Abstract
For proper chromosome segregation, sister kinetochores must interact with microtubules from opposite spindle poles (bi-orientation). To establish bi-orientation, aberrant kinetochore–microtubule attachments are disrupted (error correction) by Aurora B kinase (Ipl1 in budding yeast). Paradoxically, during this disruption, new attachments are still formed efficiently to allow fresh attempts at bi-orientation. How this is possible remains an enigma. Here we show that kinetochore attachment to the microtubule lattice (lateral attachment) is impervious to Aurora B regulation, but attachment to the microtubule plus-end (end-on attachment) is disrupted by this kinase. Thus, a new lateral attachment is formed without interference, then converted to end-on attachment and released if incorrect. This process continues until bi-orientation is established and stabilized by tension across sister kinetochores. We reveal how Aurora B specifically promotes disruption of the end-on attachment through phospho-regulation of kinetochore components Dam1 and Ndc80. Our results reveal fundamental mechanisms for promoting error correction for bi-orientation.
Introduction
Proper chromosome segregation during mitosis relies on correct kinetochore–microtubule (KT–MT) interaction 1. The KT initially interacts with the lateral surface of a single MT (lateral attachment) and is then tethered at the MT plus end (end-on attachment) 2-4. Subsequently sister KTs attach to MTs extending from the opposite spindle poles, establishing chromosome bi-orientation. If an aberrant attachment is formed (Fig 1a, left), it must be removed by Aurora B kinase (Ipl1 in budding yeast), which phosphorylates KT components and disrupts the KT–MT interaction (red arrow) 5-7. However, during this disruptive process, new KT–MT interactions can still be formed efficiently (Fig 1a, black arrows), promoting the KT–MT turnover. If bi-orientation is established and tension is applied, the KT–MT attachment is stabilized (Fig 1a, right), completing error correction. However it remains a mystery how, despite KT-MT attachments being weakened and disrupted by Aurora B (Fig 1a, red arrow), new attachments can be formed efficiently (black arrows), ensuring the KT–MT turnover. Here we address this question, using Saccharomyces cerevisiae as a model organism.
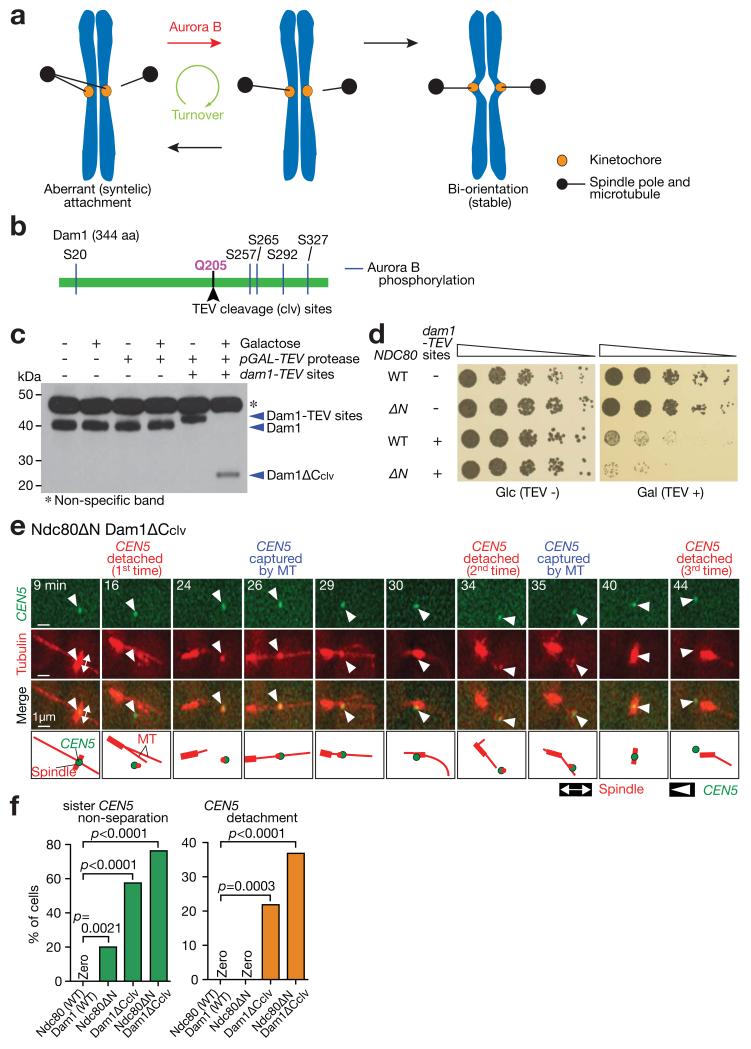
Figure 1. The KT–MT attachments are turned over repeatedly when Dam1 C-terminus and Ndc80 N-tail are deleted.
(a) Diagram shows the process of error correction. To resolve aberrant KT–MT attachment (left), it must be weakened and disrupted by Aurora B phosphorylation of KT components (red arrow). This disruption is followed by formation of new KT–MT attachment (black arrows). Such turnover (i.e. disruption followed by new formation) of KT–MT attachment continues (green arrow) until bi-orientation is established and stabilized by tension across sister KTs (right).
(b) Schematic representation of Dam1 protein showing the position of integrated TEV cleavage sites.
(c) Western blotting (with a Dam1 antibody) showing Dam1 C-terminus cleavage by TEV protease. Cells were incubated for 2 h with or without galactose. A full scan of the western blot is shown in Supplementary Fig 6.
(d) Serially diluted cells were incubated with or without TEV protease expression. Glc, glucose. Gal, galactose.
(e, f) NDC80+ DAM1+ (T7549), ndc80ΔN (T8053), dam1-TEVsites (producing Dam1ΔCclv; T8723) and ndc80ΔN dam1-TEVsites (T8725) cells with PGAL-TEV (except for T8053) CEN5-tetOs TetR-3×CFP Venus-TUB1 PMET3-CDC20 were treated with α factor for 3 h and released to fresh media with methionine (for Cdc20 depletion), in the presence of galactose (for TEV protease expression). From 70 min after the release, images were acquired every minute. e shows a representative cell with Ndc80ΔN plus Dam1ΔCclv (0 min: start of image acquisition). f shows percentage of cells that showed sister CEN5 non-separation (left) and CEN5 detachment from the spindle (usually followed by reattachment; right); n= 45, 35, 87 and 38 cells were analyzed (from left to right). The rate of CEN5 detachment was probably underestimated since it would be overlooked if CEN5 were immediately recaptured by MTs after detachment. Experiments were performed three times (statistics source data are shown in Supplementary Table 2) and a representative experiment is shown here. p-values (two tailed) were obtained by Fisher’s exact test.
The Ndc80 and Dam1 complexes (Ndc80c and Dam1c) are outer KT components comprising the KT–MT interface in budding yeast 8-11. The Ndc80c is an integral KT component since before the initial KT capture by spindle MTs, and is subsequently required for both lateral and end-on MT attachment 3. On the other hand, the Dam1c is not present on unattached KTs and shows a high accumulation at the MT end 4. When the lateral attachment is converted to an end-on attachment, the Dam1c at the MT end interacts physically with Ndc80c, forming the KT–MT interface of the end-on attachment 12-15. Nevertheless the Dam1c also localizes along the MT lattice 4,16,17 and it is unknown whether this Dam1c fraction plays any role during lateral KT–MT attachment.
The Dam1c and Ndc80c components have been identified as Aurora B substrates whose phosphorylation is important for bi-orientation 18-20. Aurora B phosphorylates Dam1 at several residues, and clustered phosphorylation at the Dam1 C-terminus is crucial for bi-orientation 18, and disrupts the Dam1c–Ndc80c and Dam1c–Dam1c interactions in vitro 12,13,17,21. The N-terminal ~100 residues (N-tail) of Ndc80, protruding from the calponin-homology domain (the essential MT-binding domain) 22,23, is also heavily phosphorylated by Aurora B, and this phosphorylation facilitates bi-orientation in both yeast and metazoan cells 19,24,25. The Ndc80 N-tail strengthens the Ndc80c–MT association, and its phosphorylation diminishes this association in vitro 15,26-28. Here we study how the Dam1 C-terminus and the Ndc80 N-tail regulate KT–MT interaction in vivo and how their phosphorylation by Aurora B promotes error correction for bi-orientation.
Results
KTs repeatedly detach from spindle MTs and are quickly recaptured by them, when the Dam1 C-terminus and Ndc80 N-tail are deleted
To investigate the roles of the Dam1 C-terminus and Ndc80 N-tail in KT–MT interaction in vivo, we generated their deletions (dam1Δ206-343 [dam1ΔC] and ndc80Δ1-112 [ndc80ΔN]). Although ndc80ΔN showed normal growth, dam1ΔC showed very slow growth, and their combination was lethal. To address the reason for this lethality, we created a conditional Dam1 C-terminus deletion by introducing TEV protease cleavage sites in the middle of Dam1 (Fig 1b). When the TEV protease was expressed, Dam1 was cleaved within two hours (Fig 1c). As expected, the Dam1 C-terminus cleavage (Dam1ΔCclv) showed very slow growth, and was lethal when combined with ndc80ΔN (Fig 1d). When a chosen CEN (CEN5) was visualized in the Dam1ΔCclv Ndc80ΔN double mutant, sister CEN5s often failed to separate on the metaphase spindle, in contrast to wild-type, implying a bi-orientation defect (Fig 1e,f). This defect was more frequent than in Ndc80ΔN and Dam1ΔCclv single mutants. Intriguingly, in the double mutant, non-separated CEN5 sisters often detached from the metaphase spindle and were quickly recaptured by spindle MTs. This cycle was repeated in many cells (Fig 1e,f). When all KTs were visualized instead of CEN5, their detachment and recapture on the spindle were observed in most of the cells having both Dam1ΔCclv and Ndc80ΔN (Supplementary Fig 1). Thus, when both the Dam1 C-terminus and Ndc80 N-tail are deleted, bi-orientation becomes defective, and centromeres show repeated detachment from, and quick reattachment to, the metaphase spindle.
KTs interact normally with the MT lateral surface, when the Dam1 C-terminus and Ndc80 N-tail are deleted
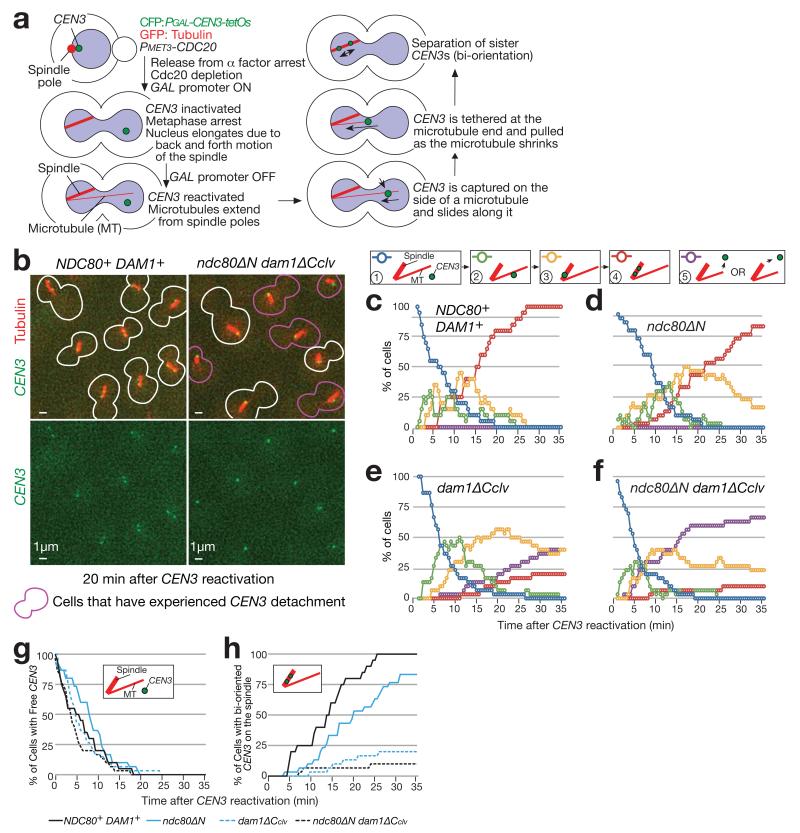
To characterize the above defects, we analysed individual KT–MT interactions, using an engineered assay system 3. In this assay, KT assembly on CEN3 was delayed by transcription from an adjacently inserted promoter 29. This increased the distance between CEN3 and the mitotic spindle 3 (Fig 2a). In wild-type control (NDC80+ DAM1+) cells, after CEN3 was reactivated for KT assembly, CEN3 was rapidly captured by the MT lateral surface (Fig 2c, decline of blue line). Soon after CEN3 reached the spindle, its sisters separated, i.e. bi-orientation was established (Fig 2b, left; c, red line). In Dam1ΔCclv Ndc80ΔN cells, the initial CEN3 capture by the MT lateral surface occurred with normal kinetics (Fig 2f, decline of blue line; g), but subsequent bi-orientation establishment was substantially delayed (Fig 2b, right; f, red line; h). Instead, CEN3 often detached from spindle MTs (Fig 2f, purple line showing cumulative percentage of detachment), followed by recapture by other MTs (not counted again as a fresh MT interaction in Fig 2f). Dam1ΔCclv cells also showed similar, but slightly milder, defects (Fig 2e,g,h). Ndc80ΔN cells showed only a slight delay in sister CEN3 separation on the spindle (Fig 2d,g,h).
Figure 2. The KT attaches normally to the MT lateral surface but detaches afterwards, when the Dam1 C-terminus and the Ndc80 N-tail are deleted.
(a) Experimental system to analyse the individual KT–MT interaction 3.
(b–h) NDC80+ DAM1+ (T9162), ndc80ΔN (T8049), dam1-TEVsites (producing Dam1ΔCclv; T8921) and ndc80ΔN dam1-TEVsites (T8965) cells with PGAL-TEV (except for T8049) PGAL-CEN3-tetOs TetR-3×CFP GFP-TUB1 PMET3-CDC20 were treated with α factor for 3 h and released to fresh media with methionine (for Cdc20 depletion), in the presence of galactose (for TEV expression and CEN3 inactivation). After 3 h, cells were suspended in medium with glucose (for CEN3 reactivation) and images were acquired every 30 sec. b shows representative images acquired at 20 min after CEN3 reactivation. Graphs show percentage of cells at each step of the KT–MT interaction (c–f); Purple lines (step 5) show cumulative percentage of CEN3 detachment; CEN3 detachment was followed by recapture by a MT, but such recapture was not included in counting for other steps. Percentage of cells at steps 1 and 4 are compared in four strains (g and h). n= 30 cells were analysed for each strain. Data represent one out of two independent experiments.
Thus, the lateral KT–MT attachment does not require the Dam1 C-terminus or the Ndc80 N-tail, although it relies on the calponin-homology domain of Ndc80 15,23 (Supplementary Fig 2). Nonetheless, when both the Dam1 C-terminus and the Ndc80 N-tail are deleted, bi-orientation becomes defective and KTs detach from spindle MTs. Subsequently, KTs can interact efficiently with the MT lateral surface, which explains the rapid recapture of KTs by MTs after their detachment from the spindle (see Fig 1e).
The Ndc80 N-tail and the Dam1 C-terminus promote distinct steps in the end-on conversion, and together prevent KT detachment from the MT end
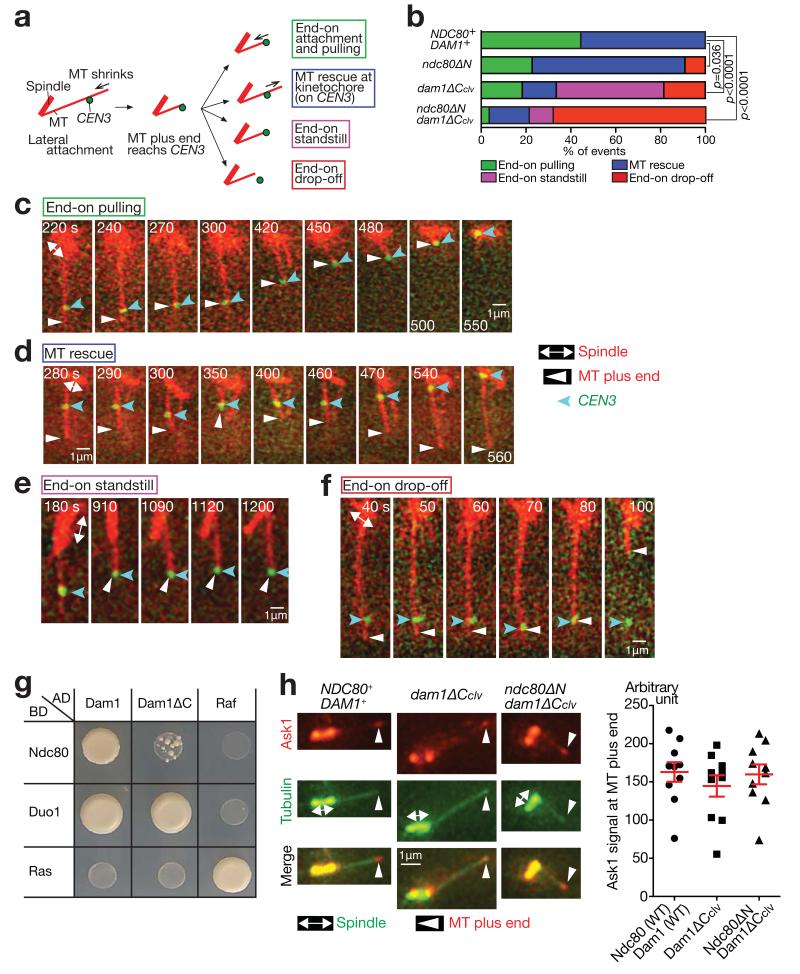
To address how Dam1ΔCclv Ndc80ΔN causes KT detachment from spindle MTs, we investigated the change in CEN3–MT interaction following lateral attachment. During lateral attachment, the plus ends of depolymerizing MTs often reached CEN3. Then, in wild-type cells, CEN3 became tethered at the MT end and was pulled towards a spindle pole as the MT depolymerized (end-on pulling), i.e. end-on attachment was established 4 (Fig 3a,b,c). However, even in wild-type cells, the conversion from the lateral to end-on attachment (end-on conversion) often failed; then, the MT grew again (MT rescue) (Fig 3a,b,d), as previously reported 30. By contrast, in Dam1ΔCclv Ndc80ΔN, after the MT plus end reached CEN3 on the MT lateral surface, the CEN3 detached from the MT end (end-on drop-off) in majority (Fig 3a,b,f). Ndc80ΔN cells showed MT rescue more frequently than wild-type (Fig 3b). In Dam1ΔCclv cells, CEN3 often localized continuously (> 5 min) at the MT end, but without subsequent MT depolymerisation (end-on standstill) (Fig 3a,b,e).
Figure 3. The KT often detaches from the MT end, when the Dam1 C-terminus and the Ndc80 N-tail are deleted.
(a) After the plus end of a shrinking MT had caught up with CEN3, four kinds of events were observed.
(b) Percentage of the events shown in a, in each strain. The four strains in Fig 2b–hwere treated as in Fig 2b–h, except that images were acquired every 10 sec. n= 27, 22, 27 and 28 events were analysed (from top to bottom). p-values (two tailed) were obtained by a chi-square test for trends. Data represent one out of two independent experiments.
(c–f) Examples of the end-on pulling (c, T9162), MT rescue (d, T8049), end-on standstill (e, T8921) and end-on drop-off (f, T8965), observed in b.
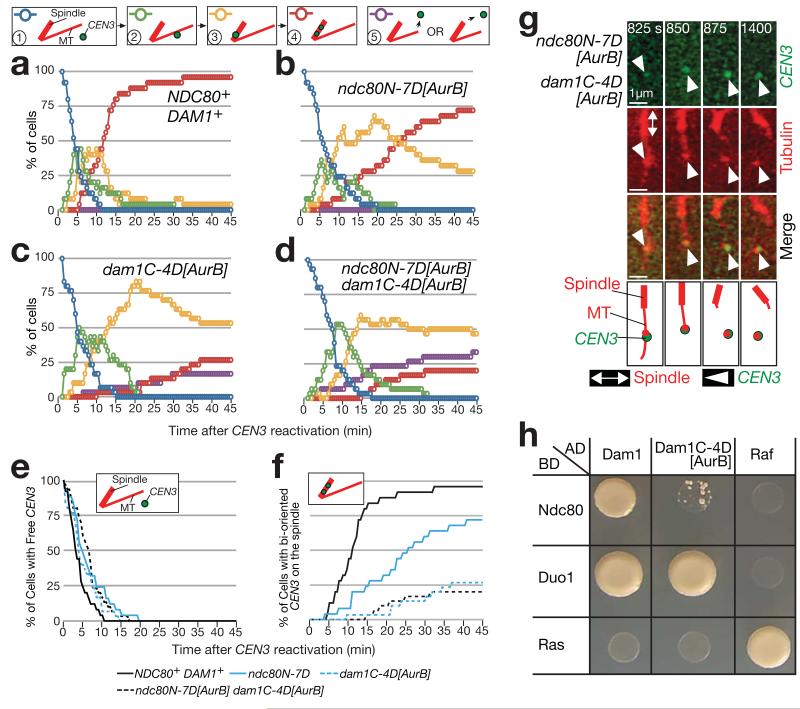
(g) Dam1 and Ndc80 interact physically in the two-hybrid assay, and this interaction requires the Dam1 C-terminus. Duo1 is a component of the Dam1 complex and serves as a control. Ras and Raf were also used as controls. AD, BD: fused with the transcription activation domain and the DNA-binding domain, respectively.
(h) The Dam1 C-terminus is dispensable for Dam1c accumulation at the MT end. NDC80+ DAM1+ (T8875), dam1-TEVsites (producing Dam1ΔCclv; T8915) and ndc80ΔN dam1-TEVsites (T8916) cells with PGAL-TEV CEN5-tetOs TetR-3×CFP ASK1-4×mCherry Venus-TUB1 were treated with α factor for 3 h and released to fresh media in the presence of galactose (for TEV protease expression). Images were acquired 70 min after release. Ask1 is a component of Dam1c. Representative cells in metaphase (left). CEN5 was on the spindle, not at the indicated MT end. Quantification of Ask1 signals at the MT ends (right). The maximum Ask1 signals at the MT ends during image acquisition were quantified in n= 10 cells of each strain. Bars represent mean ± SE. Data represent one out of two independent experiments.
The different phenotypes of the Ndc80ΔN and Dam1ΔCclv single mutants can be explained if the Ndc80 N-tail and Dam1 C-terminus promote distinct steps in the end-on conversion. The Ndc80 N-tail could be involved in an early step of the end-on conversion; i.e. tethering the KT (e.g. on CEN3) at the end of a MT (see Fig 4e). This would explain frequent MT rescue with Ndc80ΔN, as MT rescue takes place when the end-on conversion is defective 30. Meanwhile, the Dam1 C-terminus could facilitate completion of this conversion; i.e. allowing KTs tethered at the MT end to be pulled as the MT shrinks (end-on pulling; see Fig 4e). This would explain frequent end-on standstill with Dam1ΔCclv. The Dam1 C-terminus supports physical interaction with Ndc80 13 (Fig 3g); this interaction likely contributes to complete the end-on conversion 14. On the other hand, the Dam1 C-terminus was dispensable for Dam1c assembly 17 (Fig 3g) and for Dam1c accumulation at the MT end (Fig 3h).
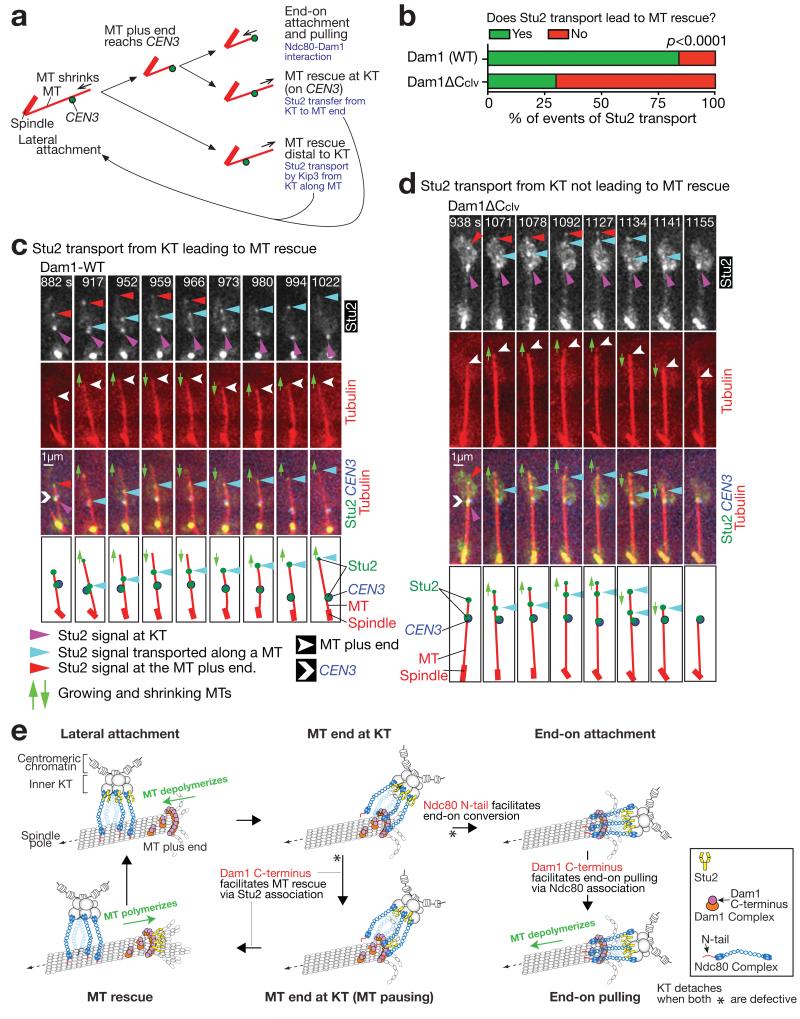
Figure 4. The Dam1 C-terminus helps Stu2 to rescue a MT.
(a) Diagrams illustrating three events following lateral KT–MT attachment; end-on attachment, MT rescue at the KT, and MT rescue distal to the KT. Relevant mechanisms are shown in pale blue 4, 30.
(b–d) Dam1 C-terminus assists Stu2 in rescuing a MT after Stu2 is transported from a KT along the MT to the MT plus end, during the lateral KT–MT interaction. DAM1+ (T9229) and dam1-TEVsites PGAL-TEV (producing Dam1ΔCclv; T9166) cells with PGAL-CEN3-tetOs TetR-3×CFP STU2-4×mCherry GFP-TUB1 PMET3-CDC20 were treated as in Fig 2b–h. Images were acquired every 7 sec. Percentage of Stu2 transport events along a MT, leading, or not leading, to MT rescue; n= 25 and 37 Stu2 transport events were analysed for T9229 and T9166, respectively (b). Representative examples of Stu2 transport along a MT, leading (c), or not leading (d), to MT rescue. In b, experiments were performed twice (statistic source data are shown in Supplementary Table 2), a representative experiment is shown here, and p-value (two tailed) was obtained by Fisher’s exact test.
(e) Diagram illustrating the roles of the Ndc80 N-tail and the Dam1 C-terminus, in the conversion to end-on attachment and in MT rescue. The diagram explains the results in Fig 3 and 4, as follows: 1) with Ndc80ΔN, the end-on conversion is not initiated efficiently, explaining frequent MT rescue; 2) with Dam1ΔCclv, the end-on conversion is initiated but not completed efficiently, i.e. end-on pulling often fails, accounting for the end-on standstill; 3) with Ndc80ΔN plus Dam1ΔCclv, the end-on conversion is not initiated efficiently and MT rescue often fails; i.e. all subsequent steps are often blocked, leading to frequent KT detachment from the MT end.
During the process towards lateral to end-on conversion, Dam1ΔCclv Ndc80ΔN cells showed frequent CEN3 detachment from the MT end. This suggests that these cells are defective in both the end-on conversion, and the failsafe mechanism that would prevent CEN detachment 30. Otherwise, when the end-on conversion fails, it should lead to MT rescue, which would maintain lateral attachment and prevent KT detachment 30 (see Fig 3d). Why is this failsafe mechanism defective in Dam1ΔCclv Ndc80ΔN cells? The following evidence suggests that the Dam1 C-terminus is required for the MT rescue upon failure in the end-on conversion (see Fig 4e).
This MT rescue is promoted by Stu2 (the MT polymerase whose vertebrate orthologues are XMAP215 and ch-TOG 31), which is transferred from the KT to the MT end 30 (Fig 4a, MT rescue at KT). We addressed whether the Dam1-C terminus could assist in this Stu2 function. Stu2 at the KT promotes MT rescue, not only upon the failure in the end-on conversion, but also during the lateral KT–MT attachment, during which Stu2 is occasionally transported from the KT along the MT, promoting MT rescue upon its arrival at the MT end 30 (Fig 4a, MT rescue distal to KT; see Fig 4c). The latter provides a convenient test for possible Stu2 assistance by Dam1, because the MT rescue happens without the KT at the MT end (which would exclude involvement of other KT components) but Dam1c is still at the MT end 4 (see Fig 3h). In Dam1 wild-type cells, Stu2 was occasionally transported from a KT along the MT and, on reaching the MT plus end, it promoted MT rescue (Fig 4b,c) 30. With Dam1ΔCclv, Stu2 was still normally transported from a KT to the MT end, but often failed to promote MT rescue on reaching the MT plus end (Fig 4b,d). Moreover, in a two-hybrid assay, Dam1 and Stu2 showed a physical interaction, relying on the Dam1 C-terminus (Supplementary Fig 3a,b). This suggests Dam1 C-terminus physically interacts with Stu2 and helps rescue a MT at its plus end. This would be the case not only for MT rescue distal to KT, but also for MT rescue at KT (see Fig 4a), as both are dependent on Stu2 accumulation at the MT end where Dam1c also accumulates4.
In summary, frequent KT detachment from the MT end in Ndc80ΔN Dam1ΔCclv is explained by distinct roles of the two domains in the end-on conversion (Fig 4e). In this mutant, the end-on conversion is not initiated efficiently but MT rescue often fails; i.e. both steps are often blocked, leading to frequent KT detachment from the MT end (Fig 4e).
Phospho-mimicking mutants of the Ndc80 N-tail and Dam1 C-terminus at Aurora B sites show similar phenotypes to their deletions
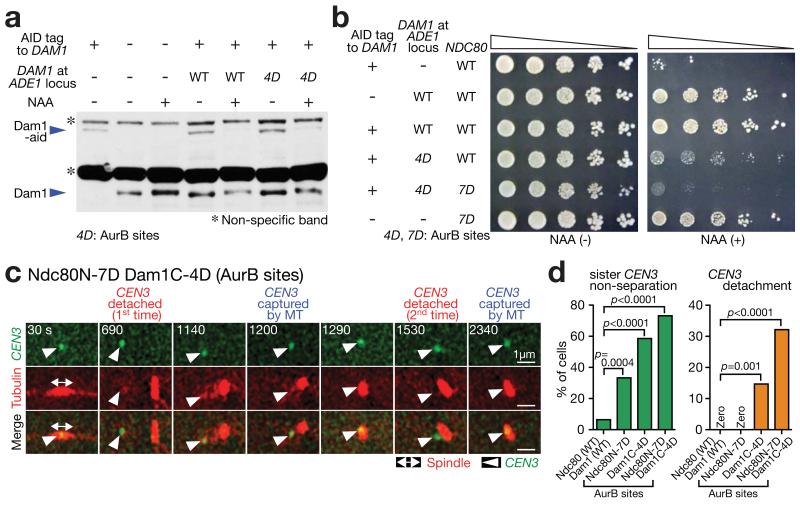
To address how Aurora B phosphorylation of the Dam1 C-terminus and the Ndc80 N-tail affects KT–MT interaction, we generated phospho-mimicking mutations at these domains (dam1C-4D[AurB] and ndc80N-7D[AurB]; see Fig 5a,b legends). dam1C-4D[AurB] showed very slow growth and dam1C-4D[AurB] ndc80N-7D[AurB] was lethal, although Dam1C-4D[AurB] was expressed similarly to Dam1 wild-type (Fig 5a). To maintain growth of these phospho-mimicking mutants, we integrated wild-type DAM1 containing an auxin-inducible degron tag (dam1-aid). With auxin, Dam1-aid was degraded (Fig 5a), dam1C-4D[AurB] cells showed very retarded growth, and dam1C-4D[AurB] ndc80N-7D[AurB] cells were lethal (Fig 5b).
Figure 5. Aurora B phospho-mimicking Dam1 and Ndc80 mutants show repeated turnover of KT–MT attachments.
(a) Western blotting (with Dam1 antibody) showing Dam1-aid depletion and Dam1 protein (wild-type or Dam1C-4D[AurB]) expression from an additional DAM1 construct. Cells were incubated with or without auxin NAA for 4 h in methionine dropout medium. In Dam1C-4D[AurB], four Aurora B target sites (S257, S265, S292 and S327 18) were replaced with aspartates at the Dam1 C-terminus. A full scan of the western blot is shown in Supplementary Fig 6.
(b) Serially diluted cells were incubated with or without auxin NAA. In Ndc80 N-7D[AurB], seven Aurora B target sites (T21, S37, T54, T71, T74, S95 and S100 19) were replaced with aspartates at the Ndc80 N-tail.
(c, d) NDC80+ DAM1+ (T9530), ndc80N-7D[AurB] (T11102), dam1C-4D[AurB] dam1-aid (T11326) and ndc80N-7D[AurB] dam1C-4D[AurB] dam1-aid (T11452) cells with TIR PGAL-CEN3-tetOs TetR-3×CFP GFP-TUB1 PMET3-CDC20 were treated with α factor for 3 h and released to fresh media with methionine (for Cdc20 depletion) in the presence of NAA (to deplete Dam1-aid) and glucose (CEN3 under the GAL promoter was always active). From 90 min after release, images were acquired every 30 sec. c shows a representative cell with ndc80N-7D[AurB] dam1C-4D[AurB] (0 sec: start of image acquisition). d displays percentage of cells showing sister CEN3 non-separation (left) and CEN3 detachment from the spindle (usually followed by reattachment; right); n= 62, 48, 75 and 56 cells were analyzed (from left to right). Experiments were performed twice (statistics source data are shown in Supplementary Table 2) and a representative experiment is shown here. p-values (two tailed) were obtained by Fisher’s exact test.
The dam1C-4D[AurB] ndc80N-7D[AurB] cells showed bi-orientation defects and repeated detachment of CEN3 from the spindle (followed by recapture) (Fig 5c,d), similarly to Dam1ΔCclv Ndc80ΔN cells (see Fig 1e). The dam1C-4D[AurB] alone also showed such phenotypes, but less frequently (Fig 5d). The Dam1 C-terminus (and perhaps the Ndc80 N-tail) is normally dephosphorylated when bi-orientation is established and tension is applied 32. However such de-phosphorylation is not recapitulated with phospho-mimicking mutations, which explains the bi-orientation defects and repeated detachment from the spindle with these mutations (Fig 5c,d).
In the assay shown in Fig 2a, dam1C-4D[AurB] ndc80N-7D[AurB] cells showed normal kinetics in lateral CEN3-MT interaction (Fig 6a,d; decline of blue line: e), but demonstrated both a delay in bi-orientation (Fig 6a,d; red line: f) and CEN3 detachment from the spindle and the MT end (Fig 6d, purple line; g), again similarly to Dam1ΔCclv Ndc80ΔN cells. dam1C-4D[AurB] made greater contribution to these defects than did ndc80N-7D[AurB], and the defects were enhanced in combination. Moreover, Dam1C-4D[AurB] showed reduced interaction with Ndc80 (Fig 6h) and with Stu2 (Supplementary Fig 3c), as did Dam1ΔC. Dam1C-4D[AurB] often failed to assist Stu2-dependent MT rescue (Supplementary Fig 3d), as did Dam1ΔC. Thus, Aurora B phospho-mimicking mutants of the Dam1 C-terminus and the Ndc80 N-tail caused CEN detachment from the MT end, without affecting CEN interaction with the MT lateral surface, probably due to suppression of functions of these Dam1 and Ndc80 domains.
Figure 6. Aurora B phospho-mimicking Dam1 and Ndc80 mutants show normal lateral KT–MT attachment, but subsequent detachment from the MT end.
(a–g) The cells shown in Fig 5c, d were treated as in Figs 2b–h and 3b (CEN3 under the GAL promoter was inactivated and then reactivated), except that NAA was added 30 min before image acquisition. a–f shows percentage of cells at each step of KT–MT interaction (as in Fig 2c–h). n= 25, 25, 30 and 30 cells were analysed in a–d, respectively. Data represent one out of two independent experiments. g shows an example of CEN3 detachment from the MT end in an ndc80N-7D[AurB] dam1C-4D[AurB] cell.
(h) The Dam1 and Ndc80 interaction, detected by the two-hybrid assay, is disrupted by Aurora B phospho-mimicking mutations at the Dam1 C-terminus (Dam1C-4D[AurB]). Duo1 is a component of the Dam1c.
In addition to Aurora B, Mps1 kinase is also required for the error correction to achieve chromosome bi-orientation 33,34. The substrates of Mps1 for this process have not yet been identified in yeast. Nevertheless Mps1 phosphorylates the Dam1 C-terminus at different sites from Aurora B phosphorylation 35. We made and tested phospho-mimicking Dam1 mutants at Mps1 sites, but they did not show the above defects found with dam1C-4D[AurB] (Supplementary Fig 4).
Physiological Aurora B activity is sufficient to promote KT detachment from the MT plus end but does not affect the lateral attachment
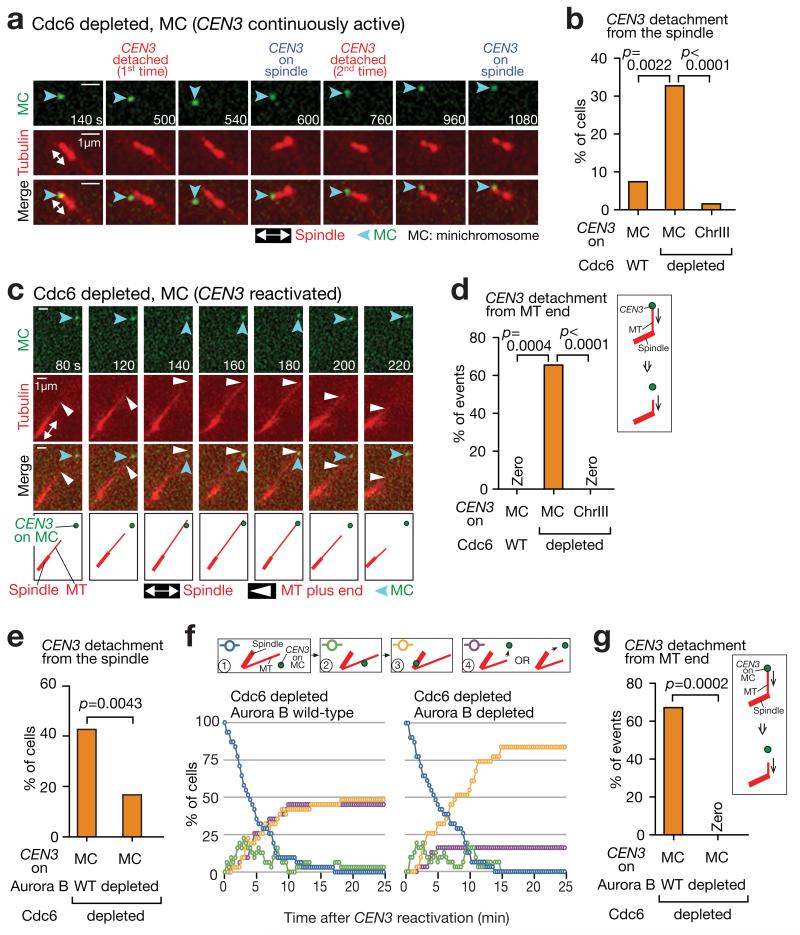
In the above experiments, we used phospho-mimicking mutations of Dam1 and Ndc80 at Aurora B phosphorylation sites. These mutations may represent their hyper-phosphorylation states. Next we aimed to study whether the physiological Aurora B activity is sufficient to cause CEN detachment from the MT end. However, CEN didn’t detach from the MT end during end-on pulling in Aurora B wild-type cells (see Fig 3c). We speculated KT detachment may occur from the MT end in Aurora B wild-type cells, but its detection may be difficult for two reasons: First, during end-on pulling, CEN is pulled towards a spindle pole with telomeres (on the same chromosome) trailing 3. Here the viscosity of the nucleoplasm may impose a weak force onto the trailing chromosome arms, generating a weak tension on the KT–MT interface. In addition, once the CEN is on the spindle, chromosome arms may collide with other chromosome arms, making the CEN less mobile after detaching from the spindle, and thus CEN detachment difficult to detect. In either case, use of a minichromosome (MC) may change the outcome, as it has minimal chromosome arms. Second, when a KT detaches from the MT end, its sister KT may still maintain MT attachment; for example, its sister KT may attach to the lateral surface of the same MT, close to the MT end. If so, we may detect detachment of unreplicated CEN from the MT end.
To test these two possibilities, we investigated the behaviour of a circular MC (~18 kb) containing CEN3, after inhibiting DNA replication initiation by Cdc6 depletion. Intriguingly, the unreplicated MC showed a higher rate of detachment from the metaphase spindle, followed by quick recapture on it, than two controls; a replicated MC, and an unreplicated CEN3 on chromosome III (Fig 7a,b). This suggests that, to detect the KT detachment with wild-type Aurora B, the KT should be on an unreplicated CEN that is on a MC. Next, to investigate MC interaction with individual MTs, we inactivated CEN3 (under the GAL promoter) on the unreplicated MC, and then reactivated it during metaphase arrest (similarly to Fig 2a). The MC was caught efficiently on the MT lateral side (see Fig 7f, Aurora B wild-type), but showed frequent detachment from the MT plus end (Fig 7c,d). To address whether Aurora B causes this detachment, we depleted Aurora B using an auxin-dependent degron tag. Indeed, Aurora B depletion led to reduction of the MC detachment from the spindle when CEN3 on the MC was always active (Fig 7e). When CEN3 on the MC was inactivated and then reactivated, Aurora B depletion did not change the kinetics in the lateral KT–MT attachment (Fig 7f, decline of blue line), but suppressed MC detachment from the MT end (Fig 7f, purple line; g). Thus, physiological Aurora B activity is sufficient to promote KT detachment from the MT end, but does not affect the lateral attachment.
Figure 7. Aurora B facilitates detachment of an unreplicated minichromosome from the MT end without changing kinetics of its lateral MT attachment.
(a, b) An unreplicated minichromosome (MC) frequently shows detachment from, and recapture on, the spindle. cdc6-anchor-away TetR-3×CFP GFP-TUB1 PMET3-CDC20 cells with pT111 (MC with PGAL-CEN3-tetOs) (T11561) or PGAL-CEN3-tetOs on chromosome III (T11629) were arrested in metaphase after Cdc6 depletion. Incubation with glucose kept CEN3 always active. Cells, like T11561, but with wild-type CDC6 (T11562) were also analyzed. Images were acquired every 10 sec. a shows a representative T11561 cell. In b, n= 51, 48 and 50 cells were analysed (left to right).
(c, d) An unreplicated MC frequently shows detachment from the MT plus end. The cells in b were treated as above, but CEN3 under the GAL promoter was inactivated and subsequently reactivated. c shows a representative T11561 cell. In d, n= 11, 21 and 26 end-on attachment events were analysed (left to right).
(e) Aurora B (Ipl1) depletion reduces detachment of an unreplicated MC from the spindle. IPL1 wild-type (T11695) and ipl1-321-aid (T11694) cells with TIR (otherwise like T11651; see a), were treated as in a (CEN3 was always active), except that NAA was added to deplete Ipl1; n= 68 and 48 cells were analysed for T11695 and T11694, respectively.
(f, g) Aurora B depletion reduces detachment of an unreplicated MC from the MT end without changing the kinetics of lateral MT attachment. T11695 and T11694 cells were treated as in c (CEN3 was inactivated and reactivated), except that NAA was added as in e. f shows percentage of cells at each step of KT–MT interaction. Purple lines show cumulative percentages of detachment from the spindle or the individual MTs (subsequent MT re-attachment was not counted). g shows percentage of MC detachment from the MT ends during end-on attachment. n= 31 and 31 cells (f) and n= 29 and 11 end-on pulling events (g) were analysed for T11695 and T11694, respectively. In b, d, e and g, experiments were performed twice (statistics source data are shown in Supplementary Table 2) and a representative experiment is shown here. p-values (two tailed) were obtained by Fisher’s exact test.
Physiological phosphorylation of the Ndc80 N-tail and Dam1 C-terminus by Aurora B promotes KT detachment from the MT plus end
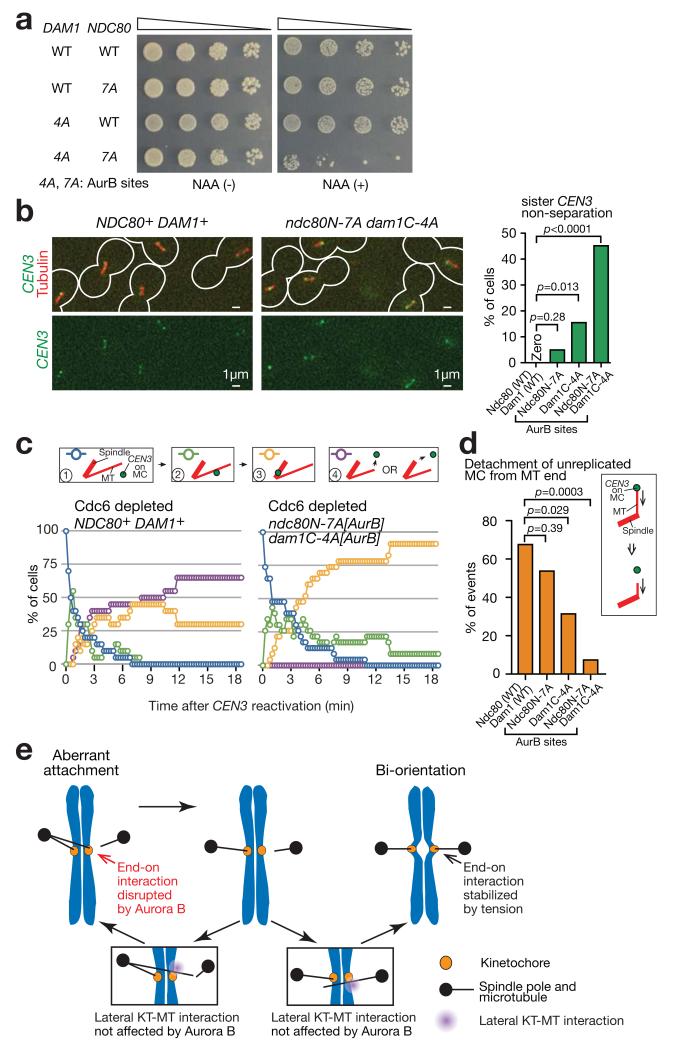
To address the effect of physiological phosphorylation of the Dam1 C-terminus and Ndc80 N-tail by Aurora B, we generated non-phosphorylatable mutations of Dam1 and Ndc80 (dam1C-4A[AurB] and ndc80N-7A[AurB]; see Fig 8a legend). The dam1C-4A[AurB] ndc80N-7A[AurB] double mutant showed a severe growth defect, when Dam1-aid was depleted in the presence of auxin (Fig 8a; dam1-aid was integrated to maintain cell viability, as in Fig 5a,b). When a chosen CEN (CEN3) was visualized, in 45 % of dam1C-4A[AurB] ndc80N-7A[AurB] cells sister CEN3s stayed together without separation in the vicinity of one spindle pole in metaphase, suggesting a bi-orientation defect (Fig 8b). Thus, Aurora B phosphorylation of the Dam1 C-terminus and Ndc80 N-tail is essential for bi-orientation.
Figure 8. Non-phosphorylatable Dam1 and Ndc80 mutants at Aurora B sites reduce the detachment rate of a KT from the MT plus end.
(a) Serially diluted cells were incubated with or without auxin NAA. In Dam1C-4A[AurB], four Aurora B target sites (S257, S265, S292 and S327 18) were replaced with alanines in the Dam1 C-terminus. In Ndc80N-7A[AurB], seven Aurora B target sites (T21, S37, T54, T71, T74, S95 and S100 19) were replaced with alanines at the Ndc80 N-tail.
(b) NDC80+ DAM1+ (T10638), ndc80N-7A[AurB] (T11813), dam1C-4A[AurB] dam1-aid (T11795) and ndc80N-7A[AurB] dam1C-4A[AurB] dam1-aid (T11794) cells with TIR (except for T11813) PGAL-CEN3-tetOs TetR-3×CFP GFP-TUB1 PMET3-CDC20 were treated and analysed as in Fig 5c; i.e. arrested in metaphase with glucose (CEN3 was always active). Representative images (of T10638 and T11794) and percentages of sister CEN3 non-separation (n= 39, 61, 45, 31 cells were analysed, respectively) are shown.
(c, d) NDC80+ DAM1+ (T11675), ndc80N-7A[AurB] (T11788), dam1C-4A[AurB] (T11790) and ndc80N-7A[AurB] dam1C-4A[AurB] (T11811) with TIR dam1-aid (except for T11788) cdc6-anchor-away TetR-3×CFP GFP-TUB1 PMET3-CDC20 and pT111 (MC with PGAL-CEN3-tetOs) were treated as in Fig 7f,g; i.e. Cdc6 was depleted, CEN3 under the GAL promoter was inactivated and subsequently reactivated, and Dam1-aid was depleted. c shows percentage of cells at each step of KT–MT interaction, as in Fig 7f. d shows percentages of MC detachment from the MT ends during end-on attachment. n= 24 and 23 cells (c) and n= 25, 24, 16 and 14 end-on pulling events (d) were analysed (left to right).
In b and d, experiments were performed three times or twice, respectively (statistics source data in Supplementary Table 2), and a representative experiment is shown here. p-values (two tailed) were obtained by Fisher’s exact test.
(e) Summary diagram. Aberrant KT–MT attachment (left) is dissolved through disruption of end-on attachment by Aurora B phosphorylation of KT components such as Dam1 and Ndc80. Subsequently lateral attachment is formed efficiently as this form of attachment is impervious to Aurora B regulation (insets). If this leads to establishment of bi-orientation (right), tension is applied across sister KTs and KT–MT attachment is stabilized. Alternatively, if that leads to aberrant attachment (left), error correction must continue to establish bi-orientation.
Next, using these mutants, we studied CEN3 interaction with individual MTs in two experiments: First, we inactivated CEN3 (using GAL promoter) on chromosome III and then reactivated it (as in Fig 2a). In dam1C-4A[AurB] ndc80N-7A[AurB] cells, CEN3 was caught on the MT lateral side with normal kinetics but subsequent establishment of bi-orientation was slower (Supplementary Fig 5). Second, as in Fig 7c, we depleted Cdc6 to inhibit DNA replication; and then inactivated CEN3 on the MC, and subsequently reactivated it, to observe interaction of an unreplicated MC with MTs. dam1C-4A[AurB] ndc80N-7A[AurB] cells showed normal kinetics of lateral KT-MT interaction (Fig 8c, decline of blue line) and suppressed MC detachment from the MT end (Fig 8c, purple line; d). dam1C-4A[AurB] cells also suppressed MC detachment, albeit modestly (Fig 8d). Thus, the physiological level of Aurora B phosphorylation of the Dam1 C-terminus and Ndc80 N-tail can cause KT detachment from the MT plus end without changing the kinetics of lateral KT–MT interaction.
Discussion
Error correction of the KT–MT attachment requires its turnover, i.e. the disruption of aberrant KT–MT attachments and formation of new ones. The disruption of KT–MT attachment is promoted by Aurora B phosphorylation of KT components that weakens KT–MT association 1,20. However, it has long been unclear how new attachments can still be formed, and errors corrected, despite attachment being weakened and disrupted by Aurora B. KT–MT interaction is initiated efficiently by lateral attachment 2,3, which presents a larger contact surface than the MT end, to the KT. It appears that this mode of attachment is impervious to Aurora B regulation (Fig 8e). Lateral attachment is converted to end-on attachment (but not vice versa) 4; end-on attachment is then weakened by Aurora B phosphorylation of KT components (unless bi-orientation is established) leading to KT detachment from the MT end. Thus, the differential regulation of the KT–MT attachment modes promotes the turnover of KT–MT interaction.
How does Aurora B specifically disrupt end-on attachment without affecting lateral attachment? The Dam1 C-terminus and the Ndc80 N-tail are known Aurora B targets whose phosphorylation is important for bi-orientation 18-20. These domains promote distinct steps in the conversion from the lateral to end-on attachment (see Fig 4e). Aurora B phosphorylation of these domains suppresses their functions, causing KT detachment from the MT plus end. The Dam1c accumulates at the MT end and this Dam1c fraction plays an important role in the end-on KT–MT interaction 4,12-15,36. The Dam1c also localizes along the MT lattice 4,16,17 but this fraction seems important neither for lateral attachment nor for Aurora B-dependent KT–MT regulation. Indeed, neither the Dam1 C-terminus nor the Ndc80 N-tail is required for the lateral KT–MT attachment. Presumably, the calponin-homology domains of Ndc80c are sufficient for lateral attachment, which is not altered by Aurora B activity.
KT detachment from the MT end was demonstrated using Dam1 and Ndc80 phospho-mimicking mutants. Such detachment was also promoted by physiological phosphorylation of Dam1 and Ndc80, but it could only be detected when the KT was on an unreplicated MC (see Fig 7 a–d). Why do we need an unreplicated MC to observe KT detachment in physiological conditions? Presumably, in physiological conditions, the level of phosphorylation by Aurora B does not reach 100%, in contrast to a phospho-mimicking state; thus, to observe KT detachment, we need conditions, such as an unreplicated MC, where a lower phosphorylation level suffices KT detachment. The level of physiological phosphorylation may be adjusted to prevent sister KTs from frequently undergoing simultaneous dissociation from the MT (a single MT in the above context and different MTs in syntelic attachment 1). If so, this would explain how cells avoid generating many unattached KTs (i.e. both sister KTs unattached to MTs) by Aurora B activity, but rather use its activity for error correction by maintaining MT attachment at least to one sister KT.
During error correction of KT–MT interaction, the lateral attachment is formed without interference. But, when it is converted to end-on attachment, the KT is released from the MT end by the action of Aurora B if tension is not applied (low-tension state). This process may repeat but, if bi-orientation is established and tension is applied across sister KTs, the end-on attachment is stabilized (high-tension state). However, a transition from low– to high–tension state still remains a mystery. For example, stable KT–MT end-on attachment and tension are mutually dependent on each other. Thus, it seems difficult to acquire tension and stable attachment when neither of them is present (initiation problem of bi-orientation) 37. Intriguingly, we find that conversion from lateral to end-on attachment may provide a solution to this problem (Zhang, et al, manuscript in preparation).
In metazoan cells, KT–MT interaction is also initiated by lateral attachment, which is then converted to end-on attachment 2. Our conclusion that differential regulation of lateral and end-on attachment promotes error correction in budding yeast may be essentially applied in metazoan cells, although the Aurora B targets are not exactly the same 20. In metazoan cells, the Ndc80 N-tail plays a more important role in facilitating the end-on attachment, and its phosphorylation seems more important for bi-orientation 23,24,27,38,39, than it is in budding yeast 19,25,40. The metazoan Ska1 complex (Ska1c), a functional equivalent to yeast Dam1c, also assists the end-on attachment, and its assembly is under negative regulation by Aurora B 41-43. Given that Ndc80c and Ska1c do not seem to be involved in lateral KT–MT attachment 42,44,45, we speculate that end-on, but not lateral, attachment is regulated by Aurora B in metazoan cells, similarly to budding yeast.
Methods
Yeast strains and cell culture
The background of yeast strains (W303; K699 and K700 from Kim Nasmyth lab), the methods for yeast culture and α factor arrest have all been described previously 4,46. Unless otherwise stated, cells were cultured at 25°C in YPA medium containing 2% glucose (YPAD). To activate the GAL promoter, cells were pre-incubated in medium containing 2% raffinose (without glucose) at least for 3 h, and subsequently incubated in medium containing both 2% galactose and 2% raffinose. To suppress the GAL promoter (without subsequent activation), cells were incubated in medium containing 2% glucose. To activate the MET3 promoter, cells were incubated in methionine drop-out media. To suppress it, 2mM methionine was added to the relevant media. Constructs of CEN5-tetOs47, PGAL-CEN3-tetOs 3,29,48, TetR-3×CFP 48, 49, PMET3-CDC20 50, Venus-TUB1 51, GFP-TUB1 52, PGAL-TEV 50 were described previously. STU2, MTW1 and ASK1 genes were tagged with 4×mCherry at their C-terminus at their original gene loci using a one-step PCR method and the pT909 plasmid 51 as a PCR template. The pT111 minichromosome was constructed by inserting tetOs (224 copies) 48 into the pGAL-CEN3 plasmid 29. The dam1 mutants and ndc80 mutants were constructed as explained in the next section. Mutants of Aurora B (ipl1 in budding yeast) and cdc6 were constructed as explained in the section after next. Two-hybrid assays, and plating after serial dilution of cells, were as described previously 51, and repeated at least twice to confirm reproducibility. Genotypes of yeast strains, used in this study, are shown in Supplementary Table 1.
Construction of ndc80 and dam1 mutants
ndc80ΔN: an ndc80 gene with deletion of the N-terminus (1–112 amino acids) was generated by a reverse PCR. A single copy of the construct was inserted at the his3 locus (while the original NDC80 gene was deleted) or it replaced the original NDC80 gene at its original locus. Ndc80ΔN protein was expressed at a similar level to that of wild-type protein.
PCUP1-ubi-DHFR-ndc80 (ndc80-td): a heat-inducible degron tag 53 was added at the N-terminus of the NDC80 gene at its original gene locus, using a one-step PCR method, as described previously 51.
ndc80-CH-K6A: an ndc80 gene, in which lysines at 122, 152, 160, 181, 192, 204 were replaced with alanines, was constructed by mutagenesis. These lysines correspond to those in the human Ndc80 calponin-homology domain, which interact with the MT surface 23. The ndc80-CH-K6A mutant was lethal. Therefore, to maintain cell viability, a single copy of the ndc80-CH-K6A construct was inserted at his3 locus while the original NDC80 gene was replaced with ndc80-td.
ndc80N-7D: an ndc80 gene, in which T21, S37, T54, T71, T74, S95 and S100 were replaced with aspartates, was constructed by gene synthesis (by DNA 2.0). These serines and threonines were the sites of phosphorylation by Aurora B 19. The mutant construct replaced the original NDC80 gene at its original locus.
dam1ΔC: the C-terminus (206–343 amino acids) of DAM1 gene was deleted at its original locus using the one-step PCR method.
dam1-aid: DAM1 gene was fused with an auxin-inducible degron tag 54 at their C-terminus at its original gene locus, using the one-step PCR method, as described previously 54. To deplete Dam1-aid, cells with the TIR gene 54 were incubated with auxin NAA (0.5 mM on plate, 1 mM in liquid media).
dam1-TEVsites: A dam1 gene with two tandem copies of TEV protease cleavage sites (ENLYFQG×2) 50 inserted between Q205 and V206 was generated by an overlap PCR. A single copy of the construct was inserted at leu2 locus while the original DAM1 was deleted.
dam1C-4D[AurB]: a dam1 gene, in which serines at 257, 265, 292 and 327 were replaced with aspartates, was constructed by gene synthesis (by DNA 2.0). S257, S265 and S292 were identified as the sites of phosphorylation by Aurora B 18 while S327 matches a consensus site of Aurora B phosphorylation 18. A single copy of the mutant construct was inserted at an auxotroph locus while the original DAM1 was replaced with dam1-aid.
dam1C-4D[Mps1], dam1C-8D[AurB+Mps1]: these dam1 mutants were generated in the same way as above, but at different phosphorylation sites. In dam1C-4D[Mps1], T217, S218, S221 and S232 (Mps1 phosphorylation sites 35) were replaced with aspartates. In dam1C-8D[AurB+Mps1], T217, S218, S221, S232, S257, S265, S292 and S327 were replaced with aspartates.
ndc80N-7A: an ndc80 gene, in which T21, S37, T54, T71, T74, S95 and S100 were replaced with alanines, was constructed by gene synthesis (by DNA 2.0). The mutant construct replaced the original NDC80 gene at its original locus.
dam1C-4A[AurB]: a dam1 gene, in which serines at 257, 265, 292 and 327 were replaced with alanines, was constructed by gene synthesis (by DNA 2.0). A single copy of the mutant construct was inserted at an auxotroph locus or at NDC80 locus (with ndc80N-7A) while the original DAM1 was replaced with dam1-aid.
Western blotting was carried out, as described previously 30, to evaluate protein expression in dam1 mutants (Figs 1c and 5a; full scans of blots are shown in Supplementary Fig 6). Dam1 proteins were detected using an anti-Dam1 affinity-purified polyclonal antibody 32. The experiment was repeated at least twice to confirm reproducibility.
Depletion of Cdc6 and Aurora B (Ipl1 in budding yeast)
cdc6 anchor away: To deplete Cdc6 protein, we used an anchor away system 55, which consists of cdc6-1-FRB, RPL13A-2×FKBP12, TOR1-1 and fpr1Δ. In the presence of rapamycin (10 μM), Cdc6 protein binds Rpl13A ribosomal protein due to the FRB-FKBP12 interaction, which leads to depletion of Cdc6 in the nucleus. We found that, by combining cdc6-1 temperature sensitive mutant with the anchor away and by exposing cells to a high temperature, Cdc6 depletion or inactivation was more thorough (even if the temperature was shifted down afterwards). In cdc6-anchor-away PMET3-CDC20 cells, Cdc6 was depleted and inactivated as follows: Cells were arrested in metaphase, and then released from it, by suppressing and reactivating the MET3 promoter. Upon release, rapamycin was added to deplete Cdc6. One hour later, the MET3 promoter was suppressed again to arrest cells in metaphase. In this procedure, cells were incubated at a high temperature (35°C) for 1.5 h (for 30 min prior to the first release from metaphase arrest and for a further 1 h after the release) and subsequently incubated at 25°C before and during image acquisition (because image quality was better at 25°C than at 35°C). A lack of appreciable DNA replication was confirmed by FACS DNA content analysis after this procedure of Cdc6 depletion and inactivation. To inactivate and then reactivate CEN3 under the GAL promoter during the procedure of Cdc6 depletion, cells were incubated with galactose after release from the first metaphase arrest, and then incubated with glucose, prior to image acquisition, after the second metaphase arrest.
ipl1-321-aid: ipl1-321 mutant gene 56 was fused with an auxin-inducible degron tag 54 at the C-terminus, in its original gene locus, using the one-step PCR method, as described previously 54. To deplete Ipl1-321-aid, cells with the TIR gene 54 were incubated with auxin NAA (0.5 mM on plate, 1 mM in liquid media). In Fig 7 e-g, NAA was added to deplete Ipl1-321-aid upon release from the first metaphase arrest (refer to the above procedure of Cdc6 depletion). Note that ipl1-321-aid showed higher sensitivity to NAA at 25°C than ipl1-aid (i.e. IPL1 wild-type gene fused with an auxin-inducible degron tag).
Live-cell imaging and image analyses
The procedures for time-lapse fluorescence microscopy were described previously57. Time-lapse images were collected at 25 °C unless otherwise stated. For image acquisition, we used a DeltaVision RT or Core microscope (Applied Precision), an UPlanSApo 100× objective lens (Olympus; NA 1.40), SoftWoRx software (Applied Precision), and either a CoolSnap HQ (Photometrics) or Cascade II 512B (Roper Scientific) CCD camera. We acquired 7–9 (0.5–0.7 μm apart) z-sections, which were subsequently deconvoluted, projected to two-dimensional images and analyzed with SoftWoRx and Volocity (Improvision) software. CFP, GFP, and mCherry signals were discriminated using the 89006 multi-band filter set (Chroma). In Fig 2c–f, Fig 6a–d, Fig 7c–d and Supplementary Fig 5, 1) cells with free CEN3 (not on MTs or the spindle) were 80–85% (Fig 2c–f), 60–70% (Fig 6a–d) and 80–90% (Fig 7c–d, Supplementary Fig 5) of the whole cell population at 0 min and we analysed what percentage of these cells showed which steps of KT–MT interaction along the time course, and 2) purple lines show cumulative percentage of CEN3 detachment and, although this CEN3 detachment was followed by recapture by a MT, such recapture was not included in counts of other steps. In Fig 8b and Supplementary Fig 5, up to 30% of T11794 cells showed two CEN3 signals, presumably due to chromosome mis-segregation in a prior cell cycle; such cells were excluded from counting. Meanwhile, the presence and absence of PGAL-TEV and TIR did not make a significant difference in NDC80+ DAM1+ and ndc80ΔN DAM1+ cells in the relevant experiments. Statistical analyses were carried out using Prism (Graph Pad) software. In Fig 7d, Fig 7g and Fig 8d, end-on attachment was analysed only when it started more than 1 μm away from a spindle pole. We used Fisher’s exact test in Figs 1f, 4b,5d, 7b, 7d, 7e, 7g, 8b and 8d, and Supplementary Figs 1, 3d and 4; and a chi-square test for trends in Fig 3b. Statistical analyses were repeated at least twice to confirm reproducibility of the results (Supplementary Table 2).
Supplementary Material
Acknowledgements
We thank M. Gierlinski and Tanaka and Novak groups for discussion, E. Griffis and L. Clayton for reading manuscript, and A. Musacchio for advising ndc80-CH-K6A mutations. This work was supported by Wellcome Trust (096535, 083524, 097945), Medical Research Council (84678), EC FP7 MitoSys (241548), ERC advanced grant (322682), Cancer Research UK (A6996) and the Human Frontier Science Program (RGP0035-2009). M.K. received BBSRC studentship. T.U.T. is Wellcome Trust Principal Research Fellow.
We thank J.-F. Maure for constructing the T7427 strain and dam1C-4D mutant; N. Kobayashi for making and testing ipl1-321-aid mutant; R. Ciosk, J.E. Haber, M. Kanemaki, K Nasmyth, K.E. Sawin, R.Y. Tsien, F. Uhlmann, EUROSCARF and Yeast Resource Centre for reagents; S. Swift and A.F.M Ibrahim for technical help.
References
- 1.Tanaka TU. Kinetochore-microtubule interactions: steps towards bi-orientation. The EMBO journal. 2010;29:4070–4082. doi: 10.1038/emboj.2010.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rieder CL, Alexander SP. Kinetochores are transported poleward along a single astral microtubule during chromosome attachment to the spindle in newt lung cells. The Journal of cell biology. 1990;110:81–95. doi: 10.1083/jcb.110.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tanaka K, et al. Molecular mechanisms of kinetochore capture by spindle microtubules. Nature. 2005;434:987–994. doi: 10.1038/nature03483. [DOI] [PubMed] [Google Scholar]
- 4.Tanaka K, Kitamura E, Kitamura Y, Tanaka TU. Molecular mechanisms of microtubule-dependent kinetochore transport toward spindle poles. The Journal of cell biology. 2007;178:269–281. doi: 10.1083/jcb.200702141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tanaka TU, et al. Evidence that the Ipl1-Sli15 (Aurora kinase-INCENP) complex promotes chromosome bi-orientation by altering kinetochore-spindle pole connections. Cell. 2002;108:317–329. doi: 10.1016/s0092-8674(02)00633-5. [DOI] [PubMed] [Google Scholar]
- 6.Hauf S, et al. The small molecule Hesperadin reveals a role for Aurora B in correcting kinetochore-microtubule attachment and in maintaining the spindle assembly checkpoint. The Journal of cell biology. 2003;161:281–294. doi: 10.1083/jcb.200208092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lampson MA, Renduchitala K, Khodjakov A, Kapoor TM. Correcting improper chromosome-spindle attachments during cell division. Nature cell biology. 2004;6:232–237. doi: 10.1038/ncb1102. [DOI] [PubMed] [Google Scholar]
- 8.Westermann S, Drubin DG, Barnes G. Structures and functions of yeast kinetochore complexes. Annual review of biochemistry. 2007;76:563–591. doi: 10.1146/annurev.biochem.76.052705.160607. [DOI] [PubMed] [Google Scholar]
- 9.Tanaka TU, Desai A. Kinetochore-microtubule interactions: the means to the end. Current opinion in cell biology. 2008;20:53–63. doi: 10.1016/j.ceb.2007.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nogales E, Ramey VH. Structure-function insights into the yeast Dam1 kinetochore complex. J Cell Sci. 2009;122:3831–3836. doi: 10.1242/jcs.004689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Biggins S. The composition, functions, and regulation of the budding yeast kinetochore. Genetics. 2013;194:817–846. doi: 10.1534/genetics.112.145276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lampert F, Hornung P, Westermann S. The Dam1 complex confers microtubule plus end-tracking activity to the Ndc80 kinetochore complex. The Journal of cell biology. 2010;189:641–649. doi: 10.1083/jcb.200912021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tien JF, et al. Cooperation of the Dam1 and Ndc80 kinetochore complexes enhances microtubule coupling and is regulated by aurora B. The Journal of cell biology. 2010;189:713–723. doi: 10.1083/jcb.200910142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maure JF, et al. The Ndc80 loop region facilitates formation of kinetochore attachment to the dynamic microtubule plus end. Current biology : CB. 2011;21:207–213. doi: 10.1016/j.cub.2010.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lampert F, Mieck C, Alushin GM, Nogales E, Westermann S. Molecular requirements for the formation of a kinetochore-microtubule interface by Dam1 and Ndc80 complexes. J Cell Biol. 2013;200:21–30. doi: 10.1083/jcb.201210091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miranda JJ, De Wulf P, Sorger PK, Harrison SC. The yeast DASH complex forms closed rings on microtubules. Nat Struct Mol Biol. 2005;12:138–143. doi: 10.1038/nsmb896. [DOI] [PubMed] [Google Scholar]
- 17.Westermann S, et al. Formation of a Dynamic Kinetochore- Microtubule Interface through Assembly of the Dam1 Ring Complex. Molecular cell. 2005;17:277–290. doi: 10.1016/j.molcel.2004.12.019. [DOI] [PubMed] [Google Scholar]
- 18.Cheeseman IM, et al. Phospho-regulation of kinetochore-microtubule attachments by the Aurora kinase Ipl1p. Cell. 2002;111:163–172. doi: 10.1016/s0092-8674(02)00973-x. [DOI] [PubMed] [Google Scholar]
- 19.Akiyoshi B, Nelson CR, Ranish JA, Biggins S. Analysis of Ipl1-mediated phosphorylation of the Ndc80 kinetochore protein in Saccharomyces cerevisiae. Genetics. 2009;183:1591–1595. doi: 10.1534/genetics.109.109041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lampson MA, Cheeseman IM. Sensing centromere tension: Aurora B and the regulation of kinetochore function. Trends Cell Biol. 2011;21:133–140. doi: 10.1016/j.tcb.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ramey VH, et al. Subunit organization in the Dam1 kinetochore complex and its ring around microtubules. Mol Biol Cell. 2011;22:4335–4342. doi: 10.1091/mbc.E11-07-0659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wei RR, Al-Bassam J, Harrison SC. The Ndc80/HEC1 complex is a contact point for kinetochore-microtubule attachment. Nat Struct Mol Biol. 2007;14:54–59. doi: 10.1038/nsmb1186. [DOI] [PubMed] [Google Scholar]
- 23.Ciferri C, et al. Implications for kinetochore-microtubule attachment from the structure of an engineered Ndc80 complex. Cell. 2008;133:427–439. doi: 10.1016/j.cell.2008.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.DeLuca JG, et al. Kinetochore microtubule dynamics and attachment stability are regulated by Hec1. Cell. 2006;127:969–982. doi: 10.1016/j.cell.2006.09.047. [DOI] [PubMed] [Google Scholar]
- 25.Demirel PB, Keyes BE, Chaterjee M, Remington CE, Burke DJ. A redundant function for the N-terminal tail of Ndc80 in kinetochore-microtubule interaction in Saccharomyces cerevisiae. Genetics. 2012;192:753–756. doi: 10.1534/genetics.112.143818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cheeseman IM, Chappie JS, Wilson-Kubalek EM, Desai A. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 2006;127:983–997. doi: 10.1016/j.cell.2006.09.039. [DOI] [PubMed] [Google Scholar]
- 27.Miller SA, Johnson ML, Stukenberg PT. Kinetochore attachments require an interaction between unstructured tails on microtubules and Ndc80(Hec1) Current biology. 2008;18:1785–1791. doi: 10.1016/j.cub.2008.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sarangapani KK, Akiyoshi B, Duggan NM, Biggins S, Asbury CL. Phosphoregulation promotes release of kinetochores from dynamic microtubules via multiple mechanisms. Proc Natl Acad Sci U S A. 2013;110:7282–7287. doi: 10.1073/pnas.1220700110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hill A, Bloom K. Genetic manipulation of centromere function. Molecular and cellular biology. 1987;7:2397–2405. doi: 10.1128/mcb.7.7.2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gandhi SR, et al. Kinetochore-dependent microtubule rescue ensures their efficient and sustained interaction in early mitosis. Developmental cell. 2011;21:920–933. doi: 10.1016/j.devcel.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Howard J, Hyman AA. Growth, fluctuation and switching at microtubule plus ends. Nature reviews. Molecular cell biology. 2009;10:569–574. doi: 10.1038/nrm2713. [DOI] [PubMed] [Google Scholar]
- 32.Keating P, Rachidi N, Tanaka TU, Stark MJ. Ipl1-dependent phosphorylation of Dam1 is reduced by tension applied on kinetochores. J Cell Sci. 2009;122:4375–4382. doi: 10.1242/jcs.055566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maure JF, Kitamura E, Tanaka TU. Mps1 kinase promotes sister-kinetochore bi-orientation by a tension-dependent mechanism. Current biology. 2007;17:2175–2182. doi: 10.1016/j.cub.2007.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jelluma N, et al. Mps1 phophorylates Borealin to control Aurora B activity and chromosome alignment. Cell. 2008;132:233–246. doi: 10.1016/j.cell.2007.11.046. [DOI] [PubMed] [Google Scholar]
- 35.Shimogawa MM, et al. Mps1 phosphorylation of Dam1 couples kinetochores to microtubule plus ends at metaphase. Current biology. 2006;16:1489–1501. doi: 10.1016/j.cub.2006.06.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Westermann S, et al. The Dam1 kinetochore ring complex moves processively on depolymerizing microtubule ends. Nature. 2006;440:565–569. doi: 10.1038/nature04409. [DOI] [PubMed] [Google Scholar]
- 37.Zhang T, Oliveira RA, Schmierer B, Novak B. Dynamical scenarios for chromosome bi-orientation. Biophys J. 2013;104:2595–2606. doi: 10.1016/j.bpj.2013.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guimaraes GJ, Dong Y, McEwen BF, Deluca JG. Kinetochore microtubule attachment relies on the disordered N-terminal tail domain of Hec1. Current biology. 2008;18:1778–1784. doi: 10.1016/j.cub.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Welburn JP, et al. Aurora B phosphorylates spatially distinct targets to differentially regulate the kinetochore-microtubule interface. Molecular cell. 2010;38:383–392. doi: 10.1016/j.molcel.2010.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kemmler S, et al. Mimicking Ndc80 phosphorylation triggers spindle assembly checkpoint signalling. The EMBO journal. 2009;28:1099–1110. doi: 10.1038/emboj.2009.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Welburn JP, et al. The human kinetochore Ska1 complex facilitates microtubule depolymerization-coupled motility. Developmental cell. 2009;16:374–385. doi: 10.1016/j.devcel.2009.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gaitanos TN, et al. Stable kinetochore-microtubule interactions depend on the Ska complex and its new component Ska3/C13Orf3. The EMBO journal. 2009;28:1442–1452. doi: 10.1038/emboj.2009.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chan YW, Jeyaprakash AA, Nigg EA, Santamaria A. Aurora B controls kinetochore-microtubule attachments by inhibiting Ska complex-KMN network interaction. J Cell Biol. 2012;196:563–571. doi: 10.1083/jcb.201109001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hanisch A, Sillje HH, Nigg EA. Timely anaphase onset requires a novel spindle and kinetochore complex comprising Ska1 and Ska2. The EMBO journal. 2006;25:5504–5515. doi: 10.1038/sj.emboj.7601426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cheerambathur DK, Gassmann R, Cook B, Oegema K, Desai A. Crosstalk between microtubule attachment complexes ensures accurate chromosome segregation. Science. 2013;342:1239–1242. doi: 10.1126/science.1246232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Amberg DC, Burke DJ, Strathern JN. Methods in yeast genetics. 2005.
- 47.Tanaka T, Fuchs J, Loidl J, Nasmyth K. Cohesin ensures bipolar attachment of microtubules to sister centromeres and resists their precocious separation. Nature cell biology. 2000;2:492–499. doi: 10.1038/35019529. [DOI] [PubMed] [Google Scholar]
- 48.Michaelis C, Ciosk R, Nasmyth K. Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell. 1997;91:35–45. doi: 10.1016/s0092-8674(01)80007-6. [DOI] [PubMed] [Google Scholar]
- 49.Bressan DA, Vazquez J, Haber JE. Mating type-dependent constraints on the mobility of the left arm of yeast chromosome III. The Journal of cell biology. 2004;164:361–371. doi: 10.1083/jcb.200311063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Uhlmann F, Wernic D, Poupart MA, Koonin EV, Nasmyth K. Cleavage of cohesin by the CD clan protease separin triggers anaphase in yeast. Cell. 2000;103:375–386. doi: 10.1016/s0092-8674(00)00130-6. [DOI] [PubMed] [Google Scholar]
- 51.Maure J-F, et al. The Ndc80 loop region facilitates formation of kinetochore attachment to the dynamic microtubule plus end. Current biology. 2011;21:207–213. doi: 10.1016/j.cub.2010.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Straight AF, Marshall WF, Sedat JW, Murray AW. Mitosis in living budding yeast: anaphase A but no metaphase plate. Science. 1997;277:574–578. doi: 10.1126/science.277.5325.574. [DOI] [PubMed] [Google Scholar]
- 53.Dohmen RJ, Wu P, Varshavsky A. Heat-inducible degron: a method for constructing temperature-sensitive mutants. Science. 1994;263:1273–1276. doi: 10.1126/science.8122109. [DOI] [PubMed] [Google Scholar]
- 54.Nishimura K, Fukagawa T, Takisawa H, Kakimoto T, Kanemaki M. An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nature methods. 2009;6:917–922. doi: 10.1038/nmeth.1401. [DOI] [PubMed] [Google Scholar]
- 55.Haruki H, Nishikawa J, Laemmli UK. The anchor-away technique: rapid, conditional establishment of yeast mutant phenotypes. Molecular cell. 2008;31:925–932. doi: 10.1016/j.molcel.2008.07.020. [DOI] [PubMed] [Google Scholar]
- 56.Biggins S, et al. The conserved protein kinase Ipl1 regulates microtubule binding to kinetochores in budding yeast. Genes & development. 1999;13:532–544. doi: 10.1101/gad.13.5.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tanaka K, Kitamura E, Tanaka TU. Live-cell analysis of kinetochore-microtubule interaction in budding yeast. Methods. 2010;51:206–213. doi: 10.1016/j.ymeth.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kitamura E, et al. Kinetochores generate microtubules with distal plus ends: their roles and limited lifetime in mitosis. Developmental cell. 2010;18:248–259. doi: 10.1016/j.devcel.2009.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.