Abstract
Antibodies to neuraminidase (NA), the second most abundant surface protein of the influenza virus, contribute to protection against influenza virus infection. Although traditional and miniaturized thiobarbituric acid (TBA) neuraminidase inhibition (NI) assays have been successfully used to characterize the antigenic properties of NA, these methods are cumbersome and not easily amendable to rapid screening. An additional difficulty of the NI assay is the interference by hemagglutinin (HA)-specific antibodies. To prevent interference of HA-specific antibodies, most NI assays are performed with recombinant viruses containing a mismatched HA. However, generation of these viruses is time consuming and unsuitable for large-scale surveillance. The feasibility of using the recently developed enzyme-linked lectin assay (ELLA) to evaluate the antigenic relatedness of NA of wild type A(H3N2) viruses was assessed. Rather than using recombinant viruses, wild type A(H3N2) viruses were used as antigen with ferret sera elicited against recombinant viruses with a mismatched HA. In this study, details of the critical steps that are needed to modify and optimize the NI ELLA in a format that is reproducible, highly sensitive, and useful for influenza virus surveillance to monitor antigenic drift of NA are provided.
Keywords: Influenza, Neuraminidase, Antigenic drift, ELLA
1. Introduction
Influenza vaccines reduce the morbidity and mortality associated with annual influenza epidemics. The seasonal influenza vaccine is designed to protect against circulating influenza A H1N1 viruses (A(H1N1)), influenza A H3N2 viruses (A(H3N2)), and influenza B viruses. The influenza virus escapes host immunity through mutations in the major surface glycoproteins hemagglutinin (HA) and neuraminidase (NA). This process is known as antigenic drift (Schild et al., 1974; Webster et al., 1982) and as a result of this drift, the influenza vaccine has to be updated frequently. In the period from 1999 to 2010, the A(H3N2) virus component was updated 6 times (Barr et al., 2010). Recently, it has been shown that only few mutations near the receptor-binding site of HA are responsible for antigenic drift of A(H3N2) viruses circulating between 1968 and 2003 (Koel et al., 2013). For NA, a number of antigenic sites have been described (Air et al., 1985). These antigenic regions surround the enzyme's active site (Colman et al., 1983, 1987) and are highly variable, most likely due to immune pressure (Laver et al., 1982; Luther et al., 1984). Influenza virus surveillance by national influenza centers is done year-round (Russell et al., 2008; Barr et al., 2010). Representatives of the predominant circulating viruses are sent to the World Health Organization (WHO) Collaborating Centers. These centers characterize the viruses by sequencing the HA and NA genes and performing hemagglutination inhibition (HI) assays (Barr et al., 2010). During vaccine strain selection, the main focus is on the genetic and antigenic characterization of HA (Fiore et al., 2010).
Influenza viruses attach to the host cell surface via binding of the HA to sialic acid-containing receptors (Sauter et al., 1989). The enzymatic activity of NA allows virus release from the cell (Palese et al., 1974; Palese and Compans, 1976; Liu et al., 1995) by cleaving the sialic acid residues from the newly formed virus particles and from the host cell (von Itzstein, 2007). Preclinical and clinical studies showed that NA-specific immunity can reduce the severity of disease (Schulman et al., 1968; Murphy et al., 1972; Couch et al., 1974; Kilbourne, 1976; Johansson et al., 1993; Brett and Johansson, 2005). Antibodies directed toward NA inhibit release and spread of newly formed virus particles from infected cells (Compans et al., 1969). The antigenic drift of NA does not closely correspond to that of HA (Schulman and Kilbourne, 1969; Kilbourne et al., 1990; Sandbulte et al, 2011). Considering these findings, investigating options to include routine analysis of NA during vaccine strain selection next to HA seems to be warranted.
Antigenic characterization of NA can be performed using NA inhibition (NI) assays to determine the extent of antibody-mediated interference with enzyme activity (Kilbourne et al., 1968). These assays rely on the enzymatic sialidase activity by measuring cleavage of sialic acid from highly glycosylated proteins such as fetuin. The NI thiobarbituric acid (TBA) assay (Warren, 1959; Webster and Laver, 1967) is based on the detection of free sialic acid. This assay is recommended by the WHO (Cox et al., 2002), but the use of large volumes of hazardous chemicals and laborious procedures has impeded antigenic characterization of NA during influenza virus surveillance. A simplified and miniaturized version of the TBA was developed (Sandbulte et al., 2009), but this assay still remains cumbersome. The recently developed enzyme-linked lectin assay (ELLA) (Lambre et al., 1991; Cate et al., 2010; Couch et al., 2012, 2013; Fritz et al., 2012; Couzens et al., 2014) also relies on the sialidase activity of NA, but instead of measuring free sialic acid, it detects the terminal galactose that becomes exposed after sialic acid cleavage by NA.
A complication of NI assays is the interference of HA-specific antibodies that block NA activity non-specifically when they bind to HA (Schulman and Kilbourne, 1969). Recombinant influenza viruses with a heterologous HA are commonly used for NI assays. Antibodies directed toward the H1 or H3 HA of A(H1N1) and A(H3N2) viruses do not cross-react with a heterologous HA (e.g. H6), and hence only the interaction between NA and NA-specific antibodies is measured (Couzens et al., 2014). However, the generation of recombinant viruses is time-consuming for large numbers of viruses and therefore this method is not suitable for large-scale surveillance of antigenic drift of circulating influenza viruses. For analysis of the antigenic drift of NA, it would be ideal to have the capability of using wild type viruses as antigen in assays that are not impacted by non-specific inhibitors, including antibodies to HA.
In this study, optimized methods to enable rapid antigenic characterization of NA, with wild type viruses as antigen, are described. Since the ELLA is less laborious and shows a good correlation to the miniaturized TBA (Fritz et al., 2012), this assay was selected as a platform. To prevent interference by antibodies directed against HA of wild type A(H3N2) viruses, ferret sera were raised against recombinant influenza A H7N2 viruses (A(H7N2)) viruses that contain the NA of various A(H3N2) viruses. Through this approach it is possible to screen wild type viruses, thus preventing the time-consuming generation of recombinant viruses or proteins for each virus of interest. Reproducibility and sensitivity of the NI assay were highest using virus concentrations that resulted in ∼50% of total NA activity of that virus. Non-specific inhibition of ferret sera was observed for some wild type viruses, especially A(H3N2) viruses, but the critical steps to overcome this non-specific inhibition and obtain reproducible and highly sensitive results are also described.
2. Materials and methods
2.1. Cells
Madin-Darby canine kidney (MDCK) cells were cultured in Eagle's minimal essential medium (EMEM, Lonza, Breda, The Netherlands) supplemented with 10% fetal bovine serum (FBS, Sigma-Aldrich, St. Louis, MO, USA), 100U/ml penicillin (Lonza), 100U/ml streptomycin (Lonza), 2mM glutamine (Lonza), 1.5mg/ml sodium bicarbonate (Lonza), 10mM HEPES (Lonza), and non-essential amino acids (MP Biomedicals, Europe, Illkirch, France).
293T cells were cultured in Dulbecco's modified Eagle's medium (DMEM, Lonza) supplemented with 10% FBS, 100U/ml penicillin, 100U/ml streptomycin, 2mM glutamine, 1mM sodium pyruvate (Life Technologies, Bleiswijk, The Netherlands), 500 μg/ml geneticin (Life Technologies) and non-essential amino acids.
2.2. Plasmids
The NA gene segments of A(H3N2) viruses A/Bilthoven/21793/ 1972 (BI/72); A/Bilthoven/1761/1976 (BI/76); A/Netherlands/233/ 1982 (NL/82); A/Netherlands/823/1992 (NL/92); A/Netherlands/ 178/1995 (NL/95); A/Netherlands/69/2009 (NL/09) and the HA and NA gene segments of A(H2N2) viruses A/Netherlands/M1/1957 (NL/57) and A/Netherlands/B1/1968 (NL/68) were amplified by reverse transcription polymerase chain reaction (RT-PCR) and cloned in a modified version of the bidirectional reverse genetics plasmid pHW2000 (Hoffmann et al., 2000; de Wit et al., 2004). To reduce pathogenicity, the multibasic cleavage site (MBCS) was removed from the bidirectional reverse genetics HA plasmid of the highly pathogenic avian A(H7N7) virus (A/Netherlands/33/03) (de Wit et al., 2010). This was done using the QuikChange multi-site-directed mutagenesis kit (Stratagene, Huissen, The Netherlands) according to the instructions of the manufacturer with specific primers (available from the authors upon request). The plasmids encoding the internal genes of A/Netherlands/219/03 (H7N7) have been described previously (de Wit et al., 2010).
The accession numbers that were used are as follows: for NA, CY112307, CY113199, CY114439, CY113735, CY116590, CY113023, KM402803, and KM402811; for HA, KM402801 and KM402809.
2.3. Viruses
All human and recombinant influenza A viruses were propagated at 37 °C and influenza B viruses at 33 °C in MDCK cells in EMEM supplemented with, 100U/ml penicillin, 100U/ml streptomycin, 2mM glutamine, 1.5 mg/mlsodium bicarbonate, 10mM HEPES, non-essential amino acids and 1 μg/ml L-1-tosylamide-2-phenylethyl chloromethyl ketone-treated trypsin (Sigma-Aldrich, Zwijndrecht, The Netherlands). Avian influenza viruses were propagated in 11-day old embryonated chicken eggs. All viruses were harvested 48 h post-inoculation and cell debris was removed by centrifugation for 15min at 3000 rpm. Supernatant was either immediately stored in suitable aliquots at -80 °C or, if needed, after concentration using an Amicon Ultra-15 Centrifugal Filter (Millipore, Amsterdam, The Netherlands).
Recombinant A(H2N2) and A(H7N2) viruses were generated by reverse genetics using transient calcium phosphate-mediated transfections of 293T cells as described previously (de Wit et al., 2004). A(H2N2) viruses were generated with plasmids carrying the HA and NA gene segments of A(H2N2) viruses (NL/57 or NL/68) and the six remaining gene segments of A/Puerto Rico/8/1934 (H1N1) under biosafety level 2 (BSL-2) conditions. A(H7N2) viruses were generated under biosafety level 3 (BSL-3) conditions with plasmids carrying the HA gene segment of A/Netherlands/33/03 (H7N7) without the MBCS, the internal gene segments of A/Netherlands/219/03 (H7N7) (de Wit et al., 2010), and one of the following NA gene segments of A(H3N2) viruses: A/Bilthoven/16190/1968 (BI/68) (Schrauwen et al., 2011); BI/72; BI/76; NL/82; NL/92; NL/95; A/Netherlands/213/2003 (NL/03) (Chutinimitkul et al., 2010); NL/09; and A(H2N2) viruses: NL/57 or NL/68. The supernatant of the transfected cells was harvested 48 h post-transfection and was passaged twice on MDCK cells. MDCK passage 2 supernatant was harvested 48 h post-inoculation and cell debris was removed by centrifugation for 15 min at 3000 rpm and immediately stored in suitable aliquots at −80 °C.
2.4. Generation of ferret sera
Male ferrets (1 year old, 0.8–1 kg) were inoculated intranasally with 1 ml of A(H7N2) virus containing MDCK passage 2 supernatant under animal BSL-3 conditions. Sera were collected 2 weeks post-inoculation. RNA was isolated from ferret sera and RT-PCR was performed as described previously (Fouchier et al., 2000) to detect A(H7N2) virus. Once absence of viral RNA was confirmed, sera were stored in suitable aliquots at −20°C. The independent animal experimentation ethical review committee “Stichting DEC Consult” approved all animal protocols (Erasmus MC permit number EMC 2617), and experiments were performed in compliance with Dutch and European guidelines for animal experimentation.
Unless noted otherwise, sera for the NI ELLA were treated with Burnet's receptor destroying enzyme (RDE) filtrated from cultures of Vibrio cholerae (Burnet and Stone, 1947). Sera were incubated overnight at 37 °C in a ratio of 1:6 with a 10-fold dilution of RDE in Dulbecco's phosphate-buffered saline (DPBS) containing 0.133 g/L CaCl2 and 0.1 g/L MgCl2 with 1% Bovine Serum Albumin (BSA) solution (Sigma–Aldrich) and 0.05% Tween 20 (Sigma–Aldrich, DPBSTBSA), followed by heat inactivation at 56 °C for 8 h.
2.5. ELLA and NI ELLA
The principals of the NI ELLA described by Lambre et al. (1991) and Cate et al. (2010) were optimized for rapid antigenic analysis of wild type viruses. Stock solutions of fetuin were made by dissolving fetuin from FBS (Sigma–Aldrich) in Coating Solution Concentrate (tebu-bio, Heerhugowaard, The Netherlands) to a concentration of 25 mg/ml. Fetuin was further diluted to a concentration of 25 ug/ml in Coating Solution Concentrate and used to coat Nunc-Immuno™ MicroWell™ 96 well solid plates (100 μl/well; Sanbio, Uden, The Netherlands) at 4 °C for at least 24 h. Aliquoted fetuin (25 mg/ml) could be stored for at least 6 months at −20 °C and coated plates could be stored for at least 2 months at 4 °C without a significant decrease in reactivity.
The ELLA was used to determine the optimal amount of each virus (antigen) for routine serology. Twofold serial dilutions of the antigens were made in DPBSTBSA. Fetuin-coated plates were washed 3 times in 0.1 M phosphate buffered saline (PBS, pH 7.4) with 0.05% Tween 20 (PBST, Sigma–Aldrich). Fifty microliter of the serial dilutions was transferred to duplicate fetuin-coated plates containing 50 μl DPBSTBSA. Plates were sealed and incubated at 37 °C for 16–18 h. The plates were subsequently washed six times with PBST, and 100 μl/well of and optimized dilution of horseradish peroxidase-conjugated peanut agglutinin lectin (PNA-HRPO, Sigma–Aldrich) in DPBS containing 0.133 g/L CaCl2 and 0.1 g/L MgCl2 with 1% BSA(DPBSBSA) was added. The optimal PNA-HRPO concentration is the concentration that gives the maximal signal in ELLA assays that allowed complete digestion of sialic acid from fetuin. PNA-HRPO could be stored at −20 °C in DPBSBSA for a maximum of one month, without loss of signal intensity. Plates were incubated at room temperature for 2 h followed by washing three times with PBST. O-Phenylenediamine dihydrochloride (OPD, Sigma–Aldrich) substrate was freshly prepared following the manufacturer's instruction and added to the plate (100 μl/well). The reaction was stopped after 10 min by the addition of stop solution (0.5 M H2SO4, 100 μl/well). The plates were read at 490 nm (OD490) for 0.25 s using an Infinite 200 96-well plate reader (Tecan, Giessen, The Netherlands). The mean background (Abkg) value was established for each plate using at least four wells that were treated identically to test wells, but in the absence of antigen. The NA activity was determined by subtracting the background value from the test well values (Atest) i.e. (Atest−Abkg). Next the NA activity was plotted against the antigen dilutions. Unless noted otherwise, the dilution of antigen that resulted in 50% of total NA activity but with an OD490 >0.75 was selected to subsequently perform the NI ELLA.
For the NI ELLA, serum samples were serially diluted (twofold) in DPBSTBSA and incubated in duplicate fetuin-coated plates with an equal volume (50 μl) of the selected antigen dilution in DPBSTBSA. These plates were subsequently sealed and incubated for 16–18h at 37 °C. Incubation with PNA-HRPO, the addition of substrate, stop buffer and subsequent absorbance measurements were performed as described above for the ELLA. The NI titers were calculated as follows, first the mean background was subtracted from the absorbance of the test wells and the wells that contained antigen but not ferret serum (positive control, Apos). Next, the percentage of NA activity was calculated by dividing the mean absorbance of test wells by the mean absorbance of positive control wells and multiplied by 100, i.e. (Atest−Abkg)/(Apos−Abkg) × 100. Finally, to determine the percentage of NA inhibition, the percentage of activity was subtracted from 100. The NI titers were defined as the reciprocal of the last dilution that resulted in at least 50% inhibition. When the duplicate titers were different, the geometric mean titers were calculated. An assay was considered valid if the NI titers of control sera run in separate plates and assays using the same conditions yielded similar titers (titer < twofold difference). The OD490 values of the duplicates did not vary more than 20%.
2.6. Hemagglutination inhibition assay
HI titers were determined using standard procedures (Hirst, 1943). In brief, antisera were pre-treated overnight with RDE at 37 °C and heat inactivated at 56 °C for 1 h. Twofold serial dilutions of RDE-treated antisera (in 50 μl), starting at a 1:20 dilution, were incubated with 25 μl of four HA units of each virus, and incubated at 37 °C for 30 min. Next, 25 μl of 1% turkey erythrocytes were added, followed by an 1 h incubation at 4 °C. The reciprocal of the last serum dilution that was able to block the agglutination of the turkey erythrocytes was recorded as the HI titer.
3. Results
3.1. Principles of the ELLA and NI ELLA
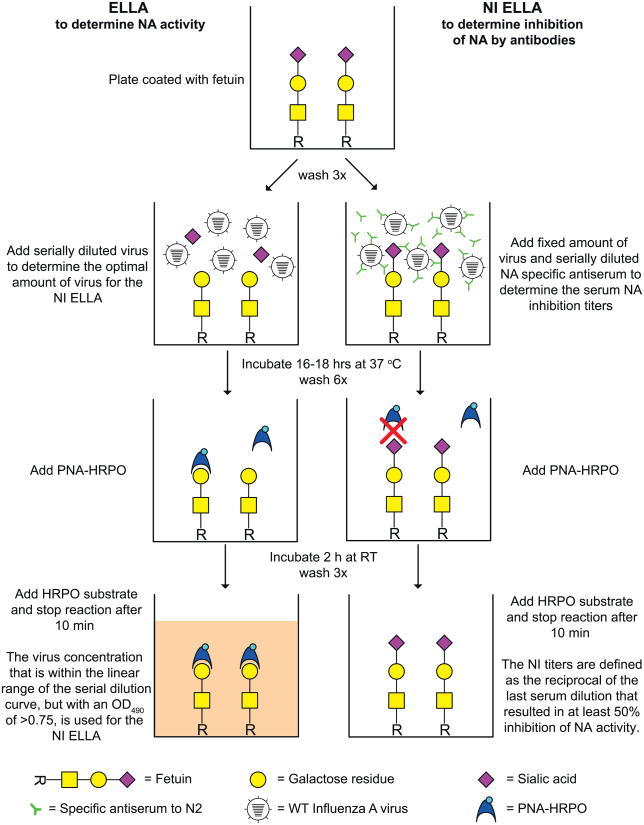
The ELLA measures the enzymatic activity of NA by detecting desialylation of the highly glycosylated fetuin by NA. PNA-HRPO is used to detect the terminal galactoses that become exposed after desialylation of fetuin by NA. The intensity of the signal after addition of the substrate is dependent on the level of desialylation and thus NA activity. In the NI ELLA, binding of NA by specific antibodies will inhibit the enzymatic function of NA and result in a reduction of desialylation and, hence, in a reduction of the signal. An illustration of the ELLA and NI ELLA principles is shown in Fig. 1. To enable rapid antigenic characterization of the NA of wild-type human A(H3N2) viruses, monospecific ferret antisera raised against recombinant viruses with a mismatched HA were generated. These recombinant A(H7N2) viruses contain the NA of A(H3N2) viruses and the HA and internal gene segments of an A(H7N7) virus. Therefore the HA-specific antibodies present in the ferret sera will not interfere with wild type A(H3N2) viruses in the NI assay. The A(H7N7) virus was selected for its ability to replicate efficiently in ferrets, thereby producing high levels of antibodies, but in principle other influenza virus subtypes can be used.
Fig. 1.
Principles of the ELLA (left) and the NI ELLA (right). After addition of influenza virus to a fetuin-coated well, the NA cleaves sialic acid from the fetuin, thus unmasking a terminal galactose residue. This terminal galactose residue is specifically recognized by PNA-HRPO. Upon addition of the peroxidase substrate, a detectable color change is produced which can be measured at OD490. By using different dilutions of virus in the ELLA, the optimal virus dilution for the NI ELLA can be determined. For the NI ELLA, serially diluted serum is added to a fixed amount of virus (OD490 >0.75). If the NA activity is neutralized by NA-specific antibodies, sialic acid will not be cleaved from the terminal galactose residue and no color change is detected. 115-21.
3.2. Assay suitability with different influenza subtypes
A panel of avian and human influenza viruses was tested in the ELLA and NI ELLA (Table 1) to evaluate the use of wild type viruses as antigens. First, the ELLA was conducted to determine the NA activity for each antigen at different concentrations. Next, the NI ELLAs were conducted using the antigen concentration that gave 50% of the maximal NA activity. However, when these viruses were tested against naïve ferret sera there was already an inhibitory effect of the sera observed for some of these viruses. This inhibitory effect was highest for human A(H3N2) and avian A(H4N2) viruses (Table 1).
Table 1.
Non-specific inhibition of avian and human influenza viruses by naïve ferret sera as measured in the NI ELLA.
| Type | Host | Subtype/lineagea | Strain | NI titer |
|---|---|---|---|---|
| A | Avian | H4N2 | A/Mallard/Netherlands/8/2007 | 1810 |
| H3N8 | A/Mallard/Netherlands/37/2010 | 28 | ||
| H3N2 | A/Mallard/Netherlands/51/2008 | 160 | ||
| H1N1 | A/Eurasian Wigeon/Netherlands/6/2007 | 14 | ||
| Human | sH1N1 | A/Netherlands/364/2006 | 20 | |
| A/Netherlands/246/2008 | 20 | |||
| pH1N1 | A/Netherlands/602/2009 | 160 | ||
| A/California/04/2009 | 113 | |||
| sH2N2b | NL/57 | 10 | ||
| NL/68 | 160 | |||
| sH3N2 | BI/68 | 640 | ||
| BI/76 | 905 | |||
| NL/82 | 640 | |||
| NL/92 | 2560 | |||
| NL/09 | 80 | |||
| B | Human | Victoria | A/Malaysia/2506/2004 | <10 |
| Yamagata | A/Florida/04/2006 | <10 |
All viruses were tested against two naïve ferret sera. These sera were not treated with RDE or heat inactivation. The geometric mean titer was calculated from these titers.
Seasonal (s) and pandemic (p).
Recombinant strains containing the internal genes of A/Puerto Rico/8/1934 (H1N1).
3.3. Treatment of serum to eliminate non-specific inhibition of NA activity
Several options were investigated to eliminate the non-specific inhibitory effect of sera used in the NI ELLA: purification of (anti)serum, sucrose-gradient purification of viruses, commercial V. cholerae neuraminidase (VCNA, Sigma–Aldrich) or RDE treatment of sera, and harvesting virus stocks at varying times after inoculation. VCNA and RDE treatment eliminated most of the non-specific inhibition, whereas the other approaches were unsuccessful (data not shown). Sera used in the HI assay are treated with RDE, followed by heat inactivation at 56 °C to inactivate non-specific inhibitors (Cox et al., 2002). Similar inhibitors may explain the inhibitory effect of naïve ferret sera in the NI ELLA.
Similar to NA, RDE is a neuraminidase and should therefore be inactivated before use in the NI assay. RDE-treated sera were heat inactivated at 56 °C for 1, 2, 4 and 8h and tested in the ELLA for residual neuraminidase activity of the RDE (data not shown). At least 4h of heat inactivation at 56 °C was required to eliminate neuraminidase activity of RDE.
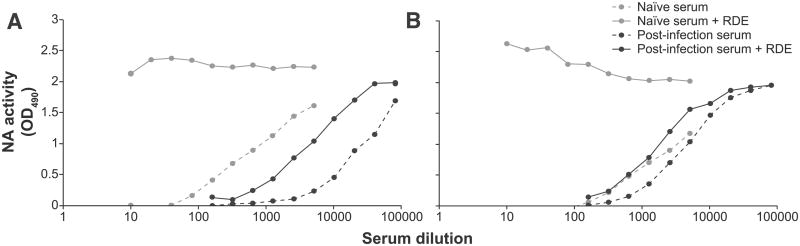
Next, untreated and RDE-treated sera were compared in the NI ELLA (Fig. 2). Although the inhibitory effect of naïve ferret serum was eliminated upon treatment with RDE for 16 h and heat inactivation at 56 °C for 8 h, the specific inhibitory effect of antisera targeted to the N2 antigen was still present. To test the effect of the duration of the heat treatment on antibodies, a panel of anti-A(H7N2) sera were treated with RDE and subjected to various durations of heat inactivation at 56 °C. These sera were tested in NI ELLA and HI assay (Table 2). Each serum was tested against the homologous recombinant A(H7N2) virus in the HI assay. The same sera were tested against their N2-homologous recombinant A(H2N2) or wild type A(H3N2) viruses in the NI ELLA. The duration of heat inactivation after the RDE treatment did not affect the HI and NI titers.
Fig. 2.
Neutralizing activity of naïve and post-infection sera after RDE treatment. Viruses BI/76 (A) and NL/92 (B) were tested against serially diluted naïve ferret sera (gray lines) and their serially diluted N2-homologous A(H7N2) ferret sera (black lines). Sera were incubated with RDE (solid lines) and without RDE (dashed lines) at 37 °C for 16 h and RDE activity was heat inactivated by incubation at 56 °C for 8 h.
Table 2.
HI and NI titers after different durations of RDE heat inactivation.
| A(H7N2) antisera | HI titersa | NI titersb | ||||||
|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||
| 1 h | 2 h | 4 h | 8 h | 1 h | 2 h | 4 h | 8 h | |
| NL/57 | 1280 | 761 | 1810 | 1280 | 5120 | 5120 | 5120 | 5120 |
| NL/68 | 640 | 538 | 640 | 640 | 2560 | 2560 | 2560 | 2560 |
| BI/68 | 640 | 453 | 640 | 640 | 20,480 | 20,480 | 20,480 | 20,480 |
| BI/76 | 269 | 320 | 320 | 320 | 5120 | 3840 | 5120 | 5120 |
| NL/82 | 640 | 538 | 640 | 640 | 160 | 320 | 226 | 226 |
| NL/92 | 538 | 453 | 453 | 538 | 1280 | 1280 | 1280 | 1280 |
| NL/09 | 160 | 160 | 160 | 160 | 2560 | 2560 | 2560 | 2560 |
A(H7N2) antisera were incubated with RDE for 16 h at 37 °C and heat inactivated at 56 °C for 1, 2, 4, or 8 h. The geometric mean titers were calculated from duplicate titers.
Recombinant A(H7N2) viruses were tested against their homologous anti-A(H7N2) ferret sera in the HI assay.
Recombinant A(H2N2) and wild type A(H3N2) viruses were tested in the NI ELLA against their N2-homologous A(H7N2) ferret sera.
Thus, RDE treatment followed by heat inactivation at 56 °C for 8 h was sufficient to inactivate RDE activity without affecting the HI and NI titers of ferret antisera. Therefore, ferret sera treated with RDE and heat-inactivated at 56 °C for 8 h was subsequently used for the NI ELLAs.
3.4. Assay sensitivity and reproducibility
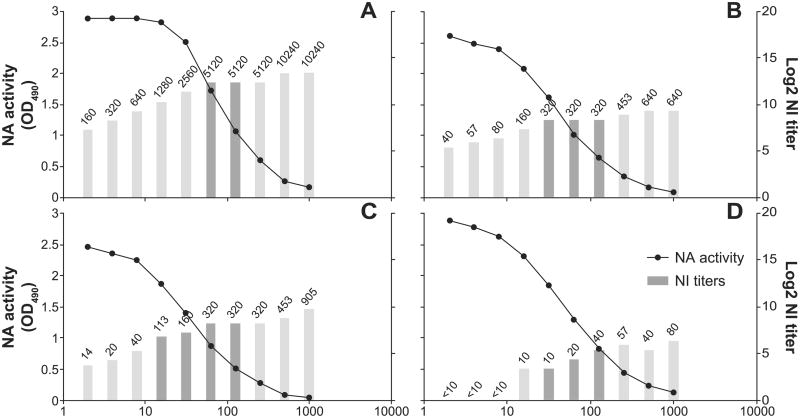
The optimal amount of antigen to use in the NI ELLA was also determined. First A(H3N2) wild type viruses were titrated and measured in the ELLA. Most of these titration curves were sigmoidal, with a maximum signal of 3.0 and background of <0.2 at OD490. Two viruses, NL92 and NL/95, were tested in the NI ELLA at concentrations that spanned the antigen titration curve. Each twofold dilution of the antigen (virus) was tested with monospecific ferret antisera directed against the homologous N2 (Fig. 3A and B) and NL/95 was also tested against two heterologous N2 (Fig. 3C and D). The NI titer was dependent on the amount of N2 antigen, with low NI titers upon use of high amounts of N2 antigen, and high NI titers upon use of low amounts of N2 antigen. However, a minimal difference in NI titer was observed when the N2 antigen concentrations were within the linear trajectory of the titration curve, which represents approximately 21–87% of total NA activity. This corresponded to an NA activity (OD490) between approximately 0.6–2.5. Therefore, to standardize the amount of N2 antigen in the assay, a dilution of virus that resulted in 50% (minimum of 25% and maximum of 75%) of total NA activity was selected for the NI ELLA. The virus dilutions that were needed to reach 50% of total NA activity ranged from 2- to 2000-fold. It should be noted that HA titers do not necessarily correlate with NA titer, and can therefore not be used as an indicator for antigen dilution.
Fig. 3.
Determination of the optimal amount of antigen for routine antigenic characterization. First, the NA activity (black circles) of serially diluted NL/95 (A, C and D) and NL/92 (B) virus was determined in the ELLA. Subsequently each of the dilutions of NL/95 and NL/92 antigens were tested against their N2-homologous (A and B, respectively) A(H7N2) ferret sera in the NI ELLA. In addition, NL/95 was tested against heterologous anti-NL/03 (C) and anti-NL/92 (D) A(H7N2) ferret sera. Absolute NI titers are shown as numbers above the log2 NI titers (bars). The dark bars represent the dilutions that resulted in 25–75% of the maximum NA activity. The assays were performed on different plates and on different days, resulting in some plate-to-plate or day-to-day variation.
For viruses that gave low total NA activity, the amount of N2 antigen within the linear trajectory but with a minimum NA activity of >0.75 (OD490) was selected. If the total NA activity was lower than 0.75 (OD490), viruses were concentrated and retested to ensure a robust measurement of NA activity inhibition.
The assay variability was assessed for NI ELLAs conducted on different plates and different days (data not shown). The results obtained on different days showed high correlation (R2 = 0.88) and the largest difference observed for individual samples was fourfold.
The titration of the viruses in the ELLA as well as the NI ELLA take 2 days to perform. Once titrated in the ELLA, the aliquoted viruses can be used in multiple NI ELLAs without the need for re-titration. A maximum of 20 plates (10 viruses measured in duplicate against eight antisera) or 60 plates (30 viruses measured in duplicate against eight antisera) can be performed by one or by two people, respectively.
3.5. Specificity of the NI ELLA
To test the specificity of the NI ELLA (Lambre et al., 1991) for antigenic characterization of wild type human A(H3N2) viruses, representative N2 antigens of the entire period of circulation of A(H2N2) and A(H3N2) viruses were selected. With these N2 antigens, recombinant A(H7N2) viruses with the internal genes of A/Netherlands/219/03 (H7N7) and the HA the A/Netherlands/33/03 (H7N7) were generated. The MBCS of the HA was removed to decrease pathogenicity. Monospecific ferret antisera elicited against these recombinant A(H7N2) viruses were obtained and tested against homologous and heterologous N2 antigens (Table 3). Homologous titers were highest for each antigen and ranged from 320 to >10,240. Heterologous titers ranged from <20 to >10,240, and showed time dependence. For example, ferret anti-NL/57 did not recognize NL/09 virus (NI titer = <20), but did recognize BI/68 virus (NI titer = 5120); ferret anti-NL/09 did not recognize NL/57 virus (NI titer = <20), but did recognize NL/03 virus (NI titer = 2560). Overall, viruses with similar isolation dates were recognized by the same antisera. Viruses with later isolation dates were not recognized by ferret sera specific for early viruses and vice versa.
Table 3.
NI titers of A(H7N2) antisera against A(H3N2) viruses.
| N2 antigens | Subtype | A(H7N2) antisera | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||
| NL/57 | NL/68 | BI/68 | BI/72 | BI/76 | NL/82 | NL/92 | NL/95 | NL/03 | NL/09 | ||
| NL/57 | H2N2 | ≥10,240 | 2560 | >10,240 | 40 | 80 | <20 | 20 | <20 | <20 | <20 |
| NL/68 | H2N2 | 10,240 | ≥10,240 | >10,240 | 40 | 80 | <20 | 160 | 320 | <20 | <20 |
| BI/68 | H3N2 | 5120 | >10,240 | ≥10,240 | 160 | 160 | 20 | 80 | 320 | 40 | <20 |
| BI/72 | H3N2 | 80 | 1280 | 20 | 5120 | 5120 | 320 | 320 | 160 | 20 | <20 |
| BI/76 | H3N2 | 40 | 640 | 20 | 2560 | 5120 | 160 | 160 | 80 | <20 | <20 |
| NL/82 | H3N2 | <20 | <20 | <20 | 40 | 160 | 320 | 160 | <20 | <20 | <20 |
| NL/92 | H3N2 | <20 | <20 | <20 | 40 | 80 | 320 | 1280 | 320 | <20 | <20 |
| NL/95 | H3N2 | <20 | 80 | 20 | <20 | 40 | <20 | 40 | 2560 | 160 | <20 |
| NL/03 | H3N2 | <20 | 40 | 20 | 20 | <20 | <20 | 20 | 160 | 640 | <20 |
| NL/09 | H3N2 | <20 | 40 | 40 | 20 | 20 | <20 | 640 | 640 | 2560 | 5120 |
The top row represents the various A(H7N2) antisera and the left column shows the N2 antigens, both ordered chronologically. N2-homologous NI titers are underscored and depicted in bold.
4. Discussion
Monitoring functional antibody responses against NA, in tandem with HA-specific antibody analysis, could enhance the antigenic characterization of influenza viruses during routine surveillance for the purpose of influenza vaccine strain selection. The NI ELLA measures functional inhibition of NA activity by antibodies, and consequently, it has clear relevance to immunity in vivo (Couch et al., 1974; Ogra et al., 1977; Beutner et al., 1979). In this study, the existing NI ELLA was optimized for rapid antigenic characterization of NA of wild type A(H3N2) viruses.
Similar to the TBA assay, the ELLA relies on the sialidase activity of NA. However, it has been demonstrated that the ELLA has a higher sensitivity compared to the TBA method (Fritz et al., 2012). The ELLA method, first described by Lambre et al. (1991), has been successfully applied in recent studies. Cate et al. (2010) showed that NI antibody seroconversion rates are greater when vaccine dose is increased. Couch et al. (2012) investigated the NA antibody response to current influenza vaccines and proved the importance of antibodies directed to the NA (Couch et al., 2013). Couzens et al. (2014) optimized the ELLA to measure influenza A virus NI titers in human sera.
NI assay results can be distorted by HA-specific antibodies interfering with substrate cleavage by NA (Schulman and Kilbourne, 1969). To avoid this possible interference, various forms of NA have been used as antigens in NI assays: purified recombinant NA (Fritz et al., 2012), recombinant A(H6N1) and A(H6N2) viruses (Sandbulte et al., 2009; Couzens et al., 2014), detergent split wild-type viruses (Cate et al., 2010), and virus-like particles (Couch et al., 2013). However, generation of these antigens is time consuming and consequently unsuitable for large scale screening of viruses for vaccine strain selection. Therefore, the usage of wild type viruses was explored in the NI ELLA in combination with sera that were raised against HA-mismatched viruses.
Upon titration of naïve ferret sera against various influenza (sub)types, a non-specific inhibitory effect of the sera was occasionally observed. Non-specific inhibitors that interfere with the HI assay have been described previously, first by Hirst (Hirst, 1942) and later by Francis (Francis, 1947). Three classes of these nonspecific inhibitors present in human and animal sera have been described: α-, β-, and γ-class (Anders et al., 1990; Ryan-Poirier and Kawaoka, 1991; Matrosovich et al., 1998). Inhibitors of the α- and γ-classes express sialic acid residues that are specifically bound by the influenza virus HA (Pritchett and Paulson, 1989; Anders et al., 1990) thereby blocking the receptor-binding sites of HA (Matrosovich et al., 1998). Inhibitors of the β-class (Anders et al., 1990; Hartley et al., 1992) are, in contrast to inhibitors of the α- and γ-classes (Krizanova and Rathova, 1969; Subbarao et al., 1992), thermolabile (Matrosovich et al., 1998). Sera used in the HI assay are treated beforehand with RDE to inactivate sialylated α and γ inhibitors (Pritchett and Paulson, 1989; Ryan-Poirier and Kawaoka, 1993), followed by incubation at 56 °C to inactivate the β inhibitors and the RDE (Konno, 1958; Cohen and Belyavin, 1959; Anders et al., 1990). The inhibitory effect of the naïve ferret sera seen in the NI ELLA may be explained by similar sialic acid expressing inhibitors that interact with the HA, the NA, or both, and thus preventing desialylation of fetuin. Since heat inactivation at 56 °C by itself did not affect the observed non-specific inhibitory effect, it indicates that the inhibitors were not β-class inhibitors (Konno, 1958) but rather α- or, most likely, γ-inhibitors. RDE treatment of serum, followed by heat inactivation, eliminated most of the non-inhibitory effect without affecting specific virus-specific NI and HI titers.
Although the NI titers showed dependence on the amount of N2 antigen used, the NI titers did not differ when N2 antigen concentrations within the linear trajectory of the titration curve were chosen. These conditions resulted in limited variability between NI ELLAs performed on different days.
To be able to use wild type viruses as antigens, recombinant viruses with a mismatched HA must be used to generate antisera. The A(H7N7) virus was selected as a backbone to generate recombinant A(H7N2) viruses, as A(H7N7) replicates well in ferrets and antibodies directed to H7 do not recognize H3. However in principle, any influenza A subtype (i.e. H9) can be selected as a backbone, providing that it replicates well in ferrets and antibodies directed to the HA do not recognize H3.
Using recombinant A(H7N2) viruses, ferret antisera yielding high NI titers without boosting or the usage of adjuvants could be generated. A panel of ferret anti-A(H7N2) sera were utilized in the NI ELLA to screen wild type A(H3N2) viruses. The observed intra-subtypic cross-reactivity of the ferret antisera permits screening of many viruses during influenza virus surveillance with a limited number of sera, while the specificity of the ferret antisera allows detection of drift variants. Although generation of the recombinant A(H7N2) viruses and anti-A(H7N2) ferret sera has to be done in a BSL-3 laboratory, sera can be used under lower biosafety conditions upon demonstration of the absence of virus and viral RNA It is likely that this strategy will also be applicable for N1 antigens to screen circulating human A(H1N1) viruses. This approach would be more difficult for influenza B viruses given that there are no different subtypes. Perhaps an early influenza B virus (i.e. B/Lee/40) could be a suitable backbone if antibodies directed to its HA do not recognize the HA of currently circulating influenza B viruses. Alternatively one could use chimeric influenza A/B viruses, with the HA and internal genes of influenza A and a NA segment with the non-coding regions of influenza A and the ORF of influenza B (Flandorfer et al., 2003; Baker et al., 2014). But these viruses are possibly too attenuated to replicate efficiently in ferrets.
In conclusion, the NI ELLA described in this study provides a practical method to evaluate the antigenic properties of NAs from different viruses, and can be used for large-scale screening of wild type influenza A viruses. The antigenic drift of HA and NA was shown to be discordant (Sandbulte et al., 2011), and NA-specific antibodies – in addition to HA-specific antibodies – contribute to protection against disease. Therefore, information regarding antigenic drift of NA may facilitate selection of viruses that are antigenically matched to circulating strains for influenza vaccine production.
Acknowledgments
This work was supported by an NWO-VICI grant and NIH contract no. HHSN266200700010C. We gratefully thank Geert van Amerongen, Sander Herfst, Eefje Schrauwen, Oanh Vuong, Martin Linster and Ruud van Beek for excellent technical assistance.
References
- Air GM, Els MC, Brown LE, Laver WG, Webster RG. Location of antigenic sites on the three-dimensional structure of the influenza N2 virus neuraminidase. Virology. 1985;145:237–248. doi: 10.1016/0042-6822(85)90157-6. [DOI] [PubMed] [Google Scholar]
- Anders EM, Hartley CA, Jackson DC. Bovine and mouse serum beta inhibitors of influenza A viruses are mannose-binding lectins. Proc Natl Acad Sci U S A. 1990;87:4485–4489. doi: 10.1073/pnas.87.12.4485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker SF, Nogales A, Finch C, Tuffy KM, Domm W, Perez DR, Topham DJ, Martinez-Sobrido L. Influenza A and B virus intertypic reassortment through compatible viral packaging signals. J Virol. 2014;88:10778–10791. doi: 10.1128/JVI.01440-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr IG, McCauley J, Cox N, Daniels R, Engelhardt OG, Fukuda K, Grohmann G, Hay A, Kelso A, Klimov A, Odagiri T, Smith D, Russell C, Tashiro M, Webby R, Wood J, Ye Z, Zhang W Writing Committee of the World Health Organization Consultation on Northern Hemisphere Influenza Vaccine Composition. Epidemiological, antigenic and genetic characteristics of seasonal influenza A(H1N1), A(H3N2) and B influenza viruses: basis for the WHO recommendation on the composition of influenza vaccines for use in the 2009-2010 Northern Hemisphere season. Vaccine. 2010;28:1156–1167. doi: 10.1016/j.vaccine.2009.11.043. [DOI] [PubMed] [Google Scholar]
- Beutner KR, Chow T, Rubi E, Strussenberg J, Clement J, Ogra PL. Evaluation of a neuraminidase-specific influenza A virus vaccine in children: antibody responses and effects on two successive outbreaks of natural infection. J Infect Dis. 1979;140:844–850. doi: 10.1093/infdis/140.6.844. [DOI] [PubMed] [Google Scholar]
- Brett IC, Johansson BE. Immunization against influenza A virus: comparison of conventional inactivated, live-attenuated and recombinant baculovirus produced purified hemagglutinin and neuraminidase vaccines in a murine model system. Virology. 2005;339:273–280. doi: 10.1016/j.virol.2005.06.006. [DOI] [PubMed] [Google Scholar]
- Burnet FM, Stone JD. The receptor-destroying enzyme of V. cholerae. Aust J Exp Biol Med Sci. 1947;25:227–233. doi: 10.1038/icb.1947.33. [DOI] [PubMed] [Google Scholar]
- Cate TR, Rayford Y, Nino D, Winokur P, Brady R, Belshe R, Chen W, Atmar RL, Couch RB. A high dosage influenza vaccine induced significantly more neuraminidase antibody than standard vaccine among elderly subjects. Vaccine. 2010;28:2076–2079. doi: 10.1016/j.vaccine.2009.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chutinimitkul S, van Riel D, Munster VJ, van den Brand JM, Rimmelzwaan GF, Kuiken T, Osterhaus AD, Fouchier RA, de Wit E. In vitro assessment of attachment pattern and replication efficiency of H5N1 influenza A viruses with altered receptor specificity. J Virol. 2010;84:6825–6833. doi: 10.1128/JVI.02737-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen A, Belyavin G. Hemagglutination inhibition of Asian influenza viruses: a new pattern of response. Virology. 1959;7:59–74. doi: 10.1016/0042-6822(59)90177-1. [DOI] [PubMed] [Google Scholar]
- Colman PM, Varghese JN, Laver WG. Structure of the catalytic and antigenic sites in influenza virus neuraminidase. Nature. 1983;303:41–44. doi: 10.1038/303041a0. [DOI] [PubMed] [Google Scholar]
- Colman PM, Laver WG, Varghese JN, Baker AT, Tulloch PA, Air GM, Webster RG. Three-dimensional structure of a complex of antibody with influenza virus neuraminidase. Nature. 1987;326:358–363. doi: 10.1038/326358a0. [DOI] [PubMed] [Google Scholar]
- Compans RW, Dimmock NJ, Meier-Ewert H. Effect of antibody to neuraminidase on the maturation and hemagglutinating activity of an influenza A2 virus. J Virol. 1969;4:528–534. doi: 10.1128/jvi.4.4.528-534.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couch RB, Kasel JA, Gerin JL, Schulman JL, Kilbourne ED. Induction of partial immunity to influenza by a neuraminidase-specific vaccine. J Infect Dis. 1974;129:411–420. doi: 10.1093/infdis/129.4.411. [DOI] [PubMed] [Google Scholar]
- Couch RB, Atmar RL, Keitel WA, Quarles JM, Wells J, Arden N, Nino D. Randomized comparative study of the serum antihemagglutinin and antineuraminidase antibody responses to six licensed trivalent influenza vaccines. Vaccine. 2012;31:190–195. doi: 10.1016/j.vaccine.2012.10.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couch RB, Atmar RL, Franco LM, Quarles JM, Wells J, Arden N, Nino D, Belmont JW. Antibody correlates and predictors of immunity to naturally occurring influenza in humans and the importance of antibody to the neuraminidase. J Infect Dis. 2013;207:974–981. doi: 10.1093/infdis/jis935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couzens L, Gao J, Westgeest K, Sandbulte M, Lugovtsev V, Fouchier R, Eichelberger M. An optimized enzyme-linked lectin assay to measure influenza A virus neuraminidase inhibition antibody titers in human sera. J Virol Methods. 2014;210C:7–14. doi: 10.1016/j.jviromet.2014.09.003. [DOI] [PubMed] [Google Scholar]
- Cox N, Webster R, Stohr K. WHO Manual on Animal Influenza Diagnosis and Surveillance. World Health Organization Department Communicable Disease Surveillance and Response 2002 [Google Scholar]
- de Wit E, Spronken MI, Bestebroer TM, Rimmelzwaan GF, Osterhaus AD, Fouchier RA. Efficient generation and growth of influenza virus A/PR/8/34 from eight cDNA fragments. Virus Res. 2004;103:155–161. doi: 10.1016/j.virusres.2004.02.028. [DOI] [PubMed] [Google Scholar]
- de Wit E, Munster VJ, van Riel D, Beyer WE, Rimmelzwaan GF, Kuiken T, Osterhaus AD, Fouchier RA. Molecular determinants of adaptation of highly pathogenic avian influenza H7N7 viruses to efficient replication in the human host. J Virol. 2010;84:1597–1606. doi: 10.1128/JVI.01783-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiore AE, Uyeki TM, Broder K, Finelli L, Euler GL, Singleton JA, Iskander JK, Wortley PM, Shay DK, Bresee JS, Cox NJ Centers for Disease Control and Prevention. Prevention and control of influenza with vaccines: recommendations of the Advisory Committee on Immunization Practices (ACIP) MMWR. Recommendations and reports: Morbidity and mortality weekly report. Recommendations and reports/Centers for Disease Control. 2010;59:1–62. [PubMed] [Google Scholar]
- Flandorfer A, Garcia-Sastre A, Basler CF, Palese P. Chimeric influenza A viruses with a functional influenza B virus neuraminidase or hemagglutinin. J Virol. 2003;77:9116–9123. doi: 10.1128/JVI.77.17.9116-9123.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouchier RA, Bestebroer TM, Herfst S, Van Der Kemp L, Rimmelzwaan GF, Osterhaus AD. Detection of influenza A viruses from different species by PCR amplification of conserved sequences in the matrix gene. J Clin Microbiol. 2000;38:4096–4101. doi: 10.1128/jcm.38.11.4096-4101.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis T. Dissociation of hemagglutinating and antibody-measuring capacities of influenza virus. J Exp Med. 1947;85:1–7. doi: 10.1084/jem.85.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz R, Sabarth N, Kiermayr S, Hohenadl C, Howard MK, Ilk R, Kistner O, Ehrlich HJ, Barrett PN, Kreil TR. A vero cell-derived whole-virus H5N1 vaccine effectively induces neuraminidase-inhibiting antibodies. J Infect Dis. 2012;205:28–34. doi: 10.1093/infdis/jir711. [DOI] [PubMed] [Google Scholar]
- Hartley CA, Jackson DC, Anders EM. Two distinct serum mannose-binding lectins function as beta inhibitors of influenza virus: identification of bovine serum beta inhibitor as conglutinin. J Virol. 1992;66:4358–4363. doi: 10.1128/jvi.66.7.4358-4363.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirst GK. The quantitative determination of influenza virus and antibodies by means of red cell agglutination. J Exp Med. 1942;75:49–64. doi: 10.1084/jem.75.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirst GK. Studies of antigenic differences among strains of influenza a by means of red cell agglutination. J Exp Med. 1943;78:407–423. doi: 10.1084/jem.78.5.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann E, Neumann G, Kawaoka Y, Hobom G, Webster RG. A DNA transfection system for generation of influenza A virus from eight plasmids. Proc Natl Acad Sci U S A. 2000;97:6108–6113. doi: 10.1073/pnas.100133697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson BE, Grajower B, Kilbourne ED. Infection-permissive immunization with influenza virus neuraminidase prevents weight loss in infected mice. Vaccine. 1993;11:1037–1039. doi: 10.1016/0264-410x(93)90130-p. [DOI] [PubMed] [Google Scholar]
- Kilbourne ED. Comparative efficacy of neuraminidase-specific and conventional influenza virus vaccines in induction of antibody to neuraminidase in humans. J Infect Dis. 1976;134:384–394. doi: 10.1093/infdis/134.4.384. [DOI] [PubMed] [Google Scholar]
- Kilbourne ED, Laver WG, Schulman JL, Webster RG. Antiviral activity of antiserum specific for an influenza virus neuraminidase. J Virol. 1968;2:281–288. doi: 10.1128/jvi.2.4.281-288.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilbourne ED, Johansson BE, Grajower B. Independent and disparate evolution in nature of influenza A virus hemagglutinin and neuraminidase glycoproteins. Proc Natl Acad Sci U S A. 1990;87:786–790. doi: 10.1073/pnas.87.2.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koel BF, Burke DF, Bestebroer TM, van der Vliet S, Zondag GC, Vervaet G, Skepner E, Lewis NS, Spronken MI, Russell CA, Eropkin MY, Hurt AC, Barr IG, de Jong JC, Rimmelzwaan GF, Osterhaus AD, Fouchier RA, Smith DJ. Substitutions near the receptor binding site determine major antigenic change during influenza virus evolution. Science. 2013;342:976–979. doi: 10.1126/science.1244730. [DOI] [PubMed] [Google Scholar]
- Konno J. Studies on several inhibitors against influenza viruses 2. beta-Inhibitor, its biological and physicochemical properties with particular emphasis on the differences from alpha-inhibitor, immune serum and properdin. Tohoku J Exp Med. 1958;67:391–405. doi: 10.1620/tjem.67.391. [DOI] [PubMed] [Google Scholar]
- Krizanova O, Rathova V. Serum inhibitors of myxoviruses. Curr Top Microbiol Immunol. 1969;47:125–151. doi: 10.1007/978-3-642-46160-6_6. [DOI] [PubMed] [Google Scholar]
- Lambre CR, Terzidis H, Greffard A, Webster RG. An enzyme-linked lectin assay for sialidase. Clin Chim Acta: Int J Clin Chem. 1991;198:183–193. doi: 10.1016/0009-8981(91)90352-d. [DOI] [PubMed] [Google Scholar]
- Laver WG, Air GM, Webster RG, Markoff LJ. Amino acid sequence changes in antigenic variants of type A influenza virus N2 neuraminidase. Virology. 1982;122:450–460. doi: 10.1016/0042-6822(82)90244-6. [DOI] [PubMed] [Google Scholar]
- Liu C, Eichelberger MC, Compans RW, Air GM. Influenza type A virus neuraminidase does not play a role in viral entry, replication, assembly, or budding. J Virol. 1995;69:1099–1106. doi: 10.1128/jvi.69.2.1099-1106.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luther P, Bergmann KC, Oxford JS. An investigation of antigenic drift of neuraminidases of influenza A (H1N1) viruses. J Hyg. 1984;92:223–229. doi: 10.1017/s002217240006424x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matrosovich M, Gao P, Kawaoka Y. Molecular mechanisms of serum resistance of human influenza H3N2 virus and their involvement in virus adaptation in a new host. J Virol. 1998;72:6373–6380. doi: 10.1128/jvi.72.8.6373-6380.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy BR, Kasel JA, Chanock RM. Association of serum anti-neuraminidase antibody with resistance to influenza in man. N Engl J Med. 1972;286:1329–1332. doi: 10.1056/NEJM197206222862502. [DOI] [PubMed] [Google Scholar]
- Ogra PL, Chow T, Beutner KR, Rubi E, Strussenberg J, DeMello S, Rizzone C. Clinical and immunologic evaluation of neuraminidase-specific influenza A virus vaccine in humans. J Infect Dis. 1977;135:499–506. doi: 10.1093/infdis/135.4.499. [DOI] [PubMed] [Google Scholar]
- Palese P, Compans RW. Inhibition of influenza virus replication in tissue culture by 2-deoxy-2,3-dehydro-N-trifluoroacetylneuraminic acid (FANA): mechanism of action. J Gen Virol. 1976;33:159–163. doi: 10.1099/0022-1317-33-1-159. [DOI] [PubMed] [Google Scholar]
- Palese P, Tobita K, Ueda M, Compans RW. Characterization of temperature sensitive influenza virus mutants defective in neuraminidase. Virology. 1974;61:397–410. doi: 10.1016/0042-6822(74)90276-1. [DOI] [PubMed] [Google Scholar]
- Pritchett TJ, Paulson JC. Basis for the potent inhibition of influenza virus infection by equine and guinea pig alpha 2-macroglobulin. J Biol Chem. 1989;264:9850–9858. [PubMed] [Google Scholar]
- Russell CA, Jones TC, Barr IG, Cox NJ, Garten RJ, Gregory V, Gust ID, Hampson AW, Hay AJ, Hurt AC, de Jong JC, Kelso A, Klimov AI, Kageyama T, Komadina N, Lapedes AS, Lin YP, Mosterin A, Obuchi M, Odagiri T, Osterhaus AD, Rimmelzwaan GF, Shaw MW, Skepner E, Stohr K, Tashiro M, Fouchier RA, Smith DJ. Influenza vaccine strain selection and recent studies on the global migration of seasonal influenza viruses. Vaccine. 2008;26(Suppl. 4):D31–D34. doi: 10.1016/j.vaccine.2008.07.078. [DOI] [PubMed] [Google Scholar]
- Ryan-Poirier KA, Kawaoka Y. Distinct glycoprotein inhibitors of influenza A virus in different animal sera. J Virol. 1991;65:389–395. doi: 10.1128/jvi.65.1.389-395.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan-Poirier KA, Kawaoka Y. Alpha 2-macroglobulin is the major neutralizing inhibitor of influenza A virus in pig serum. Virology. 1993;193:974–976. doi: 10.1006/viro.1993.1208. [DOI] [PubMed] [Google Scholar]
- Sandbulte MR, Gao J, Straight TM, Eichelberger MC. A miniaturized assay for influenza neuraminidase-inhibiting antibodies utilizing reverse genetics-derived antigens. Influenza Other Respir Viruses. 2009;3:233–240. doi: 10.1111/j.1750-2659.2009.00094.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandbulte MR, Westgeest KB, Gao J, Xu X, Klimov AI, Russell CA, Burke DF, Smith DJ, Fouchier RA, Eichelberger MC. Discordant antigenic drift of neuraminidase and hemagglutinin in H1N1 and H3N2 influenza viruses. Proc Natl Acad Sci U S A. 2011;108:20748–20753. doi: 10.1073/pnas.1113801108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauter NK, Bednarski MD, Wurzburg BA, Hanson JE, Whitesides GM, Skehel JJ, Wiley DC. Hemagglutinins from two influenza virus variants bind to sialic acid derivatives with millimolar dissociation constants: a 500-MHz proton nuclear magnetic resonance study. Biochemistry. 1989;28:8388–8396. doi: 10.1021/bi00447a018. [DOI] [PubMed] [Google Scholar]
- Schild GC, Oxford JS, Dowdle WR, Coleman M, Pereira MS, Chakraverty P. Antigenic variation in current influenza A viruses: evidence for a high frequency of antigenic ‘drift’ for the Hong Kong virus. Bull World Health Organ. 1974;51:1–11. [PMC free article] [PubMed] [Google Scholar]
- Schrauwen EJ, Bestebroer TM, Munster VJ, de Wit E, Herfst S, Rimmelzwaan GF, Osterhaus AD, Fouchier RA. Insertion of a multibasic cleavage site in the haemagglutinin of human influenza H3N2 virus does not increase pathogenicity in ferrets. J Gen Virol. 2011;92:1410–1415. doi: 10.1099/vir.0.030379-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulman JL, Kilbourne ED. Independent variation in nature of hemagglutinin and neuraminidase antigens of influenza virus: distinctiveness of hemagglutinin antigen of Hong Kong-68 virus. Proc Natl Acad Sci U S A. 1969;63:326–333. doi: 10.1073/pnas.63.2.326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulman JL, Khakpour M, Kilbourne ED. Protective effects of specific immunity to viral neuraminidase on influenza virus infection of mice. J Virol. 1968;2:778–786. doi: 10.1128/jvi.2.8.778-786.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subbarao EK, Kawaoka Y, Ryan-Poirier K, Clements ML, Murphy BR. Comparison of different approaches to measuring influenza A virus-specific hemagglutination inhibition antibodies in the presence of serum inhibitors. J Clin Microbiol. 1992;30:996–999. doi: 10.1128/jcm.30.4.996-999.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Itzstein M. The war against influenza: discovery and development of sialidase inhibitors. Nat Rev Drug Discov. 2007;6:967–974. doi: 10.1038/nrd2400. [DOI] [PubMed] [Google Scholar]
- Warren L. The thiobarbituric acid assay of sialic acids. J Biol Chem. 1959;234:1971–1975. [PubMed] [Google Scholar]
- Webster RG, Laver WG. Preparation and properties of antibody directed specifically against the neuraminidase of influenza virus. J Immunol. 1967;99:49–55. [PubMed] [Google Scholar]
- Webster RG, Laver WG, Air GM, Schild GC. Molecular mechanisms of variation in influenza viruses. Nature. 1982:296. doi: 10.1038/296115a0. [DOI] [PubMed] [Google Scholar]