Abstract
Reverse genetics systems constitute one of the most important and powerful tools to study the molecular biology of viruses. We developed a new strategy for the recovery of murine norovirus from a single plasmid in which a bacteriophage T7 RNA polymerase (T7pol) promoter for transcription and an EMCV IRES for efficient translation were engineered immediately upstream of the viral genome. Infectious noroviruses were recovered following transfection of the newly designed plasmid into nonpermissive BHK-21 and HEK293T cell lines that were engineered to express T7pol constitutively. Recovery of the virus did not require the presence of a ribozyme at the 3’-end of the virus genome. The strategy worked also for the efficient recovery of feline calicivirus in these normally nonpermissive cell types. This simplified reverse genetics approach may be broadly applicable to other caliciviruses.
Keywords: Murine norovirus, feline calicivirus, reverse genetics system, T7 RNA polymerase, EMCV IRES
The family Caliciviridae is composed of viruses with medical and veterinary importance, and is currently divided into five genera: Norovirus, Lagovirus, Vesivirus, Sapovirus and Nebovirus (Carstens, 2010). The human noroviruses (HuNV) are the major cause of gastroenteritis outbreaks worldwide (Patel et al., 2008). The study of HuNV has been restricted due to the absence of a cell culture system and an efficient small animal model. Therefore, cultivable animal caliciviruses, such as feline calicivirus (FCV) and murine norovirus (MNV), have been used as surrogate models to study the molecular biology of HuNV (Luttermann and Meyers, 2010; Wobus et al., 2006). The availability of reverse genetics system (RGS) for these animal caliciviruses, along with their well-established permissive cell culture systems, has further enhanced their importance as models (Chaudhry et al., 2007; Sosnovtsev and Green, 1995; Ward et al., 2007). Caliciviruses are non-enveloped viruses with a 6.7–8.5 kb long single-stranded, positive-sense RNA genome covalently linked to viral protein VPg at the 5’-end (Black et al., 1978; Herbert et al., 1997; Schaffer et al., 1980)The VPg is required for the infectivity of viral RNA transfected into cells, and interacts with host cellular factors involved in translation (Burroughs and Brown, 1978; Daughenbaugh et al., 2003; Dunham et al., 1998; Goodfellow et al., 2005; Herbert et al., 1997).
Capped genomic RNA synthesized in vitro from a plasmid containing a full-length (FL) cDNA of the FCV RNA genome under control of the T7pol promoter were shown to be infectious in the absence of VPg when transfected into permissive Crandell-Rees feline kidney (CRFK) cells (Sosnovtsev and Green, 1995). Infectious FCV particles could be also recovered after transfection of the plasmid into cells infected with recombinant avian vaccinia virus, MVA/T7, engineered to express T7pol (Sosnovtsev et al., 1997; Wyatt et al., 1995). Infection of cells with the MVA/T7 virus resulted in expression of the T7pol and provided the MVA virus capping enzymes necessary for calicivirus recovery. Similar approaches were employed to develop RGSs for other members of the Caliciviridae with an available cell culture system such as Porcine Enteric Calicivirus (PEC), the reference strain of genus Sapovirus (Chang et al., 2005), and Tulane virus, an unassigned calicivirus (Wei et al., 2008). Successful recovery of the rabbit hemorrhagic disease virus (RHDV), a calicivirus from the genus Lagovirus, showed that infectious virus could be generated in permissive cells transfected with a plasmid carrying the virus genome engineered downstream from the cytomegalovirus (CMV) promoter (Liu et al., 2008). Attempts to develop an RGS for the genus Norovirus were unsuccessful until the discovery of MNV (Karst et al., 2003), which is the only member of the genus that can be grown efficiently in cell culture (Wobus et al., 2004). Ward et al. (2007) developed an RGS for MNV, dependent on the co-transfection of two plasmids: one carrying the virus FL genome sequence under control of TET-O/minCMV promoter and one encoding the transcriptional transactivator TET-R/VP16 under control of a mammalian promoter. Expression of the TET-R/VP16 led to activation of MNV genome transcription and to recovery of infectious virus particles (Ward et al., 2007). Another RGS for MNV was developed using a single DNA plasmid transfection and required infection with a helper virus expressing T7pol. In the same work, co-infection with MVA/T7 blocked the replication of MNV. However, use of a different poxvirus, fowlpox virus, expressing T7pol, allowed the recovery of infectious MNV particles (Chaudhry et al., 2007). The most recently developed RGS for MNV uses in vitro transcription to generate the FL genomes capped with an optimized capping system, followed by the transfection of the capped RNAs into permissive RAW264.7 cells (Yunus et al., 2010).
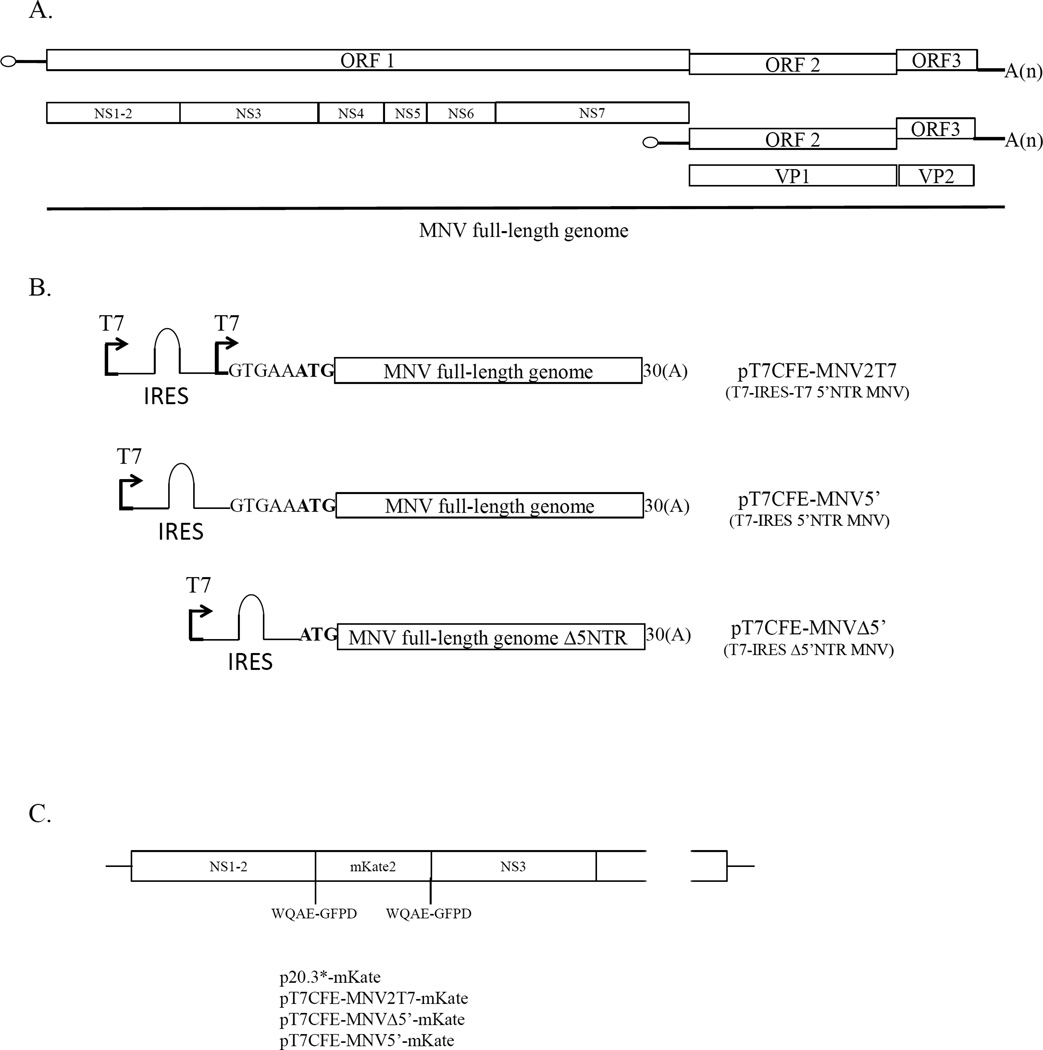
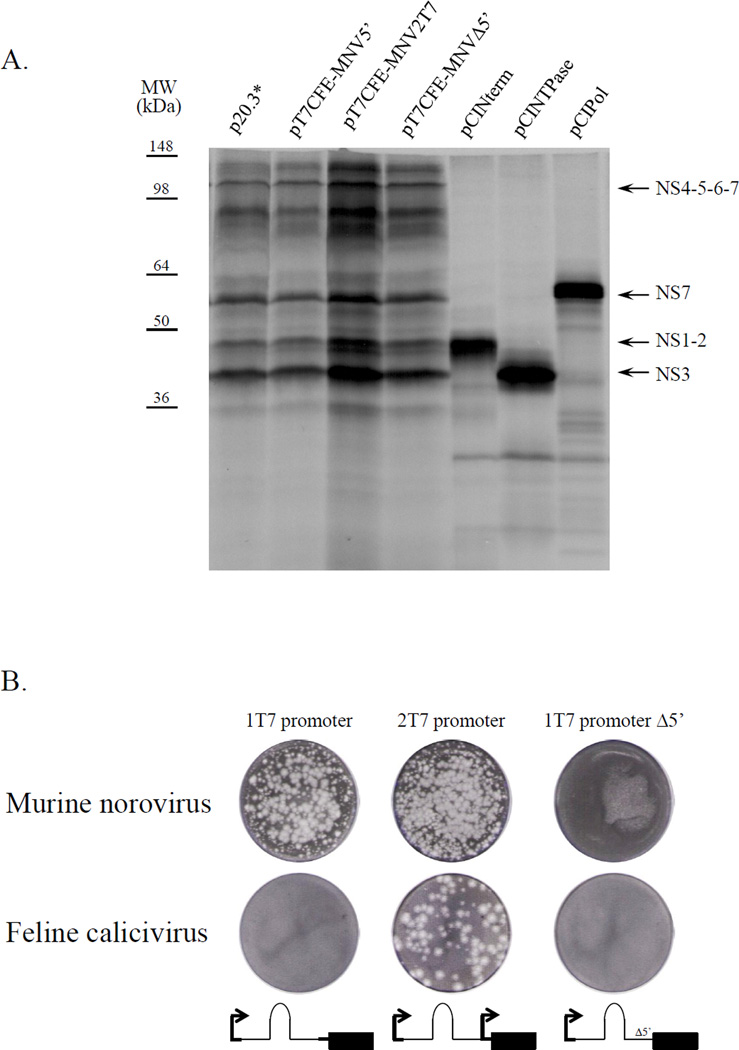
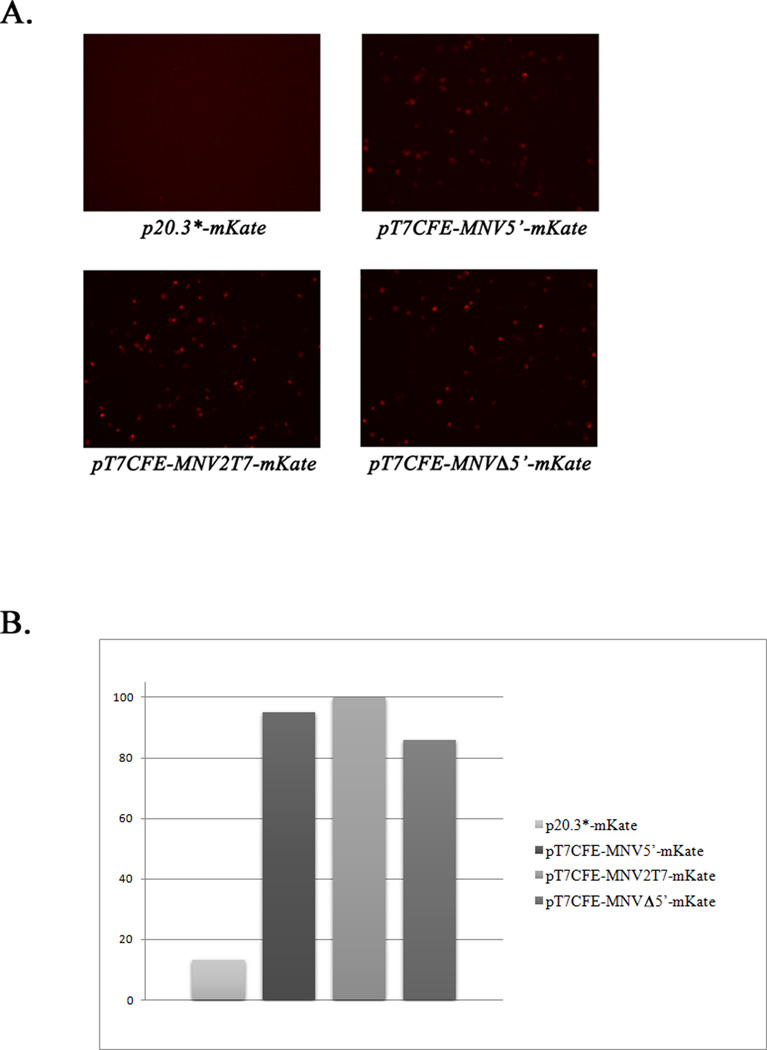
With the exception of RHDV (Liu et al., 2006), capping of transcribed FL genome RNA molecules has been required to recover infectious calicivirus particles. Because VPg was known to be essential for the infectivity of native viral RNA, it was proposed that the synthetic cap structure at the 5’-end of the transcribed calicivirus genome could substitute for the viral VPg in the initiation of translation (Sosnovtsev and Green, 1995). Although capped transcripts were infectious, a direct comparison later showed that the efficiency of translation of capped transcribed viral RNA was lower than that of the purified VPg-linked viral RNA (Chaudhry et al., 2007). Furthermore, capped RNA yielded lower virus titers in recovery experiments compared to VPg-linked RNA (Thumfart and Meyers, 2002; Yunus et al., 2010). An IRES element from the EMCV genome has been routinely engineered into calicivirus expression constructs upstream of the translated ORF to enhance viral protein expression (Pletneva et al., 1998; Sosnovtsev et al., 1998). This study explored whether IRES-driven expression from an infectious calicivirus FL clone could be used to increase the efficiency and performance of the RGS. Recombinant plasmids where FL virus (MNV or FCV) genome sequence was placed downstream of the EMCV IRES sequence were constructed. To generate the MNV constructs, the genome of the MNV-1 strain CW1P3 was amplified from p20.3 (Sosnovtsev et al., 2006) using PfuUltra II Hotstart PCR Master Mix polymerase (Agilent Technologies, Inc., Wilmington, DE) and primers listed in Table 1. Purified PCR fragments were treated with BsmBI and ligated into the pT7CFE1-His vector (Thermo Scientific, Rockford, IL) treated with BsmfI and SpeI enzymes. The resulting plasmids contained either FL (pT7CFE-MNV2T7 and pT7CFE-MNV5’) or truncated (pT7CFE-MNVΔ5’) virus genome placed downstream of the T7pol promoter and the EMCV IRES element in the pT7CFE1 (Fig. 1). The pT7CFE-MNV2T7 was engineered to bear a second T7pol promoter between the 5’-end of the MNV genome and the IRES sequence. The T7pol transcription of this plasmid would result in a synthesis of two RNA species: a full-length virus genome alone and genome fused to the IRES sequence. While the first RNA was expected to work as a template in the virus RNA replication, the second was thought to serve as template for translation of the virus nonstructural proteins (Fig. 1). All plasmids were verified by sequence analysis before evaluation in in vitro translation reactions and recovery experiments. Expression and proteolytic processing of the MNV nonstructural polyprotein were examined using an in vitro-coupled transcription-translation system (TnT T7 Coupled Reticulocyte Lysate System, Promega, Madison, WI). Briefly, 2 µg of the plasmid DNAs were added to the TnT reaction mix assembled according to the manufacturer’s instructions and reactions were performed at 30°C for 2 hrs. Proteins were radiolabeled with [S35] methionine (PerkinElmer, Waltham, MA) added to the mix at a concentration of 0.4 mCi/ml. For all tested constructs, the TnT reaction produced similar cleavage patterns yielding nonstructural proteins and precursors with molecular masses ranging from approximately 16 to 110 kDa (Fig. 2a). The identities of the proteins generated by autocatalytic processing of the MNV ORF1 polyprotein synthesized from the virus FL cDNA clone p20.3 (Fig. 2a) have been described previously (Sosnovtsev et al., 2006). The plasmids were tested next for the production of infectious virions in cells expressing T7pol. For this purpose, 1.5 × 106 BSR-T7 cells (Buchholz et al., 1999) were transfected with 4 µg of the plasmid DNA using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) according to the manufacturer’s protocol. Twenty-four hours post transfection, cells were subjected to freeze-thawing three times, and the recovery of virus was tested in the permissive RAW264.7 cells. Virus was produced only in the BSR-T7 cells transfected with the plasmids pT7CFE-MNV2T7 and pT7CFE-MNV5’. Transfection of cells with the plasmid pT7CFE-MNVΔ5’ (lacking the 5’-end of the genome) did not result in recovery of infectious virions as evidenced by the absence of cytopathic effect and plaque formation in RAW264.7 cells inoculated with the pT7CFE-MNVΔ5’ transfection lysates (Fig. 2b). Similar negative results were obtained for the FL cDNA clone of MNV genome placed directly under control of the T7pol promoter without an IRES sequence (p20.3*, data not shown). Differences in intracellular levels of expression of the ORF1 between IRES and non-IRES constructions were examined by means of a fluorescent reporter tag (mKate2, Evrogen, Moscow, Russia) inserted at the border of the NS1-2 and NS3 protein (Fig. 1C, Sandoval et al. in preparation). Transfection of the cells with p20.3*-mKate, pT7CFE-MNV2T7-mKate, pT7CFE-MNVΔ5’-mKate and pT7CFE-MNV5’-mKate (Fig. 1C) showed a significantly higher level of the ORF1-mKate protein expression in BSR-T7 cells transfected with the FL cDNA clones with IRES-mediated expression (Fig. 3). Taken together, these results suggest that the presence of the IRES element driving expression of the virus nonstructural proteins from the T7pol-transcribed virus genomic RNA and stable expression of this enzyme in BSR-T7 cells were sufficient for initiation of the virus replication in these cells.
Table 1.
PCR primers used in generation of FL MNV clones and site-directed mutagenesis of the MNV genome.
| Name | Sequence 5’-3’ |
|---|---|
| MNV sense pT7CFE2T7 | 5’ATATATATATCGTCTCTTTTGAAAAACACGATGATAATACGACTCACTATAGTGAAATGAGGATGGCAACGCC 3’ |
| MNV sense pT7CFE5 | 5’ATATATATATCGTCTCTTTTGAAAAACACGATGATAATGTGAAATGAGGATGGCAACGCCATCTTCTGC 3’ |
| MNV sense pT7CFEΔ5 | 5’ATATATATATCGTCTCTTTTGAAAAACACGATGATAATATGAGGATGGCAACGCCATCTTCTGC 3’ |
| MNV reverse | 5’ATATATATATCGTCTCACTAGTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTAAAATGCATCTAACTACCAC 3’ |
| Mutant SL1 | 5’ CGATGATAATGTGAAATGAGGATGGCAACACCTTCTTCTGCGCCCTCT GTGC 3’ |
| Mutant SL2 | 5’ GGATGGCAACGCCATCTTCTGCACCAAGTGTGCGCAACACAG AGAAACGC 3’ |
| Mutants SL1/2 | 5’ CGATGATAATGTGAAATGAGGATGGCAACACCTTCTTCT GCACCAAGTGTGCGCAACACAGAGAAACG |
The viral sequences of the oligonucleotides MNV sense pT7CFE2T7, MNV sense pT7CFE5, MNV sense pT7CFED5 and MNV reverse are shown in bold. The T7pol promoter sequence is in italics and recognition sequence of the BsmBI restriction enzyme is underlined. For oligonucleotides Mutant SL1, Mutant SL2 and Mutant SL1/2 used for site-directed mutagenesis of the MNV genome, the introduced mutations are shown in bold and underlined.
Figure 1.
Graphic representation of the MNV genome and description of the T7-IRES plasmid constructs used in this study. (A) Organization of the MNV genome showing the location of the virus nonstructural and structural protein genes. (B) Schematic diagrams of the T7-IRES clones carrying the MNV genome sequence: pT7CFE-MNV2T7 (FL MNV genome placed downstream of two T7pol promoters separated by the EMCV IRES sequence), pT7CFE-MNV5’ (FL MNV genome placed downstream of the T7pol promoter and the EMCV IRES sequence), pT7CFE-MNVΔ5’ (similar to the pT7CFE-MNV5’ construction with the MNV genome lacking the 5’-end NTR). Plasmids were engineered as described in text. First ATG codon of ORF1 is shown in bold. (C) Schematic diagram showing the site of the mKate2 gene insertion in the constructs: p20.3*-mKate, pT7CFE-MNV2T7-mKate, pT7CFE-MNVΔ5’-mKate and pT7CFE-MNV5’-mKate.
Figure 2.
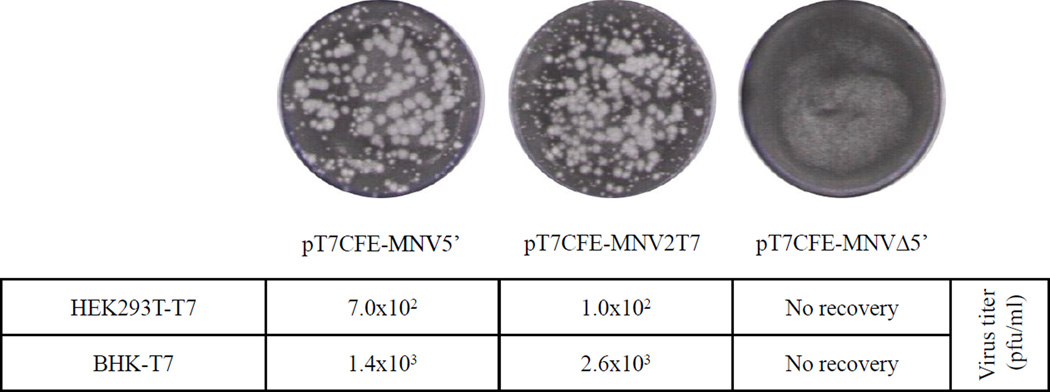
Analysis of the protein expression and virus recovery from the T7-IRES genomic clones. (A) Comparison of the in vitro translation products derived from the FL MNV genome constructs and from clones pCINterm, pCINTPase, and pCIPol encoding sequences of NS1-2, NS3 and NS7 proteins (Sosnovtsev et al., 2006). The plasmids were translated in the TnT system (Promega) in the presence of [35S]-methionine. The synthesized radiolabeled proteins were resolved in a 4–20% gradient Tris-Gly polyacrylamide gel and visualized by autoradiography. Translational products corresponding to MNV nonstructural proteins NS1-2, NS3, NS4-5-6-7, and NS7 are indicated with arrows. (B) Plaque formation assay of the recovered MNV and FCV viruses. The BHK-T7 cells were transfected with T7-IRES-FL MNV or FCV clones as described in text, and production of infectious virus particles was assayed on RAW264.7 and CRFK cells, respectively.
Figure 3.
Expression of the fluorescent protein, mKate2 in BSR-T7 cells transfected with p20.3*-mKate, pT7CFE-MNV2T7-mKate, pT7CFE-MNVΔ5’-mKate and pT7CFE-MNV5’-mKate. (A) Fluorescence microscopy analysis of the BSR-T7 cells transfected with the indicated plasmids. Images of fluorescent cells were collected 18 hrs post transfection using a Leica DMI4000 B microscope (Leica Microsystems, Buffalo Grove, IL), and a Retiga-2000R camera (QImaging, Surrey, BC, Canada). (B) Fluorescence of the transfected cells at 588/633 nm was measured and analyzed using Synergy Neo Plate Reader and Gen5 Data Analysis Software (Biotek, Winooski, VT). Fluorescence of mock-transfected cells was considered as a background. Data represent mean values of measurements (n=3) normalized to the fluorescence of pT7CFE-MNV2T7-mKate-transfected cells.
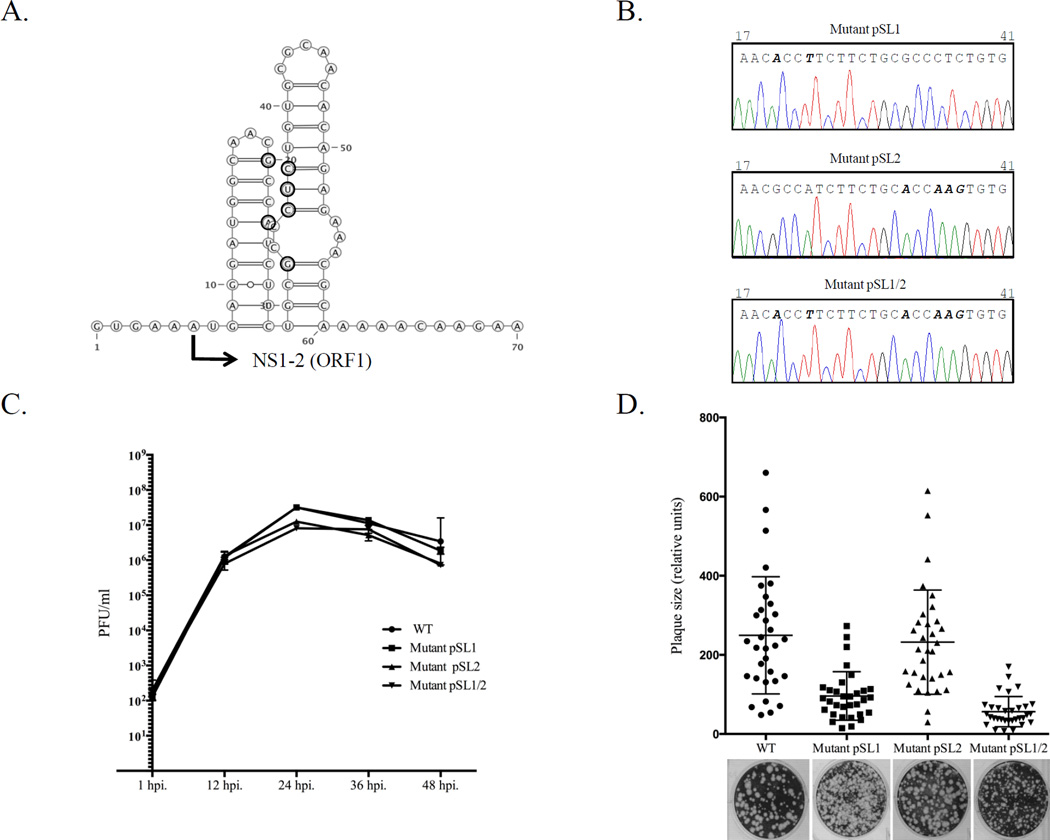
An interesting finding was recovery of viable virus from the pT7CFE-MNV5’ vector where the 5’-end of the MNV genome was fused directly to the IRES sequence (Fig. 1b). Since the only T7pol promoter in this construction was located upstream of the IRES sequence, the T7pol transcripts corresponded to the chimeric IRES-FL genome sequence (Fig. 1b). The 5’-RACE amplification (Invitrogen) and direct sequencing of the cDNA fragment corresponding to the 5’-end of the recombinant virus genome showed the identity of the recovered sequence as that of wild-type virus. This finding suggested that the MNV RNA replication machinery could regenerate the precise 5’-end of the virus genome even in the presence of additional nonviral sequences upstream of the 5’-end. To confirm that this new reverse genetics approach allowed the recovery of genetically modified virus, several silent mutations (Table 2, Fig. 4A) were introduced into predicted stem loop structures within the NS1-2 gene of the MNV genome (Simmonds et al., 2008). Briefly, the genome sequence in pT7CFE-MNV5’ was modified using the QuikChange Site-Directed mutagenesis kit (Agilent Technologies) and primers containing the desired mutations (Table 1). The resulting plasmids pSL1, pSL2 and pSL1/2 were sequenced to confirm the presence of the introduced mutations and used in recovery experiments as described above. The recovered viruses were plaque-purified and passaged in RAW264.7 cells before their NS1-2 regions were sequenced to confirm the mutants’ identities and stability of the incorporated mutations. Sequencing analysis showed that all of them retained the mutations after three passages (Fig. 4b). RNA folding analysis of the mutagenized NS1-2 sequence revealed that the suggested mutations could destabilize the stem-loop structures present in this part of the MNV genome (Simmonds et al., 2008). The functional role of these stem-loop structures in the MNV replication remains unknown; however, it has been proposed that these RNA secondary structures might be recognized by virus and cellular host proteins involved in virus replication (Carstens, 2010). To determine if the introduced mutations had an effect on virus growth, the kinetics of virus production were examined. Confluent RAW264.7 cells (approximately 1 × 106 cells) were incubated with each virus at a multiplicity of infection of 0.05 for 1 h at 37°C. The inoculum was removed, cells were washed, and fresh medium was added. Cell culture fluid was harvested 1, 12, 24, 36, and 48 h post infection, and the titer was determined by plaque assay. The mutant viruses showed growth kinetics similar to that of the wild-type virus (Fig. 4c). To compare plaque-forming characteristics of the recovered mutant viruses to that of the wild-type virus, the plaque assays were performed as described above and the plaque sizes were calculated using the image analysis software GraphClick (http://www.arizona-software.ch/graphclick). The statistical significance in the different plaque sizes was analyzed using the GraphPad Prism v.6 (GraphPad Software, La Jolla, CA). Interestingly, the mutant virus SL2 formed plaques similar in size and morphology to those of the wild-type virus, although the SL1 and SL1/2 plaques appeared to be smaller (Fig. 4d). Altogether, these results demonstrated that the new MNV RGS could be successfully used for the genetic manipulations of the virus genome and generation of recombinant viruses with altered growth properties.
Table 2.
Mutations introduced into the genome of the MNV mutants pSL1, pSL2 and pSL1/2.
| Mutant | Genome position | WT/Mut |
|---|---|---|
| SL1 | 20 | G/A |
| 23 | A/U | |
| SL2 | 32 | G/A |
| 35 | C/A | |
| 36 | U/A | |
| 37 | C/G | |
| SL1/2 | 20 | G/A |
| 23 | A/U | |
| 32 | G/A | |
| 35 | C/A | |
| 36 | U/A | |
| 37 | C/G | |
Figure 4.
Generation of genetically-modified MNV. (A) Site-directed mutagenesis was employed to introduce silent mutations in the beginning of the NS1-2 gene. The corresponding positions are indicated with bold circles and the beginning of the NS1-2 gene (ORF1) is marked with arrow. The introduced changes could alter the RNA secondary structure predicted for this part of the MNV genome (http://rna.tbi.univie.ac.at/cgi-bin/RNAfold.cgi). (B) Comparison of the genome sequence (nts 17-41) determined for the recovered SL1, SL2, and SL1/2 mutant viruses at passage 3. Mutations at positions 20 and 23 (SL1), 32 and 35–37 (SL2) and 20, 23, 32, and 35–37 (SL1/2) are shown in bold. (C) Growth properties of mutant viruses were compared with those of the wild-type (wt) virus. Confluent monolayers of RAW264.7 cells were infected with wt or mutant viruses at a multiplicity of infection of 0.05. Cell culture fluid was harvested 1, 12, 24, 36, and 48 h post infection. Virus titer was determined by end point titration in a plaque assay. (D) Recovered mutant viruses were assayed for their ability to form plaques in RAW264.7 cells. The relative size of 32 randomly selected plaques was plotted.
To determine whether the new approach could be applied to the recovery of other members of the Caliciviridae, we constructed a similar set of FL FCV cDNA clones starting with the pTMF-1 clone as template (Pletneva et al., 1998; Sosnovtsev et al., 1998). The resulting cDNA clones had an FCV FL (pFCV2T7 and pFCV5’) or 5’-end-truncated (pFCVΔ5) genome sequences engineered into the pTM-1 vector (Moss et al., 1990) downstream of the EMCV IRES element similarly to that of the MNV constructs described above (data not shown). The recombinant plasmids were verified by sequencing and by in vitro translation assay. Next, the verified plasmids were transfected into BSR-T7 cells and their infectivity was assayed in CRFK cells known to support FCV growth (Fig. 2b). The production of infectious FCV particles was observed for the construct with two T7pol promoters (FCV2T7), similar to MNV. In contrast to MNV, no FCV was recovered from the pFCV5’ vector (Fig. 2b) suggesting that FCV may differ in its ability to edit nonviral sequences upstream of the genomic 5’-end.
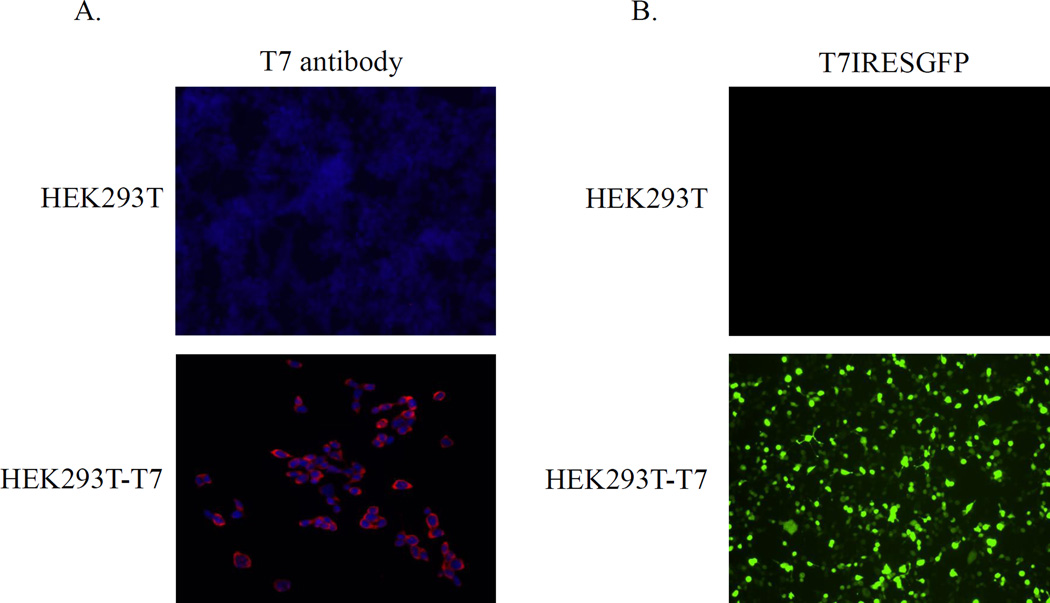
To examine if the T7-IRES-dependent system could facilitate replication of norovirus in human cells, we created HEK293T cells stably expressing T7pol. In order to make this cell line, the T7pol gene was cloned into the pIRESpuro2 vector (Clontech, Palo Alto, CA) and the resulting plasmid pIRESpuroT7pol was transfected into the HEK293T cells. Stably transfected cells were selected by the addition of 4 µg/ml puromycin (Toku-e, Bellingham, WA) to the cell culture medium, and several cell clones were isolated using a cloning discs technique (Sigma-Aldrich, St.Louis, MO). The T7pol expression in cloned cells, designated HEK293T-T7, was verified using immunofluorescence assay. Briefly, cells were fixed with cold methanol for 10 min, rinsed and then blocked with 1% normal goat serum in PBS for 1 hour at RT. Next, the fixed cells were incubated overnight with the T7pol-specific mouse monoclonal antibodies (Novagen, San Diego, CA), and bound antibodies were detected with Alexa Fluor 568-conjugated goat anti-mouse IgG (Invitrogen). Following washing, cells were visualized using a Leica DMI4000 B microscope (Leica Microsystems, Buffalo Grove, IL), and images were captured with a Retiga 2000R camera (Qimaging, Surrey, BC, Canada) and processed using iVision 4.0.14 software (BioVision, Exton, PA). The HEK293T-T7 probed with anti-T7pol antibodies showed a diffuse staining consistent with cytoplasmic localization of the expressed T7pol protein (Fig. 5a). To assess the enzymatic activity of the T7pol, the HEK293T-T7 cells were transfected with 2 ìg of pT7CFEGFP plasmid (Thermo Scientific) that encodes the GFP gene downstream of the IRES under control of the T7pol promoter. The GFP synthesis was visualized directly by fluorescence microscopy (Fig. 5b) confirming the expression of the active T7pol. Once the HEK293T-T7 cell line was established, we attempted the recovery of MNV using the FL cDNA clones described above. Transfection of the HEK293T-T7 cells with the MNV constructs pT7CFE-MNV2T7 and pT7CFE-MNV5’ plasmids resulted in recovery of infectious MNV that was confirmed by plaque formation assay in RAW264.7 cells (Fig. 6). The virus titers produced in HEK293T-T7 were comparable to those of BSR-T7 (Fig. 6).
Figure 5.
Expression of the T7pol protein in the HEK293T-T7 cells. (A) Immunofluorescence staining of the HEK293T-T7 cells was performed using a mAb specific to T7pol (red). The nuclei were counterstained with DAPI (Sigma). (B) Active T7pol drove the expression of the GFP protein when the HEK293T-T7 cells were transfected with pT7CFEGFP as described in text. Original HEK293T cells were included as a negative control.
Figure 6.
Comparative analysis of the MNV recovery in HEK293T-T7 and BHK-T7 cells. Both cells were transfected with the T7-IRES-FL MNV clones as described in text and infectious virus titers were determined at 24 h post transfection. No virus recovery was detected following transfection of pT7CFE-MNVΔ5’.
In summary, we report the development of a simplified RGS for MNV and FCV that bypasses the need for costly in vitro transcription with cap analogs, multiple plasmids, or helper viruses. The intracellular T7pol transcription of plasmids carrying the EMCV IRES sequence placed upstream of these calicivirus genomes allowed the expression of virus proteins and virion assembly in nonpermissive cells stably expressing T7pol. The recovered viruses were indistinguishable from the parental virus when analyzed in permissive cells, and silent mutations could be introduced to generate phenotypically distinct mutants. The FL constructs in our new system lacking the 5’-NTR failed to allow virus recovery, which confirms that an intact RNA template at the 5’-end is required in a calicivirus RGS. Remarkably though, MNV was able to bypass all additional sequences (up to 514 extra nucleotides) engineered upstream of the intact 5’-end (pT7CFE-MNV5’) to consistently generate progeny with authentic 5’-termini. This suggests the presence of a strong recognition site or promoter for the MNV polymerase that insures the generation of a precise 5’-end that is covalently linked to the VPg. Failure to recover FCV from a similar construct might indicate differences in the recognition and interaction of the replication machinery with the 5’-end between these two groups of caliciviruses, but additional investigation is needed.
While our manuscript was under review, we became aware of two novel RGSs developed for FCV and human norovirus (Katayama et al., 2014; Oka et al., 2014). Both systems are based on transfection of cells with plasmids encoding FL virus genomes placed under control of an EF1α promoter and, similar to the system described above, do not require the presence of helper virus or plasmid in the transfected cells. Single-step RGSs will significantly simplify functional analyses of the calicivirus genome. In addition, they will allow analysis of calicivirus individual protein functions in an environment that is helper virus free and in context of authentic virus-host interactions.
Simplified single-plasmid reverse genetics system for caliciviruses.
Recovery of caliciviruses using EMCV IRES-dependent translation of T7 RNA polymerase genomic transcripts.
Recovery of caliciviruses in nonpermissive cells expressing T7 RNA polymerase.
ACKNOWLEDGMENTS
This research was supported by the Intramural Research Program of the NIH, NIAID. We thank the editorial assistance of the NIH Fellows Editorial Board.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Black DN, Burroughs JN, Harris TJ, Brown F. The structure and replication of calicivirus RNA. Nature. 1978;274:614–615. doi: 10.1038/274614a0. [DOI] [PubMed] [Google Scholar]
- Buchholz UJ, Finke S, Conzelmann KK. Generation of bovine respiratory syncytial virus (BRSV) from cDNA: BRSV NS2 is not essential for virus replication in tissue culture, and the human RSV leader region acts as a functional BRSV genome promoter. J Virol. 1999;73:251–259. doi: 10.1128/jvi.73.1.251-259.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burroughs JN, Brown F. Presence of a covalently linked protein on calicivirus RNA. J Gen Virol. 1978;41:443–446. doi: 10.1099/0022-1317-41-2-443. [DOI] [PubMed] [Google Scholar]
- Carstens EB. Ratification vote on taxonomic proposals to the International Committee on Taxonomy of Viruses (2009) Arch Virol. 2010;155:133–146. doi: 10.1007/s00705-009-0547-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang KO, Sosnovtsev SS, Belliot G, Wang Q, Saif LJ, Green KY. Reverse genetics system for porcine enteric calicivirus, a prototype sapovirus in the Caliciviridae. J Virol. 2005;79:1409–1416. doi: 10.1128/JVI.79.3.1409-1416.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhry Y, Skinner MA, Goodfellow IG. Recovery of genetically defined murine norovirus in tissue culture by using a fowlpox virus expressing T7 RNA polymerase. J Gen Virol. 2007;88:2091–2100. doi: 10.1099/vir.0.82940-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daughenbaugh KF, Fraser CS, Hershey JW, Hardy ME. The genome-linked protein VPg of the Norwalk virus binds eIF3, suggesting its role in translation initiation complex recruitment. Embo J. 2003;22:2852–2859. doi: 10.1093/emboj/cdg251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunham DM, Jiang X, Berke T, Smith AW, Matson DO. Genomic mapping of a calicivirus VPg. Arch Virol. 1998;143:2421–2430. doi: 10.1007/s007050050471. [DOI] [PubMed] [Google Scholar]
- Goodfellow I, Chaudhry Y, Gioldasi I, Gerondopoulos A, Natoni A, Labrie L, Laliberte JF, Roberts L. Calicivirus translation initiation requires an interaction between VPg and eIF 4 E. EMBO Rep. 2005;6:968–972. doi: 10.1038/sj.embor.7400510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert TP, Brierley I, Brown TD. Identification of a protein linked to the genomic and subgenomic mRNAs of feline calicivirus and its role in translation. J Gen Virol. 1997;78:1033–1040. doi: 10.1099/0022-1317-78-5-1033. [DOI] [PubMed] [Google Scholar]
- Karst SM, Wobus CE, Lay M, Davidson J, Virgin HWt. STAT1-dependent innate immunity to a Norwalk-like virus. Science. 2003;299:1575–1578. doi: 10.1126/science.1077905. [DOI] [PubMed] [Google Scholar]
- Katayama K, Murakami K, Sharp TM, Guix S, Oka T, Takai-Todaka R, Nakanishi A, Crawford SE, Atmar RL, Estes MK. Plasmid-based human norovirus reverse genetics system produces reporter-tagged progeny virus containing infectious genomic RNA. Proc Natl Acad Sci U S A. 2014;111:E4043–E4052. doi: 10.1073/pnas.1415096111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Ni Z, Yun T, Yu B, Chen L, Zhao W, Hua J, Chen J. A DNA-launched reverse genetics system for rabbit hemorrhagic disease virus reveals that the VP2 protein is not essential for virus infectivity. J Gen Virol. 2008;89:3080–3085. doi: 10.1099/vir.0.2008/003525-0. [DOI] [PubMed] [Google Scholar]
- Liu G, Zhang Y, Ni Z, Yun T, Sheng Z, Liang H, Hua J, Li S, Du Q, Chen J. Recovery of infectious rabbit hemorrhagic disease virus from rabbits after direct inoculation with in vitro-transcribed RNA. J Virol. 2006;80:6597–6602. doi: 10.1128/JVI.02078-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luttermann C, Meyers G. Feline calicivirus. In: Hansman GS, Jiang X, Green KY, editors. Caliciviruses. Molecular and cellular virology. UK: Caister Academic Press; 2010. pp. 145–168. [Google Scholar]
- Moss B, Elroy-Stein O, Mizukami T, Alexander WA, Fuerst TR. Product review. New mammalian expression vectors. Nature. 1990;348:91–92. doi: 10.1038/348091a0. [DOI] [PubMed] [Google Scholar]
- Oka T, Takagi H, Tohya Y. Development of a novel single step reverse genetics system for feline calicivirus. J Virol Methods. 2014;207:178–181. doi: 10.1016/j.jviromet.2014.07.004. [DOI] [PubMed] [Google Scholar]
- Patel MM, Widdowson MA, Glass RI, Akazawa K, Vinje J, Parashar UD. Systematic literature review of role of noroviruses in sporadic gastroenteritis. Emerg Infect Dis. 2008;14:1224–1231. doi: 10.3201/eid1408.071114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pletneva MA, Sosnovtsev SV, Sosnovtseva SA, Green KY. Characterization of a recombinant human calicivirus capsid protein expressed in mammalian cells. Virus Res. 1998;55:129–141. doi: 10.1016/s0168-1702(98)00045-8. [DOI] [PubMed] [Google Scholar]
- Schaffer FL, Ehresmann DW, Fretz MK, Soergel MI. A protein, VPg, covalently linked to 36S calicivirus RNA. J Gen Virol. 1980;47:215–220. doi: 10.1099/0022-1317-47-1-215. [DOI] [PubMed] [Google Scholar]
- Simmonds P, Karakasiliotis I, Bailey D, Chaudhry Y, Evans DJ, Goodfellow IG. Bioinformatic and functional analysis of RNA secondary structure elements among different genera of human and animal caliciviruses. Nucleic Acids Res. 2008;36:2530–2546. doi: 10.1093/nar/gkn096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosnovtsev S, Green KY. RNA transcripts derived from a cloned full-length copy of the feline calicivirus genome do not require VPg for infectivity. Virology. 1995;210:383–390. doi: 10.1006/viro.1995.1354. [DOI] [PubMed] [Google Scholar]
- Sosnovtsev S, Sosnovtseva S, Green KY. Recovery of feline calicivirus from plasmid DNA containing a full-length copy of the genome. In: Chasey D, Gaskell RM, Clarke IN, editors. The 1st International Symposium on Caliciviruses. UK: European Society for Veterinary Virology and Central Veterinary Laboratory, Reading; 1997. pp. 125–130. [Google Scholar]
- Sosnovtsev SV, Belliot G, Chang KO, Prikhodko VG, Thackray LB, Wobus CE, Karst SM, Virgin HW, Green KY. Cleavage map and proteolytic processing of the murine norovirus nonstructural polyprotein in infected cells. J Virol. 2006;80:7816–7831. doi: 10.1128/JVI.00532-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosnovtsev SV, Sosnovtseva SA, Green KY. Cleavage of the feline calicivirus capsid precursor is mediated by a virus-encoded proteinase. J Virol. 1998;72:3051–3059. doi: 10.1128/jvi.72.4.3051-3059.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thumfart JO, Meyers G. Feline calicivirus: recovery of wild-type and recombinant viruses after transfection of cRNA or cDNA constructs. J Virol. 2002;76:6398–6407. doi: 10.1128/JVI.76.12.6398-6407.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward VK, McCormick CJ, Clarke IN, Salim O, Wobus CE, Thackray LB, Virgin HWt, Lambden PR. Recovery of infectious murine norovirus using pol II-driven expression of full-length cDNA. Proc Natl Acad Sci U S A. 2007;104:11050–11055. doi: 10.1073/pnas.0700336104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei C, Farkas T, Sestak K, Jiang X. Recovery of infectious virus by transfection of in vitro-generated RNA from tulane calicivirus cDNA. J Virol. 2008;82:11429–11436. doi: 10.1128/JVI.00696-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wobus CE, Karst SM, Thackray LB, Chang KO, Sosnovtsev SV, Belliot G, Krug A, Mackenzie JM, Green KY, Virgin HW. Replication of Norovirus in cell culture reveals a tropism for dendritic cells and macrophages. PLoS Biol. 2004;2:e432. doi: 10.1371/journal.pbio.0020432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wobus CE, Thackray LB, Virgin HWt. Murine norovirus: a model system to study norovirus biology and pathogenesis. J Virol. 2006;80:5104–5112. doi: 10.1128/JVI.02346-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt LS, Moss B, Rozenblatt S. Replication-deficient vaccinia virus encoding bacteriophage T7 RNA polymerase for transient gene expression in mammalian cells. Virology. 1995;210:202–205. doi: 10.1006/viro.1995.1332. [DOI] [PubMed] [Google Scholar]
- Yunus MA, Chung LM, Chaudhry Y, Bailey D, Goodfellow I. Development of an optimized RNA-based murine norovirus reverse genetics system. J Virol Methods. 2010;169:112–118. doi: 10.1016/j.jviromet.2010.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]