Abstract
The study of tumor metabolism has resulted in new understandings of how cancer cells modify metabolic pathways that control cellular energetics to allow increased proliferation and survival. Tumor cells have been shown to alter metabolic pathways involved in glucose, glutamine and mitochondrial metabolism to generate raw materials needed for rapid cellular proliferation, maintain favorable cellular redox environments, modify cellular epigenetics and even promote and maintain oncogenic transformation. As a consequence, there has been intense scientific and clinical interest in targeting metabolic alterations that are commonly adopted by tumor cells for therapeutic purposes. In this review, we describe common metabolic alterations seen in tumor cells and discuss how these alterations are being investigated as potential targets for pharmacological intervention in preclinical and clinical settings. We also discuss some of the challenges associated with using tumor metabolism as a therapeutic target in cancer therapy, along with potential avenues to overcome these challenges.
Keywords: Tumor metabolism, aerobic glycolysis, glutamine, IDH mutations, combination therapy
Therapeutic strategies for the treatment of cancer have undergone a revolution in recent years. In the past, frontline cancer therapy consisted largely of widely cytotoxic drugs that damaged both cancer cells and normal, healthy tissue. Over the past several decades, emphasis has been placed on identifying distinctive or preferential features of cancer cells to better target cancer yet spare normal cells. This has resulted in targeted therapies with higher treatment efficacy and improved patient outcomes. One recent focus in cancer research has been to exploit metabolic features of cancer cells that may separate them from normal tissue. The rationale behind this endeavor is that cancer cells, due to their rapid, sustained growth and proliferation and need to withstand hypoxia, must employ a metabolic program that deviates from the metabolic characteristics of normal, healthy tissue. The ability to target cancer metabolism may result in a therapeutic window where cancer cells are inhibited but normal healthy cells remain unaffected.
Metabolic Features of Cancer
Researchers and clinicians working in cancer related fields have come to understand that cancer is a heterogeneous disease. There is tremendous variation in biology between different types of cancer, different patients with the same type of cancer, and even between different cancer cells in the same tumor. Cancer metabolism also exhibits considerable heterogeneity, with tumors adopting various metabolic programs that best suit a particular microenvironment. There are, however, some commonalities in cancer metabolism that are generally applicable to a significant portion of tumors. Among these common features of cancer metabolism are alterations in glucose, glutamine and mitochondrial metabolism. These commons features may allow for the development of novel therapeutics to target a fundamental hallmark of cancer biology. Indeed, the fundamental nature of some metabolic pathways may provide a unified target to bypass and overcome the genetic heterogeneity of tumors.
Aerobic glycolysis in cancer
The metabolic needs of most normal differentiated cells are largely energetic and metabolism is oriented towards maximal ATP generation through oxidative phosphorylation. During this process, glucose is first converted through glycolysis to pyruvate in the cytosol of the cell then further metabolized in the mitochondria in the TCA cycle and electron transport to produce an electromotive force that generates large quantities of ATP. Otto Warburg first noted nearly 100 years ago, however, that cancer cells use an altered program of glucose metabolism, in which they, even under normoxic conditions, convert glucose-derived pyruvate to lactate, rather than oxidizing it in the mitochondria[1]. This metabolic program differed from conventional models at the time, where lactate was thought produced only in anaerobic conditions, and instead resembles the Crabtree effect, wherein respiration is suppressed the presence of oxygen if glucose levels are sufficiently high. Warburg termed this metabolic trait aerobic glycolysis and hypothesized it resulted from mitochondrial defects in cancer cells that rendered them unable to utilize a normal, oxidative metabolic program. While it has since been found that most cancer cells have intact mitochondria, it is certainly true that a number of oncogenic driver mutations shift cell metabolism away from oxidative phosphorylation towards glycolytic metabolism[2, 3]. The teleological reason for this metabolic reprogramming remains uncertain, but a strong consensus has emerged that glycolytic use of glucose allows cancer cells to generate biosynthetic intermediates that are necessary for cell growth and proliferation, while also avoiding the production of potentially harmful reactive oxygen species (ROS) that results from oxidative phosphorylation. This view is buttressed by findings of similar metabolisms in proliferative normal healthy cells, such as lymphocytes and endothelial cells in angiogenesis.
Since Warburg's description of aerobic glycolysis in cancer cells, key questions that have been studied are to what extent cancer cells require this metabolic program and can it be inhibited to eliminate cancer cells without excessive toxicity. There are a great number of metabolic pathways that utilize glucose or its derivatives towards the production of raw materials for biosynthesis, maintaining a favorable redox environment, and meeting the energy demands of cancer cells. Likewise amino acids can feed into a wide array of metabolic pathways. Given that core metabolic pathways are shared in nearly all cells, specificity for cancer cells and toxicity to normal cells are critical concerns. Also important is to what extent cancer cells can exert plasticity and respond to metabolic inhibition with compensatory metabolic reprogramming that may maintain cancer cell proliferation or viability.
Targeting Aerobic Glycolysis
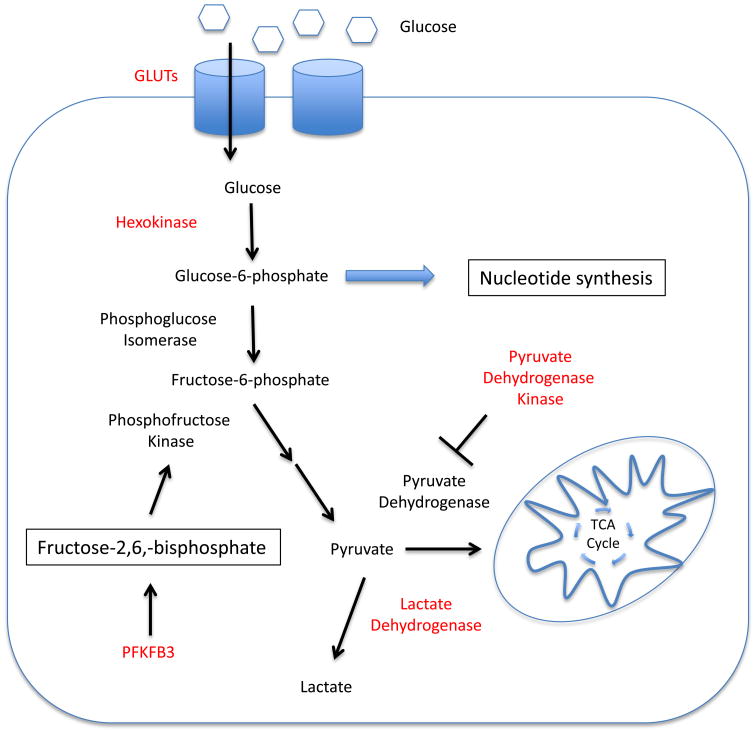
There are numerous proteins that regulate the glycolytic pathway and have been proposed as potential drug targets. Here we will discuss the potential for targeting the early stages of glycolysis, where glucose is taken up into the cancer cell and phosphorylated to trap it in the cell, and the late stages of glycolysis, at the branch point where glucose derived pyruvate is either fluxed into the TCA cycle as acetyl-CoA, or converted to lactate for export (Figure1). The first two steps of glycolysis are the uptake of glucose into the cell by glucose transporters and subsequent phosphorylation by hexokinases. In numerous types of cancer, glucose transporters[4-6] and various isoforms of hexokinase are overexpressed[7], making them tempting targets for pharmacological inhibition. Indeed, the genetic deletion of Glut1 in a mouse model of B cell acute lymphoblastic leukemia (B-ALL) greatly slowed cell proliferation and lessened disease burden[8]. Similarly, the inhibition of glucose transporters has been explored in several cancer settings. For example, small molecule based inhibition of Glut1 was found to slow the growth of non-small cell lung cancer (NSCLC)[9]and have effects against renal cell carcinoma[3]. A number of drugs in the retroviral protease inhibitor class, commonly used to treat HIV infection, have been found to also possess the off-target effect of inhibiting glucose transporters, including Glut1 and Glut4[10]. Ritonavir, a drug in this class, has been shown to have anti-proliferative effects in a mouse model of multiple myeloma through the inhibition of glucose uptake into the cells[11]. When considering glucose transporters as a potential therapeutic target for human cancer patients, it must be noted that it is unclear what toxicities would occur with potent inhibition. For instance, Glut1 is heavily expressed at the blood brain barrier[12], and inhibition may result in neurological effects, as evidenced by patients with Glut1-deficiency Syndrome[13]. Nevertheless, Glut1 inhibitors with proven clinical track records, such as Ritonavir, show that a therapeutic window of partial inhibition of glucose uptake may be present.
Figure 1. Tumor cell glucose metabolism as a therapeutic target.
The alterations to normal glucose metabolism that are exhibited in cancer cells may provide targets for therapeutic intervention. Inhibiting aerobic glycolysis in cancer cells causes slowed cell proliferation and increased cell death. Enzymes that are currently being targeted with small molecule inhibitors are indicated in red.
Hexokinase may also provide a target in cancer metabolism through isoform selective inhibition. Several different types of cancer have been shown to overexpress Hexokinase II, an isoform not expressed in most normal tissue. Multiple groups have shown that the genetic deletion of Hexokinase II is beneficial, slowing cancer progression and reducing cancer cell survival in several different types of cancer, including lung, breast[14] and brain[15, 16]. Interestingly, while germline deletion of Hexokinase II is embryonic lethal in mice, whole body knockout in adult mice was reported well tolerated[14, 16], demonstrating that cancer cells may selectively rely on this isoform that could allow therapeutic targeting of Hexokinase II in cancer. Small molecules that broadly inhibit hexokinase, such as 2-deoxyglucose (2-DG), have been shown to have activity against cancer in vitro[17-19] although in vivo efficacy of 2-DG as a single agent is modest[8, 20]. However, these compounds are not specific for a particular hexokinase isoform, and continued development of small molecules targeted at hexokinase isoforms overexpressed in cancer may provide improved specificity.
An important early step in glycolysis is the conversion of fructose-6-phosphate to fructose-1,6-bisphosphate by 6-phosphofructo-1-kinase (PFK1). This is the first committed step in glycolysis, and the activity of PFK1 is elevated in many types of cancer, allowing for increased flux of glucose into the glycolytic pathway [21, 22]. The mechanism of increased PFK1 activity in cancer relies upon the generation of an allosteric activator of PFK1. Oncogenic signaling increases the expression of an isoform of the 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase (PFKFB) family of enzymes known as PFKFB3[23, 24]. Increased PFKFB3 expression results in the production of fructose-2,6,-bisphosphate, a potent allosteric activator of PFK1[25, 26]. Studies suggest that inhibition of PFKFB3 using genetic approaches[27] and small molecule inhibition[28] results in dramatically reduced glycolytic flux and slowed cancer cell growth. Early phase clinical trials are currently underway with small molecule PFKFB3 inhibitors [29].
Another key step in glucose metabolism is the branch point at which glycolysis-derived pyruvate can either be imported into the mitochondria to be oxidized in the TCA cycle, or converted to lactate in the cytosol. The pyruvate dehydrogenase (PDH) complex, which converts pyruvate to acetyl-CoA in the mitochondria, is responsible for regulating this key junction in pyruvate fate. An important regulator of PDH activity is pyruvate dehydrogenase kinase (PDHK). PDHK reduces the activity of PDH via inhibitory phosphorylation[30], resulting in decreased flux of pyruvate into the mitochondria, and increased production of lactate[31]. Several isoforms of PDHK have been shown to be overexpressed in various cancers [32-34], and play an important role in maintaining aerobic glycolysis in tumors. Numerous studies have shown that the inhibition of PDHK through RNAi or a small molecule inhibitor, dichloroacetate (DCA), caused cancer cell death in vitro and improved outcome in in vivo models of disease[35-37]. DCA was shown to alter the energetic balance of cancer cells, promoting the oxidation of glucose and consequent production of ROS[36-39]. DCA has been utilized clinically for the treatment of lactic acidosis[40], and several clinical trials have explored DCA as an anti-cancer treatment. In a small clinical trial, DCA treatment was associated with radiological regression of glioblastoma multiforme (GBM) in some patients, along with reduced proliferation and increased apoptosis of cancer cells[38]. Targeting PDHK with DCA or other novel small molecule inhibitors may be an effective strategy for the inhibition of aerobic glycolysis.
The lactate dehydrogenase (LDH) complex also plays a key role to regulate the fate of pyruvate in cancer. LDH is responsible for the conversion of pyruvate to lactate in the cytosol of the cell, and has increased expression and activity in a variety of cancer types[41, 42]. There are two isoforms of LDH that form tetramers of mixed composition[43] and increased presence of the LDHa isoform is often implicated in contributing to aerobic glycolysis in cancer cells[41, 42, 44]. Of the isoforms of LDH, LDHa has the highest affinity for pyruvate, along with the highest Vmax for enzymatic activity[45]. Thus, LDHa is able to rapidly convert pyruvate into lactate, completing aerobic glycolysis. There are several hypothesized reasons for cancer cells to overexpress LDHa and to convert pyruvate to lactate. The reaction catalyzed by LDHa results in the production of NAD+, which is critical for maintaining the activity of other proteins in the glycolytic pathway such as GAPDH[45, 46]. Also, studies have shown that LDHa activity is critical for keeping a favorable redox environment in cancer cells[47]. Several research groups have shown that the inhibition of LDHa by small molecule inhibitors or genetic approaches results in slowed cancer cell growth and increased cell death in a variety of types of cancer settings, including hepatocellular carcinoma and breast cancer[47-49]. There have been several early stage clinical trials to evaluate the efficacy of a non-specific inhibitor of LDH, with mixed results observed[50, 51]. The pre-clinical development of inhibitors that have more specificity for LDHa is currently ongoing [52, 53].
Glutamine metabolism in cancer
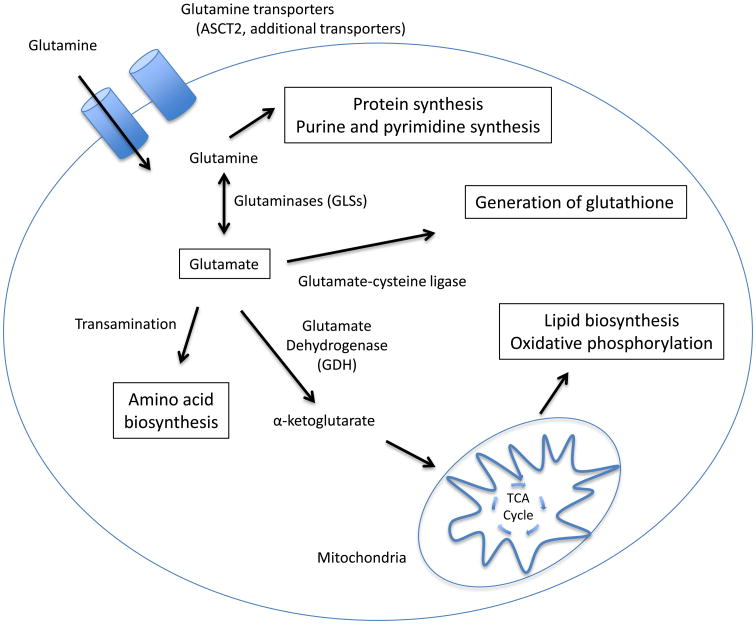
In addition to altered glucose metabolism cancer cells can have increased usage of and reliance on glutamine for cell growth and survival. Dramatically increased usage of glutamine is a metabolic phenotype often associated with oncogenic Myc signaling[54], but is also found in tumors with other driver mutations such as oncogenic KRAS[55]. Glutamine serves several purposes to tumor cells (Figure 2). The first step in glutamine metabolism is its import into the cell via glutamine transporters. There are several known transporters that are capable of taking up glutamine, with SLC1A5 (ASCT2) and LAT1 being commonly upregulated in malignancies[56-58]. Once cytosolic, glutamine can be used as a substrate for the de novo synthesis of proteins, purines and pyrimidines[59], or can be converted to glutamate by enzymes called glutaminases (GLS). After conversion, glutamine derived glutamate can then be utilized by cancer cells for a variety of important purposes[60, 61]. One of these is the generation of non-essential amino acids for growth and proliferation through transamination of glutamate. Glutamate also plays an important role as a carbon donor in cancer cell TCA flux. Glutamate can be converted to α-ketoglutarate by glutamate dehydrogenase (GDH) and then fluxed into the TCA cycle where it can be used to support oxidative phosphorylation, the production of lipids, or to replenish key intermediates such as oxaloacetate[62, 63]. Glutamate can also be used to produce reducing agents for the cell, either being converted into glutathione[64], or generating NADPH through malic enzyme[65].
Figure 2. Glutamine is used for a number of anabolic processes by cancer cells.
Glutamine is transported inside tumor cells by glutamine transporters such as ASCT2. Once inside the cell, it can be used directly as a substrate for protein and nucleotide synthesis, or converted to glutamate by glutaminases (GLS). Glutamine-derived glutamate has a number of uses for cancer cells. It may be used to generate amino acids via transamination, or used in the generation of reducing equivalents such as glutathione. Additionally, glutamate may be converted to α-ketoglutarate by glutamate dehydrogenase (GDH) and fluxed into the mitochondrial TCA cycle where it can be used to support oxidative phosphorylation, or used to generate lipids.
Targeting glutamine metabolism
The increased reliance by some cancers on glutamine metabolism provides several targets for therapeutic intervention. Several groups have explored the inhibition of glutamine transporters to limit glutamine uptake into cancer cells. LAT1 is inhibited by the small molecule inhibitor 2-amino-(2,2,1)-heptane-2-carboxylic acid (BCH), and the treatment of cancer cells in vitro and in vivo with BCH or genetic knockdown of LAT1 has been shown to slow proliferation and tumor growth[66, 67]. The inhibition of ASCT2 by RNAi or the small molecule L-γ-glutamyl-p-nitroanilide (GPNA) has also been shown to decrease pro-growth mTOR signaling and to induce autophagy in cancer cells[68]. Another study found that ASCT2 inhibition caused reduced growth and viability in several subtypes of lung cancer cells, effects that were mediated through a reduction in mTOR pathway activity [69]. Another potential therapeutic target in glutamine metabolism is GLS. A number of small molecule inhibitors of GLS have been developed, among them bis-2-(5-phenylacetamido-1,2,4-thiodiazol-2-yl)ethyl sulfide (BPTES). BPTES has been shown to successfully inhibit GLS activity in several cancer settings, resulting in slowed growth and cell death [70, 71]. There has also been interest in limiting the process of glutamine entering the TCA cycle as α-ketoglutarate by inhibiting GDH. Currently, there are no small molecule inhibitors that are specific for GDH[61]. However, non-specific inhibitors of GDH such as epigallocatechin gallate (EGCG) and aminooxyacetate (AOA) have been successfully demonstrated to be toxic to cancer cells in vitro, and to slow the growth of xenografted tumors[72, 73]. The development of more specific inhibitors of GDH may allow for more efficacious targeting of glutamine flux into the TCA cycle.
Alterations to the TCA cycle in cancer metabolism
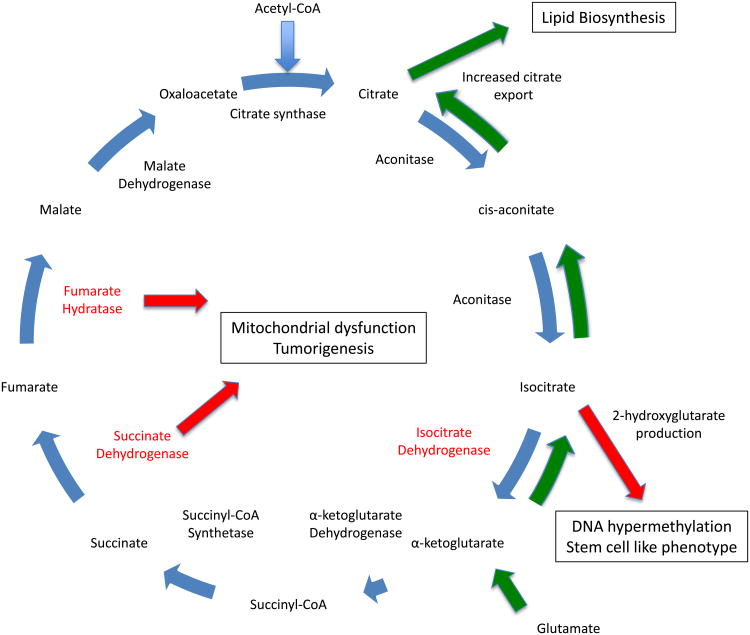
In addition to utilizing aerobic glycolysis and glutamine metabolism for proliferation and survival, it has become clear in recent years that some cancer cells extensively also alter normal TCA cycle metabolism towards these ends (Figure 3). Typically thought of as acting in support of oxidative mitochondrial metabolism, the TCA cycle can also be adapted to produce cell building blocks for proliferation. As one example, citrate produced from acetyl CoA in the TCA cycle can be exported from the mitochondria and converted into raw material for the synthesis of fatty acids that are needed for cell proliferation. The TCA cycle flow can be reversed in reductive carboxylation so that α-ketoglutarate is converted to isocitrate then citrate for lipid synthesis[74, 75]. Also, interestingly, mutations that contribute to oncogenesis and the maintenance and progression of established tumors have been identified in several TCA enzymes. To date, several mutations have been identified in TCA cycle enzymes, including succinate dehydrogenase (SDH), fumarate hydratase (FH) and isocitrate dehydrogenase (IDH). SDH and FH have come to be thought of as tumor suppressors, as mutations in either enzyme has been shown to cause sarcomas, renal cell carcinoma and other rare types of cancer[76-78]. IDH mutations are found in gliomas[79, 80] and acute myeloid leukemias[81], and evidence implicates them in other cancer settings[82-85]. These mutations result in a gain of function, allowing IDH to begin producing a new “oncometabolite” called (R)-2-hydroxyglutarate (2HG)[86, 87]. 2HG itself has been termed an oncometabolite and has the capacity to transform immortalized cells in vitro[88, 89], through mechanisms that remain somewhat unclear. Numerous studies provide evidence that increased 2HG in cells harboring IDH mutations can inhibit demethylases, leading to hypermethylated DNA and retention of a stem cell-like phenotype[89-92].
Figure 3. Cancer cells alter the TCA cycle to support proliferation and oncogenic transformation.
Tumor cells often significantly alter the flow of TCA cycle intermediates (common alterations indicated with green arrows) to increase the generation of substrates useful for cell growth and proliferation. One common alteration in TCA cycle flux is the increased export of citrate from the TCA cycle to support de novo lipid synthesis for proliferation. Along with simply increasing the amount of citrate that is exported, some cancer cells also utilize glutamine-derived glutamate to generate citrate. In this process, glutamate is converted to α-ketoglutarate and fluxed into the TCA cycle. The TCA cycle flow is then reversed, with α-ketoglutarate being converted into isocitrate and eventually citrate to yield even more substrate for lipid synthesis. Tumor cells also are known to have mutations in key TCA cycle enzymes (enzymes known to be mutated indicated in red). Isocitrate dehydrogenase mutations can result in the generation of the “oncometabolite” 2-hydroxyglutarate, which contributes the a stem cell like phenotype in tumor cells. Additional mutations have been identified in fumarate hydratase and succinate dehydrogenase. These mutations result in mitochondrial dysfunction and contribute to oncogenic transformation.
Targeting the TCA cycle in cancer metabolism
Small molecule based targeting of abnormalities in the TCA cycle has become one of the greatest successes to date in therapeutically attacking cancer metabolism. While success targeting mutant FH and SDH with small molecule inhibitors has been limited because these are loss of function mutations, novel compounds that inhibit the gain-of-function activity of mutant IDH have recently been shown to have success in preclinical and clinical settings. In preclinical studies, small molecule inhibition of mutant IDH has been shown to dramatically reduce the production of 2HG and cause cancerous cells to differentiate towards a more normal phenotype[93, 94]. Early phase clinical trials have begun with a small molecule inhibitor of mutant IDH2, AG-221.
Challenges of targeting cancer metabolism
While there are numerous potential therapeutic targets in cancer metabolism, there are profound challenges associated with utilizing metabolic inhibition as a clinical strategy. First among these challenges is the fact that it is difficult to achieve a therapeutic window in cancer metabolism, as many normal cells, especially rapidly proliferating cells of the immune system, also utilize metabolic programs similar to those utilized by cancer and toxicity could be significant by targeting some metabolic pathways. For example, effector subtypes of T cells and antibody producing B cells also rely on aerobic glycolysis[95-97] and glutamine metabolism[98, 99] to maintain immune function. Metabolic inhibition of immune cells could potentially reduce their ability to fight cancer, and further, leave patients more vulnerable to opportunistic infections. Another challenge in targeting cancer metabolism is the metabolic flexibility that many tumor cells exhibit. Except for cases where there are actual mutations in metabolic genes, cancer cells often have a remarkable ability to shift fuel sources when deprived of favored metabolic pathways[8, 73, 100, 101]. This metabolic flexibility may limit the efficacy of targeting a single pathway for therapeutic purposes.
Combination therapy as a solution
One potential way to overcome the challenges posed to successfully utilize cancer metabolism as a therapeutic target is to utilize combination therapy. There are several potential ways in which metabolic combination therapy could be used against cancer cells. Inhibiting a primary metabolic pathway, followed by the subsequent inhibition of alternate metabolic pathways used by tumor cells might be one strategy. Additionally, many groups have shown that the partial inhibition of metabolic pathways utilized by cancer cells can dramatically sensitize the cancer cells to more traditional chemotherapeutic drugs or targeted therapies[8, 102-105]. This adjuvant metabolic sensitization may allow the use of far lower doses of both metabolic inhibitors and chemotherapeutic agents to achieve greater efficacy against tumor cells and reduced off-target effects.
Conclusions
The therapeutic potential of targeting the alterations of cellular metabolism in cancer has existed since the description of aerobic glycolysis by Otto Warburg. In the decades since Warburg's observation, much progress has been made in understanding exactly how many types of cancer alter cellular energetic pathways and how these alterations may be used to design novel therapeutic strategies to combat the disease. It is clear from recent research that there are a number of potential pathways and targets that may be beneficial targets for cancer therapy. In this review, we have described potential targets in the metabolic pathways that regulate glycolysis, glutamine metabolism, and the TCA cycle. It is likely that research in the coming years will identify more potential targets for therapeutic intervention. While there are numerous challenges associated with targeting cancer metabolism, among them off-target effects of metabolic inhibitors and the suppression of immune cells, strategies such as using metabolic inhibitors in combination therapy may allow for more effective clinical use.
Acknowledgments
The authors wish to thank members of the Rathmell lab for helpful discussions during the preparation of this manuscript. This work was supported by NIH R01CA123350 (JCR), NIH R03AI106835 (JCR), Leukemia and Lymphoma Society (JCR), NIH F31CA183529 (RJK), Duke Cancer Institute Pilot Grant (JCR).
Glossary
- 2-DG
2-deoxyglucose
- 2HG
2-hydroxyglutarate
- AOA
aminoxyacetate
- ATP
adenosine triphosphate
- B-ALL
B cell acute lymphoblastic leukemia
- BCH
2-amino-(2,2,1)-heptane-2-carboxylic acid
- BPTES
bis-2-(5-phenylacetamido-1,2,4-thiodiazol-2-yl)ethyl sulfide
- DCA
dichloroacetate
- EGCG
epigallocatechin gallate
- FH
fumarate hydratase
- GAPDH
glyceraldehyde 3-phosphate dehydrogenase
- GBM
glioblastoma multiforme
- GDH
glutamate dehydrogenase
- GLS
glutaminase
- GPNA
L-γ-glutamyl-p-nitroanilide
- IDH
isocitrate dehydrogenase
- LAT1
large neutral amino acid transporter
- LDH
lactate dehydrogenase
- mTOR
mechanistic target of rapamycin
- NAD+
nicotinamide adenine dinucleotide
- NADPH
nicotinamide adenine dinucleotide phosphate
- NSCLC
non-small cell lung cancer
- PDH
pyruvate dehydrogenase
- PDHK
pyruvate dehydrogenase kinase
- PFK1
phosphofructokinase 1
- PFKFB
6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase
- RNAi
RNA interference
- ROS
reactive oxygen species
- SDH
succinate dehydrogenase
- SLC1A5 (ASCT2)
neutral amino acid transporter B(0)/ASC amino acid transporter 2
- TCA cycle
tricarboxylic acid cycle
Footnotes
Conflicts of Interest: The authors declare no conflicts of interest.
References
- 1.Warburg O. On the origin of cancer cells. Science. 1956;123(3191):309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 2.DeBerardinis RJ, et al. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008;7(1):11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 3.Jones RG, Thompson CB. Tumor suppressors and cell metabolism: a recipe for cancer growth. Genes Dev. 2009;23(5):537–48. doi: 10.1101/gad.1756509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Younes M, et al. Wide expression of the human erythrocyte glucose transporter Glut1 in human cancers. Cancer Res. 1996;56(5):1164–7. [PubMed] [Google Scholar]
- 5.Chan DA, et al. Targeting GLUT1 and the Warburg effect in renal cell carcinoma by chemical synthetic lethality. Sci Transl Med. 2011;3(94):94ra70. doi: 10.1126/scitranslmed.3002394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Flavahan WA, et al. Brain tumor initiating cells adapt to restricted nutrition through preferential glucose uptake. Nat Neurosci. 2013;16(10):1373–82. doi: 10.1038/nn.3510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith TA. Mammalian hexokinases and their abnormal expression in cancer. Br J Biomed Sci. 2000;57(2):170–8. [PubMed] [Google Scholar]
- 8.Liu T, et al. Glucose transporter 1-mediated glucose uptake is limiting for B-cell acute lymphoblastic leukemia anabolic metabolism and resistance to apoptosis. Cell Death Dis. 2014;5:e1516. doi: 10.1038/cddis.2014.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu Y, et al. A small-molecule inhibitor of glucose transporter 1 downregulates glycolysis, induces cell-cycle arrest, and inhibits cancer cell growth in vitro and in vivo. Mol Cancer Ther. 2012;11(8):1672–82. doi: 10.1158/1535-7163.MCT-12-0131. [DOI] [PubMed] [Google Scholar]
- 10.Murata H, Hruz PW, Mueckler M. The mechanism of insulin resistance caused by HIV protease inhibitor therapy. J Biol Chem. 2000;275(27):20251–4. doi: 10.1074/jbc.C000228200. [DOI] [PubMed] [Google Scholar]
- 11.McBrayer SK, et al. Multiple myeloma exhibits novel dependence on GLUT4, GLUT8, and GLUT11: implications for glucose transporter-directed therapy. Blood. 2012;119(20):4686–97. doi: 10.1182/blood-2011-09-377846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Flier JS, et al. Distribution of glucose transporter messenger RNA transcripts in tissues of rat and man. J Clin Invest. 1987;79(2):657–61. doi: 10.1172/JCI112864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klepper J, Voit T. Facilitated glucose transporter protein type 1 (GLUT1) deficiency syndrome: impaired glucose transport into brain-- a review. Eur J Pediatr. 2002;161(6):295–304. doi: 10.1007/s00431-002-0939-3. [DOI] [PubMed] [Google Scholar]
- 14.Patra KC, et al. Hexokinase 2 is required for tumor initiation and maintenance and its systemic deletion is therapeutic in mouse models of cancer. Cancer Cell. 2013;24(2):213–28. doi: 10.1016/j.ccr.2013.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wolf A, et al. Hexokinase 2 is a key mediator of aerobic glycolysis and promotes tumor growth in human glioblastoma multiforme. J Exp Med. 2011;208(2):313–26. doi: 10.1084/jem.20101470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gershon TR, et al. Hexokinase-2-mediated aerobic glycolysis is integral to cerebellar neurogenesis and pathogenesis of medulloblastoma. Cancer Metab. 2013;1(1):2. doi: 10.1186/2049-3002-1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ciavardelli D, et al. Breast cancer stem cells rely on fermentative glycolysis and are sensitive to 2-deoxyglucose treatment. Cell Death Dis. 2014;5:e1336. doi: 10.1038/cddis.2014.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang XD, et al. Effect of 2-deoxy-D-glucose on various malignant cell lines in vitro. Anticancer Res. 2006;26(5A):3561–6. [PubMed] [Google Scholar]
- 19.Coloff JL, et al. Akt-dependent glucose metabolism promotes Mcl-1 synthesis to maintain cell survival and resistance to Bcl-2 inhibition. Cancer Res. 2011;71(15):5204–13. doi: 10.1158/0008-5472.CAN-10-4531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maschek G, et al. 2-deoxy-D-glucose increases the efficacy of adriamycin and paclitaxel in human osteosarcoma and non-small cell lung cancers in vivo. Cancer Res. 2004;64(1):31–4. doi: 10.1158/0008-5472.can-03-3294. [DOI] [PubMed] [Google Scholar]
- 21.Kole HK, et al. Regulation of 6-phosphofructo-1-kinase activity in ras-transformed rat-1 fibroblasts. Arch Biochem Biophys. 1991;286(2):586–90. doi: 10.1016/0003-9861(91)90084-v. [DOI] [PubMed] [Google Scholar]
- 22.Hennipman A, et al. Glycolytic enzymes in breast cancer, benign breast disease and normal breast tissue. Tumour Biol. 1987;8(5):251–63. doi: 10.1159/000217529. [DOI] [PubMed] [Google Scholar]
- 23.Bobarykina AY, et al. Hypoxic regulation of PFKFB-3 and PFKFB-4 gene expression in gastric and pancreatic cancer cell lines and expression of PFKFB genes in gastric cancers. Acta Biochim Pol. 2006;53(4):789–99. [PubMed] [Google Scholar]
- 24.Atsumi T, et al. High expression of inducible 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase (iPFK-2; PFKFB3) in human cancers. Cancer Res. 2002;62(20):5881–7. [PubMed] [Google Scholar]
- 25.Van Schaftingen E, Hue L, Hers HG. Fructose 2,6-bisphosphate, the probably structure of the glucose- and glucagon-sensitive stimulator of phosphofructokinase. Biochem J. 1980;192(3):897–901. doi: 10.1042/bj1920897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Van Schaftingen E, et al. Control of liver 6-phosphofructokinase by fructose 2,6-bisphosphate and other effectors. Proc Natl Acad Sci U S A. 1981;78(6):3483–6. doi: 10.1073/pnas.78.6.3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Telang S, et al. Ras transformation requires metabolic control by 6-phosphofructo-2-kinase. Oncogene. 2006;25(55):7225–34. doi: 10.1038/sj.onc.1209709. [DOI] [PubMed] [Google Scholar]
- 28.Clem B, et al. Small-molecule inhibition of 6-phosphofructo-2-kinase activity suppresses glycolytic flux and tumor growth. Mol Cancer Ther. 2008;7(1):110–20. doi: 10.1158/1535-7163.MCT-07-0482. [DOI] [PubMed] [Google Scholar]
- 29.Clem BF, et al. Targeting 6-phosphofructo-2-kinase (PFKFB3) as a therapeutic strategy against cancer. Mol Cancer Ther. 2013;12(8):1461–70. doi: 10.1158/1535-7163.MCT-13-0097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yeaman SJ, et al. Sites of phosphorylation on pyruvate dehydrogenase from bovine kidney and heart. Biochemistry. 1978;17(12):2364–70. doi: 10.1021/bi00605a017. [DOI] [PubMed] [Google Scholar]
- 31.Sugden MC, Holness MJ. Recent advances in mechanisms regulating glucose oxidation at the level of the pyruvate dehydrogenase complex by PDKs. Am J Physiol Endocrinol Metab. 2003;284(5):E855–62. doi: 10.1152/ajpendo.00526.2002. [DOI] [PubMed] [Google Scholar]
- 32.Hur H, et al. Expression of pyruvate dehydrogenase kinase-1 in gastric cancer as a potential therapeutic target. Int J Oncol. 2013;42(1):44–54. doi: 10.3892/ijo.2012.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koukourakis MI, et al. Pyruvate dehydrogenase and pyruvate dehydrogenase kinase expression in non small cell lung cancer and tumor-associated stroma. Neoplasia. 2005;7(1):1–6. doi: 10.1593/neo.04373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McFate T, et al. Pyruvate dehydrogenase complex activity controls metabolic and malignant phenotype in cancer cells. J Biol Chem. 2008;283(33):22700–8. doi: 10.1074/jbc.M801765200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sanchez-Arago M, Chamorro M, Cuezva JM. Selection of cancer cells with repressed mitochondria triggers colon cancer progression. Carcinogenesis. 2010;31(4):567–76. doi: 10.1093/carcin/bgq012. [DOI] [PubMed] [Google Scholar]
- 36.Bonnet S, et al. A mitochondria-K+ channel axis is suppressed in cancer and its normalization promotes apoptosis and inhibits cancer growth. Cancer Cell. 2007;11(1):37–51. doi: 10.1016/j.ccr.2006.10.020. [DOI] [PubMed] [Google Scholar]
- 37.Sutendra G, et al. Mitochondrial activation by inhibition of PDKII suppresses HIF1a signaling and angiogenesis in cancer. Oncogene. 2013;32(13):1638–50. doi: 10.1038/onc.2012.198. [DOI] [PubMed] [Google Scholar]
- 38.Michelakis ED, et al. Metabolic modulation of glioblastoma with dichloroacetate. Sci Transl Med. 2010;2(31):31ra34. doi: 10.1126/scitranslmed.3000677. [DOI] [PubMed] [Google Scholar]
- 39.Kaplon J, et al. A key role for mitochondrial gatekeeper pyruvate dehydrogenase in oncogene-induced senescence. Nature. 2013;498(7452):109–12. doi: 10.1038/nature12154. [DOI] [PubMed] [Google Scholar]
- 40.Stacpoole PW, Nagaraja NV, Hutson AD. Efficacy of dichloroacetate as a lactate-lowering drug. J Clin Pharmacol. 2003;43(7):683–91. [PubMed] [Google Scholar]
- 41.Koukourakis MI, et al. Lactate dehydrogenase 5 expression in squamous cell head and neck cancer relates to prognosis following radical or postoperative radiotherapy. Oncology. 2009;77(5):285–92. doi: 10.1159/000259260. [DOI] [PubMed] [Google Scholar]
- 42.Koukourakis MI, et al. Lactate dehydrogenase 5 (LDH5) relates to up-regulated hypoxia inducible factor pathway and metastasis in colorectal cancer. Clin Exp Metastasis. 2005;22(1):25–30. doi: 10.1007/s10585-005-2343-7. [DOI] [PubMed] [Google Scholar]
- 43.Markert CL, Shaklee JB, Whitt GS. Evolution of a gene. Multiple genes for LDH isozymes provide a model of the evolution of gene structure, function and regulation. Science. 1975;189(4197):102–14. doi: 10.1126/science.1138367. [DOI] [PubMed] [Google Scholar]
- 44.Koukourakis MI, et al. Lactate dehydrogenase-5 (LDH-5) overexpression in non-small-cell lung cancer tissues is linked to tumour hypoxia, angiogenic factor production and poor prognosis. Br J Cancer. 2003;89(5):877–85. doi: 10.1038/sj.bjc.6601205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dawson DM, Goodfriend TL, Kaplan NO. Lactic Dehydrogenases: Functions of the Two Types Rates of Synthesis of the Two Major Forms Can Be Correlated with Metabolic Differentiation. Science. 1964;143(3609):929–33. doi: 10.1126/science.143.3609.929. [DOI] [PubMed] [Google Scholar]
- 46.Doherty JR, Cleveland JL. Targeting lactate metabolism for cancer therapeutics. J Clin Invest. 2013;123(9):3685–92. doi: 10.1172/JCI69741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Le A, et al. Inhibition of lactate dehydrogenase A induces oxidative stress and inhibits tumor progression. Proc Natl Acad Sci U S A. 2010;107(5):2037–42. doi: 10.1073/pnas.0914433107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fantin VR, St-Pierre J, Leder P. Attenuation of LDH-A expression uncovers a link between glycolysis, mitochondrial physiology, and tumor maintenance. Cancer Cell. 2006;9(6):425–34. doi: 10.1016/j.ccr.2006.04.023. [DOI] [PubMed] [Google Scholar]
- 49.Shim H, et al. c-Myc transactivation of LDH-A: implications for tumor metabolism and growth. Proc Natl Acad Sci U S A. 1997;94(13):6658–63. doi: 10.1073/pnas.94.13.6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu G, et al. An open-label, multicenter, phase I/II study of single-agent AT-101 in men with castrate-resistant prostate cancer. Clin Cancer Res. 2009;15(9):3172–6. doi: 10.1158/1078-0432.CCR-08-2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Baggstrom MQ, et al. A phase II study of AT-101 (Gossypol) in chemotherapy-sensitive recurrent extensive-stage small cell lung cancer. J Thorac Oncol. 2011;6(10):1757–60. doi: 10.1097/JTO.0b013e31822e2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Manerba M, et al. Galloflavin (CAS 568-80-9): a novel inhibitor of lactate dehydrogenase. ChemMedChem. 2012;7(2):311–7. doi: 10.1002/cmdc.201100471. [DOI] [PubMed] [Google Scholar]
- 53.Granchi C, et al. Discovery of N-hydroxyindole-based inhibitors of human lactate dehydrogenase isoform A (LDH-A) as starvation agents against cancer cells. J Med Chem. 2011;54(6):1599–612. doi: 10.1021/jm101007q. [DOI] [PubMed] [Google Scholar]
- 54.Gao P, et al. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature. 2009;458(7239):762–5. doi: 10.1038/nature07823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Son J, et al. Glutamine supports pancreatic cancer growth through a KRAS-regulated metabolic pathway. Nature. 2013;496(7443):101–5. doi: 10.1038/nature12040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fuchs BC, Bode BP. Amino acid transporters ASCT2 and LAT1 in cancer: partners in crime? Semin Cancer Biol. 2005;15(4):254–66. doi: 10.1016/j.semcancer.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 57.Witte D, et al. Overexpression of the neutral amino acid transporter ASCT2 in human colorectal adenocarcinoma. Anticancer Res. 2002;22(5):2555–7. [PubMed] [Google Scholar]
- 58.Collins CL, et al. Determinants of glutamine dependence and utilization by normal and tumor-derived breast cell lines. J Cell Physiol. 1998;176(1):166–78. doi: 10.1002/(SICI)1097-4652(199807)176:1<166::AID-JCP18>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 59.Cory JG, Cory AH. Critical roles of glutamine as nitrogen donors in purine and pyrimidine nucleotide synthesis: asparaginase treatment in childhood acute lymphoblastic leukemia. In Vivo. 2006;20(5):587–9. [PubMed] [Google Scholar]
- 60.Mates JM, et al. Glutaminase isoenzymes as key regulators in metabolic and oxidative stress against cancer. Curr Mol Med. 2013;13(4):514–34. doi: 10.2174/1566524011313040005. [DOI] [PubMed] [Google Scholar]
- 61.Hensley CT, Wasti AT, DeBerardinis RJ. Glutamine and cancer: cell biology, physiology, and clinical opportunities. J Clin Invest. 2013;123(9):3678–84. doi: 10.1172/JCI69600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Reitzer LJ, Wice BM, Kennell D. Evidence that glutamine, not sugar, is the major energy source for cultured HeLa cells. J Biol Chem. 1979;254(8):2669–76. [PubMed] [Google Scholar]
- 63.Anderson ME, Meister A. Transport and direct utilization of gamma-glutamylcyst(e)ine for glutathione synthesis. Proc Natl Acad Sci U S A. 1983;80(3):707–11. doi: 10.1073/pnas.80.3.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Meister A, Anderson ME. Glutathione. Annu Rev Biochem. 1983;52:711–60. doi: 10.1146/annurev.bi.52.070183.003431. [DOI] [PubMed] [Google Scholar]
- 65.Lyssiotis CA, et al. Pancreatic cancers rely on a novel glutamine metabolism pathway to maintain redox balance. Cell Cycle. 2013;12(13):1987–8. doi: 10.4161/cc.25307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kaira K, et al. Clinical significance of L-type amino acid transporter 1 expression as a prognostic marker and potential of new targeting therapy in biliary tract cancer. BMC Cancer. 2013;13:482. doi: 10.1186/1471-2407-13-482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang Q, et al. Targeting amino acid transport in metastatic castration-resistant prostate cancer: effects on cell cycle, cell growth, and tumor development. J Natl Cancer Inst. 2013;105(19):1463–73. doi: 10.1093/jnci/djt241. [DOI] [PubMed] [Google Scholar]
- 68.Nicklin P, et al. Bidirectional transport of amino acids regulates mTOR and autophagy. Cell. 2009;136(3):521–34. doi: 10.1016/j.cell.2008.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hassanein M, et al. SLC1A5 mediates glutamine transport required for lung cancer cell growth and survival. Clin Cancer Res. 2013;19(3):560–70. doi: 10.1158/1078-0432.CCR-12-2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Seltzer MJ, et al. Inhibition of glutaminase preferentially slows growth of glioma cells with mutant IDH1. Cancer Res. 2010;70(22):8981–7. doi: 10.1158/0008-5472.CAN-10-1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Emadi A, et al. Inhibition of glutaminase selectively suppresses the growth of primary acute myeloid leukemia cells with IDH mutations. Exp Hematol. 2014;42(4):247–51. doi: 10.1016/j.exphem.2013.12.001. [DOI] [PubMed] [Google Scholar]
- 72.Qing G, et al. ATF4 regulates MYC-mediated neuroblastoma cell death upon glutamine deprivation. Cancer Cell. 2012;22(5):631–44. doi: 10.1016/j.ccr.2012.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yang C, et al. Glioblastoma cells require glutamate dehydrogenase to survive impairments of glucose metabolism or Akt signaling. Cancer Res. 2009;69(20):7986–93. doi: 10.1158/0008-5472.CAN-09-2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Filipp FV, et al. Reverse TCA cycle flux through isocitrate dehydrogenases 1 and 2 is required for lipogenesis in hypoxic melanoma cells. Pigment Cell Melanoma Res. 2012;25(3):375–83. doi: 10.1111/j.1755-148X.2012.00989.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Scott DA, et al. Comparative metabolic flux profiling of melanoma cell lines: beyond the Warburg effect. J Biol Chem. 2011;286(49):42626–34. doi: 10.1074/jbc.M111.282046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pollard PJ, Wortham NC, Tomlinson IP. The TCA cycle and tumorigenesis: the examples of fumarate hydratase and succinate dehydrogenase. Ann Med. 2003;35(8):632–9. doi: 10.1080/07853890310018458. [DOI] [PubMed] [Google Scholar]
- 77.Gottlieb E, Tomlinson IP. Mitochondrial tumour suppressors: a genetic and biochemical update. Nat Rev Cancer. 2005;5(11):857–66. doi: 10.1038/nrc1737. [DOI] [PubMed] [Google Scholar]
- 78.Raimundo N, Baysal BE, Shadel GS. Revisiting the TCA cycle: signaling to tumor formation. Trends Mol Med. 2011;17(11):641–9. doi: 10.1016/j.molmed.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Balss J, et al. Analysis of the IDH1 codon 132 mutation in brain tumors. Acta Neuropathol. 2008;116(6):597–602. doi: 10.1007/s00401-008-0455-2. [DOI] [PubMed] [Google Scholar]
- 80.Bleeker FE, et al. IDH1 mutations at residue p.R132 (IDH1(R132)) occur frequently in high-grade gliomas but not in other solid tumors. Hum Mutat. 2009;30(1):7–11. doi: 10.1002/humu.20937. [DOI] [PubMed] [Google Scholar]
- 81.Rakheja D, et al. IDH mutations in acute myeloid leukemia. Hum Pathol. 2012;43(10):1541–51. doi: 10.1016/j.humpath.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 82.Terunuma A, et al. MYC-driven accumulation of 2-hydroxyglutarate is associated with breast cancer prognosis. J Clin Invest. 2014;124(1):398–412. doi: 10.1172/JCI71180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Amary MF, et al. IDH1 and IDH2 mutations are frequent events in central chondrosarcoma and central and periosteal chondromas but not in other mesenchymal tumours. J Pathol. 2011;224(3):334–43. doi: 10.1002/path.2913. [DOI] [PubMed] [Google Scholar]
- 84.Amary MF, et al. Ollier disease and Maffucci syndrome are caused by somatic mosaic mutations of IDH1 and IDH2. Nat Genet. 2011;43(12):1262–5. doi: 10.1038/ng.994. [DOI] [PubMed] [Google Scholar]
- 85.Borger DR, et al. Frequent mutation of isocitrate dehydrogenase (IDH)1 and IDH2 in cholangiocarcinoma identified through broad-based tumor genotyping. Oncologist. 2012;17(1):72–9. doi: 10.1634/theoncologist.2011-0386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dang L, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462(7274):739–44. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ward PS, et al. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer Cell. 2010;17(3):225–34. doi: 10.1016/j.ccr.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Losman JA, et al. (R)-2-hydroxyglutarate is sufficient to promote leukemogenesis and its effects are reversible. Science. 2013;339(6127):1621–5. doi: 10.1126/science.1231677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lu C, et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature. 2012;483(7390):474–8. doi: 10.1038/nature10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chowdhury R, et al. The oncometabolite 2-hydroxyglutarate inhibits histone lysine demethylases. EMBO Rep. 2011;12(5):463–9. doi: 10.1038/embor.2011.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Figueroa ME, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18(6):553–67. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Turcan S, et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483(7390):479–83. doi: 10.1038/nature10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Rohle D, et al. An inhibitor of mutant IDH1 delays growth and promotes differentiation of glioma cells. Science. 2013;340(6132):626–30. doi: 10.1126/science.1236062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wang F, et al. Targeted inhibition of mutant IDH2 in leukemia cells induces cellular differentiation. Science. 2013;340(6132):622–6. doi: 10.1126/science.1234769. [DOI] [PubMed] [Google Scholar]
- 95.Macintyre AN, et al. The glucose transporter Glut1 is selectively essential for CD4 T cell activation and effector function. Cell Metab. 2014;20(1):61–72. doi: 10.1016/j.cmet.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Michalek RD, et al. Cutting edge: distinct glycolytic and lipid oxidative metabolic programs are essential for effector and regulatory CD4+ T cell subsets. J Immunol. 2011;186(6):3299–303. doi: 10.4049/jimmunol.1003613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Caro-Maldonado A, et al. Metabolic reprogramming is required for antibody production that is suppressed in anergic but exaggerated in chronically BAFF-exposed B cells. J Immunol. 2014;192(8):3626–36. doi: 10.4049/jimmunol.1302062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Le A, et al. Glucose-independent glutamine metabolism via TCA cycling for proliferation and survival in B cells. Cell Metab. 2012;15(1):110–21. doi: 10.1016/j.cmet.2011.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Nakaya M, et al. Inflammatory T cell responses rely on amino acid transporter ASCT2 facilitation of glutamine uptake and mTORC1 kinase activation. Immunity. 2014;40(5):692–705. doi: 10.1016/j.immuni.2014.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Choo AY, et al. Glucose addiction of TSC null cells is caused by failed mTORC1-dependent balancing of metabolic demand with supply. Mol Cell. 2010;38(4):487–99. doi: 10.1016/j.molcel.2010.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Chen V, Shtivelman E. CC3/TIP30 regulates metabolic adaptation of tumor cells to glucose limitation. Cell Cycle. 2010;9(24):4941–53. doi: 10.4161/cc.9.24.14230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Samudio I, et al. Pharmacologic inhibition of fatty acid oxidation sensitizes human leukemia cells to apoptosis induction. J Clin Invest. 2010;120(1):142–56. doi: 10.1172/JCI38942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zhou M, et al. Warburg effect in chemosensitivity: targeting lactate dehydrogenase-A re-sensitizes taxol-resistant cancer cells to taxol. Mol Cancer. 2010;9:33. doi: 10.1186/1476-4598-9-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ayyanathan K, et al. Combination of sulindac and dichloroacetate kills cancer cells via oxidative damage. PLoS One. 2012;7(7):e39949. doi: 10.1371/journal.pone.0039949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Giacobbe A, et al. p63 regulates glutaminase 2 expression. Cell Cycle. 2013;12(9):1395–405. doi: 10.4161/cc.24478. [DOI] [PMC free article] [PubMed] [Google Scholar]