Abstract
Many cell types, including neurons, astrocytes and other cells of the central nervous system respond to changes in extracellular matrix or substrate viscoelasticity, and increased tissue stiffness is a hallmark of several disease states including fibrosis and some types of cancers. Whether the malignant tissue in brain, an organ that lacks the protein-based filamentous extracellular matrix of other organs, exhibits the same macroscopic stiffening characteristic of breast, colon, pancreatic, and other tumors is not known. In this study we show that glioma cells like normal astrocytes, respond strongly in vitro to substrate stiffness in the range of 100 to 2000 Pa, but that macroscopic (mm to cm) tissue samples isolated from human glioma tumors have elastic moduli on the order of 200 Pa that are indistinguishable from those of normal brain. However, both normal brain and glioma tissues increase their shear elastic moduli under modest uniaxial compression, and glioma tissue stiffens more strongly under compression than does normal brain. These findings suggest that local tissue stiffness has the potential to alter glial cell function, and that stiffness changes in brain tumors might arise not from increased deposition or crosslinking of collagen-rich extracellular matrix but from pressure gradients that form within the tumors in vivo.
Keywords: glioma, viscoelasticity, mechanosensing, astrocyte, brain rheology
Introduction
A common feature of many solid tumors and other diseased tissues is that they are stiffer than the normal tissue in which they arise 1–4 and often have increased interstitial fluid pressures 5, 6 and solid tissue stress 7, 8. Tissue stiffening, usually quantified as an increase in shear storage or Young's modulus, arises from multiple mechanisms including increased or chemically altered extracellular matrix production, increased matrix crosslinking due to upregulation of lysyl oxidases, and increased intercellular tensions driven by abnormal activation of acto-myosin that produces increased internal stress within the strain-stiffening matrix. Numerous recent studies have explored the hypothesis that increased matrix or stromal tissue stiffness is not simply a consequence of abnormal cell biology in cancer, fibrosis, and other diseases, but that abnormal stiffness actively contributes to disease progression by activating mechanosensitive motility, transcriptional, and proliferative pathways within the transformed cells 9. It has been hypothesized that a parallel mechanical effect derived from increased pressure within tumors potentially alters cell function and can provide a selective advantage to genetically transformed cells that adapt to increased pressure better than the surrounding normal cells 7, 8.
The hypothesis that altered extracellular matrix is essential to the increased stiffening of tumor stroma presents an interesting issue in the context of glioblastoma and other brain tumors, because brain and other CNS tissue is conspicuously devoid of the filamentous protein-based extracellular matrix (including collagens) characteristic of most mesenchymal and epidermal environments 10. Isolated glial cells and neurons are highly sensitive to substrate stiffness changes in the range from 100 to 2000 Pa 11–13, which is relevant to brain rheology, and cells are likely to respond on the time scale of seconds14, but whether the malignant tissue in brain exhibits the same macroscopic stiffening characteristic of colon, pancreatic, and other tumors is not known. To determine whether increased glioma stiffness in vivo drives the abnormal behavior of glioma cells, we evaluated the stiffness of glioma versus normal brain tissue. To our surprise, we found that isolated glioma samples have the same shear storage moduli as normal brain when measured ex vivo at low strains, but that the shear moduli of both normal and malignant brain tissue increase to the kPa range when the tissue is uniaxially compressed. We suggest that compression stiffening, which might occur with the increased vascularization and interstitial pressure gradients that are characteristic of glioma and other tumors, effectively stiffens the environment of glioma cells and that in situ, the elastic resistance these cells sense might be sufficient to trigger the same responses that are activated in vitro by increased substrate stiffness.
The mechanical properties of brain have been previously quantified by a number of shear 15, 16, tension 17, indentation 18, and compression 19 experiments in which samples were subjected to creep, stress-relaxation, frequency sweep, and strain sweep tests (reviewed in 14, 20, 21). A comprehensive characterization of brain rheology is still incomplete due to the large variability in outcomes from study to study, which may be due to differences in protocols, in vivo versus in vitro methods, and brain donor species or age 15. Values for G’ and G” found in literature, for example, range anywhere from 100-104 Pa and 20–1000 Pa, respectively (reviewed in 15, 20, 21), depending on parameters such as frequency, strain rate, and temperature. Although, the effect of compression prior to shear measurements has been noted 22, the work presented here is novel in that it demonstrates how G’ and G” change in response to both extension and compression as well as time (relaxation of both G’ and axial stress). Furthermore, in most prior work, brain rheology was studied in the context of brain injuries 16–19, whereas this study addresses possible stiffness differences between normal brain and glioma as well as understanding glial cell/neuron mechanosensing.
Materials and methods
Tissue samples
Biopsy specimens were obtained from nine patients treated for glioblastoma multiforme (GBM) at the Department of Neurosurgery, Temple University Hospital. Collection of samples was performed in accordance with an approved IRB protocol. Eight samples were frozen at −80 °C within 1–2 hours after surgery and thawed immediately before rheological testing. To determine the effect of freezing, one sample was measured within 3 hours after surgery, and its viscoelastic parameters were indistinguishable from those of the frozen samples. The patient population included both males and females with an age range of 40–75.
Normal mouse brains were obtained from wild type C57BL/6 males with an age ranging from 11 to 15 weeks. Fresh brain tissues were stored in Dulbecco`s modified Eagle medium (DMEM, Gibco, Grand Island, NY) and tested within a maximum of three hours after sacrifice.
Cells
LN229 cells were obtained from American Type Culture Collection (LN229 CRL2611; ATCC, Manassas, VA), and cultured in DMEM (Gibco, Grand Island, NY) with 10% fetal bovine serum (FBS, Gibco, Grand Island, NY), 100 U/ml penicillin and 100 µM streptomycin (SIGMA-ALDRICH, St. Louis, MO) on tissue culture plastic at 37°C and 5% CO2 in a humidified incubator. Cells were subcultured every 2–3 days. The LN229 cell line was established from cells taken from a patient with right frontal parieto-occipital glioblastoma 23. Normal astrocytes were obtained from cortices of prenatal rat embryos, removed by caesarean section from timed-pregnant Sprague-Dawley rats. Tissue samples were digested in trypsin/DNase at 37°C, centrifuged (1000 g × 5 min), and filtered to derive a cell suspension that was plated and maintained for 14 days. Neuronal cells were then removed through a series of trypsinizations as described elsewhere 12. This procedure results in cultures that are >98% astrocytes as assessed by GFAP immunocytochemistry.
Hydrogel substrate preparation and fabrication
Cells were cultured on polyacrylamide gel substrates of varying stiffness as described previously 24, 25. Briefly, different ratios of 40% acrylamide and 2% bis-acrylamide stock solutions (BIORAD Laboratories, Hercules, CA) were prepared in double distilled H20 to a total volume of 1 mL. Polymerization was initiated by addition of 1uL TEMED Electrophoresis Grade (Fisher BioReagents, Pittsburgh, PA) and 10 uL of ammonium persulfate (Thermo Fisher Scientific, Rockford, IL). A 60 uL droplet of desired solution was immediately deposited on a 15 mm glass coverslip pretreated with (3-aminopropyl)trimethoxysilane (SIGMA-ALDRICH, St. Louis, MO) and glutaraldehyde. An 18 mm chlorosilanized coverslip was placed on top of the droplet and removed after polymerization was complete. The shear modulus of each gel formulation was determined by rheological measurements conducted on a Rheometrics fluids spectrometer III (Rheometrics, Piscataway, NJ) with 8 mm parallel plate geometry using a 2% oscillatory shear strain at a frequency of 5 rad/s. The elastic moduli of different gel formulations were: 300 Pa, 1 kPa, 3.5 kPa, 4.5 kPa, 8 kPa and 14 kPa.
Because polyacrylamide gels do not adsorb proteins, UV-sensitive Sulfo-SANPAH crosslinker (Thermo Fisher Scientific, Rockford, IL) was used to covalently bind proteins to the polyacrylamide surface. After incorporation with the crosslinker, gels were incubated with 0.1 mg/ml laminin (Collaborative Biomedical, Bedford, MA) or 0.1 mg/ml collagen I (BD Bioscience, San Diego, CA) for at least 2 hours to produce a uniform coating of adhesive ligands. Previous studies, including recent ones that include HA as well as PAA gels coated with proteins 16, 18 have confirmed that gels of different stiffnesses have similar densities of adhesion proteins on their surface. The protein concentrations used to coat the surfaces or incorporate into the matrices are in the saturation range for cell adhesion 17, further supporting the conclusion that differences in viscoelasticity and not adhesion cause the effects we report here.
Rheological characterization
Macroscopic rheometry using a Rheometrics RFS3 strain controlled rheometer fitted with 8 mm diameter parallel plates was used to measure the viscoelasticity of cm sized disk shaped samples using methods previously described 26. Normal brain tissues from mice were cut into disk-shaped samples using an 8 mm diameter stainless steel punch. To avoid undesirable sample slippage during sheer deformation and to perform uniaxial extension, fibrin gel was used to glue the sample to the rheometer plate. Briefly, fibrin gels were prepared by mixing of 28 mg/ml salmon fibrinogen solution with 10 units of thrombin directly on the lower rheometer plate and the sample was immediately positioned. Subsequently, a thin layer of fibrin gel was pipetted onto the upper surface of the tissue, and the top plate was lowered until contact was made. Contact was judged by a positive normal force (ordinarily 1g) measured by a force transducer. Because the elastic modulus of 28 mg/ml fibrin is much higher than that of brain, and the height of the fibrin was negligible compared to the tissue samples, the viscoelastic response is dominated by the tissue, and no correction was needed for the presence of the fibrin glue. Control experiments in which the samples were not subjected to extensional strain showed that addition of fibrin did not alter the measured elastic modulus of brain samples. The shear modulus of the samples was measured on a strain-controlled Rheometrics fluids spectrometer III (Rheometrics, Piscataway, NJ), which can also measure normal forces simultaneously with torques arising from sample deformation 27. The dynamic shear storage modulus G’ was registered as a function of time (2% oscillatory shear strain at a frequency of 2 rad/s) and strain (with strain increasing from 1% to 50% at oscillatory shear strain at a frequency of 2 rad/s). For some experiments shown in Figure 4, the sample was first deformed uniaxially, either in compression or extension, and a sinusoidal shear strain was superimposed on the static deformation in the orthogonal direction. The force required to maintain a constant uniaxial deformation can be converted to stress by dividing the normal force by the area of the sample in contact with the rheometer plate.
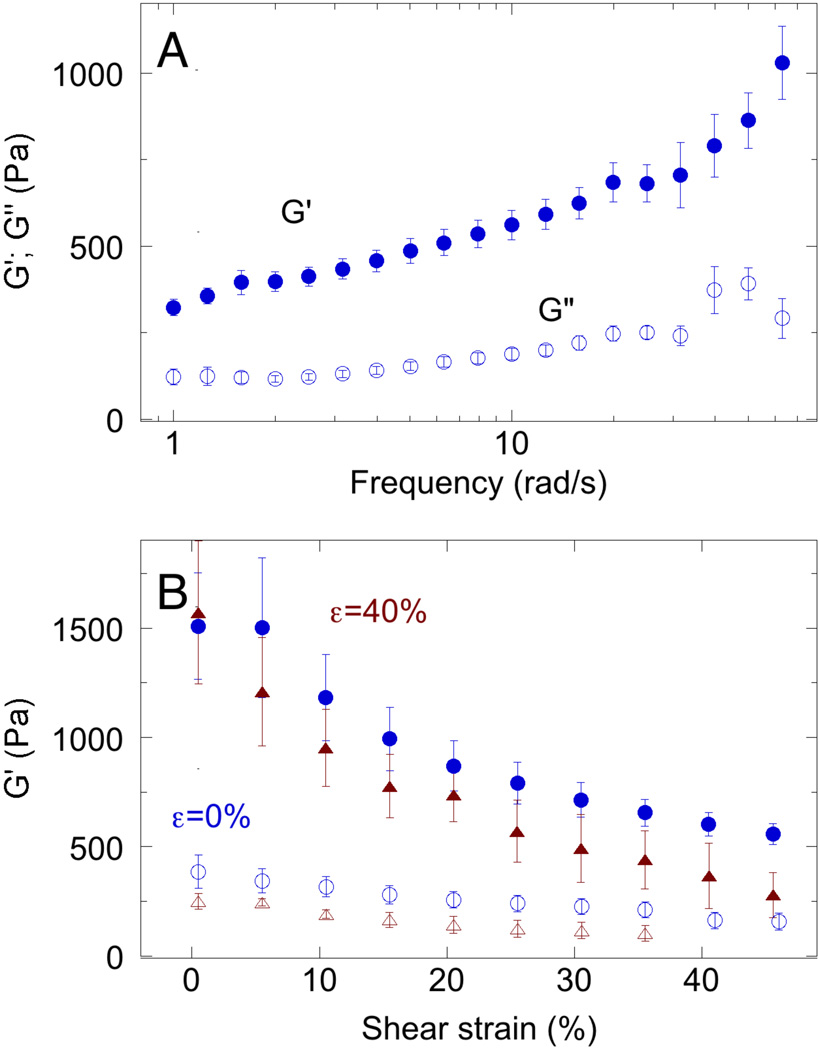
Figure 4. Frequency, temperature, and strain dependence of normal brain rheology.
A. Shear storage (G’) and loss (G") moduli measured at 2 rad/s. Error bars: standard error n=7. B Shear storage modulus (G’) measured at 2 rad/s and various levels of maximal shear strain amplitude, measured at either 23 °C (blue) or 37 °C (red) without (open symbols) or with 40% constant uniaxial compression (µ =40%; closed symbols).
Smaller millimeter-scale samples obtained from biopsy of human tumors as well as mouse brain were measured using a 300 µm diameter flat-bottomed needle indenter (Kibron, Inc., Helsinki, FI) as previously described 28, 29. Briefly, a freely hanging flat-bottomed indenter was brought in contact with either glioblastoma or brain tissue immersed in DMEM culture medium, and changes in force occurring with every 10 µm of additional displacement of the probe was registered. Correction for the deflection of the cantilever in the force probe was done by displacing the probe onto a rigid surface. All the samples were tested at 10 to 12 different locations, and measurements were conducted for no longer than one hour per sample.
Single cell and adjacent substrate stiffness was measured by indentation using atomic force microscopy as previously described 30. The DAFM-2X Bioscope (Veeco, Woodbury, NY) mounted on an Axiovert microscope (Zeiss, Thornwood, NY) was used to carry out the indentation experiments of single cells. Force indentation curves were performed as previously described 29, 31 using a silicon nitride cantilever with a spring constant of 0.06 N/m (NP-O10, Bruker, Madison, WI) and a 3.5 µm diameter polystyrene particle attached to the cantilever tip. AFM experiments were made 24h after cell plating, and cells were kept in their culture medium during experiment. Briefly, force curves were recorded on a hard surface (glass substrate) and then the substrate was replaced by the sample of interest. Indentation was carried out close to the center of the cell at three distinct spots and c.a. 100 cells were collected for each condition. The difference between the cantilever deflection on a hard surface and the cell describes the deformation of the cell under the tip load. By plotting the obtained indentation against the force, so-called force-versus-indentation curve can be plotted. To determine the Young's modulus of different cells, force-versus-indentation curves were fit to the Hertz contact model for a sphere using following formula:
Where E* is the relative Young`s modulus:
If Esample ≪ Etip (as is true for living cells), then can be simplified:
Esample is Young`s modulus of the material and μsample is the Poisson ratio of the sample, related to the compressibility of the material 32 and assumed to be 0.5, as appropriate for an incompressible material.
Microscopy and image analysis
After 24h seeding, cultures were fixed with 4% paraformaldehyde in PBS (SIGMA-ALDRICH, St. Louis, MO) at RT for 20 min. Samples were blocked and permeabilized in 10% bovine serum albumin and 0.1% Triton-X 100 for 10 and 5 minutes, respectively, and afterwards incubated for 1h with primary mouse monoclonal antibodies directed against vimentin (1:200, SIGMA-ALDRICH, St. Louis, MO). After rinsing, the following stains were used: rhodamine-phalloidin (1:500, Molecular Probes, Eugene, OR) was used to label actin filaments, Hoechst (1:1000, Molecular Probes, Eugene, OR) was used to label cell nuclei and anti-mouse Alexa Fluor 488 secondary antibody (1:500, Molecular Probes, Eugene, OR) was used for vimentin filament visualization. The cells were examined using a Leica DMIRE2 inverted microscope (Leica, Buffalo Grove, IL) and images were acquired using a Hamamatsu camera (Hamamatsu, Japan). Immunofluorescence was recorded with a 100x oil lens with a numerical aperture of 1.25. In order to quantify cell adherent area, a 40x air lens with a numerical aperture of 0.60 and phase contrast was used. For each sample, a minimum of 20 images was acquired from different locations and approximately 100 cells per substrate were analyzed. Cell area was calculated using ImageJ software by precise tracing of single cell outlines (Image J Software, NIH, Bethesda, MD) 33.
Results
Reponses of glioma cells and normal astrocytes to substrate stiffness
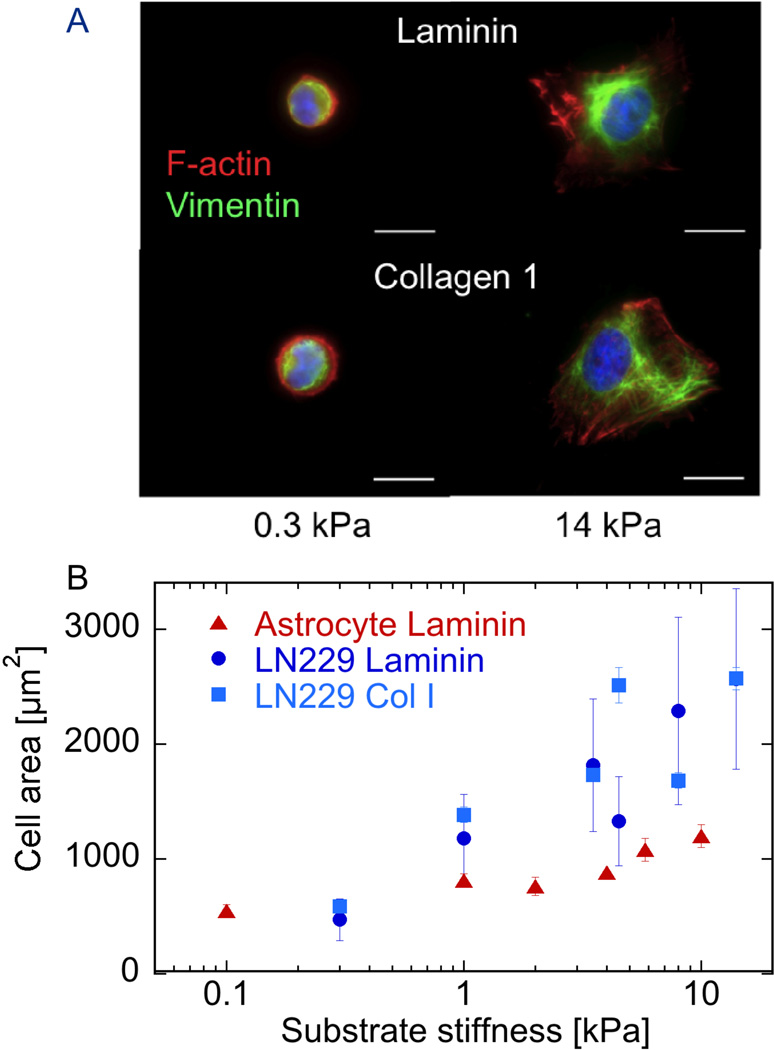
The glioma cell line LN229 responds to substrate stiffness in the range of 100 to 2000 Pa, as shown by changes in adherent area and cytoskeletal assembly depicted in Figure 1. When glioma cells are cultured on polyacrylamide gels of elastic modulus similar to that of the normal brain or on gels that are 50 times stiffer, they adhere and survive on both substrates coated with either collagen 1 or laminin, but their morphology and area are strongly dependent on substrate stiffness. Figure 1A shows that on soft substrates LN229 cells have a round morphology with relatively low adherent area with no actin-rich protrusions, whether bound to collagen 1 or laminin. These cells also expressed the intermediate filament protein vimentin in a network concentrated near the cell center. Similar to many cell types studied, glioma cells increase spread area and produce large actin-rich protrusions that also contain actin bundles on 14 kPa substrates coated with either laminin or collagen 1. The dependence of LN229 cell adherent area on substrate stiffness is quantified in Figure 1B, which shows a monotonic dependence of area on substrate shear modulus over the range from 300 to 14,000 Pa, which spans the stiffness range reported by most studies of brain viscoelasticity over a range of frequencies, strains, and brain region 14. LN229 glioma cells spread significantly more than normal astrocytes over this range of substrate stiffness, regardless of whether they bind to the surface by adhesion to laminin or collagen type I.
Figure 1. Substrate stiffness effects on cell morphology.
A. Fluorescence imaging of nucleus (blue) vimentin (red) and F-actin (green) in LN229 glioma cells cultured 24 hrs on polyacrylamide gels of two different stiffnesses coated with either laminin or collagen I. B. Adherent area of cells such as those shown in A for a range of substrate stiffnesses, compared to those of normal rat brain astrocytes adherent to laminin-coated gels. Error bars denote standard deviations for glioma cells on laminin (n = 100–143) with standard errors shown for other conditions for clarity.
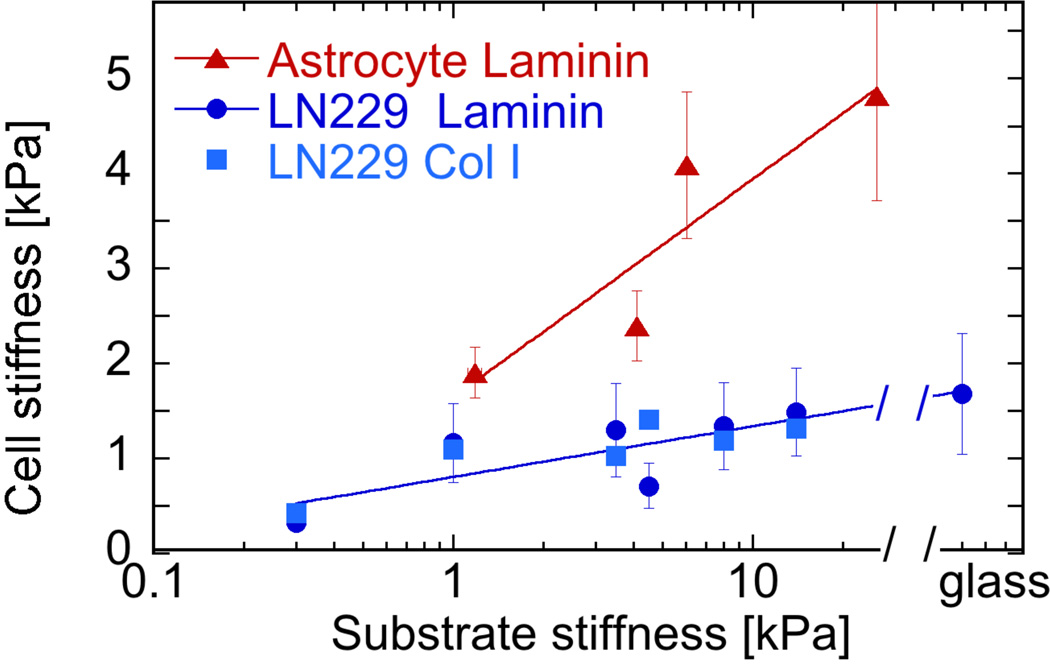
In addition to spread area, the elastic modulus of the glioma cell cortex, as measured by atomic force microscopy indentation also depends on substrate stiffness. Figure 2 shows how the glioma cell stiffness increases with substrate stiffness, reaching a maximal level on 14 kPa gels that is indistinguishable from the stiffness of glioma cells plated on glass. In contrast to these glioma cells, normal astrocytes are stiffer on all substrates studied and on glass can reach elastic moduli over 10 kPa 34.
Figure 2. Effect of substrate stiffness on cell stiffness.
Cortical cell stiffness of normal astrocytes and LN229 glioma cells cultured for 24 hrs on laminin- or collagen I-coated gels of different stiffnesses. Cell stiffness is shown as Young’s modulus derived by AFM by indentation for 700 nm into the cortical surface of cells at sites between nucleus and cell edge. Error bars denote standard deviations for glioma cells on laminin (n = 100–143) with standard errors shown for other conditions for clarity.
Viscoelasticity of glioma tissue
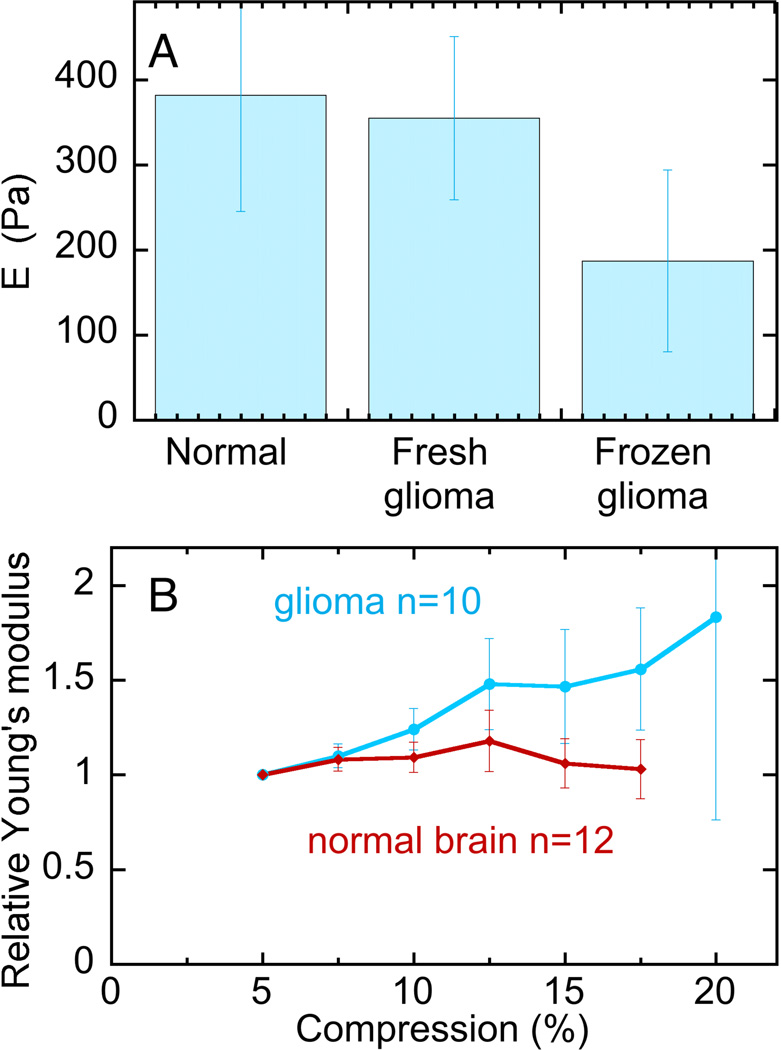
The strong dependence of glioma cell phenotype on substrate stiffness in a range that is relevant to brain rheology suggests that potential changes in tissue viscoelasticity coinciding with tumor development might contribute to progression of glioma growth, as hypothesized for breast cancers and other tumor types. Therefore, the Young's modulus of biopsy specimens from human glioma was measured and compared to that of normal mouse brain, which has an elastic modulus similar to that of human brain 14. Figure 3A shows that contrary to our expectation, the Young’s modulus of glioma tissue measured by indentation at low strain was indistinguishable from that of normal brain, at least under the minimal strain conditions by which these samples were measured ex vivo. In these low strain measurements, a 300 µm indenter was applied at various locations over the surface of 2 to 3 mm scale approximately cubic tissue samples with indentations depths of 200 µm. Although there is significant sample-to-sample variation and some spatial heterogeneity within the samples, there is no evidence from low strain measurements that glioma are stiffer than normal brain. However, as shown in Figure 3B, when indentations were made at increasing depth, corresponding to larger compressions of the tissue, the effective Young’s modulus increased for glioma samples more strongly than for normal brain.
Figure 3. Viscoelasticity of glioma tissue and normal mouse brain.
A. Young’s moduli derived from indentation at depths of 200 µm into the surface of 2 mm thick tissue samples. B. Young’s moduli normalized to their values at 100 µm indentation (5%) for compressions at different depths. The error bars represent standard deviations for 3 normal brains measured at 12 different locations, 1 fresh glioma sample measured at 12 different locations, 6 frozen glioma samples measured at 10 different locations.
Brain stiffens under uniaxial compression but not increasing shear strain
The small size of the biopsy specimens prevents a more thorough rheological characterization as can be made using cm sized samples in a strain-controlled shear rheometer, so these data are limited to tissue slices made from whole normal mouse brain. As opposed to the conditions under which the mechanical properties of excised tissues are measured in vitro, brain in vivo is confined to a specified volume within the skull, and the interstitial fluid pressure is often locally increased within the tumor and in damaged regions following traumatic brain injury. Such increased local pressure can compress the brain, and therefore the local effective mechanical environment of cells can depend on local compressions. Figure 4A shows that the shear storage modulus G' of normal brain is significantly larger than the loss modulus G" when measured at small strain. Both G' and G" increase modestly with increasing frequency of deformation in a manner consistent with numerous previous studies 14, 19, 22. When brain samples are deformed to large, but physiologically relevant strains, the effect on the shear modulus depends on whether the large strain is in shear or uniaxial compression. Figure 4B shows that G' decreases with increasing shear strain when measured at either 23 °C (blue) or 37 °C (red) in an uncompressed sample (open symbols)‥ However, the shear modulus strongly increases with uniaxial compression (closed symbols). For a simple linear viscoelastic material, shear strain is approximately independent of uniaxial compression, but nonlinear viscoelastic materials such as the composite structures of the brain and other soft tissues have more complex rheological responses. Similar compression-dependent increases in shear modulus have previously been reported for brain 26 and liver 35, but the mechanism responsible for this rheological property is not known.
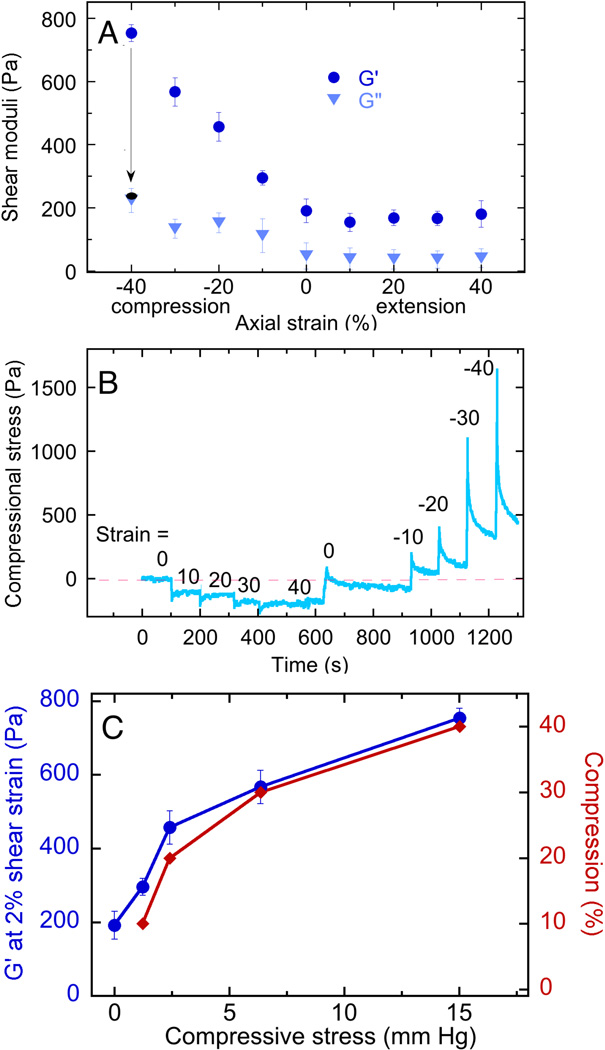
The unusual uniaxial deformation-dependent increase in the shear storage modulus occurs only in compression, but not in extension, and is fully recoverable up to compressive strains of at least 40% as shown in Figure 5. Figure 5A shows that the shear storage modulus G' increases approximately linearly with compressive strain but is nearly independent of strain in extension. The loss modulus G" is much less affected by compressive strain, confirming that the change in rheology is mainly in elasticity, and is not due to sample failure, which would lead to plastic deformation. Recoverability is also noted by the arrow from the blue to the black symbol at −40% axial strain: when a compressed sample is returned to its initial height, the shear modulus returns to its uncompressed value within the time required for the measurement (<15 s).
Figure 5. Compression-dependent stiffening of brain.
A. Shear storage (G’) and loss (G”) moduli measured at 2% shear strain and a frequency of 2 rad/sec for samples subjected to various levels of static uniaxial compression or extension. B. Normal stresses exerted by brain samples after imposition of different levels of elongation or compression. C. Shear storage modulus and compressional strain corresponding to different levels of compressional stress derived from data of Figure 5B.
By measuring the normal force exerted when samples of brain are compressed or extended, the compressive stress required to alter the shear modulus can be computed, as described in the methods section 27. Figure 5B shows that when brain samples are deformed in extension, they develop a small but sustained resisting force that is fully relieved when the sample is returned to the unstrained state. In compression, the opposing forces are much larger and show a significant but incomplete relaxation that occurs over a period of 100 s.
Figure 5C shows that the shear modulus of normal brain can be increased nearly four times by compressive stresses in the range of 3 to 15 mm Hg, as computed from the forces measured 60 seconds after an increment of strain was applied, as shown in Figure 4C. These stresses are well within the range of interstitial pressures developed in brain tumors 5, 6, 36–41.
Discussion
Like many other cell types, LN229 glioma cells respond to increased substrate stiffness by increasing their adherent area and increasing the stiffness of their cortical cytoskeleton/membrane surface. Comparison of the data in Figures 1B and 2 shows that, compared to normal astrocytes – a glial cell type derived from similar precursor cells as those from which LN229 arose by mutation – the glioma cells spread to larger cell areas, but have remarkably lower cortical elastic moduli on substrates with the same elastic modulus. This result confirms the hypothesis that at least for some types of cancer cells, their phenotypes can be characterized by a decreased cellular stiffness, a quantity that has potential for diagnostic applications 42, 43. The combination of increased spreading but decreased stiffening of glioma cells compared to normal astrocytes also has implications for developing models of mechanosensing. Some studies have suggested that cell adherent area is the major determinant of cell tension and cortical stiffness, but the data in Figures 1B and 2 are not consistent with this hypothesis. Instead, in agreement with other results 44, these data suggest that cell spreading and cell stiffening in response to substrate stiffness are independent outcomes of mechanosensing, which can in some cases change in a similar manner, but are not necessarily dependent on each other.
The response of glial cells to increased substrate stiffness suggests that if the stromal tissue within which gliomas develop is stiffer than normal brain, the increased local stiffness might contribute to increased tension, motility, or proliferation in the tumor cells, by analogy with the increased proliferation 45, tension 46 and survival 8, 47 of diseased cells documented in other malignancies such as breast cancer 48.
In most previous cases where increased stromal stiffness has been associated with cancer or fibrotic disease, the increased stiffness has been attributed to increased production of extracellular matrix components or to increased crosslinking of the fibrous matrix already formed within the tissue 49, 50. Most commonly the extracellular matrix changes have been linked to biochemical and mechanical signals arising from changes in the structure or crosslink density of collagen fibers, but the brain is almost entirely lacking in this filamentous fiber scaffold 10. There is often an upregulation of expression of various collagen types in gliomas 51, but whether this expression leads to increased tissue stiffness as it does in other tumor types is not yet known. In this study, we show that macroscopic (mm to cm) tissue samples isolated from human glioma tumors have elastic moduli on the order of 300 Pa that are indistinguishable from those of normal brain, although in vitro human glioma cells lines respond strongly to substrate stiffness changes over the range from 200 to 2000 Pa. Although glioma samples have the same shear storage moduli as normal brain when measured ex vivo at low strains, the shear moduli of both normal and malignant brain tissue increase to the kPa range when the tissue is uniaxially compressed. These results suggest that in situ, compression stiffening results in glioma cells effectively being in a stiff environment such that the elastic resistance these cells sense is sufficient to trigger the same responses that are activated in vitro by increased substrate stiffness.
Increased local pressures within the brain can arise from multiple sources such as increased vascularization within the tumor, changes in ion conductance and fluid flow across cell membranes, cytoskeletal and motor protein-dependent forces related to cell motility, and increased proliferation of cells that survive at higher homeostatic pressure, as has recently been hypothesized to be a feature of tumor growth 8. Pressure differences of 4 to 28 mmHg have been reported between probes placed at the wall of the tumor cavity and approximately 2.5 cm distal to it 6. Pressures were particularly high in glioblastomas, and the data in Figure 5C show that such pressure differences are sufficient to significantly alter the shear modulus of the tissue. In some cases tumor cells and stroma also increase deposition of abnormal extracellular matrix proteins such as collagen 1 that are characteristic of some gliomas and that can activate integrins and alter cell adhesion 51. Whether altered collagen expression affects glioma cells by biochemical or mechanical signaling is not clear, but the results in this study show that such expression is not sufficient to grossly alter the rheology of glioma samples in the same manner as it changes the rheology of fibrotic or cancerous tissues in other organs 28, 48, 50.
The mechanism by which brain increases its shear modulus when compressed but not when extended or sheared to larger strains is unknown. Increasing axial strain appears to have a different effect on the Young's modulus than on the shear modulus of normal brain, as seen by comparison of Figure 3B and Figure 4B. Figure 3B shows that the measured Young's modulus increases only slightly if at all with increasing axial strain, but Figure 4B shows that the shear modulus increases more strongly with increasing axial strain. For isotropic continuous materials, these values might be expected to change the same way as a function of axial strain. But the geometries of deformation (compression in Figure 3B; compression followed by shear strain in Figure 4B) are different, and the tissues are sufficiently heterogeneous or non-isotropic that the shear modulus is no longer simply related to the Young's modulus, once the strains are in the non-linear range.
In the experiments of this study, the compression geometry was unconfined, and at least by visual inspection the sample volumes remained constant. The compression-generated stiffening was also rapidly reversed when compression was relieved. These results strongly argue against global water flow from the sample that might increase matrix concentration or lead to increased crosslinks between network strands. Compression stiffening has also recently been reported for liver 35, but is not observed in networks formed by collagen I or other rigid or semi flexible biopolymers like fibrin or F-actin, suggesting that the compression stiffening rises either from the composite nature of the tissue in which volume-conserving cells reside within the mesh formed by the extracellular matrix or from some other feature. One possibility is the presence of large amounts of strongly charged anionic polymers such as hyaluronic acid and chondroitin sulfate in the extracellular matrix of brain and other soft tissues 10. Such strong polyelectrolyte networks might resist local deformations that alter local densities of soluble counterions if the geometry of local deformation within the tissue is different in different strain geometries.
Conclusion
Glioma cells as well as normal astrocytes are mechanosensitive to substrate stiffness differences in the range appropriate for brain. The cortical stiffness of glioma cells is significantly less than that of normal glial cells over the entire range of substrate stiffness that spans pathophysiologically relevant values, suggesting that altered matrix stiffness in brain might selectively affect glioma cells. Glioma tissue derived from human subjects is not stiffer than normal brain when measured at low strains without compression. This result is in contrast to the findings for other types of cancers that arise in tissues like breast where a filamentous protein extracellular matrix is abnormally formed, and the difference is likely related to the fact that brain does not have such a protein-based matrix. Both normal brain and glioma samples stiffen with compression, but not in elongation and increased shear strains. This outcome supports the hypothesis that compression stiffening, which might occur with increased vascularization and interstitial pressure gradients that are characteristic of tumors, effectively stiffens the environment of glioma cells, and that in situ, the elastic resistance these cells sense might be sufficient to trigger the same responses that are activated in vitro by increased substrate stiffness.
Supplementary Material
Acknowledgements
This work was supported in part by a Fulbright Junior Advanced Research Program from the Polish - US Fulbright Commission and by grants from the US National Institutes of Health (GM096971) and the US National Science Foundation (DMR-1120901).
References
- 1.Baker AM, Bird D, Lang G, Cox TR, Erler JT. Oncogene. 2013;32(14):1863–1868. doi: 10.1038/onc.2012.202. [DOI] [PubMed] [Google Scholar]
- 2.Samuel MS, Lopez JI, McGhee EJ, Croft DR, Strachan D, Timpson P, Munro J, Schroder E, Zhou J, Brunton VG, Barker N, Clevers H, Sansom OJ, Anderson KI, Weaver VM, Olson MF. Cancer Cell. 2011;19(6):776–791. doi: 10.1016/j.ccr.2011.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Olsen AL, Bloomer SA, Chan EP, Gaca MD, Georges PC, Sackey B, Uemura M, Janmey PA, Wells RG. Am J Physiol Gastrointest Liver Physiol. 2011;301(1):G110–G118. doi: 10.1152/ajpgi.00412.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kothapalli D, Liu SL, Bae YH, Monslow J, Xu T, Hawthorne EA, Byfield FJ, Castagnino P, Rao S, Rader DJ, Pure E, Phillips MC, Lund-Katz S, Janmey PA, Assoian RK. Cell Rep. 2012;2(5):1259–1271. doi: 10.1016/j.celrep.2012.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boucher Y, Salehi H, Witwer B, Harsh GRt, Jain RK. Br J Cancer. 1997;75(6):829–836. doi: 10.1038/bjc.1997.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Piek J, Plewe P, Bock WJ. Acta Neurochir (Wien) 1988;93(3–4):129–132. doi: 10.1007/BF01402894. [DOI] [PubMed] [Google Scholar]
- 7.Stylianopoulos T, Martin JD, Snuderl M, Mpekris F, Jain SR, Jain RK. Cancer Res. 2013;73(13):3833–3841. doi: 10.1158/0008-5472.CAN-12-4521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Basan M, Risler T, Joanny JF, Sastre-Garau X, Prost J. HFSP J. 2009;3(4):265–272. doi: 10.2976/1.3086732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schrader J, Gordon-Walker TT, Aucott RL, van Deemter M, Quaas A, Walsh S, Benten D, Forbes SJ, Wells RG, Iredale JP. Hepatology. 2011;53(4):1192–1205. doi: 10.1002/hep.24108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ruoslahti E. Glycobiology. 1996;6(5):489–492. doi: 10.1093/glycob/6.5.489. [DOI] [PubMed] [Google Scholar]
- 11.Flanagan LA, Ju YE, Marg B, Osterfield M, Janmey PA. Neuroreport. 2002;13(18):2411–2415. doi: 10.1097/01.wnr.0000048003.96487.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Georges PC, Miller WJ, Meaney DF, Sawyer ES, Janmey PA. Biophys J. 2006;90(8):3012–3018. doi: 10.1529/biophysj.105.073114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Betz T, Koch D, Lu YB, Franze K, Kas JA. Proc Natl Acad Sci U S A. 2011;108(33):13420–13425. doi: 10.1073/pnas.1106145108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Franze K, Janmey PA, Guck J. Annu Rev Biomed Eng. 2013;15:227–251. doi: 10.1146/annurev-bioeng-071811-150045. [DOI] [PubMed] [Google Scholar]
- 15.Hrapko M, van Dommelen JA, Peters GW, Wismans JS. J Biomech Eng. 2008;130(3):031003. doi: 10.1115/1.2907746. [DOI] [PubMed] [Google Scholar]
- 16.Nicolle S, Lounis M, Willinger R, Palierne JF. Biorheology. 2005;42(3):209–223. [PubMed] [Google Scholar]
- 17.Miller K, Chinzei K. J Biomech. 2002;35(4):483–490. doi: 10.1016/s0021-9290(01)00234-2. [DOI] [PubMed] [Google Scholar]
- 18.Finan JD, Elkin BS, Pearson EM, Kalbian IL, Morrison B., 3rd Ann Biomed Eng. 2012;40(1):70–78. doi: 10.1007/s10439-011-0394-2. [DOI] [PubMed] [Google Scholar]
- 19.Karimi A, Navidbakhsh M. J Mater Sci Mater Med. 2014 doi: 10.1007/s10856-014-5198-0. [DOI] [PubMed] [Google Scholar]
- 20.Cheng S, Clarke EC, Bilston LE. Med Eng Phys. 2008;30(10):1318–1337. doi: 10.1016/j.medengphy.2008.06.003. [DOI] [PubMed] [Google Scholar]
- 21.Chatelin S, Constantinesco A, Willinger R. Biorheology. 2010;47(5–6):255–276. doi: 10.3233/BIR-2010-0576. [DOI] [PubMed] [Google Scholar]
- 22.Hrapko M, van Dommelen JA, Peters GW, Wismans JS. Biorheology. 2008;45(6):663–676. [PubMed] [Google Scholar]
- 23.Diserens AC, de Tribolet N, Martin-Achard A, Gaide AC, Schnegg JF, Carrel S. Acta Neuropathol. 1981;53(1):21–28. doi: 10.1007/BF00697180. [DOI] [PubMed] [Google Scholar]
- 24.Pelham RJ, Jr, Wang Y. Proc Natl Acad Sci U S A. 1997;94(25):13661–13665. doi: 10.1073/pnas.94.25.13661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yeung T, Georges PC, Flanagan LA, Marg B, Ortiz M, Funaki M, Zahir N, Ming W, Weaver V, Janmey PA. Cell Motil Cytoskeleton. 2005;60(1):24–34. doi: 10.1002/cm.20041. [DOI] [PubMed] [Google Scholar]
- 26.Levental I, Georges PC, Janmey PA. Soft Matter. 2007;1:299–306. doi: 10.1039/b610522j. [DOI] [PubMed] [Google Scholar]
- 27.Janmey PA, McCormick ME, Rammensee S, Leight JL, Georges PC, MacKintosh FC. Nat Mater. 2007;6(1):48–51. doi: 10.1038/nmat1810. [DOI] [PubMed] [Google Scholar]
- 28.Levental I, Levental KR, Klein EA, Assoian R, Miller RT, Wells RG, Janmey PA. J Phys Condens Matter. 2010;22(19):194120. doi: 10.1088/0953-8984/22/19/194120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wyss HM, Henderson JM, Byfield FJ, Bruggeman LA, Ding Y, Huang C, Suh JH, Franke T, Mele E, Pollak MR, Miner JH, Janmey PA, Weitz DA, Miller RT. Am J Physiol Cell Physiol. 2011;300(3):C397–C405. doi: 10.1152/ajpcell.00438.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Byfield FJ, Wen Q, Levental I, Nordstrom K, Arratia PE, Miller RT, Janmey PA. Biophys J. 2009;96(12):5095–5102. doi: 10.1016/j.bpj.2009.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Solon J, Levental I, Sengupta K, Georges PC, Janmey PA. Biophys J. 2007;93(12):4453–4461. doi: 10.1529/biophysj.106.101386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pogoda K, Jaczewska J, Wiltowska-Zuber J, Klymenko O, Zuber K, Fornal M, Lekka M. Eur Biophys J. 2012;41(1):79–87. doi: 10.1007/s00249-011-0761-9. [DOI] [PubMed] [Google Scholar]
- 33.Murray ME, Mendez MG, Janmey PA. Mol Biol Cell. 2013 doi: 10.1091/mbc.E13-06-0326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miller WJ, Leventhal I, Scarsella D, Haydon PG, Janmey P, Meaney DF. J Neurotrauma. 2009;26(5):789–797. doi: 10.1089/neu.2008-0727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tan K, Cheng S, Juge L, Bilston LE. J Biomech. 2013;46(6):1060–1066. doi: 10.1016/j.jbiomech.2013.01.028. [DOI] [PubMed] [Google Scholar]
- 36.Whittle IR, Stavrinos N, Akil H, Yau Y, Lewis SC. Br J Neurosurg. 2010;24(4):447–453. doi: 10.3109/02688691003746290. [DOI] [PubMed] [Google Scholar]
- 37.Skjoeth J, Bjerre PK. Acta Neurol Scand. 1997;96(3):167–170. doi: 10.1111/j.1600-0404.1997.tb00261.x. [DOI] [PubMed] [Google Scholar]
- 38.Navalitloha Y, Schwartz ES, Groothuis EN, Allen CV, Levy RM, Groothuis DR. Neuro Oncol. 2006;8(3):227–233. doi: 10.1215/15228517-2006-007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Iversen BN, Rasmussen M, Cold GE. Acta Neurochir (Wien) 2008;150(4):337–344. doi: 10.1007/s00701-008-1461-1. ; discussion 344. [DOI] [PubMed] [Google Scholar]
- 40.de Castro-Costa CM, de Araujo RW, de Arruda MA, de Araujo PM, de Figueiredo EG. Arq Neuropsiquiatr. 1994;52(1):64–68. doi: 10.1590/s0004-282x1994000100011. [DOI] [PubMed] [Google Scholar]
- 41.Bruce JN, Falavigna A, Johnson JP, Hall JS, Birch BD, Yoon JT, Wu EX, Fine RL, Parsa AT. Neurosurgery. 2000;46(3):683–691. doi: 10.1097/00006123-200003000-00031. [DOI] [PubMed] [Google Scholar]
- 42.Guck J, Schinkinger S, Lincoln B, Wottawah F, Ebert S, Romeyke M, Lenz D, Erickson HM, Ananthakrishnan R, Mitchell D, Kas J, Ulvick S, Bilby C. Biophys J. 2005;88(5):3689–3698. doi: 10.1529/biophysj.104.045476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Runge J, Reichert T, Fritsch A, Kas J, Bertolini J, Remmerbach T. Oral Dis. 2013 doi: 10.1111/odi.12171. [DOI] [PubMed] [Google Scholar]
- 44.Tee SY, Fu J, Chen CS, Janmey PA. Biophys J. 2011;100(5):L25–L27. doi: 10.1016/j.bpj.2010.12.3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Klein EA, Yin L, Kothapalli D, Castagnino P, Byfield FJ, Xu T, Levental I, Hawthorne E, Janmey PA, Assoian RK. Curr Biol. 2009;19(18):1511–1518. doi: 10.1016/j.cub.2009.07.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Paszek MJ, Weaver VM. J Mammary Gland Biol Neoplasia. 2004;9(4):325–342. doi: 10.1007/s10911-004-1404-x. [DOI] [PubMed] [Google Scholar]
- 47.Montel F, Delarue M, Elgeti J, Malaquin L, Basan M, Risler T, Cabane B, Vignjevic D, Prost J, Cappello G, Joanny JF. Phys Rev Lett. 2011;107(18):188102. doi: 10.1103/PhysRevLett.107.188102. [DOI] [PubMed] [Google Scholar]
- 48.Levental KR, Yu H, Kass L, Lakins JN, Egeblad M, Erler JT, Fong SF, Csiszar K, Giaccia A, Weninger W, Yamauchi M, Gasser DL, Weaver VM. Cell. 2009;139(5):891–906. doi: 10.1016/j.cell.2009.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Barcus CE, Keely PJ, Eliceiri KW, Schuler LA. J Biol Chem. 2013;288(18):12722–12732. doi: 10.1074/jbc.M112.447631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Perepelyuk M, Terajima M, Wang AY, Georges PC, Janmey PA, Yamauchi M, Wells RG. Am J Physiol Gastrointest Liver Physiol. 2013;304(6):G605–G614. doi: 10.1152/ajpgi.00222.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Payne LS, Huang PH. Mol Cancer Res. 2013;11(10):1129–1140. doi: 10.1158/1541-7786.MCR-13-0236. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.