Abstract
Epigenetic events, including covalent post-translational modifications of histones, have been demonstrated to play critical roles in tumor development and progression. The transcriptional coactivator p300/CBP possesses both histone acetyltransferase (HAT) activity as well as scaffolding properties that directly influence the transcriptional activation of targeted genes. We have used a potent and specific inhibitor of p300/CBP HAT activity, C646, in order to evaluate the functional contributions of p300/CBP HAT to tumor development and progression. Here we report that C646 inhibits the growth of human melanoma and other tumor cells and promotes cellular senescence. Global assessment of the p300 HAT transcriptome in human melanoma identified functional roles in promoting cell cycle progression, chromatin assembly and activation of DNA repair pathways through direct transcriptional regulatory mechanisms. Additionally, C646 is shown to promote sensitivity to DNA damaging agents, leading to the enhanced apoptosis of melanoma cells following combination treatment with cisplatin. Together, our data suggest that p300 HAT activity mediates critical growth regulatory pathways in tumor cells and may serve as a potential therapeutic target for melanoma and other malignancies by promoting cellular responses to DNA damaging agents that are currently ineffective against specific cancers.
Introduction
Recent discoveries have allowed for the development of novel therapies targeting epigenetic pathways in cancers including DNA methyltransferases and histone deacetylases (Kelly, De Carvalho and Jones, 2010). Biochemical and genetic data demonstrate complex roles of HATs in determining cell fate in both normal and diseased tissues, making them a new class of potential therapeutic targets (Dekker and Haisma, 2009). The KAT3 family HAT protein p300/CBP has over 400 binding partners and at least 75 substrates (Bedford et al, 2010, Ogryzko et al, 1996). It acts as a transcriptional coactivator and a scaffold protein, and its importance to cell fate is reflected by its involvement in the regulation of cell cycle, DNA synthesis, cellular differentiation and organ development (Chan and La Thangue, 2001). P300 plays a complex role in carcinogenesis. Missense mutations and truncations of p300 have been found in solid tumors and B-cell lymphoma, suggesting that it acts as a tumor suppressor (Iyer, Ozdag and Caldas, 2004, Pasqualucci et al, 2011). In contrast, p300 is a transcriptional coactivator of known oncogenes, such as c-jun and c-fos. Furthermore, in rare cases a fusion between the MYST family HAT protein MOZ and p300 is thought to contribute to leukemia development (Chan and La Thangue, 2001, Katsumoto, Yoshida and Kitabayashi, 2008). In HCT116 colon cancer cells p300 prevents premature G1/S transitions by inhibiting RB phosphorylation while p300-null cells become sensitive to UV damage and undergo apoptosis (Iyer et al, 2004, Iyer et al, 2007).
Although mutations of p300 have not been identified in human melanoma, strong evidence has linked p300 to melanoma development. A genome-wide SAM (Significant Analysis of Microarray) analysis has shown that the EP300 gene is upregulated in melanoma cell lines (Lin et al, 2008). Additionally, p300 regulates the melanocyte lineage-specific MITF transcription factor, which is amplified in metastatic melanomas and associated with antiapoptotic and angiogenic activities (Garraway et al, 2005, Sato et al, 1997, Yajima et al, 2011). Normal human melanocytes undergo growth arrest, cyclin E repression and activation of cellular senescence following p300 HAT inhibition via a bisubstrate analog, Lys-CoA, or a dominant negative p300 transgene. This transgene was also shown to induce cellular senescence in melanomas (Bandyopadhyay et al, 2002).
Inhibitors of p300 HAT function have been derived from natural compounds that largely lack specificity (Dekker and Haisma, 2009), while bisubstrate analogs such as Lys-CoA are more selective but have limited use in biological studies. Based on the recently elucidated structure of the p300 HAT domain, a virtual ligand screen identified a potent and selective inhibitor of p300 HAT activity known as C646 (Bowers et al, 2010, Liu et al, 2008). We have demonstrated the specificity of C646 both in vitro and in culture (Bowers et al, 2010, Crump et al, 2011), and it has been used to assess p300 HAT functions in prostate cancer and leukemia (Santer et al, 2011, Wang et al, 2011). Here we evaluate the functional significance of p300 HAT activity in human melanoma and explore the global p300 HAT transcriptome using C646. We find significant effects of C646 on tumor cell growth, cellular senescence and the DNA damage response, which are mediated by direct transcriptional effects on target genes. Additionally, C646 sensitizes melanoma cells to DNA damaging agents, suggesting potential utility as a therapeutic target for this disease.
Results
Blockade of p300 HAT activity inhibits tumor cell growth
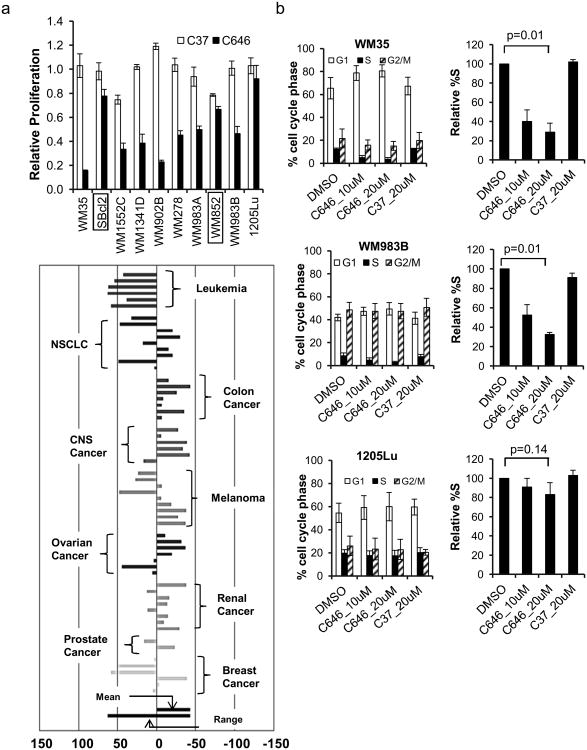
Previous data from our group and others have suggested that p300 HAT is important for tumor cell growth (Bowers et al, 2010). To evaluate the specific functional significance of p300 HAT in human melanomas, we explored the effect of p300 HAT blockade on melanoma cell proliferation using the selective inhibitor C646 in a 3H-thymidine incorporation assay. 3H-thymidine assay results were further validated using an XTT assay (Figure S3), recognizing the limitations associated with tetrazolium salt-based cell viability assays (Scudiero et al, 1988, Wang, Henning and Heber, 2010). Ten cell lines representing radial, vertical and metastatic phases of melanoma progression were evaluated with the majority demonstrating significant growth inhibition following treatment with C646 (Figure 1a) versus the non-functional control compound, C37 (Bowers et al, 2010). Furthermore, the degree of growth inhibition by C646 was not associated with tumor stage or B-raf status. Of note, previous studies from our group failed to demonstrate significant differences in EP300 transcript levels between our melanoma lines and melanocytes (Ryu et al, 2007).
Figure 1.
Effect of p300 HAT inhibition on cancer cell proliferation. (a) 3H-thymidine uptake in melanoma cells treated for 24 h with 10 μM C646, 10 μM C37, or 0.1% DMSO. Boxed: B-raf wildtype; unboxed: B-raf(V600E). The data are normalized to DMSO. N=3; error bars: standard deviations (SD). (b) Cell cycle profiles of WM35, WM983B and 1205Lu after 24 h treatment. Right panel: %S phase normalized to DMSO. N=3. P-values were calculated from Student's t-test. Representative histograms are shown in Figure S3. (c) Data from the one-dose NCI-60 screen. Bars indicate ‘Growth Percent (of each cell line) – Mean Growth Percent (of all cell lines)’; smaller numbers correspond to greater growth inhibition. See Figure S1, S2 for details.
To explore the broad spectrum of the growth inhibitory effects of C646 in cancer, we submitted C646 to the NCI Developmental Therapeutics Program NCI-60 screen (http://dip.nci.nih.gov/index.html) (Shoemaker, 2006). The data from the screen indicated that C646 inhibited proliferation of multiple cancers with IC50 values in the low μM range (Figure 1c, S1, S2). Several tumor cell lineages were particularly sensitive to C646, suggesting that p300 HAT activity may be more broadly important for cellular proliferation in these tumor types.
p300 HAT inhibition blocks S-phase progression in melanoma cells
Preliminary work from our group suggested that C646 induces a G1 cell cycle arrest in melanoma cells (Bowers et al, 2010). To more thoroughly assess the cell cycle effects of C646, melanoma lines from varying stages of progression were evaluated. We observed a statistically significant, dose-dependent decrease in %S-phase cells in C646-sensitive cell lines (Figure 1b, S4). In contrast, minimal effects on S-phase progression were seen in the C646-resistant 1205Lu cells (Figure 1b, S4).
p300 regulates genes involved in cell cycle regulation and DNA repair
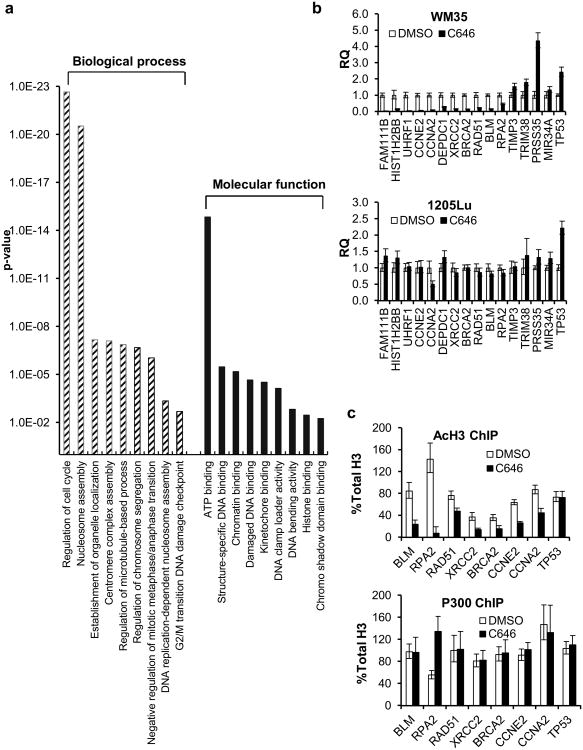
In order to evaluate the transcriptional significance of p300 HAT activity in mediating melanoma growth, we performed comprehensive gene expression profiling of the C646-sensitive cell line, WM35, following 6 h and 24 h of C646 treatment. These treatment timepoints were chosen based on previous data demonstrating significant effects on histone acetylation within hours following C646 treatment and the associated onset of cell cycle arrest at 24 h post-treatment (Bowers et al, 2010). We found few differences in the expression profiles between the C646- and DMSO-treated cells following 6 hours of treatment; however, we observed significant alterations in gene expression between the two treatment groups at 24 hours of treatment with 416 genes downregulated and 199 genes upregulated by more than 2-fold in C646-treated cells (Figure S5a). We then performed functional annotation of genes altered by more than 2-fold in the 24 h sample according to biological processes and molecular functions. While the upregulated genes could not be classified into any gene ontology (GO) categories with high significance (Figure S5b), among the downregulated genes we observed statistically significant representation of genes involved in cell cycle regulation, cell division and chromatin-associated functions such as nucleosome assembly, chromosome segregation and DNA damage checkpoint control. Of note, many of these genes also possess chromatin/DNA binding functions (Figure 2a).
Figure 2.
Gene expression profiling of WM35 cells after p300 HAT inhibition. (a) GO analysis of genes that were repressed by ≥2-fold after 24 h of treatment with 20 μM C646. The p-values represent the significance of the gene clustering. (b) qPCR validation of selected gene targets in WM35 and 1205Lu cells. The data were normalized to the DMSO control with GAPDH as the endogenous gene control. N≥3. (c) ChIP-qPCR analysis of acetylated H3 and p300 enrichment near the transcription start sites of selected genes. The data were normalized to total H3 ChIP; a GAPDH promoter sequence served as the endogenous control. N≥3.
Microarray results on selected genes of interest were validated using quantitative real time PCR (qPCR) on biological replicates of the array samples (Figure 2b, Table 1). Interestingly, the vast majority of genes whose expression was altered by C646 treatment of WM35 cells failed to be altered in the C646-insensitive 1205Lu cells, suggesting a specific correlation of the expression profile with the tumor cell growth response (Figure 2b).
Table 1.
PCR-validated gene targets of p300 HAT.The numbers indicate the relative expression level fold change in cells treated with C646 for 24 h over the DMSO control. Descriptions of gene functions are taken from PubMed gene search. “-” indicates that no broad cellular function has been associated with a particular gene.
| Gene | Fold Change | Known Function(s) | |
|---|---|---|---|
| Repressed | |||
| family with sequence similarity 111, member B | FAM111B | -42.5 | - |
| histone cluster 1, H2bb | HIST1H2BB | -29.1 | chromatin assembly |
| ubiquitin-like with PHD and ring finger domains 1 | UHRF1 | -14.5 | G1/S transition, p53-dependent DNA damage checkpoint |
| cyclin E2 | CCNE2 | -12.7 | G1/S transition |
| DEP domain containing 1 | DEPDC1 | -12.4 | Inhibition of apoptosis |
| X-ray repair complementing defective repair in Chinese hamster cells 2 | XRCC2 | -11.3 | homologous recombination |
| cyclin A2 | CCNA2 | -7.2 | G1/S and G2/M transition |
| RAD51 homolog | RAD51 | -5.6 | homologous recombination |
| Bloom syndrome, RecQ helicase-like | BLM | -5.3 | 3′-5′ helicase activity, suppression of inappropriate recombination |
| breast cancer 2, early onset | BRCA2 | -4.2 | homologous recombination |
| replication protein A2 | RPA2 | -2.6 | homologous recombination |
|
| |||
| Overexpressed | |||
| protease, serine, 35 | PRSS35 | 5.7 | - |
| tripartite motif containing 38 | TRIM38 | 4.3 | - |
| microRNA 34a | MIR34A | 2.6 | - |
| tissue inhibitor of metalloproteinase 3 | TIMP3 | 2.4 | irreversible inactivation of metalloproteinases |
| tumor protein p53 | TP53 | 1.2 | response to diverse cellular stresses, promotion of cell cycle arrest, apoptosis, senescence and DNA repair |
Inhibition of p300 HAT activity affects histone acetylation but not p300 localization at targeted gene promoters
Acetylation of promoter histones is associated with a transcriptionally permissive chromatin structure. In order to determine the mechanisms underlying p300 HAT-regulated gene expression, we first evaluated whether p300 HAT inhibition directly affects transcription by interfering with histone acetylation near gene promoters. We performed a chromatin immunoprecipitation assay followed by qPCR (ChIP-qPCR) analysis of acetylated histone H3 enrichment at the promoter regions of selected DNA repair and cell cycle regulatory genes that were identified in our microarray analyses. Notably, the genes that were repressed after C646 treatment were also depleted in acetylated H3 at the promoters (Figure 2c). TP53 expression was upregulated after C646 treatment; however, there was no increase in H3 acetylation at the tested sequence (Figure 2c).
In contrast, we did not observe changes in p300 enrichment at the promoter sites of interest following C646 treatment except for an increase seen at the RPA2 gene (Figure 2c), suggesting that specific inhibition of p300 HAT activity does not interfere with p300-chromatin interactions; rather, such inhibition targets the localization of acetylated histones to promoter sites. These findings are consistent with recent data that also found no alterations in p300 enrichment at c-fos and c-jun loci following its inhibition in vivo (Crump et al, 2011).
p300 HAT inhibition activates G1/S cell cycle arrest mechanism
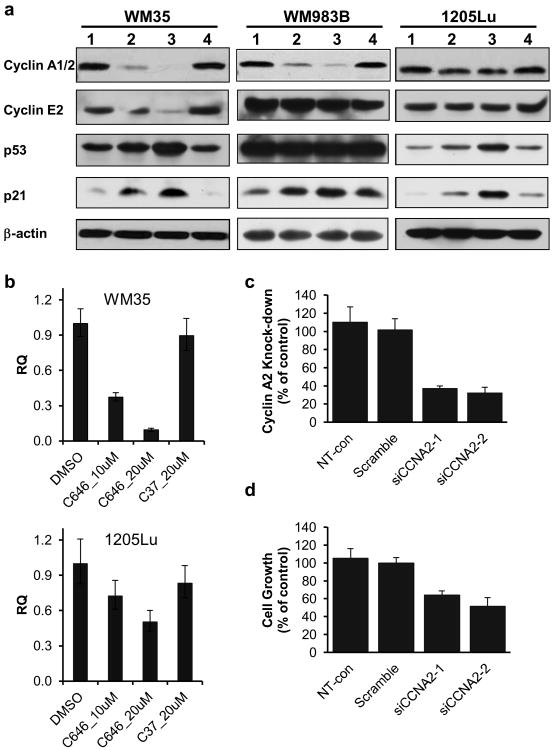
Given the results from our microarray study, we sought to further evaluate the effect of p300 HAT inhibition on cell cycle regulatory protein expression in the melanoma cell lines. We found that cyclins A2 (CCNA2) and E2 (CCNE2) were among the most downregulated genes after 24 h of C646 treatment of WM35 cells (Table 1). Remarkably, we also observed a dose-dependent decrease in the protein levels of cyclins A and E in these cells (Figure 3a). Furthermore, this decrease was accompanied by an increased expression of p53 and its downstream effector p21, a known inhibitor of cyclin-dependent kinases (Figure 3a). No changes were noted in the level of cyclin D1, which is known to be overexpressed rather than repressed in senescent cells (Burton et al, 2007, Han et al, 1999, Meyyappan et al, 1998), nor were there any changes in the levels of B-raf, Bcl-2, PARP, or PCNA (Figure S6). Interestingly, in WM983B cells, in which the TP53 gene is mutated (Weiss et al, 1994), C646 continued to induce cell cycle arrest (Figure 1b) characterized by cyclin A repression and p21 activation (Figure 3a), suggesting a p53-independent mechanism for growth arrest in these cells. In contrast to the responsive cells, the C646-resistant cell line 1205Lu showed minimal effects on cyclin A expression following C646 treatment; however, p53 and p21 were both up-regulated to a similar extent as seen in the WM35 cells (Figure 3a), suggesting the existence of a bypass mechanism preventing p21-associated growth arrest. Notably, the mRNA level of CCNA2 decreased dose-dependently in C646-treated WM35 cells; however, such effects were significantly lessened in 1205Lu cells (Figure 3b), suggesting that the extent of cyclin A attenuation in these cells may not be sufficient for cell cycle arrest. The involvement of cyclin A in melanoma cell cycle regulation was further demonstrated with the siRNA knockdown of CCNA2 mRNA, which also resulted in decreased proliferation of the C646-responsive WM35 cells (Figure 3c, d).
Figure 3.
Expression of selected growth regulatory genes in C646 sensitive and insensitive cell lines. (a) Western blot analyses on WM35, 1205Lu and WM983B cells. Lanes 1-4 represent 24 h of treatment with 0.2% DMSO, 10 μM C646, 20 μM C646 and 20 μM C37, respectively. (b) qPCR analysis of CCNA2 transcripts in WM35 and 1205Lu cells after 24 h treatment. N≥3. Please note that the Western blot antibody detects both CCNA1 and CCNA2, while the qPCR only detects CCNA2. (c) Relative expression of CCNA2 after siRNA transfection, as determined by qRT-PCR. (d) Relative cell proliferation measured by the MTT assay 96 h after plating the siRNA-transfected cells.
Based on the NCI-60 screening results and our previous data (Bowers et al, 2010), we examined cyclin A expression in lung and glioblastoma cancer lines to determine whether the growth inhibitory effects in these tumor cell lineages were also associated with altered cell cycle protein expression. Indeed, both the transcript and protein levels of cyclin A were decreased to various extents in these cancers following 24 h of C646 treatment, suggesting that p300 HAT activity plays similar functional roles in regulating cell cycle progression in these tumors (Figure S7, S8).
Prolonged p300 HAT inhibition promotes cellular senescence
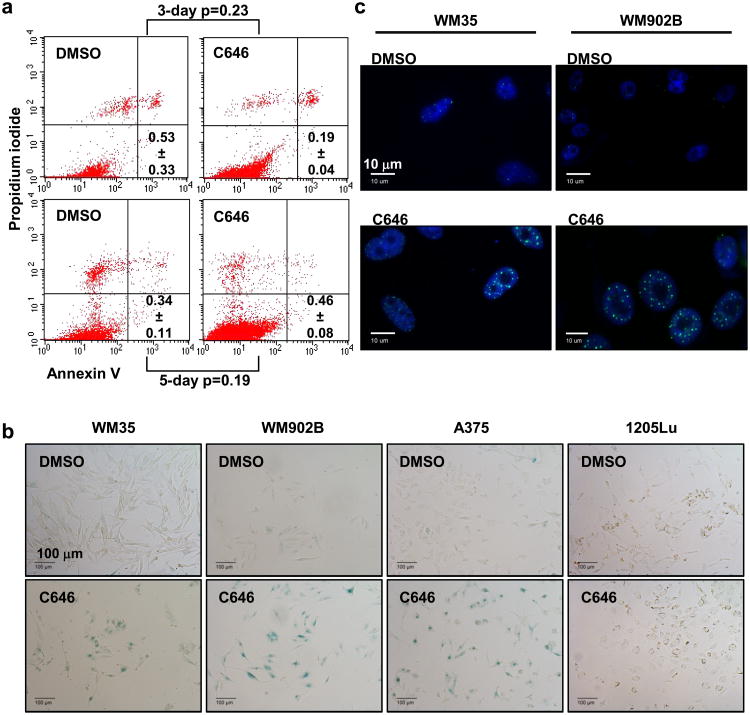
In order to determine the functional consequences of extended p300 HAT inhibition on melanoma cellular phenotypes, we evaluated cellular apoptosis following 3 and 5 days of treatment in WM35 cells and observed no statistically significant differences in the percentages of apoptotic populations between the C646-treated and DMSO-treated cells at either timepoint (p>0.05) (Figure 4a). We further sought to define the specific effects of C646 treatment on cellular senescence pathways by evaluating C646-treated cells for SA-β-gal activity and senescence-associated heterochromatin foci (SAHF) formation. We found that extended C646 treatment of multiple responsive cell lines led to increased cellular senescence versus the DMSO control (Figure 4b, c), suggesting that p300 HAT inhibition had a cytostatic rather than an apoptotic effect on melanoma cells leading to senescence-associated growth arrest. This effect was not observed in the nonresponsive 1205Lu cells (Figure 4b).
Figure 4.
Analyses of WM35 cell phenotype after extended p300 inhibition. (a) FACS analysis of apoptotic cells after 3 and 5 d of treatment with C646 (20 μM) or DMSO. The population in the lower right quadrant contains early apoptotic cells. The numbers represent the average %apoptotic cells from 3 independent experiments ± SD. P-values were derived from Student's t-test. (b) Representative images of SA-β-gal staining in WM35, WM902B, A375 and 1205Lu cells treated for 4 d. Strong perinuclear staining is considered SA-β-gal positive. Scale bars: 100 μm. (c) PML immunofluorescence staining for senescence detection of WM35 and WM902B cells. C646 treatment induced larger and flattened nucleoli as well as more SAHF formations compared to the DMSO treatment. Of note, DMSO-treated cells also showed basal levels of SAHF formation. Scale bars: 10 μm.
p300 regulates the DNA damage response in melanoma cells
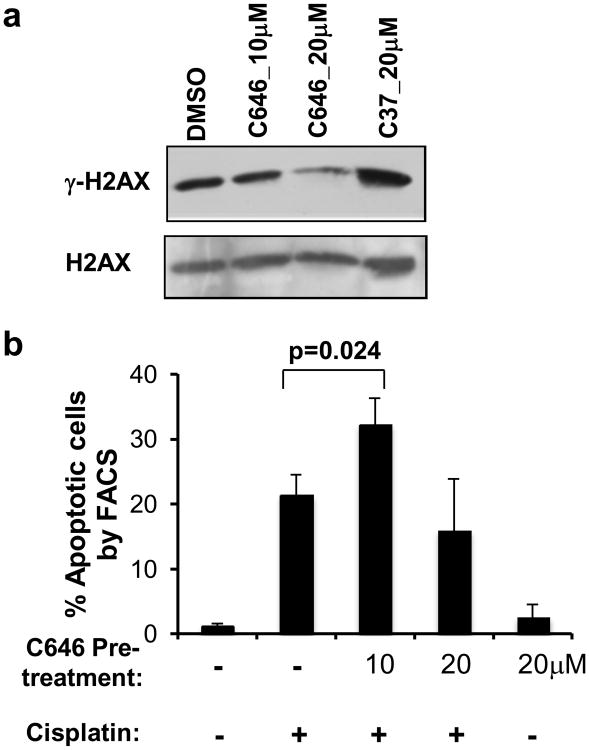
As our microarray data demonstrated the repression of DNA repair genes following p300 HAT inhibition (Table 1), we sought to explore the functional role of p300 HAT activity in the DNA damage response. The phosphorylation of the histone variant H2AX (γH2AX) occurs following DNA double strand breaks and helps to recruit repair proteins to the site of damage (Lukas, Lukas and Bartek, 2011). In cells treated with C646 for 24 h, we observed a decreased level of γH2AX protein, suggesting that tumor cells may not be able to efficiently recognize DNA damage following p300 HAT inhibition (Figure 5a).
Figure 5.
DNA damage response after p300 HAT inhibition. (a) Western blot of γH2AX in WM35 cells after 24h C646 treatment. (b) FACS analysis of TUNEL-stained cells after combination treatment with C646 and cisplatin. +/-: presence/absence of a compound. N=3.
To determine how cells responded to induced DNA damage in the presence of a HAT inhibitor, we evaluated the responses of melanoma cells to a combination treatment of C646 and cisplatin. As our expression profiling studies demonstrated a loss of DNA damage response gene expression after HAT inhibition, we anticipated that DNA damaging agents would allow for enhanced apoptotic responses in the presence of a potent, selective HAT inhibitor. Interestingly, 24 h pretreatment of melanoma cells with 10 μM C646 followed by cisplatin led to increased apoptosis in WM35 cells compared to cisplatin treatment alone; however, a 20 μM C646 pretreatment led to a reduction in apoptotic cells following cisplatin treatment (Figure 5b). Given that cisplatin efficacy is dependent on cycling cells, it is possible that the 20 μM C646-treated cells were more likely to be growth arrested than those treated at 10 μM, thereby rendering the cisplatin less effective. We conclude that p300 HAT inhibition may sensitize cancer cells to DNA damaging agents within an appropriate context.
Discussion
Epigenetic alterations in cancers have been identified as a hallmark of the malignant phenotype; however, the functional significance of particular epigenetic signatures in cancer has not been fully realized. While previous studies have sought to identify global p300 target genes and pathways, such work is unable to specifically distinguish molecular targets of p300 HAT function versus targets associated with the transcriptional adaptor function of p300. Here, we employ a specific inhibitor of p300 HAT, C646, as a tool to evaluate target genes specifically associated with the HAT activity of p300. While we previously reported the selectivity of C646 against p300 (Bowers et al, 2010), we now further demonstrate that the cellular consequences of C646 treatment are specifically linked to the loss of p300 HAT function. Our cyclin expression and senescence data are consistent with previous findings of p300 function in melanocytes. Additionally, the overall pattern of gene expression changes in C646-treated cells, which is highly skewed towards gene repression versus activation, also strongly supports targeted transcriptional effects resulting from the loss of HAT activity. Furthermore, identification of specific S-phase repression in melanoma cells and specific repression of S-phase histone HIST1H2BB (Table 1) is consistent with previous findings demonstrating the regulation of S-phase histone transcription by p300 (He et al, 2011).
Repression of cyclin A in growth-arrested melanoma cells after p300 HAT inhibition has not been previously reported; however, as the role of cyclin A in promoting G1/S and G2/M transition has previously been well established (Celis et al, 1984, Pagano et al, 1992, Shapiro and Harper, 1999), this finding is not surprising. It is notable that the degree of cyclin A repression observed is highly correlated with tumor cell responsiveness to C646, suggesting that cyclin A may serve as a useful marker for clinical response to p300 inhibition.
The microarray analysis of p300 HAT-associated transcripts and the accompanying ChIP data suggest that p300 regulates targeted gene expression through a direct transcriptional mechanism. The reduced acetylation of histones associated with C646 treatment is likely to antagonize the chromatin remodeling needed for initiation and elongation by RNA Pol II; however, we cannot exclude the possibility that direct inhibition of p300-mediated acetylation of transcription factors and coactivators by C646 may indirectly lead to subsequent inhibition of transcription. Such complexity could also account for the observed upregulation of a small set of transcripts following p300 HAT inhibition. Alternatively, such outcomes may be associated with downstream effects of direct transcriptional targets of p300 HAT.
Our previous gene expression profile study identified a set of molecular characteristics that distinguish aggressive melanoma phenotypes from less aggressive ones (Ryu et al, 2007). Such molecular characteristics of aggressive melanomas included the following: upregulation of activators of cell cycle progression, DNA replication and repair; loss of genes associated with cellular adhesion and melanocyte differentiation; and upregulation of genes associated with apoptosis resistance. The two classes of upregulated genes are, notably, also found to be significantly repressed following p300 inhibition in the current study (Table S3), suggesting that blocking p300 HAT activity may reduce the metastatic potential of melanoma cells as an alternative therapeutic approach.
One of the major obstacles in treating melanoma versus other malignancies is its resistance to DNA damage. The correlation between DNA repair gene overexpression and cancer malignancy, as determined by other studies (Kao et al, 2011, Kee and D'Andrea, 2010, Winnepenninckx et al, 2006), coupled to our finding that inhibition of p300 HAT activity results in the repression of DNA repair genes, suggests that p300 contributes to melanoma survival at least in part through the maintenance of genomic stability. While C646-induced growth arrest was found to hinder cisplatin activity at high concentrations of C646 in our study, it is yet to be studied how p300 inhibition affects the cellular response to other chemotherapeutic agents or ionizing radiation. Based on our current data, we suggest that future work should focus on the elucidation of the detailed mechanisms of p300-regulated pathways in targeted cancers and the potential role for p300 HAT inhibition as a therapeutic strategy targeting epigenetic changes in susceptible malignancies.
Materials and Methods
Detailed information on cell culture and experimental methods is provided in the supplementary material.
Cell treatment with compounds
To treat cells, C646 and C37 were dissolved in anhydrous DMSO to 10 mM and added to culture media to the desired concentration. Equal amount of DMSO was used as the vehicle control.
3H-thymidine incorporation
The use of thymidine incorporation to measure cell proliferation has been extensively described previously (Shevach, 2001). 3H-thymidine was added to cells in 96-well plates at 10 μCi/ml for 5 h and cells were collected onto a filter mat using a cell harvester (PerkinElmer). The radioactivity was measured with a MicroBeta plate reader (PerkinElmer). The samples were tested in triplicates. The data were normalized to the DMSO control.
NCI-60 Screen
Detailed methodology of the screen is described at the NCI website (http://dtp.nci.nih.gov/index.html) (Shoemaker, 2006). The initial one-dose screen used 100 μM of C646. The second stage was a 5-dose screen using 0.01-100 μM C646.
Cell cycle analysis
Cells were stained with PI according to a published protocol (Darzynkiewicz, Juan and Bedner, 2001). Data acquisition and analysis were performed on a FACSCalibur flow cytometer with the CellQuest software (BD Biosciences).
Microarray study
RNA was purified from C646- or DMSO-treated cells using the Qiagen RNAeasy Kit and submitted to the microarray core facility at JHMI for analysis on the Affymetrix GeneChip Human Exon 1.0 ST Array. The data normalization and processing were performed by the core facility. The GO analysis was performed with Spotfire (TIBCO).
qPCR
Complementary DNA was synthesized using the Superscript First Strand Synthesis System (Invitrogen). qPCR was performed using the Power SYBR Green PCR Master Mix (ABI) as described in the Supplement. The primers were designed using the NCBI PrimerBlast tool (Table S1). Gene expression values were determined with the ΔΔCt method.
ChIP-qPCR
ChIP was performed based on a published protocol (O'Geen, Frietze and Farnham, 2010). The antibodies included acetylated H3 (Upstate #06-599), total H3 (Abcam #ab1791) and p300 (SCBT #sc-585). Primers for qPCR were designed to amplify a region near transcription start (Table S2). qPCR was performed using SYBR Green.
Western blot
Whole cell lysate was prepared as described in the supplement. Western blots were performed as previously described (Cummings et al, 2008). Antibodies were obtained from the following sources: cyclin A (#sc-596), cyclin D1 (#sc-20044), PCNA (#sc-56), B-raf (#sc-166), PARP (#sc-7150) and β-actin (#sc-1616-R) were from Santa Cruz Biotechnology, Inc; cyclin E2 (#4132) and p21 (#2946) were from Cell Signaling Technology; p53 (#OP43) was from Calbiochem. The HRP-tagged secondary antibodies were from Amersham.
For the analysis of γH2AX, histones were extracted based on an established protocol with modifications as described in the supplement (Shechter et al, 2007). The antibody against γH2AX was from Abcam (#ab2893). Total H2AX (Millipore #07-627) was the loading control.
Cyclin A knockdown
The CCNA2 siRNAs (#A-003205-19-0005 and A-003205-21-0005) and the scrambled control (ON-TARGETplus Non-targeting siRNA #1, #D-001810-01) were ordered from Dharmacon (Thermo Scientific). The protocols for the knockdown and the subsequent analysis of cell proliferation are described in the supplement.
Apoptosis assay
Apoptosis was assessed with the Alexa Fluor® 488 annexin V/propidium iodide (PI) kit (Invitrogen #V13241) according to manufacturer's protocol. Approximately 105 cells per sample were analyzed by FACS.
For apoptosis analysis after C646/cisplatin co-treatment, the APO-BrdU TUNEL Assay Kit (Invitrogen A35125) was applied to paraformaldehyde-fixed cells according to the instructions. After staining, the cells were incubated with the PI/RNase solution from the kit for 30 min before FACS analysis.
Senescence detection
Senescent cells were detected by staining for lysosomal SA-β-gal activity with a commercial kit (Cell Signaling #9860) and by immunofluorescent staining of PML (promyelocytic leukemia protein) nuclear bodies using an established protocol (Vernier et al, 2011).
Supplementary Material
Acknowledgments
We thank members of the Alani, Cole and Taverna labs for their careful review of this manuscript and helpful suggestions.
Abbreviations
- HAT
histone acetyltransferase
- qPCR
quantitative real time PCR
- ChIP
chromatin immunoprecipitation
- PI
propidium iodide
- SA-β-gal
senescence activated beta galactosidase
- SAHF
senescence-associated heterochromatin foci
Footnotes
Conflict of Interest: RMA and PAC are cofounders of Acylin Therapeutics Inc. and own equity in this company, which is focused on developing HAT inhibitors as novel therapeutics.
References
- Bandyopadhyay D, Okan NA, Bales E, Nascimento L, Cole PA, Medrano EE. Down-regulation of p300/CBP histone acetyltransferase activates a senescence checkpoint in human melanocytes. Cancer Res. 2002;62:6231–6239. [PubMed] [Google Scholar]
- Bedford DC, Kasper LH, Fukuyama T, Brindle PK. Target gene context influences the transcriptional requirement for the KAT3 family of CBP and p300 histone acetyltransferases. Epigenetics. 2010;5:9–15. doi: 10.4161/epi.5.1.10449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers EM, Yan G, Mukherjee C, Orry A, Wang L, Holbert MA, et al. Virtual ligand screening of the p300/CBP histone acetyltransferase: Identification of a selective small molecule inhibitor. Chem Biol. 2010;17:471–482. doi: 10.1016/j.chembiol.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton DG, Sheerin AN, Ostler EL, Smith K, Giles PJ, Lowe J, et al. Cyclin D1 overexpression permits the reproducible detection of senescent human vascular smooth muscle cells. Ann N Y Acad Sci. 2007;1119:20–31. doi: 10.1196/annals.1404.026. [DOI] [PubMed] [Google Scholar]
- Celis JE, Bravo R, Larsen PM, Fey SJ. Cyclin: A nuclear protein whose level correlates directly with the proliferative state of normal as well as transformed cells. Leuk Res. 1984;8:143–157. doi: 10.1016/0145-2126(84)90135-8. [DOI] [PubMed] [Google Scholar]
- Chan HM, La Thangue NB. p300/CBP proteins: HATs for transcriptional bridges and scaffolds. J Cell Sci. 2001;114:2363–2373. doi: 10.1242/jcs.114.13.2363. [DOI] [PubMed] [Google Scholar]
- Crump NT, Hazzalin CA, Bowers EM, Alani RM, Cole PA, Mahadevan LC. Dynamic acetylation of all lysine-4 trimethylated histone H3 is evolutionarily conserved and mediated by p300/CBP. Proc Natl Acad Sci U S A. 2011;108:7814–7819. doi: 10.1073/pnas.1100099108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings SD, Ryu B, Samuels MA, Yu X, Meeker AK, Healey MA, et al. Id1 delays senescence of primary human melanocytes. Mol Carcinog. 2008;47:653–659. doi: 10.1002/mc.20422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darzynkiewicz Z, Juan G, Bedner E. Determining cell cycle stages by flow cytometry. Curr Protoc Cell Biol. 2001;Chapter 8 doi: 10.1002/0471143030.cb0804s01. Unit 8.4. [DOI] [PubMed] [Google Scholar]
- Dekker FJ, Haisma HJ. Histone acetyl transferases as emerging drug targets. Drug Discov Today. 2009;14:942–948. doi: 10.1016/j.drudis.2009.06.008. [DOI] [PubMed] [Google Scholar]
- Garraway LA, Widlund HR, Rubin MA, Getz G, Berger AJ, Ramaswamy S, et al. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature. 2005;436:117–122. doi: 10.1038/nature03664. [DOI] [PubMed] [Google Scholar]
- Han EK, Ng SC, Arber N, Begemann M, Weinstein IB. Roles of cyclin D1 and related genes in growth inhibition, senescence and apoptosis. Apoptosis. 1999;4:213–219. doi: 10.1023/a:1009618824145. [DOI] [PubMed] [Google Scholar]
- He H, Yu FX, Sun C, Luo Y. CBP/p300 and SIRT1 are involved in transcriptional regulation of S-phase specific histone genes. PLoS One. 2011;6:e22088. doi: 10.1371/journal.pone.0022088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer NG, Chin SF, Ozdag H, Daigo Y, Hu DE, Cariati M, et al. p300 regulates p53-dependent apoptosis after DNA damage in colorectal cancer cells by modulation of PUMA/p21 levels. Proc Natl Acad Sci U S A. 2004;101:7386–7391. doi: 10.1073/pnas.0401002101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer NG, Ozdag H, Caldas C. p300/CBP and cancer. Oncogene. 2004;23:4225–4231. doi: 10.1038/sj.onc.1207118. [DOI] [PubMed] [Google Scholar]
- Iyer NG, Xian J, Chin SF, Bannister AJ, Daigo Y, Aparicio S, et al. p300 is required for orderly G1/S transition in human cancer cells. Oncogene. 2007;26:21–29. doi: 10.1038/sj.onc.1209771. [DOI] [PubMed] [Google Scholar]
- Kao WH, Riker AI, Kushwaha DS, Ng K, Enkemann SA, Jove R, et al. Upregulation of fanconi anemia DNA repair genes in melanoma compared with non-melanoma skin cancer. J Invest Dermatol. 2011;131:2139–2142. doi: 10.1038/jid.2011.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsumoto T, Yoshida N, Kitabayashi I. Roles of the histone acetyltransferase monocytic leukemia zinc finger protein in normal and malignant hematopoiesis. Cancer Sci. 2008;99:1523–1527. doi: 10.1111/j.1349-7006.2008.00865.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kee Y, D'Andrea AD. Expanded roles of the fanconi anemia pathway in preserving genomic stability. Genes Dev. 2010;24:1680–1694. doi: 10.1101/gad.1955310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly TK, De Carvalho DD, Jones PA. Epigenetic modifications as therapeutic targets. Nat Biotechnol. 2010;28:1069–1078. doi: 10.1038/nbt.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin WM, Baker AC, Beroukhim R, Winckler W, Feng W, Marmion JM, et al. Modeling genomic diversity and tumor dependency in malignant melanoma. Cancer Res. 2008;68:664–673. doi: 10.1158/0008-5472.CAN-07-2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Wang L, Zhao K, Thompson PR, Hwang Y, Marmorstein R, et al. The structural basis of protein acetylation by the p300/CBP transcriptional coactivator. Nature. 2008;451:846–850. doi: 10.1038/nature06546. [DOI] [PubMed] [Google Scholar]
- Lukas J, Lukas C, Bartek J. More than just a focus: The chromatin response to DNA damage and its role in genome integrity maintenance. Nat Cell Biol. 2011;13:1161–1169. doi: 10.1038/ncb2344. [DOI] [PubMed] [Google Scholar]
- Meyyappan M, Wong H, Hull C, Riabowol KT. Increased expression of cyclin D2 during multiple states of growth arrest in primary and established cells. Mol Cell Biol. 1998;18:3163–3172. doi: 10.1128/mcb.18.6.3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Geen H, Frietze S, Farnham PJ. Using ChIP-seq technology to identify targets of zinc finger transcription factors. Methods Mol Biol. 2010;649:437–455. doi: 10.1007/978-1-60761-753-2_27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogryzko VV, Schiltz RL, Russanova V, Howard BH, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- Pagano M, Pepperkok R, Verde F, Ansorge W, Draetta G. Cyclin A is required at two points in the human cell cycle. EMBO J. 1992;11:961–971. doi: 10.1002/j.1460-2075.1992.tb05135.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasqualucci L, Dominguez-Sola D, Chiarenza A, Fabbri G, Grunn A, Trifonov V, et al. Inactivating mutations of acetyltransferase genes in B-cell lymphoma. Nature. 2011;471:189–195. doi: 10.1038/nature09730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu B, Kim DS, Deluca AM, Alani RM. Comprehensive expression profiling of tumor cell lines identifies molecular signatures of melanoma progression. PLoS One. 2007;2:e594. doi: 10.1371/journal.pone.0000594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santer FR, Hoschele PP, Oh SJ, Erb HH, Bouchal J, Cavarretta IT, et al. Inhibition of the acetyltransferases p300 and CBP reveals a targetable function for p300 in the survival and invasion pathways of prostate cancer cell lines. Mol Cancer Ther. 2011;10:1644–1655. doi: 10.1158/1535-7163.MCT-11-0182. [DOI] [PubMed] [Google Scholar]
- Sato S, Roberts K, Gambino G, Cook A, Kouzarides T, Goding CR. CBP/p300 as a co-factor for the microphthalmia transcription factor. Oncogene. 1997;14:3083–3092. doi: 10.1038/sj.onc.1201298. [DOI] [PubMed] [Google Scholar]
- Scudiero DA, Shoemaker RH, Paull KD, Monks A, Tierney S, Nofziger TH, et al. Evaluation of a soluble tetrazolium/formazan assay for cell growth and drug sensitivity in culture using human and other tumor cell lines. Cancer Res. 1988;48:4827–4833. [PubMed] [Google Scholar]
- Shapiro GI, Harper JW. Anticancer drug targets: Cell cycle and checkpoint control. J Clin Invest. 1999;104:1645–1653. doi: 10.1172/JCI9054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shechter D, Dormann HL, Allis CD, Hake SB. Extraction, purification and analysis of histones. Nat Protoc. 2007;2:1445–1457. doi: 10.1038/nprot.2007.202. [DOI] [PubMed] [Google Scholar]
- Shevach EM. Labeling cells in microtiter plates for determination of [3H]thymidine uptake. Curr Protoc Immunol. 2001 doi: 10.1002/0471142735.ima03ds21. Appendix 3: Appendix 3D. [DOI] [PubMed] [Google Scholar]
- Shoemaker RH. The NCI60 human tumour cell line anticancer drug screen. Nat Rev Cancer. 2006;6:813–823. doi: 10.1038/nrc1951. [DOI] [PubMed] [Google Scholar]
- Vernier M, Bourdeau V, Gaumont-Leclerc MF, Moiseeva O, Bégin V, Saad F, et al. Regulation of E2Fs and senescence by PML nuclear bodies. Genes Dev. 2011;25:41–50. doi: 10.1101/gad.1975111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Gural A, Sun XJ, Zhao X, Perna F, Huang G, et al. The leukemogenicity of AML1-ETO is dependent on site-specific lysine acetylation. Science. 2011;333:765–769. doi: 10.1126/science.1201662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Henning SM, Heber D. Limitations of MTT and MTS-based assays for measurement of antiproliferative activity of green tea polyphenols. PLoS One. 2010;5:e10202. doi: 10.1371/journal.pone.0010202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss J, Cavenee WK, Herbst RA, Jung EG, Arden KC. Point mutations and allelic loss in the TP53 locus of cutaneous malignant melanomas. Arch Dermatol Res. 1994;286:417–419. doi: 10.1007/BF00371803. [DOI] [PubMed] [Google Scholar]
- Winnepenninckx V, Lazar V, Michiels S, Dessen P, Stas M, Alonso SR, et al. Gene expression profiling of primary cutaneous melanoma and clinical outcome. J Natl Cancer Inst. 2006;98:472–482. doi: 10.1093/jnci/djj103. [DOI] [PubMed] [Google Scholar]
- Yajima I, Kumasaka MY, Thang ND, Goto Y, Takeda K, Iida M, et al. Molecular network associated with MITF in skin melanoma development and progression. J Skin Cancer. 2011;2011:730170. doi: 10.1155/2011/730170. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.