Abstract
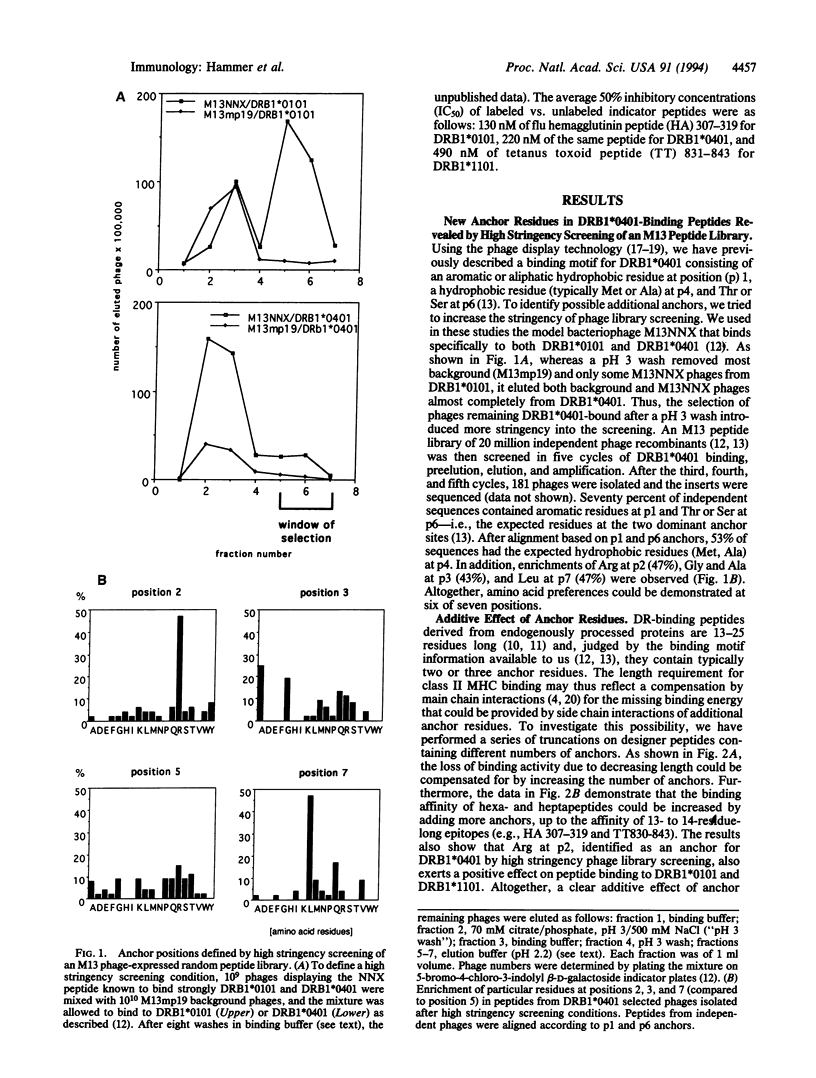
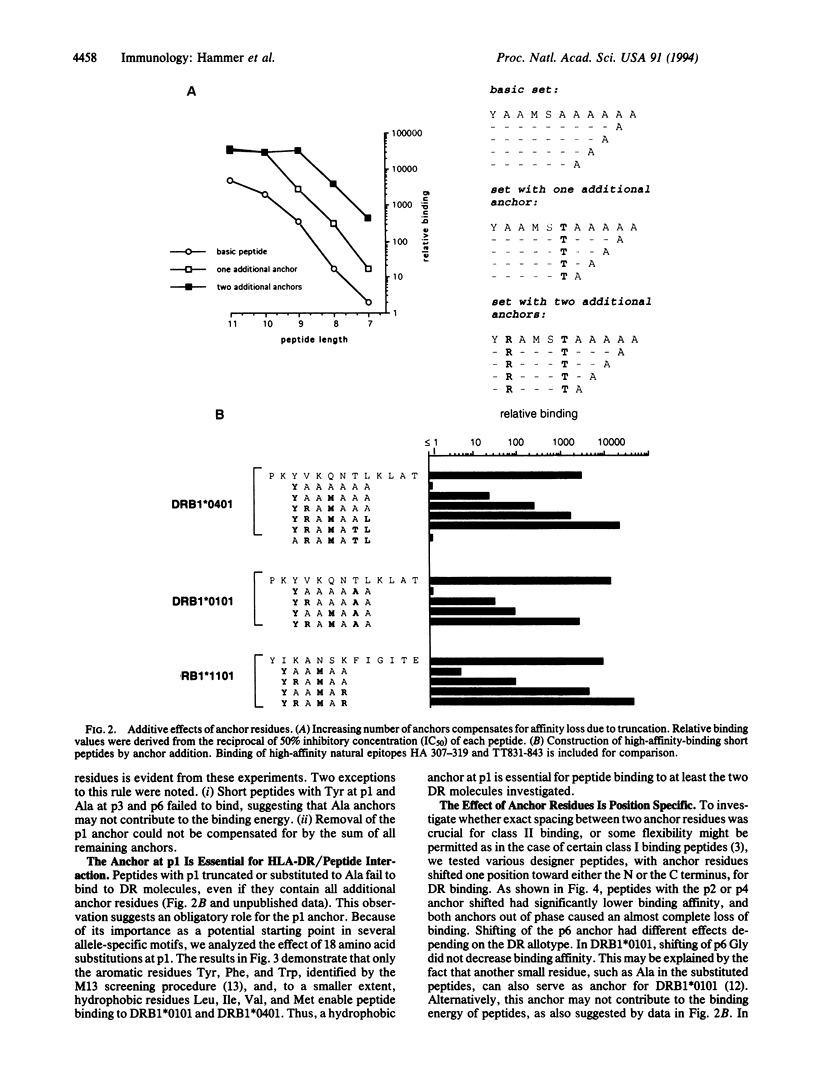
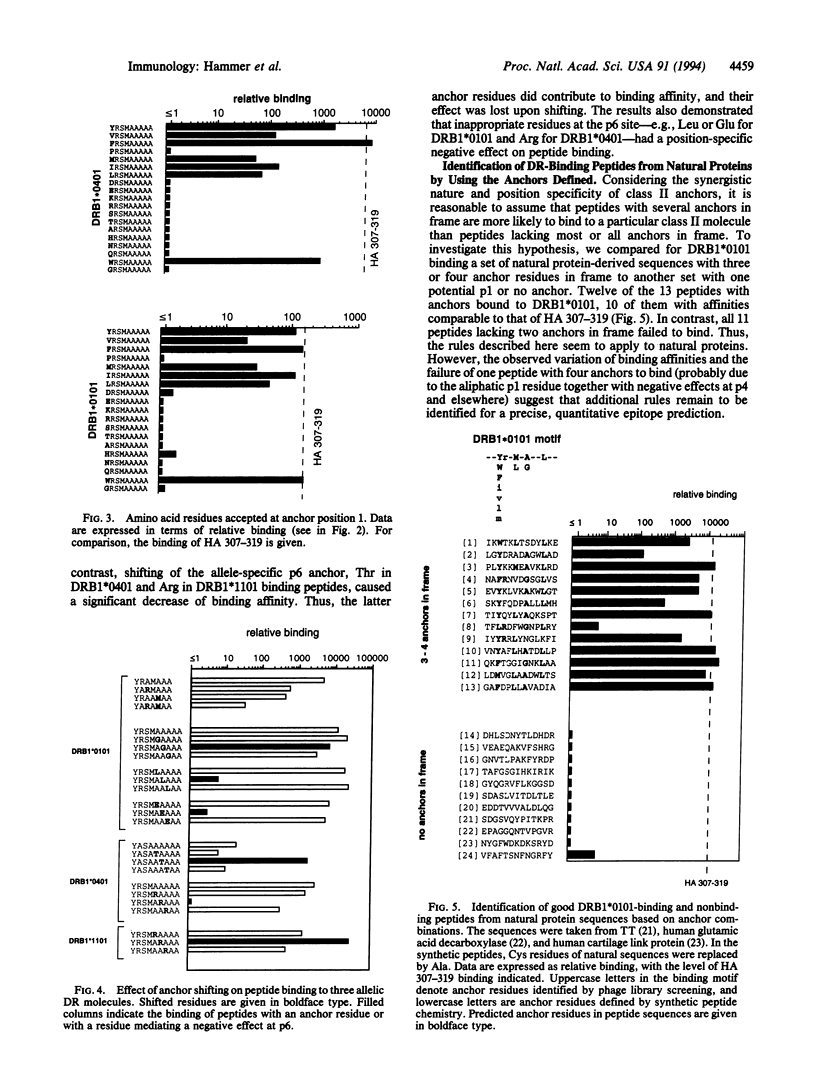
We have previously identified four anchor positions in HLA-DRB1*0101-binding peptides, and three anchors involved in peptide binding to DRB1*0401 and DRB1*1101 molecules, by screening of an M13 peptide display library (approximately 20 million independent nonapeptides) for DR-binding activity. In this study, high stringency screening of the M13 library for DRB1*0401 binding has resulted in identification of three further anchor positions. Taken together, a peptide-binding motif has been obtained, in which six of seven positions show enrichment of certain residues. We have demonstrated an additive effect of anchors in two different ways: (i) the addition of more anchors is shown to compensate for progressive truncation of designer peptides; (ii) the incorporation of an increasing number of anchors into 6- or 7-residue-long designer peptides is shown to result in a gradual increase of binding affinity to the level of 13-residue-long high-affinity epitopes. The anchor at relative position 1 seems to be obligatory, in that its substitution abrogates binding completely, whereas the elimination of other anchors results only in partial loss of binding affinity. The spacing between anchors is critical, since their effect is lost by shifting them one position toward the N or C terminus. The information born out of this study has been successfully used to identify DR-binding sequences from natural proteins.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brown J. H., Jardetzky T. S., Gorga J. C., Stern L. J., Urban R. G., Strominger J. L., Wiley D. C. Three-dimensional structure of the human class II histocompatibility antigen HLA-DR1. Nature. 1993 Jul 1;364(6432):33–39. doi: 10.1038/364033a0. [DOI] [PubMed] [Google Scholar]
- Chicz R. M., Urban R. G., Gorga J. C., Vignali D. A., Lane W. S., Strominger J. L. Specificity and promiscuity among naturally processed peptides bound to HLA-DR alleles. J Exp Med. 1993 Jul 1;178(1):27–47. doi: 10.1084/jem.178.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chicz R. M., Urban R. G., Lane W. S., Gorga J. C., Stern L. J., Vignali D. A., Strominger J. L. Predominant naturally processed peptides bound to HLA-DR1 are derived from MHC-related molecules and are heterogeneous in size. Nature. 1992 Aug 27;358(6389):764–768. doi: 10.1038/358764a0. [DOI] [PubMed] [Google Scholar]
- Cwirla S. E., Peters E. A., Barrett R. W., Dower W. J. Peptides on phage: a vast library of peptides for identifying ligands. Proc Natl Acad Sci U S A. 1990 Aug;87(16):6378–6382. doi: 10.1073/pnas.87.16.6378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin J. J., Panganiban L. C., Devlin P. E. Random peptide libraries: a source of specific protein binding molecules. Science. 1990 Jul 27;249(4967):404–406. doi: 10.1126/science.2143033. [DOI] [PubMed] [Google Scholar]
- Dudhia J., Hardingham T. E. The primary structure of human cartilage link protein. Nucleic Acids Res. 1990 Mar 11;18(5):1292–1292. doi: 10.1093/nar/18.5.1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisel U., Jarausch W., Goretzki K., Henschen A., Engels J., Weller U., Hudel M., Habermann E., Niemann H. Tetanus toxin: primary structure, expression in E. coli, and homology with botulinum toxins. EMBO J. 1986 Oct;5(10):2495–2502. doi: 10.1002/j.1460-2075.1986.tb04527.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk K., Rötzschke O., Stevanović S., Jung G., Rammensee H. G. Allele-specific motifs revealed by sequencing of self-peptides eluted from MHC molecules. Nature. 1991 May 23;351(6324):290–296. doi: 10.1038/351290a0. [DOI] [PubMed] [Google Scholar]
- Fremont D. H., Matsumura M., Stura E. A., Peterson P. A., Wilson I. A. Crystal structures of two viral peptides in complex with murine MHC class I H-2Kb. Science. 1992 Aug 14;257(5072):919–927. doi: 10.1126/science.1323877. [DOI] [PubMed] [Google Scholar]
- Hammer J., Takacs B., Sinigaglia F. Identification of a motif for HLA-DR1 binding peptides using M13 display libraries. J Exp Med. 1992 Oct 1;176(4):1007–1013. doi: 10.1084/jem.176.4.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer J., Valsasnini P., Tolba K., Bolin D., Higelin J., Takacs B., Sinigaglia F. Promiscuous and allele-specific anchors in HLA-DR-binding peptides. Cell. 1993 Jul 16;74(1):197–203. doi: 10.1016/0092-8674(93)90306-b. [DOI] [PubMed] [Google Scholar]
- Hill C. M., Hayball J. D., Allison A. A., Rothbard J. B. Conformational and structural characteristics of peptides binding to HLA-DR molecules. J Immunol. 1991 Jul 1;147(1):189–197. [PubMed] [Google Scholar]
- Hunt D. F., Henderson R. A., Shabanowitz J., Sakaguchi K., Michel H., Sevilir N., Cox A. L., Appella E., Engelhard V. H. Characterization of peptides bound to the class I MHC molecule HLA-A2.1 by mass spectrometry. Science. 1992 Mar 6;255(5049):1261–1263. doi: 10.1126/science.1546328. [DOI] [PubMed] [Google Scholar]
- Hunt D. F., Michel H., Dickinson T. A., Shabanowitz J., Cox A. L., Sakaguchi K., Appella E., Grey H. M., Sette A. Peptides presented to the immune system by the murine class II major histocompatibility complex molecule I-Ad. Science. 1992 Jun 26;256(5065):1817–1820. doi: 10.1126/science.1319610. [DOI] [PubMed] [Google Scholar]
- Jardetzky T. S., Gorga J. C., Busch R., Rothbard J., Strominger J. L., Wiley D. C. Peptide binding to HLA-DR1: a peptide with most residues substituted to alanine retains MHC binding. EMBO J. 1990 Jun;9(6):1797–1803. doi: 10.1002/j.1460-2075.1990.tb08304.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsen A. E., Hagopian W. A., Grubin C. E., Dube S., Disteche C. M., Adler D. A., Bärmeier H., Mathewes S., Grant F. J., Foster D. Cloning and primary structure of a human islet isoform of glutamic acid decarboxylase from chromosome 10. Proc Natl Acad Sci U S A. 1991 Oct 1;88(19):8337–8341. doi: 10.1073/pnas.88.19.8337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madden D. R., Gorga J. C., Strominger J. L., Wiley D. C. The structure of HLA-B27 reveals nonamer self-peptides bound in an extended conformation. Nature. 1991 Sep 26;353(6342):321–325. doi: 10.1038/353321a0. [DOI] [PubMed] [Google Scholar]
- O'Sullivan D., Arrhenius T., Sidney J., Del Guercio M. F., Albertson M., Wall M., Oseroff C., Southwood S., Colón S. M., Gaeta F. C. On the interaction of promiscuous antigenic peptides with different DR alleles. Identification of common structural motifs. J Immunol. 1991 Oct 15;147(8):2663–2669. [PubMed] [Google Scholar]
- Rudensky AYu, Preston-Hurlburt P., Hong S. C., Barlow A., Janeway C. A., Jr Sequence analysis of peptides bound to MHC class II molecules. Nature. 1991 Oct 17;353(6345):622–627. doi: 10.1038/353622a0. [DOI] [PubMed] [Google Scholar]
- Scheirle A., Takacs B., Kremer L., Marin F., Sinigaglia F. Peptide binding to soluble HLA-DR4 molecules produced by insect cells. J Immunol. 1992 Sep 15;149(6):1994–1999. [PubMed] [Google Scholar]
- Scott J. K., Smith G. P. Searching for peptide ligands with an epitope library. Science. 1990 Jul 27;249(4967):386–390. doi: 10.1126/science.1696028. [DOI] [PubMed] [Google Scholar]
- Sinigaglia F., Romagnoli P., Guttinger M., Takacs B., Pink J. R. Selection of T cell epitopes and vaccine engineering. Methods Enzymol. 1991;203:370–386. doi: 10.1016/0076-6879(91)03021-8. [DOI] [PubMed] [Google Scholar]
- Zhang W., Young A. C., Imarai M., Nathenson S. G., Sacchettini J. C. Crystal structure of the major histocompatibility complex class I H-2Kb molecule containing a single viral peptide: implications for peptide binding and T-cell receptor recognition. Proc Natl Acad Sci U S A. 1992 Sep 1;89(17):8403–8407. doi: 10.1073/pnas.89.17.8403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Bleek G. M., Nathenson S. G. The structure of the antigen-binding groove of major histocompatibility complex class I molecules determines specific selection of self-peptides. Proc Natl Acad Sci U S A. 1991 Dec 15;88(24):11032–11036. doi: 10.1073/pnas.88.24.11032. [DOI] [PMC free article] [PubMed] [Google Scholar]



