Abstract
Resistance to endocrine therapies in hormone receptor (HR)-positive breast cancer is a significant clinical problem for a considerable number of patients. The oncogenic transcription factor c-MYC (hereafter referred to as MYC), which regulates glutamine metabolism in cancer cells, has been linked to endocrine resistance. We were interested in whether MYC-mediated glutamine metabolism is also associated with aromatase inhibitor (AI) resistant breast cancer. We studied the expression and regulation of MYC and the fects of inhibition of MYC expression in both AI sensitive and resistant breast cancer cells. Considering the role of MYC in glutamine metabolism, we evaluated the contribution of glutamine to the proliferation of AI sensitive and resistant cells, and performed RNA-sequencing to investigate mechanisms of MYC-mediated glutamine utilization in AI resistance. We found that glutamine metabolism was independent of estrogen but still required ER in AI resistant breast cancer cells. The expression of MYC oncogene was up-regulated through the cross-talk between estrogen receptor (ER) and human epidermal growth factor receptor 2 (HER2) in AI resistant breast cancer cells. Moreover, the glutamine transporter solute carrier family (SLC)1A5 was significantly up-regulated in AI resistant breast cancer cells. ER down-regulator fulvestrant inhibited MYC, SLC1A5, glutaminase (GLS) and glutamine consumption in AI resistant breast cancer cells. Inhibition of MYC, SLC1A5 and GLS decreased AI resistant breast cancer cell proliferation. Our study has uncovered that MYC expression is up-regulated by the cross-talk between ER and HER2 in AI resistant breast cancer cells. MYC-mediated glutamine metabolism is associated with AI resistance of breast cancer.
Keywords: c-MYC, ER, HER2, glutamine, aromatase inhibitor resistance, breast cancer
1. Introduction
Breast cancer is the most common cancer and the second leading cause of cancer death among women worldwide [1]. Hormone receptors (HRs) – the estrogen receptor (ER) and/or the progesterone receptor (PR) are expressed in approximately 70% of breast cancer. The growth of breast cancer cells in such patients is dependent on estrogen and ER. Estrogen-mediated ER signaling can be targeted by blocking estrogen biosynthesis with aromatase inhibitors (AIs) (i.e., letrozole, anastrozole, and exemestane), antagonizing the binding of estrogens to the ER with tamoxifen, and down-regulating ER with fulvestrant. Endocrine therapies play an essential role in the treatment of patients with HR–positive breast cancer in postmenopausal women [2]. Unfortunately, a considerable number of patients have de novo resistance or ultimately develop acquired resistance to endocrine therapies [2].
Cross-talk between ER and human epidermal growth factor receptor 2 (HER2) signaling pathways has been implicated in endocrine therapy resistance [3-5]. HER2 signaling is up-regulated in breast tumors of patients treated with AIs [6]. Up-regulation of HER2 signaling pathways, including the mitogen-activated protein kinase (MAPK) and phosphatidylinositol 3′-kinase (PI3K)/AKT, can phosphorylate and activate ER in a ligand-independent manner [7, 8]. Many groups, including us, have showed that ER is up-regulated and constitutively activated in endocrine resistant breast cancer cells [9-11]. The MYC oncogene, which encodes c-MYC (MYC) protein, is a well-known ER-regulated gene [12, 13]. MYC is a transcription factor and plays a critical role in cell proliferation, growth, survival, differentiation and apoptosis [14]. Interestingly, MYC has been linked to endocrine resistance of breast cancer [15-18].
Glutamine is the most abundant amino acids in the body and plays an important role in cell proliferation. It is first converted to glutamate through the enzyme glutaminase (GLS), and then catabolized to α-ketoglutarate, an intermediate of the tricarboxylic acid (TCA) cycle [19]. Glutamine can be a source of both carbon and nitrogen for the synthesis of lipid, protein and nucleotide [20]. Although the growth and survival of most cancers depend on a high rate of aerobic glycolysis, some cancer cells cannot survive in the absence of exogenous glutamine, termed “glutamine addiction” [20]. Estrogen stimulation has been found to increase glutamine consumption in ER-positive breast cancer MCF7 cells [21], suggesting that glutamine metabolism is essential for estrogen-dependent cell proliferation. Oncogenic levels of MYC have also been linked to elevated glutamine uptake and metabolism in human cancers [22, 23].
Given the association between MYC and endocrine resistance [15, 16] as well as the regulation of glutamine metabolism by MYC in cancer cells[22, 23], we hypothesized that MYC-mediated glutamine metabolism is also associated with AI resistance. We studied the expression and regulation of MYC and the effects of inhibition of MYC expression in both AI sensitive and resistant breast cancer cells. We evaluated the contribution of glutamine to cell proliferation, and the association between glutamine consumption and hormone in breast cancer cells. Finally, we performed RNA-sequencing and investigated mechanisms of MYC-mediated glutamine metabolism in AI resistance.
2. Materials and methods
2.1. Cell culture
Human breast cancer cell line MCF7 derived cell lines MCF7aro and LTEDaro were generated in this laboratory and have been characterized and described previously [9, 24]. MCF7aro was routinely cultured in minimal Eagle's medium (MEM) supplemented with 10% fetal bovine serum (FBS), 2 mM L-glutamine, 1 mM sodium pyruvate, 100 U/mL penicillin-streptomycin, and 0.1 mg/mL G418. LTEDaro was maintained in phenol red–free MEM containing 10% charcoal/dextran-treated FBS with identical supplements as parental MCF7aro cells. For experiments under testosterone treatment, MCF7aro cells were cultured in phenol red–free MEM medium containing 10% charcoal/dextran-treated FBS for 72 hr before treatment. Experiments under deprived glutamine or glucose culture conditions were performed by using phenol red–free Dulbecco's Modified Eagle Medium (DMEM) without glucose and glutamine (GIBCO A14430-01) containing 10% charcoal/dextran-treated FBS.
2.2. Antibodies and reagents
Antihuman MYC (#5605), p-MAPK (#9101), MAPK (#9102), p-AKT (Ser473) (#9271), AKT (#9272), p-ER (Ser167) (#5587), GAPDH (#2118) antibodies were obtained from Cell Signaling Technology. Antihuman HER2 (#06-562) and p-ERα (Ser118) (ab32396) antibodies were from Abcam Inc. Antihuman ERα (HC-20) antibody (sc-543) was from Santa Cruz Biotechnology. The ER antagonist fulvestrant (#14409) and the SLC1A5 inhibitor L-Glutamic acid γ-(p-nitroanilide) (GPNA) (G6133) were obtained from Sigma-Aldrich. The glutaminase inhibitor compound 968 (#352010) was from EMD Millipore. The AKT inhibitor MK-2206 (S1078) was from Selleck Chemicals. The nontargeting control siRNA (sc-37007) and MYC siRNA (sc-29226) were obtained from Santa Cruz Biotechnology. The HER2 siRNA (L-003126-00) was from Dharmacon. The p44/42 MAPK siRNA (#6560) was from Cell Signaling Technology.
2.3. Real-time PCR
Total RNA was extracted from cells using the TRIzol reagent (Invitrogen) and quantified with a Nanodrop spectrophotometer. Reverse transcription was performed with SuperScript VILO cDNA synthesis kit (Invitrogen) from 2.5 μg total RNA. Real time PCR was performed using iQ5 multicolor real-time PCR detection system (Bio-Rad). The human MYC gene was amplified using the forward primer 5′-GGCTCCTGGCAAAAGGTCA -3′ and the reverse primer 5′- CTGCGTAGTTGTGCTGATGT -3′ (PrimerBank ID 239582723c1) [25]. The human SLC1A5 gene was amplified using the forward primer 5′-GAGCTGCTTATCCGCTTCTTC- 3′ and the reverse primer 5′- GGGGCGTACCACATGATCC-3′ (PrimerBank ID 223468565c1) [25]. The human GLS gene was amplified using the forward primer 5′- AGGGTCTGTTACCTAGCTTGG -3′ and the reverse primer 5′- ACGTTCGCAATCCTGTAGATTT -3′ (PrimerBank ID 373251163c1) [25]. The ACTB (β-actin) gene was amplified using the forward primer 5′- CACCAACTGGGACGACAT-3′ and the reverse primer 5′- GCACAGCCTGGATAGCAAC-3′. The real time PCR was established with the PerfeCTa SYBR Green SuperMix (Quanta Biosciences). The PCR results were normalized with β-actin as an internal control and then expressed as relative expression compared with reference samples. Each experiment was performed in triplicate. The data are expressed as means ± SE.
2.4. Transfection
Transfection of MCF7aro and LTEDaro with control siRNA or siRNAs against MYC, HER2 or MAPK was performed using the siPORT NeoFX transfection agent (Ambion) according to the manufacturer's protocol. For immunoblotting, transfection was performed in 60-mm dishes. For the MTT assay, 96-well plates were used. Briefly, cells were trypsinized and diluted in the medium at 1 × 105 cells/ml. siPORT NeoFX transfection agent and siRNA were diluted in OPTI-MEM I medium (Invitrogen), respectively. After being mixed and incubated for 10 min, the mixtures of siRNA and transfection agent were dispersed into a culture plate or dish. Cell suspensions (1 × 105 cells/ml) were overlaid onto the transfection complexes. The final concentration of siRNA was 30 nM.
2.5. Western blotting
Western blotting was performed as previously described [26]. In brief, cells were lysed in RIPA buffer (Cell Signaling) on ice for 5 min and then sonicated for 60 s. The protein concentration was determined by protein assay kit (Bio-Rad), and the samples were separated by 10% SDS-polyacrylamide gel electrophoresis (PAGE). After probing with a primary antibody, the membrane was incubated with a horseradish peroxidase-conjugated secondary antibody. Finally; signal intensity was determined with the SuperSignal West Pico Chemiluminescent (Thermo Scientific) substrate visualization. Relative expression of proteins was normalized to the internal control GAPDH.
2.6. Cell proliferation assay
Cell proliferation was measured by the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay. At the indicated time, the media in each well of 96-well plates were removed from the cells and replaced with 0.1 mL of fresh phenol red–free medium containing 0.5 mg/mL MTT, and then the cells were incubated at 37°C for 1 hr. After discarding the medium, the formazan dye trapped in the living cells was dissolved in 0.1 mL of DMSO and absorbance was measured at 570 nm with a SpectraMax M5 plate reader (Molecular Devices). Each experiment was performed using at least five replicates. The data are expressed as means ± SE.
2.7. Glutamine and ammonia measurements
Glutamine and ammonia concentrations were measured using a BioProfile 100 Plus analyzer (Nova Biomedical). Briefly, cells were seeded overnight and treated as indicated in replete medium. The controls were plated with an equal volume of the same medium without cells, incubated identically as the cell-containing plates. Media samples were collected for analysis immediately or stored at −20°C until the time of the assay. Glutamine and ammonia concentrations (measured in mmol/liter) were determined for each sample. Glutamine consumption was calculated by subtracting the concentration of the sample from the concentration of the medium control. Ammonia production was calculated by subtracting the concentration of the medium control from the concentration of the sample. The data are expressed as means ± SE.
2.8. RNA-sequencing and data analysis
LTEDaro cells were seeded overnight, and then cultured in phenol red-free DMEM medium containing 10% charcoal/dextran-treated FBS without glutamine, with 2mM L-glutamine, or with 2mM L-glutamine plus 100nM fulvestrant for 24 hr. Each treatment group was performed in triplicate. Total RNA was extracted from cells using the TRIzol reagent (Invitrogen). Transcriptome libraries were prepared, size selected, gel purified, and sequenced in our Integrative Genomics Core using an Illumina HiSeq 2000 system following manufacturer’s protocols (Illumina Inc.).
Reads were aligned to the human genome (build hg19) using TopHat [27]. RPKM (Reads Per Kilobase per Million mapped reads, [28]) were calculated for RefSeq [29] genes using an expectation-maximization algorithm in Partek® Genomics SuiteTM (Version 6.6, Partek, Inc., St. Louis, MO) that is based upon the method of Xing et al.[30]. RPKM values were then further normalized via log2 transformation following addition of a scaling factor of 0.1 [31]. Fold-change values were calculated on a linear scale based upon the least-squares mean, p-values were calculated 1-way ANOVA using the normalized RPKM values, and false discovery rate (FDR) values were calculated using the method of Benjamini and Hochberg [32]. Genes were defined as differentially expressed if they showed a |fold-change| value > 1.5 and FDR < 0.05.
2.9. Systems level analysis
P-values for pathway enrichment in Ingenuity Pathway Analysis (IPA, Ingenuity® Systems, www.ingenuity.com) were calculated via Fisher’s exact test. IPA was used to build networks and predict upstream regulator activity. An upstream regulator was predicted to be activated or inhibited based upon the fold-change values for known downstream targets: upstream regulators are predicted to be activated if the targets in the gene list show an activation z-score > 2 (and are predicted to be inhibited if the activation z-score < −2).
Heatmaps are displayed for standardized gene expression values. Hierarchical clustering was performed using average linkage with Euclidian distance as the distance metric. Clustering and visualization was performed in Partek® Genomics SuiteTM (Version 6.6, Partek, Inc., St. Louis, MO).
3. Results
3.1. Up-regulation of MYC in AI resistant breast cancer cells
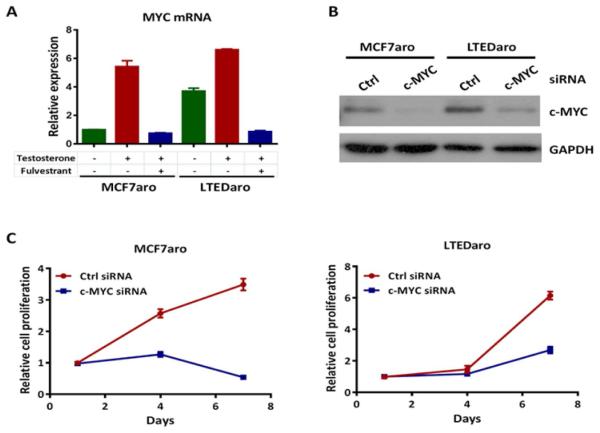
The MYC oncogene is an estrogen-dependent gene transcriptionally regulated by ER [12, 13]. ER is up-regulated and constitutively activated in AI resistant breast cancer cells [9-11]. To determine whether the regulation of MYC is through ER in AI resistant breast cancer cells, we examined the effects of testosterone and fulvestrant treatments on MYC expression in MCF7aro, an aromatase over-expressing breast cancer cell line that is sensitive to AIs, and LTEDaro, a MCF7aro derived cell line after long term estrogen deprivatioin (LTED) and a model for AI resistance. In the MCF7aro cells, we found that the MYC mRNA level was significantly elevated after treatment with testosterone, which is converted to estradiol by aromatase. Pretreatment with fulvestrant completely blocked the induction of MYC mRNA by testosterone in MCF7aro cells (Fig. 1A), confirming that MYC is an ER-regulated gene in the MCF7aro parental cells. Interestingly, the level of MYC mRNA in the LTEDaro cells was much higher than that in MCF7aro cells at baseline, and was almost compatible to the MYC mRNA level induced by testosterone treatment in MCF7aro cells. Pretreatment with fulvestrant also completely blocked the MYC mRNA expression (Fig. 1A). These findings suggest that MYC is up-regulated by constitutively activated ER in AI resistant breast cancer cells.
Fig. 1. Up-regulation of c-MYC in AI resistant breast cancer cells.
A. Real time PCR analysis of MYC expression in MCF7aro (hormone stripped for 72 hr) and LTEDaro cells treated with DMSO, testosterone (1nM) or testosterone (1nM) plus fulvestrant (100nM) for 6 hr. Values were normalized to ACTB and plotted relative to the expression of the MCF7aro cells treated with DMSO. Data are means ± SE (n=3).
B. Western blotting analysis of c-MYC expression in whole-cell lysates from MCF7aro or LTEDaro cells transfected with a nontargeting control (Ctrl) siRNA or a siRNA targeting c-MYC for 72 hr.
C. Cell proliferation of MCF7aro or LTEDaro cells transfected with a nontargeting control (Ctrl) siRNA or a siRNA targeting c-MYC. MTT was measured at the indicated time points. Data are means ± SE (n=5).
High MYC expression is a predictor of poor outcome following tamoxifen treatment in breast cancer [15, 17]. Because we found that MYC expression is up-regulated in AI resistant breast cancer cells, we sought to determine whether reduction of MYC expression by short interfering RNA (siRNA) decreases cell proliferation in MCF7aro and LTEDaro cells. We tested the knockdown efficiency of MYC expression by Western blotting. Consistent with the expression of MYC mRNA, the protein level of MYC in LTEDaro cells was higher than that in MCF7aro cells. We found a significant decrease of MYC protein levels after siRNA transfection and that the knockdown rates were similar in both cell lines (Fig. 1B). We then evaluated the effect of MYC knockdown on cell proliferation by a MTT assay. We found that cell proliferation was significantly decreased after knockdown of MYC in both the MCF7aro and LTEDaro cells (Fig. 1C), suggesting that MYC plays an important role in the proliferation of both AI sensitive and resistant breast cancer cells.
3.2. Cross-talk between ER and HER2 up-regulates MYC in AI resistant breast cancer cells
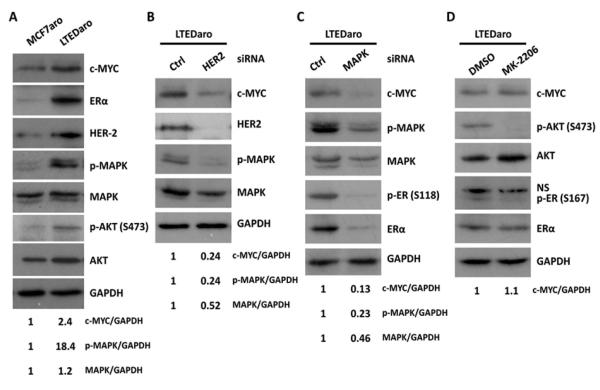
Cross-talk between ER and HER2 signaling pathways has been implicated in the endocrine therapy resistance [3-5]. We found that MYC was up-regulated in AI resistant breast cancer cells, and that up-regulation of MYC was correlated with expression levels of ERα and HER2 in LTEDaro cells (Fig. 2A). In the same set of experiments, the phosphorylation of both the MAPK and AKT was significantly increased in LTEDaro cells as compared with MCF7aro cells (Fig. 2A), confirming that HER2 signaling is up-regulated in AI resistant breast cancer cells, linking to an increased expression of MYC.
Fig. 2. Cross-talk between ER and HER2 up-regulates c-MYC in AI resistant breast cancer cells.
A. Western blotting analysis of the expression of c-MYC, ER and HER2, and the phosphorylation of MAPK and AKT in MCF7aro and LTEDaro cells cultured in normal medium. The quantification of c-MYC, p-MAPK, MAPK was normalized to the loading control GAPDH, and values were calculated relative to the expression of the MCF7aro cells.
B. c-MYC protein levels in whole-cell lysates from LTEDaro cells transfected with a nontargeting control (Ctrl) siRNA or a siRNA targeting HER2 for 72 hr. The quantification of c-MYC, p-MAPK, MAPK was normalized to GAPDH, and values were calculated relative to the expression of the LTEDaro cells treated with control siRNA.
C. c-MYC protein levels in whole-cell lysates from LTEDaro cells transfected with a nontargeting control (Ctrl) siRNA or a siRNA targeting MAPK for 72 hr. The quantification of c-MYC, p-MAPK, MAPK was normalized to GAPDH, and values were calculated relative to the expression of the LTEDaro cells treated with control siRNA.
D. c-MYC protein levels in whole-cell lysates from LTEDaro cells treated with DMSO or the AKT inhibitor MK-2206 (1μM) for 24 hr. The quantification of c-MYC was normalized to GAPDH, and values were calculated relative to the expression of the LTEDaro cells treated with DMSO.
To test whether HER2 signaling contributes to the up-regulation of MYC, we determined if reduction of HER2 expression by siRNA decreases the expression of MYC. We found that knockdown of HER2 significantly decreased MYC expression in LTEDaro cells (Fig. 2B), supporting that HER2 up-regulates MYC in AI resistant breast cancer cells.
The two major intracellular pathways activated by HER2 are the RAS-MAPK and PI3K-AKT pathways [34]. To test which pathway is responsible for the up-regulation of MYC, we treated LTEDaro cells with siRNA against MAPK and an inhibitor of AKT (MK-2206). We found that MAPK siRNA treatment significantly decreased MYC expression as well the phosphorylation of ERα (Ser118) and total ERα (Fig. 2C). In contrast, the AKT inhibitor (MK-2206) did not reduce MYC expression despite a decrease of the phosphorylation of AKT (Ser473) and ERα (Ser167) (Fig. 2D). These findings suggest that HER2 regulates MYC expression via the MAPK and constitutive activation of ER.
3.3. Glutamine consumption is independent of estrogen but requires ER in AI resistant breast cancer cells
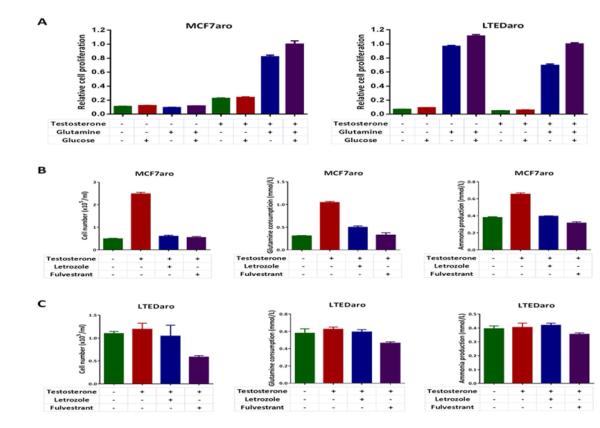
The oncogenic transcription factor MYC, which plays a role in endocrine resistance of breast cancer [15, 16], has been linked to glutamine metabolism in human cancers [22, 23]. Because glutamine is a source of nitrogen and a key source to provide metabolites to the TCA cycle for biosynthesis in proliferating cells [19], we determined the cell growth responses of MCF7aro or LTEDaro cells to glutamine and glucose deprivation in the presence or absence of testosterone. The growth of both cell lines was stimulated by glutamine, but moderately by glucose (Fig. 3A), suggesting that glutamine addiction is present in both the AI sensitive and resistant breast cancer cells. However, in the absence of testosterone, the growth of MCF7aro cells was completely diminished, whereas the growth of LTEDaro cells was not affected (Fig. 3A), indicating estrogen-independent glutamine metabolism in AI resistant breast cancer cells.
Fig. 3. Glutamine consumption is independent of estrogen but requires ER in AI resistant breast cancer cells.
A. Cell growth responses of MCF7aro cells (hormone stripped for 72 hr) to glutamine or glucose deprivation in the presence or absence of testosterone. MTT was measured at day 6. Data are means ± SE (n=6).
B. Cell growth responses of LTEDaro cells to glutamine or glucose deprivation in the presence or absence of testosterone. MTT was measured at day 6. Data are means ± SE (n=6).
C. Cell growth, glutamine consumption and ammonia production in MCF7aro cells (hormone stripped for 72 hr) treated with DMSO, testosterone (1nM), testosterone (1nM) plus letrozole (200nM), or testosterone (1nM) plus fulvestrant (100nM). Cell number, glutamine and ammonia concentrations were measured at day 3. Glutamine consumption was calculated by subtracting the concentration of the sample from the concentration of the medium control. Ammonia production was calculated by subtracting the concentration of the medium control from the concentration of the sample. Data are means ± SE.
D. Cell growth, glutamine consumption and ammonia production in LTEDaro cells treated with DMSO, testosterone (1nM), testosterone (1nM) plus letrozole (200nM), or testosterone (1nM) plus fulvestrant (100nM). Cell number, glutamine and ammonia concentrations were measured at day 3. Glutamine consumption was calculated by subtracting the concentration of the sample from the concentration of the medium control. Ammonia production was calculated by subtracting the concentration of the medium control from the concentration of the sample. Data are means ± SE.
Estrogen stimulation has been found to increase glutamine consumption in MCF7 breast cancer cells [21].To test the role of estrogen on glutamine metabolism in breast cancer cells, we determined the effects of testosterone, letrozole and fulvestrant on glutamine consumption and ammonia production in MCF7aro and LTEDaro cells. We measured cell number, glutamine and ammonia concentrations after 72 hr of treatment. We found that testosterone significantly promoted MCF7aro cell growth and increased glutamine consumption, whereas both letrozole and fulvestrant blocked the effect of testosterone treatment (Fig. 3B), showing that estrogen stimulates the consumption of glutamine through ER in AI sensitive breast cancer cells. Glutamine is the major nitrogen donor for producing ammonia in cells [35]. Consistent with the changes of glutamine consumption, ammonia levels were increased after testosterone treatment, whereas letrozole blocked the production of ammonia in MCF7aro cells (Fig. 3B). In contrast, testosterone and letrozole had no effect on the LTEDaro cell cell growth, glutamine consumption or ammonia production, whereas treatment with fulvestrant decreased cell growth and glutamine consumption (Fig. 3C), indicating that glutamine consumption becomes independent of estrogen but still requires constitutively activated ER in AI resistant breast cancer cells.
3.4. Fulvestrant suppresses glutamine-mediated cell proliferation and expression of genes in lipid and glucose metabolism in AI resistant breast cancer cells
As our results showed that ER plays a role in glutamine metabolism despite estrogen independency in AI resistant breast cancer cells, we cultured LTEDaro cells in media without glutamine (Gln−), with glutamine (Gln+), or with glutamine and fulvestrant (Gln/Fulvestrant) for 24 hr, and performed RNA-sequencing to identify the roles for ER in glutamine metabolism in LTEDaro cells.
We analyzed the molecular and cellular functions of the gene signature with the Ingenuity software. The lists of top five molecular and cellular functions with P-values are shown in Table 1. “Lipid metabolism” was the most altered function in the comparison between the Gln+ and Gln− groups, and “cellular growth and proliferation” and “cell death and survival” were in the top five lists of altered function in both comparisons of Gln+ vs. Gln− and Gln/Fulvestrant vs. Gln+.
Table 1.
Top molecular and cellular functions identified by Ingenuity Pathway Analysis (IPA)
| Category | P-value | # Molecules |
|---|---|---|
| Gln+ vs. Gln− | ||
| Lipid metabolism | 2.49E-06 - 4.85E-02 | 57 |
| Small molecule biochemistry | 2.49E-06 - 4.85E-02 | 119 |
| Vitamin and mineral metabolism | 2.49E-06 - 4.85E-02 | 23 |
| Cellular growth and proliferation | 1.37E-05 - 4.64E-02 | 213 |
| Cell death and survival | 2.56E-05 - 4.79E-02 | 206 |
| Gln/Fulvestrant vs. Gln+ | ||
| Cell cycle | 6.80E-11 – 2.73E-02 | 76 |
| DNA replication, recombination, and repair | 2.46E-09 – 2.73E-02 | 62 |
| Cellular growth and proliferation | 1.68E-05 – 2.73E-02 | 107 |
| Cell death and survival | 2.29E-05 – 2.73E-02 | 105 |
| Cellular assembly and organization | 3.53E-05 – 2.73E-02 | 22 |
| Gln/Fulvestrant vs. Gln− | ||
| Cellular growth and proliferation | 4.59E-07 – 2.98E-02 | 216 |
| Cellular assembly and organization | 2.69E-06 – 2.88E-02 | 31 |
| DNA replication, recombination, and repair | 2.69E-06 – 2.98E-02 | 26 |
| Amino acid metabolism | 8.27E-06 – 2.75E-02 | 27 |
| Molecular transport | 8.27E-06 – 2.98E-02 | 100 |
Note: P-values are calculated by the Fisher’s exact test.
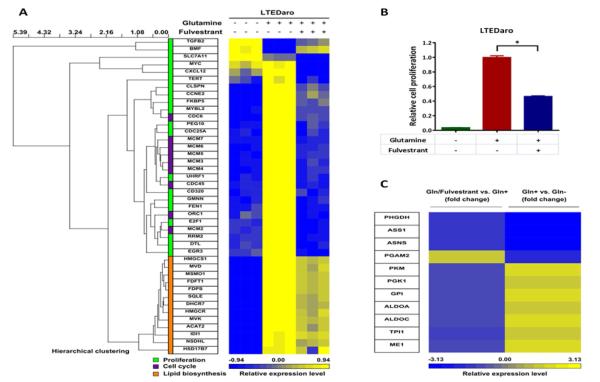
We observed increased gene expression of proliferation and cell cycle in the Gln+ group as compared with the Gln− group. The gene expression profile of proliferation and cell cycle in the Gln/Fulvestrant group approached the profile in the Gln− group (Fig. 4A). These results suggest that constitutively activated ER plays important roles in the glutamine-mediated cell proliferation. We confirmed the effect on cell proliferation by a MTT assay. We found that glutamine promoted cell proliferation, whereas fulvestrant decreased glutamine-mediated cell proliferation (Fig. 4B).
Fig. 4. Fulvestrant suppresses glutamine-mediated cell proliferation and expression of genes in lipid and glucose metabolism in AI resistant breast cancer cells.
A. The dendrogram and heat map show the hierarchical cluster analysis of gene-expression data from LTEDaro cells cultured in media without glutamine, with glutamine, or with both glutamine and fulvestrant for 24 hr. Gene-expression profiles were obtained from RNA-sequencing data. Three groups of genes were analyzed: cell proliferation, cell cycle and lipid biosynthesis. Columns represent individual genes, and rows represent individual samples. Each cell in the matrix represents the expression level of a gene in an individual sample. The scale bar indicates the level of expression; red indicates a high level of expression, and blue a low level of expression.
B. Cell proliferation of LTEDaro cells cultured in media without glutamine, with glutamine, or with both glutamine and fulvestrant. MTT was measured at day 7. Data are means ± SE (n=6). Asterisk (*) denotes P <0.05 (t test).
C. The heat map shows the gene-expression analysis of data from LTEDaro cells cultured in media without glutamine (Gln−), with glutamine (Gln+), or with both glutamine and fulvestrant (Gln/Fulvestrant) for 24 hr. Gene-expression profiles were obtained from RNA-sequencing data. Columns represent individual genes that are involved in glucose metabolism, and rows represent fold changes of Gln/Fulvestrant vs. Gln+ (left) or Gln+ vs. Gln− (right). The scale bar indicates the level of expression; red indicates a high level of expression, and blue a low level of expression.
Glutamine can serve as a source of carbon and nitrogen for the synthesis of lipid, protein and nucleotide [20]. In the RNA-sequencing data, we observed a strong transcriptional lipid biosynthesis gene signature in the Gln+ group as compared with the Gln− group. Fulvestrant treatment has less impact on the expression of genes in lipid biosynthesis (Fig. 4A), suggesting that constitutively activated ER may not be essential for glutamine-promoted lipid biosynthesis.
While the finding that glutamine stimulates lipid biosynthesis is important, we were particularly excited that glutamine promotes glycolysis (Table 2 and Fig. 4C). The latter observation supports our results in Fig. 3 that glucose is not sufficient to drive cell proliferation and glutamine is needed for the cellular utilization of glucose. Furthermore, utilization of glucose was found to be regulated by estrogen, as indicated by comparing the levels of these genes in the Gln+ group with those in the Gln/Fuvestrant group (Fig. 4C) and the review by Chen et al. [36].
Table 2.
Glutamine-mediated expression of glycolysis enzymes (Gln+ vs. Gln−) identified by Ingenuity Pathway Analysis (IPA)
| Symbol | Entrez gene name | Fold change | P-value |
|---|---|---|---|
| ALDOA | aldolase A, fructose-bisphosphate | 1.504 | 1.30E-06 |
| ALDOC | aldolase C, fructose-bisphosphate | 2.087 | 3.62E-06 |
| ENO1 | enolase 1, (alpha) | 1.553 | 4.54E-07 |
| GPI | glucose-6-phosphate isomerase | 1.665 | 1.66E-08 |
| PFKP | phosphofructokinase, platelet | 1.582 | 2.84E-07 |
| PGAM2 | phosphoglycerate mutase 2 (muscle) | -1.632 | 1.74E-03 |
| PGK1 | phosphoglycerate kinase 1 | 1.607 | 2.84E-08 |
| PKM | pyruvate kinase, muscle | 1.712 | 4.79E-07 |
| TPI1 | triosephosphate isomerase 1 | 1.568 | 3.86E-06 |
Note: P-values are calculated by the Fisher’s exact test.
3.5. Fulvestrant inhibits MYC-mediated glutamine metabolism in AI resistant breast cancer cells
MYC is an ER-regulated gene [12, 13]. As expected, expressions of ERα (encoded by ESR1) target genes, including MYC, were blocked by fulvestrant in the RNA-sequencing data (Fig. 5A). We determined the effect of fulvestrant on the protein expression of ERα and MYC by Western blotting. We found that the protein levels of ERα and MYC were significantly decreased following fulvestrant treatment (Fig. 5B).
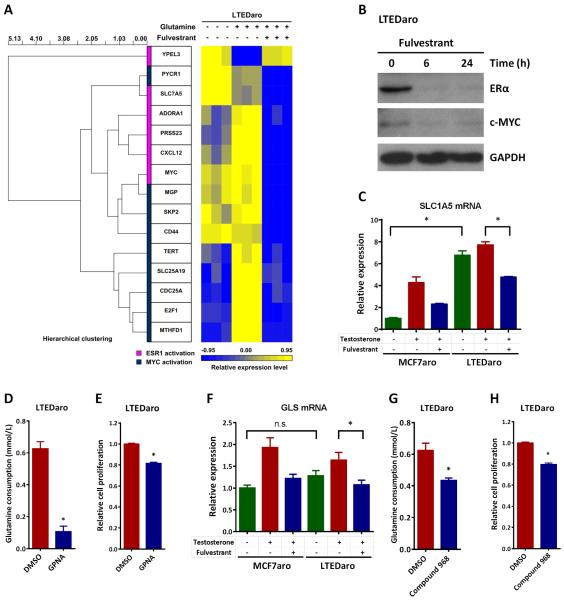
Fig. 5. Fulvestrant inhibits c-Myc, SLC1A5 and GLS in AI resistant breast cancer cells.
A. The dendrogram and heat map show the hierarchical cluster analysis of gene-expression data from LTEDaro cells cultured in media without glutamine, with glutamine, or with both glutamine and fulvestrant for 24 hr. Gene-expression profiles were obtained from RNA-sequencing data. Two groups of genes were analyzed: ESR1 activation and MYC activation. Columns represent individual genes, and rows represent individual samples. Each cell in the matrix represents the expression level of a gene in an individual sample. The scale bar indicates the level of expression; red indicates a high level of expression, and blue a low level of expression.
B. Western blotting analysis of ERα and c-MYC expression in whole-cell lysates from LTEDaro cells treated with fulvestrant (100nM) at the indicated time points.
C. Real time PCR analysis of SLC1A5 expression in MCF7aro (hormone stripped for 72 hr) and LTEDaro cells treated with DMSO, testosterone (1nM) or testosterone (1nM) plus fulvestrant (100nM) for 6 hr. Values were normalized to ACTB and plotted relative to the expression of the MCF7aro cells treated with DMSO. Data are means ± SE (n=3).
D. Glutamine consumption in LTEDaro cells treated with DMSO or the SLC1A5 inhibitor GPNA (0.5mM). Glutamine concentrations were measured at day 3. Glutamine consumption was calculated by subtracting the concentration of the sample from the concentration of the medium control. Data are means ± SE.
E. Cell proliferation of LTEDaro cells treated with DMSO or the SLC1A5 inhibitor GPNA (0.5mM). MTT was measured at day 4. Data are means ± SE (n=6).
F. Real time PCR analysis of GLS expression in MCF7aro (hormone stripped for 72 hr) and LTEDaro cells treated with DMSO, testosterone (1nM) or testosterone (1nM) plus fulvestrant (100nM) for 6 hr. Values were normalized to ACTB and plotted relative to the expression of the MCF7aro cells treated with DMSO. Data are means ± SE (n=3).
G. Glutamine consumption in LTEDaro cells treated with DMSO or the GLS inhibitor compound 968 (10μM). Glutamine concentrations were measured at day 3. Glutamine consumption was calculated by subtracting the concentration of the sample from the concentration of the medium control. Data are means ± SE.
H. Cell proliferation of LTEDaro cells treated with DMSO or the GLS inhibitor compound 968 (10μM). MTT was measured at day 4. Data are means ± SE (n=6). Asterisk (*) denotes P <0.05 (t test).
MYC enhances glutamine uptake from the extracellular space in cancer cells through up-regulation of the glutamine importer, solute carrier family (SLC)1A5 (also called ASCT2) [20]. To determine whether SLC1A5 is up-regulated in AI resistant breast cancer cells, we examined the mRNA levels of SLC1A5 in MCF7aro and LTEDaro cells at baseline and following testosterone and fulvestrant treatment. In MCF7aro cells, expression of SLC1A5 was significantly increased by testosterone treatment, and pretreatment with fulvestrant blocked the SLC1A5 expression induced by testosterone treatment, suggesting that SLC1A5 is an estrogen-inducible gene in AI sensitive breast cancer cells (Fig. 5C). Interestingly, the level of SLC1A5 mRNA in the LTEDaro cells was much higher than that in MCF7aro cells at baseline, which is consistent with the up-regulation of MYC in LTEDaro cells (Fig. 5C and Fig. 1A). Testosterone treatment did not elevate the SLC1A5 expression in LTEDaro cells, indicating that the glutamine transporter SLC1A5 is independent of estrogen in AI resistant breast cancer cells. Pretreatment with fulvestrant significantly decreased SLC1A5 expression in LTEDaro cells (Fig. 5C), suggesting that SLC1A5 is still regulated by ER in AI resistant breast cancer.
Since SLC1A5 is significantly up-regulated in LTEDaro cells, we wondered whether inhibition of SLC1A5 decreases glutamine consumption and cell proliferation. We determined the effect of GPNA, an inhibitor of SLC1A5 that suppresses SLC1A5-dependent glutamine uptake [20], on glutamine consumption and cell proliferation. We found that GPNA significantly decreased glutamine consumption and cell proliferation in LTEDaro cells (Fig. 5D-E).
Once glutamine enters the cell, it can be metabolized through glutaminolysis. GLS is the enzyme converting glutamine into glutamate and can also be regulated by MYC [20]. We thus examined the mRNA levels of GLS in MCF7aro and LTEDaro cells at baseline and following testosterone and fulvestrant treatment. In MCF7aro cells, testosterone treatment significantly elevated the GLS expression, whereas pretreatment with fulvestrant significantly blocked the effect of testosterone treatment (Fig. 5F). In contrast to SLC1A5, the expression of GLS was not significantly increased in LTEDaro cells as compared with MCF7aro cells at baseline. Testosterone treatment did not significantly increase GLS expression, but fulvestrant decreased the expression of GLS in LTEDaro cells (Fig. 5F).
To test whether inhibition of GLS affects glutamine consumption and cell proliferation in AI resistant breast cancer cells, we treated LTEDaro cells with the GLS inhibitor compound 968. We found that compound 968 significantly decreased both the glutamine consumption and cell proliferation in LTEDaro cells, indicating a role for GLS in the treatment of AI resistant breast cancer (Fig. 5G-H).
Taken together, these data suggest that fulvestrant inhibits the expression of MYC, SLC1A5 and GLS, all of which are involved in glutamine metabolism, in AI resistant breast cancer cells. Our results also suggest that the increased expression of MYC and SLC1A5 results from the constitutively activated ERα, whose expression can be inhibited by fulvestrant, in AI resistant breast cancer cells.
4. Discussion
Resistance to endocrine therapies in HR-positive breast cancer is a significant clinical problem for a considerable number of patients. In this study, we identified an association between MYC-mediated glutamine metabolism and AI resistant breast cancer. We found that the oncogenic transcription factor MYC was up-regulated through the cross-talk between ER and HER2 in AI resistant breast cancer cells. Glutamine metabolism was independent of estrogen but still required ER in AI resistant breast cancer. RNA-sequencing revealed that glutamine promoted cell proliferation, lipid biosynthesis and glycolysis, whereas the ER down-regulator fulvestrant suppressed glutamine-mediated cell proliferation and glucose metabolism in AI resistant breast cancer cells. Moreover, fulvestrant inhibited the expresion of MYC, SLC1A5 and GLS, and reduction of MYC, SLC1A5 and GLS decreased cell proliferation in AI resistant breast cancer cells.
Amplification and/or over-expression of the oncogene MYC are common in high grade breast cancer [37]. Up-regulation of MYC has been linked to endocrine resistance in breast cancer [15-18]. In our study, we demonstrated that MYC was up-regulated in AI resistant breast cancer cells. Growing evidence supports that a close interaction between the ER and HER2 signaling pathways is implicated in the endocrine therapy resistance [3-5]. We found that up-regulation of MYC was correlated with the expression levels of ER and HER2 in AI resistant breast cancer cells. In addition, we demonstrated that cross-talk between ER and HER2 regulated MYC expression in AI resistant breast cancer cells. These findings suggest that aberrant signaling through MYC regulated by ER and HER2 pathways is involved in AI resistance of breast cancer.
Oncogenic levels of MYC regulate glutamine metabolism in human cancers [22, 23]. This led us to link glutamine metabolism to AI resistance. We found that both the AI sensitive and resistant breast cancer cells were addicted to glutamine, but in an estrogen-independent manner in breast cancer cells resistant to AIs, suggesting that glutamine metabolism has been escaped from estrogen dependency in AI resistant breast cancer. Estrogen stimulation has been found to increase glutamine consumption in MCF7 breast cancer cells [21]. Consistently, we showed similar results in AI sensitive breast cancer cells. But again, glutamine consumption in AI resistant breast cancer cells was independent of estrogen. However, ER down-regulator fulvestrant decreased glutamine consumption in AI resistant breast cancer cells, suggesting that ER still plays an important role in glutamine consumption despite estrogen independency. Furthermore, our RNA-sequencing data strongly suggest that fulvestrant inhibits glutamine metabolism in AI resistant breast cancer cells. Interestingly, Shajahan-Haq et al. recently have found that MYC and glutamine metabolism are associated with antiestrogen resistance with the use of tamoxifen and fulvestrant resistant breast cancer cell lines [18]. Taken together, MYC-mediated glutamine metabolism is an emerging mechanism of breast cancer resistant to endocrine therapies.
MYC facilitates glutamine uptake through SLC1A5 and promotes glutamine metabolism (e.g. GLS that is an enzyme converting glutamine to glutamate) [20]. SLC1A5 is the transporter required for glutamine-dependent mTORC1 activation [22, 38]. Interestingly, we found that SLC1A5 was significantly up-regulated in AI resistant breast cancer cells. Expression of SLC1A5 was significantly induced by testosterone treatment in MCF7aro cells but not in LTEDaro cells, suggesting the glutamine transporter is independent of estrogen and supporting that glutamine can be consumed in the absence of estrogen in breast cancer cells resistant to AIs. Moreover, inhibition of MYC, SLC1A5 and GLS significantly decreased cell proliferation in AI resistant breast cancer cells, suggesting that targeting glutamine metabolism is a potential therapeutic strategy in the treatment of AI resistant breast cancer.
In conclusion, our work shows that MYC is up-regulated by the cross-talk between ER and HER2 in AI resistant breast cancer cells. MYC-mediated glutamine metabolism is associated with AI resistance of breast cancer.
Highlights.
Aromatase inhibitor (AI) resistant breast cancer cells are addicted to glutamine.
Glutamine consumption is independent of estrogen but still requires ER in AI resistant breast cancer cells.
c-MYC expression is up-regulated through the cross-talk between ER and HER 2 in AI resistant breast cancer cells.
Targeting c-MYC-mediated glutamine metabolism inhibits the proliferation of AI resistant breast cancer cells.
Acknowledgements
The authors would like to thank Drs. Xiwei Wu and Jinhui Wang at the City of Hope Integrative Genomics Core for assistance with RNA-sequencing. The study was supported by the National Institutes of Health (R01 ES08258 to SC) and Panda Charitable Foundation (to SC).
Abbreviations
- AI
aromatase inhibitor
- DMEM
Dulbecco's Modified Eagle Medium
- ER
estrogen receptor
- FBS
fetal bovine serum
- Gln
glutamine
- GLS
glutaminase
- GPNA
L-Glutamic acid γ-(p-nitroanilide)
- HER2
human epidermal growth factor receptor 2
- HR
hormone-receptor
- IPA
Ingenuity Pathway Analysis
- LTED
long term estrogen deprivatioin
- MAPK
mitogen-activated protein kinase
- MEM
minimal Eagle's medium
- MTT
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- PI3K
phosphatidylinositol 3'-kinase
- siRNA
short interfering RNA
- SLC1A5
solute carrier family 1A5
- TCA
tricarboxylic acid
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interest
The authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported.
References
- [1].Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- [2].Mauri D, Pavlidis N, Polyzos NP, Ioannidis JP. Survival with aromatase inhibitors and inactivators versus standard hormonal therapy in advanced breast cancer: meta-analysis. J Natl Cancer Inst. 2006;98:1285–1291. doi: 10.1093/jnci/djj357. [DOI] [PubMed] [Google Scholar]
- [3].Martin LA, Farmer I, Johnston SR, Ali S, Marshall C, Dowsett M. Enhanced estrogen receptor (ER) alpha, ERBB2, and MAPK signal transduction pathways operate during the adaptation of MCF-7 cells to long term estrogen deprivation. J Biol Chem. 2003;278:30458–30468. doi: 10.1074/jbc.M305226200. [DOI] [PubMed] [Google Scholar]
- [4].Jelovac D, Sabnis G, Long BJ, Macedo L, Goloubeva OG, Brodie AM. Activation of mitogen-activated protein kinase in xenografts and cells during prolonged treatment with aromatase inhibitor letrozole. Cancer Res. 2005;65:5380–5389. doi: 10.1158/0008-5472.CAN-04-4502. [DOI] [PubMed] [Google Scholar]
- [5].Tokunaga E, Kimura Y, Mashino K, Oki E, Kataoka A, Ohno S, Morita M, Kakeji Y, Baba H, Maehara Y. Activation of PI3K/Akt signaling and hormone resistance in breast cancer. Breast Cancer. 2006;13:137–144. doi: 10.2325/jbcs.13.137. [DOI] [PubMed] [Google Scholar]
- [6].Flageng MH, Moi LL, Dixon JM, Geisler J, Lien EA, Miller WR, Lonning PE, Mellgren G. Nuclear receptor co-activators and HER-2/neu are upregulated in breast cancer patients during neo-adjuvant treatment with aromatase inhibitors. Br J Cancer. 2009;101:1253–1260. doi: 10.1038/sj.bjc.6605324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Campbell RA, Bhat-Nakshatri P, Patel NM, Constantinidou D, Ali S, Nakshatri H. Phosphatidylinositol 3-kinase/AKT-mediated activation of estrogen receptor alpha: a new model for anti-estrogen resistance. J Biol Chem. 2001;276:9817–9824. doi: 10.1074/jbc.M010840200. [DOI] [PubMed] [Google Scholar]
- [8].Kato S, Endoh H, Masuhiro Y, Kitamoto T, Uchiyama S, Sasaki H, Masushige S, Gotoh Y, Nishida E, Kawashima H, Metzger D, Chambon P. Activation of the estrogen receptor through phosphorylation by mitogen-activated protein kinase. Science. 1995;270:1491–1494. doi: 10.1126/science.270.5241.1491. [DOI] [PubMed] [Google Scholar]
- [9].Masri S, Phung S, Wang X, Wu X, Yuan YC, Wagman L, Chen S. Genome-wide analysis of aromatase inhibitor-resistant, tamoxifen-resistant, and long-term estrogen-deprived cells reveals a role for estrogen receptor. Cancer Res. 2008;68:4910–4918. doi: 10.1158/0008-5472.CAN-08-0303. [DOI] [PubMed] [Google Scholar]
- [10].Martin LA, Farmer I, Johnston SR, Ali S, Dowsett M. Elevated ERK1/ERK2/estrogen receptor cross-talk enhances estrogen-mediated signaling during long-term estrogen deprivation. Endocr Relat Cancer. 2005;12(Suppl 1):S75–84. doi: 10.1677/erc.1.01023. [DOI] [PubMed] [Google Scholar]
- [11].Santen RJ, Song RX, Zhang Z, Kumar R, Jeng MH, Masamura A, Lawrence J, Jr., Berstein L, Yue W. Long-term estradiol deprivation in breast cancer cells up-regulates growth factor signaling and enhances estrogen sensitivity. Endocr Relat Cancer. 2005;12(Suppl 1):S61–73. doi: 10.1677/erc.1.01018. [DOI] [PubMed] [Google Scholar]
- [12].Shang Y, Hu X, DiRenzo J, Lazar MA, Brown M. Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell. 2000;103:843–852. doi: 10.1016/s0092-8674(00)00188-4. [DOI] [PubMed] [Google Scholar]
- [13].Wang C, Mayer JA, Mazumdar A, Fertuck K, Kim H, Brown M, Brown PH. Estrogen induces c-myc gene expression via an upstream enhancer activated by the estrogen receptor and the AP-1 transcription factor. Mol Endocrinol. 2011;25:1527–1538. doi: 10.1210/me.2011-1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Grandori C, Cowley SM, James LP, Eisenman RN. The Myc/Max/Mad network and the transcriptional control of cell behavior. Annual review of cell and developmental biology. 2000;16:653–699. doi: 10.1146/annurev.cellbio.16.1.653. [DOI] [PubMed] [Google Scholar]
- [15].Miller TW, Balko JM, Ghazoui Z, Dunbier A, Anderson H, Dowsett M, Gonzalez-Angulo AM, Mills GB, Miller WR, Wu H, Shyr Y, Arteaga CL. A gene expresion signature from human breast cancer cells with acquired hormone independence identifies MYC as a mediator of antiestrogen resistance. Clin Cancer Res. 2011;17:2024–2034. doi: 10.1158/1078-0432.CCR-10-2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].McNeil CM, Sergio CM, Anderson LR, Inman CK, Eggleton SA, Murphy NC, Millar EK, Crea P, Kench JG, Alles MC, Gardiner-Garden M, Ormandy CJ, Butt AJ, Henshall SM, Musgrove EA, Sutherland RL. c-Myc overexpression and endocrine resistance in breast cancer. J Steroid Biochem Mol Biol. 2006;102:147–155. doi: 10.1016/j.jsbmb.2006.09.028. [DOI] [PubMed] [Google Scholar]
- [17].Al-azawi D, Ilroy MM, Kelly G, Redmond AM, Bane FT, Cocchiglia S, Hill AD, Young LS. Ets-2 and p160 proteins collaborate to regulate c-Myc in endocrine resistant breast cancer. Oncogene. 2008;27:3021–3031. doi: 10.1038/sj.onc.1210964. [DOI] [PubMed] [Google Scholar]
- [18].Shajahan-Haq AN, Cook KL, Schwartz-Roberts JL, Eltayeb AE, Demas DM, Warri AM, Facey CO, Hilakivi-Clarke LA, Clarke R. MYC regulates the unfolded protein response and glucose and glutamine uptake in endocrine resistant breast cancer. Molecular cancer. 2014;13:239. doi: 10.1186/1476-4598-13-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell metabolism. 2008;7:11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- [20].Wise DR, Thompson CB. Glutamine addiction: a new therapeutic target in cancer. Trends in biochemical sciences. 2010;35:427–433. doi: 10.1016/j.tibs.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Forbes NS, Meadows AL, Clark DS, Blanch HW. Estradiol stimulates the biosynthetic pathways of breast cancer cells: detection by metabolic flux analysis. Metabolic engineering. 2006;8:639–652. doi: 10.1016/j.ymben.2006.06.005. [DOI] [PubMed] [Google Scholar]
- [22].Wise DR, DeBerardinis RJ, Mancuso A, Sayed N, Zhang XY, Pfeiffer HK, Nissim I, Daikhin E, Yudkoff M, McMahon SB, Thompson CB. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc Natl Acad Sci U S A. 2008;105:18782–18787. doi: 10.1073/pnas.0810199105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Gao P, Tchernyshyov I, Chang TC, Lee YS, Kita K, Ochi T, Zeller KI, De Marzo AM, Van Eyk JE, Mendell JT, Dang CV. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature. 2009;458:762–765. doi: 10.1038/nature07823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Zhou DJ, Pompon D, Chen SA. Stable expression of human aromatase complementary DNA in mammalian cells: a useful system for aromatase inhibitor screening. Cancer Res. 1990;50:6949–6954. [PubMed] [Google Scholar]
- [25].Spandidos A, Wang X, Wang H, Seed B. PrimerBank: a resource of human and mouse PCR primer pairs for gene expression detection and quantification. Nucleic Acids Res. 2010;38:D792–799. doi: 10.1093/nar/gkp1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Chen Z, Wang O, Nie M, Elison K, Zhou D, Li M, Jiang Y, Xia W, Meng X, Chen S, Xing X. Aromatase deficiency in a Chinese adult man caused by novel compound heterozygous CYP19A1 mutations: Effects of estrogen replacement therapy on the bone, lipid, liver and glucose metabolism. Mol Cell Endocrinol. 2015;399:32–42. doi: 10.1016/j.mce.2014.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Meth. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- [29].Pruitt KD, Tatusova T, Brown GR, Maglott DR. NCBI Reference Sequences (RefSeq): current status, new features and genome annotation policy. Nucleic Acids Research. 2012;40:D130–D135. doi: 10.1093/nar/gkr1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Xing Y, Yu T, Wu YN, Roy M, Kim J, Lee C. An expectation-maximization algorithm for probabilistic reconstructions of full-length isoforms from splice graphs. Nucleic Acids Research. 2006;34:3150–3160. doi: 10.1093/nar/gkl396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Warden CD, Yuan Y-C, Wu X. Optimal Calculation of RNA-Seq Fold-Change Values. Int J Comput Bioinfo In Silico Model. 2013;2:285–292. [Google Scholar]
- [32].Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society. Series B (Methodological) 1995;57:289–300. [Google Scholar]
- [33].Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene Ontology: tool for the unification of biology. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Yarden Y, Sliwkowski MX. Untangling the ErbB signalling network, Nature reviews. Molecular cell biology. 2001;2:127–137. doi: 10.1038/35052073. [DOI] [PubMed] [Google Scholar]
- [35].Shanware NP, Mullen AR, DeBerardinis RJ, Abraham RT. Glutamine: pleiotropic roles in tumor growth and stress resistance. Journal of molecular medicine. 2011;89:229–236. doi: 10.1007/s00109-011-0731-9. [DOI] [PubMed] [Google Scholar]
- [36].Chen JQ, Brown TR, Russo J. Regulation of energy metabolism pathways by estrogens and estrogenic chemicals and potential implications in obesity associated with increased exposure to endocrine disruptors. Biochim Biophys Acta. 2009;1793:1128–1143. doi: 10.1016/j.bbamcr.2009.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Blancato J, Singh B, Liu A, Liao DJ, Dickson RB. Correlation of amplification and overexpression of the c-myc oncogene in high-grade breast cancer: FISH, in situ hybridisation and immunohistochemical analyses. Br J Cancer. 2004;90:1612–1619. doi: 10.1038/sj.bjc.6601703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Nicklin P, Bergman P, Zhang B, Triantafellow E, Wang H, Nyfeler B, Yang H, Hild M, Kung C, Wilson C, Myer VE, MacKeigan JP, Porter JA, Wang YK, Cantley LC, Finan PM, Murphy LO. Bidirectional transport of amino acids regulates mTOR and autophagy. Cell. 2009;136:521–534. doi: 10.1016/j.cell.2008.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]