Abstract
J-proteins, obligate co-chaperones, provide specialization for Hsp70 function in a variety of cellular processes. Two of the 13 J-proteins of the yeast cytosol/nucleus, Zuo1 and Jjj1, are associated with 60S ribosomal subunits. Abundant Zuo1 facilitates folding of nascent polypeptides; Jjj1, of much lower abundance, functions in ribosome biogenesis. However, overexpression of Jjj1 substantially rescues growth defects of cells lacking Zuo1. We analyzed a region held in common by Zuo1 and Jjj1, outside the signature J-domain found in all J-proteins. This shared “zuotin homology domain” (ZHD) is important for ribosome association of both proteins. An N-terminal segment of Jjj1, containing the J-domain and ZHD , is ribosome-associated and, like full-length Jjj1, is competent to rescue both the cold- and cation-sensitivity of Δzuo1. However, this fragment, when expressed at normal levels, cannot rescue the cytosolic ribosome biogenesis defect of Δjjj1. Our results are consistent with a model in which the primary functions of Zuo1 and Jjj1 occur in the cytosol. In addition, our data suggest that Zuo1 and Jjj1 bind overlapping sites on ribosomes due to an interaction via their common ZHDs, but Jjj1 binds primarily to pre-60S particles and Zuo1 to mature subunits. We hypothesize that ZUO1 and JJJ1, which are conserved throughout eukaryotes, arose from an ancient duplication of a progenitor J-protein gene that encoded the ZHD ribosome-binding region; subsequently, specialized roles and additional ribosome interaction sites evolved.
Keywords: J-protein, ribosome biogenesis, Hsp70, molecular chaperone, ribosome association
1. Introduction
All Hsp70-based molecular chaperone machineries use the same fundamental biochemical mechanism of action, cycles of interaction with client proteins driven by ATP binding and hydrolysis, to function in many essential cellular processes [1, 2]. These processes range from folding of nascent chains as they emerge from ribosomes to driving protein translocation across membranes, protecting cells from heat stress, facilitating biogenesis of Fe/S clusters, and remodeling protein:protein complexes [3, 4]. Much of the capacity for such functional versatility is due to the fact that Hsp70s interact with an array of J-protein co-chaperones [5]. For example, the cytosol/nucleus of the budding yeast S. cerevisiae contains 13 J-proteins. All members of the J-protein superfamily possess a ~70 residue J-domain that binds Hsp70 and is responsible for stimulation of Hsp70’s ATPase activity, an obligatory step for stabilizing Hsp70’s interaction with client protein. However, outside their J-domains, J-proteins vary widely in sequence and structure [3]. These diverse regions often interact with client proteins, targeting them to Hsp70, or localize the J-protein to a particular site of action.
Eukaryotes contain two ribosome-associated J-proteins, called Zuo1 and Jjj1 in yeast (DNAJC2 and DNAJC21, respectively, in human cells). Both associate with the large ribosomal subunit [6-8]. Both have well-established roles: Zuo1 in chaperoning nascent chains and Jjj1 in a late step of subunit maturation, removing biogenesis factors. Zuo1 is present on approximately 1 of every 3 ribosomes [9, 10], Jjj1 is present at only about 1 per 1,000 ribosomes [10]. Cells lacking Zuo1 are slow-growing, particularly at low temperatures, cold-sensitive, and hypersensitive to cations [6, 11, 12], general defects likely reflecting the myriad of clients whose de novo folding requires ribosome-associated chaperones. As expected, loss of the ribosome-associated Hsp70:J-protein machinery results in aggregation of many newly-made polypeptides [13, 14]. Cells lacking Jjj1 are slow-growing and cold-sensitive, and exhibit hallmarks of inefficient 60S-maturation, such as decreased levels of 60S subunits and accumulation of aberrant polysomes [7, 15].
Jjj1’s role in ribosome biogenesis is an example of involvement of Hsp70/J-protein chaperone machinery in remodeling protein complexes. A few of the many factors involved in 60S subunit biogenesis transit with pre-ribosomal particles to the cytosol [16]. These shuttling factors must be removed and recycled back to the nucleus. Jjj1 is required for removal of one such shuttling factor, Arx1 [7, 15, 17]. In doing so, Jjj1 partners not only with Hsp70, but also with another 60S-biogenesis factor, Rei1. In wild-type cells, Arx1 is largely associated with nuclear pre-60S particles due to efficient removal from cytosolic 60S particles and recycling to the nucleus. In the absence of Jjj1, however, Arx1 accumulates in the cytosol.
Consistent with their different roles, many regions outside the J-domain are quite disparate [6, 8, 17-20]. In Zuo1, an N-terminal region is required for interaction with its heterodimeric partner Ssz1, a positively-charged rRNA-binding region is required for stable interaction with ribosomes, and the extreme C-terminus forms a helical bundle that may regulate ribosome association. On the other hand, the C-terminus of Jjj1 is comprised of a largely charged region flanked by C2H2 zinc fingers, which facilitates binding to Rei1. Moreover, in fungi Jjj1 and Zuo1 function with different Hsp70 partners, Jjj1 with the general Ssa class of Hsp70s, Zuo1 with the fungal-specific ribosome-associated Ssb Hsp70 [7, 21].
However, despite strong evidence that these two ribosome-associated J-proteins carry out distinct functions consistent with these sequence differences, there are intriguing hints of functional overlap. Overexpression of the relatively low-abundance Jjj1 can partially rescue the cold sensitivity and cation hyper-sensitivity of Δzuo1 [7, 22]. Here we report on our analysis of a second region of high similarity between Zuo1 and Jjj1, in addition to the J-domain, the ~80 zuotin homology domain (ZHD) [7, 18] . The ZHD is important for ribosome association of both proteins, suggesting that these proteins have overlapping ribosome-binding sites. The partial rescue of Δzuo1 phenotypes by overexpression of Jjj1 does not require its region specialized in ribosome biogenesis, suggesting that the tethering of a J-domain to an appropriate site on the 60S subunit may be sufficient for basal Zuo1-like activity.
2. Materials and methods
2.1 Yeast strains, plasmids and growth conditions
All yeast strains used in this study are isogenic with DS10, with the genotype his3-11, 15 leu2-3, 112 lys1 lys2 trp1Δ ura3-52. Deletion strains have been published as follows: Δzuo1::HIS [6], Δjjj1::TRP [7], Δarx1::KanMX [7], Δjjj1::TRP Δarx1::KanMX [7], Δjjj1::TRP ARX1-GFP::HIS+ [7]. A list of yeast plasmids used in this study is shown in Supplemental Table 1; all plasmids used are centromeric plasmids based on the pRS plasmid series [23, 24]. Substitution of codons in ZUO1 and JJJ1 and deletion of codons for residues 340-590 in JJJ1 was done by QuikChange PCR mutagenesis (Stratagene). Strains were grown in rich medium (YPD) or minimal medium as previously described [12]. For growth assays, approximately equal concentrations of cells were spotted onto minimal medium plates from 10-fold serial dilutions. Plates with paromomycin (250 μg/ml) were incubated for 3 days at 30°C, plates without paromomycin were incubated for 3 days at 23°C or for 2 days at 30°C.
2.2 Preparation of yeast extracts and analysis of ribosome association
For comparison of total protein levels, yeast cell extracts were prepared as follows. Δzuo1 or Δjjj1 cells containing the indicated plasmids were grown at 30°C in minimal medium to an OD600 of 0.4-0.5. The equivalent of 0.4 OD units of cells was harvested by centrifugation then resuspended in 50 μl of water. 50 μl of 0.2 M NaOH was added, incubated for 5 min at room temperature, then pelleted and resuspended in 50 μl of SDS sample buffer and boiled for 5 min [25]. Equivalent amounts of extract were subjected to SDS-PAGE and immunoblot analysis. Quantitation of band intensity was done using ImageJ software [26].
Lysate preparation and sucrose gradient centrifugation for polysome analysis were as follows. Δzuo1 or Δjjj1 cells containing the indicated plasmids were grown at 30°C, unless otherwise noted, to an OD600 between 0.5 and 0.7 in 50 ml of selective minimal medium, treated with 100 μg/ml of cycloheximide and harvested by centrifugation at 4°C. Cells were then washed with 10 ml of ice-cold Buffer I (20 mM Tris-HCl (pH 7.5), 50 mM KCl and 5 mM MgCl2); pelleted by centrifugation at 4°C; and resuspended in 0.75 ml of ice-cold Buffer I plus 1.5 mM pepstatin; cOmplete, Mini, EDTA-free Protease Inhibitor Cocktail (Roche) and 20 units of Recombinant RNasin Ribonuclease Inhibitor (Promega). 300 μl of ice-cold glass beads were added to the cell suspension, and lysates were prepared by vortexing at 4°C six times for 30 seconds with 30 seconds of cooling in between. Lysates were clarified by centrifugation at 14,000 rpm for 10 minutes. To fractionate polysomes, 10 OD260 units of lysate were applied to the top of a 4 ml 5–50% sucrose gradient in Buffer I and centrifuged for 80 minutes at 45,000 rpm at 4°C in a SW50.1 Ti rotor (Beckman). Gradients were monitored for absorbance at 254 nm to detect ribosomal subunits, monosomes and polysomes, and 0.4 ml fractions were collected. Proteins were precipitated by incubation with 86% acetone overnight at −20°C before SDS-PAGE and immunoblot analysis.
For RNase-treatment, lysates were prepared as above, then 250 μg/ml RNaseA was added to lysates on ice, incubated at 16° for 10 minutes, returned to ice and immediately applied to cold gradients and centrifuged, as above.
2.3 Antibodies and immunoblotting
To generate antibodies specific to the N-terminus of Jjj1, Jjj11-304 with a C-terminal 6x His tag was expressed from the pET3a vector (Novagen) in an E. coli strain, BL21 dnaK− dnaJ−, and purified. The Jjj11-304 protein was used as an immunogen to generate anti-Jjj1 polyclonal antibodies in rabbits (Harlan). Anti-Rpl3 was a gift from Jon Warner (Albert Einstein College of Medicine, Bronx, NY). Anti-Jjj1 specific to the C-terminus (raised against Jjj1304-590), anti-Zuo1 and anti-Ssc1 were produced as reported [6, 7, 27]. Immunoblot detection was done using Amersham ECL HRP-Linked Secondary Antibodies (GE Healthcare) and Western Lightning Plus ECL substrate (PerkinElmer).
2.4 Microscopy
JJJ1-encoding plasmids as indicated, as well as plasmid encoding RFP-Pus1, were transformed into Δjjj1 or Δjjj1 ARX1-GFP. Overnight cultures grown in minimal medium at 30°C were diluted into fresh minimal medium to an OD600 of 0.2 and cultured at 30°C or 23°C as indicated to an OD600 between 0.5 and 0.7, then washed with ddH2O prior to imaging. Fluorescence was visualized using a Zeiss Axio Imager.M2 epi-fluorescence microscope with a 100x oil immersion objective lens, and images were captured with an AxioCam MRm camera controlled with AxioVision software (Carl Zeiss Microscopy, LLC, Thornwood, NY).
2.5 Protein expression and purification
DNA encoding residues 166-303 of Zuo1 was amplified by PCR, and cloned into a modified pET-28a vector that contains an N-terminal His6 tag, thioredoxin (TRX) tag and a tobacco etch virus (rTEV) protease cleavage site. Two arginine mutations (RR247,251AA) were introduced into wild type ZUO1166-303 via QuikChange PCR mutagenesis (Stratagene). 15N-labeled protein expression was conducted in Rosetta 2 (DE3) pLysS E. coli cells using 1 liter of auto-induction medium [28, 29]. Cells were resuspended in lysis buffer (50 mM Tris pH 7.5, 250 mM NaCl, 10 mM imidazole) with cOmplete, EDTA-free Protease Inhibitor Cocktail (Roche), then lysed by French Press. Lysates were clarified by centrifugation. Tagged Zuo1166-303 was purified using bench top nickel column chromatography and size-exclusion chromatography. Tags were removed by proteolysis with recombinant rTEV protease, for 14-18 hrs at 4°C. Tagged protein, cleaved His6/TRX tag and His-tagged rTEV were separated from Zuo1166-303 by bench top nickel chromatography. Protein was used immediately or stored at −80°C.
2.6 NMR spectroscopy and data processing
All 2D 15N-1H Heteronuclear Single-Quantum Correlation (HSQC) spectra were recorded at 4°C using a Varian 600 MHz spectrometer. NMR data were processed using NMRPipe [30] and analyzed using Sparky [31]. All NMR experiments were conducted in 25 mM sodium phosphate (pH 7.5), 200 mM NaCl, 5mM dithiothreitol and 7% D2O.
3. Results
3.1 Two conserved ZHD region residues are important for function of both Jjj1 and Zuo1
An alignment of Zuo1 and Jjj1 from S. cerevisiae and their human orthologs, DNAJC2 and DNAJC21, was generated using ClustalW [32]. A preponderance of the conserved residues outside the J-domain were found in the ~81 residue ZHD. 15 are identical in the ZHD compared to 14 in the J-domain, with only 2 identical residues in the remainder of the alignment (Fig. 1A, Suppl. Fig. S1). Thus, the homology between the two proteins is minimal outside the J-domain and ZHD.
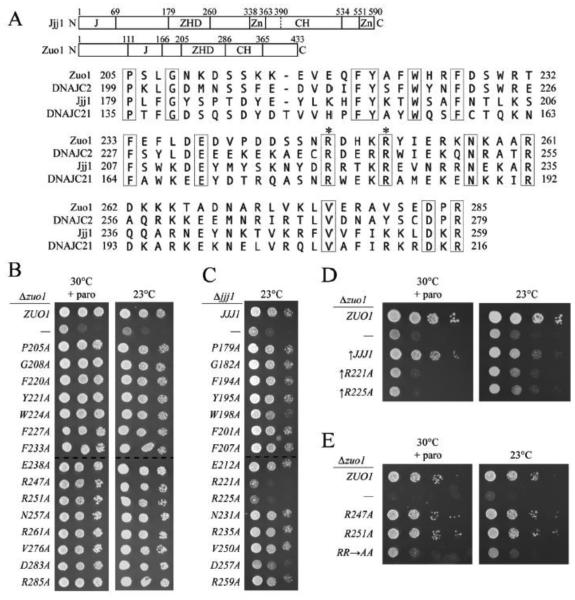
Fig. 1.
Two conserved arginines in the Zuotin homology domain (ZHD) are important for Jjj1 and Zuo1 function. A) (Upper) Diagram of Jjj1 and Zuo1 with vertical lines indicating region boundaries and numbers indicating the residue number at the boundary. J domain (J); Zuotin homology domain (ZHD); Zinc binding region (Zn); charged region (CH). (Bottom) Sequence alignment of S. cerevisiae Zuo1 and Jjj1 and their H. sapiens homologs (DNAJC2 and DNAJC21, respectively) was done with ClustalW; the ZHD sequences are shown with identical residues boxed. Residues of primary interest in this report are indicated (*). B-E) Analysis of deletion strains expressing wild-type or indicated variant proteins from the native promoter (exception noted in (D)) was as follows: strains were serially diluted, spotted on minimal medium plates and incubated at the indicated temperature for 3 days. Plates containing paromomycin (+ paro). B) Δzuo1 cells containing plasmid encoding wild-type ZUO1, no insert (—) or the indicated ZUO1 mutant. C) Δjjj1 cells containing plasmid encoding wild-type JJJ1, no insert (—) or the indicated JJJ1 mutant. D) Δzuo1 cells containing plasmid encoding wild-type ZUO1 (WT) , no insert, or WT JJJ1 or the indicated JJJ1 mutant expressed from the GPD1 promoter (↑). E) Δzuo1 cells containing plasmid encoding wild-type ZUO1, no insert (—) or the indicated ZUO1 mutant (Zuo1R247,251A indicated by RR✧AA ). Dotted lines in B-C indicate different plates from the same batch of media, analyzed at the same time.
Since no region homologous to the ZHD region was found in other yeast J-proteins, we undertook a genetic analysis to understand its function in the two ribosome-associated J-proteins. To determine which of the 15 identical ZHD region residues are most important for function, we performed an alanine mutagenesis screen. Each of the codons for the identical amino acids was singly changed to an alanine codon. The resulting ZUO1 and JJJ1 mutants were expressed from their native promoter in Δzuo1 or Δjjj1 cells, respectively. None of the cells expressing a Zuo1 single-alanine variant had an observable growth defect in the presence of cation or at 23°C (Fig. 1B). However, cells expressing Jjj1R221A or Jjj1R225A were cold-sensitive, growing as poorly as Δjjj1, even though the variants were expressed at normal levels (Fig. 1C, Suppl. Fig. S2A). Taking advantage of our previous observation that overexpression of Jjj1 partially rescues the growth defects of Δzuo1 [7], we also tested whether Jjj1R221A and Jjj1R225A could rescue Δzuo1 to the same extent. We placed JJJ1, jjj1R221A and jjj1R225A under the strong GPD1 promoter. Though all three proteins were expressed at very similar levels, only wild-type Jjj1 rescued the cation-sensitivity of Δzuo1 cells (Fig. 1D, Suppl. Fig. S2B). Since a low level of Zuo1 (<5% the level in wild-type cells) is sufficient to support growth indistinguishable from that of cells with normal Zuo1 levels [33], we considered that deleterious effects of the single-alanine substitutions in Zuo1 might be masked. To test more stringently whether the analogous arginines that are important for Jjj1 function are also important for Zuo1 function, we constructed a double substitution mutant, changing the codons for both arginine R247 and arginine R251 of ZUO1 to alanine (called zuo1RR→AA throughout). zuo1RR→AA was both cation- and cold-sensitive (Fig. 1E), though Zuo1RR→AA was expressed at a similar level to wild-type Zuo1 protein (Suppl. Fig. S2C). Together these results suggest that the analogous residues of the ZHD, R247 and R251 in Zuo1, and R221 and R225 in Jjj1, are functionally important.
3.2 ZHD residues are required for stable association of Zuo1 with ribosomes
Having identified two functionally important ZHD residues in Zuo1 and Jjj1, we next asked whether they were important for ribosome association. We first assessed ribosome association by testing for co-migration of Zuo1/Jjj1 with ribosomes. Cell lysates were centrifuged through a sucrose density gradient to separate different-sized ribosomal complexes. Zuo1 co-migrated with monosomes and polysomes, as expected [6]. However, most of Zuo1RR→AA remained near the top of the gradient (Fig. 2A), indicating destabilization of its interaction with ribosomes.
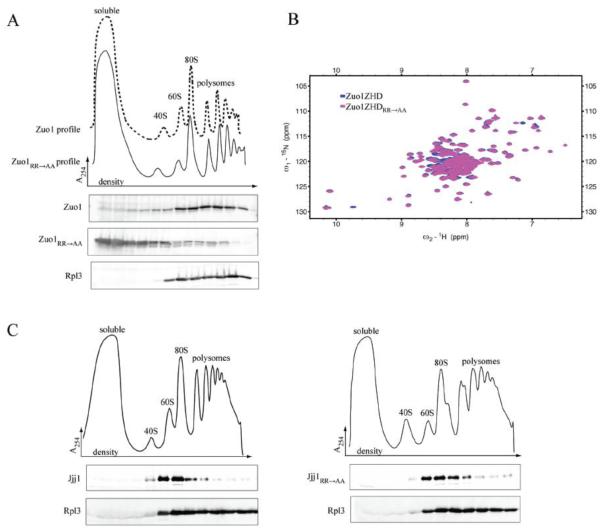
Fig. 2.
Zuo1R247,251A (Zuo1RR→AA) is largely dissociated from ribosomes. A) Lysate from Δzuo1 cells expressing wild-type Zuo1 or Zuo1RR→AA from the native promoter was centrifuged through a sucrose gradient to separate ribosomal subunits, monosomes and polysomes. Fractions were collected. Upper, absorbance at 254 nm plotted versus the relative time of fraction collection (density); dotted line (Zuo1), solid line (Zuo1RR→AA). Lower, fractions were analyzed by immunoblotting using antibodies specific for Zuo1 and Rpl3 (Rpl3 distribution in both experiments was indistinguishable, the WT sample is shown for reference). B) 2D 15N-1H Heteronuclear Single-Quantum Correlation (HSQC) NMR spectra of WT Zuo1166-303 (Zuo1ZHD), which includes the Zuotin homology domain (ZHD), and Zuo1ZHDRR→AA. C) Lysate from Δjjj1 cells expressing wild-type Jjj1 (left) or Jjj1RR→AA (right) from the native promoter was subjected to sucrose gradient analysis as in (A). Upper, absorbance at 254 nm plotted versus the relative time of fraction collection (density). Lower, fractions were analyzed by immunoblotting using antibodies specific for the C-terminus of Jjj1 and Rpl3.
We considered that the lack of association with ribosomes for Zuo1RR→AA might be due to structural changes of the ZHD, rather than loss of a specific interaction. To test this possibility we established a purification system for the ZHD of Zuo1. 15N-labeled Zuo1166-303 fragments, either with or without the RR→AA alterations, were isolated and 15N-1H HSQC NMR data collected. The 15N-1H HSQC spectra of the wild-type ZHD of Zuo1 and ZHDRR→AA fragments had very similar patterns of peak distribution, indicating that the arginine to alanine alterations did not cause a change in the overall structure of the ZHD (Fig. 2B).
3.3 In the absence of its C-terminus, Jjj1 requires ZHD residues for stable ribosome association
We next tested whether the analogous RR→AA substitutions (R221A and R225A, called Jjj1RR→AA throughout) affected Jjj1 ribosome association. As expected [7, 15], and consistent with its role in ribosome biogenesis, wild-type Jjj1 migrated at 60S in sucrose gradients. Jjj1RR→AA also migrated at 60S (Fig. 2C). We next considered that Jjj1 may interact with ribosomes via multiple domains and that destabilization of the interaction of the ZHD with the ribosome might be masked by interaction of Jjj1’s C-terminus with ribosomal complexes. Therefore, we tested whether the Jjj1 C-terminus is required for stable ribosome association. We made a construct encoding a Jjj1 variant lacking the C-terminal residues from 340-590, termed Jjj11-339. In a sucrose gradient, Jjj11-339 co-migrated not only with 60S-sized ribosome particles like Jjj1, but also with monosomes and polysomes (Fig. 3A). To determine whether this altered migration reflected association with 60S subunits or simply co-migration, cell lysates were treated with RNase to clip mRNA, thus collapsing polysomes into 80S monosomes. After RNase treatment, Jjj11-339 coincided with ribosomes, accumulating at 80S (Fig. 3B). The co-migration of Jjj11-339 with polysomes and with the 80S peak after polysome disruption strongly indicates that Jjj11-339 is ribosome-associated.
Fig. 3.
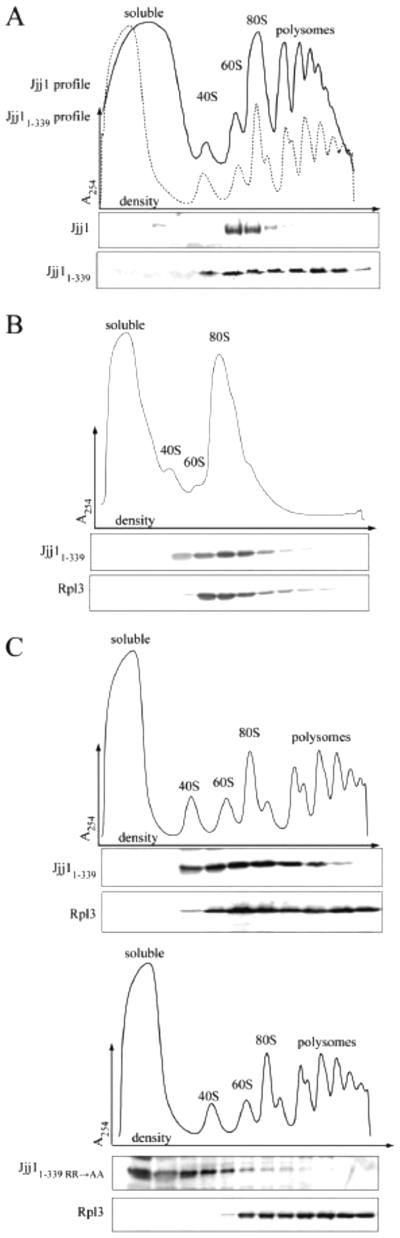
The N-terminus of Jjj1 is sufficient for ribosome association. (A-C) Lysate from Δjjj1 cells expressing wild-type Jjj1, Jjj11-339 or Jjj11-339 R221,225A (Jjj11-339 RR→AA) from the native promoter was centrifuged through a sucrose gradient. Upper, absorbance at 254 nm was monitored and plotted versus the relative time of fraction collection (density) Lower, fractions were analyzed by immunoblotting using antibodies specific for the N-terminus of Jjj1 and for Rpl3. A) Comparison of Jjj1 and Jjj11-339. Upper: dotted line (Jjj1), solid line (Jjj11-339). B) Lysate from cells expressing Jjj11-339 was briefly incubated with RNase before sucrose gradient analysis to clip the mRNA, accumulating monosomes. C) Comparison of Jjj11-339 (upper) and Jjj11-339RR->AA (lower).
Having evidence that Jjj11-339 associates with ribosomes, we tested whether the RR→AA alteration affected Jjj11-339 ribosome association. Jjj11-339/RR→AA largely remained at the top of the sucrose gradient, indicating its destabilization from ribosomes (Fig. 3C). Together with the result that the Zuo1:ribosome interaction is destabilized by the RR→AA alterations, these data indicate that the ZHD contributes to ribosome association of both Zuo1 and Jjj1.
3.4 The Jjj1 C-terminus is important for Jjj1 function in 60S ribosomal subunit maturation
The ribosome association of Jjj11-339 raises the question of what sequences are required for Jjj1’s major function in ribosome biogenesis. Therefore, we tested whether Jjj11-339 was able to rescue the effects of the absence of Jjj1, first testing for suppression of the Δjjj1 growth defect. We found that jjj11-339 is slow-growing and cold-sensitive, nearly indistinguishable from Δjjj1 (Fig. 4A, left). To facilitate accurate comparison of expression of Jjj1 and Jjj11-339, we generated polyclonal antibodies specific for residues 1-304. Using these antibodies, analysis of lysates made from cells expressing wild-type Jjj1 or Jjj11-339 revealed that Jjj11-339 was expressed at a slightly higher level than full-length Jjj1 (Fig. 4A, right), consistent with expression from a centromeric plasmid. We also inspected the cellular distribution of fluorescence signal from cells expressing Jjj1-GFP or Jjj11-339-GFP. Both Jjj1-GFP and Jjj11-339-GFP cells showed cytosolic signal (Fig. 4C), consistent both with previous reports of Jjj1 localization [7, 34] and with our observation that Jjj11-339 co-migrates with monosomes and polysomes (Fig. 3A). In some cells the fluorescence of Jjj1-GFP appeared somewhat less intense, while Jjj11-339-GFP fluorescence was consistently of similar intensity in the two compartments.
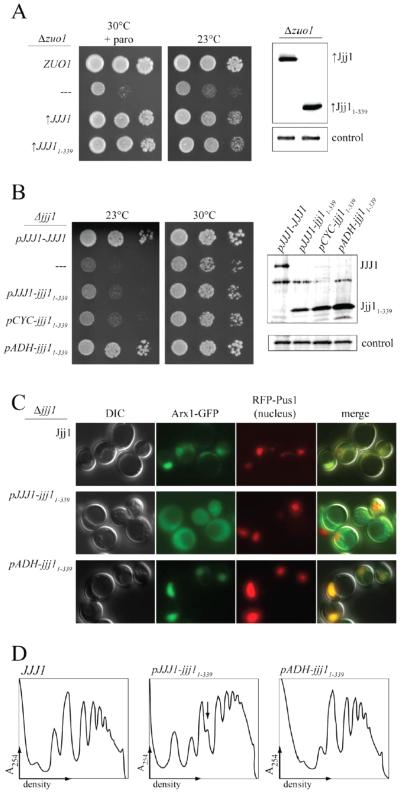
Fig. 4.
The Jjj1 C-terminus is important for the role of Jjj1 in ribosome biogenesis. A) Analysis of wild-type cells and Δjjj1 cells containing empty vector (—), or expressing Jjj1 or Jjj11-339 from the native promoter, was as follows: Left: strains were serially diluted, spotted on minimal medium plates, then incubated at 23°C for 3 days or 30°C for 2 days. Right: Cell extracts were prepared from Δjjj1+JJJ1 and Δjjj1+jjj11-339 cultures used for serial dilutions and subjected to immunoblot analysis using antibody specific to the Jjj1 N-terminus and antibody specific to Ssc1 as a loading control. B) Δjjj1 cells containing empty vector and Δjjj1 cells expressing wild-type Jjj1 or Jjj11-339 from the native promoter were grown at 23°C, lysed. The lysate was then centrifuged through a sucrose gradient. The migration of ribosomal subunits, monosomes and polysomes was monitored by absorbance at 254 nm and plotted versus the time course of fraction collection. Arrows denote half-mer polysome peaks. C) Δjjj1 cells expressing either Jjj1-GFP or Jjj11-339 -GFP from the native promoter, and the nucleus-specific protein RFP-Pus1 as a nuclear marker, were grown at 30°C prior to imaging by fluorescence and differential interference contrast (DIC) microscopy. Representative images show DIC, localization of Jjj1-GFP or Jjj11-339 -GFP as indicated, localization of RFP-Pus1, a nuclear marker, and an image overlay (merge).
A more direct indicator of inefficient 60S subunit maturation than cold-sensitivity is the presence of half-mer peaks in polysome profiles, which are interpreted as 40S ribosomal subunits stalled in an initiation complex with mRNA because of a lack of mature 60S subunits [35, 36]. Consistent with previous reports, half-mers were apparent in polysome profiles of Δjjj1, being most distinct in profiles of lysates from cells grown at a lower temperature, e.g. 23°C [7, 15]. Half-mer peaks were also present in jjj11-339 polysome profiles (Fig. 4B, also see Fig 3), consistent with a defect in 60S subunit biogenesis.
We also tested whether loss of residues 340-590 affected recycling of Arx1 from the cytosol into the nucleus. We inspected the distribution of Arx1-GFP signal between the nucleus and cytosol in wild-type, Δjjj1and jjj11-339 cells. As expected, in Δjjj1 cells Arx1-GFP accumulated in the cytosol, rather than concentrating in the nucleus as in wild-type cells [7, 15]. In jjj11-339 cells Arx1-GFP was also present present in the cytosol (Fig. 5A), indicating that the C-terminus of Jjj1 is important for efficient recycling of Arx1. Further, deletion of ARX1 has also been shown to suppress the growth phenotype of Δjjj1 [7, 15], consistent with the idea that the persistence of ARX1 in the cytosol in the absence of Jjj1 is largely responsible for the Δjjj1 growth phenotype. We found that like Δjjj1, the cold-sensitivity of jjj11-339 was suppressed by deletion of Arx1 (Fig. 5B). These results are consistent with Jjj11-339 being defective in the cytosolic function of Arx1 removal.
Fig. 5.
The Jjj1 C-terminus is important for the role of Jjj1 in Arx1 recycling. A) Δjjj1 ARX1-GFP cells containing empty vector (—), or expressing Jjj1, Jjj11-339, Jjj11-550, Jjj11-389 or Jjj11-362 from the native promoter, were grown at 23°C prior to imaging by fluorescence and differential interference contrast (DIC) microscopy. Representative images show DIC, Arx1-GFP localization, localization of a nucleus-specific RFP fusion protein, RFP-Pus1, and an image overlay (merge). B) Wild-type, Δarx1, Δjjj1, and Δjjj1 Δarx1 cells containing empty vector, and Δjjj1 and Δjjj1Δarx1 cells expressing jjj11-339 from the native promoter [(jjj11-339) and (jjj11-339 Δarx1), respectively], were serially diluted, spotted on a minimal medium plate, and incubated at 23°C for 3 days. C) Δjjj1 cells expressing either Jjj1, no Jjj1 (-) or the indicated Jjj1 truncation from the native promoter were serially diluted, spotted on a minimal medium plate, and incubated at 23°C for 2.5 days.
The C-terminal region of Jjj1 from residues 340-590 contains two zinc finger domains separated by a charged region, spanning residues 363-550, which is predominantly negative from 363-389 and predominantly positive from 390-550. To gain a better understanding of functionally important residues in the C-terminus, we analyzed the growth and Arx1-GFP fluorescence of cells expressing three truncations that removed smaller amounts of the C-terminal region than Jjj11-339, the focus of this study: Jjj11-362, Jjj11-389, and Jjj11-550 (see Fig 1A). Analysis of GFP fusions of all the truncations revealed dispersed cellular fluorescence (Suppl. Fig. 3A), indicating the presence of the Jjj1 truncations in the cytosol. The two truncations removing the smaller segments of the C-terminus, Jjj11-550 and Jjj11-389, supported growth nearly as well as full-length Jjj1, while Jjj11-362 did not (Fig. 5C). Arx1 was concentrated in the nucleus in jjj11-550 and jjj11-389, like in wild-type cells, but was dispersed in the cytosol in jjj11-362, like in jjj11-339 (Fig. 5A). Jjj11-389, the smallest C-terminal truncation that supported Jjj1 function when expressed from its native promoter, co-migrated with ribosomal subunits, consistent with a ribosome-associated function of Jjj1 (Suppl. Fig. 3B).
3.5 The C-terminus of Jjj1 is not required for rescue of Δzuo1
That Jjj11-339 is ribosome associated, but defective in its ability to carry out its ribosome biogenesis function, raises two questions: (1) is Jjj11-339, like full-length Jjj1, competent in rescuing the deleterious effects of the absence of Zuo1 when overexpressed, and (2) does Jjj11-339 retain a level of competency that enables it to carry out Jjj1’s ribosome biogenesis function if overexpressed. To answer the first question, we first placed jjj11-339 under the control of the strong GPD1 promoter, as we had done previously for full-length Jjj1 ([7] and Fig 1D). Expression of Jjj11-339 and Jjj1 to the same level rescued Δzuo1 cation- and cold-sensitivity to the same extent, indicating that the C-terminus of Jjj1 is dispensable for Δzuo1 rescue (Fig. 6A).
Fig. 6.
Effects of overexpression of jjj11-339. A) Left: Δzuo1 cells containing plasmid encoding ZUO1 under control of the native promoter (ZUO1), no insert (—), or either JJJ1 or jjj11-339 under control of the GPD1 promoter (indicated by “↑”). Strains were serially diluted, spotted on minimal medium plates and incubated at the indicated temperature for 3 days. Plate containing paromomycin is indicated (“+paro”). Right: Cell extracts were prepared from cultures used for serial dilutions and subjected to immunoblot analysis using antibody specific to the Jjj1 N-terminus and antibody specific to Ssc1, as a loading control. B) Left: Analysis of Δjjj1 cells containing plasmid encoding JJJ1 under control of the native promoter (JJJ1), no insert (—), or jjj11-339 under control of the indicated promoter: native (pJJJ1), CYC (pCYC) or ADH (pADH). Strains were serially diluted, spotted on minimal medium plates, then incubated at 23°C for 3 days or 30°C for 2 days. Right: Cell extracts were analyzed as described in (A). C) Δjjj1 ARX1-GFP cells containing a plasmid encoding RFP-Pus1 and a plasmid encoding WT JJJ1, jjj11-339 under the native promoter or jjj11-339 under control of the ADH promoter (↑jjj11-339), were grown at 23°C prior to imaging by fluorescence and differential interference contrast (DIC) microscopy. Representative images show DIC, Arx1-GFP localization, localization of a nucleus-specific RFP fusion protein, RFP-Pus1, or an image overlay (merge). D) Δjjj1 cells expressing Jjj1, Jjj11-339 or Jjj11-339 from the ADH promoter were grown at 23°C and lysed, then centrifuged through a sucrose gradient. The migration of ribosomal subunits, monosomes and polysomes was monitored by absorbance at 254 nm and plotted versus the relative time of fraction collection. Arrows denote half-mer polysome peaks.
Next we tested the ability of Jjj11-339 to rescue ribosome biogenesis defects when overexpressed. We placed jjj11-339 under promoters of varying strengths, and analyzed protein levels by immunoblotting with our antibody specific to the N-terminus of Jjj1. Modest overexpression of Jjj11-339 to 2-fold more than the normal level from the CYC1 promoter had no obvious effect on growth of Δjjj1 cells. However, approximately 8-fold overexpression from the ADH1 promoter (pADH1-jjj11-339), in which case the majority of Jjj11-339, like full-length Jjj1 co-migrated with ribosomes (Suppl. Fig. 4), resulted in significant suppression of cold-sensitivity and slow-growth (Fig. 6B).
To test whether rescue of growth by overexpression extended to rescue of Arx1 recycling, we examined Arx1-GFP localization in cells carrying pADH1-jjj11-339. Arx1-GFP is largely present in the nucleus in cells carrying pADH1-jjj11-339 (Fig. 6C), indicating that increased levels of Jjj11-339 can rescue both the growth defects and Arx1 recycling defect of Δjjj1. But this rescue requires the conserved arginines at positions 221 and 225 (Fig 6; Suppl. Fig. 4). Overexpression of Jjj11-339 also rescued the aberrant polysome profiles of Δjjj1 (Fig. 6D). Together with the evidence described above, these results suggest that the C-terminus of Jjj1 is important for its cytosolic role in Arx1 removal, though the reduced function of Jjj11-339 can be rescued by increased cellular concentration. The data also indicates that the cold sensitivity and half-mer accumulation in Jjj11-339 cells are linked to Arx1 removal defects.
4. Discussion
Overall our results indicate that the ZHD is important for the association of both Zuo1 and Jjj1 with 60S ribosomal particles, with the primary functions of these proteins ribosome-associated activities in the cytosol. However, interaction of both proteins with the ribosome involves other unrelated sequences, in addition to the ZHD.
4.1 The ZHD is important for ribosome association
That the ZHD is important for ribosome association is supported by the fact that alteration of conserved arginine residues in full-length Zuo1 results in destabilization of its interaction with the ribosome. Although the analogous alterations in Jjj1 do not cause destabilization in full-length Jjj1, ribosome association of an N-terminal fragment containing the J domain and ZHD, Jjj11-339, is destabilized. Our biophysical analysis of the ZHD fragment of Zuo1 indicates that the alterations do not significantly affect its fold. Although it is possible that these arginine alterations affect the overall structure of the protein, we think it unlikely. More likely, the ZHD of both Zuo1 and Jjj1 interact with the ribosome at the same site, as other than the J-domain, sequences outside the ZHD of the two proteins have no obvious sequence similarity. This idea is also consistent with crosslinking and cryo-electron microscopic studies that localize Zuo1 and Jjj1 to the same general area of the 60S subunit near the ribosomal exit tunnel [37, 38].
However, in addition to the ZHD interaction, both Zuo1 and Jjj1 interact with ribosome complexes via their distinct C-terminal segments. Zuo1 and Jjj1 carry out different cellular roles, Zuo1 on translating ribosomes and the much less abundant Jjj1 on Arx1-containing 60S particles. Therefore, it is easy to envision that, although they utilize a shared ribosome interaction site, additional, distinct interactions determine their affinity for specific 60S ribosomal particles, and thus the ribosome particles with which they predominately associate. Such a model would explain the predominance of Jjj1 association with subunits migrating at 60S (i.e. immature pre-60S particles). For example, Rei1, Jjj1’s partner in ribosome biogenesis with which it directly interacts, binds independently to the 60S subunit [17]. The Jjj1:Rei1 interaction could partially explain the predominant association of full-length Jjj1 with ribosomal particles migrating at 60S, compared to Jjj11-339, which associates with polysomes as well. In the case of Zuo1, recently reported cryo-electron microscopic studies [39] indicate that while the bulk of Zuo1 localizes near the exit site of the 60S subunit as does Jjj1, its extreme C-terminus, which has been shown to form a regulatory 4-helix bundle [20], interacts with the 40S subunit, thus potentially playing a role in translational regulation.
Our data also suggest that the ZHD does more than simply tether the J-proteins near the ribosome exit site. Even though Jjj1RR—>AA is ribosome-associated, it does not rescue either the cold sensitivity of Δjjj1 or the cold- or cation-sensitivity of Δzuo1 when overexpressed. In addition, that jjj1RR—>AA cells show half-mer accumulation and Arx1 mislocalization indicates that these alterations cause disruption of ribosome biogenesis. Why Jjj1RR—>AA is not functional, though ribosome-associated, is not clear. We speculate that the ZHD may play a role in positioning Jjj1 on the 60S subunit. For example, Jjj1 function requires that its J-domain interacts with its partner Hsp70. If not positioned correctly, such that this interaction can occur effectively, a null phenotype would result.
4.2 Jjj1 C-terminal regions are important for Arx1 recycling and efficient ribosome biogenesis
The results reported here indicate that the C-terminus of Jjj1 is important for Jjj1 function in ribosome biogenesis because it facilitates removal of the Arx1 biogenesis factor present on cytosolic pre-60S ribosomal subunits, thus allowing its recycling to the nucleus. However, the C-terminus is not absolutely essential for the process, as overexpression of Jjj11-339 allows removal.
The exact mechanism of Arx1 release and Jjj1’s role in it are not known. As mentioned above, however, it is known that a second cytosolic biogenesis factor, Reil is required [40, 41]. Jjj1 interacts with Rei1 via its C-terminus [17]. This interaction, though not essential for Arx1 removal, may facilitate it, thus explaining the requirement for higher levels of Jjj11-339 for function.
The alignment between growth defects and cytosolic 60S maturation defects reported here point to maturation of 60S subunits in the cytosol as the major function of Jjj1 that causes slow growth, especially at low temperatures. Both Δjjj1 and jjj11-339 displayed half-mers, cytosolic accumulation of Arx1 and suppression of the growth defect upon deletion of ARX1. However, our results do not exclude the possibility that Jjj1 also functions in the nucleus in ribosome biogenesis as previously suggested [18], as we do see some Jjj1-GFP fluorescence in the nucleus. But, our results are inconsistent with a nuclear role being Jjj1’s major function, as was concluded previously [18]. That conclusion was based in good part on the observed lack of co-migration of Jjj11-389, with ribosomes and exclusively nuclear localization of Jjj11-389-GFP. Our results also indicate that this Jjj11-389 fragment is functional, as it could both support nearly wild-type growth and nuclear localization of Arx1-GFP. However, we found it in the cytosol, co-migrating with ribosomes, like the shorter Jjj11-339 fragment, which is the focus of this report. Thus, our results continue to be consistent with the major function of Jjj1 being in the cytosol, facilitating the removal of the Arx1 ribosome biogenesis factor.
Interestingly, our analysis of a set of C-terminal truncation mutants uncovered a phenotypic difference between the jjj11-339/jjj11-362 pair and the jjj11-389/jjj11-550 pair. The intervening region between these pairs, 363-389, though part of the larger “charged” region, contains a segment that is predominately negative in charge, compared to the more C-terminal positively charged region. Thus the difference in activity between these pairs raises the possibility that this 26 residue segment is particularly important for Jjj1 function. These results are also consistent with our previous data [17] that showed a deletion of the entire charged region (363-534) resulted in growth defects, as well as Arx1 cytosolic accumulation and half-mer formation, while disruption of both zinc fingers had minimal effects.
4.3 Conclusions
The results presented here strongly suggest that the ZHD of the eukaryotic-specific J-proteins Zuo1 and Jjj1 function as a ribosome-association region for both proteins. Yet both proteins contain sequences with specialized function. For example, the C-terminus of Jjj1 is important for ribosome biogenesis, a function that appears to be unique to Jjj1. We suggest that the specialized Zuo1 and Jjj1 proteins evolved from a progenitor J-protein that evolved early in the eukaryotic lineage and was tethered near the exit tunnel of the ribosome by its ZHD. Consistent with this idea and the eukaryotic specificity of ribosome-associated J-proteins, the predicted binding site of Zuo1 and Jjj1 near the ribosomal exit tunnel is in proximity to Rpl31 and Rpl22, two eukaryotic proteins that have no homolog in bacteria [42] and reside in a region of the 60S subunit rich in eukaryote-specific rRNA expansion segments [37, 38]. It is likely that the duplication of the progenitor gene also occurred early in the lineage, as Zuo1 and Jjj1 orthologs are ubiquitous in eukaryotes.
Supplementary Material
Highlights.
J-protein Hsp70 chaperones Zuo1 and Jjj1 have conserved Zuotin Homology domain, ZHD
ZHD is important for ribosome association of both Zuo1 and Jjj1
Specialized C-terminus of Jjj1 is important for cytosolic ribosome biogenesis
Partial rescue of Δzuo1 by overexpression of Jjj1 does not require its C-terminus
ZHD likely a common ribosome association domain in specialized eukaryotic J-proteins
Acknowledgements
This work was supported by National Institutes of Health Grant GM31107 (to E. A. C.) and University of Wisconsin Molecular Biosciences Training Grant T32GM07215 from the National Institutes of Health (to L. A. K.). This study made use of the National Magnetic Resonance Facility at Madison, which is supported by NIH grant P41GM103399 (NIGMS), old number: P41RR002301. Equipment was purchased with funds from the University of Wisconsin-Madison, the NIH P41GM103399, S10RR02781, S10RR08438, S10RR023438, S10RR025062, S10RR029220), the NSF (DMB-8415048, OIA-9977486, BIR-9214394), and the USDA. We thank Jon Warner for antibody specific to Rpl3, and Symeon Siniossoglou for the pRS313-RFP-Pus1 plasmid.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Bibliography
- [1].Bukau B, Weissman J, Horwich A. Molecular chaperones and protein quality control. Cell. 2006;125:443–451. doi: 10.1016/j.cell.2006.04.014. [DOI] [PubMed] [Google Scholar]
- [2].Mayer MP. Gymnastics of molecular chaperones. Molecular cell. 2010;39:321–331. doi: 10.1016/j.molcel.2010.07.012. [DOI] [PubMed] [Google Scholar]
- [3].Kampinga HH, Craig EA. The HSP70 chaperone machinery: J proteins as drivers of functional specificity, Nature reviews. Molecular cell biology. 2010;11:579–592. doi: 10.1038/nrm2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Kim YE, Hipp MS, Bracher A, Hayer-Hartl M, Hartl FU. Molecular chaperone functions in protein folding and proteostasis. Annual review of biochemistry. 2013;82:323–355. doi: 10.1146/annurev-biochem-060208-092442. [DOI] [PubMed] [Google Scholar]
- [5].Craig EA, Huang P, Aron R, Andrew A. The diverse roles of J-proteins, the obligate Hsp70 co-chaperone. Rev Physiol Biochem Pharmacol. 2006;156:1–21. doi: 10.1007/s10254-005-0001-0. [DOI] [PubMed] [Google Scholar]
- [6].Yan W, Schilke B, Pfund C, Walter W, Kim S, Craig EA. Zuotin, a ribosome-associated DnaJ molecular chaperone. Embo J. 1998;17:4809–4817. doi: 10.1093/emboj/17.16.4809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Meyer AE, N. J. Hung,, P. Yang,, Johnson AW, Craig EA. The Specialized Cytosolic J-protein Jjj1, Functions in 60S Ribosomal Subunit Biogenesis. Proc. Natl. Acad. Sci. USA. 2007;104(5):1558–63. doi: 10.1073/pnas.0610704104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Peisker K, Braun D, Wolfle T, Hentschel J, Funfschilling U, Fischer G, Sickmann A, Rospert S. Ribosome-associated complex binds to ribosomes in close proximity of Rpl31 at the exit of the polypeptide tunnel in yeast. Molecular biology of the cell. 2008;19:5279–5288. doi: 10.1091/mbc.E08-06-0661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Raue U, Oellerer S, Rospert S. Association of protein biogenesis factors at the yeast ribosomal tunnel exit is affected by the translational status and nascent polypeptide sequence. The Journal of biological chemistry. 2007;282:7809–7816. doi: 10.1074/jbc.M611436200. [DOI] [PubMed] [Google Scholar]
- [10].Wang M, Weiss M, Simonovic M, Haertinger G, Schrimpf SP, Hengartner MO, von Mering C. PaxDb, a database of protein abundance averages across all three domains of life. Molecular & cellular proteomics : MCP. 2012;11:492–500. doi: 10.1074/mcp.O111.014704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Gautschi M, Lilie H, Funfschilling U, Mun A, Ross S, Lithgow T, Rucknagel P, Rospert S. RAC, a stable ribosome-associated complex in yeast formed by the DnaK-DnaJ homologs Ssz1p and zuotin. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:3762–3767. doi: 10.1073/pnas.071057198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Kim S, Craig E. Broad Sensitivity of Saccharomyces cerevisiae Lacking Ribosome-Associated Chaperone Ssb or Zuo1 to Cations, Including Aminoglycosides. Eukaryotic Cell. 2005;4:82–89. doi: 10.1128/EC.4.1.82-89.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Koplin A, Preissler S, Ilina Y, Koch M, Scior A, Erhardt M, Deuerling E. A dual function for chaperones SSB-RAC and the NAC nascent polypeptide-associated complex on ribosomes. The Journal of cell biology. 2010;189:57–68. doi: 10.1083/jcb.200910074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Willmund F, del Alamo M, Pechmann S, Chen T, Albanese V, Dammer EB, Peng J, Frydman J. The cotranslational function of ribosome-associated Hsp70 in eukaryotic protein homeostasis. Cell. 2013;152:196–209. doi: 10.1016/j.cell.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Demoinet E, Jacquier A, Lutfalla G, Fromont-Racine M. The Hsp40 chaperone Jjj1 is required for the nucleo-cytoplasmic recycling of preribosomal factors in Saccharomyces cerevisiae. Rna. 2007;13:1570–1581. doi: 10.1261/rna.585007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Panse VG, Johnson AW. Maturation of eukaryotic ribosomes: acquisition of functionality. Trends in biochemical sciences. 2010;35:260–266. doi: 10.1016/j.tibs.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Meyer AE, Hoover LA, Craig EA. The cytosolic J-protein, Jjj1, and Rei1 function in the removal of the pre-60 S subunit factor Arx1. The Journal of biological chemistry. 2010;285:961–968. doi: 10.1074/jbc.M109.038349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Albanese V, Reissmann S, Frydman J. A ribosome-anchored chaperone network that facilitates eukaryotic ribosome biogenesis. The Journal of cell biology. 2010;189:69–81. doi: 10.1083/jcb.201001054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Fiaux J, Horst J, Scior A, Preissler S, Koplin A, Bukau B, Deuerling E. Structural analysis of the ribosome-associated complex (RAC) reveals an unusual Hsp70/Hsp40 interaction. The Journal of biological chemistry. 2010;285:3227–3234. doi: 10.1074/jbc.M109.075804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Ducett JK, Peterson FC, Hoover LA, Prunuske AJ, Volkman BF, Craig EA. Unfolding of the C-terminal domain of the J-protein Zuo1 releases autoinhibition and activates Pdr1-dependent transcription. Journal of molecular biology. 2013;425:19–31. doi: 10.1016/j.jmb.2012.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Huang P, Gautschi M, Walter W, Rospert S, Craig EA. The Hsp70 Ssz1 modulates the function of the ribosome-associated J-protein Zuo1. Nature structural & molecular biology. 2005;12:497–504. doi: 10.1038/nsmb942. [DOI] [PubMed] [Google Scholar]
- [22].Sahi C, Craig EA. Network of general and specialty J protein chaperones of the yeast cytosol. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:7163–7168. doi: 10.1073/pnas.0702357104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Mumberg D, Muller R, Funk M. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene. 1995;156:119–122. doi: 10.1016/0378-1119(95)00037-7. [DOI] [PubMed] [Google Scholar]
- [25].Kushnirov VV. Rapid and reliable protein extraction from yeast. Yeast. 2000;16:857–860. doi: 10.1002/1097-0061(20000630)16:9<857::AID-YEA561>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- [26].Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nature methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Liu Q, D'Silva P, Walter W, Marszalek J, Craig EA. Regulated cycling of mitochondrial Hsp70 at the protein import channel. Science. 2003;300:139–141. doi: 10.1126/science.1083379. [DOI] [PubMed] [Google Scholar]
- [28].Tyler RC, Sreenath HK, Singh S, Aceti DJ, Bingman CA, Markley JL, Fox BG. Auto-induction medium for the production of [U-15N]- and [U-13C, U-15N]-labeled proteins for NMR screening and structure determination. Protein expression and purification. 2005;40:268–278. doi: 10.1016/j.pep.2004.12.024. [DOI] [PubMed] [Google Scholar]
- [29].Fox BG, Blommel PG. Coligan John E., editor. Autoinduction of protein expression. Current protocols in protein science / editorial board. doi: 10.1002/0471140864.ps0523s56. [et al.], Chapter 5 (2009) Unit 5 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. Journal of biomolecular NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- [31].Lee W, Westler WM, Bahrami A, Eghbalnia HR, Markley JL. PINE-SPARKY: graphical interface for evaluating automated probabilistic peak assignments in protein NMR spectroscopy. Bioinformatics. 2009;25:2085–2087. doi: 10.1093/bioinformatics/btp345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic acids research. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Hundley H, Eisenman H, Walter W, Evans T, Hotokezaka Y, Wiedmann M, Craig E. The in vivo function of the ribosome-associated Hsp70, Ssz1, does not require its putative peptide-binding domain. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:4203–4208. doi: 10.1073/pnas.062048399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- [35].Helser TL, Baan RA, Dahlberg AE. Characterization of a 40S ribosomal subunit complex in polyribosomes of Saccharomyces cerevisiae treated with cycloheximide. Molecular and cellular biology. 1981;1:51–57. doi: 10.1128/mcb.1.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Rotenberg MO, Moritz M, Woolford JL., Jr. Depletion of Saccharomyces cerevisiae ribosomal protein L16 causes a decrease in 60S ribosomal subunits and formation of half-mer polyribosomes. Genes & development. 1988;2:160–172. doi: 10.1101/gad.2.2.160. [DOI] [PubMed] [Google Scholar]
- [37].Greber BJ, Boehringer D, Montellese C, Ban N. Cryo-EM structures of Arx1 and maturation factors Rei1 and Jjj1 bound to the 60S ribosomal subunit. Nature structural & molecular biology. 2012;19:1228–1233. doi: 10.1038/nsmb.2425. [DOI] [PubMed] [Google Scholar]
- [38].Leidig C, Bange G, Kopp J, Amlacher S, Aravind A, Wickles S, Witte G, Hurt E, Beckmann R, Sinning I. Structural characterization of a eukaryotic chaperone--the ribosome-associated complex. Nature structural & molecular biology. 2013;20:23–28. doi: 10.1038/nsmb.2447. [DOI] [PubMed] [Google Scholar]
- [39].Zhang Y, Ma C, Yuan Y, Zhu J, Li N, Chen C, Wu S, Yu L, Lei J, Gao N. Structural basis for interaction of a cotranslational chaperone with the eukaryotic ribosome. Nature structural & molecular biology. 2014;21:1042–1046. doi: 10.1038/nsmb.2908. [DOI] [PubMed] [Google Scholar]
- [40].Hung NJ, Johnson AW. Nuclear recycling of the pre-60S ribosomal subunit-associated factor Arx1 depends on Rei1 in Saccharomyces cerevisiae. Molecular and cellular biology. 2006;26:3718–3727. doi: 10.1128/MCB.26.10.3718-3727.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Lebreton A, Saveanu C, Decourty L, Rain JC, Jacquier A, Fromont-Racine M. A functional network involved in the recycling of nucleocytoplasmic pre-60S factors. The Journal of cell biology. 2006;173:349–360. doi: 10.1083/jcb.200510080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Jenner L, Melnikov S, Garreau de Loubresse N, Ben-Shem A, Iskakova M, Urzhumtsev A, Meskauskas A, Dinman J, Yusupova G, Yusupov M. Crystal structure of the 80S yeast ribosome. Current opinion in structural biology. 2012;22:759–767. doi: 10.1016/j.sbi.2012.07.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.