, post-translational modifications of histone proteins are central to the regulation of chromatin structure, playing vital roles in regulating the activation and repression of gene transcription. In the heart, dynamic homeostasis of histone modification – driven by the actions of modifiers and recruitment of downstream effectors – is a fundamental regulator of the transcriptional reprogramming that occurs in the setting of disease-related stress. Here, we examine the growing evidence for histone modification as a key mechanism governing pathological growth and remodeling of the myocardium.
Keywords: epigenetics, chromatin, remodeling
Introduction
Eukaryotic DNA is packaged, protected, and regulated by a histone protein core, around which the DNA is wrapped. This structure, termed chromatin, can be condensed and “closed”, a state associated with relative transcriptional repression. Conversely, chromatin can be “open”, a state which allows proteins governing transcription to access the DNA and effect RNA synthesis.
Regulation of chromatin in its various active states is controlled significantly through post-translational modifications (PTMs) of the core histone proteins. Primarily targeting amino acids within the N-terminal tails of these proteins, a wide range of PTMs occurs, including phosphorylation, acetylation, methylation, ubiquitination, SUMOylation, and GlcNAcylation. Regulation of chromatin by reversible incorporation of phosphate, acetyl groups, or methyl groups within histone tails is the best understood.1
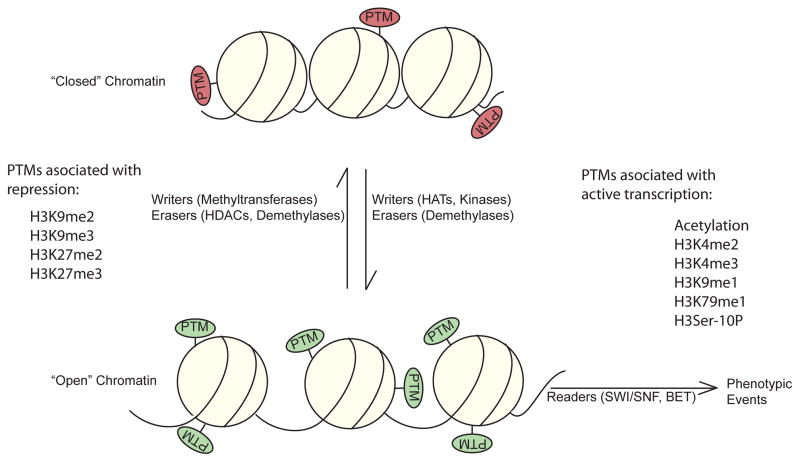
Governance of chromatin structure through histone PTMs has emerged as a key driver of transcriptional responses in numerous cells. Likewise, histone writers, erasers, and readers – the protein machinery that adds, removes or recognizes these PTMs – have become central figures in our understanding of physiological responses in many cell types (Figure 1).
Figure 1.
Post-translational modification (PTM) of histones can act to induce (green) or repress (red) the transition of chromatin to an open state. Coupled with the action of histone PTM “readers”, these changes culminate in an increase or repression of the transcription of target genes.
Writers, enzymes that add PTMs to histones, are divided into classes based on the specific PTM they effect. Similarly, erasers, enzymes which remove specific PTMs from histone substrates, are divided into PTM-specific classes. Finally, readers are dedicated protein factors that recognize either specific post-translational marks on histones or a combination of marks and histone variants to direct a particular transcriptional outcome (Table).
Table 1.
Histone modifiers and effectors active in the myocardium and their involvement in cardiac physiology.
| Writers: | Cardiac Involvement: |
|---|---|
| HATS | |
| p300/pCAF | Development28, 29; Hypertrophy 58, 63 |
| HMTs | |
| SMYD1 | Development34 |
| WHSC1 | Development35 |
| Ezh2 | Hypertrophy69 |
| SUV39h | Hypertrophy76; I/R 92 |
| DOT1L | Hypertrophy70 |
| CaMKII | Hypertrophy78, 79 |
|
| |
| Erasers: | |
| HDAC Class 1 | Hypertrophy31, 52–56; I/R 83–85 |
| HDAC1 | |
| HDAC2 | Hypertrophy59 |
| HDAC3 | Hypertrophy30, 60 |
| HDAC Class II | Development32; Hypertrophy32, 46 |
| HDAC4 | I/R84 |
| HDAC5 | |
| HDAC9 | |
| HDMs | |
| Jarid2 | Development36 |
| Jmjd1 | Hypertrophy76 |
| Jmjd2 | Hypertrophy75, 76 |
| Jmjd3 | Development37 |
| UTX | Development38 |
|
| |
| Readers: | |
| SWI/SNF | |
| Brg1 | Development26; Hypertrophy26, 42 |
| Baf60 | Development24 |
| Baf180 | Development25 |
| Baf250 | Development27 |
| BETs | Hypertrophy65, 66 |
| Brd4 | |
| 14-3-3 | Hypertrophy78 |
Writers: histone acetyltransferases (HATs)
The relationship between acetylation of histones and transcription of DNA was initially proposed in the 1960’s.2 However, it wasn’t until 1996 that the first direct link between these two events was described with the identification of histone acetyltransferase activity from a Tetrahymena thermophilia orthologue of the yeast transcription factor Gcn5.3 Identification of a number of other HATs followed, including TFIID subunit TAFII250 and p300/CBP.4–6 Mammalian cells were then found to have two distinct GCN5 homologs: PCAF and GCN5L. Dozens of HATs have been identified since, comprising roughly 5 different families, the two largest being the GNATs (Gcn5-N-acyltransferases) and MYST families, followed by the SRC (steroid receptor coactivator) family and the highly conserved ATF-2 and aforementioned TAFII250 families.7 Whereas there is little sequence homology among the families, structural similarity in the core enzymatic region exists, indicative of a mechanistic requirement of interaction with the cofactor Acetyl-CoA.8 In general, HATs function as components within a diverse set of multiprotein complexes that target to promoters and enhancers to regulate transcriptional responses.
Histone methyltransferases
A link between histone methylation and DNA transcription was first described in 1999, but unlike histone acetylation, it was quickly followed by the identification of specific histone methyltransferases.9, 10 SUV39h1, the first one identified, is now known to be part of the SET family of methyltransferases, mammalian homologs of the Drosophila suppressor of variegation 3–9.10 A number of SET domain and non-SET domain-containing methyltransferases were subsequently identified with specificity in both protein complex interactions and sites of histone methylation.11 Like histone acetylation, histone methylation occurs on multiple lysine residues, although with the potential for addition of one, two, or three methyl groups. Unlike histone acetylation, histone methylation can be linked to either repression or activation of transcription depending on the context and extent of methylation.12 Modifications associated with active transcription include di- or tri-methylation of H3K4 (H3K4me2, H3K4me3) and mono-methylation of H3K9 (H3K9me1). However di- and tri-methylation of H3K9 (H3K9me2, H3K9me3), as well as H3K27 (H3K27me2, H3K27me3), are repressive marks.
Erasers
Histone deacetylases (HDACs)
The search for HDACs, proteins capable of removing acetyl groups from histones, was an active focus of a number of groups in the 1970’s. Even so, it wasn’t until about the same time that HATs were being linked to transcription that HDAC1 was identified in mammalian cells as a homolog of the yeast transcriptional regulator Rpd3.13 Since that time, 18 mammalian HDACs have been identified and categorized into four major classes. HDACs 1, 2, 3 and 8 make up the class I HDACs. HDACs 4, 5, 7 and 9 belong to class IIa, with HDACs 6 and 10 termed class IIb. The sirtuins (SirT1-7) make up class III, with HDAC11 the sole class IV HDAC.14 Identification of specific small molecule inhibitors of HDAC activity has yielded an important tool for understanding the biology of HDAC activity and emerged as a potential therapeutic strategy for treatment of disease, including cancer, immune disorders, and heart disease.14–16
Histone demethylases
The discovery in 2004 of LSD1 as an enzyme that specifically demethylates histone H3 lysine 4 was followed by identification of a number of demethylases playing regulatory roles in transcription. These include the Jumonji C family, JHMD1, JMJD3, and JMJD2D, each of which targets specific methyllysine groups.11, 17 Much as with HDACs, small molecule inhibitors of demethylase activity are being developed for use as potential therapeutics to modulate DNA transcription in disease.18
Readers
Just as histone PTMs are accomplished by “writers” and “erasers”, their actions to govern DNA transcription are mediated by “readers.” Identification of these proteins was originally driven by use of modified histone peptides to identify proteins that recognize histone PTMs.19 This way, a number of domains have been identified that have high affinity for sites of histone methylation (e.g. PhD [plant homeodomain], chromo [chromatin organization modifier], Tudor, MBT [Malignant Brain Tumor]) or acetylation (e.g. Bromo).19
These domains are located within the chromatin modifying proteins themselves, but are also found in chromatin remodelers and adaptor proteins that respond to the histone PTMs. For example, the chromatin remodeler complex SWI/SNF (switching defective/sucrose non-fermenting) is dependent on the presence of bromodomain-containing subunits for full binding and remodeling activity.20, 21 Similarly, the BET (bromodomain and extraterminal domain) family of adaptor proteins (Brd2, Brd3, Brd4, and Brdt) employs the bromodomain to target to the modified histone and regulate transcription through interactions with the transcriptional machinery.22 These domains may also recognize PTM’s on non-histone proteins, such as transcription factors. Small molecules that disrupt the binding of these BET domains to histone PTMs are emerging as promising therapeutics in a variety of disease conditions.22
Histone PTMs and cardiac development
Changes in chromatin structure play critical roles in the differentiation of cells and the intricate orchestration of embryonic development.23 Thus, it is not surprising that loss of many of these modulators of chromatin structure elicit defects in cardiac development. Chromatin remodelers, such as the SWI/SNF subunits Brg1, Baf250, Baf180, and Baf60c are required for heart development, and loss of function in these subunits leads to embryonic lethality.24–27 Changes in expression and composition of these remodeling complexes occur at specific times during cardiac development, resulting in altered gene expression.27 Brg1, which functions within a complex that includes an HDAC (primarily HDAC2) and PARP, governs two parallel pathways to independently control myocardial growth and differentiation. Silencing of Brg1 within the myocardium results in suppression of proliferation (which can be rescued by BMP10) and premature differentiation.26
The requirement for Brg1 to maintain myocytes in an embryonic state is best characterized through its role in α/β-myosin heavy chain (MHC) switching. Brg1, as part of the BAF/HDAC/PARP complex, inhibits the expression of α-MHC and activates β-MHC expression in embryonic cells. Brg1 expression is significantly reduced in the adult cardiomyocyte, facilitating the switch to α-MHC expression.26
Histone acetylation is also critical to cardiovascular development. The HAT activity of p300 plays a role in co-activation of GATA4-dependent transcription, and genetic ablation of p300 or mutation of the HAT domain elicit defects in atrial and ventricular septum formation.28, 29 And just as acetylation is important, deacetylation is critical as well. Global deletion of HDAC1 or HDAC3 results in embryonic death at E9.5, and global silencing of HDAC2 results in morphological abnormalities in the neonatal heart and death within 24 hours of birth.30, 31 Cardiomyocyte-restricted silencing of HDAC1 or HDAC2 individually, however, has no effect on cardiac development.31 Additionally, combinatorial deletion of both of these class I HDACs in cardiac myocytes is associated with normal cardiac morphology at birth; however, these animals rapidly develop heart failure by P14, marked by lethality and dysregulation of genes involved in calcium handling and contraction.31
In contrast, loss of HDAC3 in cardiac myocytes has no immediate effect on development or function, but rather triggers cardiac hypertrophy from dysregulation of the metabolic program.30 The class II HDACs, HDAC5 and HDAC9, are functionally redundant in myocardial development with the combinatorial, double mutant manifesting incomplete penetrance of cardiac developmental defects that result in embryonic and perinatal lethality after E15.5.32 The class III HDAC, Sirt1, is similarly required for normal cardiac development, with septal and valve defects leading to lethality at birth observed in mutants.33
Finally, methylation also plays a role in cardiac development. The histone methyltransferase, SMYD1, is required for expression of a number of genes involved in myocardial growth and expression34, which may be due in part to its interaction with HDACs. Loss of SMYD1 triggers cardiac enlargement resulting in embryonic lethality.34 WHSC1 (Wolf–Hirschhorn syndrome candidate 1) is a histone methyltransferase that associates with Nkx2.5 to govern cardiovascular development.35 Jarid2, Jmjd, and UTX, enzymes within the Jumonji family of histone demethylases, also function in cardiac development, with Jarid2 involved in the formation of the trabeculated region of the developing myocardium, and Jmjd3 and UTX required for cardiovascular differentiation.36, 37,38 De-methylation of H3K27 by UTX appears to act as a signal to recruit Brg1 to cardiomyocyte-specific enhancers, thereby activating the cardiac development program.38
Histone PTM’s and cardiac hypertrophy
Stress to the heart caused by myocardial injury, hypertension, or neurohumoral activation typically triggers a hypertrophic growth response, which in the short term may be adaptive.39, 40 If the disease-related stress is not relieved, however, the heart undergoes irreversible decompensation that results in dilation of the ventricle, perturbations in contractile performance, and clinical heart failure. Many signaling pathways are involved in the development of cardiac hypertrophy, with altered intracellular Ca2+ homeostasis a central trigger.41 These multiple pathways funnel into a limited number of molecular responses, resulting in alterations in gene expression that trigger changes in cardiac structure, energy substrate utilization, and cell survival.
A key marker of pathological hypertrophy is the down-regulation of α-MHC and concurrent up-regulation of β-MHC, a reversing of the switch from fetal to adult myosin heavy chain expression seen at birth. The Brg1-Hdac-Parp chromatin remodeling complex, required for this switch, is activated during pathological stress, and loss of activity of this complex results in a diminished hypertrophic response.26 Interestingly, control of Brg1 activity occurs, in part, through expression of a cluster of long non-coding RNAs (Mhrt RNAs) that bind directly to Brg1 and decrease the helicase activity of the complex.42 Mhrt expression is abundant only in the adult cardiomyocyte and decreases in the setting of pathological stress.42 Inhibition of Mhrt expression is dependent on Brg1-Hdac-Parp complex activity, suggesting a feedback mechanism in which each complex tamps down the activity of the other. Transgenic expression in cardiomyocytes of Mhrt779 (the most abundant of these Mhrt RNAs) during pressure overload stress is protective and leads to a phenotype that is similar to that observed in Brg1 knockout mice.42
While Brg1-HDAC-Parp activity is clearly required for a robust hypertrophic response to pressure overload, the role of MHC class switching in this response is less clear. While even small shifts in isoform expression can alter cardiac output, class switching in mice in the absence of other stimuli does not result in hypertrophic growth.43, 44 Additionally, although increased β-MHC expression is associated with human heart failure, the beta isoform is the predominant MHC class in healthy human hearts.45 These facts leave open the possibility that other targets of Brg1 may impact hypertrophic growth.
The role of HDACs in pathological cardiac hypertrophy is arguably the most extensively studied context of chromatin modification in the heart. Initial work focused on class II HDACs, which are abundant in the myocardium. A number of biochemical and in vivo studies revealed that class II HDACs act as signal-responsive repressors of cardiac hypertrophy, in part through interaction with MEF2 (myocyte enhancer factor-2).32, 46
MEF2 proteins act as transcriptional switches that interact with a diverse set of cofactors to respond to specific intracellular signals to either activate or repress transcription.47 Mammals harbor four MEF2-encoding genes (Mef2a-d) that can form heterodimers, with MEF2A and MEF2D expression being highest in adult heart.48 Mice silenced for Mef2a manifest perinatal lethality and cardiac abnormalities.49 In contrast, Mef2d-null mice develop normally but manifest a blunted growth response to hypertrophic stress.50 MEF2 proteins are activated during cardiac stress, and forced expression of MEF2d in the heart is sufficient to induce a hypertrophic response.50
Class II HDACs interact directly with MEF2 to repress its transcriptional activity.51 Hypertrophic stress results in the phosphorylation of these HDACs and dissociation of the MEF2/HDAC complex.46 Mice lacking either HDAC9 or HDAC5 develop spontaneous cardiac hypertrophy as they age and elicit an exaggerated hypertrophic response to afterload stress.32, 46 Interestingly, these same mutant strains have a normal response to β-adrenergic stimulation, suggesting a difference in either the triggered pathway or extent of hypertrophic pressure under these two conditions.32
While class II HDACs appear to repress cardiac hypertrophy through multiple targets, the catalytic domain is, in part, dispensable for this activity.32 This may be due to the recruitment of other HDAC proteins to the transcriptional complex or through displacement of other activating factors, such as HATs.32
While loss of class II HDACs promotes cardiac hypertrophy, broad inhibition of the activities of both class I and class II HDACs using small molecule inhibitors, such as trichostatin A (TSA), represses hypertrophic growth in response to either pressure overload or β-adrenergic stimuli.52–54 This blunting of hypertrophy is also observed with HDAC inhibitors specific to class I isoforms.54, 55 Furthermore, treatment of mice with HDAC inhibitors after the onset of hypertrophy leads to regression of pathological growth even when the hypertrophic stress is still present.54, 56 Thus, class I HDAC activity appears dominant to class II in the balance of hypertrophic growth. The observation that in some complexes the role of the class II HDAC may not rely on catalytic activity may in part explain this differential effect.57 Additionally, there appears to be cross talk in the regulation of class I and class II HDACs with respect to their roles in hypertrophy. A recent report suggests a model in which pCAF (p300/CBP-associated factor) activates HDAC2 by direct acetylation, and HDAC5 inhibits HDAC2 by removing that acetyl moiety.58
Over-expression of HDAC2 alone in cardiac myocytes is sufficient to drive hypertrophic growth.59 However, class I HDACs appear to be functionally redundant in their ability to regulate hypertrophy, as cardiomyocyte-specific loss of either HDAC1 or HDAC2 has no impact on the response to hypertrophic stimuli.31 However, this has yet to be tested directly in adult mice. In contrast, cardiomyocyte-specific silencing of HDAC3 provokes abnormalities in myocardial energy metabolism leading to cardiac hypertrophy and failure.30, 60 Indeed, there are likely multiple pathways regulated by the activity of any given HDAC isoform, as well as redundant activities.
A complete picture of the mechanism of HDAC inhibition in blunting hypertrophy has yet to emerge. It is likely that HDAC inhibition targets a variety of pathways. HDAC2 has been shown to repress expression of the phosphatidylinositol-3,4,5-trisphosphate (PIP3) phosphatase Inpp5f, resulting in activation of the phosphatidylinositol-3-kinase- (PI3K-) Akt-Gsk3β cascade.59 Additionally, HDAC inhibition suppresses the maladaptive, disease-promoting autophagy observed under hypertrophic growth conditions.56, 61 HDAC inhibition also blunts increases in cellular protein synthesis by mechanisms that remain unclear.52 The recent finding that HDAC inhibitors can also stimulate cardiac protein SUMOylation in the absence of de novo gene transcription or protein synthesis suggests that in addition to their role in histone acetylation, acetylation of other protein targets may play a role in the regulation of hypertrophy.62
Acetyltransferases would seem to be the natural antagonist to combat the hypertrophy-promoting tendencies of HDAC activity. However, both p300 and pCAF induce hypertrophy, presumably by activating pro-growth genes.58, 63 Hypertrophic stress, or forced expression of p300, results in increased acetylation of Gata-4 and expression of Gata-4-dependent hypertrophy genes. As mentioned earlier, p300/pCAF can also activate HDAC2 by direct acetylation.58 Regardless, one would anticipate that specific HATs, yet to be identified, may be important in opposing the activity of class I HDACs. However, given that these are highly regulated, multiprotein complexes with multiple partners and targets, this expectation may be naive.
The role of readers of these hypertrophic stress-induced changes in histone acetylation is just emerging. BET proteins act to increase the occupancy of transcriptional complexes that promote chromatin remodeling, transcriptional initiation, and elongation.64 Hypertrophic stimulation triggers an increase in steady-state levels of BRD4 protein and enhanced association with target genes.65, 66 Knockdown of BRD4 in neonatal rat ventricular myocytes (NRVMs) is sufficient to inhibit PE-induced hypertrophy.65 Interestingly, the control of BRD4 abundance appears not to be transcriptional.65 Chromatin immunoprecipitation studies in TAC-treated hearts suggest a model in which BRD4, through a known interaction with the transcriptional elongation factor p-TEFb, promotes transcriptional pause release in response to pathologic stress.65
The selective bromodomain inhibitor JQ1 inhibits both PE-induced hypertrophy in vitro and pressure overload-induced cardiac hypertrophy in vivo.65 JQ1-mediated suppression affects a wide range of pathways involved in cardiac hypertrophy and failure, including cytoskeletal remodeling, cell growth signals, and pro-inflammatory signaling.65 However, despite these broad effects, JQ1 impacts only a subset of the genes up-regulated in response to hypertrophic stimuli. Specifically, JQ1 inhibition correlated strongly with calcineurin-NFAT, NFκB, and GATA4 gene signatures, but not with c-myc or E2F-regulated genes.65 Presently, it is unclear the extent to which the protective phenotype of JQ1 treatment is due specifically to blocking BRD4 activity. Conditional alleles for the BRD4 gene will allow this question to be addressed directly.
Changes in histone methylation within cardiac myocytes are observed in heart subjected to hypertrophic stress.67, 68 These include both activating (H3K9ac, H3K27ac, H3K4me3, H3K79me2) and repressive (H3K9me2, H3K9me3, H3K27me3) modifications.67 The histone methyltransferase, Ezh2, plays a critical role in repressing the hypertrophic gene program in adult cardiomyocytes by maintaining H3K4me3 modifications.69 H3K79 methylation is catalyzed by the methyltransferase Dot1L. Cardiomyocyte-specific loss of Dot1L results in decreased dystrophin expression and leads to a dilated cardiomyopathic phenotype that can be partially rescued with adenoviral delivery of the minidystrophin gene.70 Increased H3K79 methylation by Dot1L may be important for increased expression of dystrophin in the setting of pressure overload, serving to help maintain cardiac structure and function during hypertrophic growth.71, 72
The flip side of the coin is the JmjC domain-containing proteins. JMD2a is part of a family of lysine trimethyl-specific histone demethylases that target H3K9me3.73 By relieving repressive histone modifications, JMJD proteins can activate transcription through interaction with specific transcription factors, such as the androgen receptor or serum response factor.74, 75 This epigenetic control of histone methylation is critical to the cardiomyocyte transcriptional response to hypertrophic stress. Cardiomyocyte-specific silencing of Jmjd2a attenuates the myocardial response to pressure overload-induced hypertrophy.75 Conversely, transgenic expression of JMJD2A in the heart leads to an exaggerated hypertrophic response.75 This appears to occur in part through up-regulation of four-and-a-half LIM domains-1 gene expression.75
Alterations in histone methylation in the ANF and BNP promoter regions are also associated with cardiac stress, with JMJD1A and JMJD2A both bound to the promoter region of ANF.76 In NRVMs, simultaneous siRNA targeting of both of these demethylases results in decreased expression of both ANF and BNP and increased abundance of H3K9me2 and H3K9me3 in the promoter regions.76 These changes in histone methylation are correlated with the nuclear export of HDAC4 under conditions of increased hemodynamic load.76 The class II HDAC, HDAC4, is found within a complex harboring the histone methyltransferase SUV39H.76 This complex is disrupted under stress conditions by CaMKII∂B-induced phosphorylation and nuclear export of HDAC4.76 CaMKII-mediated phosphorylation of HDAC4 in NRVMs results in hypertrophic growth through de-repression of HDAC target genes.77 Once again, these data are consistent with a model in which the dynamic balance of histone acetylation/deacetylation and methylation/demethylation on promoters is regulated through interdependency of these enzymes in both complex formation and activity.
Recent studies have demonstrated that CaMKII activation under conditions of hypertrophic stress leads to hyperphosphorylation of Ser-10 on H3 at a number of hypertrophic gene loci.78, 79 Similar changes have been reported in patients with heart failure.78 Silencing of nuclear CaMKII reduces H3 Ser-10 phosphorylation, a PTM which is required for normal Mef2-dependent transcription.79 The chaperone protein 14-3-3 acts as a reader in this situation, binding to phosphorylated H3 to stimulate transcriptional elongation of hypertrophic response genes by RNA Pol II.78 Thus, in addition to the established roles of acetylation and methylation signals, histone phosphorylation plays a role in cardiac stress.
The reader/eraser paradox
Given that inhibition of readers (e.g. BET inhibitors, such as JQ1) blunts hypertrophy, it seems paradoxical that HDAC inhibitors, which would increase acetylation on histones and presumably BRD4 association with chromatin, decrease expression of hypertrophic genes. Conversely, increased HDAC activity, which would presumably promote BRD4 dissociation, increases the hypertrophic response.59 The answer to this conundrum may lie in recognizing that this biology is highly dynamic, not static. Both HATs and HDACs are targeted to transcribed regions of active genes, with the HATs activating and the HDACs resetting transcription.80 The binding of BRD4 to chromatin, the initiation of transcription, and transcript elongation appear to require differential contact with combinations of acetylated histone lysines and an exchange of Brd4-interacting partners.81 In this way, HDAC-dependent release of BRD4 from chromatin after cell stress has been proposed as a requirement for proper pTEFb recruitment, with HDAC inhibitors blocking BRD4-dependent transcriptional elongation.82
Histone PTMs and cardiac ischemia
Hearts affected by ischemic cardiomyopathy manifest a remodeling response that includes increases in fibrosis and a cellular hypertrophic response that is offset in part by cardiomyocyte death. Not surprisingly, then, many of the chromatin changes associated with the pro-hypertrophic transcriptional response are observed in failing ischemic hearts.67, 68 However, in the acute stages of ischemic cardiomyopathy, as well as during reperfusion injury, specific pathways are likely activated to limit the extent of cell death emerging from the changing oxygen environment. Interestingly, HDAC inhibitors are capable of protecting the ischemic heart from both of these insults, by limiting the extent of cell death at reperfusion and by reducing fibrosis and cardiomyocyte hypertrophy in both the border and remote zones of the infarcted heart.83–85
Treatment of Wistar rats with a daily dose of the HDAC inhibitor valproic acid (VPA) or tributyrin beginning one day after ischemic injury preserved ventricular performance, reduced fibrosis, and blunted hypertrophy in the infarcted heart.83 These results are likely a combination of cardiomyocyte-specific and nonspecific events. Class I-specific HDAC inhibitors can act directly on fibroblasts to inhibit cell cycle progression and block fibrocyte maturation, in part through the inhibition of ERK signaling.86 Additionally, HDAC inhibition has been shown to target the inflammatory cascade that accompanies heart failure.87
Perhaps the most impressive findings reported to date are the impact of HDAC inhibitors on limiting reperfusion injury. A single dose of an HDAC inhibitor administered at the time of reperfusion reduces the size of a myocardial infarct by nearly 50%.84, 85 Remarkably, this effect can occur even if the dose is delivered 45 minutes after reperfusion.84 This suggests that the effect of HDAC inhibition treatment is not on the trigger of reperfusion injury, but rather acts on the cell death/survival pathways activated by reperfusion. The mechanism of this protection is likely multifaceted; early studies suggested that it is mediated by HDAC4 and involved reduction in hypoxia-inducible factor-1a protein levels84.
Our group has demonstrated a role for induction of autophagy by HDAC inhibition in limiting myocardial reperfusion injury.85 We and others have shown that autophagic flux is repressed in cardiomyocytes during reperfusion.85, 88, 89 Short-term treatment with HDAC inhibitors restores this flux, conferring cardioprotection. This effect is in contrast to the inhibition of autophagy that occurs with long-term HDAC inhibitor treatment of hearts under hypertrophic stress.56, 85 The mechanism whereby HDACs modulate autophagy is unclear but similar activating and inhibiting effects of HDAC activity on autophagy are observed in a variety of cancer cell lines.90 Given the myriad pathways and cell types that HDACs modulate, it is not surprising that the impact of HDAC inhibition in mitigating ischemic injury is also being examined in the rescue of stroke-related injury.91
Alteration of histone methylation may also protect against I/R injury. In a model of murine diabetes, adenoviral delivery of SUV39h1 to the heart resulted in a significant decrease in infarct size after only four hours of reperfusion.92 While the interaction of SUV39h1 with HDAC4 is unknown under these specific conditions, one possibility is that over-expression of SUV39h1 compensates for the loss of HDAC4 interaction, leading to repression of HDAC4-regulated promoters.76, 84
Class III HDACs are also cardioprotective. Activation of sirtuin activity is believed to be part of the basis for the beneficial effects of resveratrol in heart failure.93 Whereas Sirt1 and Sirt3 are well established as HDACs, their non-histone protein deacetylation activities in the nucleus (Sirt1) and in mitochondria (Sirt3) are the most studied.94 Given the direct link between sirtuin activity and the energetic and redox states of the cell, it is not surprising that sirtuins play a role in the regulation of reactive oxygen species, a key driver of reperfusion injury.94 Preconditioning the heart by activation of Sirt1 can be protective against ischemia, and Sirt1 activation is believed to help preserve mitochondrial abundance and energy production in models of heart failure.95 However, transgenic expression of Sirt1 in cardiomyocytes, even at low levels, triggers a decrease in cardiac function, presumably due to dysregulation of mitochondrial function.96 Thus, the balance and timing of Sirt1 activity is clearly important.
Conclusion and perspective
Complexity in the regulation of epigenetic changes within disease-stressed heart, and their role in the underlying pathophysiology, is daunting. However, the potential of small molecules to impact the architecture of chromatin and alter the transcriptional response to stress is promising. Indeed, translation to the clinic may be in the offing. Vorinostat, a pan-HDAC inhibitor currently approved for treatment of cutaneous T cell lymphoma, has been shown to limit cardiac reperfusion injury in mice and rabbits.85 Delivered as a single dose at the time of reperfusion, infarct size is significantly blunted, a finding which is arguably closest to being tested in clinical trials. As we unveil more specifics regarding the interplay of these histone PTM pathways and the sensitivity of target genes to changes in histone PTM homeostasis, we move closer to the objective of clinically meaningful targeting of pathological cardiac remodeling.
Acknowledgments
Sources of Funding
This work was supported by grants from the NIH (HL-120732; HL-100401), AHA (14SFRN20740000), CPRIT (RP110486P3), and the Leducq Foundation (11CVD04).
Nonstandard Abbreviations and Acronyms
- PTMs
post-translational modifications
- HAT
histone acetyltransferase
- HDAC
histone deacetylase
- H3K4me3
H3-lysine-4 tri-methylation
- H3K4me2
H3-lysine-4 di-methylation
- H3K9me1
H3-lysine-mono-methylation
- H3K9me2
H3-lysine-9 di-methylation
- H3K9me3
H3-lysine-9 tri-methylation
- H3K27me2
H3-lysine-27 di-methylation
- H3K27me3
H3-lysine-27 tri-methylation
Footnotes
In January 2015, the average time from submission to first decision for all original research papers submitted to Circulation Research was 14.7 days.
Conflicts of Interest
We declare no conflicts of interest.
References
- 1.Rothbart SB, Strahl BD. Interpreting the language of histone and DNA modifications. Biochimica et biophysica acta. 2014;1839:627–643. doi: 10.1016/j.bbagrm.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allfrey VG, Faulkner R, Mirsky AE. Acetylation and methylation of histones and their possible role in the regulation of rna synthesis. Proceedings of the National Academy of Sciences of the United States of America. 1964;51:786–794. doi: 10.1073/pnas.51.5.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brownell JE, Zhou J, Ranalli T, Kobayashi R, Edmondson DG, Roth SY, Allis CD. Tetrahymena histone acetyltransferase a: A homolog to yeast gcn5p linking histone acetylation to gene activation. Cell. 1996;84:843–851. doi: 10.1016/s0092-8674(00)81063-6. [DOI] [PubMed] [Google Scholar]
- 4.Parthun MR, Widom J, Gottschling DE. The major cytoplasmic histone acetyltransferase in yeast: Links to chromatin replication and histone metabolism. Cell. 1996;87:85–94. doi: 10.1016/s0092-8674(00)81325-2. [DOI] [PubMed] [Google Scholar]
- 5.Mizzen CA, Yang XJ, Kokubo T, Brownell JE, Bannister AJ, Owen-Hughes T, Workman J, Wang L, Berger SL, Kouzarides T, Nakatani Y, Allis CD. The taf(ii)250 subunit of tfiid has histone acetyltransferase activity. Cell. 1996;87:1261–1270. doi: 10.1016/s0092-8674(00)81821-8. [DOI] [PubMed] [Google Scholar]
- 6.Bannister AJ, Kouzarides T. The cbp co-activator is a histone acetyltransferase. Nature. 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- 7.Lee KK, Workman JL. Histone acetyltransferase complexes: One size doesn’t fit all. Nature reviews Molecular cell biology. 2007;8:284–295. doi: 10.1038/nrm2145. [DOI] [PubMed] [Google Scholar]
- 8.Yuan H, Marmorstein R. Histone acetyltransferases: Rising ancient counterparts to protein kinases. Biopolymers. 2013;99:98–111. doi: 10.1002/bip.22128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Strahl BD, Ohba R, Cook RG, Allis CD. Methylation of histone h3 at lysine 4 is highly conserved and correlates with transcriptionally active nuclei in tetrahymena. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:14967–14972. doi: 10.1073/pnas.96.26.14967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rea S, Eisenhaber F, O’Carroll D, Strahl BD, Sun ZW, Schmid M, Opravil S, Mechtler K, Ponting CP, Allis CD, Jenuwein T. Regulation of chromatin structure by site-specific histone h3 methyltransferases. Nature. 2000;406:593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- 11.Del Rizzo PA, Trievel RC. Molecular basis for substrate recognition by lysine methyltransferases and demethylases. Biochimica et biophysica acta. 2014 doi: 10.1016/j.bbagrm.2014.06.008. [DOI] [PubMed] [Google Scholar]
- 12.Lyons DB, Lomvardas S. Repressive histone methylation: A case study in deterministic versus stochastic gene regulation. Biochimica et biophysica acta. 2014;1839:1373–1384. doi: 10.1016/j.bbagrm.2014.05.010. [DOI] [PubMed] [Google Scholar]
- 13.Taunton J, Hassig CA, Schreiber SL. A mammalian histone deacetylase related to the yeast transcriptional regulator rpd3p. Science. 1996;272:408–411. doi: 10.1126/science.272.5260.408. [DOI] [PubMed] [Google Scholar]
- 14.Gregoretti IV, Lee YM, Goodson HV. Molecular evolution of the histone deacetylase family: Functional implications of phylogenetic analysis. Journal of molecular biology. 2004;338:17–31. doi: 10.1016/j.jmb.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 15.Xie M, Hill JA. Hdac-dependent ventricular remodeling. Trends in cardiovascular medicine. 2013;23:229–235. doi: 10.1016/j.tcm.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Falkenberg KJ, Johnstone RW. Histone deacetylases and their inhibitors in cancer, neurological diseases and immune disorders. Nature reviews Drug discovery. 2014;13:673–691. doi: 10.1038/nrd4360. [DOI] [PubMed] [Google Scholar]
- 17.Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA, Shi Y. Histone demethylation mediated by the nuclear amine oxidase homolog lsd1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 18.Thinnes CC, England KS, Kawamura A, Chowdhury R, Schofield CJ, Hopkinson RJ. Targeting histone lysine demethylases - progress, challenges, and the future. Biochimica et biophysica acta. 2014;1839:1416–1432. doi: 10.1016/j.bbagrm.2014.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yun M, Wu J, Workman JL, Li B. Readers of histone modifications. Cell research. 2011;21:564–578. doi: 10.1038/cr.2011.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Awad S, Hassan AH. The swi2/snf2 bromodomain is important for the full binding and remodeling activity of the swi/snf complex on h3- and h4-acetylated nucleosomes. Annals of the New York Academy of Sciences. 2008;1138:366–375. doi: 10.1196/annals.1414.038. [DOI] [PubMed] [Google Scholar]
- 21.Filippakopoulos P, Knapp S. Targeting bromodomains: Epigenetic readers of lysine acetylation. Nature reviews Drug discovery. 2014;13:337–356. doi: 10.1038/nrd4286. [DOI] [PubMed] [Google Scholar]
- 22.Shi J, Vakoc CR. The mechanisms behind the therapeutic activity of bet bromodomain inhibition. Molecular cell. 2014;54:728–736. doi: 10.1016/j.molcel.2014.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Han P, Hang CT, Yang J, Chang CP. Chromatin remodeling in cardiovascular development and physiology. Circulation research. 2011;108:378–396. doi: 10.1161/CIRCRESAHA.110.224287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lickert H, Takeuchi JK, Von Both I, Walls JR, McAuliffe F, Adamson SL, Henkelman RM, Wrana JL, Rossant J, Bruneau BG. Baf60c is essential for function of baf chromatin remodelling complexes in heart development. Nature. 2004;432:107–112. doi: 10.1038/nature03071. [DOI] [PubMed] [Google Scholar]
- 25.Wang Z, Zhai W, Richardson JA, Olson EN, Meneses JJ, Firpo MT, Kang C, Skarnes WC, Tjian R. Polybromo protein baf180 functions in mammalian cardiac chamber maturation. Genes & development. 2004;18:3106–3116. doi: 10.1101/gad.1238104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hang CT, Yang J, Han P, Cheng HL, Shang C, Ashley E, Zhou B, Chang CP. Chromatin regulation by brg1 underlies heart muscle development and disease. Nature. 2010;466:62–67. doi: 10.1038/nature09130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Singh AP, Archer TK. Analysis of the swi/snf chromatin-remodeling complex during early heart development and baf250a repression cardiac gene transcription during p19 cell differentiation. Nucleic acids research. 2014;42:2958–2975. doi: 10.1093/nar/gkt1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yao TP, Oh SP, Fuchs M, Zhou ND, Ch’ng LE, Newsome D, Bronson RT, Li E, Livingston DM, Eckner R. Gene dosage-dependent embryonic development and proliferation defects in mice lacking the transcriptional integrator p300. Cell. 1998;93:361–372. doi: 10.1016/s0092-8674(00)81165-4. [DOI] [PubMed] [Google Scholar]
- 29.Shikama N, Lutz W, Kretzschmar R, Sauter N, Roth JF, Marino S, Wittwer J, Scheidweiler A, Eckner R. Essential function of p300 acetyltransferase activity in heart, lung and small intestine formation. The EMBO journal. 2003;22:5175–5185. doi: 10.1093/emboj/cdg502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Montgomery RL, Potthoff MJ, Haberland M, Qi X, Matsuzaki S, Humphries KM, Richardson JA, Bassel-Duby R, Olson EN. Maintenance of cardiac energy metabolism by histone deacetylase 3 in mice. The Journal of clinical investigation. 2008;118:3588–3597. doi: 10.1172/JCI35847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Montgomery RL, Davis CA, Potthoff MJ, Haberland M, Fielitz J, Qi X, Hill JA, Richardson JA, Olson EN. Histone deacetylases 1 and 2 redundantly regulate cardiac morphogenesis, growth, and contractility. Genes & development. 2007;21:1790–1802. doi: 10.1101/gad.1563807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chang S, McKinsey TA, Zhang CL, Richardson JA, Hill JA, Olson EN. Histone deacetylases 5 and 9 govern responsiveness of the heart to a subset of stress signals and play redundant roles in heart development. Molecular and cellular biology. 2004;24:8467–8476. doi: 10.1128/MCB.24.19.8467-8476.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cheng HL, Mostoslavsky R, Saito S, Manis JP, Gu Y, Patel P, Bronson R, Appella E, Alt FW, Chua KF. Developmental defects and p53 hyperacetylation in sir2 homolog (sirt1)-deficient mice. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:10794–10799. doi: 10.1073/pnas.1934713100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gottlieb PD, Pierce SA, Sims RJ, Yamagishi H, Weihe EK, Harriss JV, Maika SD, Kuziel WA, King HL, Olson EN, Nakagawa O, Srivastava D. Bop encodes a muscle-restricted protein containing mynd and set domains and is essential for cardiac differentiation and morphogenesis. Nature genetics. 2002;31:25–32. doi: 10.1038/ng866. [DOI] [PubMed] [Google Scholar]
- 35.Nimura K, Ura K, Shiratori H, Ikawa M, Okabe M, Schwartz RJ, Kaneda Y. A histone h3 lysine 36 trimethyltransferase links nkx2-5 to wolf-hirschhorn syndrome. Nature. 2009;460:287–291. doi: 10.1038/nature08086. [DOI] [PubMed] [Google Scholar]
- 36.Takeuchi T, Kojima M, Nakajima K, Kondo S. Jumonji gene is essential for the neurulation and cardiac development of mouse embryos with a c3h/he background. Mechanisms of development. 1999;86:29–38. doi: 10.1016/s0925-4773(99)00100-8. [DOI] [PubMed] [Google Scholar]
- 37.Ohtani K, Zhao C, Dobreva G, Manavski Y, Kluge B, Braun T, Rieger MA, Zeiher AM, Dimmeler S. Jmjd3 controls mesodermal and cardiovascular differentiation of embryonic stem cells. Circulation research. 2013;113:856–862. doi: 10.1161/CIRCRESAHA.113.302035. [DOI] [PubMed] [Google Scholar]
- 38.Lee S, Lee JW, Lee SK. Utx, a histone h3-lysine 27 demethylase, acts as a critical switch to activate the cardiac developmental program. Developmental cell. 2012;22:25–37. doi: 10.1016/j.devcel.2011.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hill JA, Olson EN. Cardiac plasticity. New Engl J Med. 2008;358:1370–1380. doi: 10.1056/NEJMra072139. [DOI] [PubMed] [Google Scholar]
- 40.Schiattarella GG, Hill JA. Inhibition of hypertrophy is a good therapeutic strategy in ventricular pressure overload. Circulation. 2015 doi: 10.1161/CIRCULATIONAHA.115.013894. In Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Burchfield JS, Xie M, Hill JA. Pathological ventricular remodeling: Mechanisms: Part 1 of 2. Circulation. 2013;128:388–400. doi: 10.1161/CIRCULATIONAHA.113.001878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Han P, Li W, Lin CH, Yang J, Shang C, Nurnberg ST, Jin KK, Xu W, Lin CY, Lin CJ, Xiong Y, Chien HC, Zhou B, Ashley E, Bernstein D, Chen PS, Chen HS, Quertermous T, Chang CP. A long noncoding rna protects the heart from pathological hypertrophy. Nature. 2014;514:102–106. doi: 10.1038/nature13596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Krenz M, Robbins J. Impact of beta-myosin heavy chain expression on cardiac function during stress. Journal of the American College of Cardiology. 2004;44:2390–2397. doi: 10.1016/j.jacc.2004.09.044. [DOI] [PubMed] [Google Scholar]
- 44.Herron TJ, McDonald KS. Small amounts of alpha-myosin heavy chain isoform expression significantly increase power output of rat cardiac myocyte fragments. Circulation research. 2002;90:1150–1152. doi: 10.1161/01.res.0000022879.57270.11. [DOI] [PubMed] [Google Scholar]
- 45.Razeghi P, Young ME, Alcorn JL, Moravec CS, Frazier OH, Taegtmeyer H. Metabolic gene expression in fetal and failing human heart. Circulation. 2001;104:2923–2931. doi: 10.1161/hc4901.100526. [DOI] [PubMed] [Google Scholar]
- 46.Zhang CL, McKinsey TA, Chang S, Antos CL, Hill JA, Olson EN. Class ii histone deacetylases act as signal-responsive repressors of cardiac hypertrophy. Cell. 2002;110:479–488. doi: 10.1016/s0092-8674(02)00861-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Black BL, Cripps RM. Myocyte enhancer factor 2 transcription factors in heart development and disease. In: Rosenthal N, Harvey RP, editors. Heart development and regeneration. Oxford: Elsevier Inc. Academic Press; 2010. pp. 673–699. [Google Scholar]
- 48.Molkentin JD, Black BL, Martin JF, Olson EN. Cooperative activation of muscle gene expression by mef2 and myogenic bhlh proteins. Cell. 1995;83:1125–1136. doi: 10.1016/0092-8674(95)90139-6. [DOI] [PubMed] [Google Scholar]
- 49.Naya FJ, Black BL, Wu H, Bassel-Duby R, Richardson JA, Hill JA, Olson EN. Mitochondrial deficiency and cardiac sudden death in mice lacking the mef2a transcription factor. Nature medicine. 2002;8:1303–1309. doi: 10.1038/nm789. [DOI] [PubMed] [Google Scholar]
- 50.Kim Y, Phan D, van Rooij E, Wang DZ, McAnally J, Qi X, Richardson JA, Hill JA, Bassel-Duby R, Olson EN. The mef2d transcription factor mediates stress-dependent cardiac remodeling in mice. The Journal of clinical investigation. 2008;118:124–132. doi: 10.1172/JCI33255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lu J, McKinsey TA, Nicol RL, Olson EN. Signal-dependent activation of the mef2 transcription factor by dissociation from histone deacetylases. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:4070–4075. doi: 10.1073/pnas.080064097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Antos CL, McKinsey TA, Dreitz M, Hollingsworth LM, Zhang CL, Schreiber K, Rindt H, Gorczynski RJ, Olson EN. Dose-dependent blockade to cardiomyocyte hypertrophy by histone deacetylase inhibitors. The Journal of biological chemistry. 2003;278:28930–28937. doi: 10.1074/jbc.M303113200. [DOI] [PubMed] [Google Scholar]
- 53.Kong Y, Tannous P, Lu G, Berenji K, Rothermel BA, Olson EN, Hill JA. Suppression of class i and ii histone deacetylases blunts pressure-overload cardiac hypertrophy. Circulation. 2006;113:2579–2588. doi: 10.1161/CIRCULATIONAHA.106.625467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kee HJ, Sohn IS, Nam KI, Park JE, Qian YR, Yin Z, Ahn Y, Jeong MH, Bang YJ, Kim N, Kim JK, Kim KK, Epstein JA, Kook H. Inhibition of histone deacetylation blocks cardiac hypertrophy induced by angiotensin ii infusion and aortic banding. Circulation. 2006;113:51–59. doi: 10.1161/CIRCULATIONAHA.105.559724. [DOI] [PubMed] [Google Scholar]
- 55.Gallo P, Latronico MV, Gallo P, Grimaldi S, Borgia F, Todaro M, Jones P, Gallinari P, De Francesco R, Ciliberto G, Steinkuhler C, Esposito G, Condorelli G. Inhibition of class i histone deacetylase with an apicidin derivative prevents cardiac hypertrophy and failure. Cardiovascular research. 2008;80:416–424. doi: 10.1093/cvr/cvn215. [DOI] [PubMed] [Google Scholar]
- 56.Cao DJ, Wang ZV, Battiprolu PK, Jiang N, Morales CR, Kong Y, Rothermel BA, Gillette TG, Hill JA. Histone deacetylase (hdac) inhibitors attenuate cardiac hypertrophy by suppressing autophagy. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:4123–4128. doi: 10.1073/pnas.1015081108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fischle W, Dequiedt F, Hendzel MJ, Guenther MG, Lazar MA, Voelter W, Verdin E. Enzymatic activity associated with class ii hdacs is dependent on a multiprotein complex containing hdac3 and smrt/n-cor. Molecular cell. 2002;9:45–57. doi: 10.1016/s1097-2765(01)00429-4. [DOI] [PubMed] [Google Scholar]
- 58.Eom GH, Nam YS, Oh JG, Choe N, Min HK, Yoo EK, Kang G, Nguyen VH, Min JJ, Kim JK, Lee IK, Bassel-Duby R, Olson EN, Park WJ, Kook H. Regulation of acetylation of histone deacetylase 2 by p300/cbp-associated factor/histone deacetylase 5 in the development of cardiac hypertrophy. Circulation research. 2014;114:1133–1143. doi: 10.1161/CIRCRESAHA.114.303429. [DOI] [PubMed] [Google Scholar]
- 59.Trivedi CM, Luo Y, Yin Z, Zhang M, Zhu W, Wang T, Floss T, Goettlicher M, Noppinger PR, Wurst W, Ferrari VA, Abrams CS, Gruber PJ, Epstein JA. Hdac2 regulates the cardiac hypertrophic response by modulating gsk3 beta activity. Nature medicine. 2007;13:324–331. doi: 10.1038/nm1552. [DOI] [PubMed] [Google Scholar]
- 60.Sun Z, Singh N, Mullican SE, Everett LJ, Li L, Yuan L, Liu X, Epstein JA, Lazar MA. Diet-induced lethality due to deletion of the hdac3 gene in heart and skeletal muscle. The Journal of biological chemistry. 2011;286:33301–33309. doi: 10.1074/jbc.M111.277707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lavandero S, Chiong M, Rothermel BA, Hill JA. Autophagy in cardiovascular biology. The Journal of clinical investigation. 2015;125:55–64. doi: 10.1172/JCI73943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Blakeslee WW, Wysoczynski CL, Fritz KS, Nyborg JK, Churchill ME, McKinsey TA. Class i hdac inhibition stimulates cardiac protein sumoylation through a post-translational mechanism. Cellular signalling. 2014;26:2912–2920. doi: 10.1016/j.cellsig.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gusterson RJ, Jazrawi E, Adcock IM, Latchman DS. The transcriptional co-activators creb-binding protein (cbp) and p300 play a critical role in cardiac hypertrophy that is dependent on their histone acetyltransferase activity. The Journal of biological chemistry. 2003;278:6838–6847. doi: 10.1074/jbc.M211762200. [DOI] [PubMed] [Google Scholar]
- 64.Dawson MA, Kouzarides T, Huntly BJ. Targeting epigenetic readers in cancer. The New England journal of medicine. 2012;367:647–657. doi: 10.1056/NEJMra1112635. [DOI] [PubMed] [Google Scholar]
- 65.Anand P, Brown JD, Lin CY, Qi J, Zhang R, Artero PC, Alaiti MA, Bullard J, Alazem K, Margulies KB, Cappola TP, Lemieux M, Plutzky J, Bradner JE, Haldar SM. Bet bromodomains mediate transcriptional pause release in heart failure. Cell. 2013;154:569–582. doi: 10.1016/j.cell.2013.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Spiltoir JI, Stratton MS, Cavasin MA, Demos-Davies K, Reid BG, Qi J, Bradner JE, McKinsey TA. Bet acetyl-lysine binding proteins control pathological cardiac hypertrophy. Journal of molecular and cellular cardiology. 2013;63:175–179. doi: 10.1016/j.yjmcc.2013.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Papait R, Cattaneo P, Kunderfranco P, Greco C, Carullo P, Guffanti A, Vigano V, Stirparo GG, Latronico MV, Hasenfuss G, Chen J, Condorelli G. Genome-wide analysis of histone marks identifying an epigenetic signature of promoters and enhancers underlying cardiac hypertrophy. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:20164–20169. doi: 10.1073/pnas.1315155110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Movassagh M, Choy MK, Knowles DA, Cordeddu L, Haider S, Down T, Siggens L, Vujic A, Simeoni I, Penkett C, Goddard M, Lio P, Bennett MR, Foo RS. Distinct epigenomic features in end-stage failing human hearts. Circulation. 2011;124:2411–2422. doi: 10.1161/CIRCULATIONAHA.111.040071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Delgado-Olguin P, Huang Y, Li X, Christodoulou D, Seidman CE, Seidman JG, Tarakhovsky A, Bruneau BG. Epigenetic repression of cardiac progenitor gene expression by ezh2 is required for postnatal cardiac homeostasis. Nature genetics. 2012;44:343–347. doi: 10.1038/ng.1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nguyen AT, Xiao B, Neppl RL, Kallin EM, Li J, Chen T, Wang DZ, Xiao X, Zhang Y. Dot1l regulates dystrophin expression and is critical for cardiac function. Genes & development. 2011;25:263–274. doi: 10.1101/gad.2018511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Maeda M, Biro S, Kamogawa Y, Hirakawa T, Setoguchi M, Tei C. Dystrophin upregulation in pressure-overloaded cardiac hypertrophy in rats. Cell motility and the cytoskeleton. 2003;55:26–35. doi: 10.1002/cm.10110. [DOI] [PubMed] [Google Scholar]
- 72.Kamogawa Y, Biro S, Maeda M, Setoguchi M, Hirakawa T, Yoshida H, Tei C. Dystrophin-deficient myocardium is vulnerable to pressure overload in vivo. Cardiovascular research. 2001;50:509–515. doi: 10.1016/s0008-6363(01)00205-x. [DOI] [PubMed] [Google Scholar]
- 73.Gray SG, Iglesias AH, Lizcano F, Villanueva R, Camelo S, Jingu H, Teh BT, Koibuchi N, Chin WW, Kokkotou E, Dangond F. Functional characterization of jmjd2a, a histone deacetylase- and retinoblastoma-binding protein. The Journal of biological chemistry. 2005;280:28507–28518. doi: 10.1074/jbc.M413687200. [DOI] [PubMed] [Google Scholar]
- 74.Shin S, Janknecht R. Activation of androgen receptor by histone demethylases jmjd2a and jmjd2d. Biochemical and biophysical research communications. 2007;359:742–746. doi: 10.1016/j.bbrc.2007.05.179. [DOI] [PubMed] [Google Scholar]
- 75.Zhang QJ, Chen HZ, Wang L, Liu DP, Hill JA, Liu ZP. The histone trimethyllysine demethylase jmjd2a promotes cardiac hypertrophy in response to hypertrophic stimuli in mice. The Journal of clinical investigation. 2011;121:2447–2456. doi: 10.1172/JCI46277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hohl M, Wagner M, Reil JC, Muller SA, Tauchnitz M, Zimmer AM, Lehmann LH, Thiel G, Bohm M, Backs J, Maack C. Hdac4 controls histone methylation in response to elevated cardiac load. The Journal of clinical investigation. 2013;123:1359–1370. doi: 10.1172/JCI61084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Backs J, Song K, Bezprozvannaya S, Chang S, Olson EN. Cam kinase ii selectively signals to histone deacetylase 4 during cardiomyocyte hypertrophy. The Journal of clinical investigation. 2006;116:1853–1864. doi: 10.1172/JCI27438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Awad S, Al-Haffar KM, Marashly Q, Quijada P, Kunhi M, Al-Yacoub N, Wade FS, Mohammed SF, Al-Dayel F, Sutherland G, Assiri A, Sussman M, Bers D, Al-Habeeb W, Poizat C. Control of histone h3 phosphorylation by camkiidelta in response to haemodynamic cardiac stress. The Journal of pathology. 2015;235:606–618. doi: 10.1002/path.4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Awad S, Kunhi M, Little GH, Bai Y, An W, Bers D, Kedes L, Poizat C. Nuclear camkii enhances histone h3 phosphorylation and remodels chromatin during cardiac hypertrophy. Nucleic acids research. 2013;41:7656–7672. doi: 10.1093/nar/gkt500. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 80.Wang Z, Zang C, Cui K, Schones DE, Barski A, Peng W, Zhao K. Genome-wide mapping of hats and hdacs reveals distinct functions in active and inactive genes. Cell. 2009;138:1019–1031. doi: 10.1016/j.cell.2009.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chiang CM. Brd4 engagement from chromatin targeting to transcriptional regulation: Selective contact with acetylated histone h3 and h4. F1000 biology reports. 2009;1:98. doi: 10.3410/B1-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ai N, Hu X, Ding F, Yu B, Wang H, Lu X, Zhang K, Li Y, Han A, Lin W, Liu R, Chen R. Signal-induced brd4 release from chromatin is essential for its role transition from chromatin targeting to transcriptional regulation. Nucleic acids research. 2011;39:9592–9604. doi: 10.1093/nar/gkr698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lee TM, Lin MS, Chang NC. Inhibition of histone deacetylase on ventricular remodeling in infarcted rats. American journal of physiology Heart and circulatory physiology. 2007;293:H968–977. doi: 10.1152/ajpheart.00891.2006. [DOI] [PubMed] [Google Scholar]
- 84.Granger A, Abdullah I, Huebner F, Stout A, Wang T, Huebner T, Epstein JA, Gruber PJ. Histone deacetylase inhibition reduces myocardial ischemia-reperfusion injury in mice. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2008;22:3549–3560. doi: 10.1096/fj.08-108548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Xie M, Kong Y, Tan W, May H, Battiprolu PK, Pedrozo Z, Wang ZV, Morales C, Luo X, Cho G, Jiang N, Jessen ME, Warner JJ, Lavandero S, Gillette TG, Turer AT, Hill JA. Histone deacetylase inhibition blunts ischemia/reperfusion injury by inducing cardiomyocyte autophagy. Circulation. 2014;129:1139–1151. doi: 10.1161/CIRCULATIONAHA.113.002416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Williams SM, Golden-Mason L, Ferguson BS, Schuetze KB, Cavasin MA, Demos-Davies K, Yeager ME, Stenmark KR, McKinsey TA. Class i hdacs regulate angiotensin ii-dependent cardiac fibrosis via fibroblasts and circulating fibrocytes. Journal of molecular and cellular cardiology. 2014;67:112–125. doi: 10.1016/j.yjmcc.2013.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.McKinsey TA. Targeting inflammation in heart failure with histone deacetylase inhibitors. Molecular medicine. 2011;17:434–441. doi: 10.2119/molmed.2011.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ma X, Liu H, Foyil SR, Godar RJ, Weinheimer CJ, Hill JA, Diwan A. Impaired autophagosome clearance contributes to cardiomyocyte death in ischemia/reperfusion injury. Circulation. 2012;125:3170–3181. doi: 10.1161/CIRCULATIONAHA.111.041814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ma X, Liu H, Foyil SR, Godar RJ, Weinheimer CJ, Diwan A. Autophagy is impaired in cardiac ischemia-reperfusion injury. Autophagy. 2012;8:1394–1396. doi: 10.4161/auto.21036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Banreti A, Sass M, Graba Y. The emerging role of acetylation in the regulation of autophagy. Autophagy. 2013;9:819–829. doi: 10.4161/auto.23908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gibson CL, Murphy SP. Benefits of histone deacetylase inhibitors for acute brain injury: A systematic review of animal studies. Journal of neurochemistry. 2010;115:806–813. doi: 10.1111/j.1471-4159.2010.06993.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yang B, Yang J, Bai J, Pu P, Liu J, Wang F, Ruan B. Suv39h1 protects from myocardial ischemia-reperfusion injury in diabetic rats. Cellular physiology and biochemistry : international journal of experimental cellular physiology, biochemistry, and pharmacology. 2014;33:1176–1185. doi: 10.1159/000358686. [DOI] [PubMed] [Google Scholar]
- 93.Howitz KT, Bitterman KJ, Cohen HY, Lamming DW, Lavu S, Wood JG, Zipkin RE, Chung P, Kisielewski A, Zhang LL, Scherer B, Sinclair DA. Small molecule activators of sirtuins extend saccharomyces cerevisiae lifespan. Nature. 2003;425:191–196. doi: 10.1038/nature01960. [DOI] [PubMed] [Google Scholar]
- 94.Michan S, Sinclair D. Sirtuins in mammals: Insights into their biological function. The Biochemical journal. 2007;404:1–13. doi: 10.1042/BJ20070140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hsu CP, Zhai P, Yamamoto T, Maejima Y, Matsushima S, Hariharan N, Shao D, Takagi H, Oka S, Sadoshima J. Silent information regulator 1 protects the heart from ischemia/reperfusion. Circulation. 2010;122:2170–2182. doi: 10.1161/CIRCULATIONAHA.110.958033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Oka S, Alcendor R, Zhai P, Park JY, Shao D, Cho J, Yamamoto T, Tian B, Sadoshima J. Pparalpha-sirt1 complex mediates cardiac hypertrophy and failure through suppression of the err transcriptional pathway. Cell metabolism. 2011;14:598–611. doi: 10.1016/j.cmet.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]