Abstract
Bacteria produce guanosine tetraphosphate and pentaphosphate, collectively named (p)ppGpp, in response to a variety of environmental stimuli. These two remarkable molecules regulate many cellular processes, including the central dogma processes and metabolism, to ensure survival and adaptation. Work in Escherichia coli laid the foundation for understanding the molecular details of (p)ppGpp and its cellular functions. As recent studies expand to other species, it is apparent that there exists considerable variation, with respect to not only (p)ppGpp metabolism, but also to its mechanism of action. From an evolutionary standpoint, this diversification is an elegant example of how different species adapt a particular regulatory network to their diverse lifestyles.
Introduction
Since their discovery over 40 years ago, the signaling nucleotides guanosine pentaphosphate and guanosine tetraphosphate, collectively named (p)ppGpp, have been shown to be critical for bacterial stress responses [1]. (p)ppGpp was first identified as a key inhibitor of stable RNA synthesis during amino acid starvation, called the stringent response. Later work expanded the role of (p)ppGpp beyond the starvation response, showing that (p)ppGpp is induced by diverse stresses, regulates many cellular targets, and exerts its influence even at much lower concentrations than those induced during the stringent response [2,3**]. Although our understanding of (p)ppGpp is becoming deeper, the mechanisms of its metabolism and action remain unclear. Much of this confusion is due to diversity in the synthesis and the action of (p)ppGpp in different bacteria and to the numerous (p)ppGpp targets even within the same bacterium. Here we summarize new mechanistic insights regarding (p)ppGpp production and regulation, in addition to the broad physiological effects and diverse targets of (p)ppGpp.
(p)ppGpp metabolism
Central to (p)ppGpp metabolism are enzymes that synthesize and degrade (p)ppGpp, which can be divided into three major groups: long RelA/SpoT homologue proteins (RSH) bearing both the synthetase and hydrolase domains, and small alarmone synthetases (SAS) and hydrolases (SAH), containing only the synthetase domain or the hydrolase domain, respectively [4]. These enzymes are widely distributed in bacteria and can coexist within one species in various combinations. For example, E. coli has two RSHs: RelA and SpoT, whereas Bacillus subtilis has one RSH and two SASs: RelA, RelP (YwaC), and RelQ (YjbM). Interestingly, genes encoding (p)ppGpp hydrolases have also been discovered in metazoans, and the gene (Mesh1) in Drosophila melanogaster was found to be important for starvation resistance [5], raising the possibility that (p)ppGpp might also function in higher organisms. An exception to the canonical RSH and SAS is a dual-function protein with both (p)ppGpp synthetase and RNase HII activities, recently identified in Mycobacterium smegmatis, suggesting crosstalk between RNA metabolism and stress signaling [6].
Differential regulation of these enzymes allows bacteria to sense various stresses. The ribosome-associated E. coli RelA senses amino acid scarcity by synthesizing (p)ppGpp, when an uncharged tRNA binds to the A-site of the ribosome. This signal is thought to be transduced by direct contact between the uncharged tRNA and RelA [7**]. Activation of RelA to synthesize (p)ppGpp may also depend on the identity of uncharged tRNAs, as those with higher affinity for the ribosome stimulate more (p)ppGpp synthesis in vitro [8]. Moreover, (p)ppGpp was shown to stimulate RelA activity in vitro, as a novel example of positive allosteric feedback [9].
In contrast, SASs seem to be activated transcriptionally, and canonical examples include RelP and RelQ, which are widely conserved in Gram-positive bacteria. The ywaC gene encoding RelP in B. subtilis is a member of the σM regulon [10] and is strongly induced by many antibiotics that target cell wall synthesis [11]. Likewise, expression of relP and relQ in Staphylococcus aureus is also induced by vancomycin and ampicillin [12].
Regulation of cellular processes by (p)ppGpp
(p)ppGpp has profound influence on bacterial physiology by directly or indirectly regulating many critical cellular processes, such as replication, transcription, translation, and metabolism. Here we focus on the molecular mechanisms by which (p)ppGpp adjusts these cellular activities to adapt cells to stresses.
Regulation of transcription initiation
(p)ppGpp has been known to regulate transcription since its discovery in E. coli [1]. Its impact on transcription was further demonstrated in recent transcriptomic analyses of several Gram-positive and Gram-negative species. These microarray-based studies showed that hundreds to thousands of genes exhibit (p)ppGpp-dependent changes [13–18]. (p)ppGpp can regulate transcription both directly and indirectly and the underlying mechanisms can vary between species. This section will center on how (p)ppGpp regulates transcription initiation of genes directing protein synthesis (downregulated) and amino acid biosynthesis (upregulated), as they are the mostly extensively studied and best understood regulations.
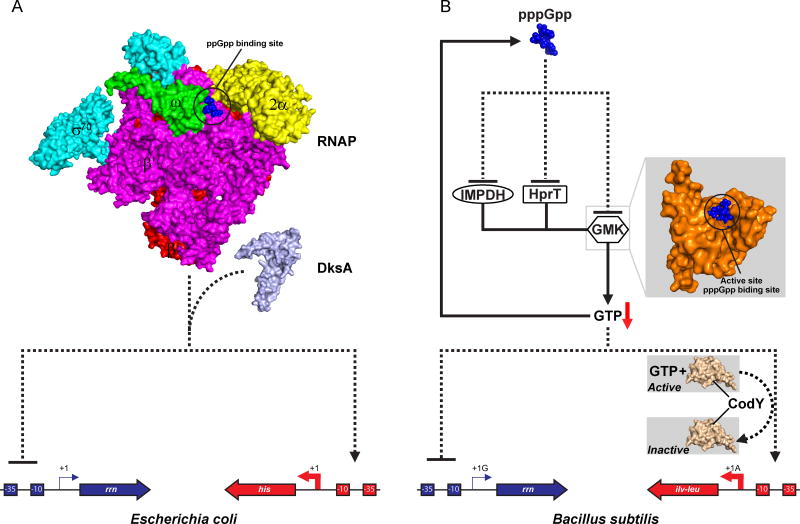
In the proteobacterium E. coli, (p)ppGpp targets RNA polymerase (RNAP) to directly regulate transcription initiation [1,19,20] (Figure 1A). It binds to an interface between the β′ and ω subunits and acts as an allosteric regulator [21**,22*,23*]. The transcription factor DksA binds the secondary channel and sensitizes RNAP to (p)ppGpp at many promoters [24,25]. Binding of (p)ppGpp and DksA to RNAP destabilizes all of the promoter open complexes examined to date, but the transcriptional outcome varies: it inhibits transcription from promoters controlling synthesis of stable RNA (rRNA and tRNA), ribosomal proteins, fatty acids, and flagella, but activates transcription from promoters governing amino acid biosynthesis [19,26–28]. A strong correlation exists between negatively regulated promoters and the short lifetime of their open complexes [19].
Figure 1. Divergent mechanisms of transcription initiation regulation by (p)ppGpp in E. coli and B. subtilis.
(A) In E. coli, this regulation is mediated by the tripartite interaction between RNAP, (p)ppGpp, and DksA. Cross-linking and crystallography suggest that ppGpp (blue spheres) binds to an interface between the β′ (magenta) and ω (green) subunits of RNA polymerase [21**,22*,23*]. This binding site is 28 Å away from the active site located between the β and β′ subunits (not visible under current view), which makes (p)ppGpp an allosteric regulator. DksA binds to the secondary channel of RNAP [24]. Classic examples of negatively and positively regulated promoters are those that direct ribosomal RNA and histidine biosynthesis, respectively. Structures of RNAP (PDB: 4JKR) and DksA (PDB: 1TJL) were used for figure preparation [22*,72]. (B) In B. subtilis, (p)ppGpp regulates GTP levels by directly inhibiting IMPDH, GMK, and HprT [3**,31], and by passively consuming GTP (GDP) during (p)ppGpp synthesis. pppGpp (blue spheres) binds the GMK active site and thus acts as a competitive inhibitor [37]. Lowering GTP levels (the downward red arrow) decreases transcription from ribosomal RNA promoters, which initiate with GTP [32], but activates transcription from amino acid biosynthesis promoters (e.g. the ilv-leu operon), in part through inactivating CodY [17,38]. The C-terminal domain of B. subtilis CodY (PDB: 2B0L) was used for figure preparation [73]. Solid lines indicate biosynthesis pathways, whereas dotted lines indicate regulation. Negatively and positively regulated promoters are colored in blue and red, respectively.
The tripartite interaction between (p)ppGpp, DksA, and RNAP is central to transcriptional regulation not only in E. coli but also very likely in species evolutionarily close to E. coli. Many proteobacteria have the E. coli ppGpp binding site well-conserved on their RNAPs [21**]. Moreover, putative dksA genes have been found in many proteobacterial genomes [29], and those of Pseudomonas aeruginosa and Rhodobacter sphaeroides have been experimentally validated [29,30]. These observations suggest that the E. coli model may be widely shared among proteobacteria.
In the distantly-related firmicute B. subtilis, (p)ppGpp does not regulate transcription initiation directly, but rather by an indirect mechanism that strongly relies on modulating intracellular GTP levels (Figure 1B). Strong (p)ppGpp induction under stresses (e.g. amino acid starvation) drastically reduces GTP levels by two mechanisms: consumption of GTP during pppGpp synthesis and inhibition of GTP biosynthesis. (p)ppGpp directly inhibits multiple enzymes in the GTP biosynthesis pathways, IMP dehydrogenase (IMPDH) [31], hypoxanthine-guanine phosphoribosyltransferase (HprT), and guanylate kinase (GMK) [3**] (Figure 1B). Lowering GTP levels affects transcription initiation directly and indirectly, depending on the promoters examined. Direct effects are observed on promoters of stable RNA synthesis, whose initiating nucleoside triphosphate is GTP [32]. These promoters display GTP concentration-dependent activities in vitro and are sensitive to intracellular GTP levels in vivo [32]. On the other hand, transcription of amino acid biosynthesis genes is indirectly regulated by GTP via several mechanisms. First, branched chain amino acid (BCAA) biosynthesis genes are repressed by the transcription factor CodY in its GTP bound form [33,34], and lowering GTP inactivates CodY and thus upregulates amino acid biosynthesis [17]. Second, decreasing GTP levels is often accompanied by a concomitant increase in ATP, which enhances transcription from BCAA promoters as they initiate with ATP and are sensitive to ATP levels [35,36]. Finally, decrease of GTP could also lead to the redistribution of RNAP from GTP-initiating promoters (e.g. those of stable RNA genes), which could contribute to the enhanced transcription of amino acid biosynthesis genes [17].
The B. subtilis mechanism appears to be conserved in Firmicutes, since many species within this group contain a (p)ppGpp-sensitive GMK [37] and they also have a CodY homologue [38]. Interestingly, species from Actinobacteria and Deinoccous-Thermus, two distantly related phyla, also exploit (p)ppGpp to inhibit GMK activity [37]. Although CodY does not appear to be conserved [38], modulation of GTP levels by (p)ppGpp may still play an important role in transcriptional regulation. In Thermus thermophilus, (p)ppGpp does not affect RNAP [39] and instead is proposed to regulate rRNA transcription by controlling GTP levels via IMPDH [40] and GMK [37].
Regulation beyond transcription initiation
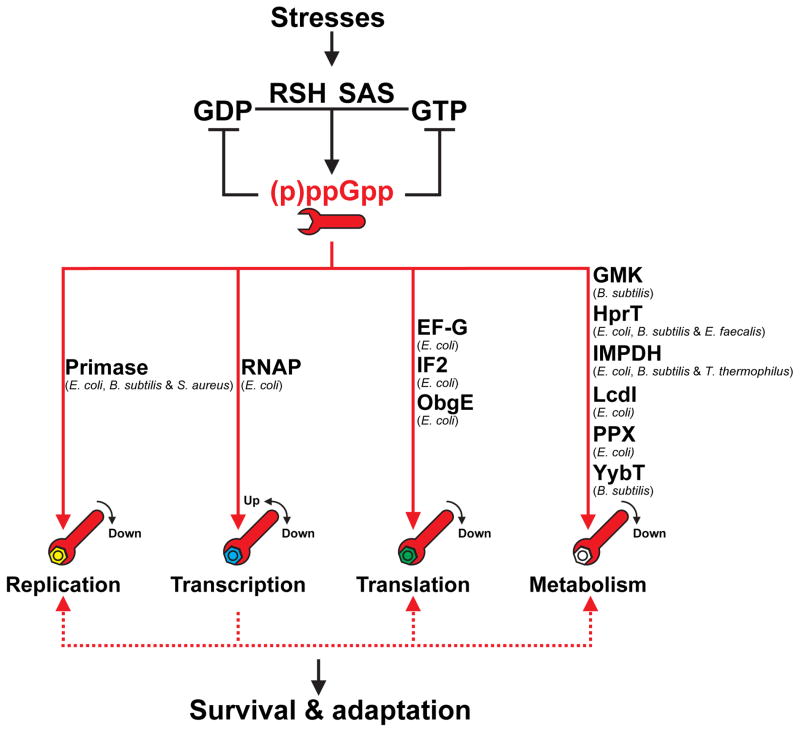
In addition to regulating transcriptional initiation, (p)ppGpp also controls many other processes, which allows it to function as a master regulator to adjust cellular physiology and to facilitate stress survival/adaptation (Figure 2).
Figure 2. (p)ppGpp regulates various important cellular processes to allow survival and adaptation to stresses.
Stresses, such as amino acid starvation and antibiotic treatment activate/upregulate (p)ppGpp synthetases (RSH and SAS) to produce (p)ppGpp (the wrench). In B. subtilis, by directly inhibiting several GTP biosynthesis enzymes, (p)ppGpp curtails production of GDP/GTP, the substrates for (p)ppGpp synthesis, thus constituting a negative feedback regulation and maintaining homeostasis of guanylate nucleotide pools [3**]. Importantly, through direct interaction with its targets, (p)ppGpp regulates replication, transcription, translation, and metabolism (differently colored bolts) to adjust the cellular physiology to survive and adapt to adverse conditions. The dotted lines indicate that (p)ppGpp also indirectly modulates replication, translation, and metabolism through its effects on transcription.
Translation
(p)ppGpp regulates translation indirectly through inhibiting transcription of ribosomal RNA and protein genes, thus curtailing production of the building blocks for ribosome assembly. In E. coli, (p)ppGpp also directly binds the translation initiation factor 2 (IF2), elongation factor G (EF-G), and the ribosome assembly factor ObgE, to regulate not only translation initiation and elongation, but also ribosome maturation during stresses [41,42].
Replication
In E. coli, (p)ppGpp indirectly inhibits replication initiation, possibly through a DNA methylation-dependent and SeqA-dependent mechanism [43]. In addition, (p)ppGpp directly binds the replication enzyme primase of S. aureus at a position overlapping the active site and interferes with its activity [44*] to slow or halt replication elongation in response to diverse stresses [45,46].
Conflicts between central dogma processes
DNA replication and transcription occur simultaneously on the same template, leading to potential conflicts between the machineries. Furthermore, translation is coupled to transcription via active ribosomes on the nascent mRNA. Amino acid starvation has the potential to exacerbate conflicts between these processes by uncoupling transcription and translation, leading to stalled transcription complexes that form barriers to replication. The (p)ppGpp cofactor DksA alleviates starvation-induced replication-transcription conflicts by preventing transcription stalling [47,48]. Intriguingly, lack of full-length IF2, a ppGpp target, sensitizes cells to DNA damaging agents, which is counteracted by increasing (p)ppGpp levels or mutations in RNAP [49]. In addition, (p)ppGpp inhibits replication, slows down transcription elongation [50], and prevents the formation of arrays of stalled RNAP [51], which may facilitate transcription-translation coupling and/or minimize transcription-replication conflicts.
Cellular metabolism
(p)ppGpp co-crystallized with the inducible lysine decarboxylase (LcdI) of E. coli and inhibits its activity in vitro and in vivo, thus regulating lysine metabolism during acid stress [52]. (p)ppGpp also inhibits exopolyphosphatase (PPX) activity to regulate metabolism of polyphosphate [53], which mediates antibiotic tolerance [54**], oxidative stress responses and general stress responses (Gray and Jakob, in this issue). Moreover, (p)ppGpp directly regulates intracellular purine nucleotide pools. Both E. coli and B. subtilis HprT and IMPDH are inhibited by (p)ppGpp [3**,55], although only the B. subtilis, but not the E. coli, GMK is sensitive to (p)ppGpp [37]. Lastly, in B. subtilis (p)ppGpp inhibits the activity of YybT, a phosphodiesterase that hydrolyzes cyclic di-AMP and cyclic di-GMP [56], suggesting crosstalk between the (p)ppGpp and c-di-AMP (or c-di-GMP) signaling pathways.
Physiological importance of (p)ppGpp
Since (p)ppGpp is involved in regulating so many essential cellular processes, it is perhaps not surprising that cells lacking (p)ppGpp, although viable, exhibit severe and pleiotropic defects during stresses.
Survival of and adaptation to amino acid starvation
The canonical phenotype of cells lacking (p)ppGpp, at least in E. coli and B. subtilis, is polyauxotrophy for amino acids [17,57,58]. In both organisms, the transcriptional regulation by (p)ppGpp is important for adapting to amino acid starvation [3**,21**]. Moreover, it appears that survival of starvation also involves a tradeoff with growth rate, at least in B. subtilis [59]. In addition, (p)ppGpp production is negatively correlated with growth rate in E. coli [57]. This suggests that in both organisms (p)ppGpp first acts as a brake on cellular processes during stresses, which could require inhibition of replication, translation, transcription of rRNA, and/or cellular metabolism. Cells would then adapt to the new conditions by modulating transcription of stress response, amino acid biosynthesis and other genes required for growth [57].
Antibiotic tolerance and resistance
Lack of (p)ppGpp often leads to impaired ability to survive antibiotic insult, suggesting a critical role of (p)ppGpp in antibiotic tolerance/resistance (for an in-depth review see [60]). In addition, strong induction of (p)ppGpp by starvation or increased basal levels of (p)ppGpp through mutation lead to enhanced antibiotic tolerance in P. aeruginosa, Enterococcus faecalis, and B. subtilis [61–64]. However, the molecular basis for tolerance/resistance in many bacterial species is still underexplored. The best understood mechanism was reported by Maisonneuve and colleagues in E. coli, who found that (p)ppGpp stochastically induces persistence through toxin-antitoxin (TA) modules [54**]. In their model, (p)ppGpp inhibits the activity of PPX to activate the Lon protease, which indirectly activates the toxin by degrading the antitoxin and thus induces persistence [54**]. Intriguingly, many toxins target translation, and at least one toxin, HipA in E. coli, increases (p)ppGpp levels by inhibiting the activity of a glutamyl-tRNA synthetase, which creates uncharged tRNAGlu that activate RelA [65**]. Thus the stochastic production of (p)ppGpp may reinforce its own synthesis through the activation of TA systems. In the closely related bacterium Salmonella Typhimurium, persistence induced by macrophage uptake also requires (p)ppGpp, Lon, and TA modules, although Lon appears to be dispensable for persistence under laboratory cultivation [66].
In addition to their implication in antibiotic tolerance, (p)ppGpp is also shown to mediate antibiotic resistance, at least in S. aureus, known for its ability to acquire resistance to multiple antibiotics. Isolates displaying high levels of methicillin resistance have point mutations in relA that lead to increased levels of (p)ppGpp [67*]. Inducing (p)ppGpp production with mupirocin also increases S. aureus resistance to β-lactam antibiotics [68,69].
Because (p)ppGpp protects cells against antibiotics, compounds that target (p)ppGpp metabolism may be viable antimicrobials when used in combination with traditional antibiotics. Recently, Relacin, a 2′-deoxyguanosine-based analogue of ppGpp, was shown to inhibit (p)ppGpp synthesis to decrease cell survival and impede biofilm formation [70*]. The peptide 1018 potently inhibits biofilm formation by directly interacting with ppGpp and promoting its degradation [71]. These studies thus may serve as models for future antimicrobial design.
Conclusions
Recent studies have highlighted conservation and divergence in (p)ppGpp metabolism and regulation. Its direct and indirect effects on multiple cellular processes and in coordinating these processes adapt cells to various stresses and maintain homeostatic growth. However, the challenge of discerning the physiologically relevant targets of (p)ppGpp under different growth and stress conditions remains.
Highlights.
(p)ppGpp metabolic enzymes sense diverse stresses
(p)ppGpp regulates transcription initiation by targeting RNAP or GTP levels
(p)ppGpp controls replication, translation, and metabolism to allow stress survival
(p)ppGpp coordinates central dogma processes to prevent conflict during stress
(p)ppGpp contributes to antibiotic tolerance and resistance
Acknowledgments
This work was supported by NIH grant GM084003 and USDA Hatch WIS01740 to JDW. We apologize to our colleagues whose work is not cited in this review due to space limitations.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Potrykus K, Cashel M. (p)ppGpp: still magical? Annu Rev Microbiol. 2008;62:35–51. doi: 10.1146/annurev.micro.62.081307.162903. [DOI] [PubMed] [Google Scholar]
- 2.Gaca AO, Kajfasz JK, Miller JH, Liu K, Wang JD, Abranches J, Lemos JA. Basal levels of (p)ppGpp in Enterococcus faecalis: the magic beyond the stringent response. MBio. 2013;4:e00646–13. doi: 10.1128/mBio.00646-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3**.Kriel A, Bittner AN, Kim SH, Liu K, Tehranchi AK, Zou WY, Rendon S, Chen R, Tu BP, Wang JD. Direct regulation of GTP homeostasis by (p)ppGpp: a critical component of viability and stress resistance. Mol Cell. 2012;48:231–241. doi: 10.1016/j.molcel.2012.08.009. Combining metabolic profiling and biochemistry, the authors found that (p)ppGpp directly inhibits the enzymatic activities of GMK and HprT to decrease GTP synthesis in B. subtilis. Furthermore, they found that GTP regulation is crucial for both survival of amino acid starvation and GTP homeostasis in unstarved conditions. They identified a negative GTP feedback loop that depends on (p)ppGpp. High GTP levels that accumulate in the absence of (p)ppGpp result in cell death even during growth in rich medium. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Atkinson GC, Tenson T, Hauryliuk V. The RelA/SpoT homolog (RSH) superfamily: distribution and functional evolution of ppGpp synthetases and hydrolases across the tree of life. PLoS One. 2011;6:e23479. doi: 10.1371/journal.pone.0023479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sun D, Lee G, Lee JH, Kim H-Y, Rhee H-W, Park S-Y, Kim K-J, Kim Y, Kim BY, Hong J-I, et al. A metazoan ortholog of SpoT hydrolyzes ppGpp and functions in starvation responses. Nat Struct Mol Biol. 2010;17:1188–1194. doi: 10.1038/nsmb.1906. [DOI] [PubMed] [Google Scholar]
- 6.Murdeshwar MS, Chatterji D. MS_RHII-RSD, a dual-function RNase HII-(p)ppGpp synthetase from Mycobacterium smegmatis. J Bacteriol. 2012;194:4003–4014. doi: 10.1128/JB.00258-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7**.Agirrezabala X, Fernández IS, Kelley AC, Cartón DG, Ramakrishnan V, Valle M. The ribosome triggers the stringent response by RelA via a highly distorted tRNA. EMBO Rep. 2013;14:811–816. doi: 10.1038/embor.2013.106. In this study, the authors obtained the first cryo-EM structure of RelA interacting with deacylated tRNA on the ribosome, which shows that RelA interacts with the acceptor arm of the tRNA toward the outer surface of the ribosome. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Payoe R, Fahlman RP. Dependence of RelA-mediated (p)ppGpp formation on tRNA identity. Biochemistry. 2011;50:3075–3083. doi: 10.1021/bi1015309. [DOI] [PubMed] [Google Scholar]
- 9.Shyp V, Tankov S, Ermakov A, Kudrin P, English BP, Ehrenberg M, Tenson T, Elf J, Hauryliuk V. Positive allosteric feedback regulation of the stringent response enzyme RelA by its product. EMBO Rep. 2012;13:835–839. doi: 10.1038/embor.2012.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eiamphungporn W, Helmann JD. The Bacillus subtilis sigma(M) regulon and its contribution to cell envelope stress responses. Mol Microbiol. 2008;67:830–848. doi: 10.1111/j.1365-2958.2007.06090.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.D’Elia Ma, Millar KE, Bhavsar AP, Tomljenovic AM, Hutter B, Schaab C, Moreno-Hagelsieb G, Brown ED. Probing teichoic acid genetics with bioactive molecules reveals new interactions among diverse processes in bacterial cell wall biogenesis. Chem Biol. 2009;16:548–556. doi: 10.1016/j.chembiol.2009.04.009. [DOI] [PubMed] [Google Scholar]
- 12.Geiger T, Kästle B, Gratani FL, Goerke C, Wolz C. Two small (p)ppGpp synthases in Staphylococcus aureus mediate tolerance against cell envelope stress conditions. J Bacteriol. 2014;196:894–902. doi: 10.1128/JB.01201-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nascimento MM, Lemos JA, Abranches J, Lin VK, Burne RA. Role of RelA of Streptococcus mutans in global control of gene expression. J Bacteriol. 2008;190:28–36. doi: 10.1128/JB.01395-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gaca AO, Abranches J, Kajfasz JK, Lemos JA. Global transcriptional analysis of the stringent response in Enterococcus faecalis. Microbiol (United Kingdom) 2012;158:1994–2004. doi: 10.1099/mic.0.060236-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Geiger T, Goerke C, Fritz M, Schäfer T, Ohlsen K, Liebeke M, Lalk M, Wolz C. Role of the (p)ppGpp synthase RSH, a RelA/SpoT homolog, in stringent response and virulence of Staphylococcus aureus. Infect Immun. 2010;78:1873–1883. doi: 10.1128/IAI.01439-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Traxler MF, Summers SM, Nguyen H-T, Zacharia VM, Hightower GA, Smith JT, Conway T. The global, ppGpp-mediated stringent response to amino acid starvation in Escherichia coli. Mol Microbiol. 2008;68:1128–1148. doi: 10.1111/j.1365-2958.2008.06229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kriel A, Brinsmade SR, Tse JL, Tehranchi AK, Bittner AN, Sonenshein AL, Wang JD. GTP dysregulation in Bacillus subtilis cells lacking (p)ppGpp results in phenotypic amino acid auxotrophy and failure to adapt to nutrient downshift and regulate biosynthesis genes. J Bacteriol. 2014;196:189–201. doi: 10.1128/JB.00918-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Durfee T, Hansen AM, Zhi H, Blattner FR, Ding JJ. Transcription profiling of the stringent response in Escherichia coli. J Bacteriol. 2008;190:1084–1096. doi: 10.1128/JB.01092-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haugen SP, Ross W, Gourse RL. Advances in bacterial promoter recognition and its control by factors that do not bind DNA. Nat Rev Microbiol. 2008;6:507–519. doi: 10.1038/nrmicro1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Srivatsan A, Wang JD. Control of bacterial transcription, translation and replication by (p)ppGpp. Curr Opin Microbiol. 2008;11:100–105. doi: 10.1016/j.mib.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 21**.Ross W, Vrentas CE, Sanchez-Vazquez P, Gaal T, Gourse RL. The magic spot: a ppGpp binding site on E. coli RNA polymerase responsible for regulation of transcription initiation. Mol Cell. 2013;50:420–429. doi: 10.1016/j.molcel.2013.03.021. This study mapped the binding site of (p)ppGpp on RNAP using crosslinking and protease mapping. It is not at the catalytic center or the nucleotide-binding site but rather the interface between the β′ and ω subunits. The authors created a mutant RNAP lacking this binding site that does not respond to (p)ppGpp in vitro. Bacteria producing mutant RNAP are defective for growth in minimal medium, thus mimicking a defect in (p)ppGpp production and providing support for the relevance of this binding site to (p)ppGpp-action. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22*.Zuo Y, Wang Y, Steitz TA. The mechanism of E. coli RNA polymerase regulation by ppGpp is suggested by the structure of their complex. Mol Cell. 2013;50:430–436. doi: 10.1016/j.molcel.2013.03.020. This study reported a crystal structure of (p)ppGpp in complex with RNAP. The binding site in this structure is on the outer surface at a position between the shelf and core modules, in agreement with Ross et al. The authors propose that binding to this site allows (p)ppGpp to inhibit closing of the active site upon nucleotide-binding, which may destabilize initiating transcription complexes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23*.Mechold U, Potrykus K, Murphy H, Murakami KS, Cashel M. Differential regulation by ppGpp versus pppGpp in Escherichia coli. Nucleic Acids Res. 2013;41:6175–6189. doi: 10.1093/nar/gkt302. The authors solved the crystal structures of RNAP in complex with ppGpp and pppGpp, which show that both molecules bind to the same site between the β′ and ω subunits of RNAP, in agreement with Ross et al. and Zuo et al. In addition, by manipulating ppGpp and pppGpp levels in vivo, the authors discovered that ppGpp more strongly affects RNAP activity than pppGpp. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lennon CW, Ross W, Martin-Tumasz S, Toulokhonov I, Vrentas CE, Rutherford ST, Lee JH, Butcher SE, Gourse RL. Direct interactions between the coiled-coil tip of DksA and the trigger loop of RNA polymerase mediate transcriptional regulation. Genes Dev. 2012;26:2634–2646. doi: 10.1101/gad.204693.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paul BJ, Barker MM, Ross W, Schneider DA, Webb C, Foster JW, Gourse RL. DksA: A critical component of the transcription initiation machinery that potentiates the regulation of rRNA promoters by ppGpp and the initiating NTP. Cell. 2004;118:311–322. doi: 10.1016/j.cell.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 26.Lemke JJ, Durfee T, Gourse RL. DksA and ppGpp directly regulate transcription of the Escherichia coli flagellar cascade. Mol Microbiol. 2009;74:1368–1379. doi: 10.1111/j.1365-2958.2009.06939.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lemke JJ, Sanchez-Vazquez P, Burgos HL, Hedberg G, Ross W, Gourse RL. Direct regulation of Escherichia coli ribosomal protein promoters by the transcription factors ppGpp and DksA. Proc Natl Acad Sci U S A. 2011;108:5712–5717. doi: 10.1073/pnas.1019383108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.My L, Rekoske B, Lemke JJ, Viala JP, Gourse RL, Bouveret E. Transcription of the Escherichia coli fatty acid synthesis operon fabHDG is directly activated by FadR and inhibited by ppGpp. J Bacteriol. 2013;195:3784–3795. doi: 10.1128/JB.00384-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blaby-Haas CE, Furman R, Rodionov DA, Artsimovitch I, De Crécy-Lagard V. Role of a Zn-independent DksA in Zn homeostasis and stringent response. Mol Microbiol. 2011;79:700–715. doi: 10.1111/j.1365-2958.2010.07475.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lennon CW, Lemmer KC, Irons JL, Sellman MI, Donohue TJ, Gourse RL, Ross W. A Rhodobacter sphaeroides protein mechanistically similar to Escherichia coli DksA regulates photosynthetic growth. MBio. 2014;5:e01105–14. doi: 10.1128/mBio.01105-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lopez JM, Dromerick A, Freese E. Response of guanosine 5′-triphosphate concentration to nutritional changes and its significance for Bacillus subtilis sporulation. J Bacteriol. 1981;146:605–613. doi: 10.1128/jb.146.2.605-613.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krásný L, Gourse RL. An alternative strategy for bacterial ribosome synthesis: Bacillus subtilis rRNA transcription regulation. EMBO J. 2004;23:4473–4483. doi: 10.1038/sj.emboj.7600423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Handke LD, Shivers RP, Sonenshein AL. Interaction of Bacillus subtilis CodY with GTP. J Bacteriol. 2008;190:798–806. doi: 10.1128/JB.01115-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brinsmade SR, Sonenshein AL. Dissecting complex metabolic integration provides direct genetic evidence for CodY activation by guanine nucleotides. J Bacteriol. 2011;193:5637–5648. doi: 10.1128/JB.05510-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krásný L, Tiserová H, Jonák J, Rejman D, Sanderová H. The identity of the transcription +1 position is crucial for changes in gene expression in response to amino acid starvation in Bacillus subtilis. Mol Microbiol. 2008;69:42–54. doi: 10.1111/j.1365-2958.2008.06256.x. [DOI] [PubMed] [Google Scholar]
- 36.Tojio S, Satomura T, Kumamoto K, Hirooka K, Fujita Y. Molecular mechanisms underlying the positive stringent response of the Bacillus subtilis ilv-leu operon, involved in the biosynthesis of branched-chain amino acids. J Bacteriol. 2008;190:6134–6147. doi: 10.1128/JB.00606-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu K, Myers AR, Pisithkul T, Claas KR, Satyshur KA, Amador-Noguez D, Keck JL, Wang JD. Molecular mechanism and evolution of guanylate kinase regulation by (p)ppGpp. Mol Cell. 2015 doi: 10.1016/j.molcel.2014.12.037. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sonenshein AL. CodY, a global regulator of stationary phase and virulence in Gram-positive bacteria. Curr Opin Microbiol. 2005;8:203–207. doi: 10.1016/j.mib.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 39.Vrentas CE, Gaal T, Berkmen MB, Rutherford ST, Haugen SP, Ross W, Gourse RL. Still looking for the magic spot: The crystallographically defined binding site for ppGpp on RNA polymerase is unlikely to be responsible for rRNA transcription regulation. J Mol Biol. 2008;377:551–564. doi: 10.1016/j.jmb.2008.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kasai K, Nishizawa T, Takahashi K, Hosaka T, Aoki H, Ochi K. Physiological analysis of the stringent response elicited in an extreme thermophilic bacterium, Thermus thermophilus. J Bacteriol. 2006;188:7111–7122. doi: 10.1128/JB.00574-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Feng B, Mandava CS, Guo Q, Wang J, Cao W, Li N, Zhang Y, Zhang Y, Wang Z, Wu J, et al. Structural and functional insights into the mode of action of a universally conserved Obg GTPase. PLoS Biol. 2014;12:e1001866. doi: 10.1371/journal.pbio.1001866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mitkevich VA, Ermakov A, Kulikova AA, Tankov S, Shyp V, Soosaar A, Tenson T, Makarov AA, Ehrenberg M, Hauryliuk V. Thermodynamic characterization of ppGpp binding to EF-G or IF2 and of initiator tRNA binding to free IF2 in the presence of GDP, GTP, or ppGpp. J Mol Biol. 2010;402:838–846. doi: 10.1016/j.jmb.2010.08.016. [DOI] [PubMed] [Google Scholar]
- 43.Ferullo DJ, Lovett ST. The stringent response and cell cycle arrest in Escherichia coli. PLoS Genet. 2008;4:e1000300. doi: 10.1371/journal.pgen.1000300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44*.Rymer RU, Solorio FA, Tehranchi AK, Chu C, Corn JE, Keck JL, Wang JD, Berger JM. Binding mechanism of metal•NTP substrates and stringent-response alarmones to bacterial DnaG-type primases. Structure. 2012;20:1478–1489. doi: 10.1016/j.str.2012.05.017. (p)ppGpp was previously found to inhibit primase activity in B. subtilis. This study reported the structures of the Staphylococcus aureus primase catalytic core either with NTP or with ppGpp. Among numerous insights obtained from the structure, the (p)ppGpp binding site was found to overlap with the NTP binding site. In addition to competing with nucleotides, (p)ppGpp also appears to destabilize the association between primase and the RNA-DNA heteroduplex. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Denapoli J, Tehranchi AK, Wang JD. Dose-dependent reduction of replication elongation rate by (p)ppGpp in Escherichia coli and Bacillus subtilis. Mol Microbiol. 2013;88:93–104. doi: 10.1111/mmi.12172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Maciąg-Dorszyńska M, Szalewska-Pałasz A, Węgrzyn G. Different effects of ppGpp on Escherichia coli DNA replication in vivo and in vitro. FEBS Open Bio. 2013;3:161–164. doi: 10.1016/j.fob.2013.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang Y, Mooney Ra, Grass Ja, Sivaramakrishnan P, Herman C, Landick R, Wang JD. DksA guards elongating RNA polymerase against ribosome-stalling-induced arrest. Mol Cell. 2014;53:766–778. doi: 10.1016/j.molcel.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tehranchi AK, Blankschien MD, Zhang Y, Halliday Ja, Srivatsan A, Peng J, Herman C, Wang JD. The transcription factor DksA prevents conflicts between DNA replication and transcription machinery. Cell. 2010;141:595–605. doi: 10.1016/j.cell.2010.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Madison KE, Jones-Foster EN, Vogt A, Kirtland Turner S, North SH, Nakai H. Stringent response processes suppress DNA damage sensitivity caused by deficiency in full-length translation initiation factor 2 or PriA helicase. Mol Microbiol. 2014;92:28–46. doi: 10.1111/mmi.12538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vogel U, Jensen KF. Effects of guanosine 3′,5′-bisdiphosphate (ppGpp) on rate of transcription elongation in isoleucine-starved Escherichia coli. J Biol Chem. 1994;269:16236–16241. [PubMed] [Google Scholar]
- 51.Trautinger BW, Jaktaji RP, Rusakova E, Lloyd RG. RNA polymerase modulators and DNA repair activities resolve conflicts between DNA replication and transcription. Mol Cell. 2005;19:247–258. doi: 10.1016/j.molcel.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 52.Kanjee U, Gutsche I, Alexopoulos E, Zhao B, El Bakkouri M, Thibault G, Liu K, Ramachandran S, Snider J, Pai EF, et al. Linkage between the bacterial acid stress and stringent responses: the structure of the inducible lysine decarboxylase. EMBO J. 2011;30:931–944. doi: 10.1038/emboj.2011.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kuroda A, Murphy H, Cashel M, Kornberg A. Guanosine tetra- and pentaphosphate promote accumulation of inorganic polyphosphate in Escherichia coli. J Biol Chem. 1997;272:21240–21243. doi: 10.1074/jbc.272.34.21240. [DOI] [PubMed] [Google Scholar]
- 54**.Maisonneuve E, Castro-Camargo M, Gerdes K. (p)ppGpp controls bacterial persistence by stochastic induction of toxin-antitoxin activity. Cell. 2013;154:1140–1150. doi: 10.1016/j.cell.2013.07.048. In this study, the authors found that stochastic production of (p)ppGpp in a subpopulation of cells slows their growth and results in persistence. The authors presented a model in which stochastic production of (p)ppGpp inhibits exopolyphosphatase, increasing inorganic polyphosphate levels and activating the Lon protease. Lon then degrades the antitoxins in toxin-antitoxin pairs, thus activating the toxins and inducing persistence. [DOI] [PubMed] [Google Scholar]
- 55.Gallant J, Irr J, Cashel M. The mechanism of amino acid control of guanylate and adenylate biosynthesis. J Biol Chem. 1971;246:5812–5816. [PubMed] [Google Scholar]
- 56.Rao F, See RY, Zhang D, Toh DC, Ji Q, Liang Z. YybT is a signaling protein that contains a cyclic dinucleotide phosphodiesterase domain and a GGDEF domain with ATPase activity. J Biol Chem. 2010;285:473–482. doi: 10.1074/jbc.M109.040238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Potrykus K, Murphy H, Philippe N, Cashel M. ppGpp is the major source of growth rate control in E. coli. Environ Microbiol. 2011;13:563–575. doi: 10.1111/j.1462-2920.2010.02357.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xiao H, Kalman M, Ikehara K, Zemel S, Glaser G, Cashel M. Residual guanosine 3′,5′-bispyrophosphate synthetic activity of relA null mutants can be eliminated by spoT null mutations. J Biol Chem. 1991;266:5980–5990. [PubMed] [Google Scholar]
- 59.Bittner AN, Kriel A, Wang JD. Lowering GTP level increases survival of amino acid starvation but slows growth rate for Bacillus subtilis cells lacking (p)ppGpp. J Bacteriol. 2014;196:2067–2076. doi: 10.1128/JB.01471-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Maisonneuve E, Gerdes K. Molecular mechanisms underlying bacterial persisters. Cell. 2014;157:539–548. doi: 10.1016/j.cell.2014.02.050. [DOI] [PubMed] [Google Scholar]
- 61.Khakimova M, Ahlgren HG, Harrison JJ, English AM, Nguyen D. The stringent response controls catalases in Pseudomonas aeruginosa and is required for hydrogen peroxide and antibiotic tolerance. J Bacteriol. 2013;195:2011–2020. doi: 10.1128/JB.02061-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Abranches J, Martinez AR, Kajfasz JK, Chávez V, Garsin Da, Lemos Ja. The molecular alarmone (p)ppGpp mediates stress responses, vancomycin tolerance, and virulence in Enterococcus faecalis. J Bacteriol. 2009;191:2248–2256. doi: 10.1128/JB.01726-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tabone M, Lioy VS, Ayora S, Machón C, Alonso JC. Role of toxin ζ and starvation responses in the sensitivity to antimicrobials. PLoS One. 2014;9:e86615. doi: 10.1371/journal.pone.0086615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nguyen D, Joshi-Datar A, Lepine F, Bauerle E, Olakanmi O, Beer K, McKay G, Siehnel R, Schafhauser J, Wang Y, et al. Active starvation responses mediate antibiotic tolerance in biofilms and nutrient-limited bacteria. Science. 2011;334:982–986. doi: 10.1126/science.1211037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65**.Germain E, Castro-Roa D, Zenkin N, Gerdes K. Molecular mechanism of bacterial persistence by HipA. Mol Cell. 2013;52:248–254. doi: 10.1016/j.molcel.2013.08.045. The toxin HipA is a key factor involved in persistence. Here the authors discovered that, rather than targeting EF-Tu, HipA phosphorylates GltX, which inhibits its aminoacyl-tRNA synthetase activity. The effect is to create “hungry” codons at the ribosomal A site, which are sensed by RelA. Thus HipA may increase persistence by indirectly stimulating (p)ppGpp production through mimicking amino acid starvation. [DOI] [PubMed] [Google Scholar]
- 66.Helaine S, Cheverton AM, Watson KG, Faure LM, Matthews Sa, Holden DW. Internalization of Salmonella by macrophages induces formation of nonreplicating persisters. Science. 2014;343:204–208. doi: 10.1126/science.1244705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67*.Mwangi MM, Kim C, Chung M, Tsai J, Vijayadamodar G, Benitez M, Jarvie TP, Du L, Tomasz A. Whole-genome sequencing reveals a link between β-lactam resistance and synthetases of the alarmone (p)ppGpp in Staphylococcus aureus. Microb Drug Resist. 2013;19:153–159. doi: 10.1089/mdr.2013.0053. In this study, whole-genome sequencing of two methicillin-resistant Staphylococcus aureus isolates identified two point mutations in relA. Cells with high levels of resistance overproduced (p)ppGpp, suggesting that the alarmone underlies the mechanism of β-lactam resistance. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kim C, Mwangi M, Chung M, Milheiriço C, Milheirço C, de Lencastre H, Tomasz A. The mechanism of heterogeneous beta-lactam resistance in MRSA: key role of the stringent stress response. PLoS One. 2013;8:e82814. doi: 10.1371/journal.pone.0082814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dordel J, Kim C, Chung M, de la Gándara MP, Holden MTJ, Parkhill J, de Lencastre H, Bentley SD, Tomasz A. Novel determinants of antibiotic resistance: Identification of mutated loci in highly methicillin-resistant subpopulations of methicillin-resistant Staphylococcus aureus. MBio. 2014;5 doi: 10.1128/mBio.01000-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70*.Wexselblatt E, Oppenheimer-Shaanan Y, Kaspy I, London N, Schueler-Furman O, Yavin E, Glaser G, Katzhendler J, Ben-Yehuda S. Relacin, a novel antibacterial agent targeting the Stringent Response. PLoS Pathog. 2012;8:e1002925. doi: 10.1371/journal.ppat.1002925. This study describes a 2′-deoxyguanosine-based analogue of (p)ppGpp, Relacin, that inhibits the (p)ppGpp synthesis activity of RelA in vivo and in vitro. Relacin inhibits growth, sporulation, and, importantly, biofilm formation. Thus Relacin may serve as a model compound for a new generation of antimicrobials. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.De la Fuente-Núñez C, Reffuveille F, Haney EF, Straus SK, Hancock REW. Broad-spectrum anti-biofilm peptide that targets a cellular stress response. PLoS Pathog. 2014;10(5):e1004152. doi: 10.1371/journal.ppat.1004152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Perederina A, Svetlov V, Vassylyeva MN, Tahirov TH, Yokoyama S, Artsimovitch I, Vassylyev DG. Regulation through the secondary channel - Structural framework for ppGpp-DksA synergism during transcription. Cell. 2004;118:297–309. doi: 10.1016/j.cell.2004.06.030. [DOI] [PubMed] [Google Scholar]
- 73.Levdikov VM, Blagova E, Joseph P, Sonenshein AL, Wilkinson AJ. The structure of CodY, a GTP- and isoleucine-responsive regulator of stationary phase and virulence in gram-positive bacteria. J Biol Chem. 2006;281:11366–11373. doi: 10.1074/jbc.M513015200. [DOI] [PubMed] [Google Scholar]