Abstract
We previously identified 9 genes and chromosomal region 3q28 as susceptibility loci for Japanese patients with myocardial infarction, ischemic stroke, or chronic kidney disease by genome-wide or candidate gene association studies. In the present study, we investigated the possible association of 13 single nucleotide polymorphisms (SNPs) at these 10 loci with the prevalence of hypertension or their association with blood pressure (BP) in community-dwelling individuals in Japan. The study subjects comprised 6,027 individuals (2,250 subjects with essential hypertension, 3,777 controls) who were recruited into the Inabe Health and Longevity Study, a longitudinal genetic epidemiological study on atherosclerotic, cardiovascular and metabolic diseases. The subjects were recruited from individuals who visited the Health Care Center of Inabe General Hospital for an annual health checkup, and they are followed up each year (mean follow-up period, 5 years). Longitudinal analysis with a generalized estimating equation and with adjustment for age, gender, body mass index and smoking status revealed that rs2116519 of family with sequence similarity 78, member B (FAM78B; P=0.0266), rs6929846 of butyrophilin, subfamily 2, member A1 (BTN2A1; P= 0.0013), rs146021107 of pancreatic and duodenal homeobox 1 (PDX1; P=0.0031) and rs1671021 of lethal giant larvae homolog 2 (Drosophila) (LLGL2; P=0.0372) were significantly (P<0.05) associated with the prevalence of hypertension. Longitudinal analysis with a generalized linear mixed-effect model and with adjustment for age, gender, body mass index and smoking status among individuals not taking anti-hypertensive medication revealed that rs6929846 of BTN2A1 was significantly associated with systolic (P=0.0017), diastolic (P=0.0008) and mean (P=0.0005) BP, and that rs2116519 of FAM78B, rs146021107 of PDX1 and rs1671021 of LLGL2 were significantly associated with diastolic (P=0.0495), systolic (P=0.0132), and both diastolic (P=0.0468) and mean (0.0471) BP, respectively. BTN2A1 may thus be a susceptibility gene for hypertension.
Keywords: hypertension, genetics, polymorphism, genetic epidemiology, longitudinal study
Introduction
Hypertension is a complex multifactorial disorder that is thought to result from an interaction between an individual’s genetic background and various lifestyle and environmental factors (1). The genetic influence on blood pressure (BP) variability has been estimated at 30–60% for a given individual (2), and the genetic heritability of hypertension estimated at 30% (3). Given that hypertension is a major risk factor for coronary artery disease, ischemic and hemorrhagic stroke, as well as chronic kidney disease (4–6), the personalized prevention of hypertension is an important public health goal.
Genome-wide association studies have identified various loci and genes associated with BP or to a predisposition to hypertension in Caucasian populations (7–11) or African Americans (12). Although the genes for adducin 2 (13) and ATPase, Ca2+ transporting, plasma membrane 1 (14) have been shown to be susceptibility loci for hypertension in Japanese individuals, the genes that confer susceptibility to this condition in Japanese individuals remain to be identified definitively.
We have previously identified 9 genes and chromosomal region 3q28 as susceptibility loci for myocardial infarction, ischemic stroke, or chronic kidney disease in Japanese individuals by genome-wide (15–17) or candidate gene (18–20) association studies. Given that hypertension is an important risk factor for these conditions (4–6), we hypothesized that certain single nucleotide polymorphisms (SNPs) at these 10 loci may contribute to their genetic susceptibility by affecting the susceptibility to hypertension. Therefore, the purpose of the present study was to examine the possible association of 13 SNPs at these 10 loci with the prevalence of essential hypertension or their association with BP in community-dwelling Japanese individuals.
Materials and methods
Study population
Study subjects comprised 6,027 community-dwelling individuals (2,250 subjects with essential hypertension and 3,777 controls) who were recruited to a population-based cohort study in Inabe City (Inabe Health and Longevity Study), Mie Prefecture, Japan. The Inabe Health and Longevity Study is a longitudinal genetic epidemiological study of atherosclerotic, cardiovascular and metabolic diseases (21–26). The subjects were recruited from individuals who visited the Health Care Center of Inabe General Hospital for an annual health checkup, and they are followed up each year. A total of 6,027 individuals was registered between March 2010 and September 2012, and genomic DNA was extracted from the venous blood cells of these subjects and stored in the genomic DNA bank of the Research Center for Genomic Medicine at Mie University. For all the participants, medical examination data obtained from April 2003 to March 2014 (11 years) were entered into a database. If individuals had a medical checkup 2 or more times per year, data from one time point for each year were entered, so that each subject had one set of health data for each year they had attended the clinic. All participants thus had undergone 1–11 medical examinations, and the average follow-up period was 5 years.
Subjects with hypertension either had a systolic BP of ≥140 mmHg or a diastolic BP of ≥90 mmHg (or both) or had taken anti-hypertensive medication. The control individuals had a systolic BP of <140 mmHg and a diastolic BP of <90 mmHg, as well as no history of hypertension or of taking any anti-hypertensive medication. BP was measured at least twice with the subjects having rested in the sitting position for >5 min; the measurements were taken by a skilled physician or nurse according to the guidelines of the American Heart Association (27). The study protocol complied with the Declaration of Helsinki and was approved by the Committees on the Ethics of Human Research of Mie University Graduate School of Medicine and Inabe General Hospital. Written informed consent was obtained from all subjects prior to enrollment in the study.
Selection and genotyping of polymorphisms
The 13 SNPs examined in the present study (Table I) were selected from our previous genome-wide (15–17) or candidate gene (18–20) association studies. Wild-type (ancestral) and variant alleles of the SNPs were determined from the dbSNP database (National Center for Biotechnology Information, Bethesda, MD, USA) (http://www.ncbi.nlm.nih.gov/SNP).
Table I.
The 13 SNPs examined in the present study.
| Chromosomal locus | Gene | dbSNP (NCBI) | Nucleotide substitution | Minor allelea |
|---|---|---|---|---|
| 1q24.1 | FAM78B | rs2116519 | C→T | C |
| 3q28 | Non-gene region | rs9846911 | A→G | G |
| 4q25 | ALPK1 | rs2074379 | G→A (Met732Ile) | G |
| 4q25 | ALPK1 | rs2074380 | G→A (Gly870Ser) | A |
| 4q25 | ALPK1 | rs2074381 | A→G (Asn916Asp) | G |
| 4q25 | ALPK1 | rs2074388 | G→A (Gly565Asp) | G |
| 6p22.1 | BTN2A1 | rs6929846 | T→C | T |
| 6q27 | THBS2 | rs8089 | T→G | G |
| 13q12.1 | PDX1 | rs146021107 | G→- (deletion) | – |
| 13q34 | F7 | rs6046 | G→A (Arg353Gln) | A |
| 17q25.1 | LLGL2 | rs1671021 | G→A (Leu479Phe) | G |
| 19p13.2 | ILF3 | rs2569512 | G→A | A |
| 22q13.3 | CELSR1 | rs6007897 | C→T (Ala2268Thr) | C |
The minor allele in Japanese individuals was determined by the allele frequency of HapMap-JPT in dbSNP. SNPs, single nucleotide polymorphisms.
Venous blood (5 ml) was collected into tubes containing 50 mmol/l ethylenediaminetetraacetic acid (disodium salt), and peripheral blood leukocytes were isolated and genomic DNA was extracted from these cells with the use of a DNA extraction kit (SMITEST EX-R&D; Medical and Biological Laboratories, Nagoya, Japan). The genotypes of the 13 SNPs were determined at G&G Science Co., Ltd. (Fukushima, Japan) by a method that combines the polymerase chain reaction and sequence-specific oligonucleotide probes with suspension array technology (Luminex, Austin, TX, USA). The primers, probes and other conditions for the genotyping of the SNPs examined in the present study are shown in Table II. Detailed genotyping methodology was as described previously (15,16,28).
Table II.
Primers, probes and other conditions for the genotyping of the 13 SNPs examined in the present study.
| Gene or locus | SNP | dbSNP | Sense primer (5′→3′) | Antisense primer (5′→3′) | Probe 1 (5′→3′) | Probe 2 (5′→3′) | Annealing | (°C) Cycles |
|---|---|---|---|---|---|---|---|---|
| FAM78B | C→T | rs2116519 | CCTGCACTGCTCTAGCTACTTC | GATCCCAATTTCAACTGTGAGATC | TCATTCCGGTCTCAGCCGCT | CCCTCATTCCGGTTTCAGCC | 60 | 50 |
| 3q28 | A→G | rs9846911 | AGTTGTGTGCCAGATTCTCCAG | TCTTCACTGAGACCTTGGGAAG | TCTCCTCTTTCAATAACAAATCTTC | AAAGTCTCCTCTTTCAGTAACAAAT | 60 | 50 |
| ALPK1 | G→A | rs2074379 | TCTGCTTCTTGGTCTTCTGATTC | AGTTGGTTTCTGGAAACTCAACAA | GAAGGATGTGTGCCTATATTCTT | GATGTGTGCCCATATTCTTGGG | 60 | 50 |
| ALPK1 | G→A | rs2074380 | CTCCACAGTGGATGAGGAGG | CTTACAGAGGAATTGGGGGTC | ACAAATGGGCACAGCTCTCATA | TATGAGAGCCGTGCCCATTTGT | 60 | 50 |
| ALPK1 | A→G | rs2074381 | AGGACTGCACTACCACAGAGG | TGATTTCAGCCACCACACTGAG | ATCAGCCTGGAAACATGCTAAAC | AGTTTAGCATGTCTCCAGGCTG | 60 | 50 |
| ALPK1 | G→A | rs2074388 | TGTGGAGACTGAGACTGAGCC | TTGCTCCAAGCACTGGAAGTC | ACTACAGCAATGATGAGGGAGC | GCTCCCTCACCATTGCTGTAG | 60 | 50 |
| BTN2A1 | T→C | rs6929846 | CCAAACATGGCGACCTAGGAGA | ATCTGCCCAGGGGCACAGGC | TTTGGGAAGGTTTGCGTCTAG | TTTGGGAAGGTTTGTGTCTAGT | 60 | 50 |
| THBS2 | T→G | rs8089 | AACCCAAGTGCCTTCAGAGGAT | CTCCACATAAAGTCTCATATATCAC | GATGTTCATCTCTGAGTTCCA | GATGTTCATCTCTGCGTTCCA | 60 | 50 |
| PDX1 | G→- | rs146021107 | TGGCTGTGGGTTCCCTCTGAG | GATTTGGCACTGTGTGGCGTTC | CGAGCAGGGGTGGCGCC | GGCGCCACCCTGCTCGCT | 60 | 50 |
| F7 | G→A | rs6046 | CGGCTACTCGGATGGCAGCA | CCAAAGTGGCCCACGGTTGC | TACCACGTGCCCCGGTAGTG | GCCACCCACTACCAGGGCA | 60 | 50 |
| LLGL2 | G→A | rs1671021 | GCTCCTGGCCTCACCTTGCG | GCTGCTCTACAAACTCAGCACTG | CTGGGCACTGAAGTTCTCGTT | CCAACGAGAACCTCAGTGCC | 60 | 50 |
| ILF3 | G→A | rs2569512 | ACCACCTCAACTGCAAGCTGAA | GGAATGATCCCTCTGGGAAGGT | GTGCAACTGCCAAAAACTGGT | GTGCAACTGCCAAGAACTGG | 60 | 50 |
| CELSR1 | C→T | rs6007897 | GGAGACGGAGGACTCCAGCTC | CTTGCTGTCGACATCTTTGACAAG | TCTTCATGGATGGCGTCGAAT | TCTTCATGGATGGTGTCGAATC | 60 | 50 |
SNPs, single nucleotide polymorphisms.
Statistical analysis
Quantitative data were compared between the subjects with hypertension and the controls with the unpaired Student’s t-test. Categorical data were compared with the χ2 test. We examined the association of the 13 SNPs with the prevalence of hypertension or their association with systolic, diastolic, or mean BP based on a 5-year longitudinal cohort study. Longitudinal changes in the prevalence of hypertension were compared between 2 groups (the dominant or recessive genetic model) with a generalized estimating equation, as previously described (29) and with adjustment for age, gender, body mass index (BMI) and smoking status. Longitudinal changes in systolic, diastolic, or mean BP in all the individuals or in the individuals not any taking anti-hypertensive medication were compared between 2 groups (the dominant or recessive model) in a generalized linear mixed-effect model, as previously described (30) with adjustment for age, gender, BMI and smoking status. The dominant or recessive model was defined as AA vs. AB + BB or AA + AB vs. BB (A, major allele; B, minor allele), respectively. Age-related changes in the prevalence of hypertension or in systolic or diastolic BP were estimated with quadratic curves controlling for the observation year. A P-value <0.05 was considered to indicate a statistically significant difference. Statistical analysis was performed using R software version 3-0-2 (the R Project for Statistical Computing) and JMP Genomics version 6.0 (SAS Institute, Cary, NC, USA).
Results
Characteristics of the 6,027 study subjects (3,352 males, 2,675 females) with regard to all measurements in a 5-year follow-up are shown in Table III. Characteristics of the subjects with hypertension and the controls according to cross-sectional analysis in March 2014 are shown in Table IV. Age, the frequency of the male gender, BMI and the prevalence of smoking were greater in the subjects with hypertension than in the controls.
Table III.
Characteristics of the study subjects: analysis of all measurements in a 5-year follow-up.
| Parameter | Malea | Femalea | Alla |
|---|---|---|---|
| No. of subjects | 3352 | 2675 | 6027 |
| Age (years) | 52.5±12.5 (15,959) | 52.5±11.9 (12,572) | 52.5±12.2 (28,531) |
| Height (cm) | 168.4±6.6 (15,550) | 155.2±5.9 (12,373) | 162.6±9.1 (27,923) |
| Weight (kg) | 67.0±11.0 (15,548) | 53.5±8.2 (12,373) | 61.0±12.0 (27,921) |
| Body mass index (kg/m2) | 23.6±3.3 (15,548) | 22.2±3.2 (12,373) | 23.0±3.3 (27,921) |
| Waist circumference (cm) | 83.2±8.7 (11,817) | 77.8±9.0 (9,541) | 80.8±9.2 (21,358) |
| Alcohol concumption (%) | 67.4 (15,959) | 26.4 (12,572) | 49.3 (28,531) |
| Current or former smoker (%) | 65.0 (15,959) | 8.5 (12,572) | 40.1 (28,531) |
| Systolic blood pressure (mmHg) | 122±16 (15,541) | 119±16 (12,370) | 121±16 (27,911) |
| Diastolic blood pressure (mmHg) | 77±12 (15,541) | 71±11 (12,370) | 75±12 (27,911) |
| Mean blood pressure (mmHg) | 92±13 (15,541) | 87±12 (12,370) | 90±13 (27,911) |
| Ocular tension (right, mmHg) | 14.0±3.0 (6,132) | 13.4±2.8 (4,886) | 13.7±3.0 (11,018) |
| Functional vital capacity (l) | 3.53±0.66 (6,173) | 2.55±0.47 (4,865) | 3.10±0.76 (11,038) |
| FEV1% (%) | 82.3±7.1 (6,168) | 84.8±6.7 (4,865) | 83.4±7.0 (11,033) |
| Serum albumin (g/l) | 44.5±2.9 (10,332) | 44.1±2.7 (8,510) | 44.3±2.8 (18,842) |
| Serum total cholesterol (mmol/l) | 5.15±0.88 (15,121) | 5.31±0.88 (11,887) | 5.22±0.89 (27,008) |
| Serum triglyceride (mmol/l) | 1.46±1.06 (15,639) | 1.01±0.58 (12,401) | 1.26±0.91 (28,040) |
| Serum HDL-cholesterol (mmol/l) | 1.47±0.39 (15,627) | 1.78±0.42 (12,378) | 1.61±0.43 (28,005) |
| Serum LDL-cholesterol (mmol/l) | 3.19±0.81 (14,997) | 3.18±0.79 (11,836) | 3.18±0.80 (26,833) |
| Fasting plasma glucose (mmol/l) | 5.82±1.27 (15,685) | 5.39±0.93 (12,395) | 5.63±1.15 (28,080) |
| Blood hemoglobin A1c (%) | 5.78±0.74 (10,849) | 5.64±0.54 (10,169) | 5.71±0.66 (21,018) |
| Blood urea nitrogen (mmol/l) | 5.61±2.86 (8,889) | 5.07±2.28 (8,162) | 5.36±2.61 (17,051) |
| Serum creatinine (μmol/l) | 88.3±116.2 (14,545) | 63.1±82.5 (11,225) | 77.3±103.6 (25,770) |
| eGFR (ml/min/1.73 m−2) | 77.2±18.0 (14,545) | 80.3±17.5 (11,225) | 78.5±17.9 (25,770) |
| Serum uric acid (μmol/l) | 372±79 (14,368) | 273±62 (10,900) | 329±87 (25,268) |
| Serum C-reactive protein (μg/l) | 1573±6428 (5,793) | 1207±4107 (4,938) | 1405±5486 (10,731) |
| White blood cells (103/μl) | 5.94±1.73 (12,521) | 5.03±1.45 (9,419) | 5.55±1.68 (21,940) |
| Red blood cells (104/μl) | 461±46 (12,651) | 415±36 (9,500) | 441±47 (22,151) |
| Hemoglobin (g/l) | 147±13 (12,651) | 127±13 (9,501) | 139±16 (22,152) |
| Hematocrit (%) | 43.3±3.7 (12,642) | 37.5±3.4 (9,497) | 40.8±4.6 (22,139) |
| Platelets (104/μl) | 23.1±5.5 (12,473) | 23.8±6.2 (9,398) | 23.4±5.8 (21,871) |
Values in parentheses indicate the numbers of measurements taken. Quantitative data are the means ± SD. FEV1%, forced expiratory volume in 1 sec percentage; HDL, high density lipoprotein; LDL, low density lipoprotein; eGFR, estimated glomerular filtration rate (ml/min/1.73 m−2) = 194 × [age (years)]−0.287 × [serum creatinine (mg/dl)]−1.094 × [0.739 if female].
Table IV.
Characteristics of subjects with hypertension and controls: cross-sectional analysis in March 2014.
| Parameter | Subjects with hypertensiona | Controlsa | P-value |
|---|---|---|---|
| No. of subjects | 2250 | 3777 | |
| Age (years) | 61.1±10.7 (2,250) | 50.1±12.4 (3,777) | <0.0001 |
| Gender (male/female, %) | 62.6/37.4 | 51.5/48.5 | <0.0001 |
| Height (cm) | 161.3±9.4 (2,207) | 163.2±9.0 (3,747) | <0.0001 |
| Weight (kg) | 63.1±12.7 (2,205) | 59.7±11.6 (3,747) | <0.0001 |
| Body mass index (kg/m2) | 24.1±3.6 (2,205) | 22.3±3.1 (3,747) | <0.0001 |
| Waist circumference (cm) | 84.0±9.5 (1,986) | 78.5±8.5 (3,619) | <0.0001 |
| Alcohol consumption (%) | 52.0 (2,250) | 46.0 (3,777) | <0.0001 |
| Current or former smoker (%) | 47.7 (2,250) | 44.5 (3,777) | 0.0147 |
| Systolic blood pressure (mmHg) | 133±15 (2,200) | 113±11 (3,745) | <0.0001 |
| Diastolic blood pressure (mmHg) | 83±12 (2,200) | 70±10 (3,745) | <0.0001 |
| Mean blood pressure (mmHg) | 99±12 (2,200) | 84±9 (3,745) | <0.0001 |
| Ocular tension (right, mmHg) | 13.9±3.0 (722) | 13.3±2.9 (1,339) | <0.0001 |
| Functional vital capacity (l) | 3.12±0.80 (768) | 3.39±0.80 (1,475) | <0.0001 |
| FEV1% (%) | 80.4±6.38 (768) | 81.7±6.6 (1,475) | <0.0001 |
| Serum albumin (g/l) | 44.5±3.0 (1,715) | 44.7±2.4 (2,497) | 0.0302 |
| Serum total cholesterol (mmol/l) | 5.19±0.90 (2,230) | 5.23±0.88 (3,720) | 0.0921 |
| Serum triglyceride (mmol/l) | 1.43±0.96 (2,215) | 1.16±0.79 (3,721) | <0.0001 |
| Serum HDL-cholesterol (mmol/l) | 1.59±0.44 (2,213) | 1.70±0.45 (3,721) | <0.0001 |
| Serum LDL-cholesterol (mmol/l) | 3.15±0.79 (2,212) | 3.19±0.81 (3,720) | 0.0632 |
| Fasting plasma glucose (mmol/l) | 5.90±1.36 (2,238) | 5.40±0.96 (3,718) | <0.0001 |
| Blood hemoglobin A1c (%) | 5.84±0.78 (1,782) | 5.59±0.59 (2,681) | <0.0001 |
| Blood urea nitrogen (mmol/l) | 5.72±2.68 (1,691) | 4.86±1.23 (2,410) | <0.0001 |
| Serum creatinine (μmol/l) | 88.5±127.4 (2,162) | 64.8±15.1 (3,414) | <0.0001 |
| eGFR (ml/min/1.73 m−2) | 71.2±18.3 (2,162) | 80.1±14.7 (3,414) | <0.0001 |
| Serum uric acid (μmol/l) | 349±88 (2,139) | 312±81 (3,392) | <0.0001 |
| Serum C-reactive protein (μg/l) | 1832±9666 (775) | 826±3359 (1,338) | 0.0005 |
| White blood cells (103/μl) | 5.51±1.74 (1,573) | 5.31±1.63 (3,034) | 0.0001 |
| Red blood cells (104/μl) | 436±48 (1,577) | 437±43 (3,046) | 0.1928 |
| Hemoglobin (g/l) | 139±16 (1,577) | 137±15 (3,046) | 0.0017 |
| Hematocrit (%) | 40.4±4.4 (1,576) | 40.1±4.2 (3,042) | 0.0186 |
| Platelets (104/μl) | 21.8±5.5 (1,557) | 22.6±5.3 (3,011) | <0.0001 |
Values in parentheses indicate the numbers of measurements taken. Quantitative data are the means ± SD. eGFR, estimated glomerular filtration rate (ml/min/1.73 m−2) = 194 × [age (years)]−0.287 × [serum creatinine (mg/dl)]−1.094 × [0.739 if female]; HDL, high density lipoprotein; LDL, low density lipoprotein.
The association of the 13 SNPs with the prevalence of hypertension was analyzed with a generalized estimating equation and with adjustment for age, gender, BMI and smoking status (Table V). The rs2116519 (C→T) SNP of the family with sequence similarity 78, member B gene (FAM78B, recessive model), rs6929846 (T→C) of the butyrophilin, subfamily 2, member A1 gene (BTN2A1, dominant model), rs146021107 (G→-) of the pancreatic and duodenal homeobox 1 gene (PDX1, dominant model) and rs1671021 (G→A) of the lethal giant larvae homolog 2 gene (LLGL2, dominant model) were significantly (P<0.05) associated with the prevalence of hypertension.
Table V.
Association of polymorphisms with hypertension analyzed for 5-year longitudinal data with a generalized estimating equation.
| Gene or locus | SNP | Genotype | Hypertensiona | Controlsa | P-value (dominant model)b | P-value (recessive model)c |
|---|---|---|---|---|---|---|
| FAM78B | rs2116519 (C→T) | TT | 1,888 (32.3) | 6,649 (30.3) | 0.3039 | 0.0266 |
| TC | 2,959 (50.7) | 11,046 (50.3) | ||||
| CC | 991 (17.0) | 4,279 (19.5) | ||||
| 3q28 | rs9846911 (A→G) | AA | 5,033 (86.2) | 19,102 (86.9) | 0.1629 | 0.1620 |
| AG | 759 (13.0) | 2,756 (12.5) | ||||
| GG | 46 (0.8) | 116 (0.5) | ||||
| ALPK1 | rs2074379 (G→A) | AA | 2,707 (46.4) | 10,004 (45.5) | 0.7330 | 0.2596 |
| AG | 2,560 (43.9) | 9,736 (44.3) | ||||
| GG | 571 (9.8) | 2,234 (10.2) | ||||
| ALPK1 | rs2074380 (G→A) | GG | 4,905 (84.0) | 18,656 (84.9) | 0.1124 | 0.1496 |
| GA | 885 (15.2) | 3,165 (14.4) | ||||
| AA | 48 (0.8) | 153 (0.7) | ||||
| ALPK1 | rs2074381 (A→G) | AA | 4,981 (85.3) | 18,815 (85.6) | 0.2390 | 0.4732 |
| AG | 821 (14.1) | 3,038 (13.8) | ||||
| GG | 36 (0.6) | 121 (0.6) | ||||
| ALPK1 | rs2074388 (G→A) | AA | 2,714 (46.5) | 10,013 (45.6) | 0.7043 | 0.2637 |
| AG | 2,552 (43.7) | 9,721 (44.2) | ||||
| GG | 572 (9.8) | 2,240 (10.2) | ||||
| BTN2A1 | rs6929846 (T→C) | CC | 4,484 (76.8) | 17,333 (78.9) | 0.0013 | 0.3602 |
| CT | 1,275 (21.8) | 4,365 (19.9) | ||||
| TT | 79 (1.4) | 276 (1.3) | ||||
| THBS2 | rs8089 (T→G) | TT | 4,895 (83.8) | 18,159 (82.6) | 0.7407 | 0.9741 |
| TG | 902 (15.5) | 3,615 (16.5) | ||||
| GG | 41 (0.7) | 200 (0.9) | ||||
| PDX1 | rs146021107 (G→-) | GG | 1,745 (29.9) | 5,983 (27.2) | 0.0031 | 0.2885 |
| G/- | 2,839 (48.6) | 11,049 (50.3) | ||||
| -/- | 1,254 (21.5) | 4,942 (22.5) | ||||
| F7 | rs6046 (G→A) | GG | 5,104 (87.4) | 19,187 (87.3) | 0.1478 | 0.8979 |
| GA | 715 (12.2) | 2,693 (12.3) | ||||
| AA | 19 (0.3) | 94 (0.4) | ||||
| LLGL2 | rs1671021 (G→A) | AA | 4,187 (71.7) | 16,353 (74.4) | 0.0372 | 0.3881 |
| AG | 1,521 (26.1) | 5,223 (23.8) | ||||
| GG | 130 (2.2) | 398 (1.8) | ||||
| ILF3 | rs2569512 (G→A) | GG | 2,563 (43.9) | 9,525 (43.3) | 0.3765 | 0.2560 |
| GA | 2,605 (44.6) | 10,180 (46.3) | ||||
| AA | 670 (11.5) | 2,269 (10.3) | ||||
| CELSR1 | rs6007897 (C→T) | TT | 5,671 (97.1) | 21,303 (96.9) | 0.6353 | not determined |
| TC | 167 (2.9) | 671 (3.1) | ||||
| CC | 0 (0) | 0 (0) |
The prevalence of hypertension was compared between 2 groups (the dominant or recessive model) for each polymorphism with adjustment for age, gender, body mass index and smoking status.
Values indicate the numbers of measurements taken, with the percentages in parentheses;
dominant model: AA vs. AB + BB (A, major allele; B, minor allele);
recessive model (AA + AB vs. BB). P-values of <0.05 are shown in bold. SNPs, single nucleotide polymorphisms.
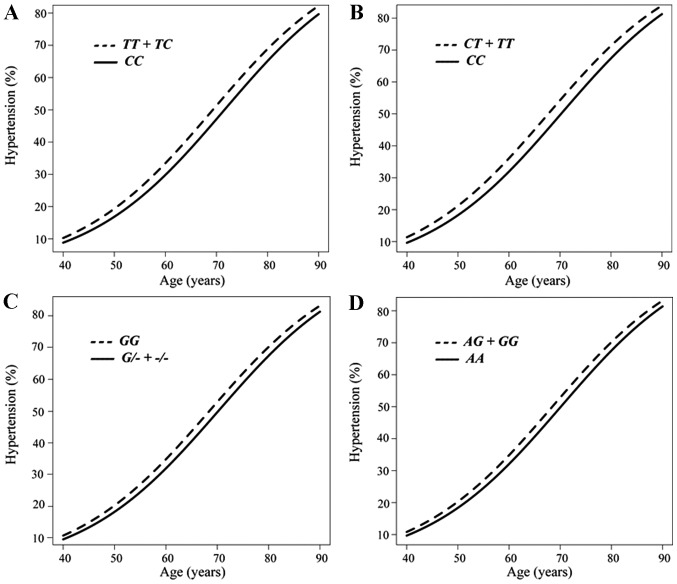
The association between the prevalence of hypertension and age analyzed longitudinally with a generalized estimating equation according to the SNP genotype is shown in Fig. 1. The prevalence of hypertension was greater in the combined group of subjects with the TT or TC genotypes of rs2116519 of FAM78B than in those with the CC genotype from 40 to 90 years of age (Fig. 1A), in the combined group of subjects with the CT or TT genotypes of rs6929846 of BTN2A1 than in those with the CC genotype (Fig. 1B), in subjects with the GG genotype of rs146021107 of PDX1 than in the combined group of subjects with the G/- or -/- genotypes (Fig. 1C), and in the combined group of subjects with the AG or GG genotypes of rs1671021 of LLGL2 than in those with the AA genotype (Fig. 1D).
Figure 1.
Longitudinal analysis with a generalized estimating equation of the association between the prevalence of hypertension and age according to the genotype for (A) rs2116519 of FAM78B (TT + TC vs. CC), (B) rs6929846 of BTN2A1 (CC vs. CT + TT) (B), (C) rs146021107 of PDX1 (GG vs. G/- + -/-), or (D) rs1671021 of LLGL2 (AA vs. AG + GG).
Given that 4 SNPs were significantly associated with hypertension, the association of these SNPs with systolic, diastolic, or mean BP in all individuals or individuals not taking any anti-hypertensive medication were analyzed with a generalized linear mixed-effect model, with adjustment for age, gender, BMI and smoking status (Table VI). The rs6929846 polymorphism of BTN2A1 was significantly associated with systolic, diastolic and mean BP in the dominant model among all individuals or individuals not taking any anti-hypertensive medication, with the T allele being associated with an increased BP. The rs146021107 SNP of PDX1 was significantly associated with systolic BP in the dominant model among all individuals or individuals not taking any anti-hypertensive medication, with the G allele being associated with an increased BP. The rs2116519 polymorphism of FAM78B was significantly associated with diastolic BP in the recessive model among individuals not taking any anti-hypertensive medication, with the T allele being associated with a high BP. The rs1671021 SNP of LLGL2 was significantly associated with diastolic and mean BP in the dominant model among individuals not taking any anti-hypertensive medication, with the G allele being associated with a high BP.
Table VI.
Association of polymorphisms with systolic, diastolic, or mean BP in all individuals or individuals not taking any anti-hypertensive medication analyzed for 5-year longitudinal data with a generalized linear mixed-effect model.
| Gene | SNP | BP (mmHg) | Dominant modela
|
P-value | Recessive modela
|
P-value | ||
|---|---|---|---|---|---|---|---|---|
| All individuals | ||||||||
| FAM78B | rs2116519 (C→T) | TT (8,537) | TC + CC (19,275) | TT + TC (2,2542) | CC (5,270) | |||
| Systolic | 121.0±16.7 | 120.4±16.1 | 0.3818 | 120.7±16.5 | 120.1±15.7 | 0.5823 | ||
| Diastolic | 74.9±12.5 | 74.6±12.1 | 0.1260 | 74.8±12.4 | 74.1±11.8 | 0.0823 | ||
| Mean | 90.2±13.1 | 89.9±12.6 | 0.1722 | 90.1±12.9 | 89.4±12.2 | 0.1814 | ||
| BTN2A1 | rs6929846 (T→C) | CC (21,817) | CT + TT (5,995) | CC + CT (27,457) | TT (355) | |||
| Systolic | 120.4±16.2 | 121.2±16.7 | 0.0061 | 120.6±16.3 | 121.4±15.4 | 0.1369 | ||
| Diastolic | 74.5±12.2 | 75.2±12.4 | 0.0023 | 74.7±12.3 | 75.2±11.0 | 0.2483 | ||
| Mean | 89.8±12.7 | 90.5±13.0 | 0.0019 | 90.0±12.8 | 90.6±11.7 | 0.1748 | ||
| PDX1 | rs146021107 (G→-) | GG (7,728) | G/- + -/- (20,084) | GG + G/- (21,616) | -/- (6,196) | |||
| Systolic | 121.1±17.1 | 120.4±16.0 | 0.0284 | 120.8±16.4 | 120.0±16.1 | 0.3884 | ||
| Diastolic | 74.5±12.8 | 74.7±12.1 | 0.2719 | 74.8±12.3 | 74.4±12.1 | 0.9222 | ||
| Mean | 90.1±13.3 | 90.0±12.5 | 0.1029 | 90.1±12.8 | 89.6±12.5 | 0.6821 | ||
| LLGL2 | rs1671021 (G→A) | AA (20,540) | AG + GG (7,272) | AA + AG (27,284) | GG (528) | |||
| Systolic | 120.4±16.2 | 121.2±16.6 | 0.1943 | 120.6±16.3 | 121.6±16.1 | 0.9056 | ||
| Diastolic | 74.5±12.2 | 75.2±12.4 | 0.1280 | 74.7±12.3 | 75.8±12.7 | 0.4665 | ||
| Mean | 89.8±12.7 | 90.5±12.9 | 0.1315 | 90.0±12.8 | 91.1±12.8 | 0.7203 | ||
| Individuals not taking any anti-hypertensive medication | ||||||||
| FAM78B | rs2116519 (C→T) | TT (8,132) | TC + CC (18,370) | TT + TC (2,1459) | CC (5,043) | |||
| Systolic | 120.5±16.7 | 119.9±16.0 | 0.2563 | 120.2±16.4 | 119.7±15.6 | 0.5041 | ||
| Diastolic | 74.6±12.5 | 74.4±12.1 | 0.2039 | 74.6±12.4 | 73.9±11.7 | 0.0495 | ||
| Mean | 89.9±13.0 | 89.6±12.6 | 0.1948 | 89.8±12.9 | 89.1±12.1 | 0.1248 | ||
| BTN2A1 | rs6929846 (T→C) | CC (20,807) | CT + TT (5,695) | CC + CT (26,163) | TT (339) | |||
| Systolic | 120.0±16.1 | 120.7±16.6 | 0.0017 | 120.1±16.3 | 120.8±15.3 | 0.1734 | ||
| Diastolic | 74.3±12.2 | 75.0±12.4 | 0.0008 | 74.4±12.3 | 75.0±10.8 | 0.2059 | ||
| Mean | 89.5±12.7 | 90.2±13.0 | 0.0005 | 89.7±12.7 | 90.3±11.5 | 0.1678 | ||
| PDX1 | rs146021107 (G→-) | GG (7,328) | G/- + -/- (19,174) | GG + G/- (20,580) | -/- (5,922) | |||
| Systolic | 120.6±17.1 | 120.0±15.9 | 0.0132 | 120.3±16.3 | 119.5±16.0 | 0.2565 | ||
| Diastolic | 74.2±12.8 | 74.5±12.0 | 0.3963 | 74.5±12.3 | 74.2±12.1 | 0.8832 | ||
| Mean | 89.7±13.3 | 89.7±12.5 | 0.1081 | 89.8±12.8 | 89.3±12.5 | 0.7018 | ||
| LLGL2 | rs1671021 (G→A) | AA (19,569) | AG + GG (6,933) | AA + AG (26,005) | GG (497) | |||
| Systolic | 119.9±16.2 | 120.7±16.5 | 0.0891 | 120.1±16.3 | 121.4±16.1 | 0.6847 | ||
| Diastolic | 74.2±12.2 | 75.0±12.4 | 0.0468 | 74.4±12.2 | 75.7±12.7 | 0.2512 | ||
| Mean | 89.5±12.7 | 90.2±12.9 | 0.0471 | 89.6±12.7 | 90.9±12.80 | 0.3889 | ||
Systolic, diastolic, or mean BP was compared between 2 groups (the dominant or recessive model) for each polymorphism with adjustment for age, gender, body mass index and smoking status.
Values in parentheses indicate the numbers of measurements taken. Data for BP are the means ± SD. P-values of <0.05 are shown in bold. BP, blood pressure. SNPs, single nucleotide polymorphisms.
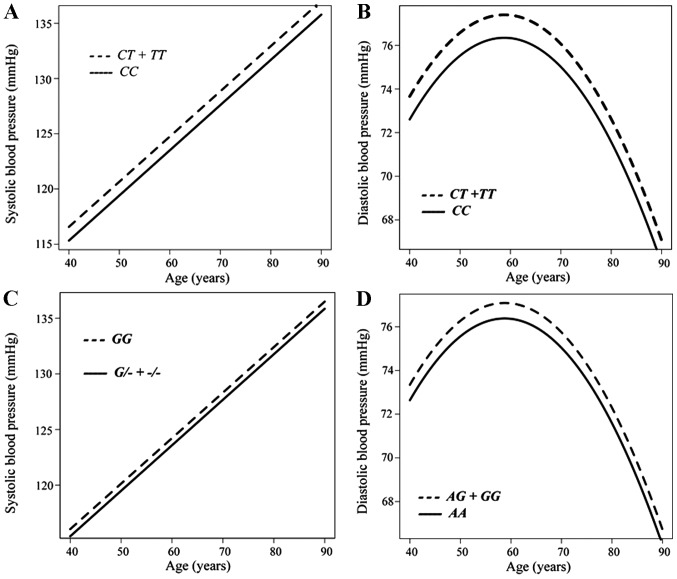
The association between systolic or diastolic BP and age in individuals not taking any anti-hypertensive medication was analyzed longitudinally according to genotype with a generalized linear mixed-effect model (Fig. 2). Systolic (Fig. 2A) and diastolic (Fig. 2B) BP were greater in the combined group of individuals with the CT or TT genotypes of rs6929846 of BTN2A1 than in those with the CC genotype from 40 to 90 years of age. Systolic BP was greater in subjects with the GG genotype of rs146021107 of PDX1 than in the combined group of individuals with the G/- or -/- genotypes (Fig. 2C). Diastolic BP was greater in the combined group of individuals with the AG or GG genotypes of rs1671021 of LLGL2 than in those with the AA genotype (Fig. 2D).
Figure 2.
Longitudinal analysis with a generalized linear mixed-effect model of the association between (A) systolic or (B) diastolic blood pressure (BP) and age according to genotype for rs6929846 of BTN2A1 (CC vs. CT + TT), (C) between systolic BP and age according to genotype for rs146021107 of PDX1 (GG vs. G/- + -/-), or (D) between diastolic BP and age according to genotype for rs1671021 of LLGL2 (AA vs. AG + GG) among individuals not taking any anti-hypertensive medication.
Discussion
Given that genetic factors, as well as interactions between multiple genes and environmental factors are important in the development of hypertension (1), the ability to predict the risk of developing hypertension on the basis of genetic variants would be beneficial for the personalized prevention of this condition. In this study, we demonstrated that rs6929846 (T→C) of BTN2A1 was significantly associated with the prevalence of hypertension and also with systolic, diastolic, and mean BP in community-dwelling Japanese individuals, with the minor T allele representing a risk factor for hypertension.
We have previously reported that rs6929846 of BTN2A1 is significantly associated with hypertension in a cross-sectional study of a different hospital-based population (31). We also observed the association of this polymorphism with hypertension in a previous cross-sectional analysis of the Inabe Health and Longevity Study (26). The results of the present longitudinal population-based study are thus consistent with these previous observations (26,31) and validate the association of rs6929846 of BTN2A1 with hypertension.
BTN2A1 is a cell-surface transmembrane glycoprotein and a member of the butyrophilin superfamily of proteins. Many of these proteins regulate immune function, and polymorphisms within the coding sequences of the corresponding genes have been associated with the predisposition to inflammatory diseases (32). We have previously demonstrated that the T allele of rs6929846 of BTN2A1 is associated with an increased risk of developing myocardial infarction and with an increased transcriptional activity of BTN2A1 (15). The serum concentration of high-sensitivity C-reactive protein was significantly greater in individuals in the combined group of CT or TT genotypes for this SNP than in those with the CC genotype among healthy subjects without neoplastic, infectious, or inflammatory disease (15,33). These observations suggest that the T allele of rs6929846 of BTN2A1 may accelerate inflammatory processes.
Previous studies have suggested that chronic vascular inflammation influences BP and vascular remodeling (34–37). Systolic and diastolic BP, as well as pulse pressure were thus found to be positively associated with the plasma concentration of interleukin-6 in healthy men (34). The plasma concentration of high-sensitivity C-reactive protein was also greater in individuals with hypertension than in the controls, and it was shown to be positively associated with systolic BP and pulse pressure (35). In addition, oxidative stress and vascular inflammation have been shown to influence BP, suggesting that chronic inflammation may play a key role in the pathogenesis of hypertension (36,37). In this study, we demonstrated that rs6929846 of BTN2A1 was significantly associated with hypertension, with the minor T allele representing a risk factor for this condition. The enhancement of chronic inflammation by the T allele of rs6929846 may account for its association with hypertension, although the molecular mechanisms underlying the effects of this polymorphism on the development of hypertension remain to be elucidated.
In a previous meta-analysis of cohort studies, a reduction of 10 mmHg in systolic or 5 mmHg in diastolic BP was estimated to result in a 22–25% decrease in the incidence of coronary artery disease and a 36–41% decrease in that of stroke (38). In our longitudinal analysis, systolic, diastolic and mean BP were each increased by 1 mmHg in individuals with the TT genotype of rs6929846 of BTN2A1 compared with those with the CC genotype. Such a difference is small at the individual level and may not have practical clinical implications. However, even small increments in BP have important effects on cardiovascular morbidity and mortality at the population level, given the high incidence of coronary artery disease, stroke and chronic kidney disease. The reduction in the mortality rate estimated for each 2-mmHg decrease in systolic BP is 4% for coronary artery disease and 6% for stroke (39). Small differences in average BP at the population level thus result in significant differences in the population mortality rate (39).
In this study, we observed that the SNPs of PDX1, LLGL2 and FAM78B were also associated with the prevalence of hypertension, as well as with systolic BP among all individuals and individuals not taking any anti-hypertensive medication (PDX1), with diastolic and mean BP among individuals without anti-hypertensive medication (LLGL2), or with diastolic BP among individuals without anti-hypertensive medication (FAM78B). FAM78B is located at 1q24.1, which has previously been suggested to harbor susceptibility loci for hypertension (40) and type 2 diabetes mellitus (41), although the function of the gene remains unclear. PDX1 is a transcriptional activator at several genes, including those for insulin, somatostatin, glucokinase, islet amyloid polypeptide and glucose transporter type 2 (NCBI Gene). It contributes to the early development of the pancreas and plays an important role in the glucose-dependent regulation of insulin gene expression (42). A rare frameshift variant of PDX1 was previously found to associated with type 2 diabetes mellitus (43). We have previously demonstrated that rs146021107 of PDX1 is significantly associated with myocardial infarction (18,20), although, to the best of our knowledge, the association of PDX1 polymorphisms with hypertension has not yet been reported. LLGL2 plays a role in asymmetric cell division, the establishment of epithelial cell polarity and cell migration (44,45). We have previously demonstrated that rs1671021 of LLGL2 is associated with ischemic stroke (16), although, to the best of our knowledge, variants of LLGL2 have not yet been associated with hypertension.
The present study had certain limitations: i) given that the study subjects comprised only Japanese individuals, further studies are required on other ethnic groups. ii) It is possible that rs6929846 of BTN2A1 is in linkage disequilibrium with other polymorphisms in BTN2A1 or in nearby genes that are actually responsible for the development of hypertension. iii) The functional relevance of rs6929846 of BTN2A1 to the pathogenesis of hypertension remains unclear.
In conclusion, the present results suggest that BTN2A1 is a susceptibility gene for essential hypertension in Japanese individuals. The determination of the genotype for rs6929846 may prove informative for the assessment of the genetic risk for hypertension in such individuals.
Acknowledgments
This study was supported by the CREST, Japan Science and Technology Agency (to Y.Y. and I.T.), and by a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan (no. 24590746 to Y.Y.).
References
- 1.Lifton RP, Gharavi AG, Geller DS. Molecular mechanisms of human hypertension. Cell. 2001;104:545–556. doi: 10.1016/S0092-8674(01)00241-0. [DOI] [PubMed] [Google Scholar]
- 2.Kupper N, Willemsen G, Riese H, Posthuma D, Boomsma DI, de Geus EJ. Heritability of daytime ambulatory blood pressure in an extended twin design. Hypertension. 2005;45:80–85. doi: 10.1161/01.HYP.0000149952.84391.54. [DOI] [PubMed] [Google Scholar]
- 3.Agarwal A, Williams GH, Fisher ND. Genetics of human hypertension. Trends Endocrinol Metab. 2005;16:127–133. doi: 10.1016/j.tem.2005.02.009. [DOI] [PubMed] [Google Scholar]
- 4.Kannel WB. Elevated systolic blood pressure as a cardiovascular risk factor. Am J Cardiol. 2000;85:251–255. doi: 10.1016/S0002-9149(99)00635-9. [DOI] [PubMed] [Google Scholar]
- 5.Sacco RL, Benjamin EJ, Broderick JP, Dyken M, Easton JD, Feinberg WM, Goldstein LB, Gorelick PB, Howard G, Kittner SJ, et al. American Heart Association Prevention Conference. IV. Prevention and Rehabilitation of Stroke. Risk factors Stroke. 1997;28:1507–1517. doi: 10.1161/01.STR.28.7.1507. [DOI] [PubMed] [Google Scholar]
- 6.Yamagata K, Ishida K, Sairenchi T, Takahashi H, Ohba S, Shiigai T, Narita M, Koyama A. Risk factors for chronic kidney disease in a community-based population: A 10-year follow-up study. Kidney Int. 2007;71:159–166. doi: 10.1038/sj.ki.5002017. [DOI] [PubMed] [Google Scholar]
- 7.Ehret GB, Munroe PB, Rice KM, Bochud M, Johnson AD, Chasman DI, Smith AV, Tobin MD, Verwoert GC, Hwang SJ, et al. CHARGE-HF consortium: Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature. 2011;478:103–109. doi: 10.1038/nature10405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wain LV, Verwoert GC, O’Reilly PF, Shi G, Johnson T, Johnson AD, Bochud M, Rice KM, Henneman P, Smith AV, et al. Genome-wide association study identifies six new loci influencing pulse pressure and mean arterial pressure. Nat Genet. 2011;43:1005–1011. doi: 10.1038/ng.922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Newton-Cheh C, Johnson T, Gateva V, Tobin MD, Bochud M, Coin L, Najjar SS, Zhao JH, Heath SC, Eyheramendy S, et al. Genome-wide association study identifies eight loci associated with blood pressure. Nat Genet. 2009;41:666–676. doi: 10.1038/ng.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Levy D, Ehret GB, Rice K, Verwoert GC, Launer LJ, Dehghan A, Glazer NL, Morrison AC, Johnson AD, Aspelund T, et al. Genome-wide association study of blood pressure and hypertension. Nat Genet. 2009;41:677–687. doi: 10.1038/ng.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Adeyemo A, Gerry N, Chen G, Herbert A, Doumatey A, Huang H, Zhou J, Lashley K, Chen Y, Christman M, et al. A genome-wide association study of hypertension and blood pressure in African Americans. PLoS Genet. 2009;5:e1000564. doi: 10.1371/journal.pgen.1000564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kato N, Miyata T, Tabara Y, Katsuya T, Yanai K, Hanada H, Kamide K, Nakura J, Kohara K, Takeuchi F, et al. High-density association study and nomination of susceptibility genes for hypertension in the Japanese National Project. Hum Mol Genet. 2008;17:617–627. doi: 10.1093/hmg/ddm335. [DOI] [PubMed] [Google Scholar]
- 14.Tabara Y, Kohara K, Kita Y, Hirawa N, Katsuya T, Ohkubo T, Hiura Y, Tajima A, Morisaki T, Miyata T, et al. Global Blood Pressure Genetics Consortium: Common variants in the ATP2B1 gene are associated with susceptibility to hypertension: The Japanese Millennium Genome Project. Hypertension. 2010;56:973–980. doi: 10.1161/HYPERTENSIONAHA.110.153429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yamada Y, Nishida T, Ichihara S, Sawabe M, Fuku N, Nishigaki Y, Aoyagi Y, Tanaka M, Fujiwara Y, Yoshida H, et al. Association of a polymorphism of BTN2A1 with myocardial infarction in East Asian populations. Atherosclerosis. 2011;215:145–152. doi: 10.1016/j.atherosclerosis.2010.12.005. [DOI] [PubMed] [Google Scholar]
- 16.Yamada Y, Fuku N, Tanaka M, Aoyagi Y, Sawabe M, Metoki N, Yoshida H, Satoh K, Kato K, Watanabe S, et al. Identification of CELSR1 as a susceptibility gene for ischemic stroke in Japanese individuals by a genome-wide association study. Atherosclerosis. 2009;207:144–149. doi: 10.1016/j.atherosclerosis.2009.03.038. [DOI] [PubMed] [Google Scholar]
- 17.Yamada Y, Nishida T, Ichihara S, Kato K, Fujimaki T, Oguri M, Horibe H, Yoshida T, Watanabe S, Satoh K, et al. Identification of chromosome 3q28 and ALPK1 as susceptibility loci for chronic kidney disease in Japanese individuals by a genome-wide association study. J Med Genet. 2013;50:410–418. doi: 10.1136/jmedgenet-2013-101518. [DOI] [PubMed] [Google Scholar]
- 18.Yamada Y, Matsuo H, Segawa T, Watanabe S, Kato K, Hibino T, Yokoi K, Ichihara S, Metoki N, Yoshida H, et al. Assessment of genetic risk for myocardial infarction. Thromb Haemost. 2006;96:220–227. [PubMed] [Google Scholar]
- 19.Fujimaki T, Kato K, Yoshida T, Oguri M, Watanabe S, Metoki N, Yoshida H, Satoh K, Aoyagi Y, Nishigaki Y, et al. Association of genetic variants with myocardial infarction in Japanese individuals with chronic kidney disease. Thromb Haemost. 2009;101:963–968. doi: 10.1160/th08-11-0761. [DOI] [PubMed] [Google Scholar]
- 20.Oguri M, Kato K, Yokoi K, Itoh T, Yoshida T, Watanabe S, Metoki N, Yoshida H, Satoh K, Aoyagi Y, et al. Association of genetic variants with myocardial infarction in Japanese individuals with metabolic syndrome. Atherosclerosis. 2009;206:486–493. doi: 10.1016/j.atherosclerosis.2009.02.037. [DOI] [PubMed] [Google Scholar]
- 21.Ueyama C, Horibe H, Fujimaki T, Oguri M, Kato K, Yamada Y. Association of genetic variants of CELSR1 and 3q28 with hypertension in community-dwelling individuals. Biomed Rep. 2013;1:840–844. doi: 10.3892/br.2013.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shimokata S, Oguri M, Fujimaki T, Horibe H, Kato K, Yamada Y. Association between polymorphisms of the α-kinase 1 gene and type 2 diabetes mellitus in community-dwelling individuals. Biomed Rep. 2013;1:940–944. doi: 10.3892/br.2013.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oguri M, Fujimaki T, Horibe H, Kato K, Ichihara S, Yamada Y. Association of a polymorphism of BTN2A1 with chronic kidney disease in community-dwelling individuals. Biomed Rep. 2013;1:868–872. doi: 10.3892/br.2013.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fujimaki T, Horibe H, Oguri M, Kato K, Yamada Y. Association of genetic variants of the α-kinase 1 gene with myocardial infarction in community-dwelling individuals. Biomed Rep. 2014;2:127–131. doi: 10.3892/br.2013.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Horibe H, Ueyama C, Fujimaki T, Oguri M, Kato K, Ichihara S, Yamada Y. Association of a polymorphism of BTN2A1 with dyslipidemia in community-dwelling individuals. Mol Med Rep. 2014;9:808–812. doi: 10.3892/mmr.2014.1902. [DOI] [PubMed] [Google Scholar]
- 26.Murakata Y, Fujimaki T, Yamada Y. Association of a butyrophilin, subfamily 2, member A1 gene polymorphism with hypertension. Biomed Rep. 2014;2:818–822. doi: 10.3892/br.2014.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Perloff D, Grim C, Flack J, Frohlich ED, Hill M, McDonald M, Morgenstern BZ. Human blood pressure determination by sphygmomanometry. Circulation. 1993;88:2460–2470. doi: 10.1161/01.CIR.88.5.2460. [DOI] [PubMed] [Google Scholar]
- 28.Itoh Y, Mizuki N, Shimada T, Azuma F, Itakura M, Kashiwase K, Kikkawa E, Kulski JK, Satake M, Inoko H. High-throughput DNA typing of HLA-A, -B, -C, and -DRB1 loci by a PCR-SSOP-Luminex method in the Japanese population. Immunogenetics. 2005;57:717–729. doi: 10.1007/s00251-005-0048-3. [DOI] [PubMed] [Google Scholar]
- 29.Hanley JA, Negassa A, Edwardes MD, Forrester JE. Statistical analysis of correlated data using generalized estimating equations: An orientation. Am J Epidemiol. 2003;157:364–375. doi: 10.1093/aje/kwf215. [DOI] [PubMed] [Google Scholar]
- 30.Dean CB, Nielsen JD. Generalized linear mixed models: A review and some extensions. Lifetime Data Anal. 2007;13:497–512. doi: 10.1007/s10985-007-9065-x. [DOI] [PubMed] [Google Scholar]
- 31.Horibe H, Kato K, Oguri M, Yoshida T, Fujimaki T, Kawamiya T, Yokoi K, Watanabe S, Satoh K, Aoyagi Y, et al. Association of a polymorphism of BTN2A1 with hypertension in Japanese individuals. Am J Hypertens. 2011;24:924–929. doi: 10.1038/ajh.2011.74. [DOI] [PubMed] [Google Scholar]
- 32.Arnett HA, Escobar SS, Viney JL. Regulation of costimulation in the era of butyrophilins. Cytokine. 2009;46:370–375. doi: 10.1016/j.cyto.2009.03.009. [DOI] [PubMed] [Google Scholar]
- 33.Oguri M, Kato K, Yoshida T, Fujimaki T, Horibe H, Yokoi K, Watanabe S, Satoh K, Aoyagi Y, Tanaka M, et al. Association of a genetic variant of BTN2A1 with metabolic syndrome in East Asian populations. J Med Genet. 2011;48:787–792. doi: 10.1136/jmg.2010.088138. [DOI] [PubMed] [Google Scholar]
- 34.Chae CU, Lee RT, Rifai N, Ridker PM. Blood pressure and inflammation in apparently healthy men. Hypertension. 2001;38:399–403. doi: 10.1161/01.HYP.38.3.399. [DOI] [PubMed] [Google Scholar]
- 35.Schillaci G, Pirro M, Gemelli F, Pasqualini L, Vaudo G, Marchesi S, Siepi D, Bagaglia F, Mannarino E. Increased C-reactive protein concentrations in never-treated hypertension: The role of systolic and pulse pressures. J Hypertens. 2003;21:1841–1846. doi: 10.1097/00004872-200310000-00010. [DOI] [PubMed] [Google Scholar]
- 36.Savoia C, Schiffrin EL. Inflammation in hypertension. Curr Opin Nephrol Hypertens. 2006;15:152–158. doi: 10.1097/01.mnh.0000203189.57513.76. [DOI] [PubMed] [Google Scholar]
- 37.Androulakis ES, Tousoulis D, Papageorgiou N, Tsioufis C, Kallikazaros I, Stefanadis C. Essential hypertension: Is there a role for inflammatory mechanisms? Cardiol Rev. 2009;17:216–221. doi: 10.1097/CRD.0b013e3181b18e03. [DOI] [PubMed] [Google Scholar]
- 38.Law MR, Morris JK, Wald NJ. Use of blood pressure lowering drugs in the prevention of cardiovascular disease: meta-analysis of 147 randomised trials in the context of expectations from prospective epidemiological studies. BMJ. 2009;338:b1665. doi: 10.1136/bmj.b1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stamler J, Rose G, Stamler R, Elliott P, Dyer A, Marmot M. INTERSALT study findings. Public health and medical care implications. Hypertension. 1989;14:570–577. doi: 10.1161/01.HYP.14.5.570. [DOI] [PubMed] [Google Scholar]
- 40.Ehret GB, O’Connor AA, Weder A, Cooper RS, Chakravarti A. Follow-up of a major linkage peak on chromosome 1 reveals suggestive QTLs associated with essential hypertension: GenNet study. Eur J Hum Genet. 2009;17:1650–1657. doi: 10.1038/ejhg.2009.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shriner D, Adeyemo A, Rotimi CN. Joint ancestry and association testing in admixed individuals. PLoS Comput Biol. 2011;7:e1002325. doi: 10.1371/journal.pcbi.1002325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stoffers DA, Zinkin NT, Stanojevic V, Clarke WL, Habener JF. Pancreatic agenesis attributable to a single nucleotide deletion in the human IPF1 gene coding sequence. Nat Genet. 1997;15:106–110. doi: 10.1038/ng0197-106. [DOI] [PubMed] [Google Scholar]
- 43.Steinthorsdottir V, Thorleifsson G, Sulem P, Helgason H, Grarup N, Sigurdsson A, Helgadottir HT, Johannsdottir H, Magnusson OT, Gudjonsson SA, et al. Identification of low-frequency and rare sequence variants associated with elevated or reduced risk of type 2 diabetes. Nat Genet. 2014;46:294–298. doi: 10.1038/ng.2882. [DOI] [PubMed] [Google Scholar]
- 44.Müsch A, Cohen D, Yeaman C, Nelson WJ, Rodriguez-Boulan E, Brennwald PJ. Mammalian homolog of Drosophila tumor suppressor lethal (2) giant larvae interacts with basolateral exocytic machinery in Madin-Darby canine kidney cells. Mol Biol Cell. 2002;13:158–168. doi: 10.1091/mbc.01-10-0496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yasumi M, Sakisaka T, Hoshino T, Kimura T, Sakamoto Y, Yamanaka T, Ohno S, Takai Y. Direct binding of Lgl2 to LGN during mitosis and its requirement for normal cell division. J Biol Chem. 2005;280:6761–6765. doi: 10.1074/jbc.C400440200. [DOI] [PubMed] [Google Scholar]