Abstract
A salient feature of the failing heart is metabolic remodeling towards predominant glucose metabolism and activation of the fetal gene program. Sunitinib is a multitargeted receptor tyrosine kinase inhibitor used for the treatment of highly vascularized tumors. In diabetic patients, sunitinib significantly decreases blood glucose. However, a considerable proportion of sunitinib-treated patients develop cardiac dysfunction or failure. We asked whether sunitinib treatment results in shift towards glycolysis in the heart. Glucose uptake by the heart was increased fivefold in mice treated with sunitinib. Transcript analysis by qPCR revealed an induction of genes associated with glycolysis and reactivation of the fetal gene program. Additionally, we observed a shift in the enzyme pyruvate kinase from the adult M1 (PKM1) isoform to the fetal M2 (PKM2) isoform, a hallmark of the Warburg Effect. This novel observation led us to examine whether a similar shift occurs in human heart failure. Examination of tissue from patients with heart failure similarly displayed an induction of PKM2. Moreover, this phenomenon was partially reversed following mechanical unloading. We propose that pyruvate kinase isoform switching represents a novel feature of the fetal gene program in the failing heart.
Keywords: heart failure, sunitinib, fetal gene program, glycolysis, PKM2, Hif1α
1. Introduction
A hallmark of the fetal heart is the use of glucose as the primary source for energy provision [1]. The greater efficiency of carbohydrate substrates per mole of oxygen allows the fetal heart to withstand its low oxygen environment, rapidly changing hemodynamic load and demand for growth [2]. Shortly after birth the heart rapidly switches to the fatty acid oxidation as the primary source of fuel. However, a salient feature of the failing heart is metabolic remodeling back towards predominant glucose metabolism. This is achieved, in part, through activation of the fetal gene program. We have previously reported that the failing human heart reverts to the fetal gene program and shows similar features of fetal cardiac metabolism [3]. We now demonstrate an additional metabolic shift related to heart failure and reflect on its potential origin and significance.
The antiangiogenic receptor tyrosine kinase inhibitor sunitinib significantly lowers blood glucose levels in both diabetic and non-diabetic cancer patients[4–6]. At the same time the incidence of left-ventricular contractile dysfunction associated with sunitinib is estimated to be between 4 and 28%[7–11]. Although the primary targets of sunitinib are well characterized, the molecular mechanism of sunitinib-induced cardiotoxicity remains unclear.
We asked whether a reversion to glycolysis occurs in the hearts from sunitinib treated mice. We observed that sunitinib treatment resulted in activation of the fetal gene program. In addition we noted the induction of the M2 isoform of pyruvate kinase (PKM2), a hallmark of the Warburg Effect. More importantly, we demonstrate induction of PKM2 in the failing human heart which is partially reversed following mechanical unloading. Potential mechanisms and consequences of this shift are discussed.
2. Materials and Methods
An expanded methods section is available in the data supplement.
2.1. Materials
Sunitinib malate was obtained from LC Laboratories (Woburn, MA). Dimethyloxalylglycine (DMOG) was obtained from Tocris Bioscience (Bristol, United Kingdom). Cobalt chloride and all other laboratory reagents were obtained from Sigma Aldrich (St. Louis, MO). Antibodies: PKM1 (SAB 42000094, Sigma Aldrich), PKM2 (#4053, Cell Signaling Technology), GAPDH (10R-G109A, Fitzgerald Industries), O-GlcNAc-CTD110.6 (9875, Cell Signaling), Hif1α (NB100-105, Novus).
2.2. Experimental Animals
All animal manipulations were approved by the Institutional Animal Care and Use Committee of the University of Texas Health Science Center at Houston. Male C57BL/6J mice (Jackson Laboratories, Bar Harbor, ME) were treated with sunitinib by oral gavage. Sunitinib (10 mg/ml) was dissolved in an oral gavage solution [polyethylene glycol 300 (10%), Tween 80 (0.5%), water, 0.1 N hydrochloric acid, pH adjusted to 3.7] as we have previously described [12].
2.3. Human Subjects
Failing Heart Samples
Failing heart muscle samples were obtained from patients referred to the Texas Heart Institute with advanced heart failure and placed on left ventricular assist device (LVAD) support. Tissue from the left ventricular apex was obtained during LVAD implantation and again at device removal. Human subjects gave informed consent and the study protocol was approved by the Committee for the Protection of Human Subjects of CHI St. Luke’s Health in Houston, Texas, and by The University of Texas Medical School at Houston.
Non-failing Heart Samples
Non-failing heart samples were obtained from the apex of the left ventricle of patients undergoing heart-lung transplantation due to pulmonary hypertension and right-ventricular failure. Left-ventricular function was normal as assessed by echocardiography. A full characterization of these patients has been published [13]. The study was approved by the Houston Methodist Hospital's institutional review board.
2.4. Hyperinsulinemic Euglycemic Clamp
The hyperinsulinemic euglycemic clamp procedure was performed by the Mouse Metabolism Core at Baylor College of Medicine and the procedure was approved by the Institutional Animal Care and Use Committee of Baylor College of Medicine.
2.5. Cell Culture
Adult mouse cardiomyocytes were isolated from the hearts of 8–12 week old C57BL/6J mice using a modified version of the protocol developed by the Simpson laboratory [14]. Experiments began 2 hours after isolation and performed in triplicate on at least three separate isolations.
2.6. RNA Extraction and qPCR
RNA was extracted using Trizol or in the case of human tissues using RNeasy miniprep kit for fibrous tissues (Qiagen, Valencia, CA). cDNA was reverse transcribed in a separate reaction. Transcript levels were evaluated using the comparative Ct method, using GAPDH as an endogenous control, and expressed as a fold change over control.
2.7. Pyruvate kinase alternative splicing
Analysis of PKM alternative splicing was performed as previously described [15] with minor changes. Alternative splicing of exon 9 and 10 of the PKM gene results in two identical length isoforms, PKM1 and PKM2. Mouse and human primer sets were designed to target exons 8 through 11. cDNA products were digested with PstI, NcoI (New England Biolabs, Ipswich, MA), both or left undigested and then ran on a 5% native polyacrylamide gel and poststained with ethidium bromide.
2.8. Statistical Analysis
Results are expressed as means ± SEM. Analysis was performed using two-tailed, unpaired Student’s t test or one-way ANOVA with Tukey post hoc test. A value of P<0.05 was considered significant.
3. Results
3.1. Sunitinib increases insulin sensitivity and peripheral glucose uptake in the mouse
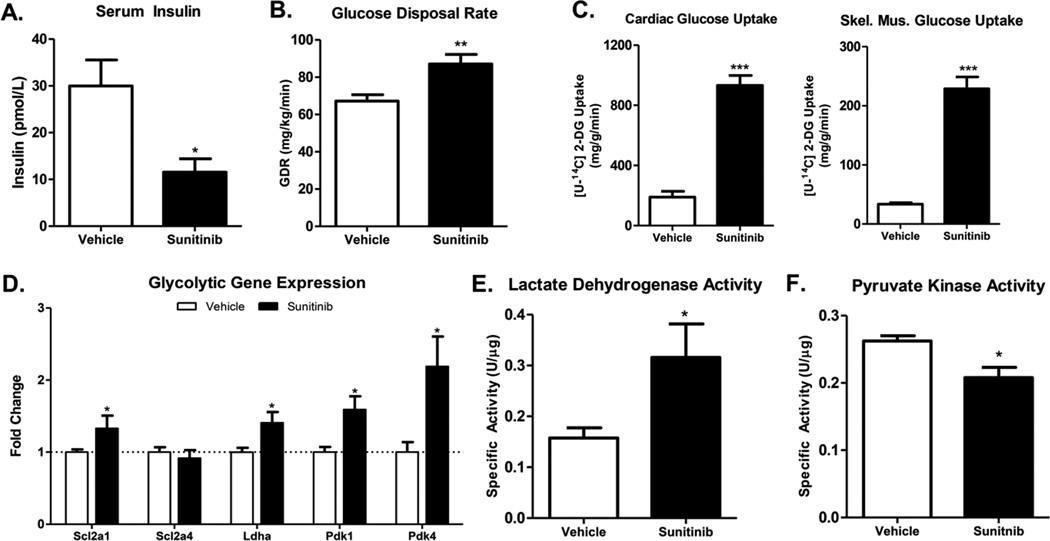
Given the number of reports demonstrating a blood glucose-lowering effect of sunitinib, we first sought to determine the primary mean by which sunitinib affects blood glucose (i.e. increases peripheral insulin sensitivity, stimulates pancreatic insulin production or decreases hepatic gluconeogenesis). Serum insulin levels were significantly lower in sunitinib treated mice, which suggested that sunitinib increases peripheral insulin sensitivity (Figure 1A). Based on this evidence, and on significantly improved insulin sensitivity and glucose clearance in oral glucose tolerance and insulin tolerance tests (data not shown), we performed hyperinsulinemic euglycemic clamp studies on mice treated with sunitinib or vehicle. Sunitinib treatment resulted in a significantly increased glucose infusion rate which equated to an increased glucose disposal rate (Figure 1B). Evaluation of tissue-specific in [U-14C]-2-deoxyglucose uptake revealed a fivefold and sevenfold rise in glucose uptake in the heart and skeletal muscle, respectively (Figure 1C).
Figure 1. Sunitinib increases insulin sensitivity in the mouse.
(A) Serum insulin levels in mice treated with sunitinib or vehicle for 21 days. (B) Glucose disposal rate and (C) Tissue-specific radiolabeled 2-deoxyglucose uptake in mice treated with sunitinib for 3 weeks before undergoing a hyperinsulinemic euglycemic clamp procedure (n=12 per group) (D) Expression level of key genes involved in glycolysis. (E) and (F) Lactate dehydrogenase and pyruvate kinase enzyme activity assays performed on cardiac lysates. Data are ± SEM with 8–12 mice per group. * P < 0.05, ** P < 0.01 and *** P < 0.001.
3.2. Sunitinib treatment increases glucose metabolism
Transcript analysis by qPCR revealed a significant increase in transcript levels of genes associated with increased glycolysis in the hearts of mice treated with sunitinib (Figure 1D). In particular, upregulation of pyruvate dehydrogenase kinase 1 and 4 (pdk1, pdk4), which inhibit the enzyme pyruvate dehydrogenase, indicated a prevention of pyruvate oxidation by the mitochondria. An increase in glycolysis was further demonstrated by increased lactate dehydrogenase activity (Figure 1E) and decreased pyruvate kinase activity (Figure 1F).
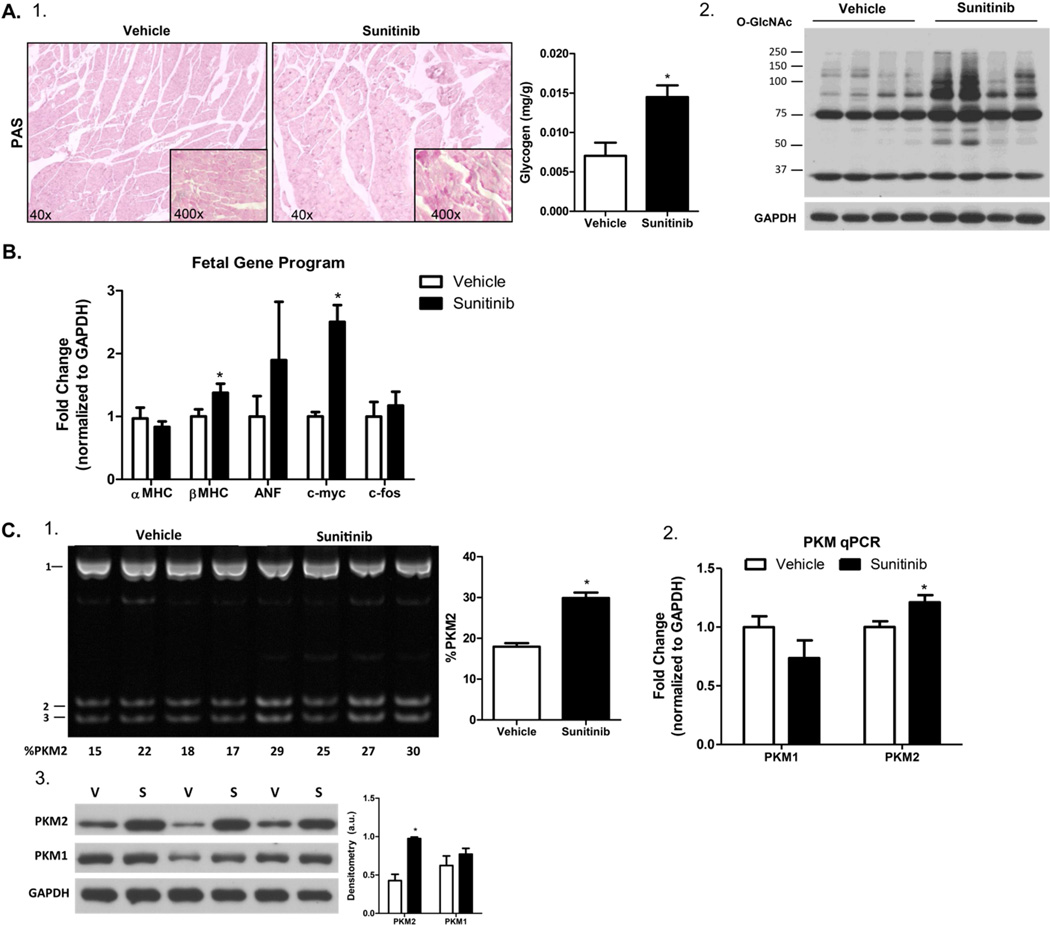
Glycogen occupies about 2% of the cell volume in the adult cardiomyocyte, however nearly a third of the cell volume in the fetal heart consists of glycogen [16]. Correspondingly, we observed a significant accumulation of glycogen (Figure 2A-1) in the hearts of sunitinib-treated mice. Consistent with the overflow of glucose into secondary pathways of metabolism, we observed an increase in proteins modified by O-linked β-N-acetylglucosamine (O-GlcNAc) in sunitinib-treated hearts (Figure 2A-2). In the failing heart, upregulation of glycolysis and glycogen accumulation is achieved through induction of the fetal gene program. This includes a switch in the contractile protein isoforms from myosin heavy chain α (MHCα) to MHCβ and increased expression of atrial naturetic factor (ANF), c-myc and c-fos [17]. These genes were also increased in the sunitinib-treated hearts, suggesting that sunitinib treatment leads to re-activation of the fetal gene program in the heart (Figure 2B).
Figure 2. PKM2 is a novel member of the fetal gene program.
(A) 1) Tissue glycogen content as assessed by periodic acid-Schiff’s base staining (pictured) and by enzyme coupled spectrophotometric quantification (right graph). 2) Western blot of O-GlcNAcylated proteins in whole tissue lysates from vehicle and sunitinib treated hearts (B) Transcript analysis by qPCR demonstrating activation of the fetal gene program. (C) Analysis of PKM2 gene expression by 1) restriction enzyme (PstI) digest of the pyruvate kinase cDNA amplicon. Data is expressed as the percentage (%) of the PKM2 isoform (bands 2 and 3) Please see the data supplement for a more detailed explanation. 2) qPCR analysis of relative mRNA expression of each pyruvate kinase isoform. 3) Protein expression of each pyruvate kinase isoforms as assessed by western blot. *P < 0.05 and **P < 0.01.
3.3. The M2 isoform of pyruvate kinase is re-expressed in sunitinib-treated hearts
Upregulation of glycolysis and decreased pyruvate kinase activity was highly reminiscent of the Warburg Effect. First described by Otto Warburg in 1924, certain cancers dramatically increase glucose uptake in order to utilize glucose-derived carbons for the synthesis of nucleotides, lipids and amino acids and sustain rapid growth [18]. A hallmark of the Warburg Effect is the induction of the fetal (M2) isoform of pyruvate kinase (PKM2) [19, 20]. The fetal heart predominately expresses PKM2 but is rapidly replaced by the adult M1 isoform (PKM1) by postnatal day 14.5 via mutually exclusive alternative splicing of exon 9 and 10 (Supplemental Figure 1) [21]. We asked whether a similar induction of PKM2 occurs in sunitinib-treated hearts. Sunitinib treatment led to increased PKM2 expression (Figure 2C-1). This was further conferred by both qPCR and western blotting (Figure 2C-2 and Figure 2C-3).
3.4. Activation of Hif1α leads to PKM2 induction
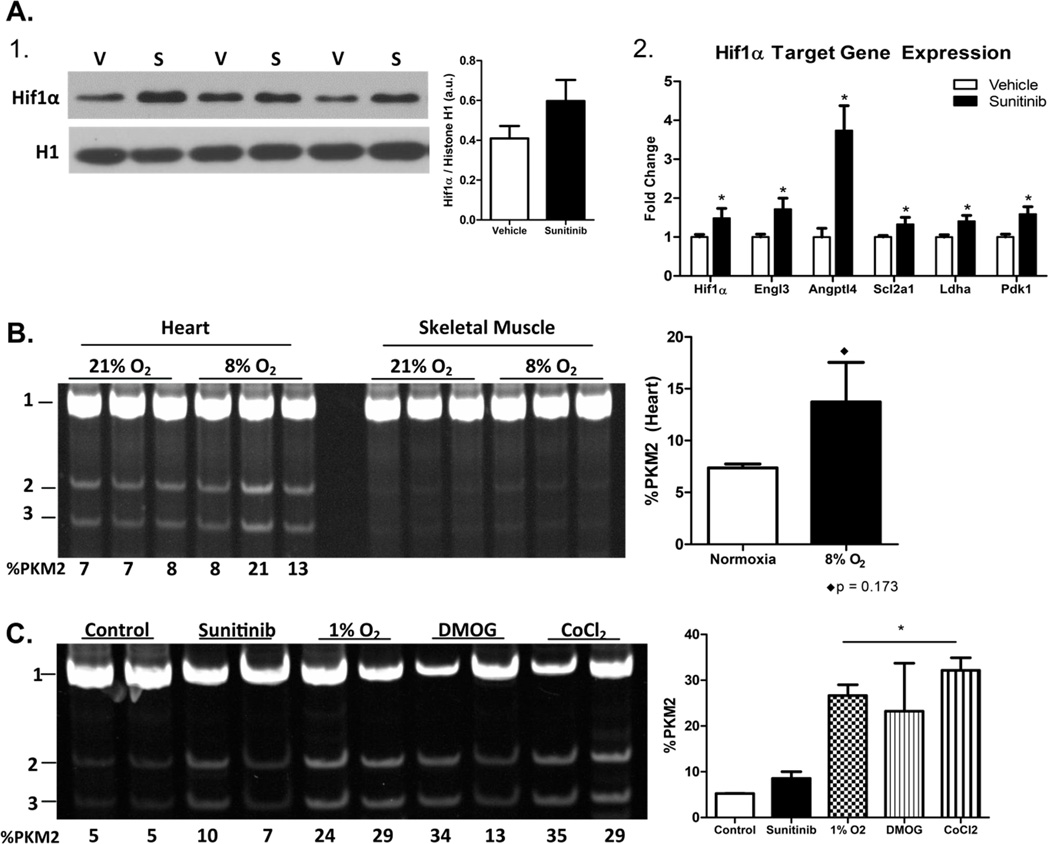
Under normoxic conditions the master hypoxia response factor, hypoxia-inducible factor 1α (Hif1α), is continually targeted for ubiquitin-mediated degradation in the cytosol. However, when oxygen levels drop, Hif1α translocates to the nucleus where it induces a host of hypoxia-response genes. In cancer, Hif1α exists in a positive feed-forward loop with hydroxylated PKM2 to promote the transcription of both and a glycolytic switch [22]. Correspondingly, nuclear content of Hif1α was increased in the sunitinib-treated hearts (Figure 3A-1). Furthermore, upregulation of Hif1α and of known Hif1α target genes was confirmed by qPCR in the hearts of mice treated with sunitinib (Figure 3A-2).
Figure 3. Activation of Hif1α promotes PKM2 induction in the heart and isolated adult mouse cardiomyocytes.
(A) 1) Western blot demonstrating increased nuclear localization of Hif1α in sunitinib treated hearts. 2) Relative expression of Hif1α and of genes known to be induced by Hif1α in the heart. (B) PstI restriction digest of PKM cDNA from wild type mice (n=3 per group) subjected to either normoxia (~21% O2) or 8% O2 for 24 hours. (C) Percent PKM2 expression in isolated adult cardiomyocytes treated with sunitinib, subjected to 1% O2 or treated with pharmacological mimetics of hypoxia, dimethyloxalylglycine (DMOG) or cobalt chloride for 24 hours. Data are ±SEM of 3–8 mice/wells per group. *P < 0.05.
We next determined if activation of Hif1α was sufficient to induce expression of PKM2 in the heart. We observed an increase in PKM2 in the heart but not in skeletal muscle (Figure 3B) of mice exposed to hypoxia (8% oxygen) for 24 hours. We then asked whether hypoxia could induce PKM2 expression in vitro. Not unexpectedly, neonatal rat ventricular myocytes (NRVMs) and the cardiac cell line H9C2 express PKM2 at high levels (Figure S1B). For this reason, we chose to use primary adult mouse cardiomyocytes. Twenty-four hour exposure of isolated adult cardiomyocytes to 1% oxygen or to the pharmacological hypoxia mimetics, dimethyloxalylglycine (DMOG) and cobalt chloride, resulted in increased PKM2 expression compared to sunitinib treatment or untreated controls (Figure 3C).
3.5. PKM2 is re-expressed in the failing human heart
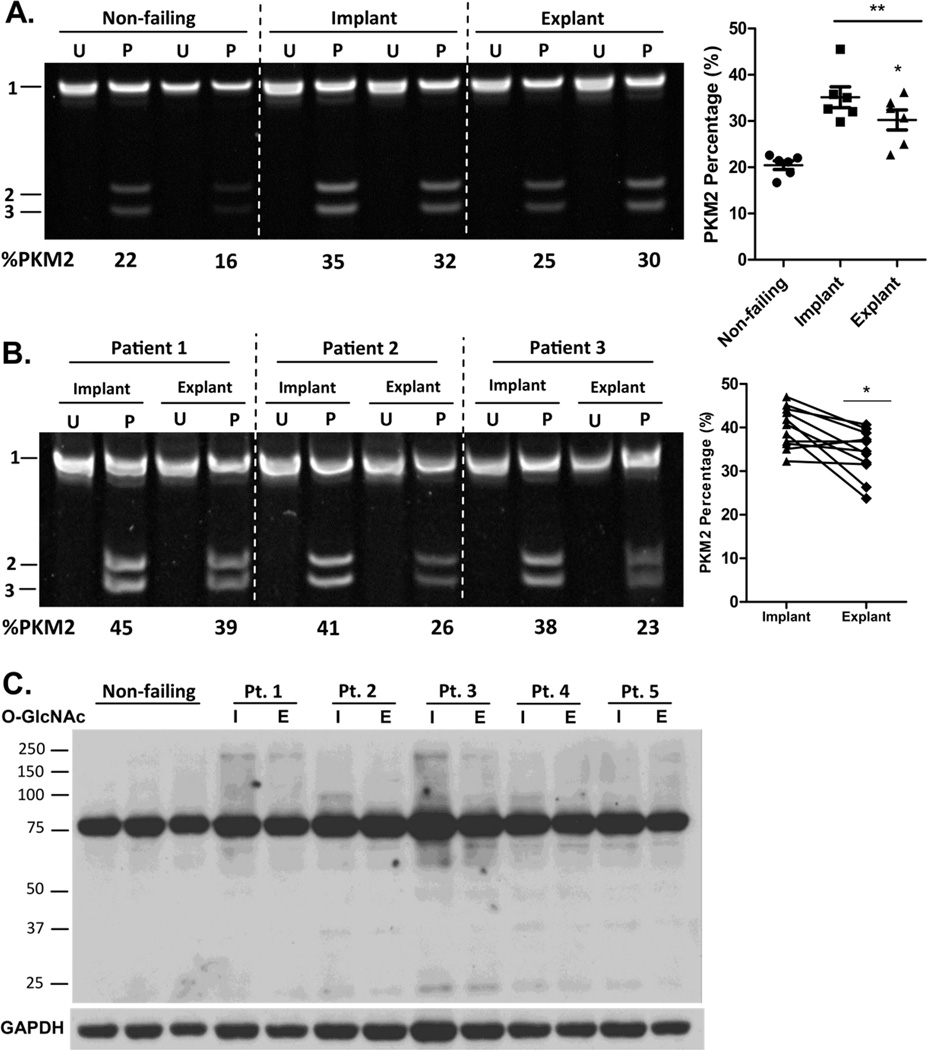
To determine the clinical relevance of our findings we evaluated PKM2 expression in heart muscle samples from non-failing and failing human hearts before and after mechanical unloading. Compared to non-failing hearts, there was a significant increase in PKM2 expression in both implant and explant failing heart samples (Figure 4A). Mechanical unloading promotes positive cardiac remodeling and functional recovery, allowing some patients to be explanted to their native heart [23]. It appeared that PKM2 was slightly decreased following mechanical unloading. A partial reversal of PKM2 induction was confirmed by comparing paired implant and explant samples from a total of 13 patients, suggesting that mechanical load may be a determinant of PKM2 expression in the heart (Figure 4B).
Figure 4. PKM2 expression is increased in the failing human heart.
(A) Uncut (U) or PstI digested (P) PKM cDNA amplicon of heart muscle samples from non-failing and failing hearts before (Implant) and after (Explant) mechanical unloading. (B) PKM2 expression in paired human samples before (implant) and after (explant) mechanical unloading. (C) Western blot of O-GlcNAcylated proteins in non-failing and failing hearts. Data are ±SEM of 6–11 human samples. *P < 0.05 and **P < 0.01.
We have previously demonstrated that glycogen accumulation in the failing human heart is partially reversed following mechanical unloading [24]. Similar to the overflow of glucose into the hexosamine biosynthetic pathway in the sunitinib treated mice, we observed an increase in O-GlcNAcylated proteins in the failing heart that was reversed following mechanical unloading, a trend that correlates with the changes in PKM2 expression in the heart.
4. Discussion
Our data suggest that sunitinib treatment leads to increased glycolysis and activation of the fetal gene program in the heart. Furthermore, activation of Hif1α leads to induction of the fetal M2 isoform of pyruvate kinase both in vivo and in vitro. Most importantly, we have revealed that induction of PKM2 is a signature of not only sunitinib cardiotoxicity but also of the stressed heart in general. To our knowledge this is the first evidence of PKM2 induction in any model of heart failure.
We propose that PKM2 is a novel member of the fetal gene program and its induction is part of the adaptive response in the stressed heart. Unlike PKM1, which exists as a near-constitutively active tetramer, PKM2 can exist as a dimer or tetramer and is uniquely subject to allosteric regulation and various post-translational modifications that determine its function as pyruvate kinase, protein kinase or transcription factor [25, 26]. Importantly, the ability of PKM2 to rapidly cycle between a tetramer and dimer could be especially advantageous to the failing heart. In its tetramer form, PKM2 acts as a canonical pyruvate kinase, allowing pyruvate oxidation and ATP generation. The low catalytic activity of the dimer form slows glycolysis and allows for the redirection of glucose-derived carbons into biosynthetic pathways, promoting cell proliferation or in the case of the heart, hypertrophy. It will be important to determine if, similar to the Warburg effect in cancer, glucose-derived carbons support cardiac hypertrophy.
It has been proposed that cellular oncogenes drive the progression to heart failure. This is based on the observation that many of the pathways driving tumor growth also drive cardiomyocyte hypertrophy [27]. Induction of PKM2 in the hypertrophying heart would fall in line with this reasoning. Included in this original hypothesis was activation of the fetal gene, c-myc. In cancer, c-myc drives the expression of three heterogeneous nuclear ribonucleoprotein (hnRNP) proteins-polypyrimidine tract binding protein (PTB, also known as hnRNPI), hnRNPA1 and hnRNPA2-that are essential for preferential splicing of PKM2 [28]. As c-myc transcription was significantly induced in the sunitinib treated hearts (Figure 2B), it is possible that it might also be important for preferential PKM2 splicing in the heart. Furthering this hypothesis, increased hemodynamic load in known to induce c-myc in the heart [29]. As PKM2 expression seemed to correlate with hemodynamic load in the failing human heart (Figure 4B), it seems likely that c-myc plays an undetermined role in promoting PKM2 induction. Hif1α and dysregulated c-myc has been shown to cooperatively promote transcription of key glycolytic genes in cancer [30]. However, Hif1α and c-myc are thought to play opposing roles in the adult heart. Whether these two factors cooperate in the failing heart or if this is an anomaly of the sunitinib-treated heart remains to be determined.
An exciting area for future investigation is the nonmetabolic functions of PKM2 in the heart. Dimeric PKM2 can translocate into the nucleus and act as a transcription factor. In cancer cells, EGFR stimulation promotes PKM2 nuclear translocation and regulated transactivation of beta-catenin [31]. PKM2 can also act as a protein kinase, using the high energy PEP to phosphorylate substrates such as stat3 [32]. As the complexity of PKM2 regulation and function continue to unfold it is increasingly clear that PKM2 is important in cell physiology beyond cancer.
Supplementary Material
Highlights.
Pyruvate kinase isoform switching represents a novel feature of the fetal gene program in the heart.
Treatment with sunitinib leads to reactivation of the fetal gene program and induction of PKM2.
PKM2 induction is also observed in failing human hearts compared to non-failing controls.
Acknowledgments
We thank Dr. Mark Entman for his help with the manuscript preparation. We also thank Drs. Pradip Saha and Lawrence Chan of the Mouse Metabolism Core at Baylor College of Medicine for aiding with the hyperinsulinemic euglycemic clamp studies; Dr. Hernan Vasquez and Sylvia Carranza for collection of the failing human heart samples; Dr. Keith Youker for the collection of the non-failing human heart samples and Drs. Anren Song and Yang Xia for help with the Hif1α experiments. These studies were supported in part by a grant from the National Heart, Lung, and Blood Institute (5R01HL061483-10). M.L.R. received a predoctoral fellowship from the National Center for Advancing Translational Sciences of the National Institutes of Health under Award Numbers TL1TR000369 and UL1TR000371.
Abbreviations
- Hif1α
hypoxia-inducible factor 1α
- LVAD
left-ventricular assist device
- PKM1
pyruvate kinase M1
- PKM2
pyruvate kinase M2
- DMOG
dimethyloxalylglycine
- O-GlcNAc
O-linked β-N-acetylglucosamine
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures
There are no financial conflicts of interest to report on behalf of the authors.
References
- 1.Stanley WC, Recchia FA, Lopaschuk GD. Myocardial Substrate Metabolism in the Normal and Failing Heart. 2005 doi: 10.1152/physrev.00006.2004. [DOI] [PubMed] [Google Scholar]
- 2.Taegtmeyer H, Sen S, Vela D. Return to the fetal gene program: a suggested metabolic link to gene expression in the heart. Ann N Y Acad Sci. 2010;1188:191–198. doi: 10.1111/j.1749-6632.2009.05100.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Razeghi P, Young ME, Alcorn JL, Moravec CS, Frazier OH, Taegtmeyer H. Metabolic gene expression in fetal and failing human heart. Circulation. 2001;104:2923–2931. doi: 10.1161/hc4901.100526. [DOI] [PubMed] [Google Scholar]
- 4.Agostino NM, Chinchilli VM, Lynch CJ, Koszyk-Szewczyk A, Gingrich R, Sivik J, Drabick JJ. Effect of the tyrosine kinase inhibitors (sunitinib, sorafenib, dasatinib, and imatinib) on blood glucose levels in diabetic and nondiabetic patients in general clinical practice. J Oncol Pharm Pract. 2011;17:197–202. doi: 10.1177/1078155210378913. [DOI] [PubMed] [Google Scholar]
- 5.Billemont B, Medioni J, Taillade L, Helley D, Meric JB, Rixe O, Oudard S. Blood glucose levels in patients with metastatic renal cell carcinoma treated with sunitinib. Br J Cancer. 2008;99:1380–1382. doi: 10.1038/sj.bjc.6604709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Templeton A, Brandle M, Cerny T, Gillessen S. Remission of diabetes while on sunitinib treatment for renal cell carcinoma. Ann Oncol. 2008;19:824–825. doi: 10.1093/annonc/mdn047. [DOI] [PubMed] [Google Scholar]
- 7.Chu TF, Rupnick MA, Kerkela R, Dallabrida SM, Zurakowski D, Nguyen L, Woulfe K, Pravda E, Cassiola F, Desai J, George S, Morgan JA, Harris DM, Ismail NS, Chen JH, Schoen FJ, Van den Abbeele AD, Demetri GD, Force T, Chen MH. Cardiotoxicity associated with tyrosine kinase inhibitor sunitinib. Lancet. 2007;370:2011–2019. doi: 10.1016/S0140-6736(07)61865-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hasinoff BB, Patel D, O'Hara KA. Mechanisms of myocyte cytotoxicity induced by the multiple receptor tyrosine kinase inhibitor sunitinib. Mol Pharmacol. 2008;74:1722–1728. doi: 10.1124/mol.108.050104. [DOI] [PubMed] [Google Scholar]
- 9.Khakoo AY, Yeh ET. Therapy insight: Management of cardiovascular disease in patients with cancer and cardiac complications of cancer therapy. Nat Clin Pract Oncol. 2008;5:655–667. doi: 10.1038/ncponc1225. [DOI] [PubMed] [Google Scholar]
- 10.Rees ML, Khakoo AY. Molecular mechanisms of hypertension and heart failure due to antiangiogenic cancer therapies. Heart Fail Clin. 2011;7:299–311. doi: 10.1016/j.hfc.2011.03.004. [DOI] [PubMed] [Google Scholar]
- 11.Rini BI, Flaherty K. Clinical effect and future considerations for molecularly-targeted therapy in renal cell carcinoma. Urol Oncol. 2008;26:543–549. doi: 10.1016/j.urolonc.2008.03.012. [DOI] [PubMed] [Google Scholar]
- 12.Chintalgattu V, Rees ML, Culver JC, Goel A, Jiffar T, Zhang J, Dunner K, Jr, Pati S, Bankson JA, Pasqualini R, Arap W, Bryan NS, Taegtmeyer H, Langley RR, Yao H, Kupferman ME, Entman ML, Dickinson ME, Khakoo AY. Coronary microvascular pericytes are the cellular target of sunitinib malate-induced cardiotoxicity. Sci Transl Med. 2013;5:187ra169. doi: 10.1126/scitranslmed.3005066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cordero-Reyes AM, Gupte AA, Youker KA, Loebe M, Hsueh WA, Torre-Amione G, Taegtmeyer H, Hamilton DJ. Freshly isolated mitochondria from failing human hearts exhibit preserved respiratory function. J Mol Cell Cardiol. 2014;68:98–105. doi: 10.1016/j.yjmcc.2013.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O'Connell TD, Rodrigo MC, Simpson PC. Isolation and culture of adult mouse cardiac myocytes. Methods Mol Biol. 2007;357:271–296. doi: 10.1385/1-59745-214-9:271. [DOI] [PubMed] [Google Scholar]
- 15.Clower CV, Chatterjee D, Wang Z, Cantley LC, Vander Heiden MG, Krainer AR. The alternative splicing repressors hnRNP A1/A2 and PTB influence pyruvate kinase isoform expression and cell metabolism. Proc Natl Acad Sci U S A. 2010;107:1894–1899. doi: 10.1073/pnas.0914845107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Navaratnam V. Heart Muscle: Ultrastructural Studies. New York: Cambridge University Press; 1987. [Google Scholar]
- 17.Rajabi M, Kassiotis C, Razeghi P, Taegtmeyer H. Return to the fetal gene program protects the stressed heart: a strong hypothesis. Heart Fail Rev. 2007;12:331–343. doi: 10.1007/s10741-007-9034-1. [DOI] [PubMed] [Google Scholar]
- 18.Warburg O. On respiratory impairment in cancer cells. Science. 1956;124:269–270. [PubMed] [Google Scholar]
- 19.Christofk HR, Vander Heiden MG, Harris MH, Ramanathan A, Gerszten RE, Wei R, Fleming MD, Schreiber SL, Cantley LC. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 2008;452:230–233. doi: 10.1038/nature06734. [DOI] [PubMed] [Google Scholar]
- 20.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao Z, Cooper TA. Reexpression of pyruvate kinase M2 in type 1 myofibers correlates with altered glucose metabolism in myotonic dystrophy. Proc Natl Acad Sci U S A. 2013;110:13570–13575. doi: 10.1073/pnas.1308806110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luo W, Hu H, Chang R, Zhong J, Knabel M, O'Meally R, Cole RN, Pandey A, Semenza GL. Pyruvate kinase M2 is a PHD3-stimulated coactivator for hypoxia-inducible factor 1. Cell. 2011;145:732–744. doi: 10.1016/j.cell.2011.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Segura AM, Dris L, Massin EK, Clubb FJ, Buja LM, Frazier OH, Taegtmeyer H. Heart failure in remission for more than 13 years after removal of a left ventricular assist device. Tex Heart Inst J. 2014;41:389–394. doi: 10.14503/THIJ-13-4029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Razeghi P, Bruckner BA, Sharma S, Youker KA, Frazier OH, Taegtmeyer H. Mechanical unloading of the failing human heart fails to activate the protein kinase B/Akt/glycogen synthase kinase-3beta survival pathway. Cardiology. 2003;100:17–22. doi: 10.1159/000072387. [DOI] [PubMed] [Google Scholar]
- 25.Wong N, De Melo J, Tang D. PKM2, a Central Point of Regulation in Cancer Metabolism. Int J Cell Biol. 2013;2013:242513. doi: 10.1155/2013/242513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anastasiou D, Yu Y, Israelsen WJ, Jiang JK, Boxer MB, Hong BS, Tempel W, Dimov S, Shen M, Jha A, Yang H, Mattaini KR, Metallo CM, Fiske BP, Courtney KD, Malstrom S, Khan TM, Kung C, Skoumbourdis AP, Veith H, Southall N, Walsh MJ, Brimacombe KR, Leister W, Lunt SY, Johnson ZR, Yen KE, Kunii K, Davidson SM, Christofk HR, Austin CP, Inglese J, Harris MH, Asara JM, Stephanopoulos G, Salituro FG, Jin S, Dang L, Auld DS, Park HW, Cantley LC, Thomas CJ, Vander Heiden MG. Pyruvate kinase M2 activators promote tetramer formation and suppress tumorigenesis. Nat Chem Biol. 2012;8:839–847. doi: 10.1038/nchembio.1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mulvagh SL, Roberts R, Schneider MD. Cellular oncogenes in cardiovascular disease. Journal of Molecular and Cellular Cardiology. 1988;20:657–662. doi: 10.1016/s0022-2828(88)80123-8. [DOI] [PubMed] [Google Scholar]
- 28.David CJ, Chen M, Assanah M, Canoll P, Manley JL. HnRNP proteins controlled by c-Myc deregulate pyruvate kinase mRNA splicing in cancer. Nature. 2010;463:364–368. doi: 10.1038/nature08697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mulvagh SL, Michael LH, Perryman MB, Roberts R, Schneider MD. A hemodynamic load in vivo induces cardiac expression of the cellular oncogene, c-myc. Biochem Biophys Res Commun. 1987;147:627–636. doi: 10.1016/0006-291x(87)90977-6. [DOI] [PubMed] [Google Scholar]
- 30.Kim JW, Gao P, Liu YC, Semenza GL, Dang CV. Hypoxia-inducible factor 1 and dysregulated c-Myc cooperatively induce vascular endothelial growth factor and metabolic switches hexokinase 2 and pyruvate dehydrogenase kinase 1. Mol Cell Biol. 2007;27:7381–7393. doi: 10.1128/MCB.00440-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang W, Xia Y, Ji H, Zheng Y, Liang J, Huang W, Gao X, Aldape K, Lu Z. Nuclear PKM2 regulates beta-catenin transactivation upon EGFR activation. Nature. 2011;480:118–122. doi: 10.1038/nature10598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gao X, Wang H, Yang JJ, Liu X, Liu ZR. Pyruvate kinase M2 regulates gene transcription by acting as a protein kinase. Mol Cell. 2012;45:598–609. doi: 10.1016/j.molcel.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.