Abstract
Cells may be captured and released using a photodegradable hydrogel (photogel) functionalized with antibodies. Photogel substrates were used to first isolate human CD4 or CD8 T-cells from a heterogeneous cell suspension and then to release desired cells or groups of cells by UV-induced photodegradation. Flow cytometry analysis of the retrieved cells revealed approximately 95% purity of CD4 and CD8 T-cells, suggesting that this substrate had excellent specificity. To demonstrate the possibility of sorting cells according to their function, photogel substrates that were functionalized with anti-CD4 and anti-TNF-α antibodies were prepared. Single cells captured and stimulated on such substrates were identified by the fluorescence “halo” after immunofluorescent staining and could be retrieved by site-specific exposure to UV light through a microscope objective. Overall, it was demonstrated that functional photodegradable hydrogels enable the capture, analysis, and sorting of live cells.
Keywords: antibodies, microarrays, micropatterning, hydrogels, single-cell detection
The presence of specific cells in bodily fluids and the function of these cells may serve as diagnostic markers of infections or malignancies.[1] This is particularly true for immune cell analysis, as the diagnosis of the acquired immune deficiency syndrome (AIDS) is based on the enumeration of CD4 T-cells, whereas latent tuberculosis (TB) is determined by identifying cytokine-producing CD4 T-cells.[2] Microengineered surfaces and microfluidic devices offer multiple advantages for the analysis of immune cells in comparison to standard immunological techniques. These advantages are related to small volumes, controlled washing conditions, miniature size of the device, and multiplexing capabilities.[3] However, the ability to capture various immune cells and analyze the function of these cells needs to be combined with the possibility of selectively retrieving cells of interest by making use of either their surface marker expression or function. For example, physicochemical stimulation (e.g., exposure to EDTA or enzymes or temperature control) can be applied to retrieve cells from the surfaces.[4] Alternatively, cells can be electrochemically released from electrodes.[5] However, these methods often threaten cell viability because of harsh and physiologically unfavorable conditions, and may be suboptimal for selective cell retrieval. Light-triggered cell retrieval is particularly promising as it may be used for the high-throughput retrieval of small groups of cells or single cells.[6] Photolabile molecules containing ortho-nitrobenzyl moieties are particularly appealing as photolabile groups because they may be degraded by 365 nm UV light and cause minimal damage to biomolecules or living cells, including T-cells, human umbilical vascular endothelial cells (HUVECs), human leukemic cells, cervical carcinoma cells, and human mesenchymal stem cells.[7] Our group has previously utilized surfaces that contain cell-capture antibodies linked to ortho-nitrobenzyl moieties for the capture and release of cells;[6a] however, we encountered non-specific binding and fouling issues with this approach. At the same time, we have been making extensive use of poly(ethylene glycol) (PEG) hydrogels as non-fouling coatings for the printing of antibody arrays and the capture of immune cells.[8] Herein, we describe the development of an antibody-functionalized photodegradable PEG hydrogel; we hypothesized that such a coating should lead to excellent non-fouling properties while additionally enabling site-specific cell release.
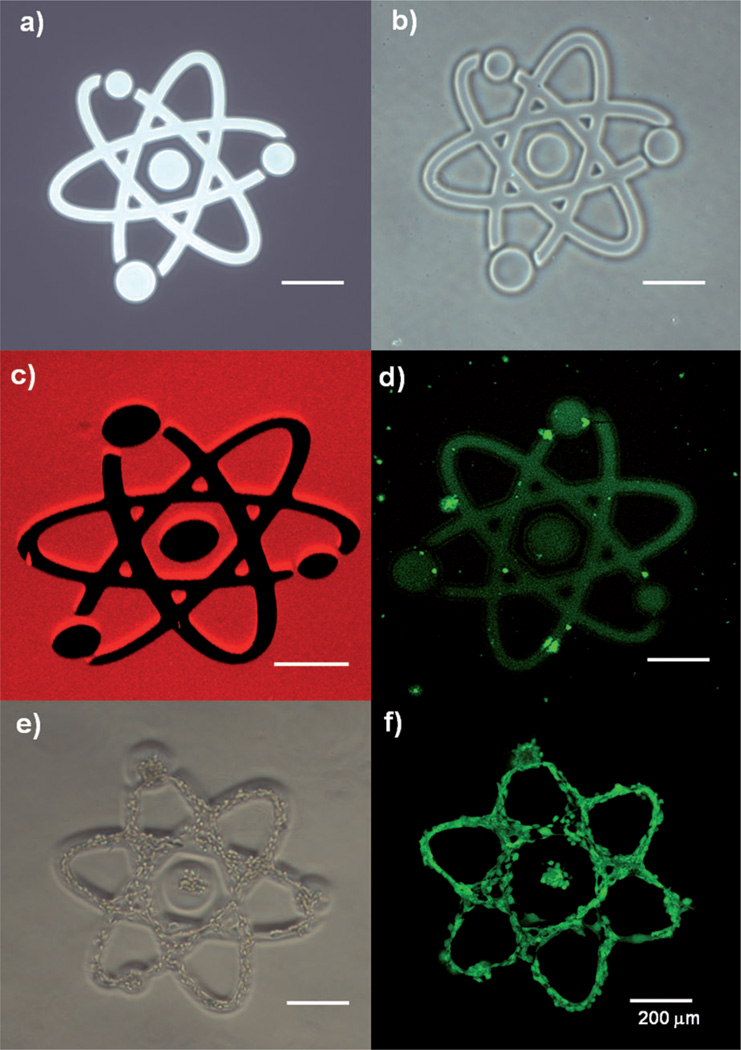
A photodegradable PEG hydrogel or “photogel” consists of PEG cross-linked by photolabile moieties (for the synthesis of photolabile cross-linkers, see the Supporting Information, Figure S1).[9] Figure 1 reveals several aspects related to the micropatterning of the photogel surfaces: 1) Pattern transfer from photomask to photogel occurred with high fidelity (Figure 1a, b). 2) The photogel was efficiently degraded and removed (see Figure 1c for an image of the fluorescently labeled gel). The dark regions, which correspond to degraded gel, and the red fluorescent regions, which correspond to retained gel, can be clearly distinguished. 3) The non-fouling properties of the photogel were confirmed by the selective adsorption of fluorescein isothiocyanate (FITC) labeled collagen I and 3T3 fibroblasts in the regions where the gel was degraded and the underlying glass substrate was exposed (Figure 1d–f). Limited adsorption of proteins or cells was seen on the photogel. Through optimization experiments that are described in detail in the Supporting Information and Figure S2, we arrived at the following procedure for gel formation and gel degradation. The photogel was grafted on acrylated glass surfaces by radical polymerization initiated by ammonium persulfate (AP) and tetramethylethylenediamine (TEMED). Regions of the photogel on the glass surface were then exposed to 365 nm UV light from a fluorescence microscope for various exposure times. The photodegradation efficiency was determined by measuring the degraded area as a function of exposure time. The photodegradation amounted to more than 95% after 20 seconds of exposure (600 mW cm−2). Importantly, a simple adjustment of microscope aperture allowed modulating the diameter of the exposure area from 50 to 200 µm, offering the possibility to address small groups of cells or single cells (Figure S3).
Figure 1.
Degradation of the photogel. a) Design of the micropattern in the chrome mask. b) Bright-field and c) Z-stacked confocal fluorescence microscopy images. d) FITC–collagen coated onto the surface after photogel removal. e) NIH-3T3 cell line seeded on the surface after photogel removal. f) Live/dead staining of the seeded cells.
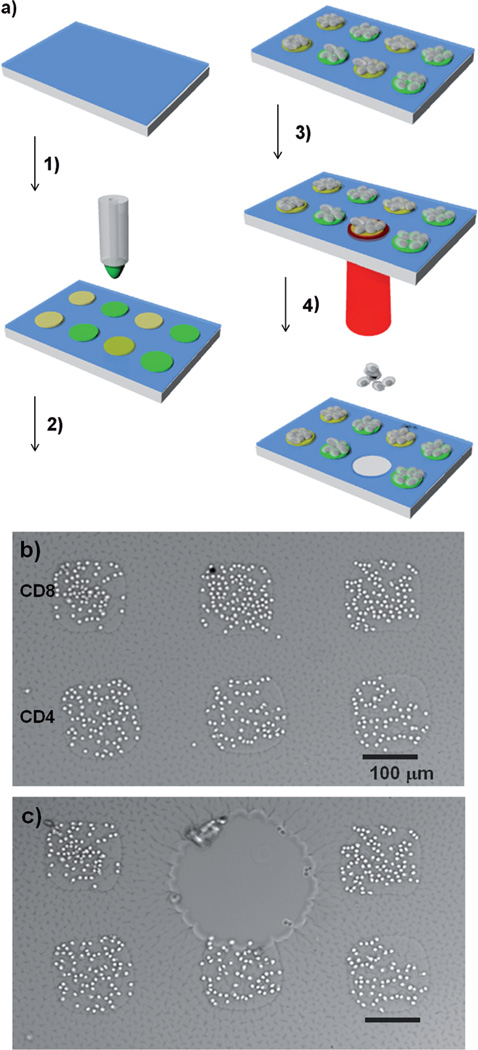
Having verified the non-fouling properties of the photogel, we used this material as a coating for printing antibody arrays and capturing T-cells from a peripheral blood mononuclear cell (PBMC) suspension. To enable the covalent linkage of antibodies, the photogel was functionalized with N-hydroxysuccinimide (NHS) ester using NHS-PEG-acrylate. Figure 2a describes the experiment procedure, according to which antibody spots specific to CD4 and CD8 T-cells were robotically printed on the hydrogels, resulting in the formation of spots with a diameter of approximately 150 µm. These patterned substrates were incorporated into microfluidic devices and perfused with a PBMC suspension, which resulted in the capture of CD4 and CD8 T-cells on their respective antibody spots. Bright-field images of the captured T-cells are shown in Figure 2b, whereas fluorescence images recorded after staining the CD3, CD4, and CD8 antigens are presented in Figure S4. The latter images demonstrate the capture of CD3+CD4+ T-cells on anti-CD4 spots and the capture of CD3+CD8+ T-cells on anti-CD8 cells (see the Supporting Information for details). Importantly, the underlying substrate could be degraded to release a specific group of cells (Figure 2c). The exposed gel region gradually degraded, and CD8 T-cells detached from the surface over the course of one hour (Figure 2b, c; for a video of the cell release, see the Supporting Information). The photodegradable substrate afforded the possibility of releasing groups of cells sequentially, one after another (Figure S5). Live/dead staining of retrieved cells showed over 97% viability, suggesting that photodegradation of the gel did not have deleterious effects on the cells (Figure S6).
Figure 2.
Capture and release of T-cells on antibody-modified surfaces. a) Cell sorting and isolation: 1) Antibody printing on the photogel layer, 2) cell capture, 3) exposure to UV light, 4) cell release. b) Bright-field image of cell capture on CD4 and CD8 antibody spots. c) Release of CD8 cells.
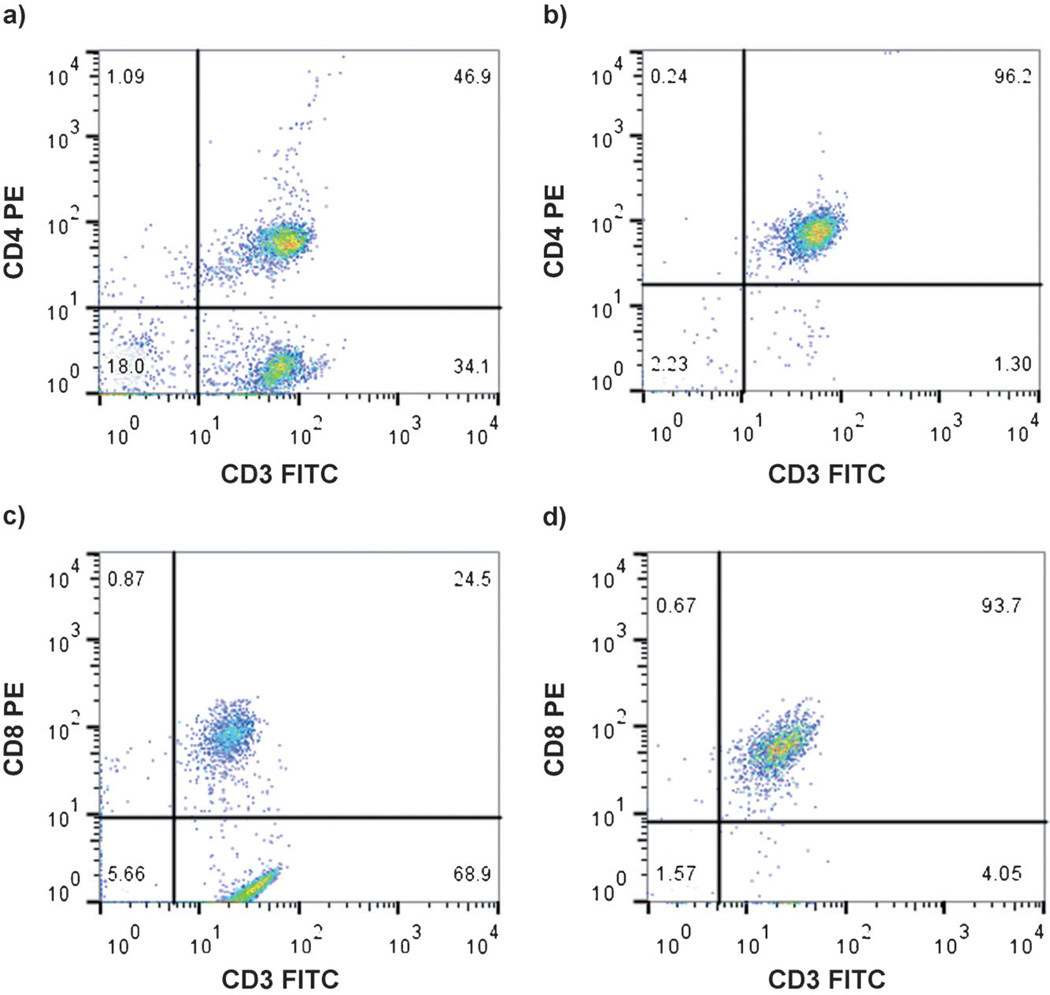
To rigorously demonstrate the purity of the T-cells, photogel surfaces were exposed to UV light to degrade the gel and collect the cells. These cells were then stained with FITC-anti-CD3 and phycoerythrin (PE)-anti-CD4 in the case of CD4 T-cells or FITC-anti-CD3 and with PE-anti-CD8 for CD8 T-cells. Flow cytometry indicated that 96% of the CD4 cells and 94% of the CD8 cells collected from the capture surface after photodegradation were CD3+CD4+ and CD3+CD8+ cells, whereas in the PMBC suspension before separation, these cells were present in 47% and 25%, respectively (Figure 3; see also Figure S7). These purity results are comparable to the cell-capture purity with non-degradable PEG hydrogel (95–97%) as well as to those obtained with commercial T-cell isolation kits (above 90%).[10]
Figure 3.
Flow-cytometry diagrams from primary cells a) before and b) after isolation from the photogel printed with anti-CD4 antibodies and c) before and d) after isolation from the photogel printed with anti-CD8 antibodies. Flow cytometry was performed after immobilized cells had been stained with FITC-anti-CD3 antibodies and biotin-anti-CD4 antibodies followed by PE-streptavidin or FITC-anti-CD3 antibodies and biotin-anti-CD8 antibodies followed by PE-streptavidin.
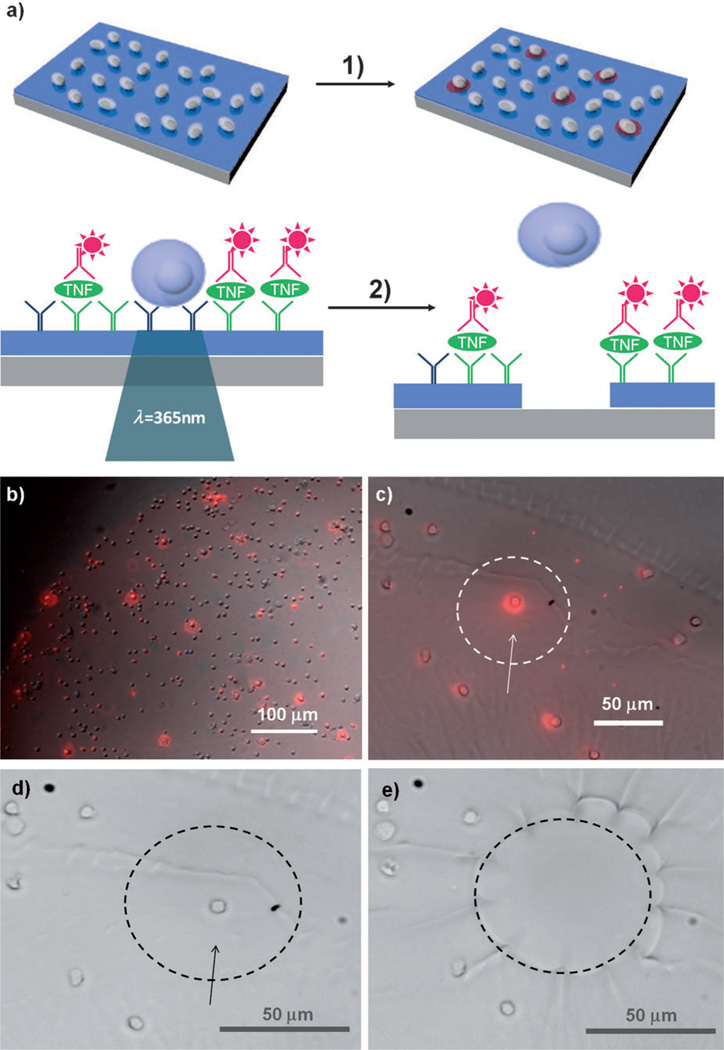
The ability to release desired cells on command is particularly appealing for single-cell analysis. Figure 4a demonstrates a method for detecting secreted molecules from single captured leukocytes. Anti-human CD4 and anti-tumor necrosis factor-α (TNF-α) antibody spots (1 mm diameter) were co-printed on top of a photodegradable gel layer. Incubation of the photogel with a PBMC suspension resulted in the selective capture of CD4-expressing cells on the antibody spots.[11] TNF-α, which was secreted upon mitogenic stimulation, was then detected by immunofluorescent staining with the secondary antibody (Figure 4b; see Figure S8 for a full image). Functional heterogeneity among the CD4 T-cell subsets was apparent, with large differences in TNF-α production from cell to cell (Figure 4b, c). An individual cell that produces a strong fluorescence signal on the photogel could then be released by site-specific UV exposure (Figure 4d, e), demonstrating that this detection and release method is a promising platform for single-cell research.[12] In fact, the approach described in Figure 4 is similar to the enzyme-linked immunosorbent spot (ELISpot) assay,[13] a well-established immunological technique, but offers several advantages: 1) Modifying a gel with cell-specific antibodies allows to capture a desired cell subset (in this case, CD4 T-cells) from a heterogeneous cell suspension. 2) Unlike ELISpot, where only a footprint of the cell is present, our approach allows to retain the cell along with its cytokine “halo”. 3) The use of a photodegradable gel allows the detection of cells of interest based on their function and the release of these cells for downstream analysis, which could entail gene expression studies or continued cultivation.
Figure 4.
Single-cell capture, cytokine detection, and release of single cells. a) Detection of TNF-α secreted from the cells. 1) CD4 cells are selectively isolated from PBMC suspensions on antibody-functionalized photogel. Immobilized cells are stimulated with mitogen to induce production of multiple inflammatory cytokines, including TNF-α. Surfaces are stained with fluorescently labelled secondary antibodies to reveal very productive cells. 2) Cells that produce TNF-α are released by exposure to UV light. b) TNF-α secretion was detected from single cells captured on the photogel. Based on the heterogeneity of TNF-α secretion within the cell population, very productive cells could be identified (bright red spots). c) Enlarged bright-field merged fluorescence TNF-α image. d, e) A single CD4 cell that produces a strong fluorescence signal (indicated with an arrow) was exposed to UV light and released.
In conclusion, we have developed a strategy for isolating and sorting cells using photogel-covered substrates. Printing leukocyte-specific antibodies enabled the capture of specific cell subsets as well as the detection of cytokines secreted by the cells. Regiospecific degradation of the photogel allowed the release of small groups of cells or single cells of interest from the capture surface. This sorting method did not compromise T-cell viability.[6,7] This strategy offers a novel means of releasing cells from culture/capture surfaces for downstream analysis and recultivation. Even though this project focused on the antibody-based capture of leukocytes, the proposed sorting strategy is broadly applicable to the immobilization of adhesive ligands, cell culturing, and cell retrieval by degradation of the underlying substrate.
Supplementary Material
Footnotes
We thank Prof. Marcu for assistance with fluorescence microscopy and Prof. Ferrara and Elizabeth Ingham at Biomedical Engineering, University of California, Davis and Bridget McLaughlin at University of California, Davis Stem Cell Program/Institute for Regenerative Cures for assistance with flow cytometry. This work was supported by NIH grants (C4EB012836 to J.S, DK073901 to A.R.) and an NSF grant (0937997 to A.R.). Additional funding came from “Research Investments in Science Engineering” from UC Davis.
Supporting information for this article is available on the WWW under http://dx.doi.org/10.1002/anie.201404323.
Contributor Information
Dr. Dong-Sik Shin, Email: dshin@ucdavis.edu, Department of Biomedical Engineering, University of California Davis, One Shields Ave, Davis, CA 95616 (USA).
Dr. Jungmok You, Department of Biomedical Engineering, University of California Davis, One Shields Ave, Davis, CA 95616 (USA)
Ali Rahimian, Department of Biomedical Engineering, University of California Davis, One Shields Ave, Davis, CA 95616 (USA).
Tam Vu, Department of Biomedical Engineering, University of California Davis, One Shields Ave, Davis, CA 95616 (USA).
Christian Siltanen, Department of Biomedical Engineering, University of California Davis, One Shields Ave, Davis, CA 95616 (USA).
Arshia Ehsanipour, Department of Biomedical Engineering, University of California Davis, One Shields Ave, Davis, CA 95616 (USA).
Dr. Gulnaz Stybayeva, Department of Biomedical Engineering, University of California Davis, One Shields Ave, Davis, CA 95616 (USA)
Prof. Julie Sutcliffe, Department of Biomedical Engineering, University of California Davis, One Shields Ave, Davis, CA 95616 (USA) Division of Hematology/Oncology, Department of Internal Medicine, Center for Molecular and Genomic Imaging, University of California, Davis, Davis, CA 95616 (USA).
Prof. Alexander Revzin, Email: arevzin@ucdavis.edu, Department of Biomedical Engineering, University of California Davis, One Shields Ave, Davis, CA 95616 (USA).
References
- 1.a) Nagrath S, Sequist LV, Maheswaran S, Bell DW, Irimia D, Ulkus L, Smith MR, Kwak EL, Digumarthy S, Muzikansky A, Ryan P, Balis UJ, Tompkins RG, Haber DA, Toner M. Nature. 2007;450:1235–1239. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Pantel K, Brakenhoff RH, Brandt B. Nat. Rev. Cancer. 2008;8:329–340. doi: 10.1038/nrc2375. [DOI] [PubMed] [Google Scholar]
- 2.a) Ewer K, Deeks J, Alvarez L, Bryant G, Waller S, Andersen P, Monk P, Lalvani A. Lancet. 2003;361:1168–1173. doi: 10.1016/S0140-6736(03)12950-9. [DOI] [PubMed] [Google Scholar]; b) Alimonti JB, Ball TB, Fowke KR. J. Gen. Virol. 2003;84:1649–1661. doi: 10.1099/vir.0.19110-0. [DOI] [PubMed] [Google Scholar]
- 3.a) Wheeler AR, Throndset WR, Whelan RJ, Leach AM, Zare RN, Liao YH, Farrell K, Manger ID, Daridon A. Anal. Chem. 2003;75:3581–3586. doi: 10.1021/ac0340758. [DOI] [PubMed] [Google Scholar]; b) Cheng X, Irimia D, Dixon M, Sekine K, Demirci U, Zamir L, Tompkins RG, Rodriguez W, Toner M. Lab Chip. 2007;7:170–178. doi: 10.1039/b612966h. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Revzin A, Maverakis E, Chang HC. Biomicrofluidics. 2012;6:021301. doi: 10.1063/1.4706845. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) You J, Shin DS, Revzin A. Methods Cell Biol. 2014;121:75–90. doi: 10.1016/B978-0-12-800281-0.00006-3. [DOI] [PubMed] [Google Scholar]
- 4.a) Plouffe BD, Brown MA, Iyer RK, Radisic M, Murthy SK. Lab Chip. 2009;9:1507–1510. doi: 10.1039/b823523f. [DOI] [PubMed] [Google Scholar]; b) Shah AM, Yu M, Nakamura Z, Ciciliano J, Ulman M, Kotz K, Stott SL, Maheswaran S, Haber DA, Toner M. Anal. Chem. 2012;84:3682–3688. doi: 10.1021/ac300190j. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Tang Z, Akiyama Y, Itoga K, Kobayashi J, Yamato M, Okano T. Biomaterials. 2012;33:7405–7411. doi: 10.1016/j.biomaterials.2012.06.077. [DOI] [PubMed] [Google Scholar]
- 5.a) Yeo WS, Yousaf MN, Mrksich M. J. Am. Chem. Soc. 2003;125:14994–4995. doi: 10.1021/ja038265b. [DOI] [PubMed] [Google Scholar]; b) Yeo WS, Mrksich M. Langmuir. 2006;22:10816–10820. doi: 10.1021/la061212y. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Zhu H, Yan J, Revzin A. Colloids Surf. B. 2008;64:260–268. doi: 10.1016/j.colsurfb.2008.02.010. [DOI] [PubMed] [Google Scholar]; d) Kim M, Lee JY, Shah SS, Tae G, Revzin A. Chem. Commun. 2009:5865–5867. doi: 10.1039/b909169f. [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Shah SS, Kim M, Foster E, Vu T, Patel D, Chen LJ, Verkhoturov SV, Schweikert E, Tae G, Revzin A. Anal. Bioanal. Chem. 2012;402:1847–1856. doi: 10.1007/s00216-011-5613-z. [DOI] [PubMed] [Google Scholar]
- 6.a) Shin DS, Seo JH, Sutcliffe JL, Revzin A. Chem. Commun. 2011;47:11942–11944. doi: 10.1039/c1cc15046d. [DOI] [PubMed] [Google Scholar]; b) Wirkner M, Alonso JM, Maus V, Salierno M, Lee TT, Garcia AJ, del Campo A. Adv. Mater. 2011;23:3907–3910. doi: 10.1002/adma.201100925. [DOI] [PubMed] [Google Scholar]; c) Byambaa B, Konno T, Ishihara K. Colloids Surf. B. 2012;99:1–6. doi: 10.1016/j.colsurfb.2011.08.029. [DOI] [PubMed] [Google Scholar]; d) Yamaguchi S, Yamahira S, Kikuchi K, Sumaru K, Kanamori T, Nagamune T. Angew. Chem. 2012;124:132–135. doi: 10.1002/anie.201106106. Angew. Chem. Int. Ed. 2012, 51, 128 – 131. [DOI] [PubMed] [Google Scholar]
- 7.a) Holmes CP. J. Org. Chem. 1997;62:2370–2380. doi: 10.1021/jo961602x. [DOI] [PubMed] [Google Scholar]; b) Pelliccioli AP, Wirz J. Photochem. Photobiol. Sci. 2002;1:441–458. doi: 10.1039/b200777k. [DOI] [PubMed] [Google Scholar]; c) Stegmaier P, Alonso JM, Campo AD. Langmuir. 2008;24:11872–11879. doi: 10.1021/la802052u. [DOI] [PubMed] [Google Scholar]; d) Kloxin AM, Kasko AM, Salinas CN, Anseth KS. Science. 2009;324:59–63. doi: 10.1126/science.1169494. [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Kloxin AM, Tibbitt MW, Anseth KS. Nat. Protoc. 2010;5:1867–1887. doi: 10.1038/nprot.2010.139. [DOI] [PMC free article] [PubMed] [Google Scholar]; f) San Miguel V, Bochet CG, del Campo A. J. Am. Chem. Soc. 2011;133:5380–5388. doi: 10.1021/ja110572j. [DOI] [PubMed] [Google Scholar]
- 8.a) Zhu H, Stybayeva G, Macal M, Ramanculov E, George MD, Dandekar S, Revzin A. Lab Chip. 2008;8:2197–2205. doi: 10.1039/b810244a. [DOI] [PubMed] [Google Scholar]; b) Stybayeva G, Kairova M, Ramanculov E, Simonian AL, Revzin A. Colloids Surf. B. 2010;80:251–255. doi: 10.1016/j.colsurfb.2010.06.015. [DOI] [PubMed] [Google Scholar]; c) Stybayeva G, Mudanyali O, Seo S, Silangcruz J, Macal M, Ramanculov E, Dandekar S, Erlinger A, Ozcan A, Revzin A. Anal. Chem. 2010;82:3736–3744. doi: 10.1021/ac100142a. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Chen A, Vu T, Stybayeva G, Pan T, Revzin A. Biomicrofluidics. 2013;7:024105. doi: 10.1063/1.4795423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Siltanen C, Shin DS, Sutcliffe J, Revzin A. Angew. Chem. 2013;125:9394–9398. doi: 10.1002/anie.201303965. Angew. Chem. Int. Ed. 2013, 52, 9224 – 9228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sekine K, Revzin A, Tompkins RG, Toner M. J. Immunol. Methods. 2006;313:96–109. doi: 10.1016/j.jim.2006.03.017. b) see the company websites for CD4 and CD8 positive isolation kits: http://www.invitrogen.com, http://www.rndsystems.com, http://www.miltenyibiotec.com, and http://www.stemcell.com.
- 11.Zhu H, Stybayeva G, Silangcruz J, Yan J, Ramanculov E, Dandekar S, George MD, Revzin A. Anal. Chem. 2009;81:8150–8156. doi: 10.1021/ac901390j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.a) Théry M, Racine V, Pépin A, Piel M, Chen Y, Sibarita JB, Bornens M. Nat. Cell Biol. 2005;7:947–953. doi: 10.1038/ncb1307. [DOI] [PubMed] [Google Scholar]; b) Newman JR, Ghaemmaghami S, Ihmels J, Breslow DK, Noble M, DeRisi JL, Weissman JS. Nature. 2006;441:840–846. doi: 10.1038/nature04785. [DOI] [PubMed] [Google Scholar]
- 13.a) Czerkinsky CC, Nilsson LA, Nygren H, Ouchterlony O, Tarkowski A. J. Immunol. Methods. 1983;65:109–121. doi: 10.1016/0022-1759(83)90308-3. [DOI] [PubMed] [Google Scholar]; b) Arend SM, van Meijgaarden KE, de Boer K, de Palou EC, van Soolingen D, Ottenhoff TH, van Dissel JT. J. Infect. Dis. 2002;186:1797–1807. doi: 10.1086/345760. [DOI] [PubMed] [Google Scholar]; c) Pathan AA, Wilkinson KA, Klenerman P, McShane H, Davidson RN, Pasvol G, Hill AV, Lalvani A. J. Immunol. 2001;167:5217–5225. doi: 10.4049/jimmunol.167.9.5217. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.