Abstract
Differential scanning calorimetry (DSC) studies of blood plasma are part of an emerging area of the clinical application of DSC to biofluid analysis. DSC analysis of plasma from healthy individuals and patients with various diseases has revealed changes in the thermal profiles of the major plasma proteins associated with the clinical status of the patient. The sensitivity of DSC to the concentration of proteins, their interactions with other proteins or ligands, or their covalent modifications underlies the potential utility of DSC analysis. A growing body of literature has demonstrated the versatility and performance of clinical DSC analysis across a range of biofluids and in a number of disease settings. The principles, practice and challenges of DSC analysis of plasma are described in this article.
Keywords: Differential scanning calorimetry, thermogram, plasma, clinical diagnostics
1. Introduction
Differential scanning calorimetry (DSC) is an established, highly sensitive thermal analysis technique. DSC has been broadly applied in the life sciences to measure the heat profiles of biomolecules with application to areas including protein engineering, biopharmaceutical formulation and the study of various biomolecular interactions such as protein-ligand and protein-protein interactions [1]. DSC normally uses highly purified biomacromolecules to provide detailed and precise thermodynamic information about their stability or reactivity. We describe here a less conventional application of DSC to the analysis of complex mixtures. DSC is sensitive to the extensive properties of a solution of macromolecules and therefore is useful for monitoring concentrations of components in mixtures. This makes possible the application of DSC to provide a signature denaturation profile (thermogram) of a complex mixture based on the thermal stability of its individual components. The DSC thermogram of a mixture provides a unique signature analogous to a spectrum of a multicomponent mixture, or to a chromatographic elution profile, but one that is based on a unique underlying physical property, thermal stability. The goal of DSC studies in such a case is not detailed thermodynamic analysis but rather a more qualitative profiling of complex samples. Thermograms of mixtures do retain quantitative features that may be used for statistical comparisons of samples. Recently DSC has been applied in this novel direction to the analysis of clinical biofluids. Table 1 shows publications in this emerging area of DSC analysis. The majority of the publications address the analysis of human blood plasma or serum with other studies exploring the use of cerebrospinal fluid and, most recently, brain tumors. The clinical areas of investigation are diverse encompassing a range of cancers, autoimmune diseases, infectious disease, chronic health conditions, healthy subjects, athletes and disease controls. These studies have shown that DSC can detect differences in thermograms of biological samples associated with health status. This presents the potential of DSC to complement existing clinical diagnostic approaches.
Table 1.
Summary of publications describing the application of DSC analysis of biofluids for clinical diagnostics
| Authors (Year) | Clinical sample |
Clinical setting | Reference |
|---|---|---|---|
| Garbett et al. (2006) | Plasma | Healthy controls, Lyme disease | [17] |
| Garbett et al. (2007) | Serum | Coronary artery disease | [8] |
| Garbett et al. (2007) | Plasma | Healthy controls, Cervical cancer, Lyme disease, Systemic lupus erythematosus, Rheumatoid arthritis | [6] |
| Garbett et al. (2008) | Plasma | Healthy controls, Lyme disease, Systemic lupus erythematosus, Rheumatoid arthritis | [5] |
| Garbett et al. (2009) | Plasma, Serum | Healthy controls, Endometrial cancer, Amyotrophic lateral sclerosis, Lung cancer, Ovarian cancer, Lyme disease, Systemic lupus erythematosus, Rheumatoid arthritis, Melanoma, Cervical cancer | [11] |
| Fish et al. (2010) | Plasma, Serum | Healthy controls, Systemic lupus erythematosus | [10] |
| Michnik et al. (2010) | Serum | Healthy controls, Chronic obstructive pulmonary disease | [18] |
| Chagovetz et al. (2011) | Cerebrospinal fluid | Glioblastoma multiforme, non-glioma neoplastic controls (carcinomatosis meningitis, CNS lymphoma or leukemia), non-neoplastic controls (head trauma, hydrocephalus, CSF leak) | [19] |
| Zapf et al. (2011) | Plasma | Healthy controls, Breast cancer | [20] |
| Ferencz et al. (2011) | Plasma | Healthy controls, Melanoma | [21] |
| Michnik (2011) | Serum | Healthy controls, Chronic obstructive pulmonary disease | [22] |
| Todinova et al. (2011) | Serum | Healthy controls, Multiple myeloma | [14] |
| Wisniewski et al. (2011) | Plasma, Serum | Healthy controls, Myositis, Rheumatoid arthritis, Systemic lupus erythematosus, Scleroderma, Cervical cancer, Melanoma, Lung cancer | [23] |
| Fekecs et al. (2012) | Plasma | Healthy controls, Melanoma | [24] |
| Todinova et al. (2012) | Plasma | Healthy controls, Colorectal cancer, Gastric cancer, non-cancerous inflammation controls | [15] |
| Mehdi et al. (2013) | Plasma | Healthy controls, Psoriasis | [25] |
| Rai et al. (2013) | Plasma | Healthy controls, Cervical cancer | [26] |
| Chagovetz et al. (2013) | Digested brain tumor tissue | Normal brain tissue controls, Brain tumor tissue | [27] |
| Krumova et al. (2013) | Serum | Schizophrenia | [4] |
| Michnik et al. (2013) | Serum | Athletes | [28] |
| Garbett et al. (2013) | Plasma | Renal function in type 1 diabetes | [13] |
| Moezzi et al. (2014) | Plasma | Healthy controls, Psoriasis | [29] |
| Garbett et al. (2014) | Plasma | Healthy controls, Cervical cancer | [3] |
DSC has a number of attributes that underlie its potential utility as a new diagnostic technology. DSC detects with high sensitivity small heat changes associated with thermally-induced events. It is a universal detector which does not rely on any form of labeling or derivatization of analytes. DSC also does not require any pre-analytical fractionation allowing the analysis of complex mixtures such as blood plasma. As was mentioned, DSC thermograms reflect the extensive properties of a sample and will therefore correspond to changes in the concentration of component proteins associated with clinical status. DSC is sensitive to any changes in the thermal stability of analytes. In the case of the plasma proteome, structural modifications to plasma proteins or changes in intermolecular interactions associated with a disease state will be sensitively detected by DSC. Changes in thermograms associated with disease conditions are typically manifested as temperature-shifted profiles. One hypothesis for disease-altered thermograms are that these could reflect the “interactome” concept [2]. This concept describes plasma as being composed of a network of protein-protein, peptide-protein and metabolite-protein interactions which one could envisage could be affected by the presence of disease biomarkers. These altered interaction networks would affect the denaturation properties of plasma and result in changes in DSC thermograms. DSC analysis of clinical samples has shown that, relative to control populations, thermograms are altered in shape and shifted to higher temperatures in a number of diseases. Further, thermograms have been shown to be sensitive to disease burden [3] as well as to patient therapy [4]. The growing body of literature applying DSC to different biological samples in an ever increasing number of diseases presents the exciting possibility that DSC might find a place as a useful clinical tool. The aim of this article is to outline the principles and practice of plasma thermogram analysis and to discuss the development of the approach in moving from basic research investigations to the clinical arena.
2. Principle of plasma thermograms
DSC is the method of choice for thermodynamic studies of protein denaturation, where temperature-induced unfolding of proteins can be directly measured. DSC monitors heat changes associated with the thermal denaturation of biomolecules providing a direct quantitative measurement of the thermostability of protein(s) in terms of a melting curve (or thermogram). DSC profiles (or thermograms) are unique for each biomolecule as a consequence of unique structural motifs and molecular forces. Under a given set of buffer conditions, every protein has a thermogram with a characteristic melting temperature (Tm) and denaturation enthalpy (ΔH) that provides a fundamental thermodynamic signature for that protein. DSC is typically applied to the analysis of a single purified biomolecule or a biomolecule with an interacting partner. Recently, the utility of DSC for the analysis of complex protein solutions has been explored in the area of clinical diagnostics. DSC thermograms are directly related to the mass of proteins present. For example, if the weight concentration of a protein is doubled, the calorimetric heat response will also double. Likewise, in a solution containing a mixture of proteins, the relative heat response will correspond to the total mass of proteins present. Fundamentally, this property constitutes the basis for DSC-based diagnostic applications since thermograms of protein mixtures can be deconvoluted into characteristic melting curves of individual protein components. In a non-interacting mixture, each protein will have a characteristic curve shape, Tm and ΔH. Thus, the thermogram observed for the mixture can be represented as the sum of all constituent individual protein thermograms weighted according to their relative molar mass and concentration. We have shown this to be the case for the plasma thermogram obtained from healthy subjects [5].
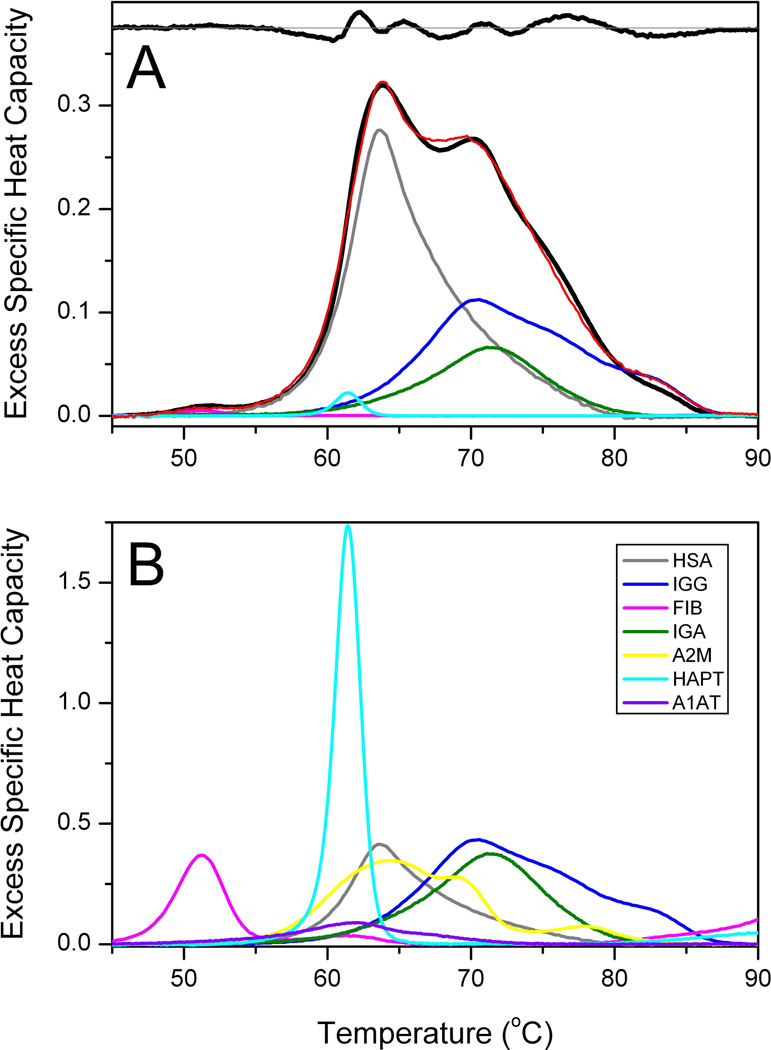
A normal thermogram and its deconvolution are shown in Figure 1. The normal thermogram in Figure 1A is an average obtained from plasma samples of 120 individuals. Figure 1B shows thermograms for seven of the most abundant pure plasma proteins, normalized on a g/L concentration scale. The observed experimental thermogram is the vector T with excess specific heat values measured over a range of temperatures. Deconvolution of T requires fitting to a vector of concentrations C using a matrix of pure reference proteins R. R is a rectangular matrix of excess specific heat capacities at each temperature for each protein component. The classic least-squares solution to this problem is given by
where R’ is the transpose of R. The experimental thermogram can be fit to a minimum of five protein components, as shown in Figure 1A. The deconvoluted contribution of human serum albumin (HSA), immunoglobulin G (IgG), fibrinogen, immunoglobulin A and haptoglobin are shown in Figure 1A, along with the residual plot inset at the top of the panel. Attempts to include more than these five proteins led to nonsensical fits characterized by negative concentration values. This indicates that only a limited number of unique component thermogram shapes can be resolved that contribute to the observed experimental thermogram. The experimental thermogram is dominated by contributions from HSA and IgG, but contributions from other components are needed to account for subtle features of the thermogram.
Figure 1.
(A) Average thermogram obtained from 120 plasma samples from healthy, normal individuals (black). The red curve is the deconvolution including five abundant plasma proteins. The residual plot (observed minus fit) is shown at the top of the panel. (B). Thermograms for seven of the most abundant pure plasma proteins. HSA, human serum albumin; IGG, immunoglobulin G; FIB, fibrinogen; IGA, immunoglobulin A; A2M, alpha-2-macroglobulin; HAPT, haptoglobin; A1AT, alpha-1-antitrypsin.
The power of DSC in characterizing binding interactions is exploited in its application to the study of the disease-state plasma interactome. Putative interaction of disease markers with the more abundant plasma proteins is akin to ligand-protein binding for which detailed protocols are firmly established for the analysis of ligand-induced shifts in denaturation thermograms. Changes in the thermogram as a result of binding interactions can be dramatic and can be related quantitatively to the binding constant and binding enthalpy of the biomarker. DSC thermograms have potential utility as a diagnostics tool because they are sensitive to changes in protein composition both in a non-interacting mixture and as a consequence of modifications or interactions with other components. An example will be shown later to illustrate this.
The aim of this article is to document necessary considerations for obtaining reproducible thermograms. An example of the utility of the approach will be provided.
3. Materials and methods
In order to obtain consistent, reproducible plasma thermogram, specific protocols must be zealously followed. A detailed, step-by-step standard operating procedure for obtaining thermograms is provided as Supporting Information. Additional comments relevant to the data presented follow.
3.1. Samples
DSC is considered a universal detector and as such is applicable to a wide array of samples. Table 1 is a testament to the versatility of DSC for the analysis of clinical samples. The majority of publications highlighted in Table 1 focused on the analysis of blood plasma or serum samples but the analysis of a range of other biological samples has been reported, including cerebrospinal fluid and digested tumor samples. This article will focus on DSC analysis of plasma/serum samples. Plasma/serum samples were purchased commercially for our studies, for example, pooled healthy control plasma was obtained from Sigma-Aldrich (St. Louis, MO) and used for the experiments examining pre-analytical and analytical factors on plasma thermograms (Section 4.1). Single donor healthy plasma was obtained from Innovative Research (Southfield, MI) and formed the majority of samples in our healthy control group. A large number of samples, including additional healthy controls as well as the majority of the disease samples we have analyzed, have been obtained through clinical collaborations. In this event, it is necessary to ensure that the studies undergo review and approval by the appropriate Institutional Review Boards before commencing and that all human subjects protocols and biosafety procedures are followed. As a result of reliance on clinical operations we have analyzed serum as well as plasma collected in the presence of different anticoagulants. We have previously shown that there is no substantial difference between the profile of serum or plasma except for fibrinogen, which is present in plasma but is removed through the clotting process for serum collection [6]. Fibrinogen can be unambiguously identified through the observation of a unique transition ~ 50 °C representing unfolding of one of its three domains.
3.2. Sample preparation
The plasma thermogram assay is simple and direct. No elaborate preparation steps are required and our standard procedure involves only four steps: (1) dialysis to normalize sample solution conditions; (2) sample filtration to remove particulates; (3) dilution to yield a suitable total protein concentration for DSC; and (4) direct analysis by DSC. Specifically, small volumes of plasma (100 microliters) were dialyzed against a standard physiological phosphate buffer (1.7 mM KH2PO4, 8.3 mM K2HPO4, 150 mM sodium chloride, 15 mM sodium citrate, pH 7.5) at 4 °C for 24 hours with four buffer changes. To effectively dialyze such small volumes of plasma we used Slide-A-Lyzer MINI dialysis devices (MWCO 3,500, 0.1 mL; Pierce, Rockford, IL) that were secured in 25-place floats, placed in a 2 liter beaker and equilibrated overnight against 1 liter of dialysis buffer in the cold room. The next morning the units were loaded with plasma samples and returned to the beaker containing dialysis buffer. Gentle stirring was employed to allow motion of the dialysis float and increase the diffusion rate during dialysis. The float was removed and placed in a beaker of fresh dialysis buffer after dialysis periods of three hours, four hours and four hours. The final dialysis period was allowed to proceed overnight before recovery of the samples and final dialysis buffer the next morning. Based on cost and reliability we routinely re-assembled washed dialysis units replacing the original dialysis membrane with cut-to-size SnakeSkin Dialysis Tubing (Pierce, Rockford, IL). Samples were filtered using a Spin-X centrifuge tube filter (0.45 µm cellulose acetate; Corning Incorporated, Corning, NY). The final dialysis buffer was filtered using 0.2 µm polyethersulfone filter disks (PALL Corporation, Port Washington, NY). Filtered samples were diluted 25-fold with final dialysis buffer to obtain a suitable concentration for DSC analysis.
3.3. Buffer exchange experiments
Buffer exchange experiments were performed using commercially obtained plasma samples. Pooled healthy control plasma was obtained from Sigma-Aldrich (St. Louis, MO) and single donor Lyme disease plasma obtained from BBI diagnostics (West Bridgewater, MA). Samples were divided into three groups: standard dialysis procedure (Section 3.2); no dialysis; and buffer exchange using Zeba™ Spin Desalting Columns (Pierce, Rockford, IL). Spin columns were prepared according to the manufacturer’s instructions for the buffer exchange procedure with 100µL of sample applied to each column. Samples were diluted 25-fold and 50-fold then analyzed in triplicate by DSC (Section 3.5). Results of a second independent experiment were found to be highly reproducible.
3.4. Equilibrium dialysis experiments
Equilibrium dialysis experiments were performed using commercially obtained plasma and HSA samples. Pooled healthy control plasma and HSA were obtained from Sigma-Aldrich (St. Louis, MO) and single donor Lyme disease plasma from BBI diagnostics (West Bridgewater, MA). Total protein concentrations of the plasma samples were measured before dialysis using the bicinchoninic acid protein assay (Section 3.5) and adjusted to equal concentrations by dilution with phosphate buffer. HSA was prepared at a concentration representing 62% of the plasma total protein concentration which equates to the typical HSA proportion in plasma. Samples were analyzed in two groups: no dialysis and dialysis using the 5-Cell Spectrum Equilibrium Dialyzer (Spectrum Labs, Los Angeles, CA) with SnakeSkin Dialysis Tubing (Pierce, Rockford, IL). Dialysis was performed in a pair-wise fashion loading 600 µL into each half-cell in the following combinations: Cell 1: Lyme vs. Buffer; Cell 2: Lyme vs. Healthy; Cell 3: Lyme vs. HSA; Cell 4: Healthy vs. Buffer; Cell 5: Healthy vs. HSA. Dialysis was performed at 4°C for 24 hours. Samples were recovered, diluted 25-fold and analyzed in duplicate by DSC (Section 3.5).
3.5. DSC measurements
DSC data have been collected with a number of different DSC systems but primarily, because of higher throughput, with the automated DSC systems: MicroCal VP-Capillary DSC (MicroCal LLC, Northampton, MA, now part of Malvern) and TA Instruments Nano DSC Autosampler System (TA Instruments, New Castle, DE). The DSC instruments used were serviced and calibrated according to the manufacturer’s procedures, for example see [3]. Plasma samples and matched dialysis buffer were transferred into 96-well plates and loaded into the instrument autosampler which was thermostated at 4 °C until loading into the DSC instrument for analysis. DSC scans were recorded from 20 °C to 110 °C at a scan rate of 1 °C/min with a pre-scan equilibration period of 900 seconds (and a mid feedback mode and filtering period of 2 s for the VP-Capillary DSC). Duplicate scans were recorded for each sample. DSC scans were processed using the instrument supplied software. First, scans were corrected for instrument baseline by subtraction of a suitable buffer reference scan. Next, corrected scans were normalized for the total protein concentration to allow direct comparison of samples. We employed a colorimetric total protein assay using the bicinchoninic acid protein assay kit and microplate procedure from Pierce (Pierce, Rockford, IL), with minor modifications to the incubation time included in the manufacturer’s protocol. Absorbance readings were taken using a Tecan Safire microplate reader (Tecan U.S., Research Triangle Park, NC). Finally, we corrected scans for non-zero baselines by application of a linear baseline fit using Origin 7 (OriginLab Corporation, Northampton, MA). Discussion of the choice of sample baseline appears in Section 4.4. Final thermograms were plotted as excess specific heat capacity (cal/°C.g) versus temperature (°C) for subsequent comparison and analysis. We have described previously our DSC procedure involving sample batching, to ensure completion of all analysis within seven days of sample thawing, and also run setup, to ensure scan reproducibility and effective cleaning of instrument chambers [3].
3.6. Data analysis
Our earlier published reports on plasma thermograms focused on qualitative comparison of profiles either through direct comparison of thermograms or through the construction of difference plots [3, 5]. We have employed another method to compare thermograms, through quantile-quantile plots [7]. To construct quantile-quantile plots, data sets were first interpolated into identical, equally spaced temperature intervals (45–90 °C at 0.1 °C intervals). Thermograms were integrated and then normalized to 1. Finally, heat capacity values were plotted in a pairwise fashion to discern differences between thermograms [13]. If there are no differences between groups then the data will fall on the 45° reference line. Deviations from this line indicate differences between groups which can be quantitated into an overall statistical measure such as a p-value. For more quantitative analysis of profiles, we have calculated a number of shape and feature metrics. Differences in thermogram metrics between clinical groups were tested for significance using the non-parametric Mann-Whitney U test for unequal medians [9]. Metrics examined were total peak area; peak height; peak width at half height; temperature of the peak maximum; heat capacity of the primary transition in the range 60–65 °C; heat capacity of the secondary transition in the range 68–72 °C; ratio of the first and second transition amplitudes; and the first moment temperature, which provides a measure of the distribution of the area of the thermogram relative to the temperature axis. The first moment temperature (TFM) was calculated as:
Another approach was to apply a general statistical methodology for quantitative analysis and classification of thermogram profiles [10]. Two parameters, a distance metric and a correlation coefficient, were combined to produce a similarity metric which was used to classify unknown thermograms against sets of reference thermograms.
4. Results and Discussion
4.1. Evaluation of pre-analytical and analytical factors on plasma thermograms
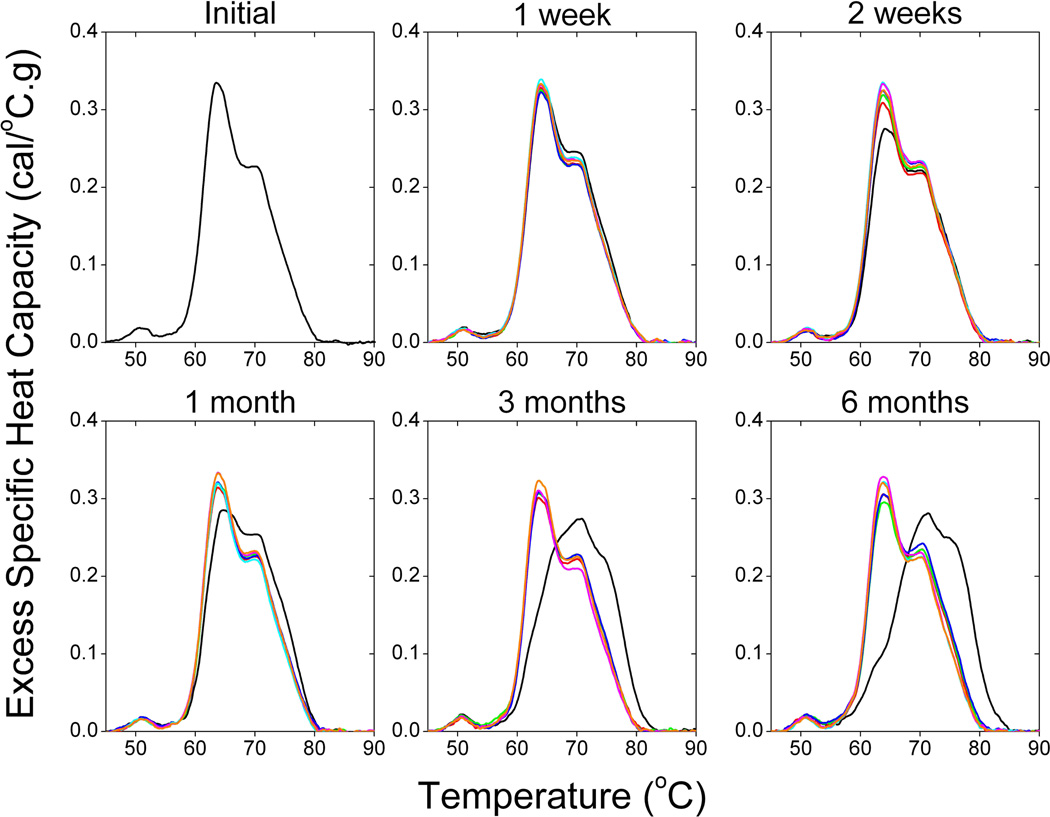
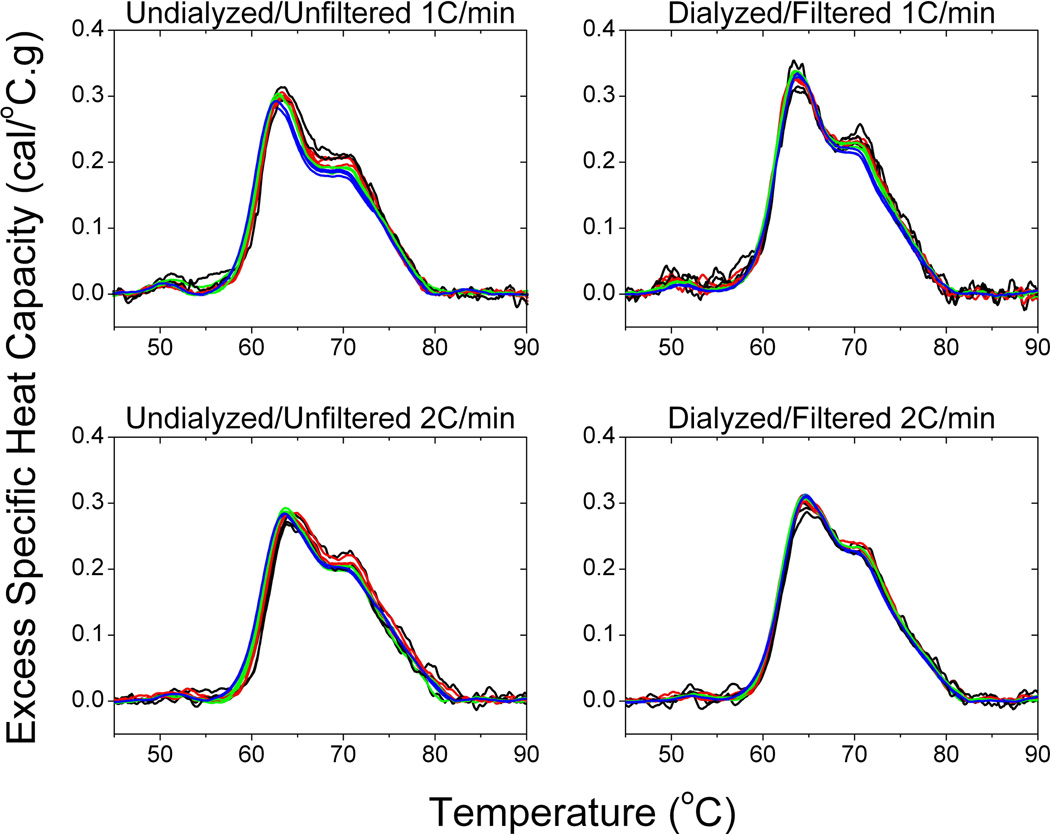
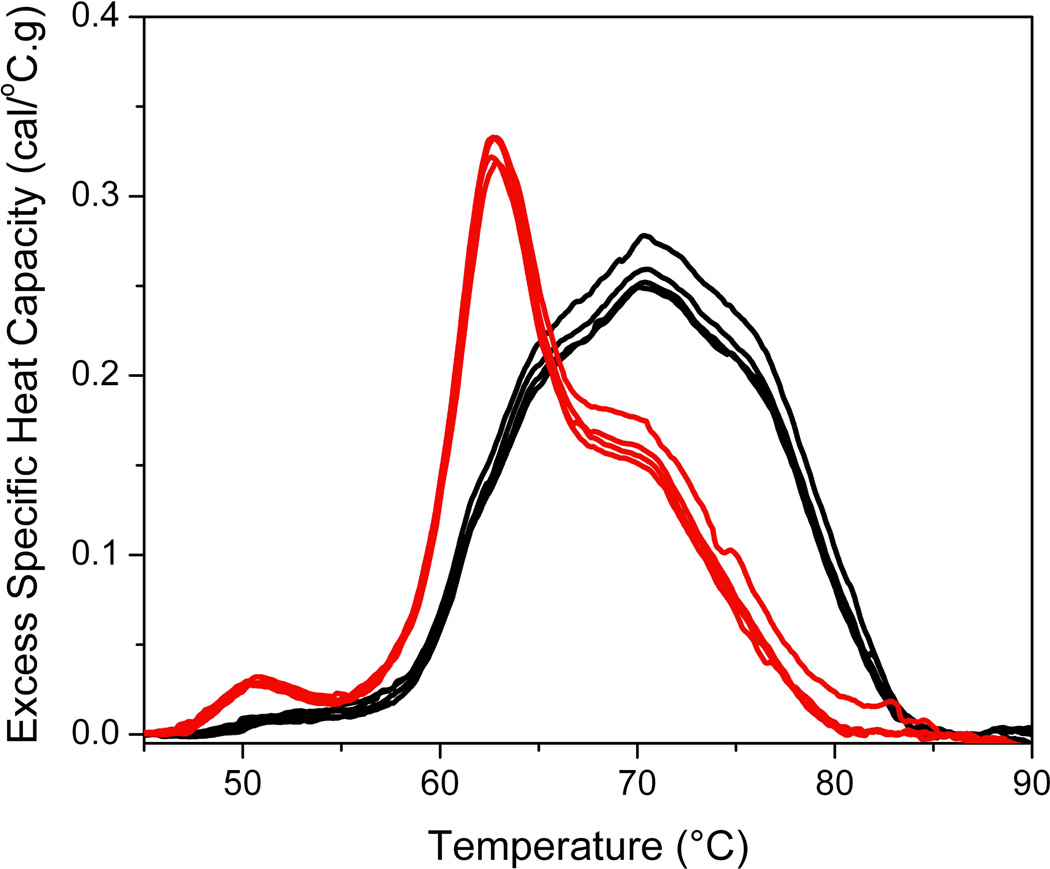
Table 1 shows a large number of studies applying plasma thermograms, as well as thermograms of other biological samples, as a complementary approach for clinical analysis. The number of studies and range of diseases examined provides substantial support of the utility of DSC for diagnostic application. To more rigorously evaluate the diagnostic potential of DSC, we have evaluated the effect of a number of analytical and pre-analytical variables on thermograms. We have previously reported that thermograms are unaffected by freeze-thaw treatment and blood collection protocol, where plasma and serum thermograms from the same individual were observed to be identical except for the absence of transitions corresponding to the denaturation of fibrinogen in the serum thermogram [6]. In an expanded study we have examined a number of pre-analytical variables: sample storage temperature (−80 °C; −20 °C; 4 °C), time of storage (no storage; 1 week; 2 weeks; 1 month; 3 months; 6 months) and freeze-thaw cycles (no freeze-thaw; 1 freeze-thaw cycle; 2 freeze-thaw cycles; 3 freeze-thaw cycles). Figure 2 shows that the thermogram profile was unaffected by these storage and handling variables with the exception of more than short term storage at 4 °C. We have also evaluated a number of analytical variables: buffer exchange method (no buffer exchange; buffer exchange column; dialysis), sample dilution (10-fold; 25-fold; 50-fold; 100-fold), instrument scan rate (1 °C/min; 2 °C/min) and analysis replicates (one scan; two scans; three scans) [Figures 3 and 4]. Sample dilution and analysis replicates gave consistent thermogram profiles well within our observed healthy thermogram standard deviation [5]. Slight changes in profile features were observed with an increase in instrument scanning rate consistent with kinetic scan broadening effects. Of most interest, the thermogram profile was unchanged for both a healthy control and disease samples with the inclusion of a buffer exchange step (Figures 3 and 4). Buffer exchange, via either dialysis or buffer exchange columns, is a routine preparation step for DSC studies deemed necessary given the high sensitivity of current instrumentation to match solvent conditions in the sample and reference instrument chambers. We employed dialysis in the plasma thermogram assay to normalize solution conditions for all biological samples examined, for example, different anticoagulants used for the preparation of blood plasma are normalized to a standard physiological buffer solution. Figure 4 shows that there is no difference in either healthy control or disease plasma thermograms in the absence of a buffer exchange step or inclusion of buffer exchange via either dialysis or buffer exchange columns. Importantly, our demonstration that there is no thermogram effect between inclusion and exclusion of dialysis has importance not only for streamlining the workflow of DSC as a diagnostic platform but also in establishing that disease modulation of the plasma proteome is robust and maintained through the dialysis step.
Figure 2.
The effect of sample storage and handling on plasma thermograms. The average of triplicate scans are shown for initial analysis then after 1 week, 2 weeks, 1 month, 3 months and 6 months of storage. Storage conditions evaluated were: 4°C (black), −20°C with 1 freeze-thaw cycle (red), −20°C with 2 freeze-thaw cycles (green), −20°C with 3 freeze-thaw cycles (blue), −80°C with 1 freeze-thaw cycle (cyan), −80°C with 2 freeze-thaw cycles (magenta), −80°C with 3 freeze-thaw cycles (orange).
Figure 3.
The effect of sample dilution, instrument scanning rate, buffer exchange method and analysis replicates on plasma thermograms. Triplicates at four sample dilutions: 100-fold dilution (black); 50-fold dilution (red); 25-fold dilution (green) and 10-fold dilution (blue) are represented in each panel.
Figure 4.
The effect of the inclusion or exclusion of a buffer exchange method on the healthy and disease plasma thermogram. Duplicate or triplicate thermograms at both 25-fold (black) and 50-fold (red) dilution are shown in each panel.
Another parameter that should be carefully considered for plasma thermogram analysis is the normalization step. Changes in both heat capacity and temperature position are observed in disease modulated thermograms and thus it is essential to include a normalization method for the diagnostic comparison of thermograms. We have adopted the bicinchoninic acid microplate assay as a widely accepted protein measurement with suitable accuracy, throughput and sample consumption. Other approaches might be considered; for example, absorbance measurements might be faster and more convenient. For any approach, it is important to have a reproducible method providing consistent normalization of thermograms and allowing for quantitative comparison and analysis.
4.2. Investigation of biomarker effects on plasma thermograms
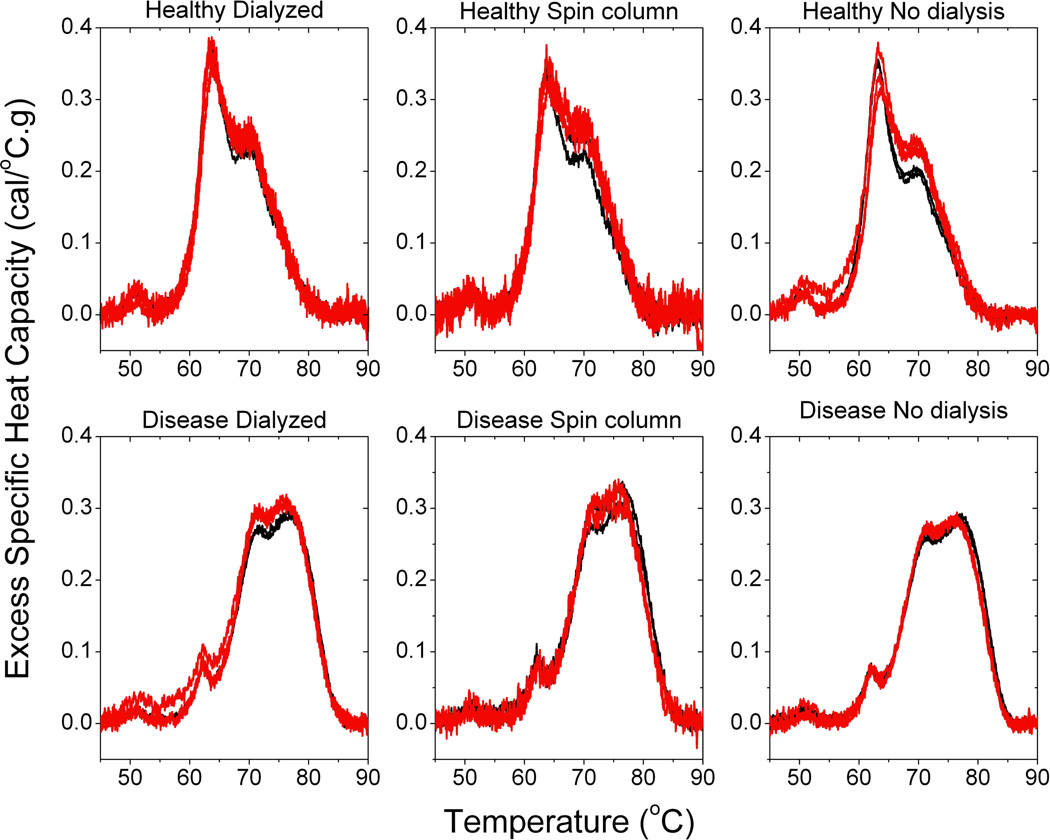
An important element to the clinical application of plasma thermograms is to understand the molecular basis of disease altered thermograms. We have begun to explore the effect of putative biomarker modifications or interactions on plasma thermograms. As a first step, we further investigated the dialysis step in the preparation of samples for DSC analysis. If biomarker interactions were responsible for the observed modulation in plasma thermogram and these interactions were labile then the thermogram might be changed as a result of the dialysis step. We employed a five-cell equilibrium dialyzer with each two-chamber cell separated by the same Snakeskin dialysis membrane we used in our general plasma preparation procedure. We charged chambers with either disease plasma, control plasma, HSA or buffer. We compared thermograms of disease plasma and healthy control in the absence of dialysis and also after dialysis against each other, against buffer or against the most abundant plasma protein, HSA. Dialysis against HSA was of interest in light of the high propensity of HSA to interact with many different molecules and its possible role in biomarker interactions within the plasma proteome, in support of the interactome concept. If biomarker interactions present in the disease plasma were labile then these biomarkers could be dialyzed out of the disease plasma chamber and into the neighboring chamber. The result would be the normalization of the disease plasma thermogram and modulation of the control plasma profile. Figure 5 shows that the disease thermogram and healthy thermogram are unchanged under all dialysis conditions examined. These results demonstrate that the modifications or interactions associated with disease plasma are robustly represented by alterations in the thermogram profile.
Figure 5.
The effect on disease and healthy thermograms in the absence of dialysis or after dialysis against each other, HSA or buffer.
In discussing the basis of biomarker effects on thermograms, our consideration of the interactome hypothesis is but one explanation for thermogram changes. The observation of disease thermogram changes employing our standard protocol of dialysis and 25-fold dilution argues against the involvement of weak interactions in thermogram modulation. Interactions involving small peptide or metabolite biomarkers might therefore be less plausible than more substantial protein-protein networks or other biomarker effects involving covalent modification of plasma proteins. While it is important to understand the origin of thermogram changes, it is not essential for use as a diagnostic indicator. As long as there is a consistent and reproducible signature for a particular disease it need not be necessary to specify the underlying mechanism that produces the signature. The mechanism is of great interest, but not essential for practical use of thermogram signatures.
4.3. Analysis of plasma thermograms
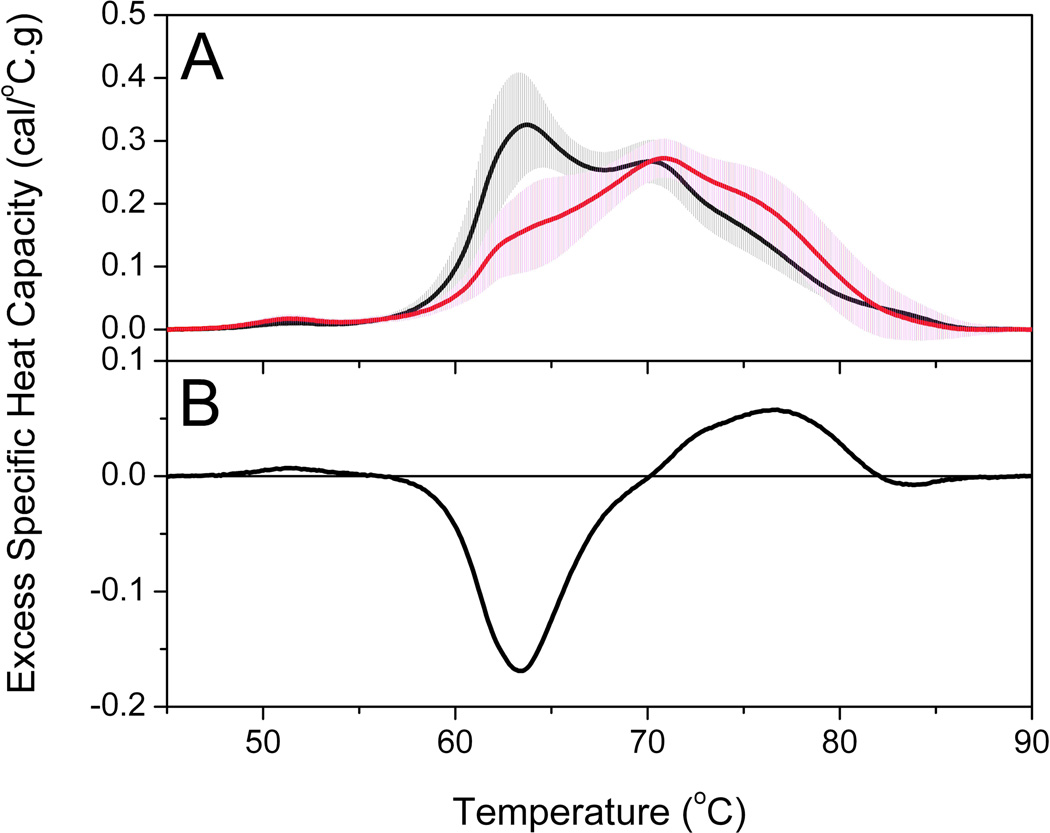
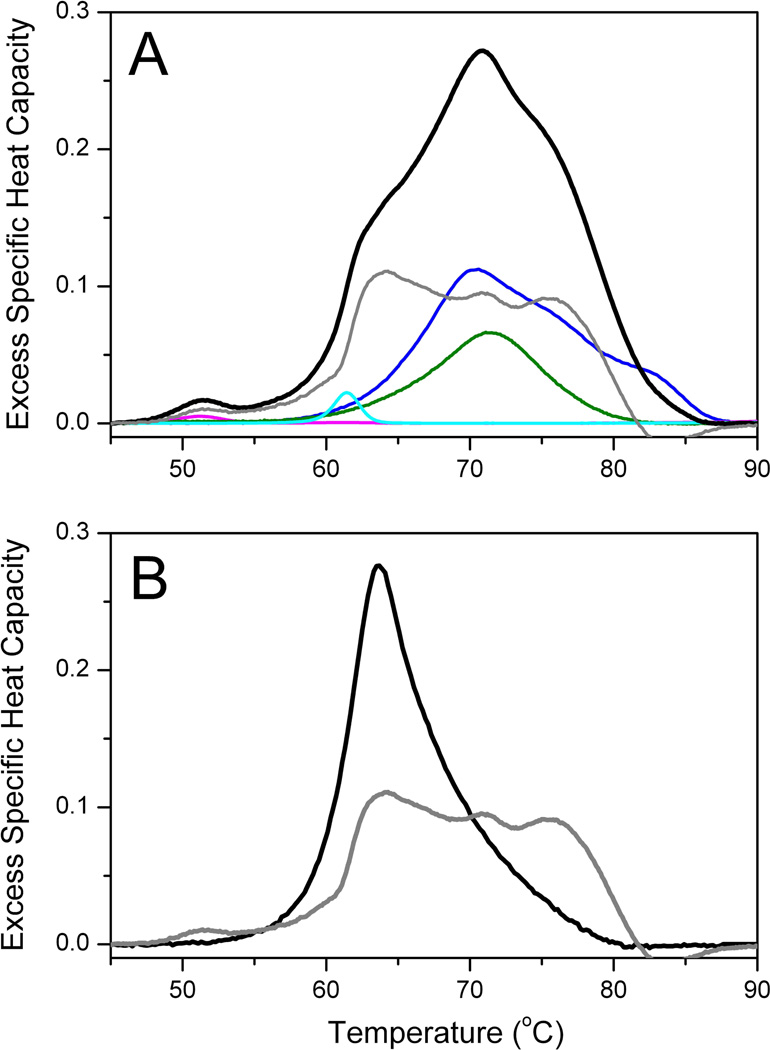
Figure 6A compares group thermograms for a healthy and lupus group. The solid lines are the average thermograms and the shaded areas represent the standard deviations of each group. The healthy control profile is the average of thermograms from 120 individuals. The average profile and standard deviation compare well with our early reports [5, 11]. A composite plot of all 120 healthy control thermograms is shown in Supplemental Material. Almost all thermograms fall within two standard deviations of the mean with the majority of thermograms clustered within one standard deviation. We have observed that the variation within a group of thermograms is much smaller than the normal clinical range for individual plasma proteins. For example, HSA, has a normal range of 35–50 g/L [12]. The lupus profile is the average from 297 individual plasma samples obtained from the Lupus Family Registry and Repository. The standard deviation is comparable to the healthy control group and similar to what we have observed for other clinical groups. It is immediately apparent that there are substantial differences between the healthy and lupus groups. The major peak ~ 63 °C is diminished in the lupus group and the thermogram is shifted to higher temperatures. Construction of a difference plot by subtracting the healthy control thermogram from the lupus thermogram emphasizes these differences (Figure 6B). There is a large negative difference peak ~ 63 °C and a smaller positive difference peak in the range 70–80 °C. These show a decrease in unfolding transition(s) ~ 63 °C and an increase in unfolding transition(s) in the range 70–80 °C. These observations are in accordance with our deconvolution analysis which we hypothesize could be associated with the change in unfolding behavior of HSA as a result of biomarker interactions or modifications. Figure 7A shows the deconvolution of the average lupus thermogram. This deconvolution shows that the altered shape arises from a shift in the denaturation of only one component, HSA. Figure 7B shows the shift in the HSA component to higher temperature and with added complexity. The multiphasic behavior of the altered HSA thermogram is fully consistent with the behavior predicted to result from protein-ligand interactions at intermediate degrees of saturation [30]. Such behavior is predicted by the interactome hypothesis.
Figure 6.
(A) Average group thermograms for healthy control and lupus groups. The solid line is the average thermogram (healthy, black line; lupus, red line) and the shaded area (healthy, gray shading; lupus, magenta shading) is the standard deviation at each temperature. (B) Difference plot between average thermograms of lupus and healthy control groups.
Figure 7.
(A) Average thermogram (black) for 297 plasma samples from lupus patients. The deconvolution using the same component protein concentrations used in Figure 1 except for an altered shape for HSA is shown. (B). Comparison of the normal and diseased HSA component shapes.
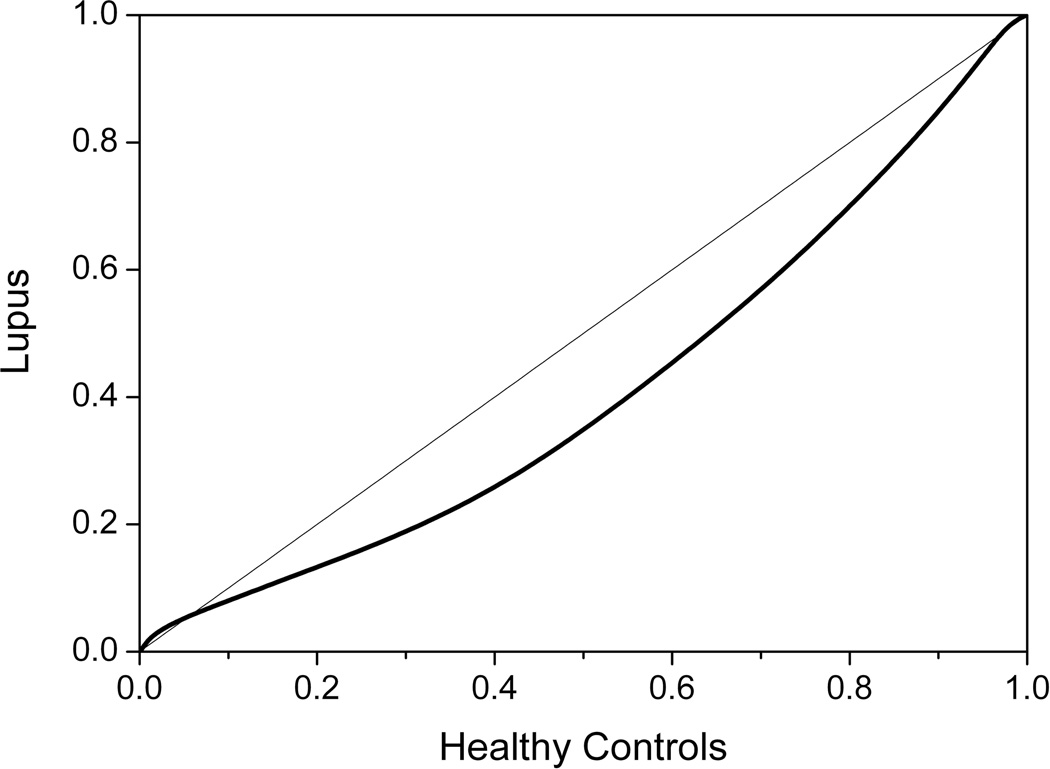
Another way to observe differences between the group thermograms and to quantify the significance of the difference is through construction of a quantile-quantile plot. To construct the quantile-quantile plot, the average thermograms are normalized to common temperature axis and total area and then plotted in a pairwise fashion against each other. If there is no deviation between groups then the data will fall on the 45° y=x line. Figure 8 shows clear deviation from this line over almost the entire data span indicating substantial differences between the healthy and lupus profiles across the entire thermogram. The greatest deviation is observed in the middle of the quantile-quantile plot, consistent with the major differences observed over the 60–80 °C envelope in Figure 6. The statistical significance of difference between thermograms is found using the Kolmogorov-Smirnov test (http://www.physics.csbsju.edu/stats/KS-test.html). For the data in Figure 8, the difference is significant at the p < 0.0001 level.
Figure 8.
Quantile-quantile plot comparing the healthy control group and the lupus group. The 45° line indicates no difference between groups.
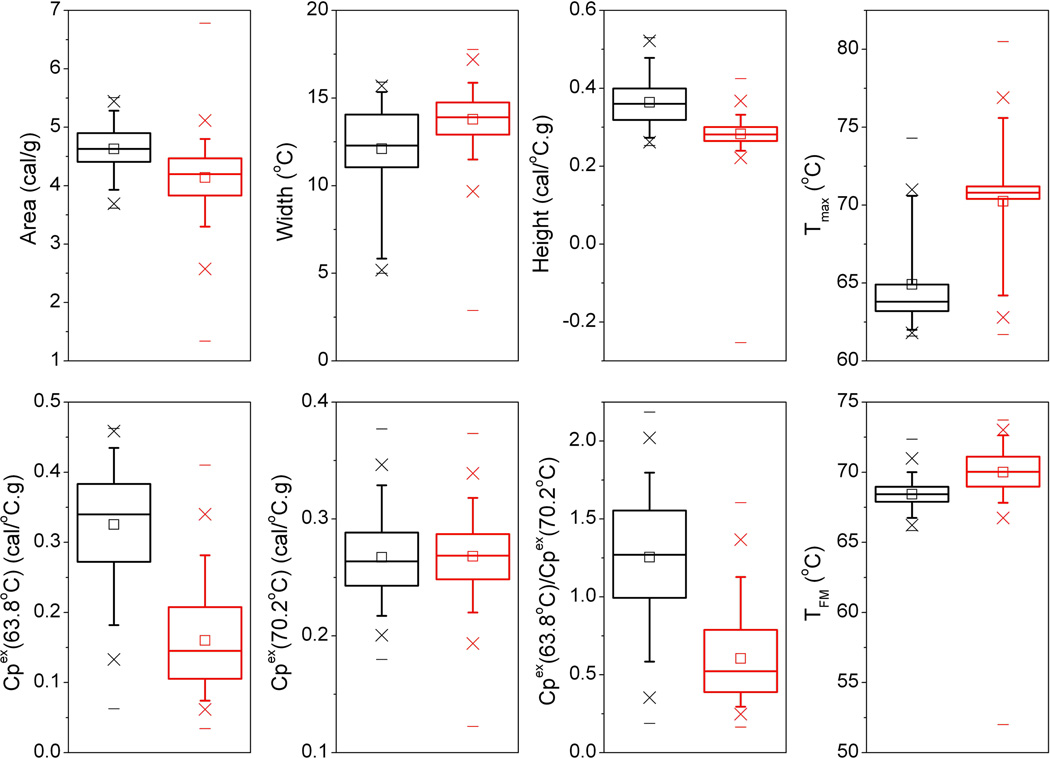
To enhance the analysis of plasma thermograms a number of other analysis procedures have been employed by our lab and others. Examining specific features of thermograms has proven useful in discriminating between clinical groups [3, 4, 13, 14, 15]. Figure 9 shows a Tukey box plot of eight thermogram metrics calculated from individual thermograms within the healthy and lupus groups. Each parameter exhibited a range of values for each group with some degree of overlap but there were clear trends in metrics between groups. Thermogram area decreased for the lupus group. Opposite trends in width and height were observed, where increased width was associated with decreased height. These parameters show that the thermogram profiles changed in distribution between the groups. The temperature of the heat capacity maximum, Tmax, increased in the lupus group but exhibited a wide spread in values as a result of variability in the shapes of thermograms. The healthy profile is well represented by a primary transition in the range of 60–65 °C and a lower amplitude secondary transition in the range 68–72 °C. However, in the lupus group, changes in the thermogram profile result in variability in the position of the major peak and hence a spread in Tmax values. We conceived the concept of calculating first moment temperature (TFM) values for thermograms in order to better represent changes in the temperature-distribution of profiles. The box plot for TFM shows a tighter distribution of values but smaller differences between groups. Other metrics of interest are the amplitudes of the primary, lower temperature transition compared with the secondary, higher temperature transition. Figure 9 shows substantial differences in the lower temperature transition with virtually no difference in the higher temperature peak. Differences in thermogram metrics between groups can be tested for significance to identify parameters which can provide useful discriminatory metrics. We applied the nonparametric Mann-Whitney U test for unequal medians to examine the statistical significance of parameter differences between the healthy and lupus groups. Table 2 shows the distribution of parameter values for the groups and the Mann-Whitney p-values. With the exception of the amplitude of the higher temperature secondary transition, all parameters showed statistically significant differences between groups. We have previously shown that the ratio of the transition amplitudes is a superior discriminator of thermograms between clinical groups [3, 13] and this might provide a simple metric to apply for thermogram analysis of clinical samples.
Figure 9.
Tukey box plots of thermogram shape and feature metrics for the healthy control (black) and lupus (red) groups. A summary of statistical analyses of these parameters is shown by Table 2. In each box plot, the median value is indicated by the horizontal line within the box; the 25th and 75th percentiles are indicated by the lower and upper box edges, respectively. The mean value is indicated by the square within the box. The upper and lower “whiskers” define the 95th and 5th percentiles, respectively. The 99th and 1st percentiles are shown by the crossed symbols and the horizontal lines denote the minimum and maximum of the data set.
Table 2.
Comparison of thermogram parameters for the healthy control and lupus groups.
| Parameter | Healthy (Mean ± SD) |
Lupus (Mean ± SD) |
Healthy (Median (LQ,UQ)) |
Lupus (Median (LQ,UQ)) |
p-value (Median) |
|---|---|---|---|---|---|
| Area (cal/g) | 4.63 ± 0.37 | 4.14 ± 0.52 | 4.63 (4.41,4.90) | 4.20 (3.83,4.47) | < 0.0001 |
| Width (°C) | 12.1 ± 2.6 | 13.8 ± 1.6 | 12.3 (11.1,14.1) | 13.9 (12.9,14.7) | < 0.0001 |
| Height (cal/°C.g) | 0.36 ± 0.06 | 0.28 ± 0.04 | 0.36 (0.32,0.40) | 0.28 (0.26,0.30) | < 0.0001 |
| Tmax (°C) | 64.9 ± 2.8 | 70.3 ± 2.8 | 63.8 (63.2,64.9) | 70.8 (70.4,71.2) | < 0.0001 |
| TFM (°C) | 68.4 ± 1.0 | 70.0 ± 1.7 | 68.4 (67.9,69.0) | 70.0 (69.0,71.1) | < 0.0001 |
| Cpex (63.8 °C) (cal/°C.g) | 0.33 ± 0.08 | 0.16 ± 0.07 | 0.34 (0.27,0.38) | 0.15 (0.11,0.21) | < 0.0001 |
| Cpex (70.2 °C) (cal/°C.g) | 0.27 ± 0.03 | 0.27 ± 0.03 | 0.26 (0.24,0.29) | 0.27 (0.24,0.29) | 0.2411 |
| Cpex (63.8 °C) / Cpex (70.2 °C) | 1.26 ± 0.38 | 0.60 ± 0.27 | 1.27 (0.99,1.55) | 0.52 (0.39,0.79) | < 0.0001 |
Parameter six in the table should appear as Cpex (63.8 °C) (cal/°C.g) Tmax, temperature of the peak maximum; TFM, first moment temperature; Cpex (63.8°C), excess specific heat capacity of the primary transition; Cpex (70.2°C), excess specific heat capacity of the secondary transition; Cpex (63.8°C)/Cpex (70.2°C), ratio of the excess specific heat capacities of the primary and secondary transitions; (Mean ± SD), mean value and standard deviation; (Median (LQ,UQ)), median value, lower quartile and upper quartile
To utilize the entire thermogram profile in distinguishing between clinical groups, we have applied a statistical methodology to compare and classify thermograms according to their similarity to reference groups [10]. In this approach, the similarity of a test thermogram and a reference set are characterized using two factors: (1) differences in vertical scaling at each temperature point and (2) similarity in shape (similar to the quantile-quantile plot). We have applied this method to characterize the similarity of the healthy and lupus thermograms. To test our approach, test healthy and lupus thermograms were compared to a healthy reference set. Substantial differences in both scaling and shape metrics were observed with relatively low similarity values obtained for both metrics for the lupus set showing dissimilarity to the healthy reference set. To further test the method, the healthy and lupus thermograms were divided into reference and test sets to allow the classification of test thermograms in both groups against group reference sets. In this way, it is possible to obtain an estimate of diagnostic performance using the classification method. Our initial results gave 82% correct classification of healthy test curves and 88% of lupus test curves. This was an encouraging result suggesting that this approach could have utility for the diagnostic application of plasma thermograms and it has subsequently been applied as a useful metric for thermogram comparison by other researchers [4, 15].
4.4. Challenges in the analysis of plasma thermograms
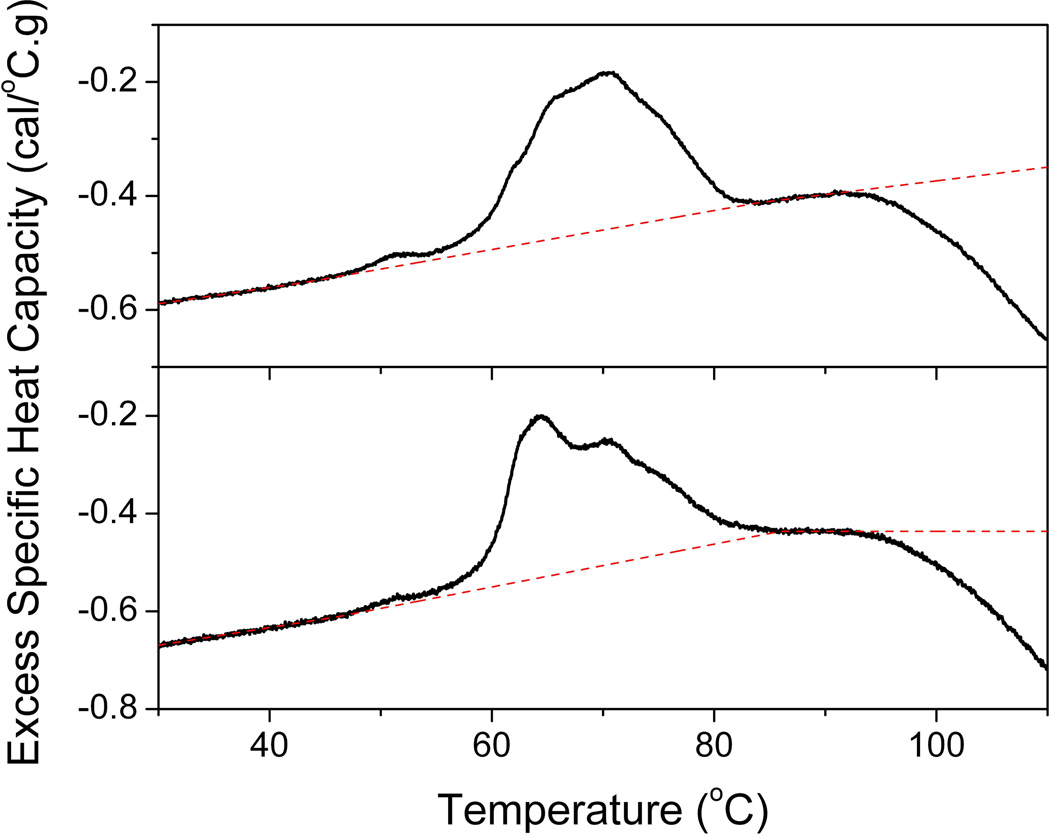
A significant challenge in the DSC analysis of proteins is the exothermic aggregation event accompanying the thermally-induced irreversible unfolding of “sticky” proteins. This is substantially reduced in capillary chamber DSC instruments but, nevertheless, is a big challenge in relatively high concentration and multi-component protein mixtures encountered in the analysis of plasma. The effects of different sample baseline selection in DSC has been previously discussed [16] with relatively small effects on unfolding temperatures but larger effects on thermogram areas. For complex protein mixtures, such as plasma, appropriate baseline selection is substantially more difficult given unfolding events and associated unfolding baselines corresponding to each of the component proteins. We have recently commented on our approach in defining sample baselines during the analysis of plasma thermograms [3]. Our approach is illustrated by Figure 10 for two example thermograms after subtraction of a buffer reference scan and normalization to the total protein concentration of the sample. The pre-transition baseline for plasma thermograms exhibits a slightly increasing linear slope for which a good fit can be obtained using a linear baseline. The post-transition is more variable. The two examples here show a post-transition baseline which follows a similar slope to the pre-transition baseline (top panel) and a horizontal post-transition baseline different in slope to the pre-transition baseline (bottom panel). These are but two examples of many hundreds of plasma thermograms collected in our laboratory. The pre-transition baseline is typically very reproducible, similar to that shown in Figure 10, whereas the post-transition baseline is highly variable and followed by a substantial “drop-off” in heat capacity corresponding to aggregation and precipitation events after protein unfolding. There is a very small area of post-transition baseline which is challenging to fit.
Figure 10.
Examples of linear baseline selection during plasma thermogram analysis.
Sample baseline fitting is a highly subjective and time-consuming part of DSC analysis in general and perhaps more so for plasma thermogram analysis. We have acknowledged that our approach in setting a linear baseline is not necessarily the most correct option but we required a method that could be most consistently applied across the large number of thermograms that we have generated. We have evaluated all common baseline setting options, linear, cubic and progressive (sigmoidal) baselines. We have found that the linear baseline method behaved the most consistently when tested on repeated measurements, different samples and by multiple users. We also wanted to employ common analysis software, Origin 7 (OriginLab Corporation, Northampton, MA), that we routinely use in the lab and could be used to analyze data sets from different instrument manufacturers from the beginnings of our thermogram research. In this analysis software, the linear baseline option could be applied without the need to truncate thermograms or set fitting limits during analysis to remove the post-transition aggregation drop-off, which was problematic for some of the other baseline fitting options we evaluated. Although our baseline fitting process is consistent and reproducible in our hands, this is the most variable part of plasma thermogram analysis. As DSC is increasingly applied in the analysis of clinical samples, particularly if DSC is to have a ultimate application in clinical diagnostics, it will be necessary to address standardization and streamlining of the baseline fitting procedure to ensure efficiency and reproducibility in analysis across multiple instruments, analysis platforms and users. This is a challenge that has yet to be addressed.
5. Conclusions
DSC is based on firm thermodynamic principles and is the method of choice for thermodynamic studies of protein denaturation. The technique has significant utility for the analysis of complex protein solutions, such as plasma, as a result of its exquisite sensitivity to the concentration, modifications and binding interactions of components. As a universal detector of heat changes associated with thermal events, DSC is applicable to the analysis of a large range of materials. These important properties have been exploited in the emerging application of DSC for clinical analysis. DSC has been applied to the direct analysis of a range of clinical samples in a number of different clinical settings. Resultant thermograms have been shown to demonstrate sensitivity to clinical status and it has been suggested that DSC could have potential utility as a complementary clinical diagnostic tool. As the field grows, challenges of clinical DSC analysis lie in demonstrating the reproducible and sensitive performance of DSC across many platforms, users and clinical settings, as well as the development of data analysis and interpretation of clinical thermograms.
Supplementary Material
Highlights.
DSC analysis of biofluids is an emerging area with application to clinical diagnostics
DSC profiles are sensitive to changes in the thermal properties of plasma proteins associated with clinical status
The principles, practice and challenges of DSC analysis of blood plasma are described
The development of approaches for thermogram analysis and interpretation are discussed
Acknowledgements
We acknowledge the immensely valuable resource of the Lupus Family Registry and Repository for generously providing access to lupus plasma samples and data. We also acknowledge C. William Helm, Lynn P. Parker, Daniel S. Metzinger and James J. Miller for generously providing clinical samples. We thank the James Graham Brown Cancer Center, the Brown Cancer Center Bio repositories and the Clinical Trials Office at the University of Louisville for support of the plasma thermogram research. Collection of plasma thermogram data was supported by use of the Biophysical Core Facility at the James Graham Brown Cancer Center and by grants P20 RR018733 and P20 GM103482 from the National Institutes of Health. Research reported in this publication was supported by the National Cancer Institute of the National Institutes of Health under Award Number R21CA187345 and by a Competitive Enhancement Grant from the Office of the Executive Vice President for Research and Innovation at the University of Louisville. The funders had no role in study design, data collection and analysis, preparation of the manuscript, or in the decision to publish.
Abbreviations
- DSC
Differential scanning calorimetry
- Tm
melting temperature
- ΔH
denaturation enthalpy
- Tmax
temperature of the peak maximum
- TFM
first moment temperature
- Cpex
excess specific heat capacity
- HSA
human serum albumin
- IgG
immunoglobulin G
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosure
NCG and JBC are co-inventors on patent applications describing the DSC plasma thermogram technology for which Louisville Bioscience, Inc. (LBI) holds an exclusive license from the University of Louisville. JBC is a founder and shareholder of LBI; NCG is a founder, shareholder and employee of LBI.
References
- 1.Bruylants G, Wouters J, Michaux C. Differential Scanning Calorimetry in Life Science: Thermodynamics, Stability, Molecular Recognition and Application in Drug Design. Curr. Med. Chem. 2005;12:2011–2020. doi: 10.2174/0929867054546564. [DOI] [PubMed] [Google Scholar]
- 2.Zhou M, Lucas DA, Chan KC, Issaq HJ, Petricoin I, E.F., Liotta LA, Veenstra TD, Conrads TP. An investigation into the human serum "interactome". Electrophoresis. 2004;25:1289–1298. doi: 10.1002/elps.200405866. [DOI] [PubMed] [Google Scholar]
- 3.Garbett NC, Merchant ML, Helm CW, Jenson AB, Klein JB, Chaires JB. Detection of Cervical Cancer Biomarker Patterns in Blood Plasma and Urine by Differential Scanning Calorimetry and Mass Spectrometry. PLOS ONE. 2014;9:e84710. doi: 10.1371/journal.pone.0084710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krumova S, Rukova B, Todinova S, Gartcheva L, Milanova V, Toncheva D, Taneva SG. Calorimetric monitoring of the serum proteome in schizophrenia patients. Thermochim. Acta. 2013;572:59–64. [Google Scholar]
- 5.Garbett NC, Miller JJ, Jenson AB, Chaires JB. Calorimetry outside the box: a new window into the plasma proteome. Biophys. J. 2008;94:1377–1383. doi: 10.1529/biophysj.107.119453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garbett NC, Miller JJ, Jenson AB, Miller DM, Chaires JB. Interrogation of The Plasma Proteome with Differential Scanning Calorimetry. Clin. Chem. 2007;53:2012–2014. doi: 10.1373/clinchem.2007.091165. Supplemental Information. [DOI] [PubMed] [Google Scholar]
- 7.Lodder RA, Hieftje GM. Quantile Analysis: A Method for Characterizing Data Distributions. Appl. Spectrosc. 1988;42:1512–1520. [Google Scholar]
- 8.Garbett NC, Miller JJ, Jenson AB, Chaires JB. Calorimetric Analysis of the Plasma Proteome. Semin. Nephrol. 2007;27:621–626. doi: 10.1016/j.semnephrol.2007.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Conover WJ. Practical Nonparametric Statistics. Third Edition ed. New York: Wiley; 1999. [Google Scholar]
- 10.Fish DJ, Brewood GP, Kim JS, Garbett NC, Chaires JB, Benight AS. Statistical analysis of plasma thermograms measured by differential scanning calorimetry. Biophys. Chem. 2010;152:184–190. doi: 10.1016/j.bpc.2010.09.007. [DOI] [PubMed] [Google Scholar]
- 11.Garbett NC, Mekmaysy CS, Helm CW, Jenson AB, Chaires JB. Differential scanning calorimetry of blood plasma for clinical diagnosis and monitoring. Exp. Mol. Pathol. 2009;86:186–191. doi: 10.1016/j.yexmp.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 12.Peters, T J. All About Albumin: Biochemistry, Genetics and Medical Applications. Academic Press; San Diego, CA: 1996. [Google Scholar]
- 13.Garbett NC, Merchant ML, Chaires JB, Klein JB. Calorimetric analysis of the plasma proteome: Identification of type 1 diabetes patients with early renal function decline. Biochim Biophys Acta. 2013;1830:4675–4680. doi: 10.1016/j.bbagen.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Todinova S, Krumova S, Gartcheva L, Robeerst C, Taneva SG. Microcalorimetry of blood serum proteome: a modified interaction network in the multiple myeloma case. Anal. Chem. 2011;83:7992–7998. doi: 10.1021/ac202055m. [DOI] [PubMed] [Google Scholar]
- 15.Todinova S, Krumova S, Kurtev P, Dimitrov V, Djongov L, Dudunkov Z, Taneva SG. Calorimetry-based profiling of blood plasma from colorectal cancer patients. Biochim. Biophys. Acta. 2012;1820:1879–1885. doi: 10.1016/j.bbagen.2012.08.001. [DOI] [PubMed] [Google Scholar]
- 16.Cooper A, Nutley MA, Wadood A. Differential scanning microcalorimetry. In: Chowdhry BZ, Harding SE, editors. Protein-Ligand Interactions: hydrodynamics and calorimetry: a practical approach. Oxford University Press; Oxford, UK: 2001. pp. 287–318. [Google Scholar]
- 17.Garbett NC, Miller JJ, Jenson AB, Chaires JB. Ligand Binding Alters the Calorimetric Thermogram of Albumin. J. Clin. Ligand Assay. 2006;29:194–197. [Google Scholar]
- 18.Michnik A, Drzazga Z, Michalik K, Barczyk A, Santura I, Sozanska E, Pierzchała W. Differential scanning calorimetry study of blood serum in chronic obstructive pulmonary disease. J. Therm. Anal. Calorim. 2010;102:57–60. [Google Scholar]
- 19.Chagovetz AA, Jensen RL, Recht L, Glantz M, Chagovetz AM. Preliminary use of differential scanning calorimetry of cerebrospinal fluid for the diagnosis of glioblastoma multiforme. J Neurooncol. 2011;105:499–506. doi: 10.1007/s11060-011-0630-5. [DOI] [PubMed] [Google Scholar]
- 20.Zapf I, Fekecs T, Ferencz A, Tizedes G, Pavlovics G, Kálmán E, Lorinczy D. DSC analysis of human plasma in breast cancer patients. Thermochim. Acta. 2011;524:88–91. [Google Scholar]
- 21.Ferencz A, Fekecs T, Lorinczy D. Differential Scanning Calorimetry, as a New Method to Monitor Human Plasma in Melanoma Patients with Regional Lymph Node or Distal Metastases. In: Xi Y, editor. Skin Cancer Overview, InTech. 2011. pp. 141–151. [Google Scholar]
- 22.Michnik A. Blood plasma, serum and serum proteins microcalorimetric studies aimed at diagnosis support. In: Lorinczy D, editor. Thermal Analysis in Medical Application. Budapest, Hungary: Akadémiai Kiadó; 2011. pp. 171–190. [Google Scholar]
- 23.Wisniewski M, Garbett NC, Fish DJ, Brewood GP, Miller JJ, Chaires JB, Benight AS. Differential Scanning Calorimetry in Molecular Diagnostics. In Vitro Diagnostic Technology. 2011;17:29–34. [Google Scholar]
- 24.Fekecs T, Zapf I, Ferencz A, Lorinczy D. Differential scanning calorimetry (DSC) analysis of human plasma in melanoma patients with or without regional lymph node metastases. J. Therm. Anal. Calorim. 2012;108:149–152. [Google Scholar]
- 25.Mehdi M, Fekecs T, Zapf I, Ferencz A, Lorinczy D. Differential scanning calorimetry (DSC) analysis of human plasma in different psoriasis stages. J. Therm. Anal. Calorim. 2013;111:1801–1804. [Google Scholar]
- 26.Rai SN, Pan J, Cambon A, Chaires JB, Garbett NC. Group Classification based on High-Dimensional Data: Application to Differential Scanning Calorimetry Plasma Thermogram Analysis of Cervical Cancer and Control Samples. Open Access Medical Statistics. 2013;3:1–9. [Google Scholar]
- 27.Chagovetz AA, Quinn C, Damarse N, Hansen LD, Chagovetz AM, Jensen RL. Differential scanning calorimetry of gliomas: a new tool in brain cancer diagnostics? Neurosurgery. 2013;73:289–295. doi: 10.1227/01.neu.0000430296.23799.cd. discussion 295. [DOI] [PubMed] [Google Scholar]
- 28.Michnik A, Drzazga Z, Poprzecki S, Czuba M, Kempa K, Sadowska-Krepa E. DSC serum profiles of sportsmen. J. Therm. Anal. Calorim. 2013;113:365–370. [Google Scholar]
- 29.Moezzi M, Ferencz A, Lorinczy D. Evaluation of blood plasma changes by differential scanning calorimetry in psoriatic patients treated with drugs. J. Therm. Anal. Calorim. 2014;116:557–562. [Google Scholar]
- 30.Brandts JF, Lin L-N. Biochemistry. 1990;29:6927–6940. doi: 10.1021/bi00481a024. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.