Abstract
Maintaining the proper lipid composition of cellular membranes is critical for numerous cellular processes but mechanisms of membrane lipid homeostasis are not well understood. There is growing evidence that membrane contact sites (MCSs), regions where two organelles come in close proximity to one another, play major roles in the regulation of intracellular lipid composition and distribution. MCSs are thought to mediate the exchange of lipids and signals between organelles. In this review, we discuss how lipid exchange occurs at MCSs and evidence for roles of MCSs in regulating lipid synthesis and degradation. We also discuss how networks of organelles connected by MCSs may modulate cellular lipid homeostasis and help determine organelle lipid composition.
Lipids are fundamental components of cellular membranes. Most eukaryotic cells contain complex mixtures of hundreds and often thousands of different lipid species [1]. The distribution of lipids among cellular compartments is tightly controlled. The lipid compositions of organelles vary dramatically. In addition, many organelles have asymmetric distributions of lipid across the two leaflets of their bilayers and lipids are often organized into domains within membranes [2]. How cells sense and regulate the complex mix and distribution of lipids in organelles is, therefore, an important and challenging question. Answering this question requires knowledge of how lipids are synthesized and degraded, how lipids traffic between cellular compartments, and how these processes are regulated.
Intracellular lipid trafficking occurs by both vesicular and nonvesicular mechanisms. There has been a growing interest in nonvesicular pathways, which are particularly important for mitochondria, plastids, and lipid droplets since they receive little or no lipid by vesicular trafficking. It is becoming increasingly clear that regions of close apposition of organelle membranes, often called membrane contact sites (MCSs), play critical roles in nonvesicular lipid trafficking [3, 4]. These zones, where organelles typically come within 30 nm of one another, are not only important for lipid trafficking but have long been known to play important roles in calcium and other signaling events. Recent findings have revealed additional functions of MCSs in organelle inheritance [5, 6], organelles division [7, 8] and autophagy [9, 10].
Here, we will focus on how MCSs affect cellular lipid homeostasis. First, we will discuss how lipid transfer at MCSs plays roles in lipid metabolism. Next, we will describe how lipid synthesis and degradation are regulated at MCSs. Finally, we will focus on lipid trafficking at MCSs between mitochondria and a variety of other organelles since recent findings suggest important new ways to think about how MCSs may participate in lipid homeostasis throughout the cell.
Lipid exchange at MCSs
The majority of nonvesicular lipid exchange between organelles probably occurs at MCSs. How might this lipid transfer occur and how would MCSs facilitated it? Lipid monomers can spontaneously move from one membrane bilayer to a second by desorbing from the first membrane, diffusing through the aqueous phase, and inserting into the second membrane. For most classes of lipids, however, the rates of spontaneous lipid exchange between membranes are much too slow to be physiologically relevant [11]. The rate-limiting step of this process is desorption of a lipid monomer from a bilayer. Lipid transport proteins (LTPs) facilitate lipid exchange between membranes in vitro by increasing rates of lipid desorption from membranes. LTPs have hydrophobic pockets or clefts that bind a lipid and often have lid-like domains that shield the bound lipid from the aqueous phase. Most lipid exchange at MCSs is probably facilitated by LTPs [12, 13].
There are a number of families of LTPs in cells. Most bind a single lipid monomer. However, it is possible that some LTPs bind more than one lipid at once. The synaptotagmin-like mitochondrial-lipid-binding protein (SMP) domain, which is found in a number of potential LTPs localized in different MCSs [14, 15], can bind two glycerolphospholipids simultaneously [•16]. The specificity of lipid binding by LTPs varies widely; some bind only a few similar lipid species while others appear to bind promiscuously to many types of lipids [13].
LTPs probably transfer lipids between membranes most efficiently at MCSs, where they have to diffuse only small distances between membranes or may even be able to transport lipids while interacting with both membranes simultaneously. Most LTPs contain at least two domains in addition to their core lipid-binding domain. These additional domains allow LTPs to interact with either proteins or lipids on the two membranes at MCSs. For example, some LTPs contain a FFAT (two phenylalanine in acid tract) motif that interacts with ER-resident proteins called VAPs (vesicle-associated membrane protein (VAMP)-associated proteins). Other LTPs are anchored in the ER by transmembrane domains. Binding of LTPs to the second organelle in a MCS is often mediated by membrane binding domains in the LTP, such as pleckstrin homology (PH) or C2 domains, or by the LTP interacting with proteins in the second membrane [17, 18]. It is likely that other targeting domains in LTPs remain to be discovered. The targeting domains of LTPs specify the MCSs where they will transfer lipids or perform other functions.
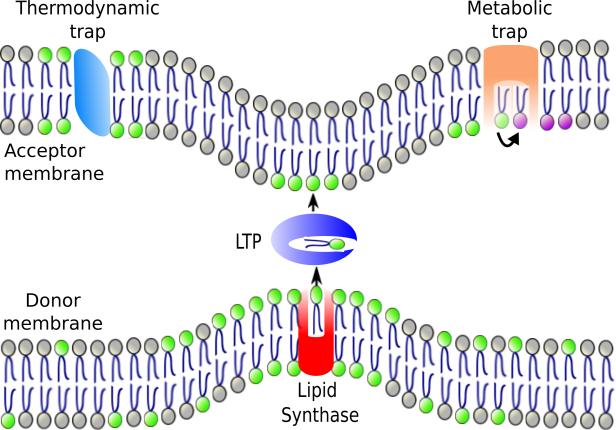
LTPs do not require energy to facilitate lipid exchange in vitro and presumably equilibrate the lipids they bind between the two membranes at MCSs [13]. This raises the question of how differences in lipid composition between the two membranes connected by MCSs could be maintained. One mechanism is trapping, in which a lipid is retained in one of the two membranes connected by MCSs (Figure 1). A lipid could be trapped when it is modified (metabolic trapping) or if the lipid has high affinity for a protein or another lipid in the membrane (thermodynamic trapping) [19]. For example, the movement of ceramide from the ER to the Golgi complex by ceramide transport protein (CERT) at contact sites is directional because ceramide is converted to sphingomyelin after it reaches the Golgi [17]. This is an example of metabolic trapping. Thermodynamic trapping probably maintains the high concentration of phosphatidylserine (PS) the inner leaflet of the plasma membrane. After PS is synthesized in the ER and transferred to the plasma membrane at MCSs by the LTPs Osh6 and Osh7 [20], it is probably held in the PM by interacting with charged proteins there.
Figure 1. Model of directional lipid exchange at an MCS.
A lipid synthase (red) localizes to an MCS where it produces a lipid (lipid with green headgroup). The enrichment of the lipid at MCS increases the likelihood that the lipid is transferred by an LTP at the MCSs. Alternatively, the synthase may hand the lipid directly to the LTP. Once the lipid reaches the second membrane it could be trapped there because of its affinity for a protein or lipid (thermodynamic trap) or by being modified (converted to a purple headgroup lipid; metabolic trap).
Recent evidence suggests another mechanism by which lipid exchange by LTPs at MCSs might be directional. A number of LTPs have the ability to sequentially bind either of two lipids in their hydrophobic pockets and could exchange the two lipids in opposite directions across a MCS [21-••23]. In this case, a difference in the concentrations of one of the lipids in the two membranes could drive the directional transport of the second lipid. For example, it has been proposed that oxysterol-binding protein (OSBP) facilitates the movement of cholesterol from the ER to the Golgi complex and phosphatidylinositol-4-phophate (PI4P) in the opposite direction at contact sites between these two organelles; the net transfer of cholesterol is driven by the high concentration of the PI4P in the Golgi complex and low concentration of PI4P in the ER [••23].
There is still much to learn about how lipids are exchanged at MCSs and how the specificity and directionality of lipid transfer at MCSs is regulated.
Role of MCSs in lipid modification and sensing
In addition to their roles in lipid trafficking, MCSs also have important functions in lipid metabolism. Although most lipid synthesis occurs in the ER, some lipid- modifying enzymes reside in other compartments [2]. As a result, the amount of lipid exchange between the ER and other organelles at MCSs could significantly affect lipid homeostasis by regulating the rate at which a lipid substrate synthesized in the ER reaches an enzyme in another compartment. An example is provided in studies on ER-plasma membrane contacts and their role in restoring phosphoinositide (PIP) levels in the plasma membrane following calcium signaling. Phosphatidylinositol (PI) is synthesized in the ER and can be converted to phosphoinositides (PIPs) in the plasma membrane. It has been found that calcium signaling induces contacts between the ER and plasma membrane, which may promote phosphatidylinositol (PI) transfer to the plasma membrane at these MCSs [24, 25]. Once the PI reaches the plasma membrane, it can be converted to PIPs. Thus, the formation of ER-plasma membrane contacts and the transfer of PI at these sites may regulate the rate of PIP formation in the plasma membrane.
Lipid synthesis at MCSs probably also has a significant effect on lipid homeostasis. Multiple studies have shown that various lipid-synthesizing enzymes are enriched at MCSs. For example PS synthase, PI synthase, and diacylglycerol cholinephosphotransferase are enriched in regions of the ER that are closely apposed to mitochondria or the plasma membrane [26-28]. Lipids produced at MCSs may be present at high concentrations at these sites, increasing the likelihood that the lipid will be transferred by an LTP to the apposing membrane (Figure 1). Indeed, it has been shown that newly synthesized PS is preferentially transferred from the ER to mitochondria where it is converted to phosphatidylethanolamine (PE) [29]. It may also be possible that lipid-synthesizing enzymes at MCSs hand lipids directly to LTPs, facilitating lipid transfer. Thus, there could be some form of substrate channeling from lipid synthesizing enzymes to LTPs at contact sites. Substrate channeling is the process by which the product of one enzyme is passed directly to a second enzyme.
Recent studies have revealed another way that lipid metabolism occurs at MCSs; enzymes located in the ER can modify lipids in the plasma membrane at regions of close contact between these organelles. In one report, the ER resident phosphatase Sac1 was found to dephosphorylate PIPs localized in the PM in yeast [30]. A second study suggested that Opi3, an enzyme in the ER that forms phosphatidylcholine (PC) from PE, can use PE in the plasma membrane as a substrate [31]. Remarkably, lipid-binding proteins localized at ER-plasma membrane MCSs regulate the activity of Sac1 and Opi3. Thus lipid sensing at MCSs may regulate plasma membrane lipid composition.
MCSs and lipid transfer to mitochondria
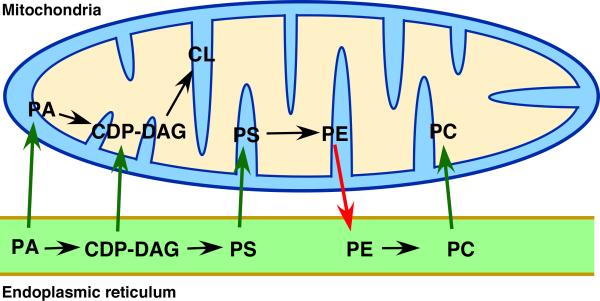
Mitochondria are thought to be endosymbiotic organelles that have evolved such that they retain the ability to synthesize some of the components necessary for their biogenesis but require the import of others from their “host” cells. Thus mitochondria have retained the ability to synthesize some lipids required for membrane biogenesis but other lipids must be imported from the rest of the cell; these include PI, PS, PC, and phosphatidic acid (Figure 2). The membrane lipids that are synthesized in mitochondria are PE, CDP-diacylglycerol, phosphatidylglycerol, and cardiolipin (CL) [32]. Some lipids synthesized in mitochondria are also transferred out of mitochondria to other organelles. For example, mitochondria are important sites of PE synthesis and this PE can be moved to the ER where it is converted to PC (Figure 2). It is thought that lipid exchange between the mitochondria and other organelles occurs at MCSs, particularly those between mitochondria and the ER, where most lipid biosynthesis occurs.
Figure 2. Lipid movement at the ER-mitochondria contacts.
Lipids that move from ER to mitochondria (green arrows) or from mitochondria to the ER (red arrows) are shown. Black arrows indicate conversion of one lipid to another. PA = phosphatidic acid; CDP-DAG = cytidine diphosphate diacylglycerol; PS = phosphatidylserine; PE = phosphatidylethanolamine; PC = phosphatidylcholine; CL = cardiolipin.
ER-mitochondria junctions are thought to be the major gateway for lipid exchange between mitochondria and the rest of the cell. We are still just beginning to understand how these close-contacts are established. A number of ER-mitochondria tethering complexes have been identified in mammalian cells but it is not currently known if they are required for lipid exchange between these organelles. An important breakthrough in our understanding of ER-mitochondrial tethering and lipid trafficking was the discovery of the ER-mitochondria encounter structure (ERMES), a complex that localizes to junctions between the ER and mitochondria [33]. ERMES is found in yeast but not in higher eukaryotes [34]. Interestingly, three of the proteins in ERMES contain lipid-binding SMP domains and thus could facilitate lipid exchange between the ER and mitochondria. Consistent with this possibility, cells lacking ERMES have reduced ER-mitochondria contacts [••35] and mitochondria from these cells have altered phospholipid composition including significantly reduced levels of PE and CL [33, 36-39]. It may be that the ERMES complex is involved in moving lipids in specific conditions, such as during mitochondrial division or mitophagy, in which the ERMES complex has been found to be involved [9, 40]. Alternatively, ERMES may not directly facilitate lipid exchange. In either case, it is clear that other complexes that tether the ER and mitochondria and facilitate lipid exchange remain to be identified.
A recent study from our group has indeed identified a second ER-mitochondria tethering complex in yeast [••35]. Mutants lacking this conserved complex, called the ER-membrane protein complex or EMC, were found in a genetic screen for strains with defects in mitochondrial PE synthesis. The EMC resides in the ER and tethers it to mitochondria by interacting with the Tom (translocase of the outer membrane) complex in the mitochondrial outer membrane. EMC mutants were also found to have reduced contact between the ER and mitochondria, altered mitochondrial lipid composition, and reduced rates of PS transfer from the ER to mitochondria. Interestingly, cells missing both the ERMES and EMC complexes were not viable and had a dramatic decrease in PS transfer to mitochondria. Both these defects were completely corrected by expressing an artificial ER-mitochondrial tether, indicating that both the ERMES and EMC together tether ER and mitochondria and mediate lipid trafficking between these organelles. How lipid exchange occurs at ER-mitochondrial junctions remains an interesting question for the future. Another important question is why multiple tethering complexes maintain ER-mitochondria contacts. Notably, work in yeast has indicated that multiple tethers also maintain ER-PM junctions [14], suggesting that multiple tethering complexes may be a common feature of many MCSs. These complexes probably have different functions, some of which may be unrelated to lipid transfer. Multiple tethers at MCSs may function like Velcro and maintain contacts by numerous weak interactions that can be rapidly formed and broken.
The finding that ER-mitochondria tethering is required for cell viability and necessary for efficient PS transfer from the ER to mitochondria is consistent with the long-held belief that lipid transfer occurs exclusively at ER-mitochondria junctions. However, this view has been challenged by two recent studies [••41, ••42]. They suggest that MCSs formed between mitochondria and vacuoles may form an alternative route for lipid transport to mitochondria. A complex named vCLAMP (vacuole and mitochondria patch) was found to tether mitochondria and vacuoles. Remarkably a mutant of missing both ERMES and vCLAMP is not viable and has a significant defect in PE and PC synthesis, suggesting that both ER-mitochondria and vacuole-mitochondria MCSs facilitate phospholipid transport to the mitochondria [••41]. As the ERMES and vCLAMP are synthetically lethal one can speculate a similar phenotype for the deletion of both EMC and vCLAMP complex as well, but this remains to be tested.
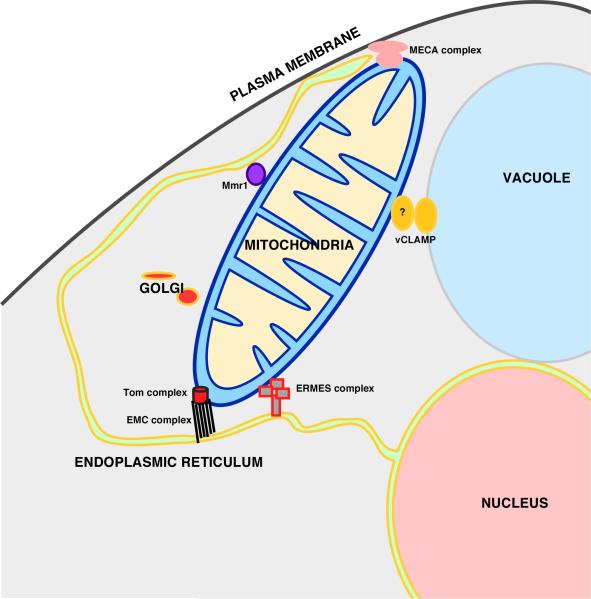
These findings raise the interesting possibility that mitochondria may exchange lipids with a number of organelles and that lipid trafficking pathways to mitochondria are redundant (Figure 3). Since mitochondria form MCSs not only with the ER and vacuole but the plasma membrane, the Golgi complex, and melanosomes [6, ••41-44], it seems possible that there is a complex network of inter-organelle lipid transfer pathways. This may well be true of other organelles as well.
Figure 3. Mitochondria centered interorganelle network in yeast.
MCSs formed between mitochondria and other organelles in yeast and the protein complexes found at these MCSs. Some of these complexes are described in the text. The other complexes are thought to tether mitochondria to various organelles but have not yet shown to mediate lipid exchange. Tthe mitochondria ER cortex anchored (MECA) complex tethers mitochondrial, plasma membrane, and the ER [6]. A complex containing Mmr1 and other unknown proteins tethers mitochondria to the ER [45].
Conclusions and future directions
A number of new components of MCSs have been identified in the last few years. However, we still lack mechanistic details about how lipids are transferred at MCSs and the roles of LTPs in these processes. It is also becoming clear that MCSs have a greater role in lipid homeostasis than simply being sites of lipid exchange. Understanding how MCSs function will require reconstitution of these sites in vitro, which will be a challenging task. Better methods of visualizing MCSs are also necessary, both for identifying new MCS components and for better understanding the regulation of MCS formation.
One of the most interesting discoveries in the last few years is that mitochondria obtain lipid not only from the ER but also from contacts with the vacuole and perhaps other organelles as well. This suggests that there may a complex network of connections between organelles by MCSs. Understanding how this network is formed and regulated will be an exciting direction for future studies.
Acknowledgements
This work was supported by the Intramural Research Program of the National Institute of Diabetes and Digestive and Kidney Diseases. We thank Orna Cohen-Fix and Amit Joshi for critically reading the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Shevchenko A, Simons K. Lipidomics: coming to grips with lipid diversity. Nat Rev Mol Cell Biol. 2010:11593–598. doi: 10.1038/nrm2934. [DOI] [PubMed] [Google Scholar]
- 2.van Meer G, Voelker DR, Feigenson GW. Membrane lipids: where they are and how they behave. Nat Rev Mol Cell Biol. 2008:9112–124. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Helle SC, Kanfer G, Kolar K, Lang A, Michel AH, Kornmann B. Organization and function of membrane contact sites. Biochim Biophys Acta. 2013:18332526–2541. doi: 10.1016/j.bbamcr.2013.01.028. [DOI] [PubMed] [Google Scholar]
- 4.Prinz WA. Bridging the gap: membrane contact sites in signaling, metabolism, and organelle dynamics. J Cell Biol. 2014:205759–769. doi: 10.1083/jcb.201401126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Knoblach B, Sun X, Coquelle N, Fagarasanu A, Poirier RL, Rachubinski RA. An ER-peroxisome tether exerts peroxisome population control in yeast. EMBO J. 2013:322439–2453. doi: 10.1038/emboj.2013.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lackner LL, Ping H, Graef M, Murley A, Nunnari J. Endoplasmic reticulum- associated mitochondria-cortex tether functions in the distribution and inheritance of mitochondria. Proc Natl Acad Sci U S A. 2013:110E458–67. doi: 10.1073/pnas.1215232110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Friedman JR, Lackner LL, West M, DiBenedetto JR, Nunnari J, Voeltz GK. ER tubules mark sites of mitochondrial division. Science. 2011:334358–362. doi: 10.1126/science.1207385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Friedman JR, Dibenedetto JR, West M, Rowland AA, Voeltz GK. Endoplasmic reticulum-endosome contact increases as endosomes traffic and mature. Mol Biol Cell. 2013:241030–1040. doi: 10.1091/mbc.E12-10-0733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bockler S, Westermann B. Mitochondrial ER contacts are crucial for mitophagy in yeast. Dev Cell. 2014:28450–458. doi: 10.1016/j.devcel.2014.01.012. [DOI] [PubMed] [Google Scholar]
- 10.Hamasaki M, Furuta N, Matsuda A, Nezu A, Yamamoto A, Fujita N, Oomori H, Noda T, Haraguchi T, Hiraoka Y, et al. Autophagosomes form at ER-mitochondria contact sites. Nature. 2013:495389–393. doi: 10.1038/nature11910. [DOI] [PubMed] [Google Scholar]
- 11.Sprong H, van der Sluijs P, van Meer G. How proteins move lipids and lipids move proteins. Nat Rev Mol Cell Biol. 2001:2504–513. doi: 10.1038/35080071. [DOI] [PubMed] [Google Scholar]
- 12.Drin G. Topological regulation of lipid balance in cells. Annu Rev Biochem. 2014:8351–77. doi: 10.1146/annurev-biochem-060713-035307. [DOI] [PubMed] [Google Scholar]
- 13.Lev S. Non-vesicular lipid transport by lipid-transfer proteins and beyond. Nat Rev Mol Cell Biol. 2010:11739–750. doi: 10.1038/nrm2971. [DOI] [PubMed] [Google Scholar]
- 14.Manford AG, Stefan CJ, Yuan HL, Macgurn JA, Emr SD. ER-to-plasma membrane tethering proteins regulate cell signaling and ER morphology. Dev Cell. 2012:231129–1140. doi: 10.1016/j.devcel.2012.11.004. [DOI] [PubMed] [Google Scholar]
- 15.Toulmay A, Prinz WA. A conserved membrane-binding domain targets proteins to organelle contact sites. J Cell Sci. 2012:12549–58. doi: 10.1242/jcs.085118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16•.Schauder CM, Wu X, Saheki Y, Narayanaswamy P, Torta F, Wenk MR, De Camilli P, Reinisch KM. Structure of a lipid-bound extended synaptotagmin indicates a role in lipid transfer. Nature. 2014:510552–555. doi: 10.1038/nature13269. [This study reveals that the SMP domain binds two lipids simultaneously in a hydrophobic tube and suggests how SMP domains can facilitate lipid exchange at MCSs.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hanada K, Kumagai K, Yasuda S, Miura Y, Kawano M, Fukasawa M, Nishijima M. Molecular machinery for non-vesicular trafficking of ceramide. Nature. 2003:426803–809. doi: 10.1038/nature02188. [DOI] [PubMed] [Google Scholar]
- 18.Schulz TA, Choi MG, Raychaudhuri S, Mears JA, Ghirlando R, Hinshaw JE, Prinz WA. Lipid-regulated sterol transfer between closely apposed membranes by oxysterol-binding protein homologues. J Cell Biol. 2009:187889–903. doi: 10.1083/jcb.200905007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holthuis JC, Menon AK. Lipid landscapes and pipelines in membrane homeostasis. Nature. 2014:51048–57. doi: 10.1038/nature13474. [DOI] [PubMed] [Google Scholar]
- 20.Maeda K, Anand K, Chiapparino A, Kumar A, Poletto M, Kaksonen M, Gavin AC. Interactome map uncovers phosphatidylserine transport by oxysterol-binding proteins. Nature. 2013:501257–261. doi: 10.1038/nature12430. [DOI] [PubMed] [Google Scholar]
- 21.de Saint-Jean M, Delfosse V, Douguet D, Chicanne G, Payrastre B, Bourguet W, Antonny B, Drin G. Osh4p exchanges sterols for phosphatidylinositol 4-phosphate between lipid bilayers. J Cell Biol. 2011:195965–978. doi: 10.1083/jcb.201104062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kono N, Ohto U, Hiramatsu T, Urabe M, Uchida Y, Satow Y, Arai H. Impaired alpha-TTP-PIPs interaction underlies familial vitamin E deficiency. Science. 2013:3401106–1110. doi: 10.1126/science.1233508. [DOI] [PubMed] [Google Scholar]
- 23••.Mesmin B, Bigay J, Moser von Filseck J, Lacas-Gervais S, Drin G, Antonny B. A four-step cycle driven by PI(4)P hydrolysis directs sterol/PI(4)P exchange by the ER-Golgi tether OSBP. Cell. 2013:155830–843. doi: 10.1016/j.cell.2013.09.056. [This study shows how OSBP can transfer PI4P and cholesterol in opposite directions at ER-Golgi contact sites and also function as a PI4P-regulated tether at these contacts.] [DOI] [PubMed] [Google Scholar]
- 24.Chang CL, Hsieh TS, Yang TT, Rothberg KG, Azizoglu DB, Volk E, Liao JC, Liou J. Feedback regulation of receptor-induced Ca2+ signaling mediated by E Syt1 and Nir2 at endoplasmic reticulum-plasma membrane junctions. Cell Rep. 2013:5813–825. doi: 10.1016/j.celrep.2013.09.038. [DOI] [PubMed] [Google Scholar]
- 25.Giordano F, Saheki Y, Idevall-Hagren O, Colombo SF, Pirruccello M, Milosevic I, Gracheva EO, Bagriantsev SN, Borgese N, De Camilli P. PI(4,5)P(2)-dependent and Ca(2+)-regulated ER-PM interactions mediated by the extended synaptotagmins. Cell. 2013:1531494–1509. doi: 10.1016/j.cell.2013.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gaigg B, Simbeni R, Hrastnik C, Paltauf F, Daum G. Characterization of a microsomal subfraction associated with mitochondria of the yeast, Saccharomyces cerevisiae. Involvement in synthesis and import of phospholipids into mitochondria. Biochim Biophys Acta. 1995:1234214–220. doi: 10.1016/0005-2736(94)00287-y. [DOI] [PubMed] [Google Scholar]
- 27.Pichler H, Gaigg B, Hrastnik C, Achleitner G, Kohlwein SD, Zellnig G, Perktold A, Daum G. A subfraction of the yeast endoplasmic reticulum associates with the plasma membrane and has a high capacity to synthesize lipids. Eur J Biochem. 2001:2682351–2361. doi: 10.1046/j.1432-1327.2001.02116.x. [DOI] [PubMed] [Google Scholar]
- 28.Stone SJ, Vance JE. Phosphatidylserine synthase-1 and -2 are localized to mitochondria-associated membranes. J Biol Chem. 2000:27534534–34540. doi: 10.1074/jbc.M002865200. [DOI] [PubMed] [Google Scholar]
- 29.Vance JE. Newly made phosphatidylserine and phosphatidylethanolamine are preferentially translocated between rat liver mitochondria and endoplasmic reticulum. J Biol Chem. 1991:26689–97. [PubMed] [Google Scholar]
- 30.Stefan CJ, Manford AG, Baird D, Yamada-Hanff J, Mao Y, Emr SD. Osh proteins regulate phosphoinositide metabolism at ER-plasma membrane contact sites. Cell. 2011:144389–401. doi: 10.1016/j.cell.2010.12.034. [DOI] [PubMed] [Google Scholar]
- 31.Tavassoli S, Chao JT, Young BP, Cox RC, Prinz WA, de Kroon AI, Loewen CJ. Plasma membrane--endoplasmic reticulum contact sites regulate phosphatidylcholine synthesis. EMBO Rep. 2013:14434–440. doi: 10.1038/embor.2013.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Horvath SE, Daum G. Lipids of mitochondria. Prog Lipid Res. 2013:52590–614. doi: 10.1016/j.plipres.2013.07.002. [DOI] [PubMed] [Google Scholar]
- 33.Kornmann B, Currie E, Collins SR, Schuldiner M, Nunnari J, Weissman JS, Walter P. An ER-mitochondria tethering complex revealed by a synthetic biology screen. Science. 2009:325477–481. doi: 10.1126/science.1175088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wideman JG, Gawryluk RM, Gray MW, Dacks JB. The ancient and widespread nature of the ER-mitochondria encounter structure. Mol Biol Evol. 2013:302044–2049. doi: 10.1093/molbev/mst120. [DOI] [PubMed] [Google Scholar]
- 35••.Lahiri S, Chao JT, Tavassoli S, Wong AKO, Choudhary V, Young BP, Loewen CJ, Prinz WA. A conserved endoplasmic reticulum membrane protein complex (EMC) facilitates phospholipid transfer from the ER to mitochondria. PLOS Biol. doi: 10.1371/journal.pbio.1001969. in press. [This study shows that a ER-mitochondrial tether formed by the interaction of the EMC complex and the Tom complex is, together with the ERMES complex, necessary for PS transfer from the ER to mitochondria.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Osman C, Merkwirth C, Langer T. Prohibitins and the functional compartmentalization of mitochondrial membranes. J Cell Sci. 2009:1223823–3830. doi: 10.1242/jcs.037655. [DOI] [PubMed] [Google Scholar]
- 37.Tamura Y, Onguka O, Hobbs AE, Jensen RE, Iijima M, Claypool SM, Sesaki H. Role for two conserved intermembrane space proteins, Ups1p and Ups2p, [corrected] in intra-mitochondrial phospholipid trafficking. J Biol Chem. 2012:28715205–15218. doi: 10.1074/jbc.M111.338665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tan T, Ozbalci C, Brugger B, Rapaport D, Dimmer KS. Mcp1 and Mcp2, two novel proteins involved in mitochondrial lipid homeostasis. J Cell Sci. 2013:1263563–3574. doi: 10.1242/jcs.121244. [DOI] [PubMed] [Google Scholar]
- 39.Voss C, Lahiri S, Young BP, Loewen CJ, Prinz WA. ER-shaping proteins facilitate lipid exchange between the ER and mitochondria in S. cerevisiae. J Cell Sci. 2012:1254791–4799. doi: 10.1242/jcs.105635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Murley A, Lackner LL, Osman C, West M, Voeltz GK, Walter P, Nunnari J. ER-associated mitochondrial division links the distribution of mitochondria and mitochondrial DNA in yeast. Elife. 2013:2e00422. doi: 10.7554/eLife.00422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41••.Elbaz-Alon Y, Rosenfeld-Gur E, Shinder V, Futerman AH, Geiger T, Schuldiner M. A dynamic interface between vacuoles and mitochondria in yeast. Dev Cell. 2014:3095–102. doi: 10.1016/j.devcel.2014.06.007. [DOI] [PubMed] [Google Scholar]
- 42••.Honscher C, Mari M, Auffarth K, Bohnert M, Griffith J, Geerts W, van der Laan M, Cabrera M, Reggiori F, Ungermann C. Cellular metabolism regulates contact sites between vacuoles and mitochondria. Dev Cell. 2014:3086–94. doi: 10.1016/j.devcel.2014.06.006. [These paper, together with ••41, shows that a complex tethers mitochondria to the vacuole in yeast and that this complex may facilitate lipid exchange and signaling between these organelles.] [DOI] [PubMed] [Google Scholar]
- 43.Daniele T, Hurbain I, Vago R, Casari G, Raposo G, Tacchetti C, Schiaffino MV. Mitochondria and melanosomes establish physical contacts modulated by Mfn2 and involved in organelle biogenesis. Curr Biol. 2014:24393–403. doi: 10.1016/j.cub.2014.01.007. [DOI] [PubMed] [Google Scholar]
- 44.Dolman NJ, Gerasimenko JV, Gerasimenko OV, Voronina SG, Petersen OH, Tepikin AV. Stable Golgi-mitochondria complexes and formation of Golgi Ca(2+) gradients in pancreatic acinar cells. J Biol Chem. 2005:28015794–15799. doi: 10.1074/jbc.M412694200. [DOI] [PubMed] [Google Scholar]
- 45.TC Swayne, Zhou C, Boldogh IR, Charalel JK, McFaline-Figueroa JR, Thoms S, Yang C, Leung G, McInnes J, Erdmann R, Pon LA. Role for cER and Mmr1p in anchorage of mitochondria at sites of polarized surface growth in budding yeast. Curr Biol. 2011:211994–1999. doi: 10.1016/j.cub.2011.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]