Abstract
Background
QT-interval (QT) prolongation is an established risk factor for ventricular tachyarrhythmia and sudden cardiac death. Previous genome-wide association studies in populations of the European descent have identified multiple genetic loci that influence QT, but few have examined these loci in ethnically diverse populations.
Methods
Here, we examine the direction, magnitude, and precision of effect sizes for 21 previously reported SNPs from 12 QT loci, in populations of European (n=16,398), African (n=5,437), American Indian (n=5,032), Hispanic (n=1,143), and Asian (n=932) descent as part of the Population Architecture using Genomics and Epidemiology (PAGE) study. Estimates obtained from linear regression models stratified by race/ethnicity were combined using inverse-variance weighted meta-analysis. Heterogeneity was evaluated using Cochran's Q test.
Results
Of 21 SNPs, seven showed consistent direction of effect across all five populations, and an additional nine had estimated effects that were consistent across four populations. Despite consistent direction of effect, nine of 16 SNPs had evidence (P < 0.05) of heterogeneity by race/ethnicity. For these 9 SNPs, linkage disequilibrium plots often indicated substantial variation in linkage disequilibrium patterns among the various racial/ethnic groups, as well as possible allelic heterogeneity.
Conclusions
These results emphasize the importance of analyzing racial/ethnic groups separately in genetic studies. Furthermore, they underscore the possible utility of trans-ethnic studies to pinpoint underlying casual variants influencing heritable traits such as QT.
Studies of the QT interval (QT), a measurement of ventricular depolarization and repolarization obtained from the electrocardiogram (ECG), have shown that QT prolongation is an established risk factor for ventricular tachyarrhythmias,1 coronary heart disease,2 sudden cardiovascular death, and all-cause mortality.2 Several correlates of QT prolongation have been identified, including structural heart disease,3 sex,4 and age.5 QT is also heritable, with estimates ranging from 35%-40%.6
Early family-based linkage studies have identified rare and highly penetrant mutations associated with long- and short-QT syndromes.7 Recent genome-wide association studies (GWAS) in large population-based studies of European descent populations also have identified several common single nucleotide polymorphisms (SNPs) associated with modest increases in QT, including NOS1AP, KCNQ1, and SCN5A,8-10 which altogether account for approximately 10% of the variance in QT.11 However, much of the variation in QT remains unexplained.
To date, the majority of published GWAS of QT have been conducted in populations of European descent.8-10 Few studies have examined the relevance of GWAS-identified QT SNPs in multi-ethnic populations,12,13 although such studies are needed to fully understand the genetic architecture underlying QT. Therefore, we examined evidence of generalizability for 21 SNPs associated with QT in previous GWAS8-10 across populations of African, American Indian, Hispanic, and Asian descent from the Population Architecture using Genomics and Epidemiology (PAGE) Study.
Methods
Study Populations
The PAGE study is a collaboration of four large, multiethnic, and deeply phenotyped consortia.14 Using the ethnically diverse populations of the participating studies, the goal of PAGE is to better understand the epidemiologic architecture of well-replicated genetic variants associated with complex disease in global populations. Three PAGE consortia contributed the following studies to this research: the National Health and Nutrition Examination Surveys (NHANES) through the Epidemiologic Architecture for Genes Linked to Environment study,15 the Women's Health Initiative Clinical Trial through the Women's Health Initiative (WHI),16 and the Causal Variants Across the Life Course Consortium which included the Atherosclerosis Risk in Communities study,17 the Cardiovascular Health Study,18 the Strong Heart Study,19 and the Strong Heart Family Study.20 Each study was approved by the Institutional Review Board at the respective sites and all participants provided written consent. Further details on each study are available in eAppendix.
A total of 42,525 participants from the above studies were genotyped in PAGE. Of these, 10,051 were excluded according to the following hierarchical criteria (eTable 1): QT information unavailable (n=5,286); poor ECG quality (grade=5, n=1,654); major conduction defects, including left- or right-bundle branch block and intraventricular conduction delay (n=1,197); QRS interval ≥ 120ms (n=662); pacemaker or defibrillator implants (n=20); atrial fibrillation or atrial flutter on baseline ECG (n=44); participants under 18 years of age (n=334); and genetic ancestry data unavailable (n=854).
QT-Interval Measurement
In each study, certified technicians recorded at baseline, resting, supine (or semi-recumbent), standard 12-lead ECGs using either Marquette MAC 12 or MAC PC machines (GE Healthcare, Milwaukee, WI, USA). Comparable procedures were used for preparing participants, placing electrodes, recording, transmitting, processing, and controlling quality of ECGs. The QT-interval was measured electronically using Marquette 12SL algorithm. ECGs from Atherosclerosis Risk in Communities study, Cardiovascular Health Study, and Women's Health Initiative were processed by the central Epidemiological Cardiology Research Center at Wake Forest University, Winston-Salem, NC, USA. Epidemiologic Architecture for Genes Linked to Environment study, Strong Heart Study and Strong Heart Family Study ECGs were read at independent ECG reading centers using comparable protocols (eTable 2).
SNP Selection and Genotyping
The 21 SNPs examined in this study were reported by previously published QT GWAS (as of January 2010). The SNPs represent 12 genetic loci, with multiple SNPs reported for NOS1AP (4 SNPs), KCNH2 (3 SNPs), KCNQ1 (3 SNPs), PLN (2 SNPs) and SCN5A (2 SNPs). The 21 SNPs examined here were either targeted for genotyping by the PAGE study or were available on previous GWAS chips. Genotyping was done separately by each study (eAppendix). Cross-study quality control was performed centrally by the PAGE Coordinating Center using 360 samples from the International HapMap Project that were genotyped by each participating study.
Statistical Analysis
Study- and race-stratified tests of association between each SNP and QT (in milliseconds [ms]) were performed using linear regression models, and assuming an additive genetic mode of inheritance. We included the following confounders: study site (where appropriate), sex, age (continuous in years), RR interval (ms) or heart rate (beats per minute) when RR interval was not measured directly, and ancestral principal components that assessed global ancestry among study participants. Results were combined by inverse-variance weighted meta-analysis using METAL,21 and heterogeneity was evaluated using Cochran's Q statistic.22 P-values were 2-sided.
Generalizability of SNPs originally identified in European descent populations was assessed by examining the direction, precision, and magnitude of estimated effects across racial/ethnic groups. SNPs were considered directionally consistent if their direction of effect was the same across populations. For each non-European descent population, we categorized SNPs as stronger than, weaker than, or equal (within 0.05 ms) to the estimates from the European-descent population, based on the absolute value of the estimated effect sizes. Precision was gauged using the 95% confidence limit difference (CLD).
Haplotype Block Analysis
Given the potential for variation in linkage disequilibrium (linkage disequilibrium) patterns between SNPs across racial/ethnic groups,23 haplotype blocks were examined using HapMap III data.24 Briefly, we calculated pairwise measures of linkage disequilibrium using Hedrick's multiallelic D′, and we generated linkage disequilibrium plots using Haploview 4.225 and dense genotype data from five International HapMap III populations: African Americans from the southwest U.S., Utah residents of northern and western European ancestry, Han Chinese from Beijing, China, Japanese from Tokyo, Japan, and a Mexican American population from Los Angeles, CA. In accordance with convention, data from the Chinese and Japanese populations were pooled before analysis and referred to as “Asian.”
Results
Study Population Characteristics
A total of 32,474 participants from the PAGE consortium were included in this analysis with the following breakdown by race/ethnicity: European American (n=18,802), African American (n=6,132), American Indian (n=5,465), Hispanic (n=1,143), and Asian (n=932) (Table 1). All studies contributed approximately equal proportions of male and female participants, with the exception of the Women's Health Initiative Clinical Trial, which enrolled only female participants. Notably, the Women's Health Initiative Clinical Trial was the only study contributing participants of Asian descent. Estimated mean age varied slightly by race/ethnicity, with the highest mean age observed among European Americans (63.8 years) and the lowest among American Indians (59.1 years). Mean QT was consistent across race/ethnicity (range = 403 ms – 405 ms).
Table 1. Descriptive Characteristics of PAGE Study Populations by Race/Ethnicity and Study.
| Study | No. | Female % |
Age (years)a Mean (SD) |
QT Interval (ms)a Mean (SD) |
|---|---|---|---|---|
| European Americans | ||||
| Total | 18,802 | 65 | 64 (3) | 405 (15) |
| ARIC | 10,926 | 53 | 54 (6) | 399 (29) |
| CHS | 2,508 | 64 | 72 (5) | 413 (32) |
| EAGLE | 1,291 | 60 | 63 (14) | 409 (33) |
| WHI CT | 4,077 | 100 | 63 (7) | 402 (29) |
| African Americans | ||||
| Total | 6,132 | 69 | 62 (3) | 403 (17) |
| ARIC | 4,020 | 63 | 54 (6) | 400 (33) |
| CHS | 492 | 67 | 72 (6) | 405 (34) |
| EAGLE | 578 | 56 | 55 (12) | 406 (33) |
| WHI CT | 1,042 | 100 | 60 (7) | 401 (34) |
| American Indians | ||||
| Total | 5,465 | 65 | 59 (5) | 404 (18) |
| SHS | 2,200 | 64 | 63 (8) | 408 (33) |
| SHFS | 3,086 | 64 | 42 (16) | 408 (33) |
| WHI CT | 179 | 100 | 59 (6) | 398 (29) |
| Hispanics | ||||
| Total | 1,143 | 71 | 60 (6) | 405 (21) |
| EAGLE | 655 | 50 | 56 (12) | 405 (30) |
| WHI CT | 488 | 100 | 61 (7) | 404 (30) |
| Asians | ||||
| WHI CT | 932 | 100 | 62 (7) | 405 (33) |
Overall mean calculated using inverse-variance weighted method
ARIC indicates Atherosclerosis Risk in Communities; CHS, Cardiovascular Health Study; EAGLE, Epidemiologic Architecture for Genes Linked to Environment study; WHI CT, Women's Health Initiative Clinical Trial; SHS, Strong Heart Study; SHFS, Strong Heart Family Study
Evidence of variation in allele frequency by race/ethnicity was observed for a majority of GWAS-identified QT SNPs (Table 2). For example, we observed mean allele frequency differences between populations greater than 20% for thirteen SNPs. The two most striking differences were for rs12053903 and rs4725982, both of which had a difference of 44% between the lowest and highest frequency; for rs12053903, this difference translated to a coded allele frequency of 33% in European Americans and 77% in African American, while for rs4725982, the largest difference was between European Americans (22%) and Asians (66%).
Table 2. GWAS-Identified QT SNP Characteristics and Allele Frequencies From 32,474 Eligible Participants From PAGE Study.
| SNP | Nearest Gene | Chr | Base Paira | CA | NCA | Coded Allele Frequency | Function | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||
| European American | African American | American Indian | Hispanic | Asian | |||||||
| rs846111 | RNF207 | 1 | 6279370 | G | C | 0.27 | 0.05 | 0.15 | 0.18 | 0.25 | Missense |
| rs12143842 | NOS1AP | 1 | 162033890 | T | C | 0.25 | 0.13 | 0.20 | 0.24 | 0.38 | Intergenic |
| rs10494366 | NOS1AP | 1 | 162085685 | T | G | 0.63 | 0.41 | 0.57 | 0.57 | 0.32 | Intronic |
| rs16857031 | NOS1AP | 1 | 162112910 | G | C | 0.14 | 0.29 | 0.06 | 0.15 | 0.15 | Intronic |
| rs12029454 | NOS1AP | 1 | 162133117 | A | G | 0.15 | 0.26 | 0.13 | 0.24 | 0.39 | Intronic |
| rs10919071 | ATP1B1 | 1 | 169099483 | G | A | 0.13 | 0.03 | 0.32 | 0.19 | 0.04 | Intronic |
| rs11129795 | SCN5A | 3 | 38589163 | A | G | 0.24 | 0.17 | 0.14 | NA | NA | Near Gene -3b |
| rs12053903 | SCN5A | 3 | 38593393 | C | T | 0.33 | 0.77 | 0.55 | 0.45 | 0.51 | Intronic |
| rs12210810 | PLN | 6 | 118653204 | C | G | 0.05 | 0.01 | 0.02 | 0.02 | 0.0005 | Intergenic |
| rs11756438 | PLN | 6 | 118993632 | A | C | 0.48 | 0.40 | 0.19 | 0.31 | 0.26 | Intronic |
| rs2968864 | KCNH2 | 7 | 150622162 | G | A | 0.24 | 0.05 | 0.07 | 0.15 | 0.05 | Intergenic |
| rs2968863 | KCNH2 | 7 | 150623137 | A | G | 0.23 | 0.05 | 0.07 | 0.17 | 0.05 | Intergenic |
| rs4725982 | KCNH2 | 7 | 150637863 | T | C | 0.22 | 0.25 | 0.46 | 0.33 | 0.66 | Intergenic |
| rs2074238 | KCNQ1 | 11 | 2484803 | T | C | 0.08 | 0.02 | 0.06 | 0.04 | 0.04 | Intronic |
| rs12296050 | KCNQ1 | 11 | 2489342 | T | C | 0.20 | 0.52 | 0.29 | 0.30 | 0.35 | Intronic |
| rs12576239 | KCNQ1 | 11 | 2502319 | T | C | 0.14 | 0.18 | 0.15 | 0.14 | 0.10 | Intronic |
| rs8049607 | LITAF | 16 | 11691753 | T | C | 0.51 | 0.46 | 0.64 | 0.57 | 0.50 | Intergenic |
| rs37062 | NDRG4 | 16 | 58567238 | G | A | 0.25 | 0.16 | 0.48 | 0.37 | 0.38 | Intronic |
| rs2074518 | LIG3, RFFL | 17 | 33324382 | A | G | 0.45 | 0.17 | 0.53 | 0.40 | 0.24 | Intronic |
| rs17779747 | KCNJ2 | 17 | 68494992 | T | G | 0.33 | 0.10 | 0.19 | 0.28 | 0.12 | Intergenic |
| rs1805128 | KCNE1 | 21 | 35821680 | A | G | 0.01 | 0.002 | 0.01 | 0.004 | 0.01 | Missense |
Base pair based on genome build 37
Indicates intergenic within 2,000 basepairs of the 3′ end of the gene
Chr indicates chromosome; CA, Coded Allele, based on allele reported in previous genome-wide association studies; NCA, non-coded allele
Summary Results Among European-Descent Populations
After meta-analysis, 20 of 21 SNPs (95%) representing all 12 genetic loci were associated with QT in European Americans, with little evidence of study heterogeneity (eTables 3 and 4). The only European ancestry-identified SNP that was not associated with QT was rs12053903 (intronic to SCN5A), which had an estimated effect size of -0.21 (95% confidence interval [CI] = -0.86 to 0.45). For the remaining 20 SNPs, the estimated direction and magnitude of effects were consistent with previously published results.
Generalizability of GWAS-Identified QT SNPs to Populations of Non-European Descent
Direction and Precision of Effect
Seven of the 21 previously reported QT SNPs (33%) had a consistent direction of effect across all populations (Figure 1). These seven SNPs (rs12143842, rs12029454, rs11129795, rs2074238, rs37062, rs2074518, and rs1805128) represented six genetic loci (NOS1AP, SCN5A, KCNQ1, NDRG4, LIG3 and KCNE1). Results were noticeably less precise for rs2074238 and rs1805128 (imprecise in all four non-European populations) than for the other five SNPs, which were precise in all five populations. Estimated imprecision for these two SNPs likely reflects the low estimated minor allele frequency across race/ethnicity (0.02 – 0.08 frequency for rs2074238; 0.002 – 0.01 for rs1805128) and a smaller sample size for rs2074238. Additionally, rs12143842, rs37062, and rs1805128 showed heterogeneity within at least one race/ethnicity (eTables 5-7).
Figure 1.
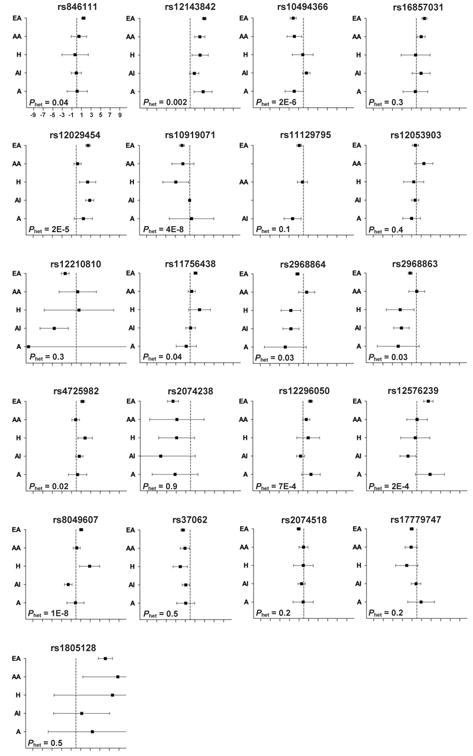
Forest Plots of Risk Variants Effect Sizes (95% Confidence Interval) for a Race/Ethnicity-Stratified Meta-Analysis of 32,474 Participants From Six Participating PAGE Studies.
Abbreviations: A, Asian; AA, African American; AI, American Indian; EA, European American; H, Hispanic; Phet, Two sided P-value for test of heterogeneity across populations;
Nine SNPs (43%) showed a consistent direction of effect in four of the five populations (Figure 1). These nine SNPs (rs17779747, rs12053903, rs11756438, rs4725982, rs2968864, rs2968863, rs10919071, rs10494366, and rs12296050) represented seven loci (KCNJ2, SCN5A, PLN, KCNH2, ATP1B1, NOS1AP, and KCNQ1). There was no race/ethnicity identified as an outlier across these nine SNPs: the directionally inconsistent effect estimate was observed in the African American, Asian, and American Indian populations four, three, and two times, respectively. Of these nine SNPs, imprecision was noted among Hispanic and Asian subpopulations for rs10919071 and rs2968863 and in Asians alone for rs2968864 and rs17779747; these groups had smaller sample sizes than the other race/ethnic groups. Additionally, two SNPs (rs4725982 and rs12296050) in African Americans and five SNPs (rs2968864, rs2968863, rs4725982, rs12296050, and rs17779747) in American Indians showed within-population heterogeneity in American Indians (eTables 5-7).
The remaining five SNPs (24%: rs12210810, rs16857031, rs846111, rs12576239, and rs8049607) showed considerable variation in direction of effect across race/ethnicity, with notable variation in precision.
Magnitude of Effect
In general, populations of non-European descent were more likely to have weaker estimated effect sizes than the European American population, although we found some variation in precision, particularly for Hispanic and Asian populations (eTable 3). Evidence of effect attenuation was particularly apparent among African American participants (18 of 21 SNPs with effects closer to the null). Notably, Hispanics were equally likely to have weaker or stronger effects compared with European-descent populations.
Heterogeneity of Estimated Effects Across Race/Ethnicity
Overall, evidence of heterogeneity was observed for 12 of the 21 examined SNPs (57%). Of the seven directionally consistent SNPs, heterogeneity of P < 0.05 among racial/ethnic groups was observed for two NOS1AP SNPs (rs12143842 [P = 2×10-3] and rs12029454 [P = 2×10-5]) (Figure 1, eTable 8). Among the nine SNPs showing a consistent direction of effect in four populations, seven demonstrated notable heterogeneity among groups: rs109109071, rs111756438, rs10494366, rs4725982, rs12296050, rs2968863, and rs2968864. For SNPs with an inconsistent direction of effect, heterogeneity of P < .05 was observed for three of these five SNPs (rs8049607 [P = 1×10-8], rs12576239 [P = 2×10-4], and rs846111 [P = 0.04]).
Haplotype Structure
Given the substantial evidence of among-race heterogeneity, we examined linkage disequilibrium patterns using data from five HapMap 3 populations to determine whether the observed heterogeneity of effect could be attributed to differences in linkage disequilibrium among racial/ethnic groups. For example, a large haplotype block surrounded rs12143842 (test for heterogeneity among racial/ethnic groups, P = 0.002) in the European ancestry (24 kilobases [kb]), Asian ancestry (14 kb), and Hispanic ancestry (10 kb) populations (Figure 2). However, for African Americans, the haplotype block containing rs12143842 was much smaller (4kb) and did not contain any SNPs downstream of rs12143842. Another example of population-specific linkage disequilibrium patterns that may help explain the observed heterogeneity was provided by rs2968864 (test for heterogeneity, P = 0.03), which exhibited marked variation in effect size and haplotype block structure by race/ethnicity. Conversely, rs17779747, which did not exhibit among-race heterogeneity (P = 0.2), showed very similar linkage disequilibrium patterns across HapMap populations.
Figure 2.
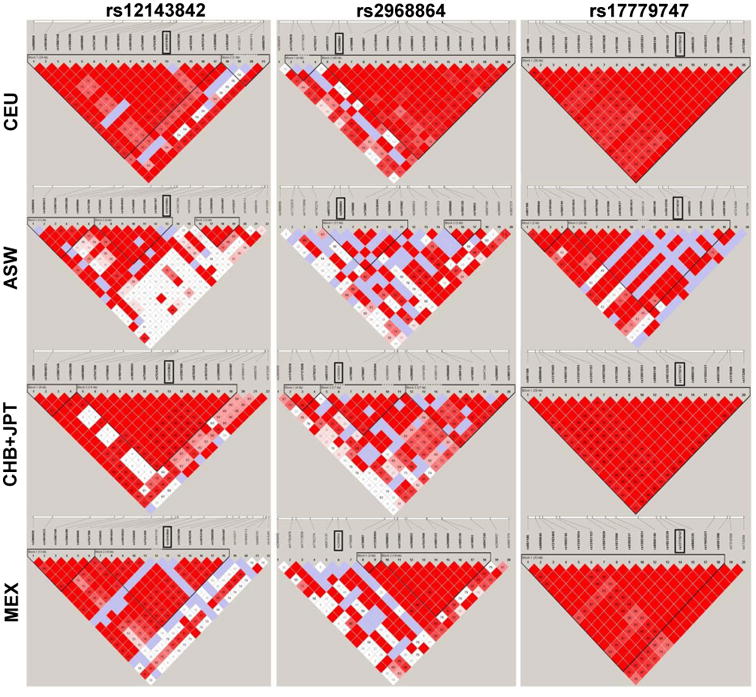
Linkage disequilibrium Plots (D′) from Haploview for Selected Risk Variants Across Four Global Populations.
All populations come from International HapMap III populations. Abbreviations: ASW, Samples of African American Ancestry; CEU, Samples of European Ancestry; CHB-JPT, Samples of Asian Ancestry; MEX, Samples of Hispanic Ancestry.
Discussion
In this study, we evaluated evidence of generalizability for 21 index QT SNPs identified by prior GWAS of European descent populations in multi-ethnic populations from the PAGE study. Evidence of heterogeneity of effect across race/ethnicity was observed for a majority of SNPs, including SNPs from NOS1AP, the most commonly identified QT locus.8-10,12,13 Analyses of HapMap 3 populations suggested that variation in estimated effect by race/ethnicity may reflect underlying variation in linkage disequilibrium among race/ethnicity. Variation in linkage disequilibrium patterns as a cause for estimated effect size variation is further supported by the weaker effects seen in African Americans, where linkage disequilibrium blocks are smaller, which suggests that the causal SNPs may not be as effectively tagged as in European Americans.
Heterogeneity of effect by race/ethnicity can reflect several phenomena, including variation in linkage disequilibrium among populations,23 allelic heterogeneity (i.e. the same locus but different causal variants influencing a trait across populations),26 or gene-gene and gene-environment interaction.27 Variation in linkage disequilibrium by race is a plausible source of the observed heterogeneity, given that several PAGE populations, particularly African Americans and Hispanics, have smaller linkage disequilibrium blocks than populations of European descent.23 For this reason, the index SNPs identified in European American populations may not tag the underlying causal SNP in populations of African or Hispanic descent because it resides in a different linkage disequilibrium block.
Another potential cause of heterogeneity of effect across race/ethnicity is allelic heterogeneity, which has been demonstrated by studies of other ECG traits, particularly the PR interval.28 In addition to the potential for race/ethnicity-specific alleles influencing QT, allelic heterogeneity is complicated by the potential for rare causal variants that can create “synthetic” signals. GWAS-identified index SNPs, including QT index SNPs, could simply represent a collection of rare causal variants, which would likely differ among populations – rather than a single causal SNP.29 As the number of rare variants increases, so too does the potential for these synthetic associations to be found.29 Thus, estimated effects would be expected to differ among racial/ethnic groups, as the SNPs evaluated above are representative of a diverse collection of rare variants.
Furthermore, heterogeneity of effect could be caused by underlying gene-gene or gene-environment interaction, especially if potential modifiers vary among populations. Previous studies have shown interaction between genetic variants and QT-altering pharmaceuticals, although only a single SNP examined in this study (rs1805128) was previously identified as a potential modifier in a large pharmacogenomics effort examining QT.30 Additionally, lifestyle factors (including physical activity, rest/sleep, and emotional stimuli) may also interact with genetic variants associated with long-QT syndromes,31 although few studies have evaluated these potential modifiers in large population-based samples. Gene-gene interactions also are biologically plausible causes of heterogeneity, given evidence of population-specific variants influencing QT.32 However, very few studies have examined the influence of gene-gene interactions with QT, given the immensity of the task of testing interactions among millions of GWAS variants.
Regardless of the source of heterogeneity, these results, as well as studies of other heritable traits including type 2 diabetes33 and obesity,34 suggest that genomic studies of ancestrally diverse populations should analyze racial/ethnic groups separately, unless strong evidence of homogeneity is observed. However, the pooling of results across these groups is common in genetic epidemiology studies, particularly when there are small samples of non-European populations.35,36 A similar practice is the meta-analysis of summary results across racial/ethnic background,37,38 although for common variants this approach is equivalent to pooling.39 Therefore, strategies that pool or meta-analyze results across race implicitly assume that the index SNP tags the causal variant across all populations; they also assume the absence of allelic heterogeneity and among-race gene-gene or gene-environment interactions. Instead, to fully understand the genetic architecture underlying disease in diverse populations, studies should allow for these potential differences across distantly related populations and analyze them independently.
Although a hurdle for researchers who wish to combine results, genetic heterogeneity can be leveraged to identify novel genes and narrow intervals flanking index SNPs.23 For example, fine-mapping studies are particularly relevant among admixed populations such as African Americans, Hispanics/Latinos, and American Indians. For QT, fine mapping has been conducted in African Americans32 to refine the region of association surrounding several genes and identify novel NOS1AP and ATP1B1 SNPs specific to African Americans. The evidence of heterogeneity identified by our study further underscores the potential utility of fine-mapping studies in other populations, which could harness among-population differences in linkage disequilibrium to refine regions of association and identify additional SNPs influencing QT.
This study had several notable limitations. First, sample sizes in non-European American populations were modest, especially for Asian Americans, and thus produced less precise estimates. Nonetheless, few studies to date have examined associations between genetic variants and QT across five distinct racial/ethnic groups. Second, SNPs were selected in 2010 and therefore did not reflect more recent publications.12 However, since SNP selection, only one new genetic locus (SLC8A1) has been identified for QT,12 ensuring that our results remain contemporary. Additionally, we did not account for potential comorbidities in our primary analysis, as was done in previous GWAS. However, our results were robust to the exclusion of participants with prevalent stroke and prevalent ischemic heart disease (eTable 9). Furthermore, variations in ECG machines, sampling rates (250 Hz vs. 500 Hz), and reading software between studies (eTable2) could have accounted for a portion of the heterogeneity observed among studies. A sensitivity analysis comparing results using two reading algorithms and sampling rates (Dalhousie at 250 Hz, Marquette 12-SL at 500 Hz) among Women's Health Initiative participants showed little change in the effect estimates between the two methods and no change in the overall conclusions, indicating that differences in ECG methods likely had little effect on the results (eTable 10). Finally, we estimated effects of small magnitude that likely explained only a small fraction of the variation in QT. However, these results have potential clinical and regulatory relevance. The Food and Drug Administration's standard for regulating QT-prolonging pharmaceuticals is a change of QT interval of 5ms,40 a threshold that is easily met when considering combinations of SNPs. Furthermore, our results suggest that the 21 SNPs are likely tag SNPs, which are expected to have smaller effects when compared to the underlying causal SNP.
In conclusion, our findings suggest the presence of considerable heterogeneity among racial/ethnic groups for previously identified QT index SNPs that may reflect several phenomena, including population-specific linkage disequilibrium patterns. More broadly, our results underscore the utility of examining heterogeneity by race/ethnicity in genetic association studies. Further characterization of these loci across multi-ethnic populations, including large-scale genotyping, is needed to provide additional insights into the genetic architecture of QT.
Supplementary Material
Acknowledgments
The data and materials included in this report result from a collaboration among the following studies:
The “Epidemiologic Architecture for Genes Linked to Environment (EAGLE)” is funded through the NHGRI PAGE program (U01HG004798-01 and its NHGRI ARRA supplement). Genotyping services for select NHANES III SNPs presented here were also provided by the Johns Hopkins University under federal contract number (N01-HV-48195) from NHLBI. The study participants derive from the National Health and Nutrition Examination Surveys (NHANES), and these studies are supported by the Centers for Disease Control and Prevention.
Funding support for the “Epidemiology of putative genetic variants: The Women's Health Initiative” study is provided through the NHGRI PAGE program (U01HG004790 and its NHGRI ARRA supplement). The WHI program is funded by the National Heart, Lung, and Blood Institute; NIH; and U.S. Department of Health and Human Services through contracts N01WH22110, 24152, 32100-2, 32105-6, 32108-9, 32111-13, 32115, 32118-32119, 32122, 42107-26, 42129-32, and 44221. The authors thank the WHI investigators and staff for their dedication, and the study participants for making the program possible. A full listing of WHI investigators can be found at: http://www.whiscience.org/publications/WHI_investigators_shortlist.pdf.
Funding support for the Genetic Epidemiology of Causal Variants Across the Life Course (CALiCo) program was provided through the NHGRI PAGE program (U01HG004803 and its NHGRI ARRA supplement). The following studies contributed to this manuscript and are funded by the following agencies: The Atherosclerosis Risk in Communities (ARIC) Study is carried out as a collaborative study supported by National Heart, Lung, and Blood Institute contracts N01-HC-55015, N01-HC-55016, N01-HC-55018, N01-HC-55019, N01-HC-55020, N01-HC-55021, N01-HC-55022. The Cardiovascular Health Study (CHS) is supported by contracts HHSN268201200036C, HHSN268200800007C, N01 HC55222, N01HC85079, N01HC85080, N01HC85081, N01HC85082, N01HC85083, N01HC85086, and grants HL080295 and HL087652 from the National Heart, Lung, and Blood Institute (NHLBI), with additional contribution from the National Institute of Neurological Disorders and Stroke (NINDS). Additional support was provided by AG023629 from the National Institute on Aging (NIA). A full list of principal CHS investigators and institutions can be found at http://www.chs-nhlbi.org/PI.htm. CHS GWAS DNA handling and genotyping at Cedars-Sinai Medical Center was supported in part by the National Center for Research Resources, grant UL1RR033176, and is now at the National Center for Advancing Translational Sciences, CTSI grant UL1TR000124; in addition the National Institute of Diabetes and Digestive and Kidney Diseases grant DK063491 to the Southern California Diabetes Endocrinology Research Center. The Strong Heart Study (SHS) is supported by NHLBI grants U01 HL65520, U01 HL41642, U01 HL41652, U01 HL41654, and U01 HL65521.
Assistance with phenotype harmonization, SNP selection and annotation, data cleaning, data management, integration and dissemination, and general study coordination was provided by the PAGE Coordinating Center (U01HG004801-01 and its NHGRI ARRA supplement). The National Institutes of Mental Health also contributes to the support for the Coordinating Center.
The PAGE consortium thanks the staff and participants of all PAGE studies for their important contributions.
Sources of Financial Support: This work was supported by the Population Architecture Using Genomics and Epidemiology (PAGE) program, which is funded by the National Human Genome Research Institute (NHGRI), supported by U01HG004803 (CALiCo), U01HG004798 (EAGLE), U01HG004802 (MEC), U01HG004790 (WHI), and U01HG004801 (Coordinating Center), and their respective NHGRI ARRA supplements. The contents of this paper are solely the responsibility of the authors and do not necessarily represent the official views of the NIH. The complete list of PAGE members can be found at http://www.pagestudy.org.
Footnotes
Conflicts of Interest: None
SDC Supplemental digital content is available through direct URL citations in the HTML and PDF versions of this article (www.epidem.com). This content is not peer-reviewed or copy-edited; it is the sole responsibility of the author.
References
- 1.Moss AJ. The QT Interval and Torsade de Pointes. Drug Safety. 1999;21(S1):5–10. doi: 10.2165/00002018-199921001-00002. [DOI] [PubMed] [Google Scholar]
- 2.Zhang Y, Post WS, Blasco-Colmenares E, Dalal D, Tomaselli GF, Guallar E. Electrocardiographic QT Interval and Mortality: A Meta-analysis. Epidemiology. 2011;22(5):660–670. doi: 10.1097/EDE.0b013e318225768b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kramer B, Brill M, Bruhn A, Kubler W. Relationship between the degree of coronary artery disease and of left ventricular function and the duration of the QT-interval in ECG. Eur Heart J. 1986;7(1):14–24. doi: 10.1093/oxfordjournals.eurheartj.a061951. [DOI] [PubMed] [Google Scholar]
- 4.Bazett HC. An Analysis of the Time-Relations of Electrocardiograms. Annals of Noninvasive Electrocardiology. 1997;2(2):177–194. [Google Scholar]
- 5.Mangoni AA, Kinirons MT, Swift CG, Jackson SH. Impact of age on QT interval and QT dispersion in healthy subjects: a regression analysis. Age Ageing. 2003;32(3):326–31. doi: 10.1093/ageing/32.3.326. [DOI] [PubMed] [Google Scholar]
- 6.Akylbekova EL, Crow RS, Johnson WD, Buxbaum SG, Njemanze S, Fox E, Sarpong DF, Taylor HA, Newton-Cheh C. Clinical correlates and heritability of QT interval duration in blacks: the Jackson Heart Study. Circ Arrhythm Electrophysiol. 2009;2(4):427–32. doi: 10.1161/CIRCEP.109.858894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shah M, Akar FG, Tomaselli GF. Molecular basis of arrhythmias. Circulation. 2005;112(16):2517–29. doi: 10.1161/CIRCULATIONAHA.104.494476. [DOI] [PubMed] [Google Scholar]
- 8.Arking DE, Pfeufer A, Post W, Kao WH, Newton-Cheh C, Ikeda M, West K, Kashuk C, Akyol M, Perz S, Jalilzadeh S, Illig T, Gieger C, Guo CY, Larson MG, Wichmann HE, Marban E, O'Donnell CJ, Hirschhorn JN, Kaab S, Spooner PM, Meitinger T, Chakravarti A. A common genetic variant in the NOS1 regulator NOS1AP modulates cardiac repolarization. Nat Genet. 2006;38(6):644–51. doi: 10.1038/ng1790. [DOI] [PubMed] [Google Scholar]
- 9.Newton-Cheh C, Eijgelsheim M, Rice KM, de Bakker PI, Yin X, Estrada K, Bis JC, Marciante K, Rivadeneira F, Noseworthy PA, Sotoodehnia N, Smith NL, Rotter JI, Kors JA, Witteman JC, Hofman A, Heckbert SR, O'Donnell CJ, Uitterlinden AG, Psaty BM, Lumley T, Larson MG, Stricker BH. Common variants at ten loci influence QT interval duration in the QTGEN Study. Nat Genet. 2009;41(4):399–406. doi: 10.1038/ng.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pfeufer A, Sanna S, Arking DE, Muller M, Gateva V, Fuchsberger C, Ehret GB, Orru M, Pattaro C, Kottgen A, Perz S, Usala G, Barbalic M, Li M, Putz B, Scuteri A, Prineas RJ, Sinner MF, Gieger C, Najjar SS, Kao WHL, Muhleisen TW, Dei M, Happle C, Mohlenkamp S, Crisponi L, Erbel R, Jockel KH, Naitza S, Steinbeck G, Marroni F, Hicks AA, Lakatta E, Muller-Myhsok B, Pramstaller PP, Wichmann HE, Schlessinger D, Boerwinkle E, Meitinger T, Uda M, Coresh J, Kaab S, Abecasis GR, Chakravarti A. Common variants at ten loci modulate the QT interval duration in the QTSCD Study. Nature Genetics. 2009;41(4):407–414. doi: 10.1038/ng.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jamshidi Y, Nolte IM, Spector TD, Snieder H. Novel genes for QTc interval. How much heritability is explained, and how much is left to find? Genome medicine. 2010;2(5):35. doi: 10.1186/gm156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim JW, Hong KW, Go MJ, Kim SS, Tabara Y, Kita Y, Tanigawa T, Cho YS, Han BG, Oh B. A common variant in SLC8A1 is associated with the duration of the electrocardiographic QT interval. Am J Hum Genet. 2012;91(1):180–4. doi: 10.1016/j.ajhg.2012.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith JG, Avery CL, Evans DS, Nalls MA, Meng YA, Smith EN, Palmer C, Tanaka T, Mehra R, Butler AM, Young T, Buxbaum SG, Kerr KF, Berenson GS, Schnabel RB, Li G, Ellinor PT, Magnani JW, Chen W, Bis JC, Curb JD, Hsueh WC, Rotter JI, Liu Y, Newman AB, Limacher MC, North KE, Reiner AP, Quibrera PM, Schork NJ, Singleton AB, Psaty BM, Soliman EZ, Solomon AJ, Srinivasan SR, Alonso A, Wallace R, Redline S, Zhang ZM, Post WS, Zonderman AB, Taylor HA, Murray SS, Ferrucci L, Arking DE, Evans MK, Fox ER, Sotoodehnia N, Heckbert SR, Whitsel EA, Newton-Cheh C on behalf of the C, consortia C. Impact of Ancestry and Common Genetic Variants on QT Interval in African Americans. Circ Cardiovasc Genet. 2012;5(6):647–655. doi: 10.1161/CIRCGENETICS.112.962787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matise TC, Ambite JL, Buyske S, Carlson CS, Cole SA, Crawford DC, Haiman CA, Heiss G, Kooperberg C, Marchand LL, Manolio TA, North KE, Peters U, Ritchie MD, Hindorff LA, Haines JL. The Next PAGE in Understanding Complex Traits: Design for the Analysis of Population Architecture Using Genetics and Epidemiology (PAGE) Study. American Journal of Epidemiology. 2011;174(7):849–859. doi: 10.1093/aje/kwr160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bush WS, Boston J, Pendergrass SA, Dumitrescu L, Goodloe R, Brown-Gentry K, Wilson S, McClellan B, Torstenson E, Basford MA, Spencer KL, Ritchie MD, Crawford DC. Enabling high-throughput genotype-phenotype associations in the Epidemiologic Architecture for Genes Linked to Environment (EAGLE) project as part of the Population Architecture using Genomics and Epidemiology (PAGE) study. Pac Symp Biocomput. 2013:373–84. [PMC free article] [PubMed] [Google Scholar]
- 16.Design of the Women's Health Initiative Clinical Trial and Observational Study. Controlled Clinical Trials. 1998;19(1):61–109. doi: 10.1016/s0197-2456(97)00078-0. [DOI] [PubMed] [Google Scholar]
- 17.Investigators TA. The Atherosclerosis Risk In Communities (ARIC) Study: Design and Objectives. American Journal of Epidemiology. 1989;129(4):687–702. [PubMed] [Google Scholar]
- 18.Fried LP, Borhani NO, Enright P, Furberg CD, Gardin JM, Kronmal RA, Kuller LH, Manolio TA, Mittelmark MB, Newman A, O'Leary DH, Psaty B, Rautaharju P, Tracy RP, Weiler PG. The cardiovascular health study: Design and rationale. Annals of Epidemiology. 1991;1(3):263–276. doi: 10.1016/1047-2797(91)90005-w. [DOI] [PubMed] [Google Scholar]
- 19.Lee ET, Welty TK, Fabsitz R, Cowan LD, Le NA, Oopik AJ, Cucchiara AJ, Savage PJ, Howard BV. The Strong Heart Study: A Study of Cardiovascular Disease in American Indians: Design and Methods. American Journal of Epidemiology. 1990;132(6):1141–1155. doi: 10.1093/oxfordjournals.aje.a115757. [DOI] [PubMed] [Google Scholar]
- 20.North KE, Howard BV, Welty TK, Best LG, Lee ET, Yeh JL, Fabsitz RR, Roman MJ, MacCluer JW. Genetic and Environmental Contributions to Cardiovascular Disease Risk in American Indians. American Journal of Epidemiology. 2003;157(4):303–314. doi: 10.1093/aje/kwf208. [DOI] [PubMed] [Google Scholar]
- 21.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26(17):2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cochran WG. The Combination of Estimates from Different Experiments. Biometrics. 1954;10(1):101–129. [Google Scholar]
- 23.Rosenberg NA, Huang L, Jewett EM, Szpiech ZA, Jankovic I, Boehnke M. Genome-wide association studies in diverse populations. Nat Rev Genet. 2010;11(5):356–366. doi: 10.1038/nrg2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.International HapMap C. Altshuler DM, Gibbs RA, Peltonen L, Altshuler DM, Gibbs RA, Peltonen L, Dermitzakis E, Schaffner SF, Yu F, Peltonen L, Dermitzakis E, Bonnen PE, Altshuler DM, Gibbs RA, de Bakker PI, Deloukas P, Gabriel SB, Gwilliam R, Hunt S, Inouye M, Jia X, Palotie A, Parkin M, Whittaker P, Yu F, Chang K, Hawes A, Lewis LR, Ren Y, Wheeler D, Gibbs RA, Muzny DM, Barnes C, Darvishi K, Hurles M, Korn JM, Kristiansson K, Lee C, McCarrol SA, Nemesh J, Dermitzakis E, Keinan A, Montgomery SB, Pollack S, Price AL, Soranzo N, Bonnen PE, Gibbs RA, Gonzaga-Jauregui C, Keinan A, Price AL, Yu F, Anttila V, Brodeur W, Daly MJ, Leslie S, McVean G, Moutsianas L, Nguyen H, Schaffner SF, Zhang Q, Ghori MJ, McGinnis R, McLaren W, Pollack S, Price AL, Schaffner SF, Takeuchi F, Grossman SR, Shlyakhter I, Hostetter EB, Sabeti PC, Adebamowo CA, Foster MW, Gordon DR, Licinio J, Manca MC, Marshall PA, Matsuda I, Ngare D, Wang VO, Reddy D, Rotimi CN, Royal CD, Sharp RR, Zeng C, Brooks LD, McEwen JE. Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467(7311):52–8. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 26.Wood AR, Hernandez DG, Nalls MA, Yaghootkar H, Gibbs JR, Harries LW, Chong S, Moore M, Weedon MN, Guralnik JM, Bandinelli S, Murray A, Ferrucci L, Singleton AB, Melzer D, Frayling TM. Allelic heterogeneity and more detailed analyses of known loci explain additional phenotypic variation and reveal complex patterns of association. Human Molecular Genetics. 2011;20(20):4082–4092. doi: 10.1093/hmg/ddr328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thomas D. Gene–environment-wide association studies: emerging approaches. Nat Rev Genet. 2010;11(4):259–272. doi: 10.1038/nrg2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith JG, Magnani JW, Palmer C, Meng YA, Soliman EZ, Musani SK, Kerr KF, Schnabel RB, Lubitz SA, Sotoodehnia N, Redline S, Pfeufer A, Muller M, Evans DS, Nalls MA, Liu Y, Newman AB, Zonderman AB, Evans MK, Deo R, Ellinor PT, Paltoo DN, Newton-Cheh C, Benjamin EJ, Mehra R, Alonso A, Heckbert SR, Fox ER Candidate-gene-Association Resource C. Genome-wide association studies of the PR interval in African Americans. PLoS Genet. 2011;7(2):e1001304. doi: 10.1371/journal.pgen.1001304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dickson SP, Wang K, Krantz I, Hakonarson H, Goldstein DB. Rare Variants Create Synthetic Genome-Wide Associations. PLoS Biol. 2010;8(1):e1000294. doi: 10.1371/journal.pbio.1000294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaab S, Crawford DC, Sinner MF, Behr ER, Kannankeril PJ, Wilde AA, Bezzina CR, Schulze-Bahr E, Guicheney P, Bishopric NH, Myerburg RJ, Schott JJ, Pfeufer A, Beckmann BM, Martens E, Zhang T, Stallmeyer B, Zumhagen S, Denjoy I, Bardai A, Van Gelder IC, Jamshidi Y, Dalageorgou C, Marshall V, Jeffery S, Shakir S, Camm AJ, Steinbeck G, Perz S, Lichtner P, Meitinger T, Peters A, Wichmann HE, Ingram C, Bradford Y, Carter S, Norris K, Ritchie MD, George AL, Jr, Roden DM. A large candidate gene survey identifies the KCNE1 D85N polymorphism as a possible modulator of drug-induced torsades de pointes. Circ Cardiovasc Genet. 2012;5(1):91–9. doi: 10.1161/CIRCGENETICS.111.960930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schwartz PJ, Priori SG, Spazzolini C, Moss AJ, Vincent GM, Napolitano C, Denjoy I, Guicheney P, Breithardt G, Keating MT, Towbin JA, Beggs AH, Brink P, Wilde AA, Toivonen L, Zareba W, Robinson JL, Timothy KW, Corfield V, Wattanasirichaigoon D, Corbett C, Haverkamp W, Schulze-Bahr E, Lehmann MH, Schwartz K, Coumel P, Bloise R. Genotype-phenotype correlation in the long-QT syndrome: gene-specific triggers for life-threatening arrhythmias. Circulation. 2001;103(1):89–95. doi: 10.1161/01.cir.103.1.89. [DOI] [PubMed] [Google Scholar]
- 32.Avery CL, Sethupathy P, Buyske S, He Q, Lin DY, Arking DE, Carty CL, Duggan D, Fesinmeyer MD, Hindorff LA, Jeff JM, Klein L, Patton KK, Peters U, Shohet RV, Sotoodehnia N, Young AM, Kooperberg C, Haiman CA, Mohlke KL, Whitsel EA, North KE. Fine-Mapping and Initial Characterization of QT Interval Loci in African Americans. PLoS Genet. 2012;8(8):e1002870. doi: 10.1371/journal.pgen.1002870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haiman CA, Fesinmeyer MD, Spencer KL, Buzkova P, Voruganti VS, Wan P, Haessler J, Franceschini N, Monroe KR, Howard BV, Jackson RD, Florez JC, Kolonel LN, Buyske S, Goodloe RJ, Liu S, Manson JE, Meigs JB, Waters K, Mukamal KJ, Pendergrass SA, Shrader P, Wilkens LR, Hindorff LA, Ambite JL, North KE, Peters U, Crawford DC, Le Marchand L, Pankow JS. Consistent directions of effect for established type 2 diabetes risk variants across populations: the population architecture using Genomics and Epidemiology (PAGE) Consortium. Diabetes. 2012;61(6):1642–7. doi: 10.2337/db11-1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fesinmeyer MD, North KE, Ritchie MD, Lim U, Franceschini N, Wilkens LR, Gross MD, Buzkova P, Glenn K, Quibrera PM, Fernandez-Rhodes L, Li Q, Fowke JH, Li R, Carlson CS, Prentice RL, Kuller LH, Manson JE, Matise TC, Cole SA, Chen CT, Howard BV, Kolonel LN, Henderson BE, Monroe KR, Crawford DC, Hindorff LA, Buyske S, Haiman CA, Le Marchand L, Peters U. Genetic Risk Factors for BMI and Obesity in an Ethnically Diverse Population: Results From the Population Architecture Using Genomics and Epidemiology (PAGE) Study. Obesity (Silver Spring) 2012 doi: 10.1002/oby.20268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marciante KD, Bis JC, Rieder MJ, Reiner AP, Lumley T, Monks SA, Kooperberg C, Carlson C, Heckbert SR, Psaty BM. Renin-Angiotensin System Haplotypes and the Risk of Myocardial Infarction and Stroke in Pharmacologically Treated Hypertensive Patients. American Journal of Epidemiology. 2007;166(1):19–27. doi: 10.1093/aje/kwm059. [DOI] [PubMed] [Google Scholar]
- 36.Olfson E, Bierut LJ. Convergence of Genome-Wide Association and Candidate Gene Studies for Alcoholism. Alcoholism: Clinical and Experimental Research. 2012;36(12):2086–2094. doi: 10.1111/j.1530-0277.2012.01843.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ober C, Nicolae DL. Meta-analysis of genome-wide association studies of asthma in ethnically diverse North American populations. Nat Genet. 2011;43(9):887–892. doi: 10.1038/ng.888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rong Y, Bao W, Rong S, Fang M, Wang D, Yao P, Hu FB, Liu L. Hemochromatosis Gene (HFE) Polymorphisms and Risk of Type 2 Diabetes Mellitus: A Meta-Analysis. American Journal of Epidemiology. 2012;176(6):461–472. doi: 10.1093/aje/kws126. [DOI] [PubMed] [Google Scholar]
- 39.Lin DY, Zeng D. Meta-analysis of genome-wide association studies: no efficiency gain in using individual participant data. Genetic Epidemiology. 2010;34(1):60–66. doi: 10.1002/gepi.20435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.FDA. Guidance for Industry: E14 Clinical Evaluation of QT/QTc Interval Prolongation and Proarrhythmic Potential for Non-Antiarrhythmic Drugs. In: Services DoHaH, editor. Guidance for Industry. 2005. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


