Abstract
Nucleotide excision repair (NER) is an evolutionarily conserved, multistep process that can detect a wide variety of DNA lesions. Transcription coupled repair (TCR) is a sub-pathway of NER that repairs the transcribed DNA strand faster than the rest of the genome. RNA polymerase (RNAP) stalled at DNA lesions mediates the recruitment of NER enzymes to the damage site. In this review we focus on a newly identified bacterial TCR pathway in which the NER enzyme UvrD, in conjunction with NusA, plays a major role in initiating the repair process. We discuss the tradeoff between the new and conventional models of TCR, how and when each pathway operates to repair DNA damage, and the necessity of pervasive transcription in maintaining genome integrity.
Introduction
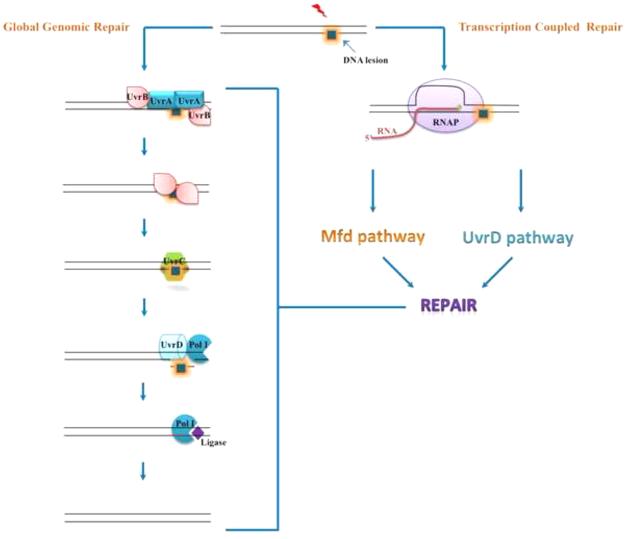
The genetic material of living organisms is under constant threat of damage by environmental agents and metabolic byproducts. NER is the most versatile DNA repair pathway that detects a wide variety of bulky damage, such as thymine dimers caused by UV light and a plethora of helix-distorting adducts formed by harmful chemical agents. NER was originally discovered in bacteria [1] and subsequently shown to be present in all domains of life. The process of NER in bacteria begins with the recognition of DNA damage by the combined action of UvrA and UvrB proteins. DNA-bound UvrA/B then recruits UvrC to the site of damage, followed by UvrA dissociation. UvrC makes incisions on both sides of the lesion and the damaged oligonucleotide is removed by UvrD and/or DNA polymerase I, with the latter of which fills the gap using the complementary strand as a template. The nick between the newly synthesized strand and the contiguous strand is then sealed by DNA ligase, completing the process of lesion removal [2,3] (Figure 1).
Figure 1. Schematics of nucleotide excision repair pathways in E. coli.
Global genomic repair (GGR): UvrA/UvrB complex recognizes DNA damage in either transcribed or non-transcribed strand and binds to the lesion site. UvrA dissociates from the complex and UvrB recruits UvrC endonuclease. UvrC makes incisions on either side of the lesion. DNA polymerase I (with or without UvrD) displaces the damaged oligonucleotide and fills the resulting single-stranded gap, using the complementary strand as a template. Finally, DNA ligase seals the nick.
Transcription coupled repair (TCR): RNAP elongation is impeded by lesions in the transcribed strand. The stalled elongation complex then triggers the repair process via Mfd- or UvrD-dependent pathway (Figure 2).
NER can be viewed as two divergent pathways: global genomic repair (GGR) and transcription coupled DNA repair (TCR) (Figure 1). GGR is DNA-strand unbiased with respect to any particular genomic location, while TCR preferentially and rapidly processes the lesions in the transcribed strand of expressed genes [4,5]. During GGR, a complex of UvrA and UvrB likely locates DNA damage via 3-D search, where the lesions are searched by random encounters. Conversely, TCR is initiated by signals from stalled RNA polymerase (RNAP), which pauses/arrests at the DNA lesions in its path [6–8]. Specialized transcription-repair coupling factors bind to and remove stalled RNAP from the site of damage. They also help to recruit NER enzymes to the exposed DNA lesion. The best-studied bacterial transcription-repair coupling factor is Mfd [9,10], which binds to the upstream edge of the stalled elongation complex [11] and pushes it forward using its ATPase dependent translocase activity [12]. Recently, an alternative, Mfd-independent, TCR pathway has been described [13]. Here we summarize and discuss the current knowledge and most recent findings pertaining to TCR.
Mfd-dependent TCR
In 1946, Evelyn Witkin discovered the rapid decrease of UV-induced suppressor mutations upon transient inhibition of protein synthesis following UV irradiation [14], a phenomenon she named mutation frequency decline (MFD) [15]. She later identified the gene responsible, naming it mfd, and showed that the process required the action of NER enzymes [15,16]. Further research by the Hanawalt group established that transcribed DNA strands were repaired preferentially compared to the entire genome and introduced the concept of TCR [5,17,18]. Sancar and colleagues expanded these pioneering studies by demonstrating that a putative transcription-repair coupling factor is a product of the mfd gene [19,20]. Their elegant biochemical work revealed that the Mfd protein was, indeed, capable of coupling transcription to DNA repair in vitro [9,10], which was later supported by in vivo studies [21].
Mfd is a monomeric, multimodular protein belonging to the DExH/D family of SF2 translocases. It contains eight domains (D1a, D1b, and D2 through D7) connected by flexible linkers [22]. Mfd binds to the arrested elongation complex through its RNAP interacting domain (D4) and makes specific contacts with the IKE motif of the N-terminal fragment of the β subunit located at the upstream edge of the elongation complex [11]. The interaction between Mfd and RNAP initiates a conformational change in Mfd [23], which, in turn, triggers its motor activity [24]. Active Mfd dislodges RNAP from the site of damage by pushing it from behind [25,26]. Mfd also recruits the UvrA protein to the damage site through its UvrB homology module (Figure 2). Interaction of Mfd with UvrA is obligatory for TCR; mutants with a modified UvrB homology module show compromised TCR activity due to defective UvrA interaction [10,23].
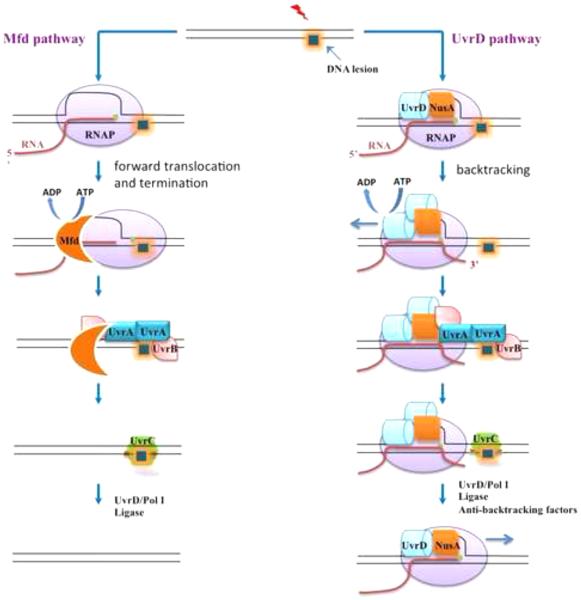
Figure 2. Schematics of alternative models of transcription coupled DNA repair.
RNAP stalls at a DNA lesion in the transcribed strand and obscures it from repair enzymes. Stalled RNAP is either dislodged (left) or forced to backtrack (right) from the site of damage by Mfd and UvrD, respectively, to initiate the process of TCR.
Mfd-dependent pathway: Mfd binds to stalled RNAP and becomes activated. Using its ATP dependent translocase activity it pushes RNAP from behind to eventually terminate the elongation complex. Mfd then recruits UvrAB to the exposed DNA lesion site. Repair then proceeds as in GGR.
UvrD-dependent pathway: Monomeric UvrD and NusA associate with RNAP throughout elongation. The SOS response triggers rapid UvrD accumulation leading to its dimerization in the elongation complex, which starts its ATP-dependent helicase activity. Activated UvrD in collaboration with NusA, pulls RNAP backwards, away from the lesion site. UvrD and NusA then recruit the UvrAB complex to the site of damage. Repair then proceeds as in GGR. In UvrD-dependent TCR transcription can resume shortly after repair has been accomplished, whereas Mfd-dependent TCR results in transcription termination.
Extensive biochemical and structural characterization of Mfd revealed the mechanistic details of its action in TCR. However, the very mild sensitivity of the mfd -deficient cells to UV radiation [15,19] and DNA damaging agents [13] raise issues regarding its precise physiological role. Indeed, TCR is only partially depends on Mfd at actively transcribed genes [27], and functions relatively slowly in vitro in terminating stalled elongation complexes [12,25,26]. It has also been reported that Mfd supports TCR on T7 RNA polymerase-transcribed genes, questioning its specificity [28]. Collectively, these observations suggested that the highly efficient TCR observed in vivo might require additional mechanisms.
Mfd-independent TCR
NusA
NusA associates with RNAP during elongation and plays an important role in transcription termination and antitermination [29]. Recently, Walker and colleagues have shown that NusA functions in DNA repair and damage tolerance pathways. NusA, while not SOS inducible, interacts physically with DNA polymerase IV (DinB) [30] and promotes DNA damage tolerance via the transcription coupled-translesion synthesis pathway [31].
It was also found that a nusA mutant (nusA11) has increased sensitivity to nitrofurazone and 4-nitroquinoline oxide – the agents causing the N2–dG adduct in DNA, independent of the Mfd and DinB pathways, as supported by epistasis analysis [32]. Proteomics and far-western blotting analysis showed that NusA also interacts with the NER enzyme UvrA, suggesting that NusA contributes to NER via an unknown TCR pathway. Furthermore, it was shown that mutations in the second largest subunit of RNAP that alter cellular resistance to nitrofurazone can be suppressed by mutations in uvrA and nusA, but not in the mfd gene [32,33].
The mechanism of NusA-dependent TCR has not been established. It has been hypothesized that RNAP arrested at DNA lesions is prone to backtracking – reverse sliding of RNAP along DNA and RNA [34] – thereby exposing the site of DNA damage [33]. In this model, the backtracked elongation complex recruits NER enzymes via the NusA-UvrA interaction.
UvrD
UvrD is a founding member of superfamily 1 (SF1) of helicases/translocases. It moves in the 3' to 5' direction on ssDNA, unwinding dsDNA in an ATP-dependent manner [35,36]. The functional state of UvrD is determined by its oligomeric composition; as a monomer, it acts as a translocase, while as a dimer it acts as a helicase [37,38]. Recently, we showed that UvrD directly couples transcription to DNA repair and proposed a new model of TCR [13]. UvrD binds RNAP tightly at near stoichiometric levels to RNAP core subunits [13,39], and likely moves with RNAP throughout elongation. Analysis of the in vivo E. coli RNAP interactome suggests that UvrD is associated with the elongation complexes at a level comparable to that of the general transcription elongation factors NusA and NusG [13]. Remarkably, UvrD can actively pull RNAP backward during RNAP pausing or stalling in vitro and in vivo, and this forced backtracking was shown to unmask the DNA lesion site for access to UvrABC in vitro [13]. Furthermore, the very high sensitivity of uvrD(-) cells to UV and various genotoxic agents can be suppressed to great extent by compromising the anti-backtracking activities of the cell [13], i.e. factors that normally inhibit backtracking of RNAP also interfere with the NER process. These factors include transcript cleavage factors GreA and GreB that catalyze reactivation of backtracked RNAP, and active ribosomes that follow elongating RNAP closely and prevent backtracking at protein coding sequences [40]. Interestingly, deletion of mfd, which also functions as an anti-backtracking factor [12], partially suppressed UV sensitivity of uvrD(-) cells [13]. These findings led us to propose a new model of TCR (Figure 2). During the course of transcription, RNAP stalls at DNA damage sites and cannot progress further, creating potential difficulties for both excision repair and replication fork progression. UvrD, which travels with the elongation complex, becomes activated during DNA damage response and pulls RNAP backwards to expose the damage site to repair enzymes. Activation may occur due to rapid accumulation of UvrD, which would favour dimerization, and thus, its helicase mode of action. In addition, UvrD may assist in recruiting repair proteins by using its inherent capability to interact with UvrB [41,42].
Crosslinking and mass spectrometry analyses established that UvrD binds to the upstream edge of the elongation complex and makes specific contacts with the β and β’ subunits of RNAP. It also interacts with the exposed portion of the non-template DNA strand close to the upstream fork of the transcription bubble [13]. Inter-protein crosslinks between RNAP and UvrD defined the UvrD-RNAP interacting surface involving the DNA binding region of UvrD and the area proximal to the RNAP β domain [13]. Notably, NusA also binds the β flap, an interaction necessary for NusA-dependent RNAP pausing [43]. Based on these observations, we proposed a mechanistic model for UvrD mediated RNAP backtracking, whereby UvrD loads onto the upstream edge of the DNA duplex junction with the transcription bubble, a natural substrate for UvrD, as it prefers to interact with such ds-ssDNA junctions [44]. Using its ATP-dependent helicase activity, UvrD unzips the upstream portion of the transcription bubble to pull RNAP backwards along from the DNA damage site, thus exposing the site to repair enzymes (Figure 2).
It has been shown that NusA renders RNAP more prone to backtracking [45] and potentiates UvrD-mediated backtracking in vitro [13]. Similar to uvrD mutants, the sensitivity of nusA mutants to DNA damaging agents can be suppressed by inactivation of anti-backtracking factors GreA/B, or by inhibition of actively translating ribosomes [13]. Thus, the role of NusA in Mfd-independent TCR is likely to be in assisting UvrD with pulling RNAP away from sites of DNA damage. Direct interactions between UvrD and UvrB [41,42] and between NusA and UvrA [32] may also help to recruit UvrAB to sites of damage.
The mechanism of UvrD/NusA-dependent TCR overcomes several apparent functional inadequacies associated with Mfd-dependent TCR. It has been reported that Mfd-dependent TCR operates only during basal levels of transcription and that Mfd is dispensable at highly induced transcription units [27]. UvrD is a SOS-inducible gene whose cellular concentration increases about three-fold soon after DNA damage occurs [46,47], resulting in about three fold molar excess of UvrD over RNAP. This should favour UvrD dimerization, which stimulates its helicase activity, thereby triggering RNAP backtracking from DNA lesion sites. This model is supported by the observation that uvrD mutant cells are defective in strand specific repair of cyclobutane pyrimidine dimers [48]. During rapid induction of transcription, stalling of RNAP at DNA lesions causes the accumulation of the arrays of elongation complexes, which cannot be readily dislodged by Mfd due to its slow enzymatic activity and difficulties accessing individual complexes packed in such arrays. During UvrD-directed TCR, each stalled RNAP can be pulled backwards by its associated UvrD dimer, and the entire array will eventually be moved away from the site of damage.
Mfd-dependent TCR results in transcription termination [19], but UvrD/NusA-dependent TCR results in backtracked RNAP [13], which can be promptly reactivated by anti-backtracking factors, including Mfd itself, shortly after DNA repair has been accomplished.
The two complementary TCR pathways may be examples of specialized systems bacteria have evolved to repair DNA damage under different circumstances. During basal levels of gene expression and normal growth conditions, Mfd-dependent TCR repairs the low-frequency lesions that occur on the transcribed strand, while UvrD/NusA-dependent TCR may be less active in this process. During genotoxic stress, however, the SOS response activates UvrD/NusA-directed TCR, which assumes the repair process, as the ability of Mfd to promptly remove RNAPs is limited. In this manner, the cell uses two different, balanced mechanisms to achieve the repair of different doses of DNA damage that occur under different growth conditions.
Mfd may also act during the post-repair, recovery phase by removing excessively backtracked RNAPs, which accumulate during DNA damage. Such complexes cause mutagenic double stranded breaks due to co-directional collisions with the replisome [34,49,50]. This hypothesis is additionally attractive as it explains the original mutation frequency decline phenotype of mfd. Indeed, by “pushing” backtracked RNAP forward to resume transcription after UV exposure, Mfd should reduce the frequency of DNA breaks, which are often repaired in an error-prone fashion. This model is consistent with the requirement of Mfd for rapid recovery of transcription after DNA damage [51], and with the lag in post UV-irradiated growth of mfd(-) cells in liquid cultures due to excessively backtracked RNAPs (Kamarthapu and Nudler, unpublished data).
RNA polymerase as a global sensor of DNA damage
The most remarkable aspect of NER is the ability of the UvrAB complex to promptly recognize DNA lesions that vary broadly in chemical structure and their impact on local DNA conformation. The number of UvrAB molecules per E. coli cell during the SOS response reaches no more than a couple of hundred [52], yet the lesions must be located rapidly and discriminated from the millions of base pairs of non-damaged DNA. In vitro, on naked DNA, a mixture of 3-D and 1-D diffusion searches can be used by these proteins [53]. However, in vivo, the search must be complicated by ubiquitous protein-DNA complexes, macromolecular crowding, and DNA compartmentalization and condensation. Unlike UvrA and UvrB, which are not processive scanners, RNAP usually moves along DNA from the beginning of an operon to its end via processive 1-D mechano-chemical tracking. Its catalytic center is finely tuned to react even to mild deviations from the canonical four bases in the template strand, leading to pauses or stalling [54]. Thus RNAP could be the most efficient DNA damage detector that continuously patrols the genome and initiates the TCR process at lesion sites. Indeed, it has become evident in recent years that pervasive transcription is conserved in all the three domains of life [55,56]. Although eukaryotic genomes contain only 1-2% protein-coding regions, more than 80% of the genome is transcribed. In prokaryotes, it was assumed that pervasive transcription is very limited because ~80% of the genome is protein-coding DNA. However, recent advances in next generation sequencing have revealed the presence of abundant ncRNA in prokaryotes as well, including substantial amounts of antisense RNA [56,57]. It is, thus, tempting to speculate that seemingly wasteful pervasive transcription provides the beneficial means to achieve global surveillance of the genome for DNA damage and to enable robust NER.
Acknowledgements
This work was supported by the NIH grant R01 GM107329 and by the Howard Hughes Medical Institute (E.N.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
*of special interest
**of outstanding interest
- 1.Setlow RB, Carrier WL. The disappearance of thymine dimers from DNA: an error-correcting mechanism. Proc. Natl. Acad. Sci. U. S. A. 1964;51:226–231. doi: 10.1073/pnas.51.2.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2*.Kisker C, Kuper J, Van Houten B. Prokaryotic nucleotide excision repair. Cold Spring Harb. Perspect. Biol. 2013;5:a012591. doi: 10.1101/cshperspect.a012591. This is a comprehensive recent review on NER. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reardon JT, Sancar A. Nucleotide excision repair. Prog. Nucleic Acid Res. Mol. Biol. 2005;79:183–235. doi: 10.1016/S0079-6603(04)79004-2. [DOI] [PubMed] [Google Scholar]
- 4.Mellon I, Spivak G, Hanawalt PC. Selective removal of transcription-blocking DNA damage from the transcribed strand of the mammalian DHFR gene. Cell. 1987;51:241–249. doi: 10.1016/0092-8674(87)90151-6. [DOI] [PubMed] [Google Scholar]
- 5.Bohr VA, Smith CA, Okumoto DS, Hanawalt PC. DNA repair in an active gene: removal of pyrimidine dimers from the DHFR gene of CHO cells is much more efficient than in the genome overall. Cell. 1985;40:359–369. doi: 10.1016/0092-8674(85)90150-3. [DOI] [PubMed] [Google Scholar]
- 6*.Ganesan A, Spivak G, Hanawalt PC. Transcription-coupled DNA repair in prokaryotes. Prog. Mol. Biol. Transl. Sci. 2012;110:25–40. doi: 10.1016/B978-0-12-387665-2.00002-X. This is a comprehesive recent review on bacterial TCR. [DOI] [PubMed] [Google Scholar]
- 7.Spivak G, Ganesan AK. The complex choreography of transcription-coupled repair. DNA Repair (Amst) 2014;19:64–70. doi: 10.1016/j.dnarep.2014.03.025. [DOI] [PubMed] [Google Scholar]
- 8.Selby CP, Sancar A. Transcription preferentially inhibits nucleotide excision repair of the template DNA strand in vitro. J. Biol. Chem. 1990;265:21330–21336. [PubMed] [Google Scholar]
- 9.Selby CP, Sancar A. Structure and function of transcription-repair coupling factor. II. Catalytic properties. J Biol Chem. 1995;270:4890–4895. doi: 10.1074/jbc.270.9.4890. [DOI] [PubMed] [Google Scholar]
- 10.Selby CP, Sancar A. Structure and function of transcription-repair coupling factor: I. Structural domains and binding properties. J. Biol. Chem. 1995;270:4882–4889. doi: 10.1074/jbc.270.9.4882. [DOI] [PubMed] [Google Scholar]
- 11.Westblade LF, Campbell EA, Pukhrambam C, Padovan JC, Nickels BE, Lamour V, Darst SA. Structural basis for the bacterial transcription-repair coupling factor/RNA polymerase interaction. Nucleic Acids Res. 2010;38:8357–8369. doi: 10.1093/nar/gkq692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Park J-S, Marr MT, Roberts JW. E. coli Transcription repair coupling factor (Mfd protein) rescues arrested complexes by promoting forward translocation. Cell. 2002;109:757–767. doi: 10.1016/s0092-8674(02)00769-9. [DOI] [PubMed] [Google Scholar]
- 13**.Epshtein V, Kamarthapu V, McGary K, Svetlov V, Ueberheide B, Proshkin S, Mironov A, Nudler E. UvrD facilitates DNA repair by pulling RNA polymerase backwards. Nature. 2014;505:372–377. doi: 10.1038/nature12928. This paper uncovers UvrD-mediated RNAP backtarcking and proposes a new TCR pathway. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Witkin EM. Time, Temperature, and Protein Synthesis: A Study of Ultraviolet-Induced Mutation in Bacteria. Cold Spring Harb. Symp. Quant. Biol. 1956;21:123–140. doi: 10.1101/sqb.1956.021.01.011. [DOI] [PubMed] [Google Scholar]
- 15.Witkin EM. Radiation-induced mutations and their repair. Science. 1966;152:1345–1353. doi: 10.1126/science.152.3727.1345. [DOI] [PubMed] [Google Scholar]
- 16.Witkin EM. Mutation frequency decline revisited. Bioessays. 1994;16:437–444. doi: 10.1002/bies.950160613. [DOI] [PubMed] [Google Scholar]
- 17.Mellon I, Hanawalt PC. Induction of the Escherichia coli lactose operon selectively increases repair of its transcribed DNA strand. Nature. 1989;342:95–98. doi: 10.1038/342095a0. [DOI] [PubMed] [Google Scholar]
- 18.Tornaletti S, Hanawalt PC. Effect of DNA lesions on transcription elongation. Biochimie. 1999;81:139–146. doi: 10.1016/s0300-9084(99)80046-7. [DOI] [PubMed] [Google Scholar]
- 19.Selby CP, Sancar A. Molecular mechanism of transcription-repair coupling. Science. 1993;260:53–58. doi: 10.1126/science.8465200. [DOI] [PubMed] [Google Scholar]
- 20.Selby CP, Witkin EM, Sancar A. Escherichia coli mfd mutant deficient in “mutation frequency decline” lacks strand-specific repair: in vitro complementation with purified coupling factor. Proc. Natl. Acad. Sci. U. S. A. 1991;88:11574–11578. doi: 10.1073/pnas.88.24.11574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mellon I, Champe GN. Products of DNA mismatch repair genes mutS and mutL are required for transcription-coupled nucleotide-excision repair of the lactose operon in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 1996;93:1292–1297. doi: 10.1073/pnas.93.3.1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deaconescu AM, Chambers AL, Smith AJ, Nickels BE, Hochschild A, Savery NJ, Darst SA. Structural basis for bacterial transcription-coupled DNA repair. Cell. 2006;124:507–520. doi: 10.1016/j.cell.2005.11.045. [DOI] [PubMed] [Google Scholar]
- 23.Deaconescu AM, Sevostyanova A, Artsimovitch I, Grigorieff N. Nucleotide excision repair (NER) machinery recruitment by the transcription-repair coupling factor involves unmasking of a conserved intramolecular interface. Proc. Natl. Acad. Sci. 2012;109:3353–3358. doi: 10.1073/pnas.1115105109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith AJ, Pernstich C, Savery NJ. Multipartite control of the DNA translocase, Mfd. Nucleic Acids Res. 2012;40:10408–10416. doi: 10.1093/nar/gks775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Howan K, Monnet J, Fan J, Strick TR. Stopped in its tracks: the RNA polymerase molecular motor as a robust sensor of DNA damage. DNA Repair (Amst) 2014;20:49–57. doi: 10.1016/j.dnarep.2014.02.018. [DOI] [PubMed] [Google Scholar]
- 26*.Howan K, Smith AJ, Westblade LF, Joly N, Grange W, Zorman S, Darst SA, Savery NJ, Strick TR. Initiation of transcription-coupled repair characterized at single-molecule resolution. Nature. 2012;490:431–434. doi: 10.1038/nature11430. This study provides a detailed mechanistical view on the Mfd action. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kunala S, Brash DE. Intragenic domains of strand-specific repair in Escherichia coli. J. Mol. Biol. 1995;246:264–272. doi: 10.1006/jmbi.1994.0082. [DOI] [PubMed] [Google Scholar]
- 28.Ganesan AK, Hanawalt PC. Transcription-coupled nucleotide excision repair of a gene transcribed by bacteriophage T7 RNA polymerase in Escherichia coli. DNA Repair (Amst) 2010;9:958–963. doi: 10.1016/j.dnarep.2010.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nudler E, Gottesman ME. Transcription termination and anti-termination in E. coli. Genes to Cells. 2002;7:755–768. doi: 10.1046/j.1365-2443.2002.00563.x. [DOI] [PubMed] [Google Scholar]
- 30.Cohen SE, Godoy VG, Walker GC. Transcriptional modulator NusA interacts with translesion DNA polymerases in Escherichia coli. J. Bacteriol. 2009;191:665–672. doi: 10.1128/JB.00941-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cohen SE, Walker GC. The transcription elongation factor NusA is required for stress-induced mutagenesis in Escherichia coli. Curr. Biol. 2010;20:80–85. doi: 10.1016/j.cub.2009.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cohen SE, Lewis CA, Mooney RA, Kohanski MA, Collins JJ, Landick R, Walker GC. Roles for the transcription elongation factor NusA in both DNA repair and damage tolerance pathways in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 2010;107:15517–15522. doi: 10.1073/pnas.1005203107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33**.Cohen SE, Walker GC. New discoveries linking transcription to DNA repair and damage tolerance pathways. Transcription. 2011;2:37–40. doi: 10.4161/trns.2.1.14228. An altearnative, Mfd-independent, model of NusA-mediated TCR is proposed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nudler E. RNA polymerase backtracking in gene regulation and genome instability. Cell. 2012;149:1438–1445. doi: 10.1016/j.cell.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matson SW. Escherichia coli helicase II (urvD gene product) translocates unidirectionally in a 3' to 5' direction. J. Biol. Chem. 1986;261:10169–10175. [PubMed] [Google Scholar]
- 36.Hickson ID, Arthur HM, Bramhill D, Emmerson PT. The E. coli uvrD gene product is DNA helicase II. MGG Mol. Gen. Genet. 1983;190:265–270. doi: 10.1007/BF00330649. [DOI] [PubMed] [Google Scholar]
- 37.Fischer CJ, Maluf NK, Lohman TM. Mechanism of ATP-dependent translocation of E. coli UvrD monomers along single-stranded DNA. J. Mol. Biol. 2004;344:1287–1309. doi: 10.1016/j.jmb.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 38.Maluf NK, Ali JA, Lohman TM. Kinetic mechanism for formation of the active, dimeric UvrD helicase-DNA complex. J. Biol. Chem. 2003;278:31930–31940. doi: 10.1074/jbc.M304223200. [DOI] [PubMed] [Google Scholar]
- 39*.Gwynn EJ, Smith AJ, Guy CP, Savery NJ, McGlynn P, Dillingham MS. The conserved C-terminus of the PcrA/UvrD helicase interacts directly with RNA polymerase. PLoS One. 2013;8:e78141. doi: 10.1371/journal.pone.0078141. Demosntartion that E. coli UvrD and its homolog PcrA from B. subtilis bind RNA polymerase. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Proshkin S, Rahmouni AR, Mironov A, Nudler E. Cooperation between translating ribosomes and RNA polymerase in transcription elongation. Science. 2010;328:504–508. doi: 10.1126/science.1184939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Manelyte L, Guy CP, Smith RM, Dillingham MS, McGlynn P, Savery NJ. The unstructured C-terminal extension of UvrD interacts with UvrB, but is dispensable for nucleotide excision repair. DNA Repair (Amst) 2009;8:1300–1310. doi: 10.1016/j.dnarep.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ahn B. A physical interaction of UvrD with nucleotide excision repair protein UvrB. Mol. Cells. 2000;10:592–597. doi: 10.1007/s10059-000-0592-5. [DOI] [PubMed] [Google Scholar]
- 43.Toulokhonov I, Artsimovitch I, Landick R. Allosteric control of RNA polymerase by a site that contacts nascent RNA hairpins. Science. 2001;292:730–733. doi: 10.1126/science.1057738. [DOI] [PubMed] [Google Scholar]
- 44.Tomko EJ, Jia H, Park J, Maluf NK, Ha T, Lohman TM. 5’-Single-stranded/duplex DNA junctions are loading sites for E. coli UvrD translocase. EMBO J. 2010;29:3826–3839. doi: 10.1038/emboj.2010.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bar-Nahum G, Epshtein V, Ruckenstein AE, Rafikov R, Mustaev A, Nudler E. A ratchet mechanism of transcription elongation and its control. Cell. 2005;120:183–193. doi: 10.1016/j.cell.2004.11.045. [DOI] [PubMed] [Google Scholar]
- 46.Siegel EC. The Escherichia coli uvrD gene is inducible by DNA damage. Mol. Gen. Genet. 1983;191:397–400. doi: 10.1007/BF00425753. [DOI] [PubMed] [Google Scholar]
- 47.Kumura K, Sekiguchi M. Identification of the uvrD gene product of Escherichia coli as DNA helicase II and its induction by DNA-damaging agents. J. Biol. Chem. 1984;259:1560–1565. [PubMed] [Google Scholar]
- 48.Crowley DJ, Hanawalt PC. The SOS-dependent upregulation of uvrD is not required for efficient nucleotide excision repair of ultraviolet light induced DNA photoproducts in Escherichia coli. Mutat. Res. 2001;485:319–329. doi: 10.1016/s0921-8777(01)00068-4. [DOI] [PubMed] [Google Scholar]
- 49.Dutta D, Shatalin K, Epshtein V, Gottesman ME, Nudler E. Linking RNA Polymerase Backtracking to Genome Instability in E. coli. Cell. 2011;146:533–543. doi: 10.1016/j.cell.2011.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Merrikh H, Machón C, Grainger WH, Grossman AD, Soultanas P. Co-directional replication-transcription conflicts lead to replication restart. Nature. 2011;470:554–557. doi: 10.1038/nature09758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schalow BJ, Courcelle CT, Courcelle J. Mfd is required for rapid recovery of transcription following UV-induced DNA damage but not oxidative DNA damage in Escherichia coli. J. Bacteriol. 2012;194:2637–2645. doi: 10.1128/JB.06725-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sancar A, Sancar GB. DNA repair enzymes. Annu. Rev. Biochem. 1988;57:29–67. doi: 10.1146/annurev.bi.57.070188.000333. [DOI] [PubMed] [Google Scholar]
- 53.Kad NM, Wang H, Kennedy GG, Warshaw DM, Van Houten B. Collaborative dynamic DNA scanning by nucleotide excision repair proteins investigated by single-molecule imaging of quantum-dot-labeled proteins. Mol. Cell. 2010;37:702–713. doi: 10.1016/j.molcel.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nudler E. RNA polymerase active center: the molecular engine of transcription. Annu. Rev. Biochem. 2009;78:335–361. doi: 10.1146/annurev.biochem.76.052705.164655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55*.Wade JT, Grainger DC. Pervasive transcription: illuminating the dark matter of bacterial transcriptomes. Nat. Rev. Microbiol. 2014;12:647–653. doi: 10.1038/nrmicro3316. A comprehensive recent review summarizing evidence of pervasive transcription in bacteria. [DOI] [PubMed] [Google Scholar]
- 56.Lybecker M, Bilusic I, Raghavan R. Pervasive transcription: detecting functional RNAs in bacteria. Transcription. 2014;5:e944039. doi: 10.4161/21541272.2014.944039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Georg J, Hess WR. cis-antisense RNA, another level of gene regulation in bacteria. Microbiol. Mol. Biol. Rev. 2011;75:286–300. doi: 10.1128/MMBR.00032-10. [DOI] [PMC free article] [PubMed] [Google Scholar]