Abstract
Long interspersed nuclear element–1 (L1) retrotransposons are mobile repetitive elements that are abundant in the human genome. L1 elements propagate through RNA intermediates. In the germ line, neighboring, nonrepetitive sequences are occasionally mobilized by the L1 machinery, a process called 3′ transduction. Because 3′ transductions are potentially mutagenic, we explored the extent to which they occur somatically during tumorigenesis. Studying cancer genomes from 244 patients, we found that tumors from 53% of the patients had somatic retrotranspositions, of which 24% were 3′ transductions. Fingerprinting of donor L1s revealed that a handful of source L1 elements in a tumor can spawn from tens to hundreds of 3′ transductions, which can themselves seed further retrotranspositions. The activity of individual L1 elements fluctuated during tumor evolution and correlated with L1 promoter hypomethylation. The 3′ transductions disseminated genes, exons, and regulatory elements to new locations, most often to heterochromatic regions of the genome.
Long interspersed nuclear element (LINE)−1 (L1) retrotransposons are widespread re-petitive elements in the human genome, composing about 17% of the entire DNA content (1). They are a remarkably successful parasitic unit, relying on the cell's transcription and translation machinery to initiate their reproduction and reinsertion, a process completed with the reverse transcriptase and integration capability of proteins encoded on the L1 transcript. In the germ line, L1s have contributed extensively to the evolution of genes and genomes by generating structural variation that can potentially affect function, shaped by ongoing mutation and natural selection across thousands of generations (2). Most of the repeat elements that reside in the human genome are inactive, because of inverted rearrangements or truncations introduced during retrotransposition or subsequent inactivating point mutations across human evolution. However, it is estimated, on the basis of full-length L1 elements with preserved open reading frames and activity in in vitro retro-transposition assays, that there are 50 to 120 currently active L1 repeats in the human genome, of which a small number are highly active, earning the moniker “hot-L1s” (3–5).
Among the many ways in which L1 retrotranspositions have shaped the human genome is the mobilization of unique DNA downstream of the LINE (6). This process, known as 3′ transduction, is likely the consequence of a weak transcription termination signal at the end of the L1, causing the transcription machinery to bypass the L1 polyadenylation signal and use another polyadenylation site downstream. The unique genomic sequence picked up in the hybrid transcript can then be reverse transcribed and reinserted by the L1 machinery, causing mobilization of up to several thousand nucleotides of nonrepetitive DNA (7). These 3′ transductions can drive the mobilization of adjacent coding regions, increasing the potential of this process to exert functional effects (6). The mobilization of unique downstream sequence effectively barcodes the source element from which a transduction arose, a property that has been exploited in germline genome analyses to track the lineages of active L1 elements (8) and that we exploit here for cancer genomes.
In somatic cells, there is increasing evidence that spontaneous L1 mobilizations occur during normal neurogenesis (9–12) and during cancer development (13–17). Similar to the germ line, this mutational process has considerable potential to restructure the cancer genome. As with all mutational processes, individual mutational events occur randomly and are then subjected to selective forces. Those events that generate functional consequences of advantage to the cell help that clone to expand over its competitors. The potential functional repertoire of somatic retro-transposition is extensive. Genes can be disrupted by insertions, leading to their inactivation and, on occasion, to their activation through disruption of cis-regulatory elements (17). Recently, processed pseudogenes, a by-product of somatic retrotrans-position occurring when mature messenger RNA transcripts are reverse transcribed and inserted in the genome by the L1 machinery, have been described in cancer genomes (18, 19). One example of a 3′ transduction has been described in a colorectal tumor (16), but this particular variation of retrotransposition has not been systematically studied in cancer genomes.
Here, we describe extensive somatic mobilization of genetic material mediated by L1 3′ transduction in many cancers, finding examples of genes, exons, and potential regulatory elements being repeatedly copied and strewn across the genome, sometimes in their tens to hundreds. With nonrepetitive sequence to fingerprint these transductions, the evidence implicates an unexpectedly small number of source elements as culpable for the vast majority of this genome shuffling. The activity of individual L1 donors may wax and wane during tumor evolution, but their progeny can themselves aspire to source element status, leading to cascading propagation of the family line.
Somatic L1 transductions can masquerade as chromosomal translocations
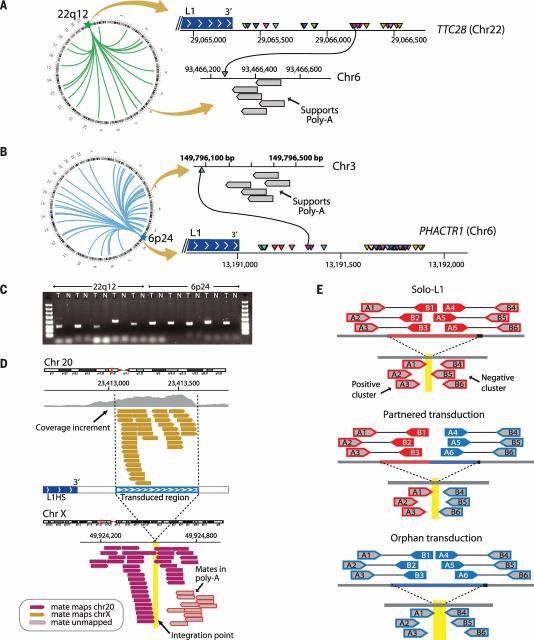
In a structural variant analysis of patients with lung cancer, we identified a patient with 22 somatically acquired interchromosomal rearrangements clustered in an intronic region of ~1700 base pairs (bp) in the gene TTC28 on chromo-some 22q12 (Fig. 1A). Preliminary analysis of the paired-end mapping reads revealed two characteristics suggestive of retrotransposition mediated by L1. First, all chromosome 22q12 breakpoints were clustered immediately after the 3′ extreme of a full-length L1 element. Second, in the relevant region of the partner chromosome, there were several reads whose unmapped mates reported a polyadenylate [poly(A)] tract inserted at the breakpoint junction. Clustered rearrangements in this same region of chromosome 22q12 were identified in two additional lung cancer patients and two lung cancer cell lines.
Fig. 1. Somatically acquired 3′ transductions are frequent in cancer genomes.
(A) Putative translocations involving 22q12 (TTC28) show characteristics suggestive of L1-mediated 3′ transduction. Breakpoints at 22q12 (triangles of different colors) are clustered immediately after a germline full-length element. On the other side of the breakpoint, there are reads whose pairs report the presence of poly(A) tails (gray boxes). (B) Breakpoints clustered in PHACTR1 just after a polymorphic L1 element not present in the reference genome. (C) PCR profiles showing the somatic acquisition of transductions from 22q12 and 6p24. T, tumor; N, normal. (D) The hallmarks of 3′ transduction. The donor-L1 locus at chromosome 20 shows coverage increment downstream of the element resulting from genome-wide amplification of the transduced material. Reads responsible for the coverage increment pair with different chromosomes (chromosome X illustrated). A cluster of reads around the breakpoint indicates the presence of a poly(A) tail. Other reads reveal the presence of target site duplication (not shown; details in table S3). (E) The strategy followed for somatic solo-L1 and transduction identification. The pipeline relies on the identification of two read clusters (i.e., positive and negative clusters) pointing to the same region of the genome where the somatic element is inserted.
In the original patient, a similar cluster of 50 rearrangements was found originating from a small intronic region in the PHACTR1 gene on chromosome 6p24.1 (Fig. 1B). Validation by polymerase chain reaction (PCR) across the breakpoint confirmed that these rearrangements were somatically acquired (Fig. 1C) and that the insertion of a poly(A) sequence was real.
The clustering of rearrangement breakpoints downstream from L1 elements coupled with poly (A) insertion suggests that these events represent L1 transductions. At first pass, it would be easy to misinterpret such events as balanced translocations because read-pairs spanning either end of the insertion appear to report reciprocal inter-chromosomal rearrangements. Indeed, retrotransductions could explain several puzzling clusters of rearrangements published by us and by others (20–24) (table S1). For example, in a colon cancer data set, 21 somatic translocations involving gene TTC28 were reported (23). We believe that these represent transductions of the same small region at chromosome 22q12 that we describe above. Similarly, in a squamous cell lung cancer data set (24), we identified that 80/592 (14%) of the reported rearrangements were located immediately downstream of germline hot-L1 loci. The distinction between balanced chromosomal translocations and retrotransductions matters, because the former can create oncogenic fusion genes whereas the latter cause insertions of up to a few kilobases and would predominantly drive oncogenesis through effects on transcription of the target gene (14, 15).
Identification of somatic transductions and retrotranspositions
We developed a bioinformatic pipeline to explore the frequency and signatures of all classes of somatic retrotransposition in cancer [TraFiC: Transposome Finder in Cancer (supplementary text)]. The pipeline is capable of detecting three different classes of retrotranspositions: solo-L1 events, in which either partial or complete LINEs are somatically retrotransposed; partnered transductions, in which a LINE and downstream nonrepetitive sequence are retrotransposed; and orphan transductions, in which only the unique sequence downstream of an active L1 is retro-transposed without the cognate LINE. We denote the L1 element from which a somatic retrotrans-position originates as the source element. The pipeline relies on the identification of genomic hallmarks of retrotransposition at both the integration point and the L1 source element locus (Fig. 1, D and E, and fig. S1).
We ran TraFiC on whole-genome sequencing data from 290 tumor and matched normal pairs (210 primary tumors, 52 metastatic tumors, and 28 cancer cell lines with matched normal cell lines) from 244 cancer patients across 12 cancer types. The analysis retrieved a total of 2756 L1 retrotranspositions that includes solo L1s and 3′ transductions (tables S2 and S3). PCR validation (fig. S2) and capillary sequencing of 308 insertions (131 solo L1s, 72 partnered transductions, 91 orphan transductions, and 14 Alu insertions from both cell lines and primary tumors) confirmed the somatic acquisition of 303 (true positive rate 98%). Mapping of at least one of the two breakpoints per insertion to base-pair resolution by capillary sequencing was obtained for 84% (259/308). To evaluate the sensitivity of TraFiC to detect somatic retrotranspositions in cancer genome sequencing data, we performed in silico simulations. We generated a mock cancer genome into which we had seeded known somatic retrotrans-position events of each type at differing levels of tumor clonality ranging from 25 to 100%. We then simulated sequencing reads from this mock cancer genome to the standard levels of coverage achieved in our cohort and ran TraFiC. The results confirmed the high specificity (>99%) of the pipeline and indicated a sensitivity ranging from 73 to 83% for tumor clonalities from 25 to 100%, respectively (table S4).
TraFiC obtained a partial reconstruction of the 5′ and 3′ extremes for 89% (2458/2756) of the total L1-mediated insertions, identified putative target site duplications for many of the insertions (table S3), and estimated an average insertion length of 1.09 kb for the insertions without 5′ inversion. Where present, target site duplications were typically in the range of 10 to 20 bp, as expected for retrotransposition. We were able to unambiguously assign 86% of somatic solo-L1 transpositions to a specific L1 subfamily; in every case, the transposable element (TE) belonged to L1Hs, based on the presence of diagnostic nucleotides (14, 25); in the remaining 14%, sequencing reads did not cover the diagnostic nucleotides. All these features are shown in table S3.
L1 3′ transductions are present in 25% of cancer genomes
Overall, 53% (129/244) of the patients have at least one somatic L1 retrotransposition event, most frequently colon cancers (93%) and lung cancers (75%) (Fig. 2A). L1-mediated transductions of downstream sequence comprise 24% (655/2756 insertions) of the total somatic L1 retrotranspositions that occurred (tables S2 and S3). This represents a substantial contribution to the mutational landscape of cancer genomes not previously documented and adds to the previous reports of somatic solo-L1 retrotransposition in cancer (13, 14, 16). Orphan transductions represent half (333/655) of all transductions, demonstrating that truncation during somatic integration is frequent. This corroborates earlier analyses of somatic solo-L1 insertions, which also showed a high rate of truncations (14). The size of the transduced regions was typically less than 1 kb (Fig. 2B), but occasional transductions were found that captured genomic sequence located up to 12 kb downstream of the L1 source element (Fig. 2C).
Fig. 2. The somatic L1 retrotransposition activity in 290 cancers.
(A) Distribution of L1 retrotransposition activity in 290 cancers. Pie charts display the proportions of analyzed cancer samples with at least one transduction (blue), no transductions but at least one solo L1 (red), and no L1 retro-transposition (white). Bars represent the somatic L1 count of each cancer sample. Horizontal black lines indicate mean somatic retrotransposition counts for each cancer type. (B) The size distribution for L1 insertions (including solo L1s and transductions), in bins corresponding to 100-nucleotide increments in insertion lengths, shows an overrepresentation of truncated L1 insertions below 2 kb (average length of insertion is ~1.1 kb). Only insertions without 5′ inversion, or with inversion when it is lower than 500 bp, are shown. There are 81 L1 elements with estimated length >5.9 kb, of which 11 are partnered transductions.These full-length insertions represent ~5% (81/1752) of the total non–5′-inverted insertions (table S3). (C) The lengths of all L1 insertions (transductions included) are illustrated as a coverage plot over the schematic representation of a canonical solo-L1 sequence (~6 kb) and its downstream sequence (~12 kb). Most somatic L1 insertions (solo L1s and partnered transductions) are truncated at the 5′ end. For insertions with 5′ extreme inversion, the insertion length estimated corresponds to the minimal size that could be recognized (table S3), so it is underestimated. Full-length transductions and orphans mobilize nonrepetitive DNA sequences up to 12 kb away from the L1 source element end. Most of the transductions correspond to DNA material located within a distance of 1 kb to the end of the L1 source element.
In numerical terms, somatic retrotransposition is an important component of the structural variation landscape in some tumors (fig. S3). In lung cancer sample PD7354k, for example, L1 activity produced 565 unique somatic events, of which 120 are transductions, compared with 142 classic genomic rearrangements. In the evolution of lung tumors PD7354 and PD7356, transductions increased the genome size by 120 and 55 kb, respectively. Somatic L1 retrotransposition activity was higher in metastatic than in primary prostate cancer (12 versus 1 retrotransposition per sample, respectively; P = 0.001), suggesting that in this disease it is a late-onset mutational process.
Few L1 loci drive 3′ transductions in cancer
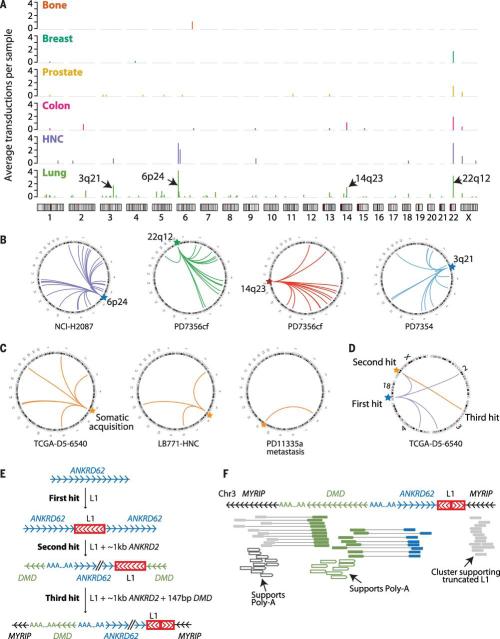
Because partnered and orphan transductions are defined by the retrotransposition of unique genomic sequence, we can unambiguously identify the L1 source element whence they derive. We find that 95% of all transductions identified can be attributed to only 72 germline L1 loci, with considerable variability in activity among them (Fig. 3A and table S5). Just two hot-L1 loci, located at chromosomes 22q12 and 6p24.1, account for more than a third of all somatic transductions we identify. Previous analyses of which L1 elements are likely to be active in the (germline) human genome have identified full-length L1 repeats with intact open reading frames found either in the reference genome or as polymorphisms in the population and studied their activity in in vitro retrotransposition assays (4, 5). Of the 72 source elements we find to be active in somatic cells, only 18 overlap with loci documented in these previous analyses (table S5). This indicates that a more extensive repertoire of L1 elements may be competent for retrotransposition, probably a reflection of events arising from intact L1 elements that are rare polymorphisms in the human population. Unfortunately, because of the short read lengths (100 bp) and library inserts (~500 bp) generated, we were unable to attribute somatic solo-L1 retrotranspositions to their germline source element and so cannot know whether the same source elements that trigger transductions also generate isolated L1 transpositions.
Fig. 3. Somatic 3′ transductions originate from a limited repertoire of L1 source elements.
(A) Rate of source element activity within and among tumor types. The y axis denotes the average number of transductions involving the given element per sample for that tumor type. (B) Individual source elements show dramatic transduction activity in some lung cancer genomes. (C) Transductions arising from somatically acquired L1 copies in a colon cancer (TCGA-D5-6540), a head and neck cancer (LB771-HNC), and a prostate cancer (PD11335a). (D) Three-hit somatic retrotransposition example. A full-length L1 element acquired somatically (first hit) generated four somatic transductions, one of which (second hit) induces further mobilization, leading to a third hit. (E) Structural configuration of the breakpoints originated in the three-hit retrotransposition example. An intact L1 is somatically retrotransposed into ANKRD62, causing further transduction of 1114 bp of ANKRD62 into DMD. A subsequent transduction picks up some of DMD together with ANKRD62 and inserts both into MYRIP on chromosome 3 (third hit). (F) Read clusters supporting breakpoints shown in (E). Paired reads are shown as boxes connected with lines, colored by the genomic region they map to.
The activity of individual L1 source elements varied across tumor types (Fig. 3A), with lung cancer showing the highest number of active copies (59 loci). In breast cancer, the source element at 22q12 is the only active L1 locus in 93% (15/17) of the breast cancer genomes where transductions occurred (fig. S4). Of particular note, some source elements, when active, can individually seed as many as 50 or more separate daughter copies in a single cancer. For example, the source element at 6p24.1 gave rise to 56 different somatic transductions during the evolution of the lung cancer PD7354 (Fig. 1B) and 25 transductions in the lung cancer cell line NCI-H2087 (Fig. 3B). Other hot-L1 elements, with at least 40 derived copies in a single sample, are located at 14q23 and 3q21 (Fig. 3B), and an additional nine source elements have shown at least 10 transductions each (table S5).
Somatically acquired L1 insertions are themselves transduction-competent
In theory, when a full-length L1 retrotransposition occurs, it takes with it all the machinery required to catalyze further retrotranspositions. By using the unique tags of downstream sequence as a marker of transduction competency, we searched for examples of somatically acquired L1 retrotranspositions that led to further dissemination from the new insertion site. We found 29 retrotranspositions from 17 L1 loci that were themselves somatically acquired insertions (Fig. 3C and table S5). The read-pair data confirmed that the de novo, somatically acquired source element was, as expected, a full-length L1 element in each example. Such somatic source elements were in general responsible for a low fraction of the transductions we observed (4.4%, 29/655) but would occasionally contribute the majority of transductions in a given sample. For instance, in colon cancer TCGA-D5-6540, four L1 insertions that were themselves somatic retrotranspositions gave rise to 12 somatic transductions, compared with 11 transductions originating from germline source elements.
In one notable colon cancer, we found a chain of three consecutive somatic retrotranspositions (Fig. 3, D to F). The first hit corresponds to the somatically acquired integration of a full-length L1 element within ANKRD62 on chromosome 18. This new somatic element itself then transduces the full-length L1 element, together with a 1114-bp region of ANKRD62, into the DMD gene on chromosome X (second hit). Last, this new element at chromosome X transduces a region of 147 bp of DMD together with the 1114 bp of ANKRD62 that was previously inserted there, integrating this new material into MYRIP on chromosome 3 (third hit). In this final transduction, the L1 element suffers an internal inversion, rendering it transduction-incompetent. This succession of events leads to the juxtaposition of intronic sequences from three different genes (MYRIP, DMD, and ANKRD62) in the final rearrangement.
Fluctuating activity of source elements during tumor evolution
To evaluate the timing of retrotransposition during cancer evolution, we analyzed eight patients with prostate cancer in whom we had sequenced multiple metastases, three patients with prostate cancer in whom we had sequenced multiple foci of the primary tumor, and three patients with squamous lung cancer in whom we had sequenced samples from two or more time points during progression from carcinoma in situ to invasive cancer. Three patients had sufficient numbers of somatic transductions for us to evaluate source element activity across the cancer's development, although we acknowledge that our sensitivity to detect early shared events is greater than for late events found in only one sample.
For one prostate cancer (Fig. 4A), we reconstructed the phylogenetic tree from 23,881 somatic substitutions across a spinal cord metastasis and three liver deposits. We mapped somatic transductions onto this phylogenetic tree. Transductions were distributed across all internal branches and all but one of the terminal branches of the tree. Four source elements (Xp22-b, 22q12, 11q14, and 13q21) gave rise to transductions on more than one branch of the phylogenetic tree, suggesting that they were repeatedly active in different clonal lineages during tumor evolution. Nonetheless, they were not universally active, nor were the activity profiles among source elements correlated.
Fig. 4. L1 source element activity waxes and wanes during tumor evolution.
(A) Evolution of prostate cancer PD11335. (Left) The phylogeny shows new somatic mobilizations in each branch of the phylogenetic tree, colored by the source element that is active on that branch. (Right) The final counts for each active source element in the sample sequenced. (B) Evolution of lung cancer PD7354.TheVenn diagram shows the number of shared and nonshared somatic L1 retrotranspositions among the four samples sequenced. Source elements at 6p22 and 22q12 differed in activity between PD7354r and PD7354h (bar graph). (C) Evolution of lung cancer PD7356, sequenced at an early carcinoma in situ phase and a late invasive cancer. Somatic retrotranspositions were classified as shared between both lesions (early) or isolated to one or other lesion alone (late). Among the events isolated to only one or other lesion, there was no overlap in source elements, indicating individual activity varied during evolution of the tumor.
Similar patterns were observed during the evolution of carcinoma in situ to invasive squamous cell lung cancer for patient PD7354. In this case, the four lesions sequenced are each complex admixtures of subclones, making reconstruction of a complete evolutionary history challenging. However, we do see differences in the activity rate of individual source elements across this time series (Fig. 4B). For example, comparing events exclusive to either PD7354h or PD7354r, the locus at 6p22 spawned six transductions in PD7354r but none in PD7354h, whereas the source element at 22q12 initiated 10 transductions in PD7354h but only two specific to PD7354r. The differences in activity across source elements between the two lesions were statistically significant (P = 0.01). Similarly, for a second patient (PD7356) in whom we sequenced a squamous carcinoma in situ and its related invasive cancer, we found no overlap in source element activity after divergence of the two clonal populations from the common ancestor (Fig. 4C).
Taken together, these data indicate that activity of individual source elements waxes and wanes during tumor evolution. We cannot know whether transposition events can occur in normal cells before oncogenic transformation—this would require single normal cell cloning and sequencing (26). These examples do show that somatic retrotransposition persists into the late stages of tumor development.
Hypomethylation activates L1 source elements in cancer genomes
DNA methylation of the L1 promoter is an important inhibitor of L1 activity (27–30). In previous analyses of methylation profiles, it had been shown that there might be genome-wide variation in methylation correlated with somatic retrotranspositions in cancer (13) and that global methylation of L1 promoters is about 20% lower in hepatocellular cancers than in normal liver (17). These assays essentially report methylation status at a global level. With transductions, because we know the individual L1 source elements that are active in any given sample, we are able to directly measure the methylation status of the given element in that sample. To do this, we combined bisulfite DNA treatment with massively parallel sequencing of the promoter region, assaying six loci in 16 samples.
We found a remarkably consistent correlation between CpG hypomethylation on the promoter of a source element and its retrotransposition activity in that sample. In every sample in which the source element had been active, the most common haplotype observed was fully unmethylated across the promoter. In contrast, samples without transductions arising from the given source element generally showed high levels of promoter methylation at that locus (Fig. 5 and fig. S5). In the source element 6p24.1, for example, in a lung cancer cell line (NCI-H2087), we found fully unmethylated CpG dinucleotides in the repeat, whereas the matched transformed lymphoblastoid cell line from the same patient (NCI-BL2087) is methylated (Fig. 5A). These correlations were also observed in primary tumors. For instance, for primary breast cancer samples encompassing the range of retrotransposition activity at source L1 locus 22q12, methylation patterns revealed by massively parallel sequencing showed a fully unmethylated haplotype (usually the most frequent one) in samples where the source element shows activity (Fig. 5B). Results were similar for the other four source elements (fig. S5) and were confirmed by capillary sequencing (fig. S6). There were occasional exceptions to this pattern; although activity was always associated with hypomethylation, sometimes hypomethylation did not indicate activity. In three samples of 66 scenarios analyzed, we found a fully unmethylated promoter at a source element from which we detected no transductions (Fig. 5 and fig. S5).
Fig. 5. Specific hypomethylation of L1 promoter of active and inactive source elements.
(A and B) After bisulfite treatment of DNA and PCR amplification of L1 promoter regions composing the six most frequently active source elements, massively parallel sequencing was undertaken.The two most commonly observed haplotypes for each sample are depicted, with open circles representing CpG dinucleotides that are unmethylated and solid circles representing methylated CpG dinucleotides. The fraction of reads reporting each haplotype is shown on the right. Green circles on the left indicate which samples showed transductions derived from that source element; red circles indicate samples without activity of that source element. Asterisks after the sample name indicate tumor samples; those without asterisks are matched normal samples.
To determine how far the hypomethylation at active L1 elements extended, we analyzed Illumina (Illumina, Incorporated, San Diego, California) methylation array data available for 19 colon cancer samples from the Cancer Genome Atlas (TCGA) (23). We did not find any correlation between source element activity and hypomethylation in the 5 Mb surrounding the L1 locus (P values ranging from 0.4 to 0.9, t test) (fig. S7). This suggests that L1 activation in cancer genomes is more likely the consequence of the hypomethylation of a highly localized DNA region surrounding the source element (or restricted to each specific L1 locus) rather than hypomethylation of larger chromosomal regions. These results reveal that, although global hypomethylation is a factor predisposing to L1 activation (13) in cancer, individual L1 source elements become active only when hypomethylation occurs in the promoter region of that element.
Transduction of genes, exons, and regulatory elements in cancer genomes
When an L1 source element is located near a coding region, transductions can cause mobilization of exons (6, 31). We found capture of adjacent coding regions to be relatively common in somatic transductions (Fig. 6A), with about 2.3% of events (15/655) distributing a proximal exon or complete gene elsewhere in the genome.
Fig. 6. Somatic shuffling of coding and regulatory regions mediated by L1 transductions.
(A) Somatic transductions can mobilize coding sequences. Gray rectangles represent each L1 source element (LINE1 master), and white boxes at 5′ represent the L1 promoter. The x axis shows the distance downstream of the source element. Exons are represented by green rectangles. Blue lines represent the region transduced elsewhere in the genome. (B) In PD7354, the Circos (http://circos.ca) plot shows transductions mediated by the source element at chromosome 7q34 involved in the somatic amplification of OR9A4. (C) Coverage increment demonstrating amplification of OR9A4 in different samples of tumor PD7354. (D) Read clusters supporting the integration of STK31 exon into chromosome 14, with the sequence of events shown in (E). (E) Structural configuration of breakpoints involved in the STK31 exon shuffling mediated by a somatic L1 element. An intact, transduction-competent L1 element inserts somatically immediately downstream of an exon of STK31. A further partnered transduction event occurs in which the exon of STK31 and a portion of the somatic L1 element retrotranspose to an intron of NRXN3. (F) Somatic transductions frequently mobilize DNA sequences with regulatory potential. Gray rectangles represent the 3′ end of the L1 source elements. The x axis shows the distance downstream of each source element. Green rectangles represent DNAse-I–hypersensitive sites, and horizontal blue lines represent transcription factor binding sites. Every vertical red line represents the end point of a somatic transduction event.
One lung carcinoma in situ, PD7354, demonstrates recurrent transduction of the single-exon gene OR9A4, an olfactory receptor, originating from a germline L1 element just 862 bp downstream of the gene (Fig. 6, B and C). Among 14 different somatic transductions, we found four complete copies of the OR9A4 gene and five partial copies distributed across the genome, often inserted into footprints of other genes. Similarly, we identified duplication of the third exon of the TPST1 gene in two primary lung cancers, TCGA-60-2722 and PD7356 (fig. S8). There, the exon is mobilized by the action of a germline L1 element located 145 bp upstream.
Somatic retrotranspositions can mobilize coding sequence when they themselves propagate. In one lung cancer cell line, NCI-H2087, a full-length L1 element had integrated into the kinase gene STK31 as a somatically acquired retrotrans-position (Fig. 6, D and E, and fig. S9). This new somatic L1 insertion was transduction-competent: In one event, it picked up the whole of exon 18 of STK31 located 169 bp downstream, catalyzing its insertion into the footprint of another gene, NRXN3. Such secondary transductions mean that the range of genomic elements that could potentially be targeted by transduction is considerably larger than just those near germline transduction-competent L1 elements.
Transductions could also potentially mediate distribution of sequences with regulatory potential around the genome (Fig. 6F). We screened the somatic transductions identified in our cohort for mobilization of deoxyribonuclease I (DNAse-I) hypersensitive sites or transcription factor binding sites defined by the Encyclopedia of DNA Elements (ENCODE) project (32). We found 43 transductions that mobilized a total of 54 full-length DNAse-I hypersensitive sites. Similarly, 86 transductions led to 251 transcription factor binding sites being copied and inserted elsewhere in the genome, although we recognize that ENCODE used a variety of normal cell types to define these sites. In the absence of tumor-specific epigenetic profiling data, it is difficult to know what the effects of mobilizing any one of these putative regulatory elements would be, but our data do show that somatic transductions would theoretically have the potential to shuffle such elements around the genome and near key genes.
Chromatin organization influences regional rates of L1 retrotransposon insertion
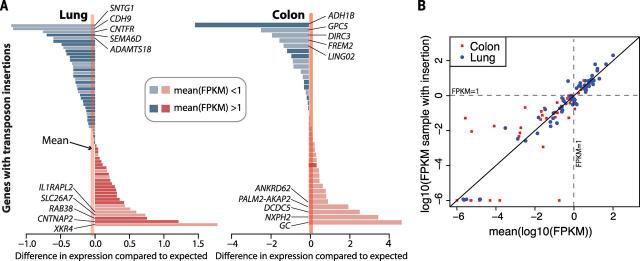
Recent work has provided evidence that a small, but important, fraction of the L1 insertions in a cancer genome may influence gene function (14). To assess this, we analyzed RNA-sequencing data available from TCGA samples of 13 lung and 11 colon cancers. We did not find any strong evidence that retrotransposon insertion altered expression levels generally for the 66 and 39 genes, respectively, with somatic L1 insertions (Fig. 7). This finding differs from a previous analysis that suggested L1 insertions caused frequent down-regulation of gene expression (14), a discrepancy we believe to be due to skewness in the distribution of gene expression levels (discussed extensively in the materials and methods and figs. S10 and S11). In addition, we did not find any evidence for aberrant fusion transcripts arising from inclusion of transduced sequence in the target gene. In total, 16 insertions occurred in footprints of genes identified as recurrently mutated (33), mostly intronic, and probably of minimal importance (table S6). There was one somatic insertion, of an Alu element into the promoter of NFE2L2 in a squamous lung cancer, of uncertain importance.
Fig. 7. Gene expression effects associated with L1 insertions.
(A) For lung and colon cancers, each bar represents the difference between the log10 (FPKM) for the target gene in the relevant sample compared to the average log10(FPKM) for other samples of that tumor type. FPKM, fragments per kilobase of transcript per million mapped reads. Genes with FPKM > 1 average expression have darker bars. (B) Scatter plot showing the data in (A).The y axis shows the log10(FPKM) for the target gene in the relevant sample, and the x axis shows the average log10(FPKM) for that gene for other samples of that tumor type. In expressed genes [mean log10(FPKM) > 0], the expression level in the affected sample is very close to the overall expression level of the gene in the corresponding tissue. Most large expression level differences occur at unex-pressed genes [mean log10(FPKM) < 0].
To understand the properties of the insertion points of retrotranspositions, we analyzed the genome-wide distribution of 2850 somatic TEs (including 2756 L1 and 94 non-L1 insertions). At a macroscopic scale, we found significant heterogeneity across the genome in the density of insertion points (Fig. 8A). This is particularly evident in the first 30 Mb of chromosome 5p, where we identified no fewer than 93 separate insertions. Beyond this, we also found evidence for more-localized clusters of integration sites, identifying four particular hotspots, three of which are located in the subtelomeric region of 5p. Each of these hotspots is 1 to 1.5 Mb in size and contains 10 insertions from our data set (Fig. 8A).
Fig 8. Somatic L1 insertions favor heterochromatin.
(A) Bars show number of elements per 10-Mb window. Red bars represent the 13 regions with overrepresentation of elements. Asterisk represents hotspots of TEs in the cancer genome (i.e., 10 or more elements are clustered together within a region of 1 to 1.5 Mb). (B) Somatic integrations of TEs are more abundant far away from the transcription start site of the nearest gene.The x axis shows the rate of observed versus expected somatic insertions.The y axis shows the distance to the transcription start site of the nearest gene. (C) Somatic integrations of TEs are more frequently associated with exon-poor regions of the cancer genome.The x axis shows the number of somaticTEs in windows of 3 Mb of the genome, whereas the y axis shows the density of exons.Windows at chromosome 5p, which showed the highest somaticTE insertion rates in the cancer genome, are highlighted. (D) TEs are enriched in lowly expressed genes (<3 FPKM) relative to highly expressed genes. (E) Overall, TEs are overrepresented in transcriptionally repressed regions of the genome (most likely heterochromatic), similar to previous observations of point mutations in cancer (37). The relative abundance of insertions in repressed chromatin is 4.55 times higher than in transcriptionally active regions of the genome. R, repressed; T, transcriptionally active; CTCF, CCCTC binding factor–enriched element; E/WE, enhancer or weak enhancer regions;TSS/PF, promoters and flanking regions. Error bars reflect Poisson confidence intervals. (F) Average rate of TE insertions and synonymous point mutations in repressed and active chromatin.The difference in mutation rate between repressed and active chromatin is much larger in TE insertions relative to point mutations.
Somatic retrotranspositions are more likely to insert in intergenic or heterochromatic regions of the cancer genome than expected by chance. First, higher frequency of TE integration correlates with greater distance to the transcription start site of the nearest gene (Fig. 8B), indicating that most of the integration events occur at intergenic regions. Second, somatic insertions are more frequently observed in regions of the genome where exon density is lower (Fig. 8C). Third, TEs more frequently insert into lowly expressed genes compared with those that are highly expressed (P = 2 × 10−24), even after correction for the length of the gene (P = 5 × 10−4) (Fig. 8D). Fourth, the rate of somatic insertion is five times higher in repressed or low-activity regions of the genome predicted by ENCODE than in transcribed regions (Fig. 8E). This difference in mutation rates between heterochromatin and euchromatin is much larger for retrotransposon insertions (4.55 times higher rate in heterochromatin) than for somatic substitutions (1.54 times higher) (Fig. 8F). When we repeated this analysis for polymorphic germline retrotranspositions (that must have occurred recently in evolutionary time) identified from three different databases, we find a very similar enrichment for heterochromatin insertion as for somatic events (P = 0.6; fig. S12).
Two possible explanations, not necessarily mutually exclusive, could underpin the apparent enrichment of somatic retrotransposition into heterochromatin. First, insertions into gene footprints could be deleterious to the cancer clone and therefore subject to negative selection, or, second, the insertion machinery could preferentially target heterochromatin. We cannot formally distinguish between these possibilities on the basis of our data but believe the latter to be the more dominant because most of the insertions in active chromatin do not have an impact on gene expression and because negative selection is less powerful in somatic cells than in the germ line.
The majority of these retrotransposition events are likely to be passenger mutations. This is no different from any other mutational process—kataegis, the POLE hypermutator phenotype in endometrial cancer, and the tandem duplicator phenotype in breast and ovarian cancer (34–36), for example. Nonetheless, generation of genomic variability is the first stage of Darwinian evolution, and it is a key aim of cancer genomics to describe mutational processes in full, multidimensional detail. This includes information about the patterns and frequency of the mutational process, its genome-wide distribution, its distribution across tumor types, its timing during cancer evolution, and its correlation with other processes generating genomic instability. To understand how 3′ transductions can generate functional and tumorigenic consequences, we will need to survey the topography of somatic retrotransposition on a considerably larger scale, across thousands of cancer genomes, integrated with other mutational processes and transcriptional data.
Supplementary Material
ACKNOWLEDGMENTS
J.M.C.T. is supported by a Marie Curie Fellowship FP7-PEOPLE-2012-IEF (project number 328264) and thanks H. Kazazian and S. Solyom for providing with L1Hs specific oligonucleotide primer sequences. We thank the TCGA project team and their specimen donors for providing sequencing data. This work was supported by the Wellcome Trust, the Chordoma Foundation, and the Skeletal Cancer Action Trust. Y.S.J. and I.M. are supported by European Molecular Biology Organization long-term fellowships (LTF 1203-2012 and 1287-2012). P.J.C. and S.M.J. are Wellcome Trust Senior Clinical Fellows. P.V.L. is a postdoctoral researcher of the Research Foundation Flanders (FWO). S.N.-Z. is funded by a Wellcome Trust Intermediate Clinical Research Fellowship (WT100183MA). U.M. is supported by a CRUK Clinician Scientist fellowship. Support was provided to A.M.F. by the National Institute for Health Research, the University College London Hospitals Biomedical Research Centre, and the Cancer Research UK University College London Experimental Cancer Centre. C.D. is supported by a grant from the Brussels Region–Impulse Programme Life Sciences. Samples from Addenbrooke's Hospital were collected with support from the NIHR Cambridge Biomedical Resource Centre. The ICGC Breast Cancer Consortium was supported by a grant from the European Union (BASIS) and the Wellcome Trust. The ICGC Prostate UK Cancer Consortium is funded by CRUK grant C5047/A14835, by the Dallaglio Foundation, and the Wellcome Trust. We also acknowledge support from the Bob Champion Trust, the Orchid Cancer appeal, the RoseTrees Trust, the North West Cancer Research Fund, Big C, the Grand Charity of Freemasons, and the Research Foundation Flanders (FWO). We acknowledge the Biomedical Research Centre at the Institute of Cancer Research and the Royal Marsden NHS Foundation Trust. We thank the NIHR, Hutchison Whampoa Limited, Human Research Tissue Bank (Addenbrookes Hospital), CRUK Cambridge Research Institute Histopathology, In-situ Hybridisation Core Facility, Genomics Core Facility Cambridge, and Cambridge University Hospitals Media Studio. P.J.C. holds equity in, and is a paid consultant for, 14M Genomics Limited.
Footnotes
SUPPLEMENTARY MATERIALS
REFERENCES AND NOTES
- 1.Lander ES, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. doi: 10.1038/ 35057062; pmid: 11237011. [DOI] [PubMed] [Google Scholar]
- 2.Kazazian HH., Jr Mobile elements: Drivers of genome evolution. Science. 2004;303:1626–1632. doi: 10.1126/science.1089670. doi: 10.1126/ science.1089670; pmid: 15016989. [DOI] [PubMed] [Google Scholar]
- 3.Sassaman DM, et al. Many human L1 elements are capable of retrotransposition. Nat. Genet. 1997;16:37–43. doi: 10.1038/ng0597-37. doi: 10.1038/ng0597-37; pmid: 9140393. [DOI] [PubMed] [Google Scholar]
- 4.Brouha B, et al. Hot L1s account for the bulk of retrotransposition in the human population. Proc. Natl. Acad. Sci. U.S.A. 2003;100:5280–5285. doi: 10.1073/pnas.0831042100. doi: 10.1073/ pnas.0831042100; pmid: 12682288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beck CR, et al. LINE-1 retrotransposition activity in human genomes. Cell. 2010;141:1159–1170. doi: 10.1016/j.cell.2010.05.021. doi: 10.1016/ j.cell.2010.05.021; pmid: 20602998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moran JV, DeBerardinis RJ, Kazazian HH., Jr Exon shuffling by L1 retrotransposition. Science. 1999;283:1530–1534. doi: 10.1126/science.283.5407.1530. doi: 10.1126/science.283.5407.1530; pmid: 10066175. [DOI] [PubMed] [Google Scholar]
- 7.Szak ST, Pickeral OK, Landsman D, Boeke JD. Identifying related L1 retrotransposons by analyzing 3′ transduced sequences. Genome Biol. 2003;4:R30. doi: 10.1186/gb-2003-4-5-r30. doi: 10.1186/ gb-2003-4-5-r30; pmid: 12734010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Macfarlane CM, et al. Transduction-specific ATLAS reveals a cohort of highly active L1 retrotransposons in human populations. Hum. Mutat. 2013;34:974–985. doi: 10.1002/humu.22327. doi: 10.1002/ humu.22327; pmid: 23553801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coufal NG, et al. L1 retrotransposition in human neural progenitor cells. Nature. 2009;460:1127–1131. doi: 10.1038/nature08248. doi: 10.1038/nature08248; pmid: 19657334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Muotri AR, et al. L1 retrotransposition in neurons is modulated by MeCP2. Nature. 2010;468:443–446. doi: 10.1038/nature09544. doi: 10.1038/nature09544; pmid: 21085180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Evrony GD, et al. Single-neuron sequencing analysis of L1 retrotransposition and somatic mutation in the human brain. Cell. 2012;151:483–496. doi: 10.1016/j.cell.2012.09.035. doi: 10.1016/j.cell.2012.09.035; pmid: 23101622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baillie JK, et al. Somatic retrotransposition alters the genetic landscape of the human brain. Nature. 2011;479:534–537. doi: 10.1038/nature10531. doi: 10.1038/nature10531; pmid: 22037309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iskow RC, et al. Natural mutagenesis of human genomes by endogenous retrotransposons. Cell. 2010;141:1253–1261. doi: 10.1016/j.cell.2010.05.020. doi: 10.1016/j.cell.2010.05.020; pmid: 20603005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee E, et al. Landscape of somatic retrotransposition in human cancers. Science. 2012;337:967–971. doi: 10.1126/science.1222077. doi: 10.1126/ science.1222077; pmid: 22745252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miki Y, et al. Disruption of the APC gene by a retrotransposal insertion of L1 sequence in a colon cancer. Cancer Res. 1992;52:643–645. pmid: 1310068. [PubMed] [Google Scholar]
- 16.Solyom S, et al. Extensive somatic L1 retrotransposition in colorectal tumors. Genome Res. 2012;22:2328–2338. doi: 10.1101/gr.145235.112. doi: 10.1101/gr.145235.112; pmid: 22968929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shukla R, et al. Endogenous retrotransposition activates oncogenic pathways in hepatocellular carcinoma. Cell. 2013;153:101–111. doi: 10.1016/j.cell.2013.02.032. doi: 10.1016/j.cell.2013.02.032; pmid: 23540693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cooke SL, et al. Processed pseudogenes acquired somatically during cancer development. Nat. Commun. 2014;5:3644. doi: 10.1038/ncomms4644. doi: 10.1038/ncomms4644; pmid: 24714652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ewing AD, et al. Retrotransposition of gene transcripts leads to structural variation in mammalian genomes. Genome Biol. 2013;14:R22. doi: 10.1186/gb-2013-14-3-r22. doi: 10.1186/gb-2013-14-3-r22; pmid: 23497673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baca SC, et al. Punctuated evolution of prostate cancer genomes. Cell. 2013;153:666–677. doi: 10.1016/j.cell.2013.03.021. doi: 10.1016/j.cell.2013.03.021; pmid: 23622249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Banerji S, et al. Sequence analysis of mutations and translocations across breast cancer subtypes. Nature. 2012;486:405–409. doi: 10.1038/nature11154. doi: 10.1038/nature11154; pmid: 22722202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Campbell PJ, et al. The patterns and dynamics of genomic instability in metastatic pancreatic cancer. Nature. 2010;467:1109–1113. doi: 10.1038/nature09460. doi: 10.1038/nature09460; pmid: 20981101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Muzny DM, et al. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. doi: 10.1038/nature11252; pmid: 22810696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hammerman PS, et al. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489:519–525. doi: 10.1038/nature11404. doi: 10.1038/nature11404; pmid: 22960745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ewing AD, Kazazian HH., Jr High-throughput sequencing reveals extensive variation in human-specific L1 content in individual human genomes. Genome Res. 2010;20:1262–1270. doi: 10.1101/gr.106419.110. doi: 10.1101/gr.106419.110; pmid: 20488934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Welch JS, et al. The origin and evolution of mutations in acute myeloid leukemia. Cell. 2012;150:264–278. doi: 10.1016/j.cell.2012.06.023. doi: 10.1016/j.cell.2012.06.023; pmid: 22817890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bourc'his D, Bestor TH. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature. 2004;431:96–99. doi: 10.1038/nature02886. doi: 10.1038/nature02886; pmid: 15318244. [DOI] [PubMed] [Google Scholar]
- 28.Menendez L, Benigno BB, McDonald JF. L1 and HERV-W retrotransposons are hypomethylated in human ovarian carcinomas. Mol. Cancer. 2004;3:12. doi: 10.1186/1476-4598-3-12. doi: 10.1186/ 1476-4598-3-12; pmid: 15109395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yoder JA, Walsh CP, Bestor TH. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 1997;13:335–340. doi: 10.1016/s0168-9525(97)01181-5. doi: 10.1016/S0168-9525(97)01181-5; pmid: 9260521. [DOI] [PubMed] [Google Scholar]
- 30.Hur K, et al. Hypomethylation of long interspersed nuclear element-1 (LINE-1) leads to activation of proto-oncogenes in human colorectal cancer metastasis. Gut. 2014;63:635–646. doi: 10.1136/gutjnl-2012-304219. doi: 10.1136/gutjnl-2012-304219; pmid: 23704319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ejima Y, Yang L. Trans mobilization of genomic DNA as a mechanism for retrotransposon-mediated exon shuffling. Hum. Mol. Genet. 2003;12:1321–1328. doi: 10.1093/hmg/ddg138. doi: 10.1093/hmg/ddg138; pmid: 12761047. [DOI] [PubMed] [Google Scholar]
- 32.ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. doi: 10.1038/nature11247; pmid: 22955616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lawrence MS, et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505:495–501. doi: 10.1038/nature12912. doi: 10.1038/nature12912; pmid: 24390350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nik-Zainal S, et al. Mutational processes molding the genomes of 21 breast cancers. Cell. 2012;149:979–993. doi: 10.1016/j.cell.2012.04.024. doi: 10.1016/j.cell.2012.04.024; pmid: 22608084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McBride DJ, et al. Tandem duplication of chromosomal segments is common in ovarian and breast cancer genomes. J. Pathol. 2012;227:446–455. doi: 10.1002/path.4042. doi: 10.1002/path.4042; pmid: 22514011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Palles C, et al. Germline mutations affecting the proofreading domains of POLE and POLD1 predispose to colorectal adenomas and carcinomas. Nat. Genet. 2013;45:136–144. doi: 10.1038/ng.2503. doi: 10.1038/ng.2503; pmid: 23263490. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.