Abstract
Acetyl-CoA represents a key node in metabolism due to its intersection with many metabolic pathways and transformations. Emerging evidence reveals that cells monitor the levels of acetyl-CoA as a key indicator of their metabolic state, through distinctive protein acetylation modifications dependent on this metabolite. We offer the following conceptual model for understanding the role of this sentinel metabolite in metabolic regulation. High nucleocytosolic acetyl-CoA amounts are a signature of a “growth” or “fed” state and promote its utilization for lipid synthesis and histone acetylation. In contrast, under “survival” or “fasted” states, acetyl-CoA is preferentially directed into the mitochondria to promote mitochondrial-dependent activities such as the synthesis of ATP and ketone bodies. Fluctuations in acetyl-CoA within these subcellular compartments enable the substrate-level regulation of acetylation modifications, but also necessitates the function of sirtuin deacetylases to catalyze removal of spontaneous modifications that might be unintended. Thus, understanding the sources, fates, and consequences of acetyl-CoA as a carrier of two-carbon units has started to reveal its underappreciated but profound influence on the regulation of numerous life processes.
Introduction
In response to a dynamic nutrient environment, cells must assess their metabolic state to decide whether to grow, survive, or die. It has become evident that metabolites themselves must feed back to regulate gene expression, signal transduction, and various protein activities in cellular decision-making processes [1,2]. These small molecule metabolites play critical roles in relaying metabolic information to their protein and nucleic acid counterparts. However, despite increased recognition of such reciprocal interplay, many aspects of the mechanisms through which metabolites exert their influence on cellular regulatory mechanisms are still being unraveled.
Amongst the thousands of metabolites present in the cellular milieu at any given time, which might represent the “sentinel” metabolites that signify cellular metabolic state? One well-known signature of metabolic state is AMP, which indicates cellular energy charge and accumulates upon ATP insufficiency. AMP regulates the activity of the AMP-activated protein kinase (AMPK), which phosphorylates many proteins involved in cellular energy homeostasis [3]. Another example is NAD+, which indicates the cellular redox status as a ratio of NAD+ to NADH [4,5]. Herein, we discuss the hypothesis that acetyl-CoA represents an additional prominent gauge of the cell’s metabolic state with substantial influence on numerous biological regulatory mechanisms.
Growth or Fed State - High acetyl-CoA in cytosol/nucleus
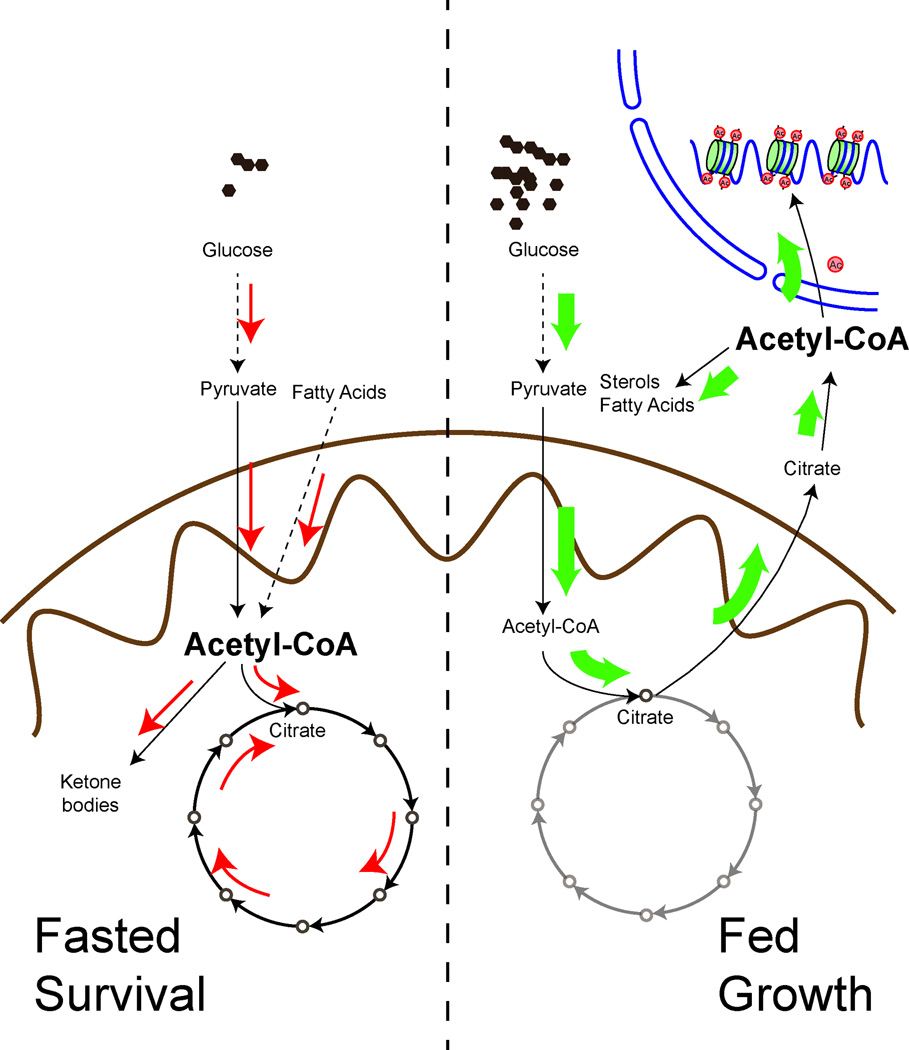
Acetyl-CoA is a metabolite derived from glucose, fatty acid, and amino acid catabolism. During glycolysis, glucose is broken down into two three-carbon molecules of pyruvate. The mitochondrial pyruvate dehydrogenase complex then catalyzes the oxidative decarboxylation of pyruvate to produce acetyl-CoA, a two-carbon acetyl unit that is ligated to the acyl-group carrier, CoA [6]. In the mitochondria, citrate synthase then catalyzes the condensation of the acetyl moiety of acetyl-CoA with oxaloacetate to yield a six-carbon citrate molecule. Citrate can proceed to be oxidized via the TCA cycle, or alternatively it can be transported to the cytosol as a substrate for the enzyme ATP citrate lyase, which cleaves citrate to regenerate acetyl-CoA and oxaloacetate [7] (Fig. 1). Under conditions of carbohydrate or glucose excess, the function of this pathway is to direct acetyl-CoA away from the mitochondria and back to the cytosol for the synthesis of fatty acids and sterols [8]. As such, cells can store excess carbohydrates as fat. Thus, the function of the ATP citrate lyase enzyme offers a clue to the logic and direction of carbon flow – acetyl-CoA units are shipped out of the mitochondria in the form of citrate when carbon sources are abundant, indicating a favorable nutrient state.
Figure 1. Schematic model proposing a general logic of acetyl-CoA utilization under fed versus fasted or growth versus survival states.
Under fed or growth states, acetyl-CoA is directed out of the mitochondria and to the cytosol and nucleus for use in lipid synthesis or histone acetylation. Nucleocytosolic amounts of acetyl-CoA increase relative to mitochondrial amounts. Under fasted or survival states, acetyl-CoA is channeled into the mitochondria for synthesis of ATP and ketone bodies. Mitochondrial amounts of acetyl-CoA increase relative to nucleocytosolic amounts. Fatty acid oxidation significantly increases mitochondrial acetyl-CoA.
Nucleocytosolic pools of acetyl-CoA are also utilized for histone acetylation and the activation of gene expression. ATP citrate lyase was shown to provide a source of acetyl-CoA for histone acetylation in mammalian cells [9]. The budding yeast Saccharomyces cerevisiae, which lacks ATP citrate lyase, relies on acetyl-CoA synthetase enzymes to supply acetyl-CoA for histone acetylation [10]. Moreover, a special cohort of yeast genes important for growth, such as those required for ribosome biogenesis and the G1 cyclin CLN3, are especially dependent on histone acetylation for their activation [11,12]. As such, the expression of these growth genes is closely coupled to acetyl-CoA as an indicator of the cell’s nutritional state. Thus, when carbon sources are abundant, nucleocytosolic amounts of acetyl-CoA accumulate and facilitate the processes of lipid synthesis and histone acetylation (Fig. 1).
Survival or Fasted State - High acetyl-CoA in mitochondria
During starvation, cells must typically shift from growth to survival mode and alter metabolism towards functions important for viability. Instead of shipping acetyl units out to the cytosol, there is now a greater requirement for acetyl-CoA to be oxidized in the mitochondria for ATP synthesis (Fig. 1). Under such conditions, nucleocytosolic acetyl-CoA levels therefore decrease. Fatty acids are a significant source of this mitochondrial acetyl-CoA pool [13]. CoA synthesis is induced to activate fatty acids as fatty acyl-CoAs [14,15], which can then be transported into mitochondria via the carnitine shuttle for β-oxidation. As a result, acetyl-CoA is generated in the mitochondria for oxidation or other possible fates. In the liver, mitochondrial acetyl-CoA is used to synthesize ketone bodies (acetoacetate and β-hydroxybutyrate) as alternative fuel sources for the brain and heart under conditions of carbohydrate scarcity [13,16]. Under such conditions, lower nucleocytosolic acetyl-CoA will also limit fatty acid synthesis, histone acetylation, and other growth-related processes. ATP citrate lyase is inhibited under these situations at both the transcriptional and post-translational levels [17,18].
Depletion of nucleocytosolic acetyl-CoA also represents a cue to induce autophagy [19,20]. In yeast, the expression of a core autophagy gene (ATG7) is repressed by acetyl-CoA [19]. During the yeast metabolic cycle (YMC), many ATG genes (including ATG1 and ATG8) are repressed during growth phases when acetyl-CoA levels rise, and are activated only during the stress/survival phases when acetyl-CoA levels fall [21,22]. More generally, many other genes with functions involved in stress and survival are induced concomitantly with core autophagy genes [21,23,24]. These genes tend to be less dependent on histone acetylation for their activation [11,25,26], perhaps due to reduced availability of acetyl-CoA. In mammalian cells, autophagy regulation by acetyl-CoA occurs in a manner dependent on the p300 acetyltransferase [20]. Thus, the regulation of autophagy by acetyl-CoA may occur primarily at the level of transcriptional control of core autophagy genes.
Taken together, under fasted or carbon-poor states, nucleocytosolic amounts of acetyl-CoA decrease in cells, while mechanisms to channel acetyl-CoA into the mitochondria are engaged. These considerations support a model in which the subcellular compartmentalization of acetyl-CoA units undergoes a major shift during starvation, and the utilization of these acetyl units is re-purposed to support survival strategies (Fig. 1).
Sensing of acetyl-CoA through protein acetylation modifications
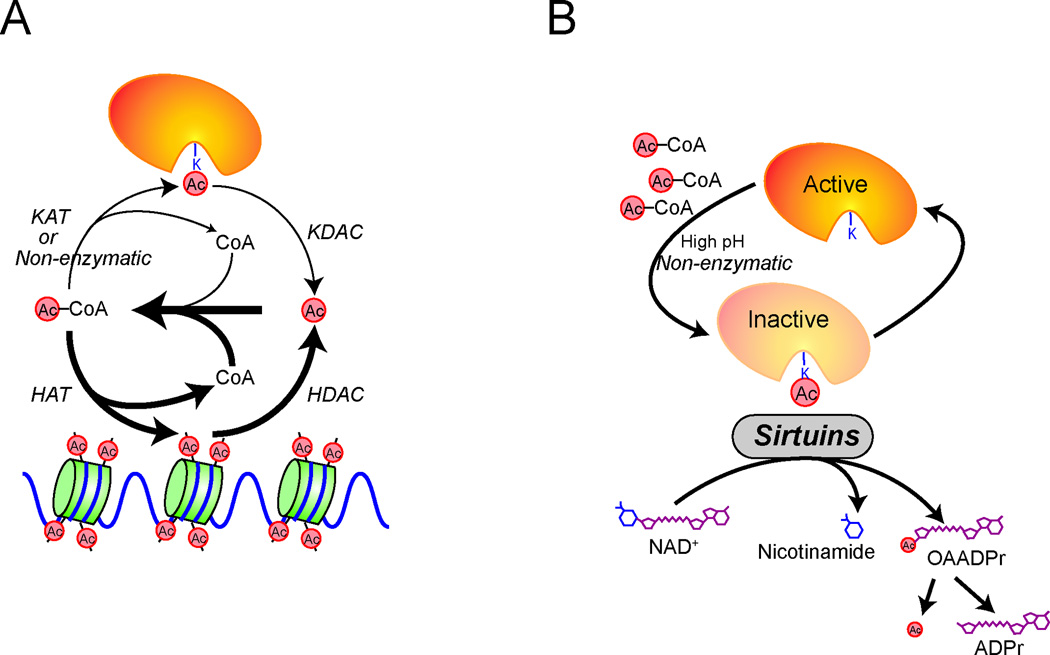
How might cells actually sense the abundance of acetyl-CoA? It is perhaps no coincidence that acetyl-CoA doubles as the acetyl donor for protein acetylation modifications (including histone acetylation) (Fig. 2). The abundance of protein acetylation modifications could therefore reflect the cell’s metabolic state to regulate various protein activities. Studies performed under carbon-rich conditions where acetyl-CoA synthesis is not limiting may mask the contributions of this metabolite in cellular regulation. However, most organisms, as well as particular tissue microenvironments in vivo experience challenges in the nutrient environment that might limit acetyl-CoA biosynthesis or availability (e.g., carbon starvation or hypoxia). Recent studies have begun to provide compelling evidence that many protein acetylation modifications are indeed modulated by acetyl-CoA availability [27,28].
Figure 2. Dynamic acetylation and deacetylation of proteins.
(A) The acetylation of proteins may be catalyzed by acetyltransferase enzymes or can occur spontaneously through reaction with acetyl-CoA directly. Deacetylase enzymes catalyze the removal of acetylation modifications. Liberated acetate can be converted back to acetyl-CoA. (B) Sirtuins utilize NAD+ to catalyze protein deacetylation, yielding nicotinamide and O-acetyl-ADP-ribose (OAADPr). The removal of aberrant acetylation or acylation modifications may restore protein function. Abbreviations: HAT (histone acetyltransferase), KAT (lysine acetyltransferase), HDAC (histone deacetylase), KDAC (lysine deacetylase), Ac (acetate), CoA (Coenzyme A).
Besides histones, the acetyl-CoA synthetase family of enzymes was also identified to be regulated by reversible acetylation [29–31]. The acetylation of an active site lysine residue was observed to inhibit the activity of acetyl-CoA synthetase as a mechanism of feedback inhibition in response to high acetyl-CoA [32–34]. The deacetylation of these enzymes, catalyzed by sirtuins, restores their activity [32–34]. Subsequent mass spectrometry surveys have now revealed that thousands of other proteins, including many other metabolic enzymes, can be acetylated [35–38]. In some cases, every enzyme in a particular biochemical pathway was found to be acetylated [39]. Although the majority of these modifications were found to be inhibitory, several were reported to be activating [40]. In some instances, the acetylation of particular metabolic enzymes was responsive to glucose levels in the media, suggesting that they could be linked to intracellular acetyl-CoA abundance. Whether specific acetyltransferase enzymes catalyze the majority of these acetylation modifications present on metabolic enzymes is not yet clear.
The yeast metabolic cycle (YMC) offers a system to investigate whether particular acetylation modifications might be coupled to acetyl-CoA itself. Studies of yeast cells undergoing the YMC during continuous, glucose-limited growth in a chemostat have revealed periodic changes in intracellular acetyl-CoA amounts as yeast cells alternate between growth and quiescent-like phases [22]. Several proteins are dynamically acetylated precisely in phase with the observed acetyl-CoA oscillations [11]. These include histones, several components of the transcriptional coactivator SAGA, a subunit of the SWI/SNF chromatin remodeling complex Snf2p, and a transcriptional coactivator of ribosomal subunit gene expression Ifh1p [11,41]. Interestingly, the dynamic acetylation of all of these proteins is dependent on the acetyltransferase Gcn5p, suggesting this enzyme has the capability of acetylating its substrates in tune with acetyl-CoA fluctuations in vivo. Consistent with this hypothesis, mutations within Gcn5p slow growth, disrupt the yeast metabolic cycle, or alter the cell’s responsiveness to acetate [11,12]. Moreover, acetylation of SAGA subunits appears to aid its recruitment to growth genes [11]. A brief survey of other acetylated proteins that are not known to be Gcn5p substrates showed they are not dynamically acetylated across the YMC [11]. An analysis of the genomic regions bound by these acetylated histones revealed that several marks, in particular H3K9Ac, were present predominantly at growth genes, specifically during the growth phase of the YMC when acetyl-CoA levels rise [11,25]. These considerations suggest that the acetylation of these nuclear-localized proteins collectively functions to promote the activation of growth genes in response to a burst of nucleocytosolic acetyl-CoA.
Stoichiometry of acetylation modifications
Given the thousands of newly identified acetylated proteins, a pertinent question is what proportion of each protein is acetylated? Recent studies aiming to determine the stoichiometry of acetylated sites estimate that for many proteins, only a small fraction of the peptides are actually acetylated [42,43]. However, nuclear proteins, including histones and transcription factors, were estimated to be acetylated at much higher stoichiometry [43]. Conventional shotgun detection of peptides by mass spectrometry is biased towards abundant proteins, so perhaps it is unsurprising that a small fraction of a very abundant protein that is acetylated could be scored as a positive. Moreover, lysine residues on proteins can react spontaneously with thioesters such as acetyl-CoA or other acyl-CoA metabolites, resulting in non-enzymatic acetylation or acylation [44–48]. Non-enzymatic acetylation or acylation may be especially prominent within the mitochondria [43,46,48,49], which is thought to have higher acetyl-CoA concentrations and higher pH, thereby increasing the nucleophilicity of lysyl side chains. Thus, while some non-enzymatic acetylation or acylation events could have evolved to be regulatory, the possibility also exists that many of these modifications could be spurious.
These considerations must be taken into account when determining the physiological significance of any detected acetylation site. Moreover, there are limitations to mutation of a lysine residue to either arginine or glutamine. These mutations are not always accurate acetylated or deacetylated lysine mimics, and could perturb protein function independent of site-specific acetylation. As such, it can be challenging to demonstrate whether a particular acetylation modification is functionally important in vivo. To help address these issues, methods for site-specific incorporation of acetyllysine [50], as well as better acetylated or deacetylated lysine mimics, have been developed [51,52]. The use of these and other methods will help clarify the extent through which protein acetylation modifications are responsive to acetyl-CoA fluctuations in a regulatory manner, either enzymatically or non-enzymatically.
Implications for sirtuin function
The accumulation of acetyl-CoA in subcellular compartments may also necessitate the activity of deacetylase enzymes to remove non-enzymatic acetylation modifications that could intentionally or unintentionally compromise protein function [28,53,54]. Such a “repair” or “detoxification” role may be fulfilled by the sirtuin family of protein deacylases (Fig. 2). Consistent with this idea, hyperacetylation of mitochondrial enzymes occurs in the absence of mitochondrial SIRT3 [55–57], and deacetylation of these enzymes typically increases their activity [53]. Moreover, the expression of SIRT3 is increased specifically under fasting states, in response to high-fat diets, or during exercise - conditions that all promote increased mitochondrial acetyl-CoA [53]. Likewise, the potential of proteins to be modified by other acyl-CoA metabolites besides acetyl-CoA is supported by the discovery of a wide variety of acylation modifications present on proteins, along with associated sirtuins that preferentially catalyze their removal [58–61]. Evidence that sirtuins evolved specifically to remove non-enzymatic protein acylation as a form of protein quality control has been summarized in a recent review [54]. In this model, failure of sirtuins to remove aberrant acylation modifications would hinder the function of effected proteins and consequently lead to dysfunctions in metabolism and susceptibility to disease [47,55,57].
The sirtuins utilize the cofactor NAD+ to catalyze protein deacylation. However, deacylation of a lysine residue can also be executed using water as a nucleophile without a requirement for NAD+, a mechanism employed by many histone deacetylases (HDACs) [62]. The dependency of sirtuins on NAD+ have led to the hypothesis that their activity could be regulated by fluctuations in NAD+ concentrations [63,64]. However, such dependency on NAD+ may serve an additional purpose and enable the removal of the acyl group via covalent attachment to ADP-ribose, to produce O-acyl-ADP-ribose metabolites, which themselves may have biological functions [65]. Moreover, if the acyl group were liberated as a free carboxylate, then the respective acyl-CoA synthetase enzymes could potentially convert these free carboxylates back to acyl-CoA metabolites, facilitating re-acylation and thus leading to a futile cycle (Fig. 2).
Summary and perspective
In summary, there is now compelling evidence that acetyl-CoA represents a fundamental gauge of cellular metabolic state that is monitored by the cell by way of distinctive protein acetylation modifications. Perhaps it is no coincidence that nature chose to carefully monitor the abundance of acetyl-CoA as a proxy for its metabolic state due to its requirement in the biosynthesis of cellular building blocks, two carbons at a time. Some of these acetyl-CoA-responsive modifications may be established by a delicate balance between the opposing activities of acetyltransferase and deacetylase enzymes, while others could be set in a non-enzymatic manner. Understanding the sources and fates, as well as the movement of acetyl groups between subcellular compartments reveals an underlying logic to metabolic strategies employed under growth versus survival, fed versus fasted, or normal versus tumorigenic metabolic states. Based on recent stoichiometry studies, it is tempting to speculate that acetylation originally evolved as a means to link nuclear activities with acetyl-CoA equivalents produced in the mitochondria. However, a consequence of using electrophilic acyl-CoA metabolites in cellular metabolism and regulation is their tendency to react spontaneously with nucleophilic moieties on proteins such as lysine residues. As such, the accumulation of particular acyl-CoA metabolites in various cellular compartments may have necessitated a mechanism to remove unintended acylation modifications on proteins. Future studies will continue to reveal the mechanisms and consequences of employing acetyl-CoA and other acyl-CoAs in cellular metabolism - their reciprocal influence on metabolism and cell regulation should no longer be overlooked.
Acknowledgments
The authors acknowledge support from NIH grant R01GM094314. Text and citation restrictions prevented us from discussing many other interesting and important studies in this field.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Literature Cited
• of special interest
- 1.Wellen KE, Thompson CB. A two-way street: reciprocal regulation of metabolism and signalling. Nat Rev Mol Cell Biol. 2012;13:270–276. doi: 10.1038/nrm3305. [DOI] [PubMed] [Google Scholar]
- 2.Kaelin WG, Jr, McKnight SL. Influence of metabolism on epigenetics and disease. Cell. 2013;153:56–69. doi: 10.1016/j.cell.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hardie DG. AMPK: positive and negative regulation, and its role in whole-body energy homeostasis. Curr Opin Cell Biol. 2014;33C:1–7. doi: 10.1016/j.ceb.2014.09.004. [DOI] [PubMed] [Google Scholar]
- 4.Canto C, Auwerx J. NAD+ as a signaling molecule modulating metabolism. Cold Spring Harb Symp Quant Biol. 2011;76:291–298. doi: 10.1101/sqb.2012.76.010439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Houtkooper RH, Canto C, Wanders RJ, Auwerx J. The secret life of NAD+: an old metabolite controlling new metabolic signaling pathways. Endocr Rev. 2010;31:194–223. doi: 10.1210/er.2009-0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Theodoulou FL, Sibon OC, Jackowski S, Gout I. Coenzyme A and its derivatives: renaissance of a textbook classic. Biochem Soc Trans. 2014;42:1025–1032. doi: 10.1042/BST20140176. [DOI] [PubMed] [Google Scholar]
- 7.Srere PA. The citrate cleavage enzyme. I. Distribution and purification. J Biol Chem. 1959;234:2544–2547. [PubMed] [Google Scholar]
- 8.Srere PA. The molecular physiology of citrate. Nature. 1965;205:766–770. [Google Scholar]
- 9.Wellen KE, Hatzivassiliou G, Sachdeva UM, Bui TV, Cross JR, Thompson CB. ATP-citrate lyase links cellular metabolism to histone acetylation. Science. 2009;324:1076–1080. doi: 10.1126/science.1164097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takahashi H, McCaffery JM, Irizarry RA, Boeke JD. Nucleocytosolic acetyl-coenzyme a synthetase is required for histone acetylation and global transcription. Mol Cell. 2006;23:207–217. doi: 10.1016/j.molcel.2006.05.040. [DOI] [PubMed] [Google Scholar]
- 11. Cai L, Sutter BM, Li B, Tu BP. Acetyl-CoA induces cell growth and proliferation by promoting the acetylation of histones at growth genes. Mol Cell. 2011;42:426–437. doi: 10.1016/j.molcel.2011.05.004. Study showing how and why some acetylation modifications are coupled to acetyl-CoA fluctuations in yeast.
- 12.Shi L, Tu BP. Acetyl-CoA induces transcription of the key G1 cyclin CLN3 to promote entry into the cell division cycle in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2013;110:7318–7323. doi: 10.1073/pnas.1302490110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McGarry JD, Foster DW. Regulation of hepatic fatty acid oxidation and ketone body production. Annu Rev Biochem. 1980;49:395–420. doi: 10.1146/annurev.bi.49.070180.002143. [DOI] [PubMed] [Google Scholar]
- 14.Jackowski S, Leonardi R. Deregulated coenzyme A loss of metabolic flexibility and diabetes. Biochem Soc Trans. 2014;42:1118–1122. doi: 10.1042/BST20140156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leonardi R, Rehg JE, Rock CO, Jackowski S. Pantothenate kinase 1 is required to support the metabolic transition from the fed to the fasted state. PLoS One. 2010;5:e11107. doi: 10.1371/journal.pone.0011107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Robinson AM, Williamson DH. Physiological roles of ketone bodies as substrates and signals in mammalian tissues. Physiol Rev. 1980;60:143–187. doi: 10.1152/physrev.1980.60.1.143. [DOI] [PubMed] [Google Scholar]
- 17.Lin R, Tao R, Gao X, Li T, Zhou X, Guan KL, Xiong Y, Lei QY. Acetylation stabilizes ATP-citrate lyase to promote lipid biosynthesis and tumor growth. Mol Cell. 2013;51:506–518. doi: 10.1016/j.molcel.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Uyeda K, Repa JJ. Carbohydrate response element binding protein, ChREBP, a transcription factor coupling hepatic glucose utilization and lipid synthesis. Cell Metab. 2006;4:107–110. doi: 10.1016/j.cmet.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 19. Eisenberg T, Schroeder S, Andryushkova A, Pendl T, Kuttner V, Bhukel A, Marino G, Pietrocola F, Harger A, Zimmermann A, et al. Nucleocytosolic depletion of the energy metabolite acetyl-coenzyme a stimulates autophagy and prolongs lifespan. Cell Metab. 2014;19:431–444. doi: 10.1016/j.cmet.2014.02.010. One of two studies identifying acetyl-CoA as a repressor of autophagy. The authors demonstrated nucleocytosolic acetyl-CoA repressed autophagy and ATG7 gene expression in yeast.
- 20.Marino G, Pietrocola F, Eisenberg T, Kong Y, Malik SA, Andryushkova A, Schroeder S, Pendl T, Harger A, Niso-Santano M, et al. Regulation of autophagy by cytosolic acetyl-coenzyme A. Mol Cell. 2014;53:710–725. doi: 10.1016/j.molcel.2014.01.016. [DOI] [PubMed] [Google Scholar]
- 21.Tu BP, Kudlicki A, Rowicka M, McKnight SL. Logic of the yeast metabolic cycle: temporal compartmentalization of cellular processes. Science. 2005;310:1152–1158. doi: 10.1126/science.1120499. [DOI] [PubMed] [Google Scholar]
- 22.Tu BP, Mohler RE, Liu JC, Dombek KM, Young ET, Synovec RE, McKnight SL. Cyclic changes in metabolic state during the life of a yeast cell. Proc Natl Acad Sci U S A. 2007;104:16886–16891. doi: 10.1073/pnas.0708365104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, Brown PO. Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brauer MJ, Huttenhower C, Airoldi EM, Rosenstein R, Matese JC, Gresham D, Boer VM, Troyanskaya OG, Botstein D. Coordination of growth rate, cell cycle, stress response, and metabolic activity in yeast. Mol Biol Cell. 2008;19:352–367. doi: 10.1091/mbc.E07-08-0779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuang Z, Cai L, Zhang X, Ji H, Tu BP, Boeke JD. High-temporal-resolution view of transcription and chromatin states across distinct metabolic states in budding yeast. Nat Struct Mol Biol. 2014;21:854–863. doi: 10.1038/nsmb.2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mehrotra S, Galdieri L, Zhang T, Zhang M, Pemberton LF, Vancura A. Histone hypoacetylation-activated genes are repressed by acetyl-CoA- and chromatin-mediated mechanism. Biochim Biophys Acta. 2014;1839:751–763. doi: 10.1016/j.bbagrm.2014.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shi L, Tu BP. Protein acetylation as a means to regulate protein function in tune with metabolic state. Biochem Soc Trans. 2014;42:1037–1042. doi: 10.1042/BST20140135. [DOI] [PubMed] [Google Scholar]
- 28.Cai L, Tu BP. On Acetyl-CoA as a Gauge of Cellular Metabolic State. Cold Spring Harb Symp Quant Biol. 2011;76:195–202. doi: 10.1101/sqb.2011.76.010769. [DOI] [PubMed] [Google Scholar]
- 29.Lipmann F, Tuttle LC. The detection of activated carboxyl groups with hydroxylamine as interceptor. J Biol Chem. 1945;161:415. [PubMed] [Google Scholar]
- 30.Jones ME, Lipmann F, Hilz H, Lynen F. On the enzymatic mechanism of Coenzyme A acetylation with adenosine triphosphate and acetate. J Am Chem Soc. 1953;75:3285–3286. [Google Scholar]
- 31.Berg P. Acyl adenylates, an enzymatic mechanism of acetate activation. J Biol Chem. 1956;222:991–1013. [PubMed] [Google Scholar]
- 32.Starai VJ, Celic I, Cole RN, Boeke JD, Escalante-Semerena JC. Sir2-dependent activation of acetyl-CoA synthetase by deacetylation of active lysine. Science. 2002;298:2390–2392. doi: 10.1126/science.1077650. [DOI] [PubMed] [Google Scholar]
- 33.Hallows WC, Lee S, Denu JM. Sirtuins deacetylate and activate mammalian acetyl-CoA synthetases. Proc Natl Acad Sci U S A. 2006;103:10230–10235. doi: 10.1073/pnas.0604392103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schwer B, Bunkenborg J, Verdin RO, Andersen JS, Verdin E. Reversible lysine acetylation controls the activity of the mitochondrial enzyme acetyl-CoA synthetase 2. Proc Natl Acad Sci U S A. 2006;103:10224–10229. doi: 10.1073/pnas.0603968103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kim SC, Sprung R, Chen Y, Xu Y, Ball H, Pei J, Cheng T, Kho Y, Xiao H, Xiao L, et al. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol Cell. 2006;23:607–618. doi: 10.1016/j.molcel.2006.06.026. The first proteomic survey demonstrating that acetylation can occur on a variety of proteins, especially mitochondrial proteins.
- 36.Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 37.Wang Q, Zhang Y, Yang C, Xiong H, Lin Y, Yao J, Li H, Xie L, Zhao W, Yao Y, et al. Acetylation of metabolic enzymes coordinates carbon source utilization and metabolic flux. Science. 2010;327:1004–1007. doi: 10.1126/science.1179687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao S, Xu W, Jiang W, Yu W, Lin Y, Zhang T, Yao J, Zhou L, Zeng Y, Li H, et al. Regulation of cellular metabolism by protein lysine acetylation. Science. 2010;327:1000–1004. doi: 10.1126/science.1179689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guan KL, Xiong Y. Regulation of intermediary metabolism by protein acetylation. Trends Biochem Sci. 2011;36:108–116. doi: 10.1016/j.tibs.2010.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hallows WC, Yu W, Denu JM. Regulation of glycolytic enzyme phosphoglycerate mutase-1 by Sirt1 protein-mediated deacetylation. J Biol Chem. 2012;287:3850–3858. doi: 10.1074/jbc.M111.317404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cai L, McCormick MA, Kennedy BK, Tu BP. Integration of multiple nutrient cues and regulation of lifespan by ribosomal transcription factor Ifh1. Cell Rep. 2013;4:1063–1071. doi: 10.1016/j.celrep.2013.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baeza J, Dowell JA, Smallegan MJ, Fan J, Amador-Noguez D, Khan Z, Denu JM. Stoichiometry of site-specific lysine acetylation in an entire proteome. J Biol Chem. 2014;289:21326–21338. doi: 10.1074/jbc.M114.581843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weinert BT, Iesmantavicius V, Moustafa T, Scholz C, Wagner SA, Magnes C, Zechner R, Choudhary C. Acetylation dynamics and stoichiometry in Saccharomyces cerevisiae. Mol Syst Biol. 2014;10:716. doi: 10.1002/msb.134766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Paik WK, Pearson D, Lee HW, Kim S. Nonenzymatic acetylation of histones with acetyl-CoA. Biochim Biophys Acta. 1970;213:513–522. doi: 10.1016/0005-2787(70)90058-4. [DOI] [PubMed] [Google Scholar]
- 45.Berndsen CE, Denu JM. Assays for mechanistic investigations of protein/histone acetyltransferases. Methods. 2005;36:321–331. doi: 10.1016/j.ymeth.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 46.Pougovkina O, te Brinke H, Ofman R, van Cruchten AG, Kulik W, Wanders RJ, Houten SM, de Boer VC. Mitochondrial protein acetylation is driven by acetyl-CoA from fatty acid oxidation. Hum Mol Genet. 2014;23:3513–3522. doi: 10.1093/hmg/ddu059. [DOI] [PubMed] [Google Scholar]
- 47. Pougovkina O, Te Brinke H, Wanders RJ, Houten SM, de Boer VC. Aberrant protein acylation is a common observation in inborn errors of acyl-CoA metabolism. J Inherit Metab Dis. 2014;37:709–714. doi: 10.1007/s10545-014-9684-9. Study showing aberrant protein acylation in several models of inborn errors of metabolism associated with accumulation of particular acyl-CoA metabolites.
- 48.Wagner GR, Payne RM. Widespread and enzyme-independent Nepsilon-acetylation and Nepsilon-succinylation of proteins in the chemical conditions of the mitochondrial matrix. J Biol Chem. 2013;288:29036–29045. doi: 10.1074/jbc.M113.486753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ghanta S, Grossmann RE, Brenner C. Mitochondrial protein acetylation as a cell-intrinsic, evolutionary driver of fat storage: chemical and metabolic logic of acetyl-lysine modifications. Crit Rev Biochem Mol Biol. 2013;48:561–574. doi: 10.3109/10409238.2013.838204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Neumann H, Peak-Chew SY, Chin JW. Genetically encoding N(epsilon)-acetyllysine in recombinant proteins. Nat Chem Biol. 2008;4:232–234. doi: 10.1038/nchembio.73. [DOI] [PubMed] [Google Scholar]
- 51.Huang R, Holbert MA, Tarrant MK, Curtet S, Colquhoun DR, Dancy BM, Dancy BC, Hwang Y, Tang Y, Meeth K, et al. Site-specific introduction of an acetyl-lysine mimic into peptides and proteins by cysteine alkylation. J Am Chem Soc. 2010;132:9986–9987. doi: 10.1021/ja103954u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dancy BC, Ming SA, Papazyan R, Jelinek CA, Majumdar A, Sun Y, Dancy BM, Drury WJ, 3rd, Cotter RJ, Taverna SD, et al. Azalysine analogues as probes for protein lysine deacetylation and demethylation. J Am Chem Soc. 2012;134:5138–5148. doi: 10.1021/ja209574z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.He W, Newman JC, Wang MZ, Ho L, Verdin E. Mitochondrial sirtuins: regulators of protein acylation and metabolism. Trends Endocrinol Metab. 2012;23:467–476. doi: 10.1016/j.tem.2012.07.004. [DOI] [PubMed] [Google Scholar]
- 54. Wagner GR, Hirschey MD. Nonenzymatic protein acylation as a carbon stress regulated by sirtuin deacylases. Mol Cell. 2014;54:5–16. doi: 10.1016/j.molcel.2014.03.027. Review discussing evidence supporting a role for sirtuins in protein quality control and the removal of nonenzymatic protein acylation modifications.
- 55.Hirschey MD, Shimazu T, Goetzman E, Jing E, Schwer B, Lombard DB, Grueter CA, Harris C, Biddinger S, Ilkayeva OR, et al. SIRT3 regulates mitochondrial fatty-acid oxidation by reversible enzyme deacetylation. Nature. 2010;464:121–125. doi: 10.1038/nature08778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lombard DB, Alt FW, Cheng HL, Bunkenborg J, Streeper RS, Mostoslavsky R, Kim J, Yancopoulos G, Valenzuela D, Murphy A, et al. Mammalian Sir2 homolog SIRT3 regulates global mitochondrial lysine acetylation. Mol Cell Biol. 2007;27:8807–8814. doi: 10.1128/MCB.01636-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ahn BH, Kim HS, Song S, Lee IH, Liu J, Vassilopoulos A, Deng CX, Finkel T. A role for the mitochondrial deacetylase Sirt3 in regulating energy homeostasis. Proc Natl Acad Sci U S A. 2008;105:14447–14452. doi: 10.1073/pnas.0803790105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Du J, Zhou Y, Su X, Yu JJ, Khan S, Jiang H, Kim J, Woo J, Kim JH, Choi BH, et al. Sirt5 is a NAD-dependent protein lysine demalonylase and desuccinylase. Science. 2011;334:806–809. doi: 10.1126/science.1207861. Inspired by protein structural analysis, the authors discovered the lysine demalonylase and desuccinylase activity of SIRT5. This study explains why SIRT5 did not exhibit deacetylase activity and implicates other sirtuin members in regulating various protein acylation modifications other than acetylation.
- 59. Jiang H, Khan S, Wang Y, Charron G, He B, Sebastian C, Du J, Kim R, Ge E, Mostoslavsky R, et al. SIRT6 regulates TNF-alpha secretion through hydrolysis of long-chain fatty acyl lysine. Nature. 2013;496:110–113. doi: 10.1038/nature12038. The authors discovered the specific role of SIRT6 in antagonizing protein lysine long chain fatty acylation by enzymology and structural analysis. This study resolves how SIRT6 regulates TNFα.
- 60.Tan M, Peng C, Anderson KA, Chhoy P, Xie Z, Dai L, Park J, Chen Y, Huang H, Zhang Y, et al. Lysine glutarylation is a protein posttranslational modification regulated by SIRT5. Cell Metab. 2014;19:605–617. doi: 10.1016/j.cmet.2014.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Feldman JL, Baeza J, Denu JM. Activation of the protein deacetylase SIRT6 by long-chain fatty acids and widespread deacylation by mammalian sirtuins. J Biol Chem. 2013;288:31350–31356. doi: 10.1074/jbc.C113.511261. The authors profiled distinct but overlapping deacylation specificities of sirtuins and suggested potential regulation of SIRT6 by long chain fatty acids. The results reveal distinct regulations and roles for each sirtuin in response to cellular metabolic state.
- 62.de Ruijter AJ, van Gennip AH, Caron HN, Kemp S, van Kuilenburg AB. Histone deacetylases (HDACs): characterization of the classical HDAC family. Biochem J. 2003;370:737–749. doi: 10.1042/BJ20021321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Imai S, Guarente L. NAD+ and sirtuins in aging and disease. Trends Cell Biol. 2014;24:464–471. doi: 10.1016/j.tcb.2014.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Houtkooper RH, Pirinen E, Auwerx J. Sirtuins as regulators of metabolism and healthspan. Nat Rev Mol Cell Biol. 2012;13:225–238. doi: 10.1038/nrm3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tong L, Denu JM. Function and metabolism of sirtuin metabolite O-acetyl-ADP-ribose. Biochim Biophys Acta. 2010;1804:1617–1625. doi: 10.1016/j.bbapap.2010.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]