Abstract
In chronic wounds, biofilm infects host tissue for extended periods of time. This work establishes the first chronic pre-clinical model of wound biofilm infection aimed at addressing long-term host response. Although biofilm infected wounds did not show marked differences in wound closure, the repaired skin demonstrated compromised barrier function. This observation is clinically significant because it leads to the notion that even if a biofilm infected wound is closed as observed visually, it may be complicated by the presence of failed skin which is likely to be infected and or further complicated post-closure. Study of underlying mechanisms recognized for the first time biofilm-inducible miR-146a and miR-106b in the host skin wound-edge tissue. These miRs silenced ZO-1 and ZO-2 to compromise tight junction function resulting in leaky skin as measured by transepidermal water loss. Intervention strategies aimed at inhibiting biofilm-inducible miRNAs may be productive in restoring barrier function of host skin.
Keywords: Wound biofilm, porcine burn wounds, microRNA, TEWL, transepidermal water loss, Mixed species biofilm
INTRODUCTION
In the United States of America, 6.5 million patients are affected by chronic wounds. The national annual healthcare cost of treating chronic wounds is estimated at US$25 billion annually and this burden is rapidly growing due to increasing health care costs, an aging population and a sharp rise in the incidence of diabetes and obesity worldwide [1]. Wound infections are not only the most expensive complications following surgery but a major source of bacteria that drive the nosocomial infection rates in hospitals. Bacteria may establish social networking and structurally organize in aggregates known as biofilms. In the biofilm form, bacteria are encased within an extracellular polymeric substance (EPS) and become recalcitrant to antimicrobials and host defenses, posing a rapidly escalating threat to human health [2]. Biofilms are estimated to account for 60% of chronic wound infections [3].
Current understanding of wound biofilm biology, including bacterial mechanisms of pathogenesis and host responses, is limited by the availability of appropriate chronic models of wound biofilm infection where cascading mechanisms may be studied longitudinally over time. Biofilms have been studied for decades under in vitro conditions [4]. In vivo animal models of biofilm infection have been of short-term acute phase studies lasting from 2-16 days [5-8]. In the case of skin wounds, the porcine model is in agreement with human studies 78% of the time compared to 53% for the small mammal studies [9]. With respect to translational value, the Wound Healing Society (WHS) recommends the use of pig as the preclinical model for wound studies [10]. Preclinical models refer to experimental systems that attempt to most closely mimic a human pathological condition of interest [11]. Wound biofilm has been mostly studied in flies, rodents and rabbits. The porcine model has been also examined albeit for a short term of 2-7 days following induced infection [12]. Host response to biofilm infection is typically chronic and governed by micro-environmental cues present at site of infection [13]. Our interest to understand the host response to chronic infection necessitated development of a wound infection model that is sustained, in this case eight weeks long.
Current knowledge supports the notion that the biology of pathological biofilms is driven not only by interactions between different microorganisms at the site of infection but also by bacteria-host interactions which play a critical role in defining the human disease conditions. Thus, we sought to establish a mixed bacterial species biofilm infection (using Psedumonas aeruginosa PAO1 and Acinetobacter baumannii 19606) on full thickness burn wounds. These microorganisms are known to be associated with battle burn wound casualties [14] contributing to chronicity [15]. This novel experimental system led to the observation that although effects of chronic biofilm infection on wound closure may be marginal over a period of two months, such infection results in failure of the skin barrier function at the site of healing. While wound appears closed by visual assessment, the current standard of care, high epidermal water loss resulting from malfunctioning tight junctions was identified at the site of biofilm infection. Interestingly this effect was only evident after a month of biofilm infection, i.e, when the wound has achieved chronicity [16]. This work reports the first evidence of biofilm-inducible microRNAs that specifically silence tight junction proteins. Observations in this study underscore the clinical need to utilize objective measures of skin function when evaluating healing outcomes of biofilm-infected wounds.
MATERIALS AND METHODS
Ethics Statement
All animal (mouse and pig) experiments were approved by the Ohio State University Institutional Laboratory Animal Care and Use Committee (ILACUC) under protocols 2008A0012-R1 (pig) and 2009A0214-R1 (mouse).
Bacterial Isolates
Pseudomonas aeruginosa strain PAO1[17] with constitutive expression of GFP was obtained from the Mathew Parsek laboratory and Pseudomonas aeruginosa strain Δpsl PAO1 were grown on Pseudomonas Isolation Agar (PIA) plates or Luria broth without sodium chloride (LBNS) at 37°C. Acinetobacter baumannii strain 19606 with spontaneous rifampicin mutation was grown on Luria agar (LA) plates with 100μg/mL rifampicin or LBNS at 37°C.
Porcine Full Thickness Burns and Bacterial Inoculation
A total of 64 domestic Yorkshire pigs were used in this study. See supplemental text for procedural description.
Histology and Imaging
Histopathology was performed as described [18]. In brief, formalin-fixed paraffin-embedded wound sections were deparaffinized and stained with hematoxylin & eosin (H&E), Mason Trichrome and Gram/Twort stain (Newcomer Supply Inc.), using standard procedures. Immunohistochemical staining of OCT embedded frozen- sections were performed as described earlier [18] using the following primary antibodies: anti-Pseudomonas antibody (custom developed by Covance, Denver, PA), anti-Acinetobacter antibody (custom developed by Covance, Denver, PA) and ZO-1 and ZO-2 (Invitrogen). Because of the high auto-fluorescence in porcine tissue and weak GFP signal emitted from our PAO1::gfp strain, we chose to visualize P. aeruginosa with anti-Pseudomonas antibody rather than relying on the GFP signal. Mosaic images were collected using a Zeiss Axiovert 200 inverted fluorescence microscope supported by an AxioCam digital camera, a motorized stage, and guided by Axiovision software (Zeiss). Each mosaic image was generated by combining a minimum of 100 images. Confocal Laser Scanning Microscopy (CLSM) was performed using a Fluoview FV1000 spectral confocal microscope (Olympus, Pittsburgh, PA) equipped with an argon laser. Z-stack images were created by merging serial scans of tissue sections (20-50 μm).
Scanning Electron Microscope Imaging
Samples processing and imaging was performed as described [19]. In brief, following glutaraldehyde fixation and dehydration with graded ethanol, the samples are treated with hexamethyldisilazane (HMDS, Ted Pella Inc.) and left overnight for drying. Before scanning, samples were mounted and coated with gold. Imaging of the samples was done by using a FEI™ NOVA nanoSEM scanning electron microscope (FEI™, Hillsboro, OR) equipped with a field-emission gun electron source.
Surgical Debridement and Laser Doppler Blood Flow Imaging
Debridement was performed using a 0.12 inch weck blade to remove necrotic and infected tissues until bleeding healthy tissue was exposed. Debridement was performed as per standard of care in a clinical setting. The MoorLDI-Mark 2 laser Doppler blood perfusion imager (Moor Instruments Ltd., UK) was used to map tissue blood flow in debrided wound as described previously [18].
Trans-epidermal Water Loss (TEWL)
DermaLab TEWL Probe (cyberDERM inc., Broomall, PA) was used to measure the trans-epidermal water loss from the wounds. TEWL was expressed in g/m2/h.
Laser Capture microdissection (LCM) & Biofilm Gene Expression Analysis
Serial sections were obtained from OCT embedded frozen wound sections, one of the serial sections was stained with anti-Pseudomonas antibody as described in an earlier section for visualization of the biofilm infected area. Other serial sections (10 μm) were stained using a modified hematoxylin QS procedure. The sections were mounted on a PEN (polyethylene napthalate) membrane glass slides (P.A.L.M. Microlaser Technologies AG Germany), that had been RNAsin (Ambion) and UV treated, for cutting and catapulting as described by our group [20]. Corresponding P. aeruginosa stained areas were captured in chaotropic RNA lysis solution [20]. The RNA was extracted using TRIzol® Max™ Bacterial RNA Isolation Kit (Life Technologies) as per manufacturer’s instructions. Reverse transcription and mRNA quantitation using real-time PCR were performed as described [20].
Cell Culture and Transfection
Human immortalized keratinocytes (HaCaT) were grown under standard culture conditions (at 37°C in a humidified atmosphere consisting of 95% air and 5% CO2) in DMEM growth medium supplemented with 10% FBS, 100 IU/ml penicillin, 0.1 mg/ml streptomycin, 10 mmol/l L-glutamine as described previously [21].
In Vitro Static Biofilm Co-culture
Bacterial biofilms were grown on HaCaT cells using a co-culture model system adapted from Anderson et al., 2008 [22]. In brief, the confluent cultures of HaCaT cells were inoculated with bacterial cultures of P. aeruginosa (105 CFU/ml) and A. baumannii (106 CFU/ml) in antibiotic free culture media. The plates were incubated for 1 h at 37°C and 5% CO2. After 1 h, the supernatant was replaced with fresh DMEM (without antibiotics) supplemented with 0.4% arginine. In vitro biofilm culture on polycarbonate membrane was adapted from Zhao et al., 2010 [23]. The cells were further incubated in regular culture conditions for specified time as described in figures.
Western Blot Analyses
Western blot was performed using antibodies against ZO1 (Invitrogen), ZO2 (Invitrogen) and β- actin (Sigma) as described previously [21].
RNA Isolation and Quantitative Real Time PCR
Total RNA, including the miRNA fraction, was isolated using mirVana miRNA isolation kit, according to the manufacturer’s protocol (Ambion). Specific Taqman assays for miRNA (Applied Biosystems) and mirVana qRT-PCR miRNA RT Kit (Applied Biosystems) were used with real-time PCR system and Taqman universal master mix. Levels of miRNA were quantified with the relative quantification method using U6 small nuclear RNA as the housekeeping. The transcription levels of ZO1 and ZO2 and house-keeping control GAPDH was quantified using SYBR green-I(Applied Biosystems). Expression levels of miRNA and mRNA were quantified employing the 2(−ΔΔct) relative quantification method [21]. Primer sequences have been provided as supplementary information.
miR-Target 3′-UTR Luciferase Reporter Assay
miRIDIAN mimic-miR-146a or −106b were transfected to HaCaT cells followed by transfection with with pmiR Target-ZO-1-3′-UTR or pmiR Target-ZO-2-3′-UTR plasmids. Luciferase assay were performed using the dual-luciferase reporter assay system (Promega) as described. Normalization was achieved by co-transfection with Renilla plasmid [24]. Data are presented as ratio of firefly to Renilla luciferase.
Immunocytochemistry
Cells were fixed with IC fixation buffer (eBioscience), blocked and incubated with primary antibody against ZO1 (1: 100) or ZO2 (1:100) overnight at 4°C. Signal was visualized using FITC-tagged α-rabbit,1:200 (Invitrogen) and counterstained with DAPI (Invitrogen 1:10,000) [25].
miR Mimic Delivery on Mice Skin
Male C57BL/6 mice (age, 8 weeks old) were used. miRNA mimics, 3 μg per 5μl (miR-146a mimic, miR-106b mimic and non-targeting miRNA mimic C.elegans miR-67 as negative control, Dharmacon) were incubated with 5 μl of siPort transfection reagent (Ambion) for 30 min and mixed with 10 μl cream. Composition of the cream was as described [26]. The miRNA mix was rubbed onto a marked (8mmX16mm) and naired dorsal area. The mimics were applied daily for a period of ten days. Before the application of the miRNA mimics, TEWL measurements of the mice skin were taken using Dermalab Series Skinlab Combo (Cortex Technologies). The animals were sacrificed at day 11 and the skin where the mimic was applied was harvested. All of the animal studies were performed in accordance with protocols approved by the Laboratory Animal Care and Use Committee of the Ohio State University.
Statistics
In vitro data are reported as mean ± SD of 3-6 experiments as indicated in respective figure legends. Comparisons among multiple groups were tested using analysis of variance (ANOVA). p<0.05 was considered statistically significant. For animal studies, data are reported as mean ± SD of at least 3-4 animals as indicated. Given the small sample size, Mann–Whitney or Kruskal–Wallis one-way analysis of variance tests was performed to test significance (p<0.05) of difference between means.
RESULTS
Establishment and characterization of mixed-species full-thickness wound biofilm model
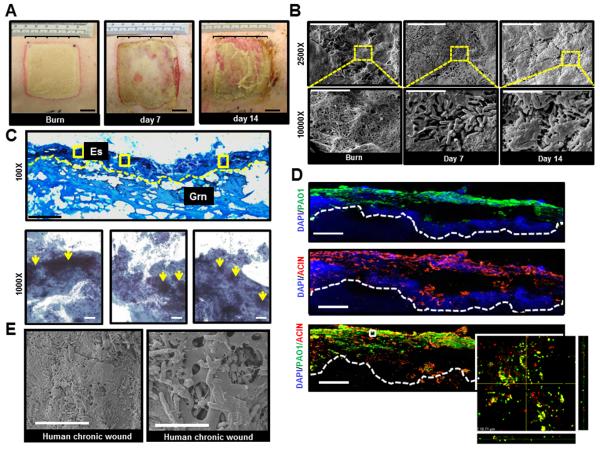
The first objective of this work was to develop a pre-clinical, large animal model that would enable our understanding of underlying impaired host tissue responses implicated in healing of biofilm infected burn wounds. We developed a microprocessor controlled electrically heated burning device to create uniform, full-thickness thermal injuries of a defined size and depth (Fig S1). A mixed bacterial species infection approach was adopted, combining Acinetobacter baumannii 19606 and Pseudomonas aeruginosa PAO1 to co-infect burn wounds and establish a clinically relevant mixed-species biofilm infection (Fig 1). These microorganisms are highly relevant as they are typically associated with battle burn wound casualties [14]. Following induced infection (II group, representing clinically infected wounds; spontaneously infected wounds or SI group served as controls; see Methods), inspection of the wounds revealed yellowish green discoloration with discharge that increased with time (Fig 1A). Histological and microbiological analyses confirmed bacterial colonization of the wound bed (Fig 1C). Direct microscopic visualization of A. baumannii and P. aeruginosa colonies revealed the presence of both organisms in burn wounds seven days post-inoculation. Occasionally, co-localization of both organisms in the wound bed was noted supporting the establishment of mixed species biofilm infection (Fig 1D). Imaging of burn wound biopsies with scanning electron microscope (SEM) demonstrated aggregates of bacteria attached to the surface of the burn wounds that were embedded in an EPS indicating biofilm matrix (Fig 1B) similar to that in clinical pressure ulcer wound SEM micrographs (Fig 1E). Thus, our newly developed preclinical mixed infection biofilm model replicates key histologic characteristics when compared to human clinical chronic wound samples.
Figure 1. Establishment of mixed-species biofilm infection in a full thickness porcine burn wound model.
Six 2 × 2 sq inch size burn wounds were created with a burning device (Fig S1). Co- infection of the burn wounds was performed day 3 post-burn with P. aeruginosa PAO1 and A. baumannii 19606. A, representative digital photograph of wounds on the day of burn, days 7 and 14 post-infection. Note signs of active infection with localized erythema, yellowish exudates, and friable wound edges on days 7 and 14 post-infection. B, scanning electron microscope (SEM) images of biopsies collected from the wounds on the day of burn and days 7 & 14 post-infection. The images clearly demonstrate a mix of extracellular tissue matrix, fibers, and red blood cells over the surface of burn wounds before bacterial inoculation. Large aggregates of rods attached to the wound surface encased in a layer of extracellular amorphous material was noted on days 7 & 14 post-infection. Upper panel, scale bar = 20 μm, 2500× magnification. Lower panel, magnification of the red dashed boxes in the upper panels. Scale bar = 5 μm, 10000× magnification. C, Gram stained images of inoculated wounds shows presence of bacterial aggregates. Upper panel, representative mosaic image of a day 7 post infection wound. Images collected under 400× magnification using microscope supported with a motorized stage. Scale bar = 200 μm. Es= eschar, Grn= granulation tissue. Lower panels, Zoom of three boxed area in upper panel image showing Gram negative bacilli & coccobacilli over the surface of the burn wound while large Gram negative clumps colonizing the wound tissues (yellow arrows). Scale bar = 50 μm. D, both P. aeruginosa and A. baumannii on the burn wounds were visualized using anti-pseudomonas (green) and anti-acinetobactor (red) antibody and confocal laser scanning (CLSM) microscopy. Merged (red & green) immunofluorescence images of day 7 (post-infection) wound biopsies show heavy colonization of wound tissues with both strains. Mosaic images were collected under 400× magnification using fluorescent microscope supported with a motorized stage. Scale bar = 100 μm. Z-stack images of boxed areas of the lower panel surface of burn wound tissues. Inset, zoom of the boxed area in merged image. The image was created by merging serial scans of thick tissue section (20 μm), viewed under 600× magnification in a confocal laser scanning microscope (CLSM). Dense micro-colonies of P. aeruginosa and A. baumannii with some co-localization in the wound tissues were noted. x/z and y/z planes display the thickness of the microbial clumps within the tissue section. E, representative SEM image of wound biopsies from human chronic pressure ulcers showing that the bacterial colonization and biofilm established in experimental porcine biofilm model is comparable to that of human chronic wounds. left panel, scale bar = 20 μm, 2500× magnification. Right panel, scale bar = 5 μm, 10000× magnification.
In addition to bacteria embedded in EPS, criteria for defining biofilm infection include: i) adherence to a surface or each other; ii) persistent and localized infection; and iii) resistance to anti-microbial treatments [27]. Data that support these criteria are indicated here: i) Adherence to surface. A scrubbing technique was used to test adherence of biofilm to wound surface [12, 28]. This technique is known to remove free (planktonic) bacteria from burn wounds. Scrubbing was performed with double open ended sterile plastic tubes; sterile saline was added thrice followed by once with a detergent (4% Tween 80 v/v). Microbiology analyses revealed that bacterial counts did not significantly change before or after scrubbing suggesting strong bacterial adherence to the wound (not shown). This, combined with direct visualization of the biofilms by SEM and confocal microscopy (Fig 1) support this criteria ii) Persistent and localized infection. Bacterial infection was present in the burn wounds until days 14 and 35 post inoculation, indicating persistence (Fig S2). Localized infection was established with negative blood cultures and lack of systemic signs of infection (data not shown). iii) Resistance to anti-microbial treatments. Silver dressing Acticoat 7™ represents standard of care for the management of infected wounds [29]. In vitro, incubation of Acticoat 7™ with planktonic cultures of A. baumannii and P. aeruginosa effectively eradicated both bacterial species (Fig 2A). However, Acticoat 7™ dressing was ineffective against bacterial biofilm in burn wounds (Fig 2B-C).
Figure 2. Resistance of mixed species biofilm to antimicrobial and debridement.
A-C, antimicrobial Acticoat 7™ effectively kills bacteria in planktonic phase while ineffective against wound biofilm. A, planktonic cultures of P. aeruginosa and A. baumannii were incubated in presence or absence of Acticoat 7™ for 24h followed by determination of bacterial CFU. The treatment was highly effective in killing the bacteria. nd, not detectable. Data are mean ±SD (n=4), *, p<0.05. B, images of wounds covered with Tegaderm™ or Acticoat 7™ .C, Microbiology analysis of porcine burn wound were covered with Tegaderm™ or Acticoat 7™ on days 7 or 14 post-inoculation. Acticoat 7™ was ineffective in attenuating any bacterial load from wounds suggesting that the bacteria in biofilm were resistant to anti-microbial treatment. Data are mean ± SD, n=3. D-H, debridement was performed using a 0.12 inch Weck Blade by removing necrotic and infected tissues until bleeding healthy tissue is exposed. D, digital images of burn wound pre debridement (pre-D), immediate post-debridement (post-D, 0h) or 48h (post-D, 48h) after debridement. E, microbiology analysis reveals a significant decrease in bacterial burden after debridement. The bacterial burden for P. aeruginosa was restored to almost to the initial level 48h post-D suggesting debridement alone does not eradicate biofilm. Data are mean ± SD. * p<0.05 (n=3); F, laser doppler imaging was performed to demonstrate that there was no blood flow (blue color) in pre debrided burn wounds while the healthy well perfused (red) tissue is exposed immediately after debridement. G, bar graph represents quantitation of the laser Doppler blood flow imaging. H, Immuno-histochemical staining of P. aeruginosa (green) and A. baumannii (red) burn wounds following debridement demonstrate that the bacterial biofilm (micro colonies) are restored in deeper layers of the wounds on days 14 post-debridement. White dashed line represents the line of debridement. Scale bar = 100μM.
Mixed species wound biofilm is resistant to debridement
Debridement of the biofilm infected burn wound was performed by a plastic surgeon wound care specialist using a Weck Blade to remove necrotic and infected tissues until bleeding healthy tissue was exposed as documented using Doppler blood flow analysis (Fig 2D & F-G). CFU analyses revealed significant but transient lowering of bacterial burden following debridement. However, the P.aeruginosa bacterial burden was restored to almost initial levels within 48h demonstrating that debridement alone was not effective to eradicate biofilm infection (Fig 2E). IHC staining of A. baumannii and P. aeruginosa indicated micro colonies of both bacteria in deeper tissue of the debrided wound bed cautioning that debridement may actually promote deep tissue infection (Fig 2H).
Induction of biofilm specific genes in infected wound tissue elements
It is well established that in a sessile biofilm lifestyle, bacteria acquire unique phenotypes by modulating gene expression that supports biofilm biology [30]. While there is no known “biofilm biomarker gene” identified for P. aeruginosa, we evaluated expression patterns of some genes previously studied under biofilm growth conditions. These included rpoS, which is implicated in the morphology and antibiotic resistance of biofilms [30], and rhlR/aprA previously linked to quorum sensing and biofilms [31, 32]. The expression of rhlR, rpoS and arpR were significantly upregulated in biofilm bacteria laser captured from burn wounds compared to those of planktonic bacteria obtained from wound scrubbed fluid (Fig 3A-C). These observations further support biofilm formation in the model reported herein.
Figure 3. Abundant expression of biofilm specific genes in biofilm tissue elements captured using LCM.
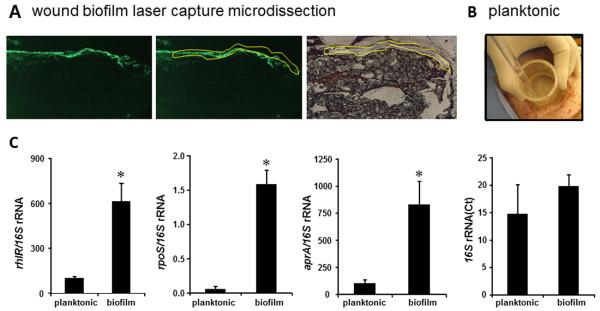
A, frozen serial sections (10 μm) from biofilm infected wound tissue were stained with anti-PAO1 (green) antibody. The next serial sections were stained with hematoxylin to visualize corresponding biofilm affected area. Images are representative (left) anti-PAO1 stained image; (middle) anti-PAO1 stained tissue marked to delineate the biofilm affected area; and (right) the area of biofilm was cut/captured using laser capture microdissection (LCM) from corresponding hematoxylin stained section. B, to collect planktonic bacteria from wounds, a sterile double open-ended plastic tube was placed on a wound followed by washing once with 4ml of PBS and collected. The wash suspension was collected and spun down to retrieve bacterial pellets. This procedure results in removal of loosely adhered or planktonic form of bacteria. C, LCM captured tissue was used for quantification of mRNA levels of biofilm specific mRNA using real-time PCR and normalized against 16S rRNA expression. 16S rRNA level have been presented to demonstrate comparable levels in both plankonic and captured biofilm groups. Data are mean ±SD (n=4); * p<0.05 compared to planktonic.
Biofilm infections compromise skin barrier function
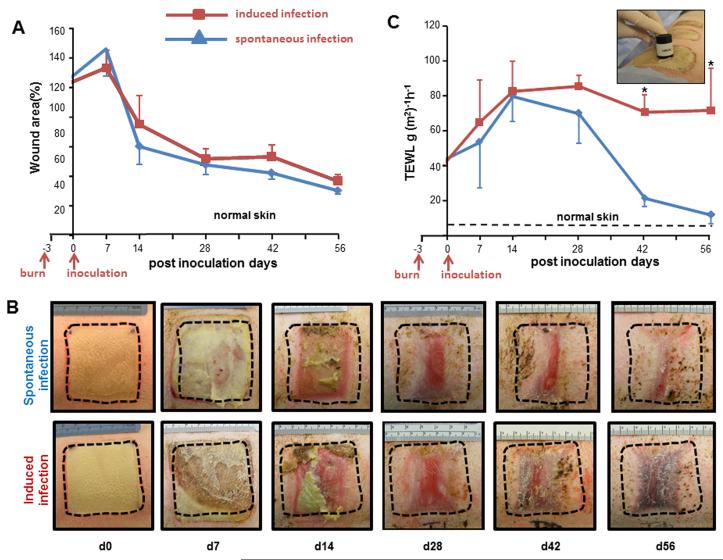
Development of the wound biofilm model was aimed at elucidating novel aspects of host response that may only be studied in a long-term experimental system. For the purposes of studying host response to induced biofilm formation, controls were considered burn wounds not exposed to bacterial inoculation (sterile, PBS mock inoculated). Despite standard wound care management, control wounds occasionally developed spontaneous infection from skin micro-flora. During a long-term experimentation setting we observed that inoculation of some wounds on a pig resulted in colonization of all wounds by the inoculated bacteria. Thus, pigs bearing control wounds were separately housed and wholly dedicated for that purpose. In the current standard of clinical care, wound closure is determined based on visual inspection by the wound care physician. Closure, as appreciated by visual inspection, of biofilm infected wounds was comparable to that of non-inoculated control wounds (Fig. 4A-B). When skin is injured, its barrier function is impaired resulting in higher transepidermal water loss (TEWL). TEWL is an index to assess skin barrier function in vivo [33]. Therefore, in addition to visual documentation of wound closure, a TEWL assay was performed to quantitatively determine the re-establishment of skin barrier function following burn wound. Interestingly, although visual wound closure was unaffected by biofilm infection, marked impairment in the restoration of skin barrier function was noted in biofilm-infected wounds compared to non-inoculated control wounds (Fig 4C).
Figure 4. Biofilms infection compromised re-establishment of skin barrier function.
A, wound closure in spontaneous infection (blue line, control) or induced infection (red line) wounds. Data have been presented as percentage of the initial wound area. Data are mean ± SD (n=4). B, representative digital images from spontaneous infection or induced infection wounds burn wounds. C, trans epidermal water loss (TEWL) analysis from spontaneous infection and induced infection burn wounds. Inset presents the DermaLab TEWL Probe used for the measurement of the trans-epidermal water loss from the wounds. TEWL was expressed in g/m2/h. Data are mean ±SD (n=4); * p<0.05 compared to spontaneous infection.
Silencing of tight junction proteins by biofilm infection
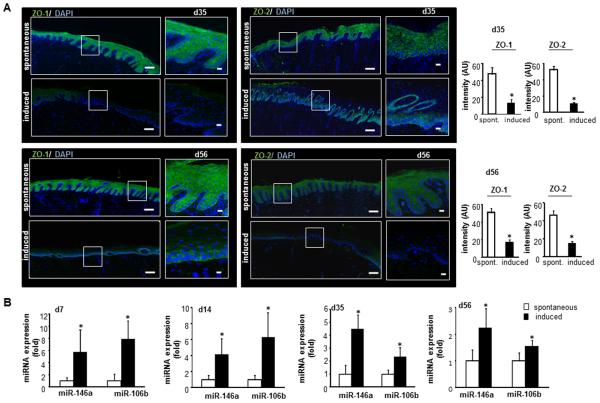
The barrier function of the skin is maintained by integrity of adhesive interaction at the epithelial apical junction complex comprised mainly of tight junctions, adherens junctions, desmosomes and gap junction proteins [34]. Zona occludens-1 (ZO-1) and zona occludens-2 (ZO-2) represent two key tight junction proteins that were found to be significantly down regulated in biofilm infected burn wounds (Fig. 5A).
Figure 5. Silencing of tight junction proteins by biofilm infection.
Wound biopsies were collected at specified time-points after inoculation from spontaneous infected (spont, no inoculation, control) or induced infected (induced), inoculated with A. baumannii and P. aeruginosa. A, representative mosaic (scale bar=200 μm) and corresponding zoomed (scale bar=50 μm) images of ZO-1 and ZO-2 stained sections on days 35 & 56 post inoculation demonstrating reduced expression of the proteins following induced infection. OCT embedded frozen sections (10 μm) and stained using anti-ZO-1 (green) or anti-ZO-2 (green). The sections were counterstained using DAPI. Bar graphs present quantitation of ZO-1 and ZO-2 signal intensity. Data are presented as mean ± SD (n=3), * p<0.05 compared to spontaneous. B, expression of miR-146a and miR-106b in wound biopsies collected on days 7-35 post inoculation from spontaneous or induced burn wounds. Data presented as mean ± SD (n=3), * p<0.05 compared to spontaneous infection.
Biofilm inducible cutaneous miRNA silence ZO-1 and ZO-2 and increases trans-epidermal water loss (TEWL)
In our search for mechanisms silencing ZO-1 and ZO-2, bioinformatic analyses recognized miR-146a and miR106b with potential binding sites on ZO-1 and ZO-2 mRNA (Fig S4). Interestingly, expression of miR-146a and miR-106b were significantly induced in response to biofilm infection of wounds when compared with uninfected controls (Fig 5B). Does biofilm infection indeed induce miR-146a and miR-106b? Are ZO-1 and ZO-2 biologically validated targets of miR-146a and miR-106b? To answer these two questions we relied on an in vitro model involving static biofilms grown on human keratinocyte monolayers [22]. Exposure of keratinocytes to mixed-species biofilm silenced ZO-1/ZO-2 (Fig 6A) and induced miR-146a/miR106b (Fig 6B) consistent with in vivo findings (Fig 5). While Pseudomonas monospecies biofilm induced both miR-146a and miR-106b, Acenitobacter biofilm induced miR-106b but not miR-146a. Biofilm featuring co-presence of Pseudomonas and Acenitobacter demonstrated additive effect in inducing miR-106b (Fig 6B). The use of Δpsl poor biofilm producing strain of P.aeruginosa PAO1 failed to induce both miRs pointing towards the significance of biofilm infection in inducing miR-106b and miR-146a (Fig 6C). The observation that conditioned media from biofilm cultures, not planktonic cultures, were able to induce miR-146a and miR-106b indicate that the miR regulatory factor may be secreted (Fig 6D).
Figure 6. Biofilm inducible miRNA silence ZO-1 and ZO-2.
Human keratinocyte (HaCaT) cells were infected with a static biofilm infection as described in methods. A, Western blot of ZO-1 and ZO-2 expression following 12h of infection as compared to non-infected cells (control). Data are mean ± SD (n=3), * p<0.05 compared to control. B, expression of miR-146a and miR-106b in keratinocytes following 12h of P.aeruginosa PAO1 (Pa), A. baumannii 19606 (Ab) mixed (both Pa and Ab) infection compared to non-infected control. Data represented as mean ± SD (n=4), * p<0.05 compared to control, † p<0.05 compared to A. baumannii and ‡ p<0.05 compared P.aeruginosa. C, expression of miR-146a and miR-106b in keratinocytes following 12h of poor-biofilm forming P.aeruginosa Δpsl PAO1 (Pa), wild type biofilm forming P.aeruginosa PAO1 (Pa) infection compared to non infected control. Data represented as mean ± SD (n=4), * p<0.05 compared to control. D, expression of miR-146a and miR-106b in keratinocytes following 12h of incubation with planktonic bacterial media and biofilm bacterial medium compared to non infected control media. Data represented as mean ± SD (n=4), * p<0.05 compared to control media and † p<0.05 compared to planktonic control media. E, human keratinocytes were transfected with miR-146a a miR-106b mimics for 48h followed by Immunocytochemistry. Images of HaCaT stained with anti- ZO-1 or anti-ZO-2 (green) and counter stained with DAPI (nuclear, blue). The quantification of ZO-1 and ZO-2 has been provided in Fig S4. Scale bar =10 μm. F, expression of ZO-1 and ZO-2 expression in HaCaT cells transfected with either miRIDIAN hsa-miR-146a or hsa-miR-106b mimic or control-mimic for 48h. Data represented as mean ± SD (n=4), * p<0.05 compared to control miRNA mimic. G, expression of ZO-1 and ZO-2 in presence of anti-miR to miR-146a and miR-106b inhibitors. Data represented as mean ± SD (n=4), * p<0.05 compared to control anti-miR. H, to test if ZO-1 or ZO-2 are direct targets of miR-146a or miR-106b keratinocytes were transfected with pmiR Target-ZO1-3′-UTR or pmiR Target-ZO2-3′-UTR firefly luciferase expression constructs and co-transfected with pRL-TK Renilla luciferase expression construct along with either miR-146a, miR106b or control mimics. I, expression of ZO-1 and ZO-2 mRNA in presence of miR-146a and miR-106b mimics. Data represent mean ± SD (n=3). *, p<0.05 compared to control transfected cells.
To directly connect these miRs to ZO-1/ZO-2, immunocytochemistry studies using miRidian miR-146a and miR-106b mimics were performed (Fig 6E). hsa-miR-146a or hsa-miR-106b mimics or their corresponding inhibitors transfection showed that both miRNAs were inversely associated with expression of corresponding ZO proteins (Fig 6F-G,I). Using ZO-1 3′-UTR and ZO-2 3′-UTR firefly luciferase expression constructs the effect of the said miRs on ZO transcription was tested (Fig 6H).
Findings of this work show that ZO-1 and ZO-2 are direct targets of miR146a. While miR-106 expression is co-regulated with ZO-1/ZO-2, these proteins are only targeted by miR146a (Fig 6H). Finally, to determine if miR-146a and miR-106b may directly influence skin barrier function, miR-146a mimic, miR-106b mimic and non-targeting miRNA mimic were delivered to the dorsal skin of mice via a cream-based vehicle. Delivery of miR mimics silenced ZO-1 and ZO-2 expression and compromised skin barrier function as manifested by increased TEWL (Fig 7A-C).
Figure 7. miRNA 146a and miR106b disrupts tight junction proteins in epidermis and increases trans epidermal water loss (TEWL) in skin.
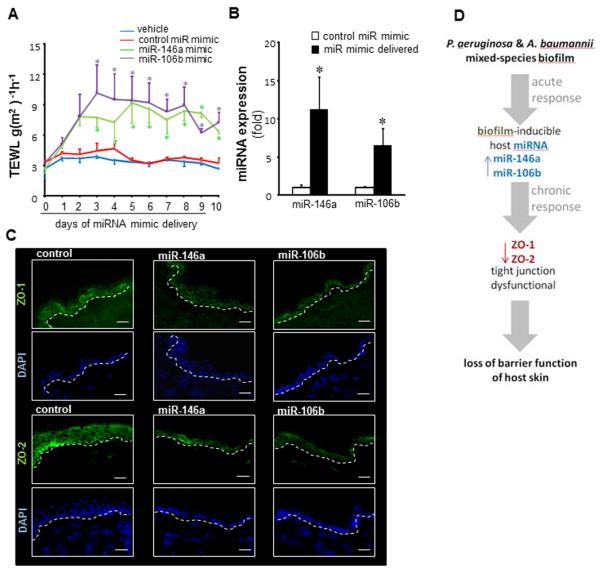
MiR-146a mimic, miR-106b mimic and non-targeting miRNA mimic (C. elegans miR-67 as negative control) were mixed in a cream and applied on a marked area on the dorsal skin of mice for 10 days. TEWL measurements were taken daily using Dermalab Series Skinlab Combo. The animals were sacrificed at day 11 and the skin where the mimic was applied was harvested for miRNA and immunohistochemistry (IHC) analysis. A, TEWL analysis of mice skin following delivery of miR-146a and miR-106b or control mimics. Data are mean ± SD (n=3), * p<0.05 compared to mice applied control miRNA mimic. B, quantitative Real Time PCR of skin where miR-146a and miR-106b mimic was applied. Data are mean ± SD (n=3), *p<0.05 compared to mice applied with control miRNA mimic. C, IHC images of mouse skin delivered with miR-146a and miR-106b mimic stained with tight Junction proteins ZO-1 and ZO-2 (green) and counterstained with DAPI (blue). The quantification of ZO-1 and ZO-2 has been provided in Fig S5. D, summary illustration depicting acute phase induction of biofilm-inducible miRs followed by silencing of ZO-1 & ZO-2 resulting in longer-term compromise of skin barrier function.
DISCUSSION
The skin is the largest organ of the human body, which provides an effective barrier between the organism and the environment. When barrier is breached as in chronic wounds, biofilm are often present in the host tissue for extended periods during which time they may compromise the host response to injury. Presence of biofilm has been reported in ischemic wounds, including burn [35, 36]. The pig is widely acknowledged as the animal of choice to serve as a pre-clinical model of skin wound healing [9, 18]. At present all experimental systems addressing skin wound biofilm in pigs are short in duration and therefore inadequate to understand long-term clinically relevant host responses to wound biofilm [12, 37]. This is the first work studying mechanisms underlying the wound biofilm over an eight week period. The burn wound biofilm developed herein satisfied the criteria as an established biofilm by being surface adherent, persistent and localized over four weeks, and resistant to Ag dressing and debridement used as a standard of care to manage infected wounds. We found that debridement promoted deep tissue colonization of bacterial biofilms. Though, colonization of bacteria to deeper tissue had been reported [38], no clear evidence was provided linking debridement with deep tissue infection.
Transepidermal water loss is a reliable measure of skin barrier function [33]. The nucleated epidermis contributes to this barrier primarily via tight, gap and adherens junctions. The absence of the tight junction proteins [39] in mice, results in lethal skin-barrier function failure [40]. ZO proteins are ubiquitous scaffolding proteins that enable assembly of multi-protein complexes at the cytoplasmic surface of the plasma membrane and link transmembrane proteins to the filamentous cytoskeleton. ZO-1 acts as a scaffolding molecule during formation of junctional complexes such as tight junctions, gap junctions, and adherens junctions [41]. ZO-2 is present at the tight junction of epithelial cells regulating barrier function [42]. Both ZO-1 and ZO-2 deficient mice are embryonic lethal [43, 44]. ZO-2 deficient mice show overt signs of epithelial barrier defect leading to death [44]. Bacteria are known to perturb tight junctions. Several bacterial pathogens actively spread within human tissues by the pathogen-induced recruitment of host filamentous (F)-actin. F-actin forms a tail behind the microbe, propelling it through the cytoplasm and compromising tight junction function [45]. Specifically, P. aeruginosa type III secreted toxins disrupt epithelial barrier in the lung by downregulating tight junction proteins including ZO-1 [46]. Consistent with these observations, this work presents the first evidence demonstrating that skin barrier function is compromised by mixed species biofilm infection. Primary targets in such pathology are ZO-1 and ZO-2, both of which are significantly downregulated in response to biofilm infection. This observation has outstanding clinical significance because it raises the possibility that although biofilm infected wounds may visually appear to be closed, the current standard for clinical care, they may actually be open because of the presence of functionally compromised leaky skin.
miRNA regulate epithelial barrier function. Gene ablation of Dicer1 demonstrated that miRNAs play a vital role in the differentiation and function of the intestinal epithelium [47]. Specifically, TNF-α regulates intestinal permeability by inducing miR-122a-mediated degradation of occludin mRNA [48]. More recently, overexpression of miR-21 in patients with ulcerative colitis has been shown to impair intestinal epithelial barrier function through targeting the Rho GTPase RhoB [49]. None of these mechanisms implicate miR-dependent silencing of ZO proteins in compromising epithelial barrier function in any tissue system. As of now, the only miR known to directly silence ZO proteins is miR-212 [50]. The current work validates both ZO-1 as well as ZO-2 as direct targets of miR-146a. miR-146a is induced in skin in response to inflammation [51]. In addition, miR-146a expression is elevated by microenvironmental signals in the epidermis, rendering Langerhans cells less susceptible to inappropriate activation by commensal bacteria, triggering TLR2 [52]. In the skin, miR-146a may also function as a negative regulator of TGF-β1-induced myofibroblast transdifferentiation by targeting SMAD4 [53]. In non-infected murine diabetic wounds, low expression of injury-inducible miR-146a is claimed to be responsible for impaired wound healing [54]. The current report presents first evidence demonstrating that biofilm infection may induce miRNAs in the host tissue. Both miR-146a and miR-106b were recognized as being biofilm inducible in the skin. Although the 3′ UTR regions of ZO-1 and ZO-2 were recognized to be a direct target of miR-146a, that was not the case for miR-106b. miR-106b did not target the 3′ UTR region but did suppress gene expression by possibly targeting the exonic or 5′ UTR regions of the mRNA [55, 56]. The significance of miR-106b is even more poorly understood than that of miR-146a. Recently miR-106b has been causally linked to mitochondrial dysfunction [57], a response that is also known to be implicated in epithelial barrier disruption [58]. Findings of this study lead to the notion that topical delivery of miR-146a and miR-106b inhibitors to the functionally compromised skin produced from biofilm infected wounds may restore barrier function and secure functional closure of the wound.
When bacteria form a biofilm in the human host, the infection often becomes very resistant to treatment and can contribute to the development of chronic disease. The current literature is equivocal on whether biofilm may compromise wound closure [36, 59]. Our model of host response in a chronic wound has enabled the observation that following weeks of biofilm infection, skin barrier function may be compromised. This observation is particularly clinically significant because it shows that even if a biofilm infected wound visually appeared to be closed, its barrier function may be compromised. Such failed or leaky skin may serve as an open portal allowing further infection and wound healing complications. Thus, reliance on functional measures of skin barrier function as a measure in addition to conventional wound inspection seems prudent. Furthermore, wound care may be enhanced by restoration of barrier function of incompletely closed wounds. Another highlight of this work is the recognition of wound biofilm inducible miR-146a and miR-106b. The observation that wound biofilm may induce host skin miRNAs is novel and particularly significant because these miRNAs target the same tight junction proteins of the ZO family which are known to be critically required for skin barrier function. The field of post-transcriptional gene silencing of ZO proteins is in its infancy with miR-212 being the only miR to silence ZO-1. The current report identifies two miRs that silence both ZO-1 and ZO-2, the most critical ZOs of the three-member ZO family. The observation that biofilm may induce host skin miRNAs to compromise skin function is novel and sets the stage for intervention strategies aimed at inhibiting these miRNAs with the goal to restore skin barrier function.
Supplementary Material
Acknowledgment
Supported by NIH grant GM077185, GM069589 and DOD W81XWH-11-2-0142 to CKS, NIH NR013898 to CKS and DJW and in part by NIH DK076566 to SR and NIH R01AI097511 to DJW. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Author contributions: SR, HE, DJW, CHC, GMG and CKS conceived and designed the work. CKS, SR, HE, DJW, HMP, CHC, GMG and VB wrote the manuscript. SR, HE, KG, MS, CM, SK, EM, SC, VB and HMP collected and analyzed data for this work and participated in the preparation of the manuscript.
Conflict of Interest: None for any of the authors.
REFERENCES
- 1.Sen CK, Gordillo GM, Roy S, et al. Human skin wounds: a major and snowballing threat to public health and the economy. Wound Repair Regen. 2009;17:763–771. doi: 10.1111/j.1524-475X.2009.00543.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lopez D, Vlamakis H, Kolter R. Generation of multiple cell types in Bacillus subtilis. FEMS Microbiol Rev. 2009;33:152–163. doi: 10.1111/j.1574-6976.2008.00148.x. [DOI] [PubMed] [Google Scholar]
- 3.James GA, Swogger E, Wolcott R, et al. Biofilms in chronic wounds. Wound Repair Regen. 2008;16:37–44. doi: 10.1111/j.1524-475X.2007.00321.x. [DOI] [PubMed] [Google Scholar]
- 4.Hall-Stoodley L, Costerton JW, Stoodley P. Bacterial biofilms: from the natural environment to infectious diseases. Nat Rev Microbiol. 2004;2:95–108. doi: 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- 5.Seth AK, Geringer MR, Hong SJ, et al. In vivo modeling of biofilm-infected wounds: a review. J Surg Res. 2012;178:330–338. doi: 10.1016/j.jss.2012.06.048. [DOI] [PubMed] [Google Scholar]
- 6.Watters C, DeLeon K, Trivedi U, et al. Pseudomonas aeruginosa biofilms perturb wound resolution and antibiotic tolerance in diabetic mice. Med Microbiol Immunol. 2013;202:131–141. doi: 10.1007/s00430-012-0277-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Watters C, Everett JA, Haley C, et al. Insulin treatment modulates the host immune system to enhance Pseudomonas aeruginosa wound biofilms. Infect Immun. 2014;82:92–100. doi: 10.1128/IAI.00651-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dalton T, Dowd SE, Wolcott RD, et al. An in vivo polymicrobial biofilm wound infection model to study interspecies interactions. PLoS One. 2011;6:e27317. doi: 10.1371/journal.pone.0027317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sullivan TP, Eaglstein WH, Davis SC, et al. The pig as a model for human wound healing. Wound Repair Regen. 2001;9:66–76. doi: 10.1046/j.1524-475x.2001.00066.x. [DOI] [PubMed] [Google Scholar]
- 10.Gordillo GM, Bernatchez SF, Diegelmann R, et al. Preclinical Models of Wound Healing: Is Man the Model? Proceedings of the Wound Healing Society Symposium. Adv Wound Care (New Rochelle) 2013;2:1–4. doi: 10.1089/wound.2012.0367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Geyer M, Markou A. The Role of preclinical models in the development of psychotropic drugs. In: Davis KL, editor. In Neuropsychopharmacology: The Fifth Generation of Progress. 2002. pp. 445–454. [Google Scholar]
- 12.Davis SC, Ricotti C, Cazzaniga A, et al. Microscopic and physiologic evidence for biofilm-associated wound colonization in vivo. Wound Repair Regen. 2008;16:23–29. doi: 10.1111/j.1524-475X.2007.00303.x. [DOI] [PubMed] [Google Scholar]
- 13.Bjarnsholt T. The role of bacterial biofilms in chronic infections. APMIS Suppl. 2013:1–51. doi: 10.1111/apm.12099. [DOI] [PubMed] [Google Scholar]
- 14.Calhoun JH, Murray CK, Manring MM. Multidrug-resistant organisms in military wounds from Iraq and Afghanistan. Clin Orthop Relat Res. 2008;466:1356–1362. doi: 10.1007/s11999-008-0212-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dallo SF, Weitao T. Insights into acinetobacter war-wound infections, biofilms, and control. Adv Skin Wound Care. 2010;23:169–174. doi: 10.1097/01.ASW.0000363527.08501.a3. [DOI] [PubMed] [Google Scholar]
- 16.CMS Decision Memo for Electrostimulation for Wounds (CAG-00068N) 2002 [Google Scholar]
- 17.Holloway BW, Krishnapillai V, Morgan AF. Chromosomal genetics of Pseudomonas. Microbiol Rev. 1979;43:73–102. doi: 10.1128/mr.43.1.73-102.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roy S, Biswas S, Khanna S, et al. Characterization of a preclinical model of chronic ischemic wound. Physiol Genomics. 2009;37:211–224. doi: 10.1152/physiolgenomics.90362.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elgharably H, Mann E, Awad H, et al. First Evidence of Sternal Wound Biofilm following Cardiac Surgery. PLoS One. 2013;8:e70360. doi: 10.1371/journal.pone.0070360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roy S, Patel D, Khanna S, et al. Transcriptome-wide analysis of blood vessels laser captured from human skin and chronic wound-edge tissue. Proc Natl Acad Sci U S A. 2007;104:14472–14477. doi: 10.1073/pnas.0706793104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Biswas S, Roy S, Banerjee J, et al. Hypoxia inducible microRNA 210 attenuates keratinocyte proliferation and impairs closure in a murine model of ischemic wounds. Proc Natl Acad Sci U S A. 2010;107:6976–6981. doi: 10.1073/pnas.1001653107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Anderson GG, Moreau-Marquis S, Stanton BA, et al. In vitro analysis of tobramycin-treated Pseudomonas aeruginosa biofilms on cystic fibrosis-derived airway epithelial cells. Infect Immun. 2008;76:1423–1433. doi: 10.1128/IAI.01373-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao G, Hochwalt PC, Usui ML, et al. Delayed wound healing in diabetic (db/db) mice with Pseudomonas aeruginosa biofilm challenge: a model for the study of chronic wounds. Wound Repair Regen. 2010;18:467–477. doi: 10.1111/j.1524-475X.2010.00608.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chan YC, Roy S, Khanna S, et al. Downregulation of endothelial microRNA-200b supports cutaneous wound angiogenesis by desilencing GATA binding protein 2 and vascular endothelial growth factor receptor 2. Arterioscler Thromb Vasc Biol. 2012;32:1372–1382. doi: 10.1161/ATVBAHA.112.248583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roy S, Khanna S, Azad A, et al. Fra-2 mediates oxygen-sensitive induction of transforming growth factor beta in cardiac fibroblasts. Cardiovasc Res. 2010;87:647–655. doi: 10.1093/cvr/cvq123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mehta RC, Stecker KK, Cooper SR, et al. Intercellular adhesion molecule-1 suppression in skin by topical delivery of anti-sense oligonucleotides. J Invest Dermatol. 2000;115:805–812. doi: 10.1046/j.1523-1747.2000.00143.x. [DOI] [PubMed] [Google Scholar]
- 27.Parsek MR, Singh PK. Bacterial biofilms: an emerging link to disease pathogenesis. Annu Rev Microbiol. 2003;57:677–701. doi: 10.1146/annurev.micro.57.030502.090720. [DOI] [PubMed] [Google Scholar]
- 28.Martineau L, Davis SC, Peng HT, et al. Controlling methicillin resistant Staphyloccocus aureus and Pseudomonas aeruginosa wound infections with a novel biomaterial. J Invest Surg. 2007;20:217–227. doi: 10.1080/10717540701481275. [DOI] [PubMed] [Google Scholar]
- 29.Tredget EE, Shankowsky HA, Groeneveld A, et al. A matched-pair, randomized study evaluating the efficacy and safety of Acticoat silver-coated dressing for the treatment of burn wounds. J Burn Care Rehabil. 1998;19:531–537. doi: 10.1097/00004630-199811000-00013. [DOI] [PubMed] [Google Scholar]
- 30.Whiteley M, Bangera MG, Bumgarner RE, et al. Gene expression in Pseudomonas aeruginosa biofilms. Nature. 2001;413:860–864. doi: 10.1038/35101627. [DOI] [PubMed] [Google Scholar]
- 31.Perez-Osorio AC, Williamson KS, Franklin MJ. Heterogeneous rpoS and rhlR mRNA levels and 16S rRNA/rDNA (rRNA gene) ratios within Pseudomonas aeruginosa biofilms, sampled by laser capture microdissection. J Bacteriol. 2010;192:2991–3000. doi: 10.1128/JB.01598-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lenz AP, Williamson KS, Pitts B, et al. Localized gene expression in Pseudomonas aeruginosa biofilms. Appl Environ Microbiol. 2008;74:4463–4471. doi: 10.1128/AEM.00710-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pinnagoda J, Tupker RA, Agner T, et al. Guidelines for transepidermal water loss (TEWL) measurement. A report from the Standardization Group of the European Society of Contact Dermatitis. Contact Dermatitis. 1990;22:164–178. doi: 10.1111/j.1600-0536.1990.tb01553.x. [DOI] [PubMed] [Google Scholar]
- 34.Pummi K, Malminen M, Aho H, et al. Epidermal tight junctions: ZO-1 and occludin are expressed in mature, developing, and affected skin and in vitro differentiating keratinocytes. J Invest Dermatol. 2001;117:1050–1058. doi: 10.1046/j.0022-202x.2001.01493.x. [DOI] [PubMed] [Google Scholar]
- 35.Kennedy P, Brammah S, Wills E. Burns, biofilm and a new appraisal of burn wound sepsis. Burns. 2010;36:49–56. doi: 10.1016/j.burns.2009.02.017. [DOI] [PubMed] [Google Scholar]
- 36.Wolcott RD, Rhoads DD. A study of biofilm-based wound management in subjects with critical limb ischaemia. J Wound Care. 2008;17:145–148. 150–142, 154–145. doi: 10.12968/jowc.2008.17.4.28835. [DOI] [PubMed] [Google Scholar]
- 37.Gurjala AN, Geringer MR, Seth AK, et al. Development of a novel, highly quantitative in vivo model for the study of biofilm-impaired cutaneous wound healing. Wound Repair Regen. 2011;19:400–410. doi: 10.1111/j.1524-475X.2011.00690.x. [DOI] [PubMed] [Google Scholar]
- 38.Kirketerp-Moller K, Jensen PO, Fazli M, et al. Distribution, organization, and ecology of bacteria in chronic wounds. J Clin Microbiol. 2008;46:2717–2722. doi: 10.1128/JCM.00501-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Furuse M. Molecular basis of the core structure of tight junctions. Cold Spring Harb Perspect Biol. 2010;2:a002907. doi: 10.1101/cshperspect.a002907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kirschner N, Rosenthal R, Gunzel D, et al. Tight junctions and differentiation--a chicken or the egg question? Exp Dermatol. 2012;21:171–175. doi: 10.1111/j.1600-0625.2011.01431.x. [DOI] [PubMed] [Google Scholar]
- 41.Hartsock A, Nelson WJ. Adherens and tight junctions: structure, function and connections to the actin cytoskeleton. Biochim Biophys Acta. 2008;1778:660–669. doi: 10.1016/j.bbamem.2007.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Monteiro AC, Sumagin R, Rankin CR, et al. JAM-A associates with ZO-2, Afadin and PDZ-GEF1 to activate Rap2c and regulate epithelial barrier function. Mol Biol Cell. 2013 doi: 10.1091/mbc.E13-06-0298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Katsuno T, Umeda K, Matsui T, et al. Deficiency of zonula occludens-1 causes embryonic lethal phenotype associated with defected yolk sac angiogenesis and apoptosis of embryonic cells. Mol Biol Cell. 2008;19:2465–2475. doi: 10.1091/mbc.E07-12-1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xu J, Kausalya PJ, Phua DC, et al. Early embryonic lethality of mice lacking ZO-2, but Not ZO-3, reveals critical and nonredundant roles for individual zonula occludens proteins in mammalian development. Mol Cell Biol. 2008;28:1669–1678. doi: 10.1128/MCB.00891-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ireton K. Molecular mechanisms of cell-cell spread of intracellular bacterial pathogens. Open Biol. 2013;3:130079. doi: 10.1098/rsob.130079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Clark CA, Thomas LK, Azghani AO. Inhibition of protein kinase C attenuates Pseudomonas aeruginosa elastase-induced epithelial barrier disruption. Am J Respir Cell Mol Biol. 2011;45:1263–1271. doi: 10.1165/rcmb.2010-0459OC. [DOI] [PubMed] [Google Scholar]
- 47.Yi R, O’Carroll D, Pasolli HA, et al. Morphogenesis in skin is governed by discrete sets of differentially expressed microRNAs. Nat Genet. 2006;38:356–362. doi: 10.1038/ng1744. [DOI] [PubMed] [Google Scholar]
- 48.Ye D, Guo S, Al-Sadi R, et al. MicroRNA regulation of intestinal epithelial tight junction permeability. Gastroenterology. 2011;141:1323–1333. doi: 10.1053/j.gastro.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang Y, Ma Y, Shi C, et al. Overexpression of miR-21 in patients with ulcerative colitis impairs intestinal epithelial barrier function through targeting the Rho GTPase RhoB. Biochem Biophys Res Commun. 2013;434:746–752. doi: 10.1016/j.bbrc.2013.03.122. [DOI] [PubMed] [Google Scholar]
- 50.Tang Y, Banan A, Forsyth CB, et al. Effect of alcohol on miR-212 expression in intestinal epithelial cells and its potential role in alcoholic liver disease. Alcohol Clin Exp Res. 2008;32:355–364. doi: 10.1111/j.1530-0277.2007.00584.x. [DOI] [PubMed] [Google Scholar]
- 51.Sonkoly E, Stahle M, Pivarcsi A. MicroRNAs and immunity: novel players in the regulation of normal immune function and inflammation. Semin Cancer Biol. 2008;18:131–140. doi: 10.1016/j.semcancer.2008.01.005. DOI 10.1016/j.semcancer.2008.01.005. [DOI] [PubMed] [Google Scholar]
- 52.Jurkin J, Schichl YM, Koeffel R, et al. miR-146a is differentially expressed by myeloid dendritic cell subsets and desensitizes cells to TLR2-dependent activation. J Immunol. 2010;184:4955–4965. doi: 10.4049/jimmunol.0903021. [DOI] [PubMed] [Google Scholar]
- 53.Liu Z, Lu CL, Cui LP, et al. MicroRNA-146a modulates TGF-beta1-induced phenotypic differentiation in human dermal fibroblasts by targeting SMAD4. Arch Dermatol Res. 2012;304:195–202. doi: 10.1007/s00403-011-1178-0. [DOI] [PubMed] [Google Scholar]
- 54.Xu J, Wu W, Zhang L, et al. The role of microRNA-146a in the pathogenesis of the diabetic wound-healing impairment: correction with mesenchymal stem cell treatment. Diabetes. 2012;61:2906–291. doi: 10.2337/db12-0145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Forman JJ, Coller HA. The code within the code: microRNAs target coding regions. Cell Cycle. 2010;9:1533–1541. doi: 10.4161/cc.9.8.11202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang WX, Wilfred BR, Xie K, et al. Individual microRNAs (miRNAs) display distinct mRNA targeting “rules”. RNA Biol. 2010;7:373–380. doi: 10.4161/rna.7.3.11693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang Y, Yang L, Gao YF, et al. MiR-106b induces mitochondrial dysfunction and insulin resistance in C2C12 myotubes by targeting mitofusin-2. Mol Cell Endocrinol. 2013 doi: 10.1016/j.mce.2013.08.004. [DOI] [PubMed] [Google Scholar]
- 58.Ma C, Wickham ME, Guttman JA, et al. Citrobacter rodentium infection causes both mitochondrial dysfunction and intestinal epithelial barrier disruption in vivo: role of mitochondrial associated protein (Map) Cell Microbiol. 2006;8:1669–1686. doi: 10.1111/j.1462-5822.2006.00741.x. [DOI] [PubMed] [Google Scholar]
- 59.Thomson CH. Biofilms: do they affect wound healing? Int Wound J. 2011;8:63–67. doi: 10.1111/j.1742-481X.2010.00749.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.