SUMMARY
Oncogenic mutations of FLT3 and KIT receptors are associated with poor survival in patients with acute myeloid leukemia (AML) and myeloproliferative neoplasms (MPN) and currently available drugs are largely ineffective. Although Stat5 has been implicated in regulating several myeloid and lymphoid malignancies, how precisely Stat5 regulates leukemogenesis, including its nuclear translocation to induce gene transcription is poorly understood. In leukemic cells, we show constitutive activation of focal adhesion kinase (FAK), whose inhibition represses leukemogenesis. Downstream of FAK, activation of Rac1 is regulated by RacGEF Tiam1, whose inhibition prolongs the survival of leukemic mice. Inhibition of the Rac1 effector PAK1 prolongs the survival of leukemic mice in part by inhibiting the nuclear translocation of Stat5. These results reveal a leukemic pathway involving FAK/Tiam1/Rac1/PAK1 and demonstrate an essential role for these signaling molecules in regulating the nuclear translocation of Stat5 in leukemogenesis.
INTRODUCTION
Acute myeloid leukemia (AML) is a lethal disease characterized by uncontrolled growth of myeloid cells and is predominantly a disease of the elderly. Little progress has been made in terms of standard of care treatment for AML, which has essentially remained the same over decades. Long-term survival is observed in ~ 30% of younger patients and ~5% of older patients greater than 60 years of age. Internal tandem duplications (ITD), in-frame insertions or duplication of amino acids near the juxtamembrane domain of FLT3, have been observed in ~25 to 30% of all AML patients and confer a poor prognosis (Kottaridis et al., 2001). Likewise, gleevec resistant activation loop mutations of KIT (KITD816V) are found in a number of patients with core binding factor (CBF)-AML and ~95% patients with systemic mastocytosis (SM) and confer poor overall survival (Beghini et al., 2004). Both FLT3ITD and KITD816V receptors are constitutively phosphorylated (Kiyoi et al., 2002; Spiekermann et al., 2003) and induce growth in a ligand-independent manner. While effort has been devoted to the development of FLT3 and KIT inhibitors; as single agents, the efficacy of these inhibitors is limited and in some cases results in drug resistance (Smith et al., 2012). Given that direct targeting of FLT3ITD or KITD816V has met with little success, signaling pathways downstream from FLT3ITD/KITD816V provide attractive alternate targets for treating hematologic malignancies involving these receptors.
Overexpression of focal adhesion kinase (FAK) in up to 50% of AML patient derived cells but not in normal cells has been observed, and FAK is hyper-phosphorylated on Y397 in a number of patients. FAK+ AML cells show greater migration and resistance to daunorubicin compared to FAK- cells and FAK expression correlates with high blast cell counts, early death and shorter survival rate (Despeaux et al., 2011; Recher et al., 2004) (Li and Hua, 2008). Presence of phosphorylated (p) pStat5 in newly diagnosed AML patients is also associated with poor overall survival (Brady et al., 2012). Constitutive activation of pStat5 is observed in 100% of systemic mastocytosis (SM) patients bearing the KITD816V mutation (Baumgartner et al., 2009). A strong correlation between the presence of pStat5 and FLT3ITD mutations is seen in AML patients and FLT3ITD expression results in constitutive Stat5 phosphorylation (Obermann et al., 2010) (Spiekermann et al., 2003), (Choudhary et al., 2007; Choudhary et al., 2005). Mutating the binding sites for Stat5 in the FLT3ITD abrogates the development of MPN (Rocnik et al., 2006). Taken together, studies suggest that FLT3ITD/KITD814V, FAK and Stat5 may be involved in regulating a critical pathway in AML and MPNs; however, the relationship between these signaling molecules in the context of leukemogenesis is not fully understood. Importantly, although Stat5 has been implicated in regulating several hematologic malignancies; how precisely activation of Stat5 is regulated in the cytosol or in the nucleus of leukemic cells and what are the signaling molecules involved in its nuclear import in the context of AML or MPN remains unclear. Here we reveal a leukemic pathway involving FAK/Tiam1/Rac1/PAK1 and demonstrate an essential role for these signaling molecules in regulating the nuclear translocation of Stat5 in leukemogenesis.
RESULTS
FAK is constitutively phosphorylated in FLT3ITD and KITD814V expressing cells
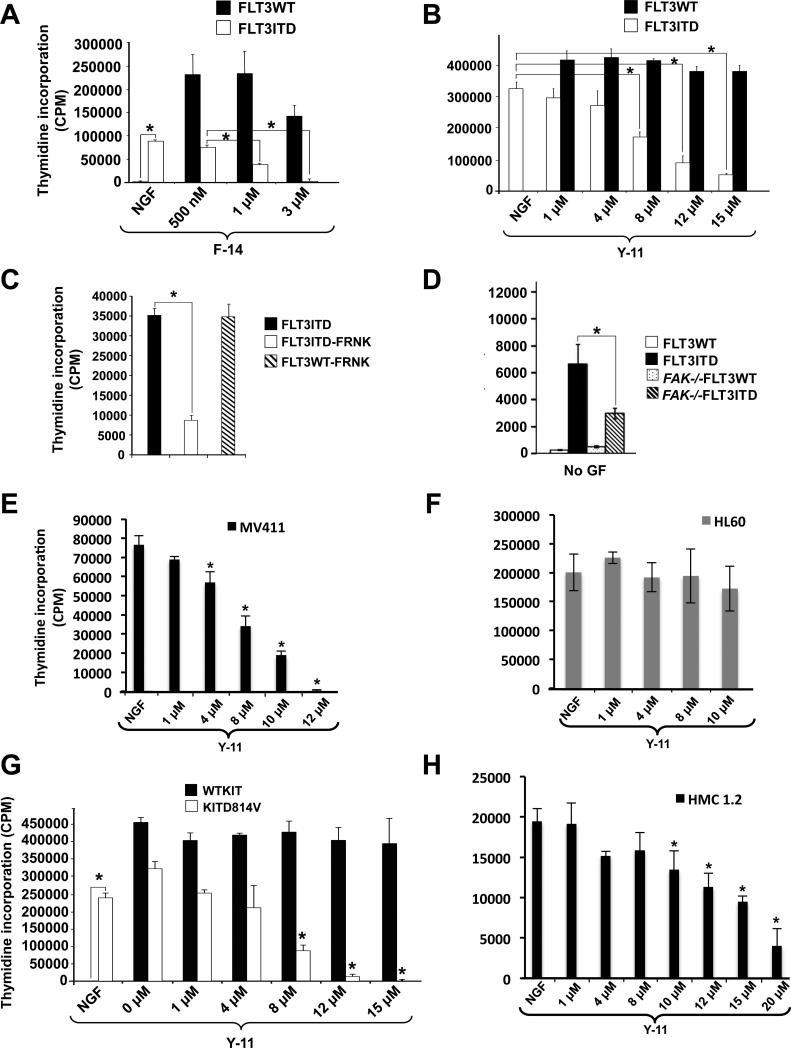
32D cells expressing the WT FLT3 or KIT receptor (FLT3WT or KITWT) or its oncogenic version (FLT3ITD or KITD814V) were starved and treated with a FAK specific inhibitor F-14 (Golubovskaya et al., 2008). Enhanced activation of FAK was observed in FLT3ITD bearing cells compared to controls (Fig.1A, lane1 vs 5), which was inhibited in the presence of F-14 (Fig.1A, lane 6 vs 5). Similar results were observed in cells expressing KITWT and KITD814V receptors (Fig.1B). To assess if activation of FAK was restricted to oncogenic FLT3 and KIT receptor expressing cells, same cells were stimulated with IL-3 to activate the IL-3 receptor, and analyzed for FAK activation. As seen in Figure 1A (lane 3), IL-3 stimulation also resulted in activation of FAK, which was inhibited in the presence of F-14 (Fig.1A, lane 4). Similar results were observed upon treatment of cells with FLT3 ligand (FL) (Fig.1C, lane 1 vs 3). To assess the direct involvement of FLT3ITD in FAK activation, cells were treated with AC220, a potent FLT3ITD inhibitor (Smith et al., 2012). Treatment of FLT3ITD cells with AC220 inhibited the activation of FLT3ITD (Fig.1D, lane 3 vs 4) and also resulted in reduced FAK activation (Fig.1E, lane 1 vs 3). To rule out the non-specific effects of F-14 on FAK inhibition, we utilized a genetic approach. WT BM cells expressing KITD814V demonstrated increased levels of active FAK compared to controls, while FAK−/− BM cells showed absence of FAK expression (Fig.1F). FLT3ITD+ve AML patient derived cells also demonstrated constitutive FAK activation, which was inhibited in the presence of F-14 (Fig.1G). Figures 1D and S8D show the expression of total FLT3 and KIT receptors. We also performed intracellular staining to determine the effect of Y-11; another FAK specific inhibitor (Golubovskaya et al., 2012) on FLT3ITD mediated repression of FAK. As seen in Figure 1H, the percentage of cells showing activated FAK was significantly higher in FLT3ITD expressing cells (83.1%, middle panel) as compared to WT (19.2%) (left vs middle panel). Treatment of FLT3ITD cells with Y-11 inhibited the activation of FAK (69.7%, right panel), and correspondingly increased the levels of un-phosphorylated FAK (30.3%) compared to vehicle treated (16.9%) (right panel vs middle panel). These results suggest that FAK is hyperactive in FLT3 and KIT oncogene bearing cells and pharmacologic inhibition or genetic loss of FAK can repress the activation of FAK in these cells.
Fig.1. FAK is constitutively phosphorylated in FLT3 and activating KITD814V oncogene bearing cells.
(A) Serum starved 32D cells expressing FLT3ITD and FLT3WT were treated with DMSO (lanes 1, 5); F-14 (lanes 2, 6); IL-3 (lanes 3, 7) or F-14 followed by IL-3 (lanes 4, 8). An equal amount of protein was subjected to western blot analysis and probed with phospho-FAK (Y397) antibody (n=3). ‘MK’ denotes lane with protein ladder. (B) 32D cells expressing KITWT or KITD814V were treated with F-14 (n=2) and analyzed as described in (A). (C) 32D cells expressing FLT3WT were treated with F-14 (lane 2) or stimulated with FLT3 ligand (FL) (lane 3), and analyzed as described in (A). (D) 32D cells expressing FLT3WT or FLT3ITD were treated with FLT3ITD inhibitor AC220 and analyzed for activated FLT3 (pY589/591). (E) 32D cells expressing FLT3ITD were treated with F-14 (lane 2) or with AC220 (lane 3) (n=2) and analyzed as above. (F) Lysates from primary FAK−/− deficient or WT BM cells expressing KITD814V or empty vector were analyzed for activated FAK. (G) FLT3ITD+ve AML patient sample was analyzed for activated FAK (n=2). (H) 32D cells expressing FLT3ITD were treated with Y-11 and subjected to flow cytometry analysis. The percentage of cells show activated FAK under basal conditions in FLT3WT (i), FLT3ITD vehicle treated cells (ii), and FLT3ITD treated with Y-11 (iii). n=2.
Inhibition of FAK suppresses the constitutive growth of oncogenic FLT3 and KIT expressing cells
We assessed the functional significance of constitutive activation of FAK in FLT3ITD expressing cells. As seen in Fig.2A, treatment of BaF3 cells with F-14 significantly repressed the ligand independent growth of FLT3ITD bearing cells in a dose dependent manner, with minimal effect on FLT3WT expressing cells. Similar growth repression was seen in 32D cells treated with Y-11 (Fig.2B). Expression of FRNK, a dominant negative version of FAK (Zhao and Guan, 2009), also repressed the ligand independent growth of FLT3ITD expressing cells (Fig.2C), which was partly a result of reduced survival (Fig.S1A,B). Expression of FRNK in FLT3 cells is shown in Fig.S8D. Although these results suggest an essential role for FAK in ligand independent growth of FLT3ITD expressing cells, a direct role of FAK was ascertained by complete ablation of FAK. As seen in Fig.2D, expression of FLT3ITD in WT BM cells (FAK+/+) demonstrated ligand independent growth, which was significantly repressed in FAK−/− cells. We also assessed whether targeting hyperactive FAK in cell lines derived from human leukemic patients, shows similar effects. We used MV4-11 and HL60 cells that express the FLT3ITD and WT receptors, respectively. Treatment of MV4-11 cells with Y-11 showed a dose dependent repression of constitutive growth (Fig.2E), while no such effect was observed in HL60 cells (Fig.2F).
Fig.2. Inhibition of FAK suppresses the constitutive growth of oncogenic FLT3 and KIT bearing cells.
(A) BaF3 or (B) 32D cells expressing FLT3WT or FLT3ITD were cultured for 48 hours in the presence or absence of F-14 or Y-11 in replicates of four and subjected to a thymidine incorporation assay. (C) BaF3 cells co-expressing FRNK and either FLT3ITD or FLT3WT were subjected to thymidine incorporation assay as in (A) & (B). (D) WT or FAK−/− BM cells expressing FLT3ITD or FLT3WT were subjected to proliferation assay in the absence of growth factors as described in (A) & (B). (E) MV4-11 cells expressing endogenous levels of FLT3ITD or (F) HL60 cells harboring FLT3WT were subjected to thymidine incorporation assay in presence of Y-11. (G) 32D cells expressing KITD814V or WTKIT and (H) HMC1.2 human leukemic cells line harboring KIT (D816V + G560V) mutations were cultured in the absence or presence of Y-11, and subjected to thymidine incorporation assay. Thymidine incorporation is depicted on y-axis as mean ± SD, *p<0.05. NGF/ No GF= cells grown in presence of no growth factors/cytokines. Data are representative of at least 3 independent experiments.
To assess whether FAK plays a similar role in cells bearing an oncogenic form of KIT (KITD816V in humans and KITD814V in mouse); we used 32D cells expressing WTKIT or KITD814V. As seen in Fig.2G, treatment of these cells with Y-11 resulted in growth repression of KITD814V expressing cells. Similar results were observed upon Y-11 treatment of HMC1.2 cells derived from a human mastocytosis patient bearing the activating KIT mutation (Fig.2H). These results show that FAK plays an essential role in supporting the constitutive growth of FLT3 and KIT oncogene bearing hematopoietic cells which is modulated by pharmacologic or genetic inhibition of FAK.
AC220 resistant “driver” mutations of FLT3 are sensitive to FAK inhibition
Recent translational studies validated FLT3ITD mutations in AML to function as “driver” but not “passenger” mutations (Smith et al., 2012). Smith et al. demonstrated the presence of point mutations at three residues within the kinase domain of FLT3ITD that conferred resistance to AC220, an inhibitor of FLT3 and KIT. Acquisition of AC220 resistant substitutions at two of these residues was observed in all FLT3ITD+ AML patients with acquired resistance to AC220, thus validating FLT3ITD to function as a driver mutation and a critical therapeutic target in AML. We assessed whether these mutants were sensitive to FAK inhibition. As seen in Fig. 3A, AC220 resistant FLT3 kinase domain mutant (D835Y, F691L and D835V) induced growth is inhibited by F-14.
Fig.3. AC220 resistant FLT3 mutations, primary AML FLT3ITD+ cells or KITD816V+ SM cells are sensitive to FAK inhibition.
(A) BaF3 cells bearing FLT3ITD or FLT3 receptors with acquired AC220 resistant mutations in the kinase domain (D835Y, F691L and D835V) were subjected to proliferation assay as described in Fig 2, *p<0.05. (B) Primary AML patient cells positive for FLT3ITD mutation (AML#1-4) or (C) primary KITD816V(+) or KITD816V(-) SM cells were treated with indicated concentrations of F-14 or Y-11. After 48 hours proliferation assay was performed. Bars denote mean ± SD, *p<0.05.
AML FLT3ITD+ and KITD816V+ SM patient derived cells are sensitive to FAK inhibition
We next assessed whether inhibition of FAK in primary FLT3ITD+ AML cells inhibits their growth. We examined cells derived from 16 independent patients. Data from four representative patients is shown. In Figure 3B, a dose dependent reduction in the growth of all FLT3ITD+ AML cells was observed in the presence of F-14 and Y-11. Likewise, SM patient derived cells positive for KITD816V mutation also demonstrated significantly greater growth reduction relative to patients lacking the expression of KITD816V (Fig. 3C). These results demonstrate that FAK is indeed hyperactive in FLT3ITD and KITD816V expressing cells and its inhibition is associated with enhanced apoptosis and growth repression.
FAK and Rac1 modulate the nuclear translocation of active Stat5 in FLT3 and KIT oncogene expressing cells
In an effort to identify downstream targets of FAK that might contribute to FLT3ITD induced growth and enhanced survival, we examined the activation of PI3Kinase, ERK MAP Kinase as well as RacGTPases. In non-hematopoietic cell types, all three pathways have been shown to be regulated by FAK (Gabarra-Niecko et al., 2003; Yin, 2011). We found constitutive and enhanced activation of Rac1 in FLT3ITD bearing cells relative to FLT3WT bearing cells (Fig.4A; lanes 1 & 2 vs 4 in control [CT] panel). Expression of FRNK in FLT3ITD expressing cells significantly inhibited the constitutive activation of Rac (Fig.4A; lane 3, control [CT] panel). Furthermore, treatment of these cells with F-14 abolished Rac activation (lanes 1-4 upper control [CT] panel vs. lanes 1-4 lower [F-14] panel) (Fig.4A). A similar reduction in Rac activation was also observed in FLT3ITD+ve human leukemic MV4-11 cells and FLT3ITD+ AML patient derived cells, respectively (Figs. 4B, 4C); as well as in FAK−/−deficient cells expressing FLT3ITD (Fig.4D). shRNA mediated down-regulation of FAK in FLT3ITD bearing cells also showed similar results (Fig.4E). These results suggest that FAK plays a role in the activation of Rac1 in FLT3ITD bearing cells. Consistent with these observations, treatment of cells with a Rac1 inhibitor NSC23766 repressed the constitutive growth of FLT3ITD+ AML patient derived cells and of MV4-11 leukemic cells expressing the FLT3ITD receptor (Fig.S1C,D). HL60 cells that express the FLT3WT receptor were used as a negative control (Fig.S1E). Similar findings were observed in FLT3ITD bearing cells expressing a dominant negative version of Rac (RacN17) (Fig.S1F).
Fig.4. Rac1 is a downstream effector of FAK in FLT3ITD bearing oncogenic pathway.
(A) BaF3 (lane 1) or 32D (lane 2) cells expressing the FLT3WT receptor or FLT3ITD both alone or in combination with FRNK were starved and subjected to a Rac activation assay. These cells were either vehicle treated alone (upper panel [CT]) or with F-14 (lower panel) (n=2). (B) MV4-11 cells expressing endogenous FLT3ITD, and (C) AML patient FLT3ITD+ cells were subjected to Rac activation assay as described in (A). (D) FLT3ITD bearing BM cells in the setting of FAK deficiency were subjected to Rac activation assay as in (A) (n=2). (E) 32D FLT3ITD cells expressing shRNA's targeting FAK (lane 2, 3) and control shRNA (lane 1) were subjected to Rac1 activation assay as described in (A).
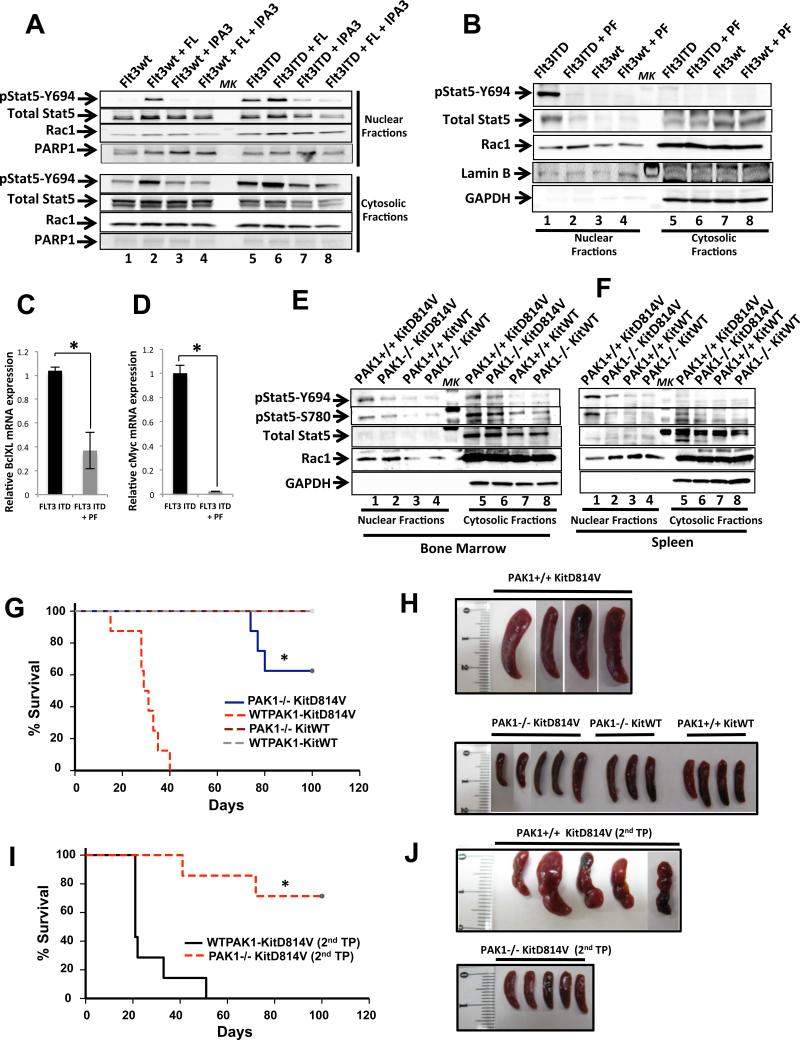
Active Stat5 is thought to play an essential role in regulating the transformation of FLT3 and KIT oncogene bearing cells (Baumgartner et al., 2009; Brady et al., 2012). Although Stat5 is a transcription factor, mechanism(s) involved in the transport of this molecule in and out of the nucleus in oncogene bearing cells remains poorly understood. We therefore performed cellular fractionation assays to determine the mechanism(s) behind active Stat5 translocation into the nucleus mediated by Rac1 and its upstream activator FAK in oncogene bearing cells. We found active Stat5 in the nuclear fractions of FLT3ITD bearing cells, which was associated with enhanced presence of Rac1, as compared with cells expressing FLT3WT (Fig.5A, lanes 1 vs 3). To determine whether nuclear translocation of Stat5 could be mimicked in FLT3WT expressing cells upon cytokine stimulation, cells were treated with IL-3 and analyzed for Stat5 and Rac1 localization. As seen in Figure 5A (lane 2), addition of IL-3 resulted in activation of FLT3WT mediated Stat5-Rac1 nuclear localization, which was inhibited in the presence of F-14 (Fig.5B, lane 1 vs 2). Next, we assessed if the effect of FAK inhibition could be overcome after ligand stimulation. FLT3ITD expressing cells were treated with FLT3 ligand, followed by treatment with F-14. As seen in Figure 5B, stimulation of FLT3ITD with its ligand FL resulted in a modest increase in nuclear localized active Stat5 and Rac1 (Fig.5B, lane 1 vs 3) (Zheng et al., 2011), which was repressed in the presence of F-14 (Fig.5B, lane 4). A similar reduction in the activation of Stat5, and accumulation of Rac1, respectively was noted in the nuclear fractions of primary FLT3ITD+ AML (Fig.S2A), KITD816V+ mastocytosis (HMC1.2) (Fig.5C), and AML patient derived cells (MV4-11) treated with F-14 (Fig.5D). HL60 cells that express the FLT3WT receptor served as a negative control (Fig.5D, right panels). AC220 resistant “driver” mutations of FLT3 also demonstrated reduced nuclear accumulation of active Stat5 and total Rac1, respectively when treated with F-14 (Fig. S2B). Although we did not observe inhibition in the activation of MAPKinase or PI3Kinase/Akt pathway in FLT3ITD bearing cells lacking FAK or in which FAK was inhibited, we nonetheless directly examined the contribution of these pathways in the nuclear translocation of Stat5 and total Rac1. We performed a fractionation assay in the presence of MEK (PD98059) or Raf (PLX4720) or Akt (124005) inhibitor and observed no significant inhibition of Rac1 or active Stat5 nuclear translocation (Fig.S2C). Raf and MEK inhibitors also failed to repress the constitutive growth of FLT3ITD bearing cells (Figs.S2D, E).
Fig.5. FAK regulates the translocation of active Stat5 and Rac1 to the nucleus in FLT3ITD and KITD816V expressing cells.
(A) 32D cells expressing FLT3ITD or FLT3WT were serum starved and subjected to fractionation assays, and nuclear and cytosolic fractions analyzed for levels of active Stat5 (pY694), total Stat5 and Rac1, and (B) 32D cells expressing FLT3ITD were subjected to fractionation assays after treatment with DMSO control, F-14, FLT3 ligand (FL) or with F-14 followed by FL (n=3). (C) HMC1.2 cells bearing KITD816V+G560V mutations, (D) MV4-11 and HL60 cells derived from leukemia patients harboring endogenous FLT3ITD and FLT3WT mutations were subjected to fractionation assay in presence of F-14 or Y-11 as in (A). Fractionation assays were performed in BM cells harvested from primary transplanted mice cohorts transplanted with KITD814V in a wild type FAK (FAK+/+) or FAK deficient (FAK−/−) background (n=2) (E). Fractionation assay from BM cells harvested from F-14 or DMSO (vehicle) treated primary transplant mice cohorts (F). The level of Stat5 phosphorylation/expression and Rac1 expression in the nuclear and cytosolic fractions is indicated. Expression of GAPDH was used as an indicator of cytosolic marker and loading control. ‘MK’ denotes lane with protein ladder. qRT-PCR analysis of relative mRNA expression levels of Stat5 responsive genes c-Myc (G) and BclXL (H) in FLT3ITD cells treated with F-14 or vehicle (DMSO) (n=2), *p<0.05.
A direct role for FAK in regulating the translocation of active Stat5 can be seen in Fig.S2F. Expression of FLT3ITD failed to translocate active Stat5 and total Rac1 to the nucleus in FAK−/− deficient BM cells (Fig.S2F, lane 1 vs 2). The expression of FLT3 receptor in WT (FAK+/+) and FAK−/− BM cells is shown in Fig.S8E. To further understand the mechanism behind FAK regulated Stat5 translocation in FLT3 and KIT oncogene bearing cells in an in vivo setting, we used cells derived directly from mice that were transplanted with BM from WT (FAK+/+) and FAK deficient (FAK−/−) mice expressing KITD814V. Mice transplanted with WTFAK BM cells expressing KITD814V came down with disease significantly earlier than FAK−/− KITD814V mice (Fig.7G), at which point time-point matched control mice (FAK−/− KITD814V) were euthanized and BM cells harvested from both groups of mice and subjected to cellular fractionation analysis. As seen in Fig.5E (lane 1 vs 2), and consistent with our in vitro findings (Fig.S2F), genetic ablation of FAK in vivo inhibited the nuclear translocation of active Stat5 and Rac1 in KITD814V bearing BM derived leukemic cells. Furthermore, similar results were also observed after F-14 treatment of leukemic mice transplanted with BM cells expressing KITD814V. As seen in Figure 5F, BM cells derived from leukemic mice bearing the KITD814V mutation that were treated with F-14 also showed a reduction in Stat5 activation and nuclear accumulation of Rac1, as compared to cells derived from vehicle (DMSO) treated mice (Fig.5F, lane 1 vs 2).
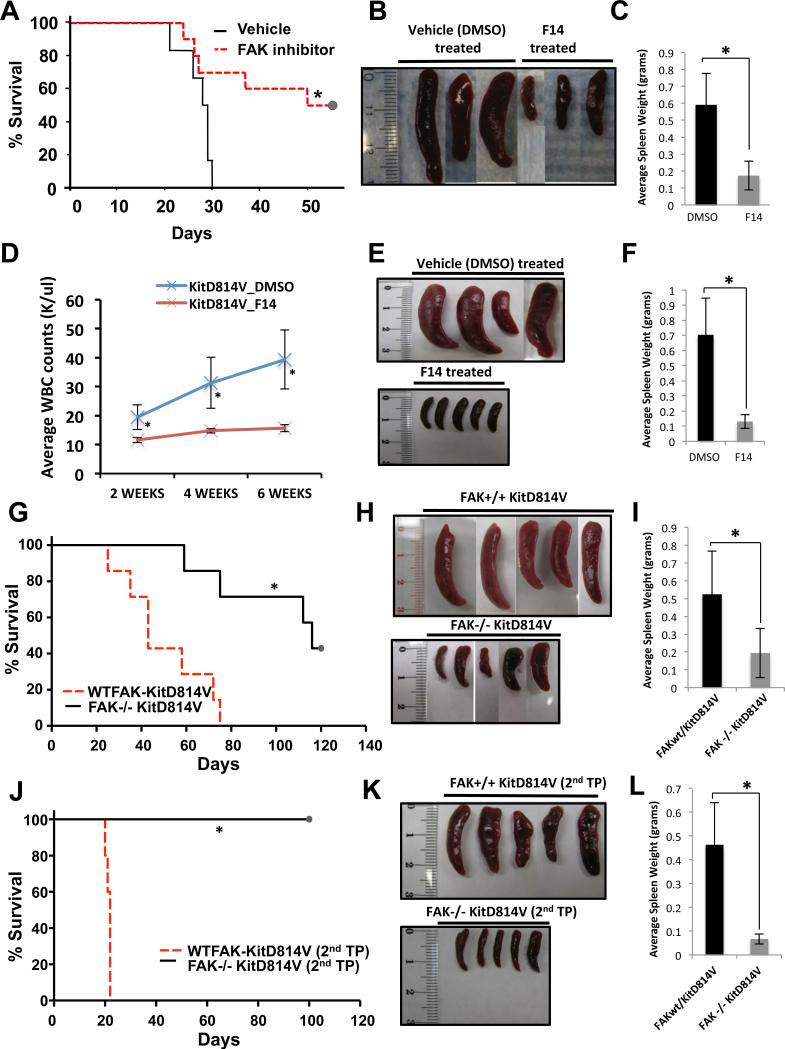
Fig.7. In vivo inhibition of FAK delays the development of MPN in mice transplanted with FLT3ITD and KITD814V bearing cells.
(A) C3H/HeJ mice were transplanted with 32D cells bearing FLT3ITD, and treated with 20 mg/kg body weight F-14 for 28 days. Kaplan-Meier survival analysis of vehicle (n=14) vs. F-14 (n=15) treated mice showed significant increase in overall survival (*p<0.02), and significant reduction of spleen size and weight (B,C) in F-14 treated mice as compared to vehicle (DMSO) control treated mice. (D) BoyJ mice were irradiated and transplanted with 5-FU treated BM cells expressing KITD814V. Mice were randomly divided into 2 groups and treated with vehicle (DMSO) (n=7) or F-14 (n=7) for 3 weeks post-transplantation. Peripheral blood from mice was analyzed at intervals of 2, 4 and 6 weeks (D). After 6 weeks mice were harvested to determine spleen size (E) and weight (F). (G) 5-FU treated BM cells from WTFAK or FAK−/− mice expressing KITD814V were transplanted into lethally irradiated C57BL/6 mice. Kaplan-Meier survival analysis of FAK+/+ KITD814V (n=9) vs. FAK−/− KITD814V (n=9) mice, spleen size (H) and weight (I) is shown (*p<0.002). (J) Secondary transplants were performed using BM from FAK+/+KITD814V and FAK−/−KITD814V primary recipients. Kaplan-Meier survival analysis of FAK+/+ KITD814V (n=5) vs. FAK−/− KITD814V (n=5) is shown (*p<0.003), and spleen size (K) and weight (L) (*p<0.05).
Active Stat5 downstream of FLT3ITD translocates to the nucleus to bind DNA and express Stat5 responsive genes like c-Myc and BclXL that play a crucial role in leukemogenesis (Li et al., 2007; Zhang et al., 2000). We next performed qRT-PCR analysis to determine the relative expression of c-Myc and BclXL genes in FLT3ITD cells treated with F-14. As seen in Figures 5G and 5H, a significant reduction in the expression of Stat5 responsive genes c-Myc and BclXL was observed upon FAK inhibition. Similar results were observed in FAK−/− FLT3ITD cells (Fig.S3A, B). To ascertain whether FAK also regulates the nuclear association between Rac1 and Stat5, besides directly activating Rac1, F-14 treated nuclear fractions were subjected to Rac1 immunoprecipitation assay. As seen in Fig.6A, the amount of active Stat5 that interacts with Rac1 was significantly reduced upon treatment of FLT3ITD cells with F-14 (Fig.6A, lane 1 vs 2). These results explain our initial observation demonstrating that reduced levels of total Rac1 and active Stat5 in nuclear fractions of FLT3ITD bearing cells in which FAK activation was repressed, and is directly a consequence of FAK's role in the activation of Rac1, and more importantly its association and subsequent translocation with active Stat5 into the nucleus. To further assess whether FAK regulates the association between active Rac1 and active Stat5, we performed active Rac1 pull down assay from WT and FAK−/− BM cells expressing FLT3ITD and analyzed for active Stat5 binding. As seen in Fig.6B, increased levels of active and total Stat5 protein was bound to activated Rac1 in WT (FAK+/+) BM cells (lane 1), which was reduced in FAK deficient (FAK−/−) FLT3ITD bearing cells (lane 2). Taken together, these findings suggest that first, FAK regulates the formation of an active Rac1-active Stat5 complex and second, it modulates the translocation of Rac1-Stat5 complex into the nuclear compartment.
Fig.6. Downstream of FAK, Tiam1 regulates the activation of Rac1 and subsequent translocation of active Stat5 to the nuclear compartment to develop leukemia in mice.
(A) Nuclear fractions from F-14 treated FLT3ITD and FLT3WT bearing cells were subjected to Rac1 immunoprecipitation assay to assess the level of Rac1 binding to active Stat5. Level of active Stat5 (pY694), total Stat5 and Rac1 were analyzed. (B) Active Rac1 fractions from WT and FAK−/− deficient BM cells expressing FLT3ITD was determined, along with levels of active and total Stat5. Total Rac1 levels are shown in the lowermost panel (n=2). (C) Fractionation assay was performed using WT and Rac1−/− BM cells expressing FLT3ITD or FLT3WT receptors. Nuclear and cytosolic fractions were analyzed as described above. GAPDH was used as a loading control and cytosolic marker (n=3). ‘MK’ denotes lane with protein ladder. (D) 32D FLT3ITD and FLT3WT cells were treated with or without F-14 and subjected to Tiam1 activation assay (n=2). (E) 32D FLT3ITD and FLT3WT cells were subjected to Tiam1 IP n presence or absence of F-14. Samples were analyzed for amount of Rac1 binding Tiam1 (IP:Tiam1 panels). Lower two panels (Lysate (input)) depict the total protein levels of Rac1 and Tiam1 (n=2). (F) 32D cells co-expressing FLT3ITD and Tiam1 shRNA or scrambled shRNA were subjected to cellular fractionation assay and nuclear and cytosolic fractions were analyzed for the levels of active Stat5 (pY694), total Stat5 and Rac1, and nuclear marker/loading control PARP-1. (G) 32D cells co-expressing FLT3ITD and Tiam1 shRNA's (lanes 2-4) or scrambled shRNA (lane 1) were subjected to active Rac1 pull-down assay. The amount of active and total Rac1 are shown in the upper and lower panels, respectively. (H) Kaplan-Meier survival curve of mice transplanted with 32D cells co-expressing FLT3ITD and Tiam1 shRNA (n=5) or scrambled shRNA (n=5), (*p<0.01).
Since Rac1 contains a functional nuclear localization signal (NLS), and also forms a complex with active Stat5 in the nucleus, we next investigated the role of Rac1 in nuclear translocation of active Stat5 in FLT3ITD-bearing cells. Cells were treated with or without Rac inhibitor NSC23766, and subjected to a fractionation assay. Levels of active Stat5, along with total Rac1, were reduced in FLT3ITD cells treated with NSC23766 (Fig.S3C). Likewise, cells co-expressing FLT3ITD along with a dominant negative form of Rac1 (Rac1N17) showed reduced levels of active Stat5 in the nuclear fractions (Fig.S3D). Equal expression levels of GFP-Rac1N17 in FLT3ITD and WT cells can be seen in Fig.S3E,F. As seen in Fig.6C (lane 1 vs 2), robust levels of active Stat5 were present in nuclear fractions of FLT3ITD expressing Rac1+/+ BM cells, while a significant reduction in the levels of nuclear localized active Stat5 was observed in Rac1−/− BM cells, with no detectable levels of Stat5 activation in FLT3WT bearing cells. These results demonstrate an essential role for Rac1 in the translocation of active Stat5 into the nucleus.
RacGEF Tiam1 is essential for FLT3ITD induced leukemic development in mice
To identify RacGEFs involved downstream of FAK in activating Rac1 in FLT3ITD bearing cells, we analyzed the role of Tiam1. We ascertained whether Tiam1 is active in cells bearing FLT3ITD. As seen in Figure 6D, increased levels of active Tiam1 were observed in FLT3ITD bearing cells compared to FLT3WT expressing cells (lane 1 vs 3), which was attenuated upon treating these cells with F-14 (lane 1 vs 2). Further, F-14 treatment perturbed the interaction between Rac1 and Tiam1 (Fig.6E). Having observed the presence of Tiam1 in active Rac1 complex in FLT3ITD bearing cells, and the perturbation of this association upon FAK inhibition, we next examined the functional significance of Tiam1 in FLT3ITD induced transformation. We knocked down Tiam1 using shRNA in cells bearing FLT3ITD (Fig. S3G). Cells bearing FLT3ITD and Tiam1 shRNA showed significant reduction in constitutive growth in comparison to cells bearing scrambled shRNA (Fig.S3H). Moreover, when FLT3ITD and Tiam1shRNA co-expressing cells were subjected to a fractionation assay, the levels of active Stat5 and total Rac1 in nuclear fractions were significantly reduced compared to scrambled shRNA expressing cells (Fig.6F, lane 1 vs 2-5). As shown in Fig.6G (lane 1 vs 2-4), knockdown of Tiam1 significantly inhibited the activation of Rac1 compared to cells co-expressing FLT3ITD and scrambled shRNA. These results indicate that Tiam1 plays an important role in the activation of Rac1 in FLT3ITD expressing cells, which in-turn regulates the nuclear translocation of the Rac1/Stat5 complex. To further investigate the role of Tiam1 in FLT3ITD induced leukemogenesis, we performed transplantation experiments. Fig.6H shows that mice transplanted with FLT3ITD and Tiam1 shRNA survived significantly longer compared to mice transplanted with FLT3ITD and scrambled vector (*p<0.01).
Inhibition of FAK delays the onset of FLT3ITD and KITD814V induced MPN development and prolongs the survival of mice
We next examined the in vivo impact of FAK inhibition on FLT3ITD and KITD814V induced leukemogenesis. Although mice bearing FLT3ITD cells treated with DMSO died within 30 days post-transplant, mice treated with F-14 showed significantly prolonged survival (Fig.7A). F-14 treated mice showed significantly reduced spleen weight (Fig.7B,C), and demonstrated absence of lesions in lungs compared to DMSO treated mice (Fig.S4A,B). Histopathologic analysis showed leukemic infiltration of myeloid cells and destruction of alveolar architecture in lungs and of normal architecture in spleens of DMSO treated mice but significant improvement in the F-14 treated mice (Fig.S4C,D). Moreover, F-14 treated mice also showed reduced percentage of leukemic cells in tissues (peripheral blood and spleen) as determined by the presence of GFP positive cells (Fig.S4E,F), relative to vehicle treated mice. Likewise, F-14 treatment of mice transplanted with cells expressing KITD814V survived significantly longer than vehicle treated mice (Fig.S5A, *p<0.01). We next assessed these findings in mice transplanted with primary BM cells expressing KITD814V. One cohort of mice was treated with F-14 and the other with vehicle (DMSO). As seen in Figure 7D, WBC counts remained constant over the entire duration of F-14 treatment in KITD814V bearing mice (6 weeks), while KITD814V bearing mice treated with vehicle demonstrated a steady rise in WBC counts over time, a hallmark of MPN development and progression. At the end of 6 weeks, all mice were euthanized and analyzed. As seen in Fig. 7E, F, vehicle treated mice demonstrated significant enlargement of spleen compared to F-14 treated mice. Collectively, these data supports the observation that targeting FAK rescues the development of FLT3ITD and KITD814V induced MPN in vivo. To further investigate the role of FAK in FLT3ITD induced MPN, we knocked down FAK expression using shRNA in cells bearing FLT3ITD and transplanted into mice as described in Fig.7A. Knockdown of FAK not only repressed the constitutive growth of FLT3ITD bearing cells (Fig.S5B), but more importantly mice transplanted with cells co-infected with FLT3ITD and FAK shRNA survived significantly longer compared to mice transplanted with FLT3ITD and scrambled vector (Fig.S5C, *p<0.025). To rule out non-specific effects of F-14, and diminished but not absolute effects of shRNA mediated knockdown of FAK, we performed transplantation studies using primary BM cells from WT (FAK+/+) and FAK−/− deficient mice expressing the oncogenic KITD814V receptor. As seen in Figure 7G, genetic ablation of FAK significantly prolonged the survival of leukemic mice (FAK−/− KITD814V) compared to controls (FAK+/+ KITD814V). Leukemic mice harboring the KITD814V oncogene in the FAK−/− background demonstrated reduced spleen size (Fig.7H, I) and WBC counts relative to controls (Fig.S5D). To further ascertain whether targeting FAK in the context of KITD814V induced MPN inhibits the growth of cells that give rise to leukemia; we performed secondary transplants using BM cells derived from the primary cohorts. As seen in Figure 7J, mice transplanted with BM cells from primary donor harboring KITD814V in a FAK deficient background (FAK−/− KITD814V) survived significantly longer than mice transplanted with BM from WT background (FAK+/+ KITD814V). The prolonged survival of these mice correlated with reduced splenomegaly (Figs. 7K, L). Importantly, loss of FAK in HSCs did not impair the engraftment or the self-renewal of these cells (Lu et al., 2012).
Targeting PAK1 inhibits the nuclear translocation of active Stat5
To determine the functional role of p21-activated kinase (PAK), a downstream effector of Rac1, in Stat5 regulation and leukemogenesis, we utilized a recently described allosteric PAK inhibitor, IPA-3 (Deacon et al., 2008). As seen in Fig.8A (lane 5 vs 7), IPA-3 treatment significantly inhibited the presence of activated nuclear Stat5 in FLT3ITD expressing cells. As seen in Fig.8A (lane 1 vs 2), addition of ligand also resulted in activation of the Stat5 oncogenic pathway in FLT3WT cells, which was also inhibited by IPA-3 (Fig.8A, lane 4). Next, we used another PAK inhibitor PF-3758309 (PF) (Murray et al., 2010). As seen in Figure 8B (lane 1 vs 2), similar results were observed upon treating FLT3ITD cells with PF-3758309 resulting in repression of nuclear translocation of active Stat5. Consistent with these results, expression of a dominant negative form of PAK1 (K299R), in FLT3ITD expressing cells (Fig.S5F) demonstrated reduced tyrosine phosphorylated Stat5 (Fig.S5E). These results suggest that downstream from Rac1, PAK contributes to the translocation of active Stat5 in the nucleus in FLT3ITD bearing cells. To determine whether PAK is active in FLT3ITD and KITD814V oncogene bearing cells, whole cell lysates were analyzed for PAK1 activity. As seen in Figure S6A, cells expressing KITD814V showed increased levels of constitutive PAK1 activation (pPAK1) compared to KITWT cells (lane 1 vs 5), which was readily inhibited upon treatment with F-14 (lane 2, 6). Similar results were seen in FLT3ITD expressing cells treated with F-14 (Fig.S6B, lane 1 vs 2). Inhibition of active PAK1 levels were also observed in FLT3ITD expressing cells treated with PAK inhibitors IPA-3 and PF-3758309 (Fig.S6C, lane 1 vs 2-3). These results suggest that PAK1 is hyperactive downstream of a FAK/Rac1 signaling pathway in FLT3ITD and KITD814V oncogene bearing cells.
Fig.8. In vivo inhibition of PAK1 delays the onset of MPN in mice transplanted with FLT3ITD bearing cells and inhibits the growth of leukemic patient cells.
(A) 32D cells expressing FLT3ITD or FLT3WT were starved of serum and treated with the PAK inhibitor IPA-3 (lanes 3,7) alone, with FLT3 ligand (FL) (lanes 2,6), or with IPA-3 followed by FL (lanes 4,8) as indicated, and subjected to cellular fractionation assay. Nuclear and cytosolic fractions were quantitated and equal lysates were loaded on a gel and probed with the indicated antibodies. Arrows indicate the activation/expression of the labeled molecules in nuclear as well as in cytosolic fractions of FLT3ITD and FLT3WT bearing cells. Expression of PARP-1 was used as an indicator of nuclear loading (n=2). (B) 32D FLT3ITD and FLT3WT were serum starved and treated with the PAK inhibitor PF-3758309 (PF) and analyzed as described in (A). qRT-PCR analysis of relative mRNA expression levels of Stat5 responsive genes BclXL (C) and c-Myc (D) in FLT3ITD cells treated with PAK inhibitor PF-3758309 (n=2), *p<0.05. Fractionation assays were performed in BM cells (E) and splenocytes (F). Mice cohorts transplanted with KITD814V in a WT PAK1 (PAK1+/+KIT814V) or PAK1 deficient (PAK1−/−KITD814V) cells (n=2). The level of phospho-Stat5, total Stat5 and Rac1 in the nuclear and cytosolic fractions is indicated. Expression of GAPDH was used as an indicator of cytosolic marker and loading control. ‘MK’ denotes lane with protein ladder. (G) Primary transplant were carried out using 5-FU treated BM cells from WTPAK1 or PAK1−/− mice transduced with KITD814V or KITWT, and transplanted into lethally irradiated C57BL/6 mice. Four groups of mice were used: WTPAK1 KITD814V (n=8), WTPAK1 KITWT (n=5), PAK1−/− KITD814V (n=8), and PAK1−/− KITDWT (n=5). Kaplan-Meier survival analysis of PAK1+/+ KITD814V vs PAK1−/− KITD814V, WTPAK1 KITWT, PAK1−/−KITWT mice showed significant overall survival (*p<0.0003, Hom-Sidak Method), and significant reduction of spleen size (H). (I) Secondary transplants were performed using BM cells from PAK1+/+KITD814V and PAK1−/−KITD814V mice and transplanted into irradiated C57BL/6 mice. Kaplan-Meier survival analysis of PAK1+/+ KITD814V (n=5) vs PAK1−/− KITD814V (n=5) mice showed significant overall survival (*p<0.001), and significant reduction of spleen size (J).
Group I family of PAKs consist of three members including PAK1, PAK2 and PAK3. While PAK1 and PAK2 are ubiquitously expressed, PAK3's expression is predominantly restricted to the brain (Ye and Field, 2012). We assessed the role of PAK1 and PAK2 in oncogene induced transformation by examining Stat5 activation in FLT3ITD cells in which the expression of these two isoforms was knocked down (Fig.S6E). As seen in Fig.S6D, knockdown of PAK1 (lane 1 vs 2-3) but not PAK2 (lane 1 vs 4-6) significantly reduced the nuclear accumulation of active Stat5 in FLT3ITD expressing cells. Consistent with these findings, expression of FLT3ITD in PAK1 deficient BM (PAK1−/−) cells also impaired the nuclear translocation of active Stat5 (Fig.S6I, lane 1 vs 2). Furthermore, loss of active Stat5 nuclear import in PAK1−/− FLT3ITD cells resulted in reduced expression of Stat5 target genes including Bcl-xL and c-Myc compared to controls (Fig.S6F), and also reduced relative mRNA levels of c-Myc and BclXL in FLT3ITD cells treated with PAK inhibitor PF-3758309 (Fig. 8C,D).
Inhibition of PAK1 delays the onset of FLT3ITD and KITD814V induced MPN and prolongs the survival of mice
To determine the role of PAK isoforms in FLT3ITD and KITD814V mediated MPN development, BM transplant studies were performed. Transplantation studies utilizing myeloid cells bearing FLT3ITD in the context of PAK2 knockdown did not prolong the survival of leukemic mice compared to controls (Fig.S6G). In contrast, PAK1 knockdown in the context of FLT3ITD expression significantly enhanced the life span of leukemic mice (Fig.S6H). To further confirm PAK1's involvement in KITD814V mediated leukemogenesis, we transplanted mice with KITD814V bearing PAK1−/− BM cells or WT (PAK1+/+) controls. Mice transplanted with WTPAK1 BM cells expressing KITD814V came down with disease significantly earlier than PAK1−/− KITD814V mice (Fig.8G), at which juncture time-point matched control mice (PAK1−/− KITD814V, PAK1+/+ KITWT, PAK1−/− KITWT) were euthanized and BM (Fig.8E) and spleen (Fig.8F) cells harvested and subjected to cellular fractionation analysis. As seen in Figure 8E (lane 1 vs 2) and 8F (lane 1 vs 2), similar to our data using cell lines and PAK inhibitors, genetic ablation of PAK1 in vivo abrogated the nuclear translocation of active Stat5 in KITD814V bearing BM derived cells. As seen in Figure 8G, genetic ablation of PAK1 significantly prolonged the survival of leukemic mice (PAK1−/− KITD814V) compared to controls (PAK1+/+ KITD814V), and modulated the development of MPN in mice as shown by reduced splenomegaly and WBC counts (Figs.8H, S6J,K). Mice transplanted with WTKIT did not demonstrate any signs of MPN, and showed normal survival and spleen size (Figs.8H, S6J). To further ascertain whether targeting PAK1 in the context of KITD814V induced MPN, selectively impacts the leukemia initiating cell ‘LIC’ population; we performed secondary transplants using BM cells from primary recipients. As seen in Figure 8I, mice transplanted with BM cells from primary donor harboring KITD814V in a PAK1 deficient background (PAK1−/− KITD814V) survived significantly longer than mice transplanted with BM from WT background (PAK1+/+ KITD814V). The survival of mice correlated with correction in spleen size (Figs. 8I,J, S6L), similar to primary transplants described above.
Inhibition of PAK inhibits the constitutive growth of FLT3ITD+ AML cells and KITD816V (+) SM patient derived cells
To assess the functional consequence(s) of PAK1 repression on the growth and transforming ability of FLT3ITD and KITD814V bearing cells, we performed a proliferation assay using cells expressing FLT3 and KIT receptors treated with or without PF-3758309. A dose dependent reduction in the growth of FLT3ITD and KITD814V bearing cells was observed but not that of FLT3WT and KITWT bearing cells (Fig.S7A,B and S7C,D, respectively). Similar results were observed for FLT3ITD cells treated with the PAK inhibitor IPA-3 (Fig.S7E). To further validate these observations, we performed similar studies in FLT3ITD bearing cells co-expressing a dominant negative version of PAK1 (K299R) (Fig.S7F) as well as in BM cells expressing FLT3ITD in the setting of PAK1 deficiency (Fig.S7G). A significant inhibition in the growth of FLT3ITD bearing cells was noted in the background of dominant negative or genetic ablation of PAK1. A similar inhibition in the growth of AML patient cells (Fig.S7I) and KITD816V (+) SM patient derived cells (Fig.S7K) was observed in the presence of IPA-3. Lastly, we assessed the role of PAK1 overexpression on the rescue of ligand-independent growth of BM cells expressing KITD814V in PAK1 deficient (PAK1−/−) background. An activated version of PAK1 (PAK1T423E) (Schurmann et al., 2000) was co-expressed with KITD814V in BM cells derived from PAK1−/− mice. As seen in Figure S7H we observed rescue of ligand-independent growth of cells over-expressing PAK1T423E, as compared to vector control.
DISCUSSION
Presence of active Stat5 in the nucleus, and the subsequent expression of Stat5 dependent pro-survival and anti-apoptotic genes, plays a key role in the transformation of cells bearing oncogenic forms of FLT3 and KIT (Benekli et al., 2003; Choudhary et al., 2007; Tse et al., 2000). However, the mechanism(s) involved in regulating the active form of nuclear Stat5 remain poorly understood. To this end, a role for Rac1GTPase/MgcRacGAP complex in the translocation of active Stat5 into the nucleus has been suggested; however the upstream and downstream signaling proteins from Rac1 involved in this process have not been identified and the extent to which these proteins contribute to leukemogenesis is unknown (Sallmyr et al., 2008). Using pharmacologic, biochemical and genetic approaches, we demonstrate that the FAK/Rac1-Tiam1/PAK1 axis plays a crucial role in the transformation induced by oncogenic forms of FLT3 (FLT3ITD) and KIT (KITD816V). Targeting FAK, Tiam1, and PAK1 in oncogene bearing cells in vitro or in vivo inhibits the presence of active Stat5 in the nuclear compartment, which profoundly delays the onset of leukemia by repressing the expression of Stat5 responsive genes. These findings were validated in both murine and human models of AML and MPN and suggest that the signaling axis we have identified is highly conserved across species. More importantly, we show that this axis is active in leukemia initiating cells as well as in leukemic cells that acquire AC220 resistant mutations of FLT3.
We and others have shown that FAK may contribute to Rac activation in hematopoietic cells as well as in other heterologous cell systems (Chang et al., 2006; Elias et al., 2010; Vemula et al., 2010) and that FLT3ITD can activate Rac1 and regulate the production of reactive oxygen species (ROS) via its association with Stat5 (Sallmyr et al., 2008). Furthermore, active Rac1 can also induce the activation and nuclear translocation of Stat3 (Simon et al., 2000). FLT3ITD can phosphorylate Stat5, independent of Jak kinase family members (Choudhary et al., 2007), while Rac1 mediated ROS production can induce the activation of Jak kinases and Stats downstream of G-protein coupled receptors (Pelletier et al., 2003). Although significant work has been done in identifying the above described linkages; how these molecules connect and what is their relationship in regulating transformation via oncogenic forms of KIT and FLT3 has never been described. Our findings provide insight into how FAK-Rac1-Tiam1 and PAK1 axis contributes to leukemic transformation in part by regulating active nuclear Stat5.
The role of GEFs such as Vav1 and Vav2 is largely considered promiscuous, as they regulate the activity of all three members of the RhoGTPase family including Rac, Rho and Cdc42 (Schmidt, 2002). In contrast, GEF Tiam1 is highly specific for Rac1 in vivo, and has been implicated in activating Rac1 to mediate Stat3 activation and its subsequent nuclear localization in COS-1 cells (Simon et al., 2000). We demonstrate that FAK activates Rac1 via RacGEF Tiam1 in FLT3ITD bearing cells, and targeting FAK and Tiam1 results in inhibition of Rac1. We also show that shRNA mediated knockdown of Tiam1 prolongs the survival of FLT3ITD bearing leukemic mice and genetic and pharmacologic inhibition of FAK and Tiam1 results in failure of active Stat5 to be expressed in the nucleus, along with Rac1. While Tiam1 regulates epithelial cancers such as carcinomas of breast and colon (Bourguignon et al., 2000; Buongiorno et al., 2008); we show its role in regulating hematologic malignancies.
Although evidence in this study and reported earlier (Sallmyr et al., 2008) suggests a role of Rac1 in translocating Stat5 into the nucleus, the relationship between Stat5 and RacGTPases in the context of FLT3 and KIT oncogenic mutations is unclear. An indirect role of PAK1 in nuclear shuttling of Stat5 has been suggested, where PAK1 plays a role in ‘switching’ in occupancy of the same promoter region between BCL6 and Stat5. In colorectal cancer PAK1, activated via Rac1, translocates into the nucleus and phosphorylates chromatin-bound BCL6, leading to its dissociation from the promoter, thereby allowing active Stat5 that is already present in the nucleus via a Rac1/MgcRacGAP dependent mechanism, to bind to the same promoter regions (Barros et al., 2012). In line with reported findings by Barros et al., we have observed a reduction in BCL6 activation and its mRNA expression levels upon treatment of FLT3ITD cells with PAK1 inhibitor (Fig.S8A, B). This data demonstrates that in the FLT3 and KIT oncogenic pathway the FAK/Tiam1/Rac1 signaling axis activates PAK1, which in-turn inhibits the transcriptional repressor BCL6, while correspondingly activating Stat5 to mediate leukemic transformation.
In BCR-ABL induced CML, majority of Stat5 is persistently active and retained in the cytoplasmic compartment, primarily via an association of active Stat5 with Gab2 and PI3k/Akt, subsequently leading to leukemogenesis (Nyga et al., 2005). FLT3ITD also interacts with Gab2 and results in the activation of PI3K/Akt, Stat5 and Gab2 mediated recruitment of Src kinases can also result in the activation of Stat5. Thus, while FLT3ITD can directly activate Stat5 (Choudhary et al., 2007), other tyrosine kinases present in complexes with FLT3ITD may also be involved in regulating Stat5 activation including Src kinases, as mutating Src kinase binding sites Y589 and Y591 in FLT3 receptor inhibits Stat5 activation (Hayakawa et al., 2000; Rocnik, 2006). Our data using FLT3ITD specific inhibitor AC220 not only shows inhibition in the phosphorylation of FLT3ITD on Y589/591, but also downstream inhibition of activating residue Y397 on FAK, indicating that oncogenic FLT3 mediates direct activation of FAK. In breast cancer cells, Prolactin (PRL) induced activation of FAK and Stat5 is mediated via Src family kinases. Upon phosphorylation by Src, phosphorylated FAK recruits Grb2/Gab2 to mediate activation of Ras/MAPK signaling pathway. The involvement of the PI3K/Rac/Pak pathway in PRL-induced activation of Erk has also been suggested. These results demonstrate a complex cross-talk between various signaling pathways involved in breast cancer metastasis (Aksamitiene et al., 2011). Future studies will determine whether Gab2 or other Src kinases play a role in activating FAK downstream of FLT3ITD and KITD814V receptors, which results in oncogenic transformation mediated by the subsequent nuclear translocation of active Stat5.
EXPERIMENTAL PROCEDURES
Antibodies and Reagents
Phycoerythrin (PE)-conjugated annexin V antibody and 7-amino actinomycin D (7-AAD) were purchased from BD Biosciences Pharmingen (San Jose, CA). Rabbit anti-phospho-PAK1, anti-PAK1, anti-phospho-Stat5 (Y694), anti-Stat5 antibodies, rabbit anti-phospho-FAK (Y397) and anti-FAK antibodies were purchased from Cell Signaling Technology (Beverly, MA). Anti-rabbit IgG DyLight 649 from Biolegend (San Deigo, CA), Anti-actin and GAPDH antibodies were purchased from Sigma (St. Louis, MO). Tiam1 activation kit was purchased from Cell BioLabs (San Diego, CA). Anti-mouse-HRP, anti-rabbit-HRP, anti-goat-HRP, anti-phospho-Stat5 (S780), anti-Tiam1, PARP-1, LaminB, BclXL and cMyc antibodies were purchased from Santa Cruz Biotechnology (Dallas,TX). FAK inhibitors F-14 and Y-11, PAK inhibitors IPA-3 and PF-3758309, and Rac inhibitor NSC23766 were purchased from R&D Systems Inc. (Minneapolis, MN). Lumina Forte Western HRP Substrate, Chemiluminescent Blocker (bløk-CH), anti-phospho-FAK (Y397) rabbit polyclonal, anti-Rac1 (23A8) and Rac1 activation kit were purchased from Millipore Corporation (Billerica, MA). Recombinant murine and human IL-3, Flt3, GM-CSF, SCF, IL-6, and Tpo were purchased from Peprotech (Rocky Hill, NJ). Retronectin was obtained from Takara (Madison, WI). Iscove's modified Dulbecco's medium (IMDM) was purchased from Invitrogen (Carlsbad, CA). Monothioglycerol was purchased from Sigma (St. Louis, MO). [3H] Thymidine was purchased from PerkinElmer (Boston, MA). Protein A– Sepharose beads were purchased from Amersham Biosciences (Piscataway, NJ). MKK/MEK inhibitor PD98059 was purchased from Cell Signaling Technology (Danvers, MA), Raf inhibitor PLX4720 from Selleckchem (Houston,TX) and Akt inhibitor 124005 from Millipore corporation (Billerica, MA).
Mice
C57BL/6 and C3H/HeJ mice were purchased from Jackson Laboratory (Bar Harbor, ME). FAK, Rac1 and PAK1 deficient mice have been previously described (Martin et al., 2013; McDaniel et al., 2008; Vemula et al., 2010). All mice used in this study were between 6 to 12 weeks of age and were maintained under specific pathogen-free conditions at the Indiana University Laboratory Animal Research Center (Indianapolis, IN). The studies were approved by the Institutional Animal Care and Use Committee (IACUC) of the Indiana University School of Medicine.
Patient Samples
Peripheral blood mononuclear cells from patients with AML were obtained at the time of diagnostic testing after informed consent. Approval was obtained from the institutional review boards of Indiana University School of Medicine. The buoyant fraction was isolated over Ficoll-Hypaque, and then washed with phosphate-buffered saline (PBS) before processing as described previously (Hartman et al., 2006). KITD816V positive or negative SM patient derived cells were obtained as described (Traina et al., 2012).
Cells
Primary low density mononuclear cells (LDMNC) were harvested as described earlier and used in the study (Mali et al., 2011). The murine IL-3 dependent myeloid cell line 32D cells bearing FLT3, FLT3ITD (N51), MIEG3 vector, KIT, or KITD814V have been described (Mali et al., 2011). Puromycin resistant BaF3 cells bearing the AC220 resistant mutants (Flt3ITD+TKD_D835Y/F, F691L) have been described (Smith et al., 2012). The human mast cell leukemia line, bearing the KITV560G as well as KITD816V mutations, HMC1.2 and acute myeloid leukemia (AML) cell line, bearing the FLT3ITD mutation, MV4-11 have been described (Butterfield et al., 1988; Lange et al., 1987).
Expression of WT and oncogenic receptors
Transduction of 32D and primary BM derived hematopoietic stem and progenitor cells was performed as described previously (Mali et al., 2011).
shRNA silencing of FAK, Tiam1 and PAK
FAK, Tiam1 or PAK-specific shRNA expression plasmids were purchased from OriGene Technologies (Rockville, MD). Purified and sequence verified plasmid containing a non-effective 29-mer sh GFP cassette (Scrambled vector) was used as a negative control. Cells were transduced with scrambled vector or shRNA plasmid and grown in the presence of puromycin (10ng/mL) to select for the transduced cells.
Proliferation and Apoptosis assays
Proliferation assays were performed as previously described (Mali et al., 2011).
Cytoplasm and Nuclear extraction
To extract nuclear and cytosolic fractions, the NE-PER Nuclear and Cytoplasmic Extraction Kit (Thermo Scientific) was used as per the manufacturer's instructions.
Immunoprecipitation (IP) and Western blot analysis (WB)
Immunoprecipitation and western blot analysis was performed as described previously (Mali et al., 2011).
qRT-PCR
Total RNA was isolated from 5×106 cells using RNeasy Plus Mini Kit (Qiagen) according to the manufacturer's instructions. cDNA was generated using random hexamer primers and Superscript II reverse transcriptase (Invitrogen). qRT-PCR was performed using FastStart Universal SYBR Green master mix (Roche) and a Applied Biosystem 7500 Real Time PCR system. ß-actin amplification was used to normalize sample RNA content.
Mouse leukemia induction and in vivo drug treatment
Mouse leukemia induction and in vivo drug treatments were performed as described previously (Mali et al., 2011). 1×106 32D cells bearing FLT3ITD in 200 μL PBS was injected into C3H/HeJ mice intravenously. After 48 hours of transplantation, mice were treated with vehicle (PBS/DMSO) or FAK inhibitor F-14 (25 mg/kg body weight) by intraperitoneal injection at 24 hour interval for 21 days. Mice were closely monitored for MPN development and harvested at moribund. For F-14 treatment in a primary transplant model, irradiated BoyJ mice were transplanted with 2.0 ×106 GFP-KITD814V cells and 0.1 ×106 supporting BM cells. Three weeks post-transplant mice were treated with 10 mg/kg body weight F-14 for 5 days a week, for 6 weeks. All mice were harvested and peripheral blood (PB) counts were monitored using a Hemavet 950 (Drew Scientific). PB, BM and spleens were analyzed for GFP expression. BM, spleen and lungs were also fixed in 10% buffered formalin to perform histopathologic analysis by hematoxylin and eosin (H&E) staining.
Statistics
All graphical data was evaluated by paired Student t- test (2-tailed) and results were considered significantly different with p-value <0.05. All data are represented as mean values ± standard deviations (SD). Survival probability of transplanted mice groups was compared using a Kaplan-Meier Survival Analysis in which statistical significance was determined as p-values <0.05 by log rank test.
Supplementary Material
ACKNOWLEDGMENTS
We thank Ms. Marilyn Wales for providing administrative support, Dr. D. Wade Clapp and Dr. Jonathan Chernoff (Fox Chase Cancer Center, Philadelphia, PA) for providing PAK1−/− mice, members of Dr. Christie M. Orschell's laboratory and Dr. James Henderson (Millipore). This work was supported in part by grants from National Institutes of Health (R01HL077177 to RK; R01HL081111 to RK, R01CA173852 to R.K., and R01CA134777 to RC and RK), and Riley Children's Foundation. A.C. is an American Cancer Society post doctoral Fellow supported by PF13-065-01, and by T32HL007910 from National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
A.C. conceived, designed, performed, analyzed experiments and wrote manuscript; J.G. designed, performed, analyzed experiments and edited manuscript; B.R., R.S.M., H.M., M.K., S.V., V.H.C., E.R.W. performed experiments; V.V., R.V.T., C.C.M. provided reagents; N.S., K.D.B., H.S.B., Y.L., R.J.C. provided expertise and reagents; R.K. conceived, designed, analyzed experiments and wrote manuscript.
SUPPLEMENTAL INFORMATION
Supplemental information includes eight figures and associated figure legends.
REFERENCES
- Aksamitiene E, Achanta S, Kolch W, Kholodenko BN, Hoek JB, Kiyatkin A. Prolactin-stimulated activation of ERK1/2 mitogen-activated protein kinases is controlled by PI3-kinase/Rac/PAK signaling pathway in breast cancer cells. Cell Signal. 2011;23:1794–1805. doi: 10.1016/j.cellsig.2011.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barros P, Lam EW, Jordan P, Matos P. Rac1 signalling modulates a STAT5/BCL-6 transcriptional switch on cell-cycle-associated target gene promoters. Nucleic acids research. 2012;40:7776–7787. doi: 10.1093/nar/gks571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgartner C, Cerny-Reiterer S, Sonneck K, Mayerhofer M, Gleixner KV, Fritz R, Kerenyi M, Boudot C, Gouilleux F, Kornfeld JW, et al. Expression of activated STAT5 in neoplastic mast cells in systemic mastocytosis: subcellular distribution and role of the transforming oncoprotein KIT D816V. The American journal of pathology. 2009;175:2416–2429. doi: 10.2353/ajpath.2009.080953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beghini A, Ripamonti CB, Cairoli R, Cazzaniga G, Colapietro P, Elice F, Nadali G, Grillo G, Haas OA, Biondi A, et al. KIT activating mutations: incidence in adult and pediatric acute myeloid leukemia, and identification of an internal tandem duplication. Haematologica. 2004;89:920–925. [PubMed] [Google Scholar]
- Benekli M, Baer MR, Baumann H, Wetzler M. Signal transducer and activator of transcription proteins in leukemias. Blood. 2003;101:2940–2954. doi: 10.1182/blood-2002-04-1204. [DOI] [PubMed] [Google Scholar]
- Bourguignon LY, Zhu H, Shao L, Chen YW. CD44 interaction with tiam1 promotes Rac1 signaling and hyaluronic acid-mediated breast tumor cell migration. J Biol Chem. 2000;275:1829–1838. doi: 10.1074/jbc.275.3.1829. [DOI] [PubMed] [Google Scholar]
- Brady A, Gibson S, Rybicki L, Hsi E, Saunthararajah Y, Sekeres MA, Tiu R, Copelan E, Kalaycio M, Sobecks R, et al. Expression of phosphorylated signal transducer and activator of transcription 5 is associated with an increased risk of death in acute myeloid leukemia. Eur J Haematol. 2012;89:288–293. doi: 10.1111/j.1600-0609.2012.01825.x. [DOI] [PubMed] [Google Scholar]
- Buongiorno P, Pethe VV, Charames GS, Esufali S, Bapat B. Rac1 GTPase and the Rac1 exchange factor Tiam1 associate with Wnt-responsive promoters to enhance beta-catenin/TCF-dependent transcription in colorectal cancer cells. Molecular cancer. 2008;7:73. doi: 10.1186/1476-4598-7-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butterfield JH, Weiler D, Dewald G, Gleich GJ. Establishment of an immature mast cell line from a patient with mast cell leukemia. Leuk Res. 1988;12:345–355. doi: 10.1016/0145-2126(88)90050-1. [DOI] [PubMed] [Google Scholar]
- Chang F, Lemmon CA, Park D, Romer LH. FAK Potentiates Rac1 Activation and Localization to Matrix Adhesion Sites: A Role for betaPIX. Molecular Biology of the Cell. 2006;18:253–264. doi: 10.1091/mbc.E06-03-0207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary C, Brandts C, Schwable J, Tickenbrock L, Sargin B, Ueker A, Bohmer FD, Berdel WE, Muller-Tidow C, Serve H. Activation mechanisms of STAT5 by oncogenic Flt3-ITD. Blood. 2007;110:370–374. doi: 10.1182/blood-2006-05-024018. [DOI] [PubMed] [Google Scholar]
- Choudhary C, Muller-Tidow C, Berdel WE, Serve H. Signal transduction of oncogenic Flt3. Int J Hematol. 2005;82:93–99. doi: 10.1532/IJH97.05090. [DOI] [PubMed] [Google Scholar]
- Deacon SW, Beeser A, Fukui JA, Rennefahrt UE, Myers C, Chernoff J, Peterson JR. An isoform-selective, small-molecule inhibitor targets the autoregulatory mechanism of p21-activated kinase. Chem Biol. 2008;15:322–331. doi: 10.1016/j.chembiol.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Despeaux M, Labat E, Gadelorge M, Prade N, Bertrand J, Demur C, Recher C, Bonnevialle P, Payrastre B, Bourin P, et al. Critical features of FAK-expressing AML bone marrow microenvironment through leukemia stem cell hijacking of mesenchymal stromal cells. Leukemia. 2011;25:1789–1793. doi: 10.1038/leu.2011.145. [DOI] [PubMed] [Google Scholar]
- Elias BC, Bhattacharya S, Ray RM, Johnson LR. Polyamine-dependent activation of Rac1 is stimulated by focal adhesion-mediated Tiam1 activation. Cell Adh Migr. 2010;4:419–430. doi: 10.4161/cam.4.3.12043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabarra-Niecko V, Schaller MD, Dunty JM. FAK regulates biological processes important for the pathogenesis of cancer. Cancer metastasis reviews. 2003;22:359–374. doi: 10.1023/a:1023725029589. [DOI] [PubMed] [Google Scholar]
- Golubovskaya VM, Nyberg C, Zheng M, Kweh F, Magis A, Ostrov D, Cance WG. A small molecule inhibitor, 1,2,4,5-benzenetetraamine tetrahydrochloride, targeting the y397 site of focal adhesion kinase decreases tumor growth. J Med Chem. 2008;51:7405–7416. doi: 10.1021/jm800483v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman AD, Wilson-Weekes A, Suvannasankha A, Burgess GS, Phillips CA, Hincher KJ, Cripe LD, Boswell HS. Constitutive c-jun N-terminal kinase activity in acute myeloid leukemia derives from Flt3 and affects survival and proliferation. Exp Hematol. 2006;34:1360–1376. doi: 10.1016/j.exphem.2006.05.019. [DOI] [PubMed] [Google Scholar]
- Hayakawa F, Towatari M, Kiyoi H, Tanimoto M, Kitamura T, Saito H, Naoe T. Tandem-duplicated Flt3 constitutively activates STAT5 and MAP kinase and introduces autonomous cell growth in IL-3-dependent cell lines. Oncogene. 2000;19:624–631. doi: 10.1038/sj.onc.1203354. [DOI] [PubMed] [Google Scholar]
- Kiyoi H, Ohno R, Ueda R, Saito H, Naoe T. Mechanism of constitutive activation of FLT3 with internal tandem duplication in the juxtamembrane domain. Oncogene. 2002;21:2555–2563. doi: 10.1038/sj.onc.1205332. [DOI] [PubMed] [Google Scholar]
- Kottaridis PD, Gale RE, Frew ME, Harrison G, Langabeer SE, Belton AA, Walker H, Wheatley K, Bowen DT, Burnett AK, et al. The presence of a FLT3 internal tandem duplication in patients with acute myeloid leukemia (AML) adds important prognostic information to cytogenetic risk group and response to the first cycle of chemotherapy: analysis of 854 patients from the United Kingdom Medical Research Council AML 10 and 12 trials. Blood. 2001;98:1752–1759. doi: 10.1182/blood.v98.6.1752. [DOI] [PubMed] [Google Scholar]
- Lange B, Valtieri M, Santoli D, Caracciolo D, Mavilio F, Gemperlein I, Griffin C, Emanuel B, Finan J, Nowell P, et al. Growth factor requirements of childhood acute leukemia: establishment of GM-CSF-dependent cell lines. Blood. 1987;70:192–199. [PubMed] [Google Scholar]
- Li L, Piloto O, Kim KT, Ye Z, Nguyen HB, Yu X, Levis M, Cheng L, Small D. FLT3/ITD expression increases expansion, survival and entry into cell cycle of human haematopoietic stem/progenitor cells. Br J Haematol. 2007;137:64–75. doi: 10.1111/j.1365-2141.2007.06525.x. [DOI] [PubMed] [Google Scholar]
- Li S, Hua ZC. FAK expression regulation and therapeutic potential. Adv Cancer Res. 2008;101:45–61. doi: 10.1016/S0065-230X(08)00403-X. [DOI] [PubMed] [Google Scholar]
- Lu J, Sun Y, Nombela-Arrieta C, Du KP, Park SY, Chai L, Walkley C, Luo HR, Silberstein LE. Fak depletion in both hematopoietic and nonhematopoietic niche cells leads to hematopoietic stem cell expansion. Exp Hematol. 2012;40:307–317. e303. doi: 10.1016/j.exphem.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali RS, Ramdas B, Ma P, Shi J, Munugalavadla V, Sims E, Wei L, Vemula S, Nabinger SC, Goodwin CB, et al. Rho Kinase Regulates the Survival and Transformation of Cells Bearing Oncogenic Forms of KIT, FLT3, and BCR-ABL. Cancer Cell. 2011;20:357–369. doi: 10.1016/j.ccr.2011.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin H, Mali RS, Ma P, Chatterjee A, Ramdas B, Sims E, Munugalavadla V, Ghosh J, Mattingly RR, Visconte V, et al. Pak and Rac GTPases promote oncogenic KIT-induced neoplasms. J Clin Invest. 2013;123:4449–4463. doi: 10.1172/JCI67509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDaniel AS, Allen JD, Park SJ, Jaffer ZM, Michels EG, Burgin SJ, Chen S, Bessler WK, Hofmann C, Ingram DA, et al. Pak1 regulates multiple c-Kit mediated Ras-MAPK gain-in-function phenotypes in Nf1+/− mast cells. Blood. 2008;112:4646–4654. doi: 10.1182/blood-2008-04-155085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray BW, Guo C, Piraino J, Westwick JK, Zhang C, Lamerdin J, Dagostino E, Knighton D, Loi CM, Zager M, et al. Small-molecule p21-activated kinase inhibitor PF-3758309 is a potent inhibitor of oncogenic signaling and tumor growth. Proc Natl Acad Sci U S A. 2010;107:9446–9451. doi: 10.1073/pnas.0911863107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyga R, Pecquet C, Harir N, Gu H, Dhennin-Duthille I, Regnier A, Gouilleux-Gruart V, Lassoued K, Gouilleux F. Activated STAT5 proteins induce activation of the PI 3-kinase/Akt and Ras/MAPK pathways via the Gab2 scaffolding adapter. The Biochemical journal. 2005;390:359–366. doi: 10.1042/BJ20041523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obermann EC, Arber C, Jotterand M, Tichelli A, Hirschmann P, Tzankov A. Expression of pSTAT5 predicts FLT3 internal tandem duplications in acute myeloid leukemia. Ann Hematol. 2010;89:663–669. doi: 10.1007/s00277-009-0890-8. [DOI] [PubMed] [Google Scholar]
- Pelletier S, Duhamel F, Coulombe P, Popoff MR, Meloche S. Rho family GTPases are required for activation of Jak/STAT signaling by G protein-coupled receptors. Mol Cell Biol. 2003;23:1316–1333. doi: 10.1128/MCB.23.4.1316-1333.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Recher C, Ysebaert L, Beyne-Rauzy O, Mansat-De Mas V, Ruidavets JB, Cariven P, Demur C, Payrastre B, Laurent G, Racaud-Sultan C. Expression of focal adhesion kinase in acute myeloid leukemia is associated with enhanced blast migration, increased cellularity, and poor prognosis. Cancer Res. 2004;64:3191–3197. doi: 10.1158/0008-5472.can-03-3005. [DOI] [PubMed] [Google Scholar]
- Rocnik JL. Roles of tyrosine 589 and 591 in STAT5 activation and transformation mediated by FLT3-ITD. Blood. 2006;108:1339–1345. doi: 10.1182/blood-2005-11-011429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocnik JL, Okabe R, Yu JC, Lee BH, Giese N, Schenkein DP, Gilliland DG. Roles of tyrosine 589 and 591 in STAT5 activation and transformation mediated by FLT3-ITD. Blood. 2006;108:1339–1345. doi: 10.1182/blood-2005-11-011429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sallmyr A, Fan J, Datta K, Kim KT, Grosu D, Shapiro P, Small D, Rassool F. Internal tandem duplication of FLT3 (FLT3/ITD) induces increased ROS production, DNA damage, and misrepair: implications for poor prognosis in AML. Blood. 2008;111:3173–3182. doi: 10.1182/blood-2007-05-092510. [DOI] [PubMed] [Google Scholar]
- Schmidt A. Guanine nucleotide exchange factors for Rho GTPases: turning on the switch. Genes & Development. 2002;16:1587–1609. doi: 10.1101/gad.1003302. [DOI] [PubMed] [Google Scholar]
- Schurmann A, Mooney AF, Sanders LC, Sells MA, Wang HG, Reed JC, Bokoch GM. p21-activated kinase 1 phosphorylates the death agonist bad and protects cells from apoptosis. Mol Cell Biol. 2000;20:453–461. doi: 10.1128/mcb.20.2.453-461.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon AR, Vikis HG, Stewart S, Fanburg BL, Cochran BH, Guan KL. Regulation of STAT3 by direct binding to the Rac1 GTPase. Science. 2000;290:144–147. doi: 10.1126/science.290.5489.144. [DOI] [PubMed] [Google Scholar]
- Smith CC, Wang Q, Chin CS, Salerno S, Damon LE, Levis MJ, Perl AE, Travers KJ, Wang S, Hunt JP, et al. Validation of ITD mutations in FLT3 as a therapeutic target in human acute myeloid leukaemia. Nature. 2012;485:260–263. doi: 10.1038/nature11016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiekermann K, Bagrintseva K, Schwab R, Schmieja K, Hiddemann W. Overexpression and constitutive activation of FLT3 induces STAT5 activation in primary acute myeloid leukemia blast cells. Clin Cancer Res. 2003;9:2140–2150. [PubMed] [Google Scholar]
- Traina F, Visconte V, Jankowska AM, Makishima H, O'Keefe CL, Elson P, Han Y, Hsieh FH, Sekeres MA, Mali RS, et al. Single nucleotide polymorphism array lesions, TET2, DNMT3A, ASXL1 and CBL mutations are present in systemic mastocytosis. PLoS One. 2012;7:e43090. doi: 10.1371/journal.pone.0043090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tse KF, Mukherjee G, Small D. Constitutive activation of FLT3 stimulates multiple intracellular signal transducers and results in transformation. Leukemia. 2000;14:1766–1776. doi: 10.1038/sj.leu.2401905. [DOI] [PubMed] [Google Scholar]
- Vemula S, Ramdas B, Hanneman P, Martin J, Beggs HE, Kapur R. Essential role for focal adhesion kinase in regulating stress hematopoiesis. Blood. 2010;116:4103–4115. doi: 10.1182/blood-2010-01-262790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye DZ, Field J. PAK signaling in cancer. Cell Logist. 2012;2:105–116. doi: 10.4161/cl.21882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin B. Focal adhesion kinase as a target in the treatment of hematological malignancies. Leuk Res. 2011;35:1416–1418. doi: 10.1016/j.leukres.2011.04.017. [DOI] [PubMed] [Google Scholar]
- Zhang S, Fukuda S, Lee Y, Hangoc G, Cooper S, Spolski R, Leonard WJ, Broxmeyer HE. Essential role of signal transducer and activator of transcription (Stat)5a but not Stat5b for Flt3-dependent signaling. J Exp Med. 2000;192:719–728. doi: 10.1084/jem.192.5.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Guan JL. Signal transduction by focal adhesion kinase in cancer. Cancer Metastasis Rev. 2009;28:35–49. doi: 10.1007/s10555-008-9165-4. [DOI] [PubMed] [Google Scholar]
- Zheng R, Bailey E, Nguyen B, Yang X, Piloto O, Levis M, Small D. Further activation of FLT3 mutants by FLT3 ligand. Oncogene. 2011;30:4004–4014. doi: 10.1038/onc.2011.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.