Abstract
Heterologous expression of secondary metabolic pathways is a promising approach for the discovery and characterization of bioactive natural products. Herein we report the first heterologous expression of a natural product from the model marine actinomycete genus Salinispora. Using the recently developed method of yeast-mediated transformation-associated recombination for natural product gene clusters, we captured a type II polyketide synthase pathway from Salinispora pacifica with high homology to the enterocin pathway from Streptomyces maritimus and successfully produced enterocin in two different Streptomyces host strains. This result paves the way for the systematic interrogation of Salinispora’s promising secondary metabolome.
With the increased ease in obtaining next-generation DNA sequencing information, bioinformatic analyses and genome mining methods have become important modern tools in the discovery of new microbial natural products.1 A recent survey of all publicly available genomes revealed over 30 000 biosynthetic gene clusters that potentially allow the production of novel bioactive small molecules.2 This genetic reservoir represents a readily available resource for biosynthetic discovery and inspiration. However, the identification and characterization of the associated compounds can be challenging due to immense variations in microbial culturing and secondary metabolite production under normal laboratory conditions. A number of analytical methods have been developed in recent years to overcome these vexing problems and efficiently connect genes and gene clusters to small molecules.3−6 One of the most promising approaches involves the heterologous expression of entire biosynthetic gene clusters in a reliable and genetically tractable host organism. Heterologous expression not only allows the production of small molecules independent of growth rate and culturability of the natural producer, but also enables genetic manipulations to enhance compound production levels, the discovery of new biosynthetic enzymes, or engineering of unnatural biosynthetic products.7
So far, no compatible heterologous host system has been reported for the obligate marine actinomycete Salinispora.(8) This genus is known to produce a wealth of bioactive natural products of often unparalleled chemical structures9,10 and has been found to remarkably devote approximately 10% of its genome to their production.11 Recent bioinformatic efforts allowed the detection of more than 100 orphan pathways associated with polyketide and nonribosomal peptide biosynthesis in 75 newly sequenced Salinispora strains.12 Previous genome mining studies in Salinispora connected orphan gene clusters to salinilactam A,11 salinosporamide K,13 and the lomaiviticins,14 yet in each of these cases, production was limited to the native strain. Thus, the goal of this study was to develop an efficient heterologous expression system to provide a key tool for interrogating the biosynthesis of new natural products by these chemically prolific marine actinobacteria.
Common genetic tools and methods used with terrestrial actinomycetes work well for in vivo manipulations in Salinispora species.15,16 The 70% average GC content of Salinispora genomes is very similar to the well-established Streptomyces superhosts widely used for the expression of actinomycete pathways.17 Therefore, we decided to test the suitability of two common Streptomyces host strains, the genome-minimized Streptomyces coelicolor M1146 and Streptomyces lividans TK23,18,19 for the production of Salinispora compounds. Of the more than 100 orphan biosynthetic pathways identified in an earlier study,12 we selected an 18 kb type II PKS gene cluster from Salinispora pacifica CNT-150 that showed high homology to the enterocin locus in Streptomyces maritimus for our proof of principle study (Table 1).20
Table 1. Enterocin Gene Cluster in Salinispora pacifica CNT-150.
| JGI gene locus | gene product | homologue | identity | accession no. | |
|---|---|---|---|---|---|
| 1 | B174DRAFT_04104 | hypothetical protein | EncG, S. maritimus | 59% | AAF81722.1 |
| 2 | B174DRAFT_04105 | methyltransferase | EncK, S. maritimus | 61% | AAF81726.1 |
| 3 | B174DRAFT_04106 | ketoreductase | EncD, S. maritimus | 66% | AAF81727.1 |
| 4 | B174DRAFT_04107 | 3-oxoacyl-(acyl-carrier-protein) synthase, α subunit | EncA, S. maritimus | 72% | AAF81728.1 |
| 5 | B174DRAFT_04108 | 3-oxoacyl-(acyl-carrier-protein) synthase, β subunit | EncB, S. maritimus | 62% | AAF81729.1 |
| 6 | B174DRAFT_04109 | acyl carrier protein | EncC, S. maritimus | 37% | AAF81730.1 |
| 7 | B174DRAFT_04110 | acyl transferase | EncL, S. maritimus | 51% | AAF81731.1 |
| 8 | B174DRAFT_04111 | FAD-dependent favorskiiase | EncM, S. maritimus | 64% | AAF81732.1 |
| 9 | B174DRAFT_04112 | cytochrome P450, monooxygenase | EncR, S. maritimus | 49% | AAF81737.1 |
| 10 | B174DRAFT_04113 | phenylalanine ammonia-lyase | EncP, S. maritimus | 64% | AAF81735.1 |
| 11 | B174DRAFT_04114 | hypothetical protein | EncO, S. maritimus | 44% | AAF81734.1 |
| 12 | B174DRAFT_04115 | benzoyl-CoA ligase | EncN, S. maritimus | 46% | AAF81733.1 |
| 13 | B174DRAFT_04117 | drug resistance transporter, EmrB/QacA subfamily | EncT, S. maritimus | 57% | AAF81738.1 |
| 14 | B174DRAFT_04118 | transcriptional regulator | EncS, S. maritimus | 58% | AAF81739.1 |
| 15 | B174DRAFT_04119 | transcriptional activator | hypothetical protein [S. sp. MspMP-M5] | 37% | WP_018542724.1 |
| 16 | B174DRAFT_04120 | acyl-CoA synthetase (AMP-forming)/AMP-acid ligase II | long-chain-fatty-acid-CoA ligase [B. saxobsidens DD2] | 49% | YP_005329444.1 |
Enterocin (1) is a bacteriostatic polyketide with a highly unusual structure containing a caged tricyclic, nonaromatic core whose formation involves a Favorskii-like oxidative rearrangement of a reactive poly(β-carbonyl) substrate catalyzed by the unique flavoenzyme EncM.21 Previously, enterocin and its structural analogues were heterologously expressed in S. lividans, thereby confirming the function and entirety of the enc locus in S. maritimus.22
Sequence comparisons of both enc gene clusters revealed that all essential enzymes involved in enterocin biosynthesis in S. maritimus are also present in the S. pacifica pathway,23 although with somewhat different gene arrangement (Figure 1, Table 1). Genes missing from the S. pacifica enc locus are encH–J, which encode beta-oxidation enzymes involved in starter unit processing, and encQ, which codes for a ferredoxin cofactor, all of which were previously shown to be complemented by primary metabolic enzymes.24 Two additional genes flanking the S. pacifica enc cluster, enc15 and enc16, showed no homology to those in S. maritimus and are predicted to code for a transcription factor and an acyl-CoA synthetase, respectively.
Figure 1.
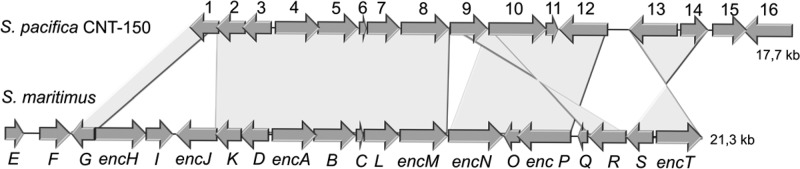
Comparison of the enterocin (enc) gene clusters of Streptomyces maritimus and Salinispora pacifica CNT-150. Numbered genes in S. pacifica are further described in Table 1.
To test whether the enterocin gene cluster in S. pacifica CNT-150 is active under laboratory conditions, extracts from 7-day-old cultures were tested by HR-LCMS and compared to an enterocin standard (Figure 2). Mass and UV data of one of the prominent peaks in the tested extracts indeed fully matched enterocin, which was furthermore verified by NMR analysis of the purified compound (Figure S1). S. pacifica CNT-150 is thus the first bacterium outside the Streptomyces genus shown to produce enterocin.
Figure 2.
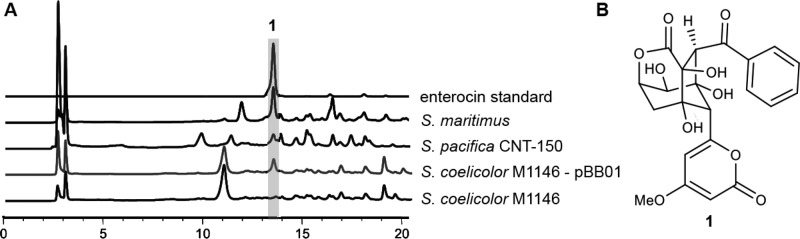
Heterologous expression of enterocin. (A) HPLC chromatograms of extracts of the heterologous host Streptomyces coelicolor M1146 with and without the expression plasmid pBB01 as well as the natural producer strains Streptomyces maritimus and Salinispora pacifica CNT-150. Detection at 254 nm. (B) Structure of enterocin.
To express a selected microbial metabolite such as enterocin in a Streptomyces host, it is necessary to clone the gene cluster in an appropriate integrative vector, a process that usually involves time-consuming genomic library and cloning efforts. To apply a more direct cloning method, we employed the pCAP01 vector that we recently designed for the transformation-associated recombination (TAR) of biosynthetic pathways in yeast directly from genomic DNA.25,26 The method was used to capture and express marinopyrrole from Streptomyces sp. CNQ-418 and taromycin A from Saccharomonospora sp. CNQ-490 in a S. coelicolor host strain. Hence we reasoned that this method seemed well suited for Salinispora as well as other actinomycete genera.
We captured by TAR a 21.3 kb region of genomic DNA containing the enterocin gene cluster from S. pacifica CNT-150 and subsequently transferred the vector through yeast and Escherichia coli to the Streptomyces host strains. Extracts of positive clones were tested for enterocin production after 6 days of growth by HR-LCMS (Figure 2). Indeed, enterocin production could be detected in all tested Streptomyces clones containing the pathway. No significant differences in production levels for both Streptomyces host species were detected, proving that the genus is principally well suited for the heterologous expression of Salinispora biosynthetic pathways (Figure S5).
The enterocin gene cluster in S. maritimus has been shown to be a highly versatile biosynthetic system that produces a large series of metabolites, including 5-deoxyenterocin, 3-epi-5-deoxyenterocin, and different wailupemycin analogues.20 The latter are assumed to arise from spontaneous cyclization of the extremely unstable EncM substrate, which competes with the enzymatic Favorskii rearrangement. Further enterocin analogues have been isolated from marine ascidians.27 The lipoesters enterocin-arachidate and -behenate were speculated to be produced by uncultivated bacterial symbionts of a Western Australian Didemnum species. To test the spectrum of enterocin analogues produced by the S. pacifica cluster, we employed mass spectral molecular networking28 to compare the metabolome of both natural producers and one heterologous host (Figure S6). Although mass spectra of known analogues such as 5′-deoxyenterocin could be observed, no new enterocin or wailupemycin derivatives were detected in the network. Notably, wailupemycins were completely absent in extracts of both S. pacifica CNT-150 and the heterologous host strain. Why these spontaneous cyclization products are not produced by S. pacifica CNT-150 can only be speculated and might suggest a more controlled substrate channeling by the early enterocin biosynthetic machinery (EncABCDM). Gene deletion experiments of the superfluous acyl-CoA synthetase gene enc16 in the heterologous producer S. coelicolor M1146 showed no effects on the production of enterocin and related compounds, thereby implying that the enzyme is not involved in the biosynthesis of alternative derivatives.
Of the 75 sequenced Salinispora genomes presently available in public databases, only two additional strains contain the enterocin pathway, the Hawaiian isolates S. pacifica CNT-851 and S. pacifica CNT-796. Although these strains belong to a different phylotype29 and originate from a different location than S. pacifica CNT-150, which was isolated in Fiji, the nucleotide sequence is nearly identical in all three strains. This relationship suggests a very recent event of horizontal gene transfer, as has been demonstrated for the majority of secondary metabolite pathways in this highly diverse species.12
In conclusion, in silico analyses and comparative genomics have shown that the marine actinomycete genus Salinispora contains among the highest diversity of secondary metabolite classes within the biosynthetic rich group of actinomycetes.30 The recent genome sequencing of 75 Salinispora genomes furthermore revealed a high number and variety of orphan pathways in closely related strains,12 suggesting an enormous untapped potential of novel small molecules. With this study we show for the first time the heterologous expression of a Salinispora natural product pathway. Because heterologous expression and genetic manipulations are well established in Streptomyces, this study sets the stage for a more systematic approach to access the available genetic potential of Salinispora. Using TAR cloning to directly capture the gene clusters from genomic DNA offers a fast and efficient way for production and genetic manipulations, thus allowing rapid and detailed investigation of promising biosynthetic pathways.
Experimental Section
Bacterial Strains, Plasmids, and Culture Conditions
All strains used are listed in Table S1. E. coli cultures were grown in LB liquid media or on LB agar plates with antibiotics when necessary (50 μg/mL kanamycin, 50 μg/mL apramycin, 25 to 50 μg/mL chloramphenicol, 50 μg/mL carbenicillin) at 37 °C with shaking. Streptomyces liquid precultures were grown in tryptic soy broth with the appropriate antibiotics (50 μg/mL kanamycin, 50 μg/mL apramycin, 25 μg/mL nalidixic acid), inoculated into SSM media (1% soytone, 1% soluble starch, 2% maltose, 0.5% trace element solution, pH 5.7) and grown at 30 °C with shaking. Streptomyces were also grown on mannitol soya flour medium plates (2.0% agar, 2.0% mannitol, 2.0% soya flour) with appropriate antibiotics (50 μg/mL kanamycin, 50 μg/mL apramycin, 25 μg/mL nalidixic acid). Salinispora were grown in A1 media (1.0% starch, 0.4% yeast extract, 0.2% peptone, and 2.8% sea salt) at 30 °C with shaking.
TAR Capture of the Salinispora pacifica enc Gene Cluster
The method of TAR capturing used in this study has been previously reported.25 The isolation of high-quality genomic DNA from Salinispora was performed according to standard procedures.31 The capture vector, pCAP01, containing yeast ARSH4/CEN6 and TRP1 marker, E. coli pUC ori, Streptomyces φC31 integrase gene(int), its attachment site (attP) and origin of DNA transfer (oriT), aph(3)II gene (Kan/Neo resistance), and two 1 kb regions flanking the gene cluster on either side was used to capture and propagate the enterocin gene cluster. The vector pCAP01 with the captured enc gene cluster from S. pacifica CNT-150 was named pBB01.
Heterologous Expression, Extraction, and HPLC Analysis
The plasmid pBB01 and its derivatives were conjugated into S. lividans TK23 and S. coelicolor M1146 using a standard triparental mating method with E. coli ET 12567/pBB01 and E. coli ET 12567/pUB307.31 After 6 days of growth in SSM media, the cultures were extracted with four volumes of EtOAc and concentrated in vacuo. The extracts were then dissolved in 100 μL of MeCN and analyzed by HPLC using a Luna 5u C18 column (150 mm × 4.60 mm, 5 μm beads) with a linear gradient of 2–100% MeCN in water with 0.1% TFA over 30 min with a flow rate of 0.7 mL/min.
HPLC-MS/MS and Molecular Networking
HPLC-HR-ESI-MSMS analysis of all extracts was carried out on an Agilent 1290 Q-TOF (200–2000 m/z, 20 keV). The data were subjected to the molecular networking workflow and analyzed as described previously.36
Gene Deletion Experiment
The inactivation of the acyl-CoA synthetase encoding gene, enc16, in the heterologous host strain S. coelicolor M1146-pBB01 was done using λ-Red recombination with an apramycin-resistant marker (aac(3)IV) as described previously.32
Acknowledgments
We thank P. R. Jensen and M. J. Bibb for providing bacterial strains S. pacifica CNT-150 and S. coelicolor M1146, respectively. We also thank B. M. Duggan for assistance with NMR, and X. Tang and P. C. Dorrestein for helpful discussions about λ-Red recombination techniques and molecular networking. This work was supported by NIH grants AI-47818 and GM-85770 to B.S.M. and DFG postdoctoral fellowships to R.T. and M.C.
Supporting Information Available
NMR spectra for 1, molecular network of Salinispora pacifica, Streptomyces maritimus, and Streptomyces coelicolor extracts, and detailed information about the strains and plasmids used in this study. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
Dedication
Dedicated to Dr. William Fenical of Scripps Institution of Oceanography, University of California–San Diego, for his pioneering work on bioactive natural products.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Jensen P. R.; Chavarria K. L.; Fenical W.; Moore B. S.; Ziemert N. J. Ind. Microbiol. Biotechnol. 2014, 41, 203–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimermancic P.; Medema M. H.; Claesen J.; Kurita K.; Wieland Brown L. C.; Mavrommatis K.; Pati A.; Godfrey P. A.; Koehrsen M.; Clardy J.; Birren B. W.; Takano E.; Sali A.; Linington R. G.; Fischbach M. A. Cell 2014, 158, 412–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersten R. D.; Yang Y.-L.; Xu Y.; Cimermancic P.; Nam S.-J.; Fenical W.; Fischbach M. A.; Moore B. S.; Dorrestein P. C. Nat. Chem. Biol. 2011, 7, 794–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersten R. D.; Ziemert N.; Gonzalez D. J.; Duggan B. M.; Nizet V.; Dorrestein P. C.; Moore B. S. Proc. Natl. Acad. Sci. U.S.A. 2013, 110, E4407–4416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross H.; Stockwell V. O.; Henkels M. D.; Nowak-Thompson B.; Loper J. E.; Gerwick W. H. Chem. Biol. 2007, 14, 53–63. [DOI] [PubMed] [Google Scholar]
- Winter J. M.; Behnken S.; Hertweck C. Curr. Opin. Chem. Biol. 2011, 15, 22–31. [DOI] [PubMed] [Google Scholar]
- Ongley S. E.; Bian X.; Neilan B. A.; Müller R. Nat. Prod. Rep. 2013, 30, 1121–1138. [DOI] [PubMed] [Google Scholar]
- Maldonado L. A.; Fenical W.; Jensen P. R.; Kauffman C. A.; Mincer T. J.; Ward A. C.; Bull A. T.; Goodfellow M. Int. J. Syst. Evol. Microbiol. 2005, 55, 1759–1766. [DOI] [PubMed] [Google Scholar]
- Feling R. H.; Buchanan G. O.; Mincer T. J.; Kauffman C. A.; Jensen P. R.; Fenical W. Angew. Chem., Int. Ed. 2003, 42, 355–357. [DOI] [PubMed] [Google Scholar]
- Jensen P. R.; Williams P. G.; Oh D. C.; Zeigler L.; Fenical W. Appl. Environ. Microbiol. 2007, 73, 1146–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udwary D. W.; Zeigler L.; Asolkar R. N.; Singan V.; Lapidus A.; Fenical W.; Jensen P. R.; Moore B. S. Proc. Natl. Acad. Sci. U.S.A. 2007, 104, 10376–10381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziemert N.; Lechner A.; Wietz M.; Millán-Aguiñaga N.; Chavarria K. L.; Jensen P. R. Proc. Natl. Acad. Sci. U.S.A. 2014, 111, E1130–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eustáquio A. S.; Nam S. J.; Penn K.; Lechner A.; Wilson M. C.; Fenical W.; Jensen P. R.; Moore B. S. ChemBioChem 2011, 12, 61–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersten R. D.; Lane A. L.; Nett M.; Richter T. K. S.; Duggan B. M.; Dorrestein P. C.; Moore B. S. ChemBioChem 2013, 14, 955–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eustáquio A. S.; Pojer F.; Noel J. P.; Moore B. S. Nat. Chem. Biol. 2008, 4, 69–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechner A.; Eustáquio A. S.; Gulder T. A. M.; Hafner M.; Moore B. S. Chem. Biol. 2011, 18, 1527–1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltz R. H. J. Ind. Microbiol. Biotechnol. 2010, 37, 759–772. [DOI] [PubMed] [Google Scholar]
- Gomez-Escribano J. P.; Bibb M. J. Microb. Biotechnol. 2011, 4, 207–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penn J.; Li X.; Whiting A.; Latif M.; Gibson T.; Silva C. J.; Brian P.; Davies J.; Miao V.; Wrigley S. K.; Baltz R. H. J. Ind. Microbiol. Biotechnol. 2006, 33, 121–128. [DOI] [PubMed] [Google Scholar]
- Piel J.; Hertweck C.; Shipley P. R.; Hunt D. M.; Newman M. S.; Moore B. S. Chem. Biol. 2000, 7, 943–955. [DOI] [PubMed] [Google Scholar]
- Teufel R.; Miyanaga A.; Michaudel Q.; Stull F.; Louie G.; Noel J. P.; Baran P. S.; Palfey B.; Moore B. S. Nature 2013, 503, 552–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piel J.; Hoang K.; Moore B. S. J. Am. Chem. Soc. 2000, 122, 5415–5416. [Google Scholar]
- Cheng Q.; Xiang L.; Izumikawa M.; Meluzzi D.; Moore B. S. Nat. Chem. Biol. 2007, 3, 557–558. [DOI] [PubMed] [Google Scholar]
- Xiang L.; Moore B. S. J. Bacteriol. 2003, 185, 399–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka K.; Reynolds K. A.; Kersten R. D.; Ryan K. S.; Gonzalez D. J.; Nizet V.; Dorrestein P. C.; Moore B. S. Proc. Natl. Acad. Sci. U.S.A. 2014, 111, 1957–1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross A. C.; Gulland L. E. S.; Dorrestein P. C.; Moore B. S. ACS Synth. Biol. 2014, 10.1021/sb500280q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H.; Jensen P. R.; Fenical W.. J. Org. Chem. 1996, 61, 1543–1546. [Google Scholar]
- Watrous J.; Roach P.; Alexandrov T.; Heath B. S.; Yang J. Y.; Kersten R. D.; van der Voort M.; Pogliano K.; Gross H.; Raaijmakers J. M.; Moore B. S.; Laskin J.; Bandeira N.; Dorrestein P. C. Proc. Natl. Acad. Sci. U.S.A. 2012, 109, E1743–E1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freel K. C.; Millán-Aguiñaga N.; Jensen P. R. Appl. Environ. Microbiol. 2013, 79, 5997–6005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nett M.; Ikeda H.; Moore B. S. Nat. Prod. Rep. 2009, 26, 1362–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieser T.; Bibb M. J.; Buttner M. J.; Chater K. F.; Hopwood D. A.. Practical Streptomyces Genetics; The John Innes Foundation: Norwich, UK, 2000. [Google Scholar]
- Gust B.; Challis G. L.; Fowler K.; Kieser T.; Chater K. F. Proc. Natl. Acad. Sci. U.S.A. 2003, 100, 1541–1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


