The localization of MyBP-C within cardiac muscle cells provides efficient mechanochemical excitation-contraction coupling.
Keywords: myosin-binding protein C, native thick filaments, native thin filaments, calcium dynamics, phosphorylation, muscle activation, muscle regulation, cardiac excitation-contraction coupling
Abstract
The beating heart exhibits remarkable contractile fidelity over a lifetime, which reflects the tight coupling of electrical, chemical, and mechanical elements within the sarcomere, the elementary contractile unit. On a beat-to-beat basis, calcium is released from the ends of the sarcomere and must diffuse toward the sarcomere center to fully activate the myosin- and actin-based contractile proteins. The resultant spatial and temporal gradient in free calcium across the sarcomere should lead to nonuniform and inefficient activation of contraction. We show that myosin-binding protein C (MyBP-C), through its positioning on the myosin thick filaments, corrects this nonuniformity in calcium activation by exquisitely sensitizing the contractile apparatus to calcium in a manner that precisely counterbalances the calcium gradient. Thus, the presence and correct localization of MyBP-C within the sarcomere is critically important for normal cardiac function, and any disturbance of MyBP-C localization or function will contribute to the consequent cardiac pathologies.
INTRODUCTION
Ventricular power generation involves the rapid release of calcium from the sarcoplasmic reticulum (SR) and diffusion of these ions through the sarcomere to activate the contractile machinery (Fig. 1A). Each heartbeat is initiated by an action potential that triggers the opening of ryanodine receptors (1) (calcium release channels in the terminal cisternae of the SR) that are located within nanometers of the Z-lines of each sarcomere (Fig. 1A). Calcium then binds to the troponin-tropomyosin regulatory complex to activate the actin-based thin filament so that myosin molecular motors, comprising the thick filaments, can interact with the thin filaments (Fig. 1A) to generate force and motion (2, 3). Because of the spatial organization of the calcium release channels and diffusion rates in the sarcomere, molecular modeling (4) and experimental evidence from elongated frog skeletal muscle (5) suggest that a spatiotemporal calcium gradient should exist along the length of each cardiac sarcomere that could lead to heterogeneity in excitation-contraction (E-C) coupling. Cardiac myosin-binding protein C (MyBP-C) (Fig. 1B), which is considered to be a potential activator of the thin filament (6–9), is located in a region of the sarcomere (that is, C-zone; Fig. 1A) that would experience such temporal delays in calcium concentrations. Therefore, we hypothesize that localization of MyBP-C to the C-zone may mitigate potential inhomogeneities in calcium activation.
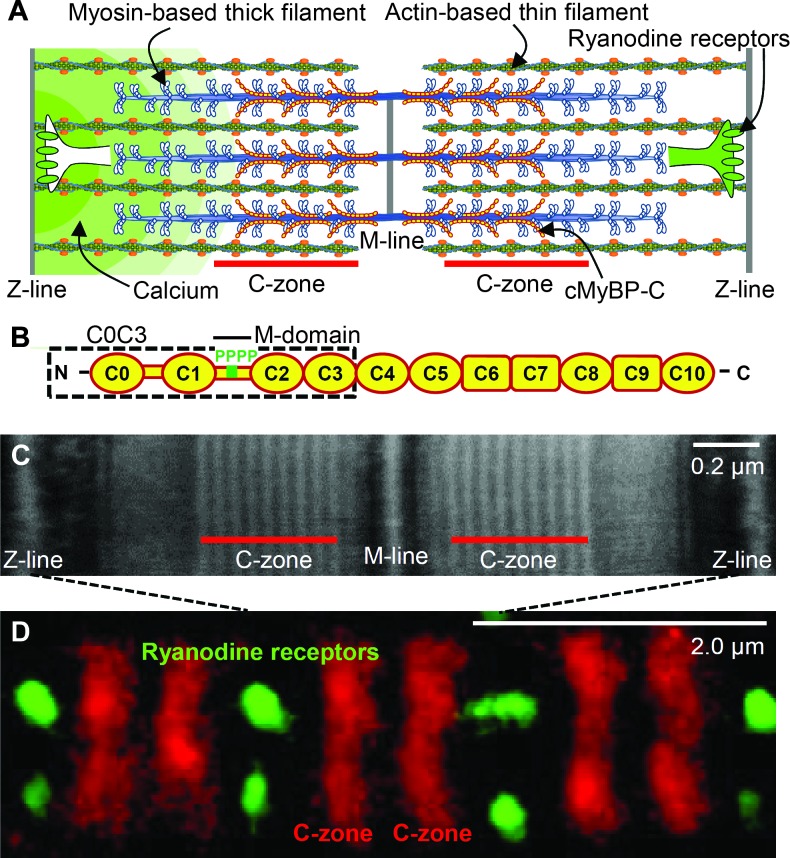
Fig. 1. Sarcomeric organization and MyBP-C.
(A) Cardiac muscle sarcomere with interdigitating thick and thin filaments. MyBP-C localized to the C-zone, whereas the ryanodine receptors are localized in puncta (CRUs) along the Z-lines, forming the boundaries of each sarcomere. (B) Schematic diagram of cardiac MyBP-C’s Ig-like (oval) and fibronectin-like (rectangular) domains with four phosphorylation sites (P) in the M-domain and C0C3 fragment (dashed box) used in the 3D EM and in vitro motility experiments. (C) Negatively stained EM image of a sarcomere within a mouse ventricular myocyte labeled with antibodies to MyBP-C. (D) Two-color dSTORM super-resolution image of sarcomeres as in (C), labeled with Alexa Fluor–conjugated antibodies to MyBP-C and ryanodine receptors.
Cardiac MyBP-C is a 140-kD immunoglobulin (Ig) protein superfamily member (Fig. 1B) that is strongly bound to the thick filament by its C terminus, whereas its N-terminal domains extend radially (10) to allow their binding to the thin filament and the myosin head region that emanates from the thick filament backbone (11). In the normal heart, MyBP-C’s M-domain contains four highly phosphorylated cardiac-specific serine residues (Fig. 1B) (12–14). Although the role of MyBP-C in vivo is unclear, its functional importance is evidenced by its genetic mutations being a leading cause of hypertrophic cardiomyopathy (15) and its decreased phosphorylation being a hallmark of the failing heart (12, 13). Here, we have used advanced multidimensional light microscopy techniques and analytic modeling to quantify a calcium transient across a single sarcomere during the initial phase of cardiac muscle contraction that would lead to a gradient in thin filament activation. We then combined our recently developed single-particle thick filament–based analytic platform (14) and three-dimensional (3D) electron microscopy (EM) to determine that MyBP-C’s N-terminal domains can sensitize thin filaments to calcium within the C-zone. Thus, we propose that MyBP-C effectively normalizes an intrinsic inhomogeneity in E-C coupling that exists because of the sarcomeric calcium gradient.
RESULTS
A transient calcium gradient is observed within single sarcomeres
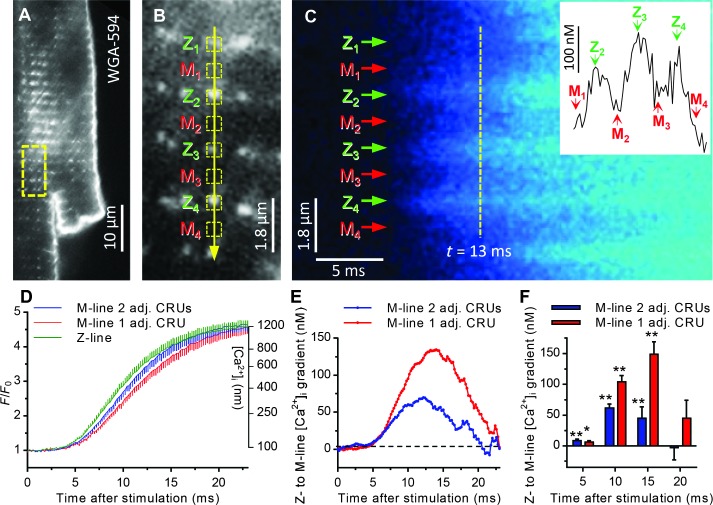
The locations of MyBP-C and the ryanodine receptors within isolated mouse cardiac myocytes were visualized by EM and two-color direct stochastic optical reconstruction microscopy (dSTORM) (16) with nanometer spatial resolution (Fig. 1, C and D, and fig. S1). Punctate clusters of ryanodine receptors form calcium release units (CRUs) that reside along the Z-lines of each sarcomere to which the thin filaments are anchored (Fig. 1D and fig. S1), whereas MyBP-C is localized to nine distinct stripes within each C-zone, flanking the M-line and centered ~650 nm from the Z-line (Fig. 1, C and D). We used high-speed laser scanning confocal microscopy (Fig. 2, A to F) to monitor the electrically stimulated increase in cytosolic calcium [[Ca2+]i (intracellular calcium concentration)] in 500-nm regions (Fig. 2B) encompassing either CRUs at the Z-lines (ends of the sarcomere) or MyBP-C near the M-line (center of the sarcomere) with 200-μs temporal resolution (16). We identified the locations of CRUs through live cell labeling with a fiduciary transverse tubule (t-tubule)/Z-line marker (Fig. 2, A and B) and placed our confocal scan line directly over several confirmed CRUs and their neighboring M-lines in series (Fig. 2B). In relaxed myocytes, the basal level of calcium within both the Z- and M-line regions was 90 to 105 nM [that is, −log calcium (pCa), 7.05 to 6.98], reaching 1 to 1.4 μM (pCa 6.00 to 5.85) at the peak of contraction. Excitation resulted in the triggering of calcium release from most, but not all, of the CRUs on any given beat, as previously suggested (17). Although calcium is being released from CRUs in three dimensions, we are only able to resolve these events in the visual plane, and for simplicity, we refer to the events as originating from one or both CRUs flanking a single sarcomere. We thus separately analyzed the time course of [Ca2+]i from sarcomeres where only one CRU was triggered versus where two CRUs were triggered on opposing ends of an individual sarcomere. Under physiological circumstances, a sarcomere would be activated by only one CRU between 25 and 50% of the time, which would also be influenced by factors such as sarcomeric calcium load, ryanodine receptor sensitivity, and any CRU structural remodeling (18). Within 5 ms of stimulation, calcium concentrations began to increase at both the Z- and M-line regions, but with a temporal delay at the M-line, being distant from the CRUs (Fig. 2D). This delay led to an average calcium gradient of 60 to 140 nM from the Z- to the M-line, with the magnitude dependent on whether calcium was released from one or two CRUs flanking a given M-line. The calcium gradient peaked roughly 13 ms after electrical stimulation and persisted for 20 ms before collapsing because of diffusion (Fig. 2, E and F). Notably, contraction was detected 8 to 10 ms after electrical stimulation.
Fig. 2. Sarcomeric calcium gradients in ventricular myocytes.
(A) Single cardiac myocyte labeled with a lipophilic wheat germ agglutinin conjugated to Alexa Fluor 594 to identify t-tubule CRUs. Yellow box indicates area expanded in (B). (B) Line of yellow arrow represents confocal line scan across several CRUs. Only confirmed CRUs and their adjacent M-lines were scanned. Yellow boxes represent 5-pixel regions of interest (ROIs) (500 nm) over the Z- and M-line regions. (C) Rising phase of the calcium transient ([Ca2+]i) from scanned region depicted in (B) immediately after stimulation of ventricular myocyte loaded with fluorescent calcium indicator. Certain M-lines are flanked by two adjacent CRUs that release calcium immediately upon stimulation (for example, M2 and M3), whereas others are adjacent to only one CRU that is triggered (for example, M1 and M4). The inset depicts the spatial profile of [Ca2+]i 13 ms after stimulation (vertical yellow line). (D) Average [Ca2+]i transient ± error bars (SEM) rising phase time course from 59 Z-line CRUs (green, n = 12 cells) and their adjacent (adj.) M-lines when flanked by two triggered CRUs (blue, n = 59 sites, 12 cells) and a single triggered CRU (red, n = 53 sites, 12 cells). (E) Z- to M-line [Ca2+]i gradient calculated by subtracting M-line from Z-line traces in (D). (F) Z- to M-line [Ca2+]i gradient ± SEM 5, 10, 15, and 20 ms after stimulation in sarcomeres. *P < 0.01, **P < 0.001, one-sample t test.
Is this gradient intrinsic to the diffusion of calcium within the sarcomere or an artifact of sarcomere movement? To control for potential artifacts from myocyte contraction, we repeated these experiments in the presence of the contractile inhibitor blebbistatin. Blebbistatin blocked myocyte contraction without qualitatively altering the sarcomeric calcium gradient (fig. S2). The persistence of the gradient can be explained by calcium release flux from each CRU remaining significantly elevated for 10 to 20 ms after stimulation (19) and is considerably longer-lasting than would be predicted for a calcium gradient in skeletal muscle, where rapid channel inactivation limits release flux to ~2 ms (20).
Native thin filaments are activated by calcium
Could such temporal differences in calcium distribution within the sarcomere create spatial gradients in contractile activation along the length of the calcium-regulated thin filament? In a simplified model system of cardiac contractility, we determined the concentrations over which calcium regulates the sliding of fluorescently labeled, native mouse cardiac thin filaments on a surface of monomeric mouse α-cardiac myosin [25 mM KCl, 100 μM ATP (adenosine triphosphate), 22°C] (16). On the basis of this in vitro motility assay, thin filaments are highly sensitive to calcium over the range of concentrations measured in the sarcomere after excitation (pCa 7.05 to 5.85; Fig. 2D), as evidenced by the sigmoidal increases in both the sliding velocities (fig. S3A) and the fraction of thin filaments moving (fig. S3B and Fig. 3E). Graded responses in both calcium-activated thin filament sliding velocities and fraction of thin filaments moving reflect the increased engagement of force-generating actomyosin cross-bridges (21). However, both velocity and fraction of thin filaments moving may not be directly proportional to the number of attached, force-generating cross-bridges because few cross-bridges are required to propel a native thin filament (21). Regardless, on the basis of the fraction of filaments moving (Fig. 3E), these data suggest that the observed gradient in free calcium between the Z- and M-lines (Fig. 2, E and F) would result in thin filaments being up to ~22% less activated near the center of the sarcomere 10 ms after excitation. This inhomogeneity in E-C coupling could compromise mechanical efficiency in the heart that must repetitively contract and relax in a synchronous manner.
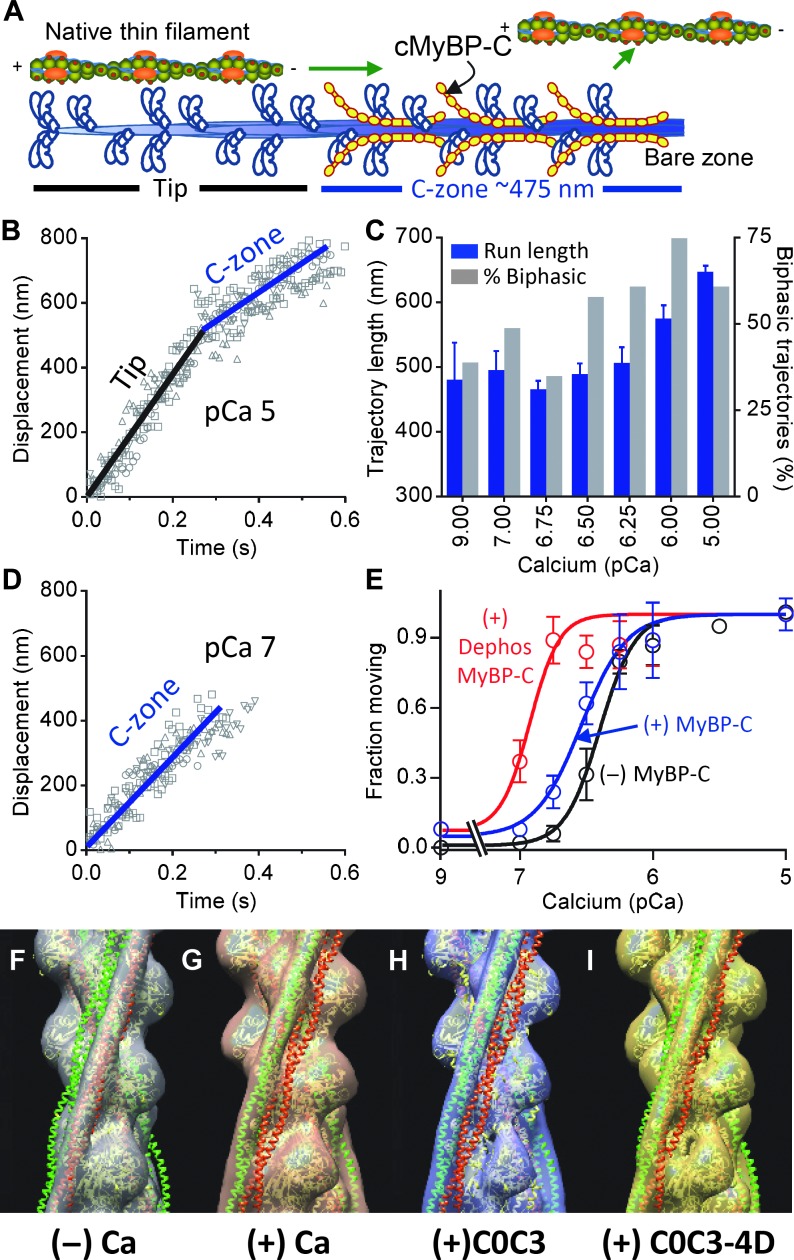
Fig. 3. The effects of MyBP-C on native thin filament activation.
(A) Illustration of a thin-filament shard traveling over one-half of a native thick filament, as in TIRFM experiments. (B) Displacement-time plots for five thin filaments on wild-type thick filaments at pCa 5 demonstrating two phases of velocity (black, fast; blue, slow). (C) Trajectory length ± SEM–pCa and percent trajectories with biphasic velocities (as in B)–pCa histograms for thin filaments on wild-type thick filaments. (D) As in (B) for five filaments at pCa 7 demonstrating movement only within the thick filament C-zone. (E) Fraction of thin filaments moving ± SEM–pCa plots for thin filament motion in the absence of (black) and presence of wild-type (blue) and λ-phosphatase–treated MyBP-C (red). Data fitted with sigmoidal dose-response curves with differing pCa50 (P < 0.001, extra sum-squares F test). Dephos, dephosphorylated with λ-phosphatase. (F and G) 3D structure of native thin filaments with tropomyosin (Tm) in the (F) “blocked” position in the absence of and (G) “closed” position in the presence of calcium. Atomic structures of actin with Tm in the “blocked” position (red) and “closed” position (green) were fitted to each 3D structure. (H and I) 3D structure of native thin filaments decorated with MyBP-C N-terminal fragments in the absence of calcium. Position of tropomyosin with addition of (H) C0C3 and (I) phosphomimetic C0C3-4D fragments. The 3D structure fitted with atomic structures showed Tm’s movement in thin filament decorated with C0C3 from “blocked” to “closed” position. In contrast, the fitted structures for the thin filament decorated with C0C3-4D showed only partial movement of Tm toward the “closed” position.
MyBP-C enhances thin filament activation within the C-zone
Could MyBP-C residing within the C-zone near the sarcomere center eliminate this potential gradient in E-C coupling by effectively sensitizing the thin filament to calcium? To answer this question, we isolated native thick filaments from wild-type mouse hearts. Electron microscopy, Western blotting, and quantitative mass spectrometry were previously used to show that these native thick filaments retained MyBP-C’s in vivo stoichiometry and spatial orientation within the thick filament C-zone (Fig. 3A) and that there was a high level of phosphorylation (~65%) on each of MyBP-C’s four phosphorylatable serines (14), as seen in healthy human hearts (12, 13). Using total internal reflectance microscopy with high temporal (8.3 ms) and spatial (30 nm) resolution, we observed the calcium-dependent sliding of short ~250-nm native thin filament shards over native thick filaments (pCa 9 to 5; 25 mM KCl, 100 μM ATP, 22°C) (16). This assay preserves to a large extent the spatial orientations and relationships that exist between thin and thick filaments in vivo. Thus, the use of the short thin filaments allows for the observation of actomyosin interactions in regions of the thick filament with (that is, C-zone) and without MyBP-C. As with unregulated bare actin filaments (14), at pCa 5 when thin filaments are fully calcium-activated (fig. S3), the thin filaments slide for 643 ± 14 nm (n = 112) along the thick filaments with the majority (60%, n = 67) sliding with an initial fast phase of velocity, which is then slowed by MyBP-C as the thin filament enters the C-zone (Fig. 3, A to C). Finally, the thin filament detaches once in the bare zone, where the thick filament is devoid of myosin heads (Fig. 3, A to C). At calcium concentrations leading to submaximal thin filament activation (pCa > 5), the fractions of runs demonstrating biphasic velocities (Fig. 3B) were reduced (Fig. 3C), and at the lowest calcium concentrations (pCa ≥ 7) where thin filaments should be inactive, runs were observed of constant, slow velocity (Fig. 3D), with short run lengths (Fig. 3C) nearly equivalent to that of the C-zone (~475 nm; Fig. 3A). When compared to the calcium-dependent motion of thin filaments observed in the in vitro motility assay using depolymerized mouse cardiac myosin in the absence of MyBP-C (Fig. 3E, black, and fig. S3B), the fraction of filaments moving over wild-type native thick filaments (Fig. 3E, blue) demonstrated a sigmoidal relationship that was more sensitive to calcium (pCa50, 6.5 ± 0.04 versus 6.4 ± 0.02; P < 0.05). These data suggest that MyBP-C increases the number of force-generating cross-bridges at low calcium concentrations to enhance the thin filament’s effective calcium sensitivity, as supported by previous in vitro (8, 9) and muscle fiber studies (6, 7), but only within the thick filament C-zone where MyBP-C resides.
MyBP-C’s ability to activate the thin filament is enhanced by dephosphorylation
Because thick filaments are highly phosphorylated in the normal heart and become dephosphorylated during heart failure (12, 13), does MyBP-C phosphorylation tune MyBP-C’s thin filament activating potential? To test this hypothesis, we treated thick filaments using λ-phosphatase, which dephosphorylated MyBP-C to ~22% (14). Whereas the trajectory run length distributions were similar to the untreated thick filaments (Fig. 3C and fig. S4), the pCa50 for fraction of thin filaments moving (Fig. 3E; 6.9 ± 0.06 versus 6.5 ± 0.04) increased markedly (P < 0.001), suggesting that dephosphorylation enhances MyBP-C’s capacity to effectively sensitize the thin filament to calcium. Thus, MyBP-C phosphorylation appears to modulate MyBP-C function. One potential molecular mechanism is that phosphorylation alters how MyBP-C N-terminal domains interact with and activate the thin filament, which we next characterized using EM.
MyBP-C’s N-terminal domains displace tropomyosin in a phosphorylation-dependent manner
Conventional models of thin filament activation suggest that calcium binding to troponin shifts tropomyosin azimuthally from the “blocked” to the “closed” position on actin (2, 3), allowing myosin access to the thin filament. With EM and generated 3D reconstructions (16), we visualized this calcium-induced shift in tropomyosin position on native thin filaments (Fig. 3, F and G). An expressed (dephosphorylated) MyBP-C N-terminal fragment (C0C3) binds to the thin filament and shifts tropomyosin to the myosin-accessible, “closed” position even in the absence of calcium (Fig. 3H and figs. S5 and S6), as recently observed for the shorter C0C2 fragment (9). In contrast, a C0C3 fragment with its four phosphorylatable serines replaced by aspartic acid residues (D) to mimic 100% phosphorylation (C0C3-4D; Fig. 1B) was still capable of displacing tropomyosin in the absence of calcium but only partially toward the “closed” position (Fig. 3I and figs. S5 and S6). Although these data suggest that phosphorylation of all four endogenously phosphorylated sites alters MyBP-C’s ability to modulate tropomyosin’s position on the thin filament, the impact of phosphorylation in vivo may be more complex given that one or more phosphorylatable sites may affect MyBP-C’s molecular function differentially (22, 23). The ability of MyBP-C phosphorylation to modulate the extent of tropomyosin displacement (Fig. 3, H and I) provides a molecular mechanism to tune MyBP-C’s activating potential in the native thick filament (Fig. 3E) or when the N-terminal domains are added to the in vitro motility assay (fig. S7) (16). MyBP-C appears to be a tunable mechanical element within the thick filament that acts synergistically with calcium through shifts in tropomyosin (Fig. 3, H and I) to modulate levels of thin filament activation (Fig. 3E). The localization of MyBP-C in the C-zone of the sarcomere could thus impose a countergradient in activation to effectively sensitize the thin filament to calcium where the spatiotemporal gradient of free calcium is greatest (Fig. 2, D to F). Although both calcium and MyBP-C binding independently move tropomyosin into the “closed” position, myosin strong-binding is required to displace tropomyosin to the “open” position to cooperatively amplify and turn on the thin filament for additional myosin binding (2, 3). Experimental evidence suggests that MyBP-C can also bind to the S2-domain of the myosin head in a phosphorylation-dependent manner (6). Such S2 binding affects the positioning of the myosin head relative to the thick filament (24, 25), which in turn can influence the attachment and detachment kinetics of myosin to the thin filament (6, 26). Therefore, MyBP-C binding to the myosin S2-domain may also effectively sensitize the thin filament to calcium and contribute to mitigating the functional impact of the calcium gradient. Therefore, the timing and distance over which myosin strong-binding to the thin filament can affect thin filament activation both within and outside the C-zone requires further experimental consideration.
DISCUSSION
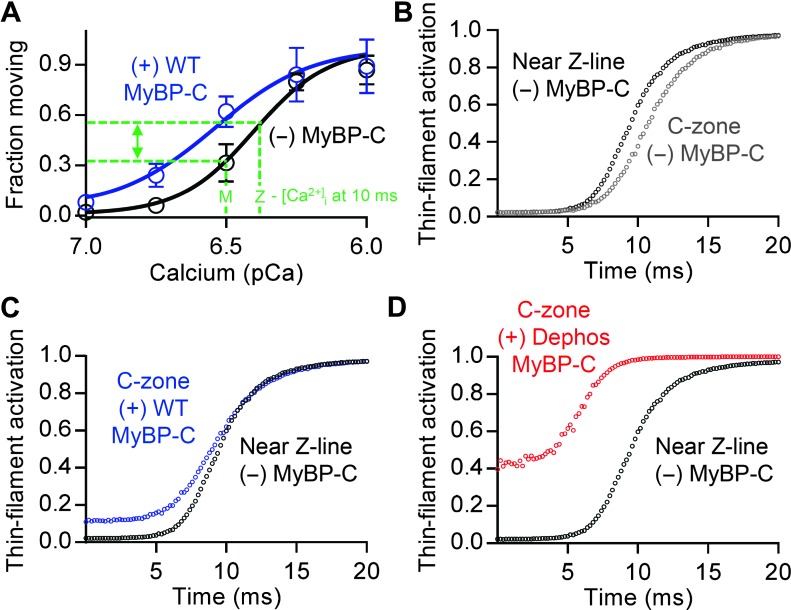
To appreciate the potential spatial effect of both this transient difference in calcium across the sarcomere and MyBP-C on the activation state of the thin filament, we developed an analytical model that uses our data to approximate the thin filament’s activation state over the length of the thin filament after myocyte stimulation (16). Therefore, we spatially and temporally modeled the thin filament activation state by assuming that the fraction of filaments moving was a sensitive indicator of the thin filament’s activation state rather than velocity, because changes in thin filament sliding velocities due to MyBP-C may occur through mechanisms other than changes in the number of attached force-generating cross-bridges (27–30). We then transformed the [Ca2+]i time course at the Z-line in which the thin filaments insert and at the M-line adjacent to the C-zone where MyBP-C resides (Fig. 2D) into time courses of relative thin filament activation (that is, fraction of filaments moving) using the sigmoidal-fitted relationship for the fraction of thin filaments moving versus pCa (Fig. 4A). On the basis of these data, within the initial 15 ms of electrical stimulation when the calcium gradient is present and at its greatest, the activation state within the C-zone would be up to 22% less than the region of the thin filament closest to the calcium release sites near the Z-line (Fig. 4, A and B). Given the activating potential of natively phosphorylated MyBP-C (Fig. 3E), MyBP-C within the C-zone is predicted to mitigate the inhomogeneity in E-C coupling due to the calcium gradient (Fig. 4B) and thus normalize E-C coupling (Fig. 4C) to ensure rapid and uniform force development within the sarcomere during the early portion of the systolic phase of cardiac contraction. Both this critical role for MyBP-C and its slowing or “breaking” action (Fig. 3B) would be reduced or lost in hypertrophic cardiomyopathy patients with MyBP-C haploinsufficiency (15). In contrast, MyBP-C dephosphorylation, as observed in failing human hearts (12, 13), is predicted to hypersensitize the thin filament’s activation state by as much as 39% in the C-zone over the range of calcium we measured (Fig. 4D). This would cause cardiac cells to maintain considerable levels of activation even during diastole, resulting in impaired relaxation and decreased ventricular filling—hallmarks of the failing heart. Reduced diastolic filling limits ventricular stretch between heartbeats, decreasing normal gains in force generation that occur through the Frank-Starling mechanism. Thus, our findings provide a mechanistic explanation for how dephosphorylated MyBP-C contributes to impaired relaxation and contractile dysfunction (31, 32). In conclusion, MyBP-C should be considered a potential link in the heart’s E-C coupling system that may mechanically compensate for an intrinsic spatial nonuniformity in calcium release to ensure rapid and efficient activation of the sarcomere.
Fig. 4. Effect of [Ca2+]i on thin filament activation in the absence of MyBP-C near the Z-line and presence of MyBP-C within the C-zone.
(A) Fraction of thin filaments moving ± SEM–pCa plots within the Z- and M-line regions of the sarcomere in the (black) absence and (blue) presence of wild-type (WT) MyBP-C. [Ca2+]i in the Z- and M-line regions 10 ms after cardiac excitation indicated (green dashed lines). (B to D) Model generated estimates of thin filament activation after calcium release. Thin filament activation–time plot generated for regions of the thin filaments near the Z-lines (black) and within the C-zone, near the center of the sarcomere. (B) Model assumes that the C-zone lacks MyBP-C and thus predicts that thin filament activation in the C-zone (gray) would lag behind the Z-line. (C) Model assumes normally phosphorylated wild-type MyBP-C in C-zone (blue) and predicts that C-zone activation level would no longer lag behind the Z-line. (D) Model assumes dephosphorylated MyBP-C within C-zone (red), resulting in a sensitized C-zone that would contribute to significant activation even during relaxation.
MATERIALS AND METHODS
Experimental design
The objective of our study was to test the hypothesis that MyBP-C is localized to a defined and limited region of the sarcomere, the elementary force-generating unit of the heart, to correct an intrinsic inhomogeneity in cardiac E-C coupling. We established a collaborative multidisciplinary investigative team with expertise in intracellular cardiac calcium signaling (Lederer Lab), contractile protein molecular biophysics (Warshaw Lab), and contractile protein structural biology (Craig Lab) to address this question at the subcellular level using state-of-the-art molecular techniques. We then attempted to develop an analytical model to illustrate the gross level impact of MyBP-C in the sarcomere on contractile function in response to the calcium gradient.
Myofibril isolation and EM imaging
Myofibrils were isolated from the left ventricle of adult 129 SVE mice for EM imaging of MyBP-C within the sarcomere. The tissue was cut into small slices in rigor solution containing 100 mM NaCl, 3 mM MgCl2, 2 mM EGTA, 1 mM NaN3, and 5 mM PIPES (pH 7.0). The slices were skinned in rigor solution containing 0.1% w/v saponin with agitation (3 hours, 4°C) and then homogenized with a Polytron homogenizer (Brinkmann Instruments) in rigor solution. Myofibrils were placed on EM grids, blocked with 1% bovine serum albumin (BSA) in phosphate-buffered saline (PBS), and then incubated with a cMyBP-C antibody in PBS (gift from S. Harris). For negative staining, grids were fixed with 2.5% glutaraldehyde (buffered in rigor solution, pH 7.0), washed with 0.1 M ammonium acetate, and negatively stained with 0.5% ammonium molybdate. Images were taken on a Tecnai Spirit transmission electron microscope (FEI) with an Erlangshen CCD (charge-coupled device) camera (Gatan Inc.) at 80 kV.
Cardiomyocyte isolation
Cardiomyocytes were prepared from the left ventricle of adult C57BL/6 mice for dSTORM and confocal imaging as described (33). Cardiomyocytes were stored in a normal Tyrode’s solution containing 140 mM NaCl, 5 mM KCl, 1.8 mM CaCl2, 0.5 mM MgCl2, 5 mM Hepes, 5 mM glucose, and 0.33 mM NaH2PO4 (pH 7.4).
Two-color dSTORM super-resolution imaging
dSTORM (34) was performed on murine ventricular myocytes. Immunolabeling was performed as previously described (35). Cells were probed with anti–N-terminal MyBP-C (1:100 dilution, rabbit polyclonal; sc-67353, Santa Cruz Biotechnology) and anti-RyR2 (1:100 dilution, mouse monoclonal; C3-33, Pierce) antibodies, followed by labeling with secondary antibodies (1:500 dilution, goat anti-mouse Alexa Fluor 488 or 647, goat anti-rabbit Alexa Fluor 647, Life Technologies). Images were captured on a Nikon Eclipse Ti-E inverted wide-field/total internal reflection fluorescence microscope (TIRFM) using a 100× 1.49 N.A. (numerical aperture) oil immersion lens. Two-color acquisition was performed in an interlaced fashion, with images collected at 55 frames/s on an 897 Ultra EMCCD camera (Andor Technology). A minimum of 40,000 single-molecule events were detected per field of view, and super-resolution images were reconstructed from Gaussian fits to these single-molecule events.
T-tubule and calcium imaging
Cells were loaded with Fluo-4 by 15-min incubation with 3 μM Fluo-4–acetoxymethyl ester (Life Technologies) and 0.01% Pluronic F127 (a poloxamer made by BASF Corporation) and allowed an additional 10 min for de-esterification. Cells were additionally stained with the lipophilic t-tubule marker wheat germ agglutinin conjugated to Alexa Fluor 594 (WGA-594, Life Technologies). All experiments were performed in custom, rotatable glass-bottomed cell chambers (Four Hour Day Foundation) equipped with local and bath perfusion, and platinum electrodes for field stimulation at 1 Hz. Cells were imaged on a LSM 5 LIVE inverted ultrahigh-speed confocal microscope using a 63× 1.4 N.A. water immersion objective (Zeiss). Loading of indicators and experiments was performed at room temperature (22 ± 1°C). First, WGA-594 was excited and imaged using a 563-nm diode laser. After t-tubule CRUs were located, a confocal line scan was placed over 7 to 15 CRUs in series. This region of the cell was then scanned using a 488-nm argon ion laser in confocal line scan mode at 200 μs per line. This scan speed and chemical indicator combination was found to produce the best signal/noise and necessary temporal resolution to resolve sarcomeric calcium gradients. Pixel size was 0.1 μm, and 500-nm (5-pixel) ROIs were placed over the known CRUs at the Z-line or their directly adjacent M-lines, and the fluorescence time course after field stimulation was calculated for each ROI. Fluorescence time courses were converted to F/F0, with F0 the resting (before stimulation) fluorescence for each ROI to control for dye loading and potential compartmentalization. The calibration of the fluorescence signal (F) to [Ca2+]i from a single wavelength indicator was done using the following equation: [Ca2+] = Kd(F − Fmin)/(Fmax − F) as described (36), where Fmin is the fluorescence intensity in the absence of Ca2+ and Fmax is the fluorescence in the presence of saturating Ca2+. Fmin was achieved by perfusing the cell with a normal Tyrode’s solution lacking CaCl2 with the following additions: 25 μM cytochalasin D (to inhibit contraction), 2 μM calcium ionophore ionomycin, and 10 mM EGTA. Fmax was achieved by perfusing the cell with normal Tyrode’s solution with 25 μM cytochalasin D, 2 μM ionomyocin, and 10 mM CaCl2. A Kd (dissociation constant) of 891 nM was used as recently measured for Fluo-4 in the presence of protein (37).
Protein preparation
Native thick and thin filaments were isolated for from the apex of adult FVB mouse hearts as described (9, 14). Briefly, sarcomeric proteins were purified by serial mechanochemical extractions. The native thick filaments were liberated from 5 to 10 small bundles of muscle strips by digestion in a 10-μl drop containing 0.2 U/μl of calpain-1 from porcine erythrocytes (Calbiochem). The thick filaments were stored in 250 μl of chilled 75 mM NaCl, 5 mM MgCl2, 2 mM EGTA, 1 mM DTT (dithiothreitol), 7 mM phosphate buffer (pH 7), 10 mM creatine phosphate, and 2.5 mM ATP. Although MyBP-C is a target for calpain-induced degradation, this procedure results in 79% of the MyBP-C being intact (14). For dephosphorylation experiments, 50-μl aliquots of thick filament samples were incubated with 5 μl (2000 U) of λ-phosphatase from Escherichia coli (New England BioLabs) and 6.5 μl of 10 mM MnCl2 (1 hour, 30°C) to decrease the phosphorylation of the endogenous MyBP-C to 22% (14). Native thin filaments were liberated from muscle strips by serial extraction through homogenization and centrifugation. The thin filaments were stored in chilled 100 mM KCl, 5 mM MgCl2, 1 mM EGTA, 25 mM imidazole (pH 6.45), and 10 mM DTT. The thin filaments were fluorescently labeled with tetramethyl rhodamine phalloidin and diluted in actin buffer [25 mM KCl, 1 mM EGTA, 10 mM DTT, 25 mM imidazole, and 4 mM MgCl2, adjusted to pH 7.4; containing an oxygen scavenging system [glucose oxidase (0.1 μg/ml), catalase (0.018 μg/ml), and glucose (2.3 μg/ml)]. Before use, 1.5-ml tubes containing 1 ml of 0.01 μM TRITC (tetramethyl rhodamine isothiocyanate)–labeled thin filaments in actin buffer were sonicated for 20, 1-s pulses using a probe sonicator (Model 300, Fisher) to produce ~250-nm thin filament shards (14).
For the in vitro motility assays, monomeric myosin was freshly isolated from the ventricles of FVB mouse hearts as described (9, 14). Cardiac MyBP-C N-terminal C0C3 (amino acids 1 to 539) and phosphomimetic C0C3-4D fragments were bacterially expressed as described (9, 14). The C0C3-4D fragment contained phosphomimetic aspartic acid residues in place of the four phosphorylatable serines (S273, S282, S302, and S307) located within the M-domain.
Fluorescence image acquisition
All native thin filament sliding assays were performed at room temperature (22 ± 1°C) using a Nikon Eclipse Ti-U inverted microscope equipped with a Plan Apo 100× 1.49 N.A. oil emersion lens for epifluorescence or through-the-objective total internal reflectance microscopy (TIRFM) using a custom “Micro Optic Fiber Launch TIRFM” (patent pending) as described (14). Briefly, either a Lumen 200-W metal arc lamp (Prior Scientific) or 532-nm diode-pumped, solid-state, 50-mW laser (Lasever Inc.) was used for excitation. Images were obtained using an intensified high-resolution 10-bit digital camera (XR/Turbo-Z running Piper Control v2.6.09 software, Stanford Photonics).
In vitro motility assay
To determine the calcium sensitivity of the native thin filaments, in vitro motility assays were performed on the surface of a flow cell coated with mouse cardiac myosin (100 μg/ml) with the calcium-dependent motion of the filaments observed by epifluorescence microscopy as described (9). Motility buffer (actin buffer with 100 μM ATP, 0.5% methyl cellulous, and calcium chloride ranging from pCa 4 to 9) was added to the flow cell, and thin filament motion was observed after 3 min of incubation. Calcium levels were determined using MaxChelator software (38) to account for the 1 mM EGTA present in the motility buffer. Images were captured at 10 frames/s and down-sampled to 2 frames/s using ImageJ 1.43u (National Institutes of Health), and DiaTrack 3.03 software for Windows (Semasopht) was used to determine their velocity and fraction of moving thin filaments. The mean velocity and fraction moving ± SEM were determined from the average of four movies for each experimental condition from a minimum of three independent myosin and native thin filament preparations. The velocity and fraction of filaments moving were plotted with respect to pCa (negative log of the calcium concentration) and fitted with sigmoidal dose-response curves with variable slopes using GraphPad Prism 6 (GraphPad Software). To determine the effect of MyBP-C’s N-terminal C0C3 and phosphomimetic C0C3-4D domains on thin filament activation and inhibition of maximal sliding velocities, we added the fragment into the motility buffer, over a range of fragment concentrations.
Thick filament assay
The movement of ~250-nm TRITC-labeled native thin filament shards was observed on native thick filaments from three independent thick and thin filament preparations using TIRFM as described for bare actin filaments (14). Thick filaments were incubated in a Sigmacote-treated glass flow cell (20 min). The surface of the flow cell was blocked with BSA (1 mg/ml) for 3 min and then washed with actin buffer containing 1 mM ATP. A 30-μl aliquot of 0.01 μM TRITC-labeled thin filament shards in actin buffer without ATP or calcium was infused into the flow cell, incubated for 1 min, and then washed with actin buffer. The flow cell was oil-coupled to the microscope objective, and the TRITC-labeled thin filaments were visualized using TIRFM. Next, three 30-μl aliquots of 0.01 μM TRITC-labeled actin shards in actin buffer with 100 μM ATP and calcium chloride ranging from pCa 5 to 9 were sequentially added to the flow cell in a random order. Thin filament motility on thick filaments was recorded (120 frames/s, 1000 frames) at each pCa in multiple areas on the flow cell. The image stacks were processed using ImageJ SpotTracker 2D with spatial resolution being 30 nm as reported (14). The X- and Y-positions of linear tracks less than 800 nm in length were recorded in Excel (Microsoft). The instantaneous velocity of each thin filament was plotted using GraphPad Prism 6 (GraphPad Software) and fitted with a segmental linear regression to determine if there was one or two phases of velocity as described (14). The fraction of filaments moving from pCa 6 to 9 in each 1000 frame acquisition was determined relative to the average number of filaments moving at pCa 5. The average percent moving ± SEM was reported. The data were fitted with a sigmoidal dose-response curve with variable slopes (GraphPad Prism 6) to determine the effective pCa50. Statistical significance between fitted data was determined by the extra sum-squares F test.
3D electron micrograph reconstructions
EM and 3D reconstructions were carried out on native thin filaments as described (9). In brief, for control measurements of tropomyosin in the “blocked” position, native thin filaments were diluted in low-Ca2+ buffer [100 mM K acetate, 2 mM MgCl2, 0.2 mM EGTA, 1 mM DTT, and 10 mM MOPS (pH 7.0)]. For high-Ca2+ controls with tropomyosin in the “closed” position, 100 mM CaCl2 was added to the buffer (to total 0.33 mM Ca2+). For decoration with MyBP-C N-terminal fragments, 2 μM native thin filaments were mixed with 6 μM C0C3 (actin subunits/C0C3, 1:3) in low-Ca2+ buffer. After mixing, solutions were incubated at room temperature for 30 min, applied to EM grids, and negatively stained with 1% uranyl acetate (39). Thin filaments were observed with a Philips CM120 electron microscope (FEI) at 80 KV under low-dose conditions, with images acquired at a pixel size of 0.35 nm, using a 2 K × 2 K CCD camera (F224HD, TVIPS GmbH). Not all filaments were well decorated by C0C3. The most strongly decorated were selected on the basis of the increase in their diameter due to bound C0C3 (9). Long, relatively straight filaments were unbent using ImageJ (National Institutes of Health) and converted to SPIDER format (EM2EM; Image Science and Imperial College, London). A density projection along the filament axis was obtained using the SPIDER (v11.2) software package (40) to determine the diameter calculated from the distance between the minima in the projection profile. Selected filaments were cut into segments in SPIDER (40), and Iterative Helical Real Space Reconstruction (IHRSR) (41) was carried out using SPIDER (40). An F-actin model (no tropomyosin) was used as an initial reference for the first round of IHRSR so that the model would not bias the position of tropomyosin in the reconstruction. UCSF Chimera (42) was used for visualization, analysis, and atomic fitting of 3D volumes. Accurate matching of 3D reconstructions was carried out by fitting each reconstruction to the same-reference F-actin model; comparison of different reconstructions, all fitted to the same-reference F-actin model, revealed any changes in tropomyosin position (9). The reconstructions were fitted with atomic models of F-actin and/or F-actin–tropomyosin determined by x-ray diffraction and cryo-EM (43, 44).
Modeling
An analytic model was developed to predict the spatial impact of the calcium gradient on the activation state of the thin filament in regions of the sarcomere with and without MyBP-C and on MyBP-C’s phosphorylation state (Fig. 4). To model the relative levels of thin filament activation as a function of time after stimulation near the Z-line (region lacking MyBP-C) and within the C-zone (where MyBP-C resides), we used the [Ca2+]i time course at the Z-line and M-line, respectively (Fig. 2D), and plotted the fraction of filaments moving at each concentration using the sigmoidal-fitted fraction of thin filaments moving–pCa curves (Fig. 4A) as a measure for activation. The resultant predicted levels of thin filament activation within these two sarcomeric regions were plotted with respect to time (Fig. 4, B to D).
Statistical analysis
Average [Ca2+]i transients were determined from 59 Z-line and M-line regions flanked by two CRUs and 53 Z-line and M-line regions flanks by one CRU originating from 12 cardiac myocytes (Fig. 2E). Z- to M-line [Ca2+]i differences were determined by subtracting 5, 10, 15, and 20 ms after stimulation in each trace (Fig. 2F). Statistical differences were determined by a one-sample t test.
The average percentage of thin filaments moving with respect to calcium was determined in the absence of MyBP-C in the in vitro motility assay in four movies at each calcium concentration from three independent protein preparations and normalized to 100% of the filaments moving at pCa 4 to 5. The average fraction of filaments moving on native thick filaments and λ-phosphatase–treated thick filaments was determined from five movies at each calcium concentration relative to the movement at pCa 5. Each data set was fitted with a sigmoidal dose-response curve with variable slopes (GraphPad Prism 6). Statistical significance between the effective pCa50 for each curve was determined by the extra sum-squares F test.
Supplementary Material
Acknowledgments
We thank B. Palmer and Y. Wang for mouse colony management; M. Redmond for technical assistance; G. Kennedy, from the University of Vermont Instrumentation and Model Facility, for imaging expertise; S. P. Harris for the MyBP-C antibody; and M. T. Nelson for helpful comments on the manuscript. Funding: NIH funds supported M.J.P., J.Y.M., K.L., J.G., J.R., R.C., and D.M.W. (grant HL059408); M.J.P. (grant HL124041); R.C. (grant AR034711); B.L.P. (grant HL114879); and W.J.L. (grants HL106059 and HL116321). Author contributions: M.J.P. performed all of the thick and thin filament functional assays with help from S.B.P. for protein isolation and microscopy and S.B.P. and D.M.W. for data analysis. B.L.P. isolated cardiac myocytes for dSTORM with help from M.J.P. for antibody labeling and confocal/calcium measurements with help from W.J.L. for data analysis. J.G. and J.R. expressed the N-terminal protein fragments for in vitro motility assays and EM experiments. J.Y.M. performed all of the thin filament EM experiments with help from S.B.P. for protein isolation and carried out the 3D reconstructions with help from R.C. for data analysis. K.L. isolated cardiac myofibrils and performed antibody-labeling EM experiments. M.J.P., B.L.P., J.Y.M., J.R., R.C., W.J.L., and D.M.W. designed the experiments, interpreted the results, and wrote the manuscript. Competing interests: The authors declare that they have no competing financial interests. Data and materials availability: Data described in the paper are presented in the Supplementary Materials.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/ content/full/1/1/e1400205/DC1
Materials and Methods
Fig. S1. Two-color dSTORM super-resolution imaging of MyBP-C and ryanodine receptors (RyR2).
Fig. S2. Treatment with the contractile inhibitor blebbistatin does not obscure sarcomeric calcium gradients.
Fig. S3. Calcium-dependent regulation of native thin filament motion.
Fig. S4. Histograms for thin filaments on λ-phosphatase–treated thick filaments containing dephosphorylated MyBP-C (trajectory length ± SEM and percent trajectories with biphasic velocities versus pCa).
Fig. S5. 3D reconstructions of low-Ca2+ native thin filaments decorated with C0C3 (actin/C0C3, 1:3) in which the four phosphorylatable serines have been replaced with alanines (fully dephosphorylated, C0C3-4A) or with aspartic acid residues (phosphomimetic, C0C3-4D).
Fig. S6. Transverse sections of the respective low-Ca2+ reconstructions in fig. S5, in which native thin filaments have been decorated with C0C3-4A and C0C3-4D.
Fig. S7. Phosphorylation of MyBP-C N-terminal domains modulates their ability to activate native thin filament motion in the in vitro motility assay on a bed of monomeric myosin.
REFERENCES AND NOTES
- 1.Bers D. M., Cardiac excitation–contraction coupling. Nature 415, 198–205 (2002). [DOI] [PubMed] [Google Scholar]
- 2.McKillop D. F., Geeves M. A., Regulation of the interaction between actin and myosin subfragment 1: Evidence for three states of the thin filament. Biophys. J. 65, 693–701 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vibert P., Craig R., Lehman W., Steric-model for activation of muscle thin filaments. J. Mol. Biol. 266, 8–14 (1997). [DOI] [PubMed] [Google Scholar]
- 4.Cannell M. B., Allen D. G., Model of calcium movements during activation in the sarcomere of frog skeletal muscle. Biophys. J. 45, 913–925 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hollingworth S., Soeller C., Baylor S. M., Cannell M. B., Sarcomeric Ca2+ gradients during activation of frog skeletal muscle fibres imaged with confocal and two-photon microscopy. J. Physiol. 526, 551–560 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kunst G., Kress K. R., Gruen M., Uttenweiler D., Gautel M., Fink R. H., Myosin binding protein C, a phosphorylation-dependent force regulator in muscle that controls the attachment of myosin heads by its interaction with myosin S2. Circ. Res. 86, 51–58 (2000). [DOI] [PubMed] [Google Scholar]
- 7.Harris S. P., Rostkova E., Gautel M., Moss R. L., Binding of myosin binding protein-C to myosin subfragment S2 affects contractility independent of a tether mechanism. Circ. Res. 95, 930–936 (2004). [DOI] [PubMed] [Google Scholar]
- 8.Razumova M. V., Shaffer J. F., Tu A. Y., Flint G. V., Regnier M., Harris S. P., Effects of the N-terminal domains of myosin binding protein-C in an in vitro motility assay: Evidence for long-lived cross-bridges. J. Biol. Chem. 281, 35846–35854 (2006). [DOI] [PubMed] [Google Scholar]
- 9.Mun J. Y., Previs M. J., Yu H. Y., Gulick J., Tobacman L. S., Beck Previs S., Robbins J., Warshaw D. M., Craig R., Myosin-binding protein C displaces tropomyosin to activate cardiac thin filaments and governs their speed by an independent mechanism. Proc. Natl. Acad. Sci. U.S.A. 111, 2170–2175 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luther P. K., Winkler H., Taylor K., Zoghbi M. E., Craig R., Padrón R., Squire J. M., Liu J., Direct visualization of myosin-binding protein C bridging myosin and actin filaments in intact muscle. Proc. Natl. Acad. Sci. U.S.A. 108, 11423–11428 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pfuhl M., Gautel M., Structure, interactions and function of the N-terminus of cardiac myosin binding protein C (MyBP-C): Who does what, with what, and to whom? J. Muscle Res. Cell Motil. 33, 83–94 (2012). [DOI] [PubMed] [Google Scholar]
- 12.Copeland O., Sadayappan S., Messer A. E., Steinen G. J., van der Velden J., Marston S. B., Analysis of cardiac myosin binding protein-C phosphorylation in human heart muscle. J. Mol. Cell. Cardiol. 49, 1003–1011 (2010). [DOI] [PubMed] [Google Scholar]
- 13.Jacques A. M., Copeland O., Messer A. E., Gallon C. E., King K., McKenna W. J., Tsang V. T., Marston S. B., Myosin binding protein C phosphorylation in normal, hypertrophic and failing human heart muscle. J. Mol. Cell. Cardiol. 45, 209–216 (2008). [DOI] [PubMed] [Google Scholar]
- 14.Previs M. J., Beck Previs S., Gulick J., Robbins J., Warshaw D. M., Molecular mechanics of cardiac myosin-binding protein C in native thick filaments. Science 337, 1215–1218 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harris S. P., Lyons R. G., Bezold K. L., In the thick of it: HCM-causing mutations in myosin binding proteins of the thick filament. Circ. Res. 108, 751–764 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Materials and methods are available as supplementary materials on Science Online.
- 17.Bridge J. H., Ershler P. R., Cannell M. B., Properties of Ca2+ sparks evoked by action potentials in mouse ventricular myocytes. J. Physiol. 518, 469–478 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Prosser B. L., Ward C. W., Lederer W. J., Subcellular Ca2+ signaling in the heart: The role of ryanodine receptor sensitivity. J. Gen. Physiol. 136, 135–142 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Williams G. S., Chikando A. C., Tuan H. T., Sobie E. A., Lederer W. J., Jafri M. S., Dynamics of calcium sparks and calcium leak in the heart. Biophys. J. 101, 1287–1296 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baylor S. M., Hollingworth S., Sarcoplasmic reticulum calcium release compared in slow-twitch and fast-twitch fibres of mouse muscle. J. Physiol. 551, 125–138 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gordon A. M., LaMadrid M. A., Chen Y., Luo Z., Chase P. B., Calcium regulation of skeletal muscle thin filament motility in vitro. Biophys. J. 72, 1295–1307 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gresham K. S., Mamidi R., Stelzer J. E., The contribution of cardiac myosin binding protein-c Ser282 phosphorylation to the rate of force generation and in vivo cardiac contractility. J. Physiol. 592, 3747–3765 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang L., Ji X., Barefield D., Sadayappan S., Kawai M., Phosphorylation of cMyBP-C affects contractile mechanisms in a site-specific manner. Biophys. J. 106, 1112–1122 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Colson B. A., Bekyarova T., Fitzsimons D. P., Irving T. C., Moss R. L., Radial displacement of myosin cross-bridges in mouse myocardium due to ablation of myosin binding protein-C. J. Mol. Biol. 367, 36–41 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Colson B. A., Bekyarova T., Locher M. R., Fitzsimons D. P., Irving T. C., Moss R. L., Protein kinase A-mediated phosphorylation of cMyBP-C increases proximity of myosin heads to actin in resting myocardium. Circ. Res. 103, 244–251 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Lange W. J., Grimes A. C., Hegge L. F., Ralphe J. C., Ablation of cardiac myosin–binding protein-C accelerates contractile kinetics in engineered cardiac tissue. J. Gen. Physiol. 141, 73–84 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saber W., Begin K. J., Warshaw D. M., VanBuren P., Cardiac myosin binding protein-C modulates actomyosin binding and kinetics in the in vitro motility assay. J. Mol. Cell. Cardiol. 44, 1053–1061 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weith A., Sadayappan S., Gulick J., Previs M. J., Vanburen P., Robbins J., Warshaw D. M., Unique single molecule binding of cardiac myosin binding protein-C to actin and phosphorylation-dependent inhibition of actomyosin motility requires 17 amino acids of the motif domain. J. Mol. Cell. Cardiol. 52, 219–227 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weith A. E., Previs M. J., Hoeprich G. J., Previs S. B., Gulick J., Robbins J., Warshaw D. M., The extent of cardiac myosin binding protein-C phosphorylation modulates actomyosin function in a graded manner. J. Muscle Res. Cell Motil. 33, 449–459 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Walcott S., Docken S., Harris S. P., Effects of cardiac myosin binding protein-C on actin motility are explained with a drag-activation-competition model. Biophys. J. 108, 10–13 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sadayappan S., Gulick J., Osinska H., Martin L. A., Hahn H. S., Dorn G. W. Jr., Klevitsky R., Seidman C. E., Seidman J. G., Robbins J., Cardiac myosin-binding protein-C phosphorylation and cardiac function. Circ. Res. 97, 1156–1163 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tong C. W., Stelzer J. E., Greaser M. L., Powers P. A., Moss R. L., Acceleration of crossbridge kinetics by protein kinase A phosphorylation of cardiac myosin binding protein C modulates cardiac function. Circ. Res. 103, 974–982 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mitra R., Morad M., A uniform enzymatic method for dissociation of myocytes from hearts and stomachs of vertebrates. Am. J. Physiol. 249, H1056–H1060 (1985). [DOI] [PubMed] [Google Scholar]
- 34.Heilemann M., van de Linde S., Schüttpelz M., Kasper R., Seefeldt B., Mukherjee A., Tinnefeld P., Sauer M., Subdiffraction-resolution fluorescence imaging with conventional fluorescent probes. Angew. Chem. Int. Ed. Engl. 47, 6172–6176 (2008). [DOI] [PubMed] [Google Scholar]
- 35.Prosser B. L., Ward C. W., Lederer W. J., X-ROS signaling: Rapid mechano-chemo transduction in heart. Science 333, 1440–1445 (2011). [DOI] [PubMed] [Google Scholar]
- 36.Cheng H., Lederer W. J., Cannell M. B., Calcium sparks: Elementary events underlying excitation-contraction coupling in heart muscle. Science 262, 740–744 (1993). [DOI] [PubMed] [Google Scholar]
- 37.Hagen B. M., Boyman L., Kao J. P., Lederer W. J., A comparative assessment of fluo Ca2+ indicators in rat ventricular myocytes. Cell Calcium 52, 170–181 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Patton C., Thompson S., Epel D., Some precautions in using chelators to buffer metals in biological solutions. Cell Calcium 35, 427–431 (2004). [DOI] [PubMed] [Google Scholar]
- 39.Craig R., Lehman W., Crossbridge and tropomyosin positions observed in native, interacting thick and thin filaments. J. Mol. Biol. 311, 1027–1036 (2001). [DOI] [PubMed] [Google Scholar]
- 40.Frank J., Radermacher M., Penczek P., Zhu J., Li Y., Ladjadj M., Leith A., SPIDER and WEB: Processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 116, 190–199 (1996). [DOI] [PubMed] [Google Scholar]
- 41.Egelman E. H., A robust algorithm for the reconstruction of helical filaments using single-particle methods. Ultramicroscopy 85, 225–234 (2000). [DOI] [PubMed] [Google Scholar]
- 42.Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E., UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 43.Holmes K. C., Angert I., Kull F. J., Jahn W., Schröder R. R., Electron cryo-microscopy shows how strong binding of myosin to actin releases nucleotide. Nature 425, 423–427 (2003). [DOI] [PubMed] [Google Scholar]
- 44.Poole K. J., Lorenz M., Evans G., Rosenbaum G., Pirani A., Craig R., Tobacman L. S., Lehman W., Holmes K. C., A comparison of muscle thin filament models obtained from electron microscopy reconstructions and low-angle x-ray fibre diagrams from non-overlap muscle. J. Struct. Biol. 155, 273–284 (2006). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/ content/full/1/1/e1400205/DC1
Materials and Methods
Fig. S1. Two-color dSTORM super-resolution imaging of MyBP-C and ryanodine receptors (RyR2).
Fig. S2. Treatment with the contractile inhibitor blebbistatin does not obscure sarcomeric calcium gradients.
Fig. S3. Calcium-dependent regulation of native thin filament motion.
Fig. S4. Histograms for thin filaments on λ-phosphatase–treated thick filaments containing dephosphorylated MyBP-C (trajectory length ± SEM and percent trajectories with biphasic velocities versus pCa).
Fig. S5. 3D reconstructions of low-Ca2+ native thin filaments decorated with C0C3 (actin/C0C3, 1:3) in which the four phosphorylatable serines have been replaced with alanines (fully dephosphorylated, C0C3-4A) or with aspartic acid residues (phosphomimetic, C0C3-4D).
Fig. S6. Transverse sections of the respective low-Ca2+ reconstructions in fig. S5, in which native thin filaments have been decorated with C0C3-4A and C0C3-4D.
Fig. S7. Phosphorylation of MyBP-C N-terminal domains modulates their ability to activate native thin filament motion in the in vitro motility assay on a bed of monomeric myosin.