Abstract
Combination therapy concurrently targeting PD1 and CTLA4 immune checkpoints leads to remarkable anti-tumor effects. While both PD1 and CTLA4 dampen the T cell activation, the in vivo effects of these drugs in humans remain to be clearly defined. In order to better understand biologic effects of therapy, we analyzed blood/tumor tissue from 45 patients undergoing single or combination immune checkpoint blockade. We show that blockade of CTLA4, PD1 or combination of the two leads to distinct genomic and functional signatures in vivo in purified human T cells and monocytes. Therapy-induced changes are more prominent in T cells than in monocytes and involve largely non-overlapping changes in coding genes including alternatively-spliced transcripts, and non-coding RNAs. Pathway analysis revealed that CTLA4-blockade induces a proliferative signature predominantly in a subset of transitional memory T cells, while PD1-blockade instead leads to changes in genes implicated in cytolysis and natural killer cell function. Combination blockade leads to non-overlapping changes in gene expression including proliferation-associated and chemokine genes. These therapies also have differential effects on plasma levels of CXCL10, sIL2R and IL1α. Importantly, PD1 receptor occupancy following anti-PD1 therapy may be incomplete in the tumor T cells even in the setting of complete receptor occupancy in circulating T cells. These data demonstrate that in spite of shared property of checkpoint blockade, antibodies against PD1, CTLA4 alone or in combination have distinct immunologic effects in vivo. Improved understanding of pharmacodynamic effects of these agentsin patients will support rational development of immune-based combinations against cancer.
Introduction
Antigen-specific T cell activation is regulated by a balance of co-stimulatory and co-inhibitory signals, such as those mediated by inhibitory receptors CTLA4 and PD1(1). Antibodies that block these inhibitory receptors or their ligands such as PDL1 have led to impressive anti-tumor effects in several cancers(2, 3). CTLA4 blockade with Ipilimumab (Yervoy; Bristol-Myers Squibb, Princeton, NJ) was the first treatment demonstrated to improve survival of patients with stage IV melanoma in randomized trials(4). Clinical trials with anti-PD1 antibody (such as Nivolumab) have demonstrated promising clinical activity in diverse tumor types including melanoma, renal cell carcinoma and lung cancer(5–8). In preclinical models, combined blockade of both PD1 and CTLA4 led to greater anti-tumor effects than either therapy alone, and in a recent clinical trial, the combination of Nivolumab and Ipilimumab led to a distinct pattern of anti-tumor activity, with rapid and deep tumor regressions in a substantial proportion of melanoma patients(9, 10).
Clinical studies of PD1 and CTLA4 blockade in cancer patients have shown that the two therapies have clear differences in the frequency and pattern of immune-related adverse events (3, 11). While the preclinical models of PD1 or CTLA4 blockade have to date been poorly predictive of the pattern of immune-related adverse events observed in the clinic, genetic deletion of PD1 or CTLA4 leads to very different effects in mice. CTLA4 knockout mice suffer from a lethal lymphoproliferative disease, while deficiency of PD1 leads to less severe phenotype with strain-specific autoimmunity(12–16). Although both PD1 and CTLA4 act to dampen T cell activation via shared signaling pathways, differences in sites of action have been proposed to help understand the differences in patterns of autoimmunity as well as anti-tumor effects with PD1 and CTLA4 blockade. For example, the effects of CTLA4 may be mostly in lymphoid tissue, while PD1 interactions may primarily occur in the periphery. Inhibitory signaling via both PD1 and CTLA4 in human T cells in culture was shown to converge on certain nodes such as inhibition of Akt phosphorylation, although the proximate events may differ, such as the involvement of phosphatase PP2A with CTLA4 but not PD1(17–19). Improved understanding of the changes in gene expression in vivo in humans using genome wide approaches in specific immune cells in response to checkpoint blockade therapy may provide new insights into the mechanisms of anti-tumor and autoimmune effects with these agents. In particular, it is important to understand whether combination checkpoint blockade in vivo leads to distinct or synergistic biologic effects compared to blockade of individual checkpoints in humans.
Materials and Methods
Patients
Peripheral blood and tumor tissue was obtained from patients (n=45)undergoing immune checkpoint blockade after obtaining informed consent under a separate protocol for the collection of research samples approved by the Yale University IRB. This included patients receiving anti-PD1 (n=24), anti-CTLA4 (n=9) or combination therapy (n=12; 9 concurrent, 3 sequential).
Cell separation for gene expression analysis
Peripheral blood mononuclear cells (PBMCs) were obtained by density gradient centrifugation process using FicollPaque™ Plus (GE Health care Life Sciences, UK). Monocytes were sorted from PBMCs using anti-human CD14 microbeads (MiltenyiBiotec, Germany) using the manufacturers protocol(20). Sample separation was performed using MACS-LS columns (Miltenyi). The CD14-fraction was further subjected to a second round of separation for T cells using the Human Pan T Cell Isolation Kit I (MiltenyiBiotec). Purity of sorted populations was monitored by flow cytometry (Supplementary Figure1A). Monocytes and T cells obtained by MACS bead separation were pelleted suspended in RLT buffer (Qiagen) and stored at −80 for RNA isolation.
Gene expression analysis of purified monocytes and T cells
RNA was extracted from purified monocytes and T cells using the RNeasy® Mini kit from Qiagen. We employed Affymetrix GeneChip® Human Transcriptome 2.0 (HTA2) microarrays for gene expression profiling to allow analysis of coding as well as non-coding and alternatively spliced transcripts. Paired pre and post-therapy samples from each patient for each cell type (monocyte or T cells) were compared directly to evaluate therapy-induced changes. We used Genespring GX 12.5 platform to analyze the changes in coding genes, exon workflow of the Partek GS 6.6 to analyze the alternatively spliced exons 2.0 genechip, as described by Whistler et al(21, 22)and Partek GS 6.6 platform to analyze changes in the non-coding genes.
Analysis of coding genes
Data on coding genes was analyzed using Genespring GX 12.5 platform(20). Data was imported via exon expression workflow employing RMA16 or PLIER16 (normalization) for analyzing the coding genes using the specific annotation support file for human transcriptome array 2.0 (as provided by Genespring GX). Experiment grouping for each treatment cohort (anti-PD1 alone, anti-CTLA4 alone, concurrent anti-PD1 + anti-CTLA4; combination and PD1 following CTLA4; Sequential) was created for both T cells and monocytes, and interpretation for the post treatment compared to pre-treatment samples was generated. QC analysis was using PCA analysis and for probe hybridization intensities. For the identification of the differentially regulated coding genes between post vs pre-treatment samples for all treatment groups under each cell type, the locus filter was set on coding only genes (HTA gene chip 2.0 covers 44,699 genes/transcript clusters). We applied a T test with a p value cut-off set at 0.05 followed by the unsupervised clustering analysis using Genespring clustering workflow (Euclidean distance metric and Ward’s linkage rule). Further statistical analysis involved application of a fold-change threshold of 1.3; differentially regulated genes were then manually curated to include coding genes only.
Pathway analysis
Pathway analysis of differentially expressed genes was performed using the Metacore pathway analysis platform(20). Differentially regulated gene lists with p<0.05 and fold change of +/−1.3 were used from each treatment group as input for the Metacore pathway platform and differentially regulated pathways maps and GO terms were generated.
Analysis of alternatively spliced genes
Analysis to detect the potentially alternatively spliced genes was performed using the Exon workflow of the Partek GS 6.6 by taking into account exon probes from Affymetrix HTA 2.0 genechip, as described by Whistler et al(21, 22). Exons with a p value <0.05 were included for analysis. We obtained number of exon probe-sets in each transcript cluster (with corresponding transcript_cluster_id derived from the meta-probeset file) and discarded clusters with greater than 100 probe-sets or less than 10 probe-sets and included exon clusters with alt-splice p value<0.00001. The list of alternatively spliced genes was filtered on gene clusters with p values <0.05 and a fold change of +/− 1.3 between pre therapy and post therapy samples to obtain splice variants that were also differentially regulated at the gene level.
Analysis of non-coding genes
For analyzing the non-coding data we used the Partek GS 6.6, post importing data and we employed the filter on non-coding genomic loci (22,829) to retain only the non-coding probe sets on the HTA 2.0 genechip from all samples. Statistical analysis involved one-way ANOVA, followed by application of a fold change threshold of 1.3 to generate non-coding transcript lists (unadjusted p value, 0.05). All the non-coding probe-sets/transcripts lists were further curated manually (via UCSC, Ensembl). Venn diagrams were generated between different treatment groups under each cell type.
Q-PCR
RNA from T cells isolated from patients pre- and post-therapy was used to validate the microarray data for the expression of Ki-67 and ICOS by Q-PCR using the assays on demand primer probes (Applied Biosciences). Expression of GAPDH was monitored as a housekeeping gene. Reactions were set up in triplicates using EZ PCR Core reagents (Applied Biosciences) according to manufacturer’s protocol. Relative expression of target genes was calculated using comparative threshold cycle method.
Immunoassay for detecting plasma levels of various cytokines
Plasma collected from patient samples before and after therapy (ipilimumab alone, n=5; nivolumab alone, n=20; concurrent ipilimumab+nivolumab (Combo), n=6; and sequential nivolumab in patients with prior ipilimumab (Seq), n=3) and stored at −20°C was thawed and used for the assay. Samples were used undiluted and in duplicate. Milliplex® MAP Human Cytokine/Chemokine Magnetic Bead Panel kit (MPXHCYTO-60K-PMX39; EMD Millipore, USA) for 96 well plate assay was used for the simultaneous quantification of 39 human cytokines and chemokines (EGF, Eotaxin, FGF-2, Flt-3 Ligand, Fractalkine, G-CSF, GM-CSF, GRO, IFNα2, IFNγ, IL-1α, IL-1β, IL-1rα, IL-2, IL-3, IL-4, IL-5, IL-6, IL-7, IL-8, IL-9, IL-10, IL-12 (p40), IL-12 (p70), IL-13, IL-15, IL-17A, IP-10, MCP-1, MCP-3, MDC, MIP-1α, MIP-1β, sCD40L, sIl-2Rα, TGFα, TNFα, TNFβ, VEGF) using the protocol provided by the manufacturer. xPONENT® software (Luminex Corp., USA) was used to detect, quantitate and analyze the samples on the Luminex 100™ instrument.
Detection of cytokines secreted by tumor infiltrating T cells
Tumors were processed into single cell suspension by manual dissociation and either left untreated or treated with anti-CD3/CD28 beads for 48 hrs in 96 well round bottom plates in 5%PHS. Cell supernatant was collected at 48 hrs and analyzed for cytokines and chemokines using the Luminex assay as described above.
Immunophenotyping of PBMCs
Cryopreserved patient pre and post samples PBMCs were thawed togetherand stained with dead cell exclusion dye and Fluorochrome conjugated anti-human antibodies CD3, CD4 and CD8 (all from BD Pharmingen) and CD56 (BioLegend Inc.), CD25 (clone 4E3, MiltenyiBiotec GmbH), CD45RO (BD Horizon) as well as PD-1 (clone J105; eBioscience Inc). For some samples, cells were fixed and permeabilized. After permeabilization of cells, fluorochrome conjugated antibodies against human Granzyme B, (BD Biosciences) and Ki-67(eBiosciences), were used to stain and detect the respective intracellular molecules. For detection of cytokine production, cells were rested overnight after thawing and then stimulated with PMA (Phorbol 12-myristate 13-acetate) and Ionomycin, both at 500ng/ml in the presence of protein transport inhibitor BD Golgi Stop™ (0.7ul/ml). After 5 hours of stimulation, the cells were stained with the dead cell exclusion dye, fixed, permeabilized and stained with fluorochrome-conjugated antibodies against human CD3, CD4, CD8, IFN-γ (all from BD Biosciences).. All live cell stains were acquired on BD-LSR Fortessa™ and the data was analyzed using Flowjo v9.7.5 software (Tree Star Inc). Intracellular staining samples were acquired on BD™LSR II and the data analysis was done using Flowjo software.
Immunophenotyping by Mass Cytometry
Cryopreserved patient PBMCs were thawed in warm media containing Benzonase® Nuclease (25U/ml; Sigma) and washed twice. Pre and post therapy samples were thawed and stained at the same time. Cells were suspended at up to 10million/ml in 1X PBS for viability staining by Cell-ID™ Cisplatin (final concentration of 5µM, Fluidigm Sciences). Cells were mixed well and incubated for 5 minutes at room temperature. The staining was quenched with MaxPar® Cell staining buffer and washed twice before proceeding to the usual procedure of surface and intracellular staining as per manufacturers protocol. MaxPar® Human T Cell Phenotyping Panel Kit (CD11a-142Nd, CD4-145Nd, CD8a-146Nd, CD16-148Nd, CD25-149Sm, CD45-154Sm, CCR7-159Tb, CD69-162Dy, CD45RO-165Ho, CD44-166Er, CD27-167Er, CD45RA-169Tm, CD3-170Er, CD57-172Yb, HLA-DR-174Yb and CD127-176Yb) was used along with CD28-160Gd (CD28.2), CD117-143Nd (104D2), CD95-164Dy (DX2) for surface staining. 3million PBMCs were incubated in a volume of 100µl of cell staining buffer with antibodies in a polystyrene tube for 30 minutes at room temperature. After staining, cells were washed twice with buffer before fixing with BD Cytofix™ fixation buffer (100µl/million cells) and permeabilizing with BD Perm/Wash™ buffer. Fixed and permeabilized cells were stained with anti-human Ki-67-151Eu (B56; BD Pharmingen antibody conjugated with lanthanide MaxPar® Europium Chloride 151Eu using the MaxPar® X8 Antibody Labeling kit) for 30 minutes at room temperature. Cells were washed twice with buffer and suspended in 1ml intercalation solution containing MaxPar Intercalator-Ir in MaxPar Fix and Perm buffer at final conc. of 125nM. Cells were left overnight in the intercalator solution, washed with staining buffer and finally suspended in 500 µl of MaxPar Water before acquiring on CyTOF® 2 instrument (DVS, Fluidigm Sciences Inc.). All data were analyzed and graphs generated using the DVS Cytobank software (Cytobank Inc.).
Results
In order to better understand the effects of combination versus individual PD1/CTLA4 checkpoint blockade on human T cells and monocytes in vivo using a genome-wide approach, we initially analyzed gene expression profiles of purified CD3+ T cells and CD14+ monocytes in 20 patients before and 3 weeks after receiving checkpoint blockade therapy with ipilimumab (anti-CTLA4;n=5), nivolumab (anti-PD1; n=6) concurrent ipilimumab and nivolumab (Combo; n=6) or sequential nivolumab in patients with prior ipilimumab (Seq; n=3) using an exon expression array (Human HT2.0, Affymetrix). Therapy-induced changes in gene expression were analyzed utilizing baseline sample from each patient as its own control. Unsupervised cluster analysis revealed that the samples largely clustered according to pre or post therapy status indicating that the impact of therapy was more dominant than the baseline variability between samples (Supplementary Figure 1B). The use of exon expression array allowed us to examine changes in the expression of coding genes and splice variants as well as non-coding genes.
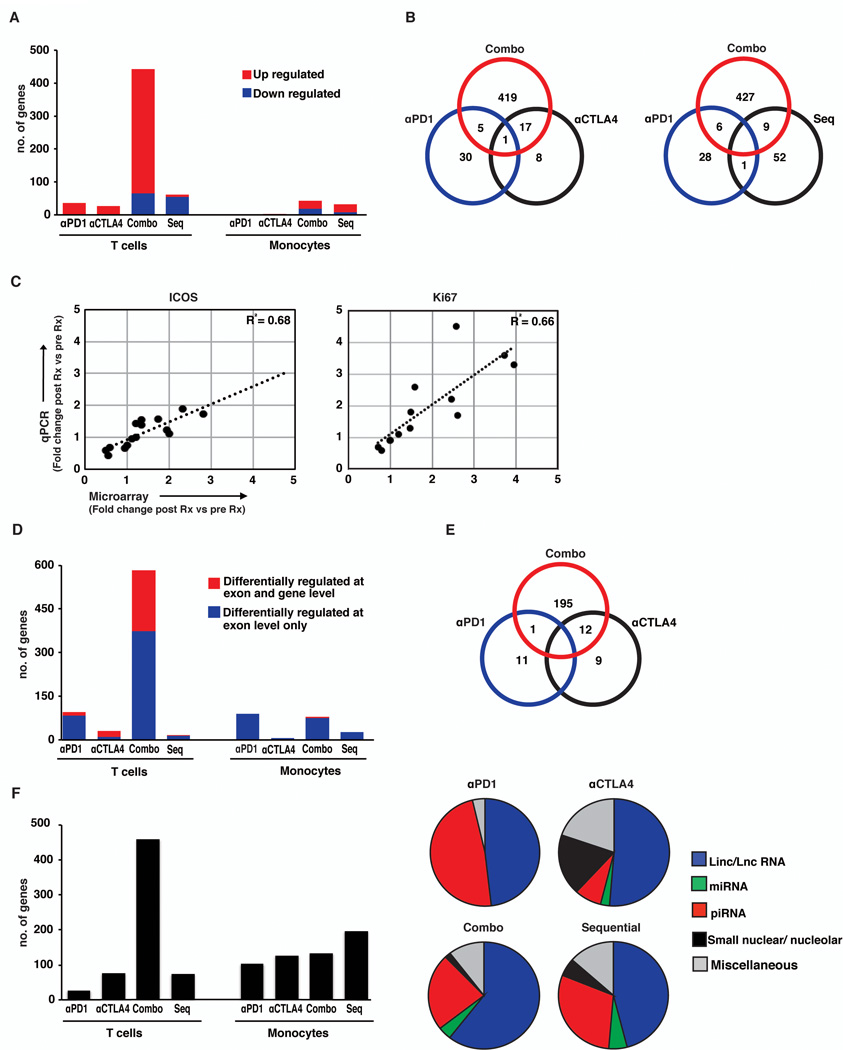
We first focused on therapy-induced changes in coding genes. Concurrent combination therapy with both anti-CTLA4 and anti-PD1 led to greater number of differentially expressed T cell genes compared to either agent alone or both agents given sequentially (Combination=442 genes; anti-CTLA4=26 genes; anti-PD1=36 genes; sequential anti-PD1 in patients with prior anti-CTLA4=62 genes) (Figure 1A). Therapy-induced changes in gene expression included genes unique to each treatment cohort, including combination therapy, with only minor overlap (Figure 1B). The only up regulated gene shared between all 3 cohorts is interferon-γ. Interestingly, genomic changes following anti-PD1 therapy showed little overlap with therapy-induced changes when anti-PD1 therapy was applied as a sequential strategy in patients with prior anti-CTLA4 therapy (Figure 1B). Some of the notable genes induced in vivo following checkpoint blockade were Ki-67 (anti-CTLA4 and combination), granzyme A/B (anti-PD1 and combination), FCRL3 and KLRF1(anti-PD1), CTLA4 and ICOS (anti-CTLA4), IL8 and HLADR (Combination) and IFNγ (all 3 cohorts) (Table 1). Data for changes in selected genes (ICOS and Ki67) was validated by Q-PCR (Figure 1C). In contrast to T cells, relatively few genes were differentially up regulated in monocytes following checkpoint blockade. This included increase in TNF following anti-CTLA4 therapy.
Figure 1.
RNA extracted from freshly isolated monocytes and T cells from peripheral blood of patients treated with either anti-PD1 (n=6), anti-CTLA4 (n=5), combination therapy with anti-PD1 and anti-CTLA4 concurrently (Combo, n=6), and sequential anti-PD1 in patients with prior anti-CTLA4 (Seq, n=3) was analyzed using the Affymetrix HTA2.0 exon array.
(A)The expression data for coding genes was analyzed using genespring 12.5platform.Coding genes were identified using the locus type filter on coding followed by manual curation of the list obtained. Differentially regulated genes were obtained by using p<0.05 and FC+/− 1.3 fold in post therapy samples compared to pre therapy samples. Changes in expression of coding genes in peripheral blood T cells and monocytes of patients treated with anti-PD1, anti-CTLA4, Combination (Combo) or Sequential (Seq) therapy. Figure shows genes differently regulated (p<0.05, fold change ≥ ±1.3) in samples obtained 3 weeks post therapy compared to baseline prior to starting therapy. The genes that were upregulated are in red and those that were downregulated are in blue.
(B). Venn diagrams showing differentially regulated T cell coding genes that were shared between patients treated with anti-PD1 (αPD1), anti-CTLA4 (αCTLA4), Combination (Combo) and Sequential therapy (Seq). The figure shows that majority of the genes were unique to each specific treatment group.
(C). Q-PCR was performed to confirm expression of ICOS and Ki67 as determined by microarray (n=15). Figure shows correlation between levels obtained by Q-PCR versus expression levels as determined by gene array on the same patients.
(D) Expression data was analyzed to detect alternatively spliced genes using exon flow workflow of Partek GS 6.6. Exons with probe sets between 10 and 100, a p value of <0.05 at exon level and <0.00001 at the exon cluster level were included in the analysis. Figure shows alternatively spliced genes in peripheral T cells and monocytes of patients treated with anti-PD1, anti-CTLA4, Combination and Sequential therapy. Alternatively spliced genes differentially regulated at the exon level only are in blue, and genes differentially regulated at both the exon and gene level are represented in red.
(E) Venn diagram showing differentially regulated alternatively spliced genes in T cells shared between patients treated with anti-PD1, anti-CTLA4 and Combination therapy.
(F) Expression data was analyzed for changes in non-coding genes using Partek GS 6.6. The bar graph shows non-coding genes that are differentially regulated (p<0.05 and fold change of +/− 1.3) in peripheral blood T cells and monocytes of patients receiving therapy with checkpoint blockade inhibitors. The pie charts show the type of non- coding genes (ielinc/LncRNA, miRNA, piRNAetc) that are differentially regulated in the T cells of the patients. Miscellaneous group includes anti-sense, transfer, ribosomal and Y-RNA.
Table 1. Selected list of coding genes differentially regulated in T cells of patients treated with anti-CTLA4, anti-PD1, combination therapy or sequential therapy.
Selected list of coding genes from the top 30 differentially regulated genes in T cells of patients treated with anti-CTLA4, anti-PD1, combination therapy (concurrent anti-CTLA4 and anti-PD1) or sequential therapy (anti-PD1 following anti-CTLA4 therapy).
| TRANSCRIPT ID |
P VALUE | FOLD CHANGE |
GENESYMBOL | GENEDESCRIPTION |
|---|---|---|---|---|
| Combo Therapy (Anti-CTLA4+ Anti-PD1) | ||||
| 18876628 | 0.01 | 3.17 | MKI67 | antigen identified by monoclonal antibody Ki-67 |
| 18906869 | 0.01 | 2.84 | IFNG | interferon gamma |
| 19224629 | 0.02 | 2.77 | HIST1H3B | histone cluster 1, H3b |
| 18764511 | 0.05 | 2.77 | IL8 | interleukin 8 |
| 18775469 | 0.03 | 2.65 | FGFBP2 | fibroblast growth factor binding protein 2 |
| 18772965 | 0.02 | 2.64 | CCNA2 | cyclin A2 |
| 18949520 | 0.02 | 2.45 | SLC7A5 | solute carrier family 7,member 5 |
| 18966489 | 0.001 | 2.39 | KPNA2 | karyopherin alpha 2 |
| 18923230 | 0.04 | 2.29 | GZMB | granzyme B |
| 18961872 | 0.02 | 2.13 | TOP2A | topoisomerase (DNA)II alpha 170kDa |
| Anti-CTLA4 Therapy | ||||
| 18689122 | 0.021 | 2.33 | FAM46C | family with sequence similarity 46, member C |
| 18876628 | 2.5E-04 | 2.18 | MKI67 | antigen identified by monoclonal antibody Ki-67 |
| 18961872 | 0.002 | 1.59 | TOP2A | topoisomerase(DNA)II alpha 170kDa |
| 18724103 | 0.035 | 1.58 | CTLA4 | cytotoxic T-lymphocyte-associated protein 4 |
| 18797874 | 0.012 | 1.55 | STX11 | syntaxin 11 |
| 19224662 | 0.003 | 1.54 | HIST1H3F | histone cluster1, H3f |
| 18772965 | 0.007 | 1.49 | CCNA2 | cyclin A2 |
| 18906869 | 0.020 | 1.42 | IFNG | interferon gamma |
| 18988349 | 0.001 | 1.42 | TPX2 | TPX2 (microtubule associated) |
| 18724111 | 0.06 | 1.57 | ICOS* | InducibleT-cell co-stimulator |
| Anti-PD1 Therapy | ||||
| 18895449 | 0.028 | 2.67 | KLRF1 | killer cell lectin-like receptor subfamily F, member 1 |
| 18707750 | 0.048 | 2.51 | SH2D1B | SH2 domain containing 1B |
| 18971449 | 0.027 | 2.23 | KIR2DL2 | killer cell immunoglobulin-like receptor, two domains, long cytoplasmic tail, 2 |
| 18777270 | 0.021 | 2.07 | GZMA | granzyme A |
| 18988173 | 0.013 | 1.97 | CST7 | cystatin F (leukocystatin) |
| 18718479 | 0.019 | 1.93 | GNLY | granulysin |
| 18707252 | 0.004 | 1.77 | FCRL3 | Fc receptor-like 3 |
| 19011576 | 0.025 | 1.60 | HLA-DMA | major histocompatibility complex, class II, DM alpha |
| 18978382 | 0.039 | 1.53 | NCR1 | natural cytotoxicity triggering receptor 1 |
| 18906869 | 0.030 | 1.52 | IFNG | interferon gamma |
| Sequential Therapy | ||||
| 18896172 | 0.043 | 2.01 | FAR2 | fatty acyl CoA reductase 2 |
| 18698492 | 0.010 | 1.46 | PADI2 | peptidyl arginine deiminase, type II |
| 19576522 | 0.008 | 1.43 | ZFP57 | zinc finger protein 57 homolog (mouse) |
| 18851261 | 0.000 | 1.35 | IL3RA | interleukin 3 receptor, alpha (low affinity) |
| 18892472 | 0.030 | 1.35 | CD3D | CD3D molecule, delta(CD3-TCR complex) |
| 18968132 | 0.029 | 1.35 | HRH4 | histamine receptor H4 |
| 18882589 | 0.007 | 1.34 | FOLR3 | folate receptor 3 (gamma) |
| 18728884 | 0.031 | −1.30 | PRKD3 | protein kinase D3 |
| 18872598 | 0.021 | −1.30 | TIMM23 | translocase of inner mitochondrial membrane 23 homolog (yeast) |
| 18891105 | 0.039 | −1.30 | RAB30 | |
Similar patterns emerged when exon level data were analyzed for evaluation of alternate splicing (Figure 1D–E). The differentially spliced genes included both differentially regulated genes as well as genes that were not differentially regulated with therapy. The proportion of genes with altered exon usage that were also differentially regulated at the gene level was 34% with combination therapy, 67% with anti-CTLA4, 13% with anti-PD1 therapy and 7% with sequential therapy. The sites of altered exon usage were similar for genes shared between CTLA4 and combination treated patients (data not shown). Similar to the pattern with the coding genes, combination therapy led to greatest changes in non-coding genes in T cells. Majority of the changes in non-coding genes in T cells were observed in long non-coding RNAs and piwi RNAs (Figure 1F) and there was little overlap between differentially regulated non-coding genes between the three treatment groups (data not shown). Together these data demonstrate that each of the checkpoint blockade strategies leads to distinct and largely non-overlapping effects on the transcriptome of human T cells in vivo.
Pathway analysis of differentially expressed coding transcripts revealed that the dominant pathway in the setting of CTLA4 blockade and combination therapy was cell cycle / proliferation (Table 2). In contrast to CTLA4/combination blockade, PD1-regulated genes did not include a proliferation signature and were instead enriched for genes implicated in cytolytic function and regulation of effector T and natural killer cell function.
Table 2. Metacore pathway analysis on differentially regulated T cell genes (p<0.05, FC±1.3).
Metacore pathway analysis was used to analyze coding genes that were differentially regulated (p<0.05, FC±1.3) between pre and post therapy T cells obtained from patients treated with either anti-PD1 alone, anti-CTLA4 alone as well as anti-CTLA4 and anti-PD1 in combination either concurrently (combination therapy) or sequentially (Sequential therapy). Table shows pathways that were differentially regulated in each group.
| Pathway | P value (FDR) |
|
|---|---|---|
| Anti PD1 therapy | ||
| 1 | Immune response: Role of DAP12 receptors in NK cells | 2.2E-13 |
| 2 | Immune response: T regulatory cell-mediated modulation of effector T cell and NK cell functions | 8.5E-03 |
| 3 | Immune response: IL-27 signaling pathway | 1.9E-02 |
| 4 | Immune response: IL-12-induced IFN-gamma production | 4.2E-02 |
| 5 | Immune response: Differentiation and clonal expansion of CD8+ T cells | 4.2E-02 |
| Anti CTLA4 therapy | ||
| 1 | Cell cycle: Transition and termination of DNA replication | 5.5E-03 |
| 2 | Cell cycle: Spindle assembly and chromosome separation | 5.5E-03 |
| 3 | Cell cycle: Nucleocytoplasmic transport of CDK/Cyclins | 2.4E-02 |
| 4 | Cell cycle: Chromosome condensation in prometaphase | 3.0E-02 |
| Combination Therapy (PD1+ CTLA4) | ||
| 1 | Cell cycle: Chromosome condensation in prometaphase | 6.4E-04 |
| 2 | DNA damage: ATM/ATR regulation of G1/S checkpoint | 1.4E-06 |
| 3 | DNA damage: ATM / ATR regulation of G2 / M checkpoint | 1.4E-06 |
| 4 | Cell cycle: Transition and termination of DNA replication | 1.4E-06 |
| 10 | Immune response: Differentiation and clonal expansion of CD8+ T cells | 0.059 |
| Sequential Therapy | ||
| 1 | Immune response: T regulatory cell-mediated modulation of effector T cell and NK cell functions | 3.7E-02 |
| 2 | Apoptosis and survival: Endoplasmic reticulum stress response pathway | 4.3E-02 |
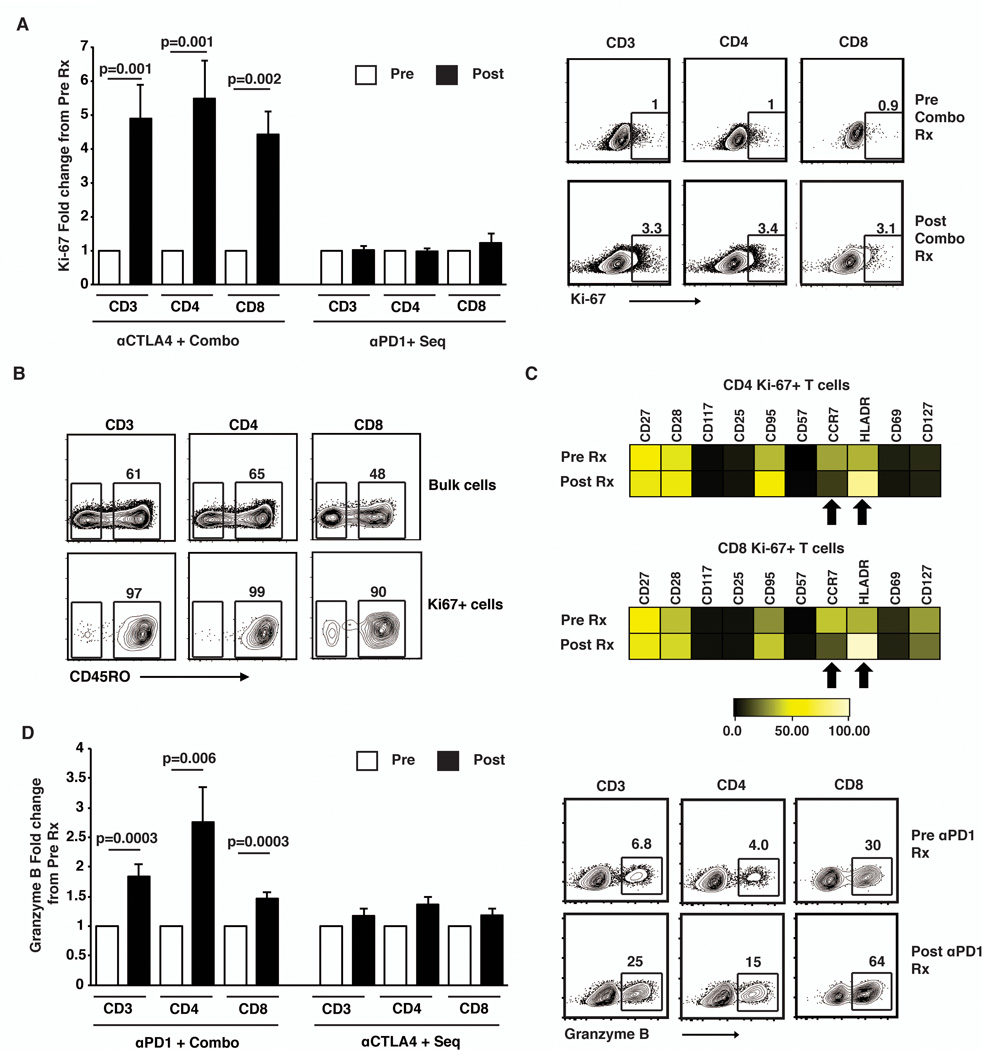
Next, we analyzed the expression of Ki-67 and granzyme in T cells by flow cytometry, as these represented two distinct aspects of T cell function (proliferation and cytolytic function) that appear to be the major functional pathways differentially activated by CTLA4 and PD1 blockade respectively in vivo in human T cells. Flow cytometry of paired pre and post therapy samples (n=34 patients) confirmed the microarray data for the induction of Ki-67 following combination and CTLA4 blockade. Ki67 was induced after therapy with anti-CTLA4 or combination in both CD4 and CD8 T cells (Fig 2A) and the Ki67+ cells had a CD45RO+ memory phenotype (Fig 2B). In order to further dissect the phenotype of Ki-67+ cells following checkpoint blockade, these cells were analyzed by single cell mass cytometry. The data revealed that the Ki-67+ cells increasing after combination checkpoint blockade have a phenotype of CD45RO+, CCR7-CD27+CD28+CD95+, consistent with transitional memory T cells (Fig 2C)(23). The proliferating cells up regulated HLADR. Anti-PD1 as well as combination therapy led to an increase in Granzyme B, while this was not observed in the cohort that received anti-CTLA4 or sequential therapy (Fig 2D). Together these data demonstrate that each of the therapeutic approaches at immune checkpoint blockade leads to a distinct, but largely non-overlapping signature of changes in gene expression in T cells in vivo, which represent distinct aspects of T cell function and can be readily detected in freshly isolated circulating immune cells.
Figure 2.
Changes in T cell proliferation and cytolytic function following therapy with checkpoint blockade inhibitors.
Freshly collected frozen samples were used to for these assays. All pre and post therapy samples from the same patient were thawed at the same time, stained together using the same antibody cocktail and analyzed at the same time.
(A) Bar graph shows expression of Ki67 (mean and standard error of mean) in peripheral blood (CD3, CD4 and CD8 T cells) of patients (n=34) before (Pre-white bars) and after (Post-Black bars) therapy with either anti-CTLA4 and Combo therapy (n=15) or anti-PD1 and Sequential therapy (n=19). Flow cytometry plots on the right show a representative patient with increase in Ki67+ cells after combination therapy.
(B) Expression of T cell memory marker (CD45RO) in bulk T cells as well as Ki67 positive T cells after therapy in the same patient as in figure 2A.
(C) Single cell mass cytometry (CyTOF) analysis for expression of surface markers on CD3+CD4+ Ki67+ as well as CD3+CD8+ Ki67+ cells before (Pre Rx) and 3 weeks after (Post Rx) starting combination therapy with anti-CTLA4 and anti-PD1. The figure shows median MFI. Plot is representative of 3 similar patients (anti-CTLA4=2 and combination treated patients n=1).
(D) Expression of Granzyme B (mean and standard error of mean) in CD3 and CD8 T cells in peripheral blood of patients (n=34) before(pre) and after(post) therapy with anti-PD1 and Combo therapy (n=23) and those treated with anti-CTLA4 and Sequential therapy(n=11). The flow cytometry plot shows a representative patient.
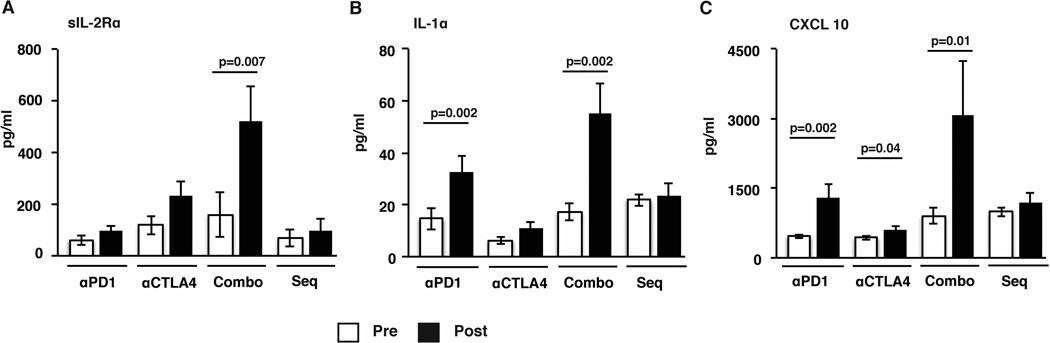
As checkpoint blockade therapy led to changes that were detected in circulating immune cells, we hypothesized that these therapies would also lead to detectable changes in systemic levels of cytokines. Accordingly, we evaluated changes in the plasma levels of a panel of 39 cytokines/chemokines/growth factors before/after therapy in these patients using Luminex analysis. Of the analytes tested, sIL2R was increased following combination therapy, while the levels of IL1α were increased following anti-PD1 and combination blockade and CXCL10 levels were increased following anti-PD1, anti-CTLA4 and combination blockade (Fig 3A–C). Together these data demonstrate that each form of immune checkpoint blockade is associated with a distinct pattern of systemic changes in cytokines.
Figure 3.
Changes in plasma chemokine and cytokines of patients treated with checkpoint blockade inhibitors.
Plasma collected before and after therapy with either anti-PD1, anti-CTLA4, combination therapy (Combo) as well as sequential therapy (Seq) was analyzed for presence of cytokines and chemokines using 39-plex luminex assay. All samples were tested in duplicate. Figure shows data for levels of cytokines and chemokines (mean and standard error of mean) that were differentially secreted. (A) sIL2Rα levels (B) IL1α levels and (C) CXCL10/IP10 levels in plasma of patients pre and post therapy.
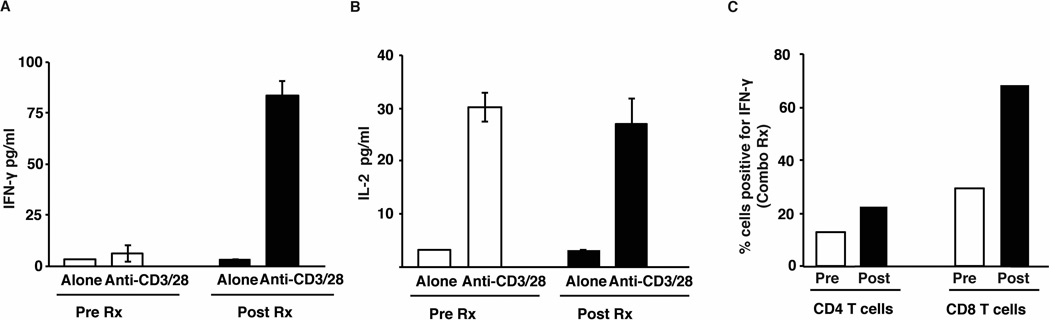
In contrast to single agent CTLA4 or PD1 blockade, less is known about the effects of combination checkpoint blockade on T cells infiltrating human tumors. An impressive aspect of such therapy is the rapidity and depth of clinical response. Therefore in order to characterize early changes in tumor- infiltrating immune T cell function following such therapy, we analyzed serial biopsies before and after initiation of therapy in a patient treated with combination checkpoint blockade. When compared to baseline, freshly isolated tumor-infiltrating T cells from just 3 weeks post therapy demonstrated increased production of interferon-gamma, indicating rapid induction of effector T cell function in the tumor bed (Fig 4A–B). Analysis of circulating T cells obtained at the same time as the tumor biopsies also showed an increase in IFNγ+ T cells following combination therapy indicating that these early changes are also detectable in the peripheral blood (Fig 4C).
Figure 4.
Early effects of combination blockade on cytokine secretion by tumor infiltrating lymphocytes
Tumor biopsy and peripheral blood were obtained from a patient before and 3 weeks after starting combination therapy with ipilimumab and nivolumab. Tumor infiltrating lymphocytes were either cultured alone (Alone) or with anti-CD3/CD28 beads (Anti-CD3/28). Culture supernatant obtained at 48 hours was subjected to luminex assay. Peripheral blood mononuclear cells obtained at the same time were stimulated with PMA and ionomycin and intracellular flow cytometry was performed for the detection of IFNγ.
(A) Secretion of IFNγ by tumor infiltrating lymphocytes before and after therapy.
(B) Secretion of IL2 by the tumor infiltrating lymphocytes.
(C) Percent of IFNγ positive CD4 and CD8 T cells in the peripheral blood obtained pre and post therapy from the same patient.
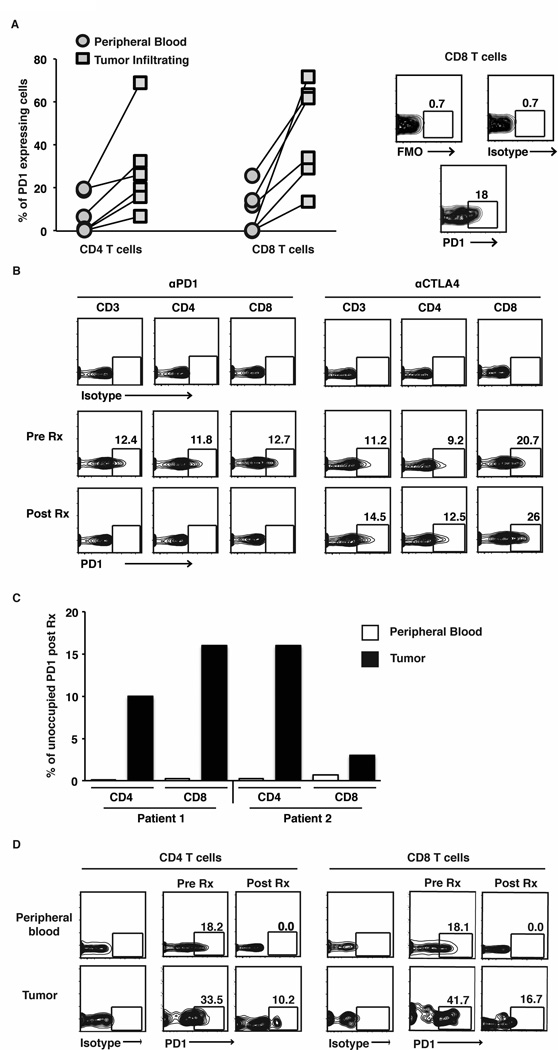
Current dosing strategies for Nivolumab in the clinic have been guided by prior studies demonstrating that doses of Nivolumab as low as 1 mg/kg lead to complete saturation of PD1 receptor on circulating T cells(8). As in prior studies, tumor infiltrating T cells expressed higher levels of PD1 on their surface compared to peripheral blood T cells (Fig 5A). Interestingly, surface staining for PD1(clone J105, e- Biosciences) was abrogated in circulating T cells from patients treated with anti-PD1 antibody, but in not those treated with anti-CTLA4 antibody, consistent with treatment specific staining interference likely due to receptor occupancy in vivo following anti-PD1 therapy (8) (Fig 5B). However at least in two patients with available biopsies before and after therapywith anti-PD1 (either alone or in combination with CTLA4), the blockade of PD1 on tumor infiltrating lymphocytes was incomplete, even in the setting of complete blockade of PD1 on both CD4 and CD8 T cells in the circulation (Fig 5C–D).
Figure 5.
PD1 expression and occupancy in patients treated with checkpoint blockade inhibitors.
PD1 surface expression was determined on tumor infiltrating lymphocytes and blood from patients (n=6 different patients). Cells were obtained from the tumor sample by manual dissociation. PBMCs obtained from the blood and the tumor tissue were stained and analyzed together.
(A) Graph in the left panel shows expression of PD1 on CD4 and CD8 T cells in the tumor as well as peripheral blood T cells drawn at the same time. Panel on the right is a representative flow cytometry plot showing controls (Fluorensence minus one; FMO and Isotype) as well as PD1 staining on CD8 T cells.
(B) Expression of PD1 on CD3, CD4 and CD8 T cells in peripheral blood of representative patients before and after treatment with anti-PD1 and anti-CTLA4.
(C). Bar graph shows percent PD1 on CD4 and CD8 T cells that was unoccupied after therapy with anti-PD1 compared to that prior to therapy in 2 separate patients. Bar in white shows data from peripheral blood T cells and bar in black shows data from tumor infiltrating T cells.
(D) Expression of PD1 on peripheral blood and tumor infiltrating T cells in a patient before and after combination therapy with anti-CTLA4 and anti-PD1 therapy. Data are representative of similar findings on two patients.
Discussion
These studies provide the first comparison of in vivo changes in purified human immune cells following combination checkpoint blockade versus individual CTLA4 or PD1 checkpoint blockade in human cancer. An important aspect of this study is that the changes in gene expression were analyzed in highly purified immune cells and that both T cells and monocytes were purified from fresh blood mononuclear cells immediately after blood draw, without a potential artifact of in-vitro culture, shipping, or cryopreservation. The data clearly demonstrate that the effects of CTLA4, PD1 or combination blockade can be readily detected in circulating T cells and that each therapy leads to distinct patterns of immune activation in vivo, in spite of known convergence of signaling pathways downstream of the inhibitory receptors (17). Notably, the effects of combination blockade are distinct from that of individual checkpoints, which may help the differences in clinical effects of these therapies.
These findings have several potential implications for understanding the anti-tumor and autoimmune effects observed with these therapies in the clinic and the development of new combinations. While CTLA4 blockade induces a robust proliferation signature in T cells, this was not observed following PD1 blockade indicating that the two modes of “checkpoint blockade” therapies have very different effects on human T cells in vivo. In contrast to CTLA4, PD1 blockade is associated with the induction of several cytolysis and natural killer-associated genes. For example, in addition to the well documented roles of granzyme and granulysin, majority of the top PD1-regulated genes such as cystatin F and CD244 have also been implicated in the regulation of lytic function or secretory granules (24–27). The finding that PD1 blockade particularly leads to the expression of several NK-associated genes on T cells should encourage further exploration of the possible effects of PD1 blockade on NKT cells as well as combinations targeting innate cells with PD1 blockade(28–30). Genomic signature of CTLA4 blockade includes the induction of several cell cycle-associated genes, best exemplified by up-regulation of Ki-67 on a subset of transitional memory T cells, which is consistent with preclinical studies showing CTLA4-mediated dampening of proliferation and enhanced memory following CTLA4 blockade in mice(31–33). It is notable that the CTLA4-regulated genes (such as Ki-67) described here bear a strong similarity to findings in another study analyzing changes in gene expression in purified T cells post-ipilimumab with a different GEP platform, which provides additional validation for our findings(34). To our knowledge, this is the first study to compare GEP of purified immune cells in vivo following combination and PD1 blockade therapies. In contrast to single agent therapies, combination therapy leads to the induction of much larger number of genes, involving greater increase in genes induced by single agents, but also distinct genes observed only with combination blockade. Interestingly, the latter set of genes includes potent chemokines (such as IL8), which may increase immune infiltration and may help explain why the clinical response to the combination therapy was found to be independent of preexisting immune infiltration and PDL1 expression in the tumor(9, 10).
In addition to the coding genes, we also identified differential regulation of several non-coding genes as well as altered exon splicing. Most of these have not yet been studied in the context of human immune system, although the importance of RNA splicing and non-coding RNAs in immune regulation is increasingly appreciated. It is notable that some of the coding as well as non-coding genes (such as linc RNAs) identified here, have prominent species-specific effects, imploring the need to directly study patients treated with these agents. For example, FCRL3 induced following PD1 blockade is a human-specific T/NK-associated gene strongly implicated by genetic studies in human autoimmunity (35, 36). In contrast to some prior studies, purification of immune cells allowed us to specifically study genomic changes in T cells and monocytes(37). While the immune signatures that we observed are quite robust, one of the limitations of this study is the small numbers of patients studied. Further studies with additional patients are needed to better understand the correlation between genomic signatures and other aspects of immune response with clinical outcome following checkpoint blockade (8, 38).
These data may also have possible implications for optimal management of therapy-induced autoimmunity. Currently, immune-related adverse events following checkpoint blockade are clinically managed with steroids, with TNF blockade reserved for refractory cases. The use of TNF blockade in severe colitis associated with CTLA4 blockade is also supported by our finding of TNF as the top differentially regulated gene in myeloid cells of anti-CTLA4 treated patients(39). However, differences in genomic and cytokine profiles following CTLA4 and PD1 blockade as shown here raise the possibility that optimal management of autoimmune events in the two settings may differ. For example the finding that PD1(but not CTLA4) blockade leads to an increase in plasma IL1α suggests that some cases of PD1-induced auto-inflammation may benefit from consideration of such as IL1 blockade already in the clinic, but yet unexplored in this setting(40).
Recent clinical success of PD1/CTLA4 combination therapy has led to much excitement in cancer immunotherapy (10). When designing this combination, the dose of anti-PD1 was not escalated beyond 3 mg/kg, due to dose-limiting toxicity and prior data about saturation of the receptor in circulating T cells at much lower doses(8, 10). Our data demonstrate that such a strategy might need to be reevaluated and monitoring changes in tumor infiltrating lymphocytes including receptor occupancy in individual patients may allow for optimal dosing of anti-PD1 antibody as a part of the combination. Optimal development of checkpoint blockade combinations in the clinic will require careful evaluation of pharmacodynamics effects in the tumor bed in patients undergoing these combination therapies.
Supplementary Material
Acknowledgments
Authors acknowledge Matthew Burke and Antonella Bacchiocchi for help with patient consents and sample collection, Anumeha Shah and Lin Zhang for technical support and Xiting Yan for help with pathway analysis.
1Funding: This work is supported in part by funds from the National Institutes of Health – RO1-AI0792222 (KMD), CA106802 (MVD), CA135110 (MVD), K24CA172123 (HK), P50-CA121974 (RH, KMD), Dana Foundation and Hyundai Hope on Wheels (KMD).
References
- 1.Krummel MF, Allison JP. CD28 and CTLA-4 have opposing effects on the response of T cells to stimulation. J.Exp.Med. 1995;182:459–465. doi: 10.1084/jem.182.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer. 2012;12:252–264. doi: 10.1038/nrc3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sznol M, Chen L. Antagonist Antibodies to PD-1 and B7-H1 (PD-L1) in the Treatment of Advanced Human Cancer. Clin Cancer Res. 2013;19:1021–1034. doi: 10.1158/1078-0432.CCR-12-2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hodi FS, O'Day SJ, McDermott DF, Weber RW, Sosman JA, Haanen JB, Gonzalez R, Robert C, Schadendorf D, Hassel JC, Akerley W, van den Eertwegh AJ, Lutzky J, Lorigan P, Vaubel JM, Linette GP, Hogg D, Ottensmeier CH, Lebbe C, Peschel C, Quirt I, Clark JI, Wolchok JD, Weber JS, Tian J, Yellin MJ, Nichol GM, Hoos A, Urba WJ. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med. 2010;363:711–723. doi: 10.1056/NEJMoa1003466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hamid O, Robert C, Daud A, Hodi FS, Hwu WJ, Kefford R, Wolchok JD, Hersey P, Joseph RW, Weber JS, Dronca R, Gangadhar TC, Patnaik A, Zarour H, Joshua AM, Gergich K, Elassaiss-Schaap J, Algazi A, Mateus C, Boasberg P, Tumeh PC, Chmielowski B, Ebbinghaus SW, Li XN, Kang SP, Ribas A. Safety and tumor responses with lambrolizumab (anti-PD-1) in melanoma. N Engl J Med. 2013;369:134–144. doi: 10.1056/NEJMoa1305133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Topalian SL, Sznol M, McDermott DF, Kluger HM, Carvajal RD, Sharfman WH, Brahmer JR, Lawrence DP, Atkins MB, Powderly JD, Leming PD, Lipson EJ, Puzanov I, Smith DC, Taube JM, Wigginton JM, Kollia GD, Gupta A, Pardoll DM, Sosman JA, Hodi FS. Survival, durable tumor remission, and long-term safety in patients with advanced melanoma receiving nivolumab. J Clin Oncol. 2014;32:1020–1030. doi: 10.1200/JCO.2013.53.0105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brahmer JR, Tykodi SS, Chow LQ, Hwu WJ, Topalian SL, Hwu P, Drake CG, Camacho LH, Kauh J, Odunsi K, Pitot HC, Hamid O, Bhatia S, Martins R, Eaton K, Chen S, Salay TM, Alaparthy S, Grosso JF, Korman AJ, Parker SM, Agrawal S, Goldberg SM, Pardoll DM, Gupta A, Wigginton JM. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N Engl J Med. 2012;366:2455–2465. doi: 10.1056/NEJMoa1200694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Topalian SL, Hodi FS, Brahmer JR, Gettinger SN, Smith DC, McDermott DF, Powderly JD, Carvajal RD, Sosman JA, Atkins MB, Leming PD, Spigel DR, Antonia SJ, Horn L, Drake CG, Pardoll DM, Chen L, Sharfman WH, Anders RA, Taube JM, McMiller TL, Xu H, Korman AJ, Jure-Kunkel M, Agrawal S, McDonald D, Kollia GD, Gupta A, Wigginton JM, Sznol M. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med. 2012;366:2443–2454. doi: 10.1056/NEJMoa1200690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Curran MA, Montalvo W, Yagita H, Allison JP. PD-1 and CTLA-4 combination blockade expands infiltrating T cells and reduces regulatory T and myeloid cells within B16 melanoma tumors. Proc Natl Acad Sci U S A. 2010;107:4275–4280. doi: 10.1073/pnas.0915174107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wolchok JD, Kluger H, Callahan MK, Postow MA, Rizvi NA, Lesokhin AM, Segal NH, Ariyan CE, Gordon RA, Reed K, Burke MM, Caldwell A, Kronenberg SA, Agunwamba BU, Zhang X, Lowy I, Inzunza HD, Feely W, Horak CE, Hong Q, Korman AJ, Wigginton JM, Gupta A, Sznol M. Nivolumab plus ipilimumab in advanced melanoma. N Engl J Med. 2013;369:122–133. doi: 10.1056/NEJMoa1302369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Page DB, Postow MA, Callahan MK, Allison JP, Wolchok JD. Immune modulation in cancer with antibodies. Annu Rev Med. 2014;65:185–202. doi: 10.1146/annurev-med-092012-112807. [DOI] [PubMed] [Google Scholar]
- 12.Chambers CA, Sullivan TJ, Allison JP. Lymphoproliferation in CTLA-4-deficient mice is mediated by costimulation-dependent activation of CD4+ T cells. Immun. 1998;7:885–895. doi: 10.1016/s1074-7613(00)80406-9. [DOI] [PubMed] [Google Scholar]
- 13.Waterhouse P, Penninger JM, Timms E, Wakeham A, Shahinian A, Lee KP, Thompson CB, Griesser H, Mak TW. Lymphoproliferative disorders with early lethality in mice deficient in Ctla-4. Science. 1995;270:985–988. doi: 10.1126/science.270.5238.985. [DOI] [PubMed] [Google Scholar]
- 14.Nishimura H, Minato N, Nakano T, Honjo T. Immunological studies on PD-1 deficient mice: implication of PD-1 as a negative regulator for B cell responses. Int Immunol. 1998;10:1563–1572. doi: 10.1093/intimm/10.10.1563. [DOI] [PubMed] [Google Scholar]
- 15.Tivol EA, Borriello F, Schweitzer AN, Lynch WP, Bluestone JA, Sharpe AH. Loss of CTLA-4 leads to massive lymphoproliferation and fatal multiorgan tissue destruction, revealing a critical negative regulatory role of CTLA-4. Immun. 1995;3:541–547. doi: 10.1016/1074-7613(95)90125-6. [DOI] [PubMed] [Google Scholar]
- 16.Nishimura H, Okazaki T, Tanaka Y, Nakatani K, Hara M, Matsumori A, Sasayama S, Mizoguchi A, Hiai H, Minato N, Honjo T. Autoimmune dilated cardiomyopathy in PD-1 receptor-deficient mice. Science. 2001;291:319–322. doi: 10.1126/science.291.5502.319. [DOI] [PubMed] [Google Scholar]
- 17.Parry RV, Chemnitz JM, Frauwirth KA, Lanfranco AR, Braunstein I, Kobayashi SV, Linsley PS, Thompson CB, Riley JL. CTLA-4 and PD-1 receptors inhibit T-cell activation by distinct mechanisms. Mol Cell Biol. 2005;25:9543–9553. doi: 10.1128/MCB.25.21.9543-9553.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Riley JL, Mao M, Kobayashi S, Biery M, Burchard J, Cavet G, Gregson BP, June CH, Linsley PS. Modulation of TCR-induced transcriptional profiles by ligation of CD28, ICOS, and CTLA-4 receptors. Proc Natl Acad Sci U S A. 2002;99:11790–11795. doi: 10.1073/pnas.162359999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Okazaki T, Maeda A, Nishimura H, Kurosaki T, Honjo T. PD-1 immunoreceptor inhibits B cell receptor-mediated signaling by recruiting src homology 2-domain-containing tyrosine phosphatase 2 to phosphotyrosine. Proc Natl Acad Sci U S A. 2001;98:13866–13871. doi: 10.1073/pnas.231486598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sehgal K, Guo X, Koduru S, Shah A, Lin A, Yan X, Dhodapkar KM. Plasmacytoid dendritic cells, interferon signaling, and FcgammaR contribute to pathogenesis and therapeutic response in childhood immune thrombocytopenia. Sci Transl Med. 2013;5:193ra189. doi: 10.1126/scitranslmed.3006277. [DOI] [PubMed] [Google Scholar]
- 21.Furney SJ, Pedersen M, Gentien D, Dumont AG, Rapinat A, Desjardins L, Turajlic S, Piperno-Neumann S, de la Grange P, Roman-Roman S, Stern MH, Marais R. SF3B1 mutations are associated with alternative splicing in uveal melanoma. Cancer discovery. 2013;3:1122–1129. doi: 10.1158/2159-8290.CD-13-0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Whistler T, Chiang CF, Lonergan W, Hollier M, Unger ER. Implementation of exon arrays: alternative splicing during T-cell proliferation as determined by whole genome analysis. BMC Genomics. 2010;11:496. doi: 10.1186/1471-2164-11-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fritsch RD, Shen X, Sims GP, Hathcock KS, Hodes RJ, Lipsky PE. Stepwise differentiation of CD4 memory T cells defined by expression of CCR7 and CD27. J Immunol. 2005;175:6489–6497. doi: 10.4049/jimmunol.175.10.6489. [DOI] [PubMed] [Google Scholar]
- 24.Sivori S, Parolini S, Falco M, Marcenaro E, Biassoni R, Bottino C, Moretta L, Moretta A. 2B4 functions as a co-receptor in human NK cell activation. Eur J Immunol. 2000;30:787–793. doi: 10.1002/1521-4141(200003)30:3<787::AID-IMMU787>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 25.Hamilton G, Colbert JD, Schuettelkopf AW, Watts C. Cystatin F is a cathepsin C-directed protease inhibitor regulated by proteolysis. EMBO J. 2008;27:499–508. doi: 10.1038/sj.emboj.7601979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Clark R, Griffiths GM. Lytic granules, secretory lysosomes and disease. Curr Opin Immunol. 2003;15:516–521. doi: 10.1016/s0952-7915(03)00113-4. [DOI] [PubMed] [Google Scholar]
- 27.Lieberman J. The ABCs of granule-mediated cytotoxicity: new weapons in the arsenal. Nat Rev Immunol. 2003;3:361–370. doi: 10.1038/nri1083. [DOI] [PubMed] [Google Scholar]
- 28.Parekh VV, Lalani S, Kim S, Halder R, Azuma M, Yagita H, Kumar V, Wu L, Kaer LV. PD-1/PD-L blockade prevents anergy induction and enhances the anti-tumor activities of glycolipid-activated invariant NKT cells. J Immunol. 2009;182:2816–2826. doi: 10.4049/jimmunol.0803648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Benson DM, Jr, Bakan CE, Mishra A, Hofmeister CC, Efebera Y, Becknell B, Baiocchi RA, Zhang J, Yu J, Smith MK, Greenfield CN, Porcu P, Devine SM, Rotem-Yehudar R, Lozanski G, Byrd JC, Caligiuri MA. The PD-1/PD-L1 axis modulates the natural killer cell versus multiple myeloma effect: a therapeutic target for CT-011, a novel monoclonal anti-PD-1 antibody. Blood. 2010;116:2286–2294. doi: 10.1182/blood-2010-02-271874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moll M, Kuylenstierna C, Gonzalez VD, Andersson SK, Bosnjak L, Sonnerborg A, Quigley MF, Sandberg JK. Severe functional impairment and elevated PD-1 expression in CD1d-restricted NKT cells retained during chronic HIV-1 infection. Eur J Immunol. 2009;39:902–911. doi: 10.1002/eji.200838780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pedicord VA, Montalvo W, Leiner IM, Allison JP. Single dose of anti-CTLA-4 enhances CD8+ T-cell memory formation, function, and maintenance. Proc Natl Acad Sci U S A. 2011;108:266–271. doi: 10.1073/pnas.1016791108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krummel MF, Allison JP. CTLA-4 engagement inhibits IL-2 accumulation and cell cycle progression upon activation of resting T cells. J.Exp.Med. 1996;183:2533–2540. doi: 10.1084/jem.183.6.2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Walunas TL, Bakker CY, Bluestone JA. CTLA-4 ligation blocks CD28-dependent T cell activation. J.Exp.Med. 1996;183:2541–2550. doi: 10.1084/jem.183.6.2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang W, Yu D, Sarnaik AA, Yu B, Hall M, Morelli D, Zhang Y, Zhao X, Weber JS. Biomarkers on melanoma patient T cells associated with ipilimumab treatment. J Transl Med. 2012;10:146. doi: 10.1186/1479-5876-10-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nagata S, Ise T, Pastan I. Fc receptor-like 3 protein expressed on IL-2 nonresponsive subset of human regulatory T cells. J Immunol. 2009;182:7518–7526. doi: 10.4049/jimmunol.0802230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kochi Y, Yamada R, Suzuki A, Harley JB, Shirasawa S, Sawada T, Bae SC, Tokuhiro S, Chang X, Sekine A, Takahashi A, Tsunoda T, Ohnishi Y, Kaufman KM, Kang CP, Kang C, Otsubo S, Yumura W, Mimori A, Koike T, Nakamura Y, Sasazuki T, Yamamoto K. A functional variant in FCRL3, encoding Fc receptor-like 3, is associated with rheumatoid arthritis and several autoimmunities. Nat Genet. 2005;37:478–485. doi: 10.1038/ng1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ribas A, Comin-Anduix B, Chmielowski B, Jalil J, de la Rocha P, McCannel TA, Ochoa MT, Seja E, Villanueva A, Oseguera DK, Straatsma BR, Cochran AJ, Glaspy JA, Hui L, Marincola FM, Wang E, Economou JS, Gomez-Navarro J. Dendritic cell vaccination combined with CTLA4 blockade in patients with metastatic melanoma. Clin Cancer Res. 2009;15:6267–6276. doi: 10.1158/1078-0432.CCR-09-1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dhodapkar KM, Gettinger SN, Das R, Zebroski H, Dhodapkar MV. SOX2-specific adaptive immunity and response to immunotherapy in non-small cell lung cancer. Oncoimmunology. 2013;2:e25205. doi: 10.4161/onci.25205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Beck KE, Blansfield JA, Tran KQ, Feldman AL, Hughes MS, Royal RE, Kammula US, Topalian SL, Sherry RM, Kleiner D, Quezado M, Lowy I, Yellin M, Rosenberg SA, Yang JC. Enterocolitis in patients with cancer after antibody blockade of cytotoxic T-lymphocyte-associated antigen 4. J Clin Oncol. 2006;24:2283–2289. doi: 10.1200/JCO.2005.04.5716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dinarello CA, van der Meer JW. Treating inflammation by blocking interleukin-1 in humans. Semin Immunol. 2013;25:469–484. doi: 10.1016/j.smim.2013.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.