Abstract
Although activation of the AMP-activated protein kinase (AMPK) as well as of ubiquitin/proteasome degradative pathways play an essential role in the preservation of metabolic homeostasis, little is known concerning interactions between protein turnover and AMPK activity. In the present studies, we found that inhibition of the 26S proteasome resulted in rapid activation of AMPK in macrophages, epithelial and endothelial cells. This was associated with increased levels of non-degraded Ub-protein conjugates, in both cytosolic and mitochondrial fractions. Selective inhibitors of ubiquitination or siRNA-dependent knockdown of Ub-ligase E1 diminished AMPK activation in cells treated with MG132, a 26S proteasome inhibitor. In addition to inhibition of AMPK activation by Ub-ligase E1 inhibitors, deficiency in Park2 mitochondria-associated Ub-ligase E3 also reduced AMPK activation upon dissipation of mitochondrial membrane potential (Δψm). Accumulation of Ub-proteins was correlated with decreases in cellular bioenergetics, including mitochondria oxidative phosphorylation, and an increase in ROS formation. Antioxidants, such as N-acetyl-L-cysteine or mitochondria-targeted MitoTEMPO, effectively diminished MG132-induced AMPK activation. Glucose-dependent regulation of AMPK or AMPK-mediated autophagy was modulated by alterations in intracellular levels of Ub-protein conjugates. Our results indicate that accumulation of ubiquitinated proteins alter cellular bioenergetics and redox status, leading to AMPK activation.
Keywords: AMP kinase, bioenergetics, proteasome, ubiquitination, autophagy, PARK2
1. Introduction
The ubiquitin-proteasome system (UPS) is a major protein degradative pathway involved in the preservation of cellular structure and function [1, 2]. While the 20S proteasome is involved in direct protein hydrolysis, degradation of ubiquitinated proteins by the 26S proteasome is a relatively more important process in protein turnover [3–5]. In addition to facilitating protein turnover and removal of damaged proteins, ubiquitination and deubiquitination serve important roles signaling/regulatory pathways [6–8]. In addition to proteasome function, autophagy is important degradative/regenerative system that involves lysosome-dependent degradation of bulk proteins and cellular components, including mitochondrial quality control system we known as mitophagy [9].
Ubiquitination of proteins designed for degradation is an ATP-dependent process and involves cooperation of three ubiquitin ligase enzymes. In particular, the ubiquitin moiety is transferred by Ubiquitin-activating enzyme E1 to the Ubiquitin-conjugating enzyme E2 followed by formation of ubiquitin chain ligation on target proteins by a substrate specific E3 ubiquitin ligase [10]. Selected components of the 26S cap proteins are involved in recognition and transport of ubiquitinated proteins for degradation by the 26S proteasome [11, 12]. Impairment of ubiquitin ligases is typically associated with the appearance of excess and misfolded proteins, whereas inhibition of the 26S proteasome results in accumulation of non-degraded ubiquitinated proteins. Prolonged proteasomal inactivation may lead to activation of alternative degradative pathways, including autophagy and mitophagy, or result in cell death [13–15]. Pathologic conditions associated with dysregulation of ubiquitin/proteasome degradative capacity are implicated in diabetes and obesity, inflammation, neurodegeneration and aging [16–19]. For example, Ub-ligase E3 MG53 deficiency can trigger insulin resistance and cardiovascular complications associated with metabolic stress in animal models [20]. Aberrant function of E3 ubiquitin ligases, such as BRCA1, MDM2 or FANC, is associated with diminished DNA repair and increased risk of malignancy [21–24]. Overall decline in proteasome/ubiquitin protein degradative function is a hallmark of aging [17].
The AMP-activated protein kinase (AMPK), a serine/threonine protein kinase, is a heterotrimer that consists of regulatory β and γ subunits, and one α catalytic subunit [25]. AMPK activation is associated with alterations in cellular bioenergetics and redox status, particularly relating to glycolysis and mitochondrial function [26]. Crystal structure analysis revealed that binding of AMP and ADP to the AMPK γ subunit induces allosteric domain rearrangement [27], that allows for phosphorylation and optimal activation of AMPK by upstream kinases [28, 29]. In addition to being activated through interactions with AMP and ADP, AMPK can be also activated by reactive oxygen species, particularly oxidative modification (glutathionylation) of specific cysteine thiols in the α and β subunits [30, 31]. Previous studies indicate that mitochondrial ROS can activate AMPK in many cell types, including lung fibroblast, epithelial and endothelial cells, and macrophages [32–34]. Once activated, AMPK switches on catabolic pathways to promote ATP synthesis and switches off biosynthetic pathways, thereby limiting energy expenditure and providing alternative sources of ATP production.
Although AMPK is a central energetic sensor and regulator of lipid and carbohydrate metabolism [26, 35], AMPK also improved outcomes from cardiovascular complications, as well as diminished liver, kidney and lung injury, in murine models of acute inflammation [36, 37]. AMPK activators, including metformin, prevented disruption of vascular integrity in preclinical models of acute lung injury and airway remodeling in asthma [38, 39]. The AMPK/bioenergetics axis is linked to regulation of autophagy and mitochondrial biogenesis, as well as cellular redox status [9, 34, 40, 41].
Although alterations in cellular bioenergetics and redox status can trigger AMPK activation, little is known about upstream signaling events responsible for the ability of mitochondria to modulate AMPK activity. One potential mechanism may be associated with imbalance in protein ubiquitination and degradation, including turnover of mitochondria-associated proteins. In the present experiments, we examined the hypothesis that protein ubiquitination and degradation plays an important role in regulating the mitochondria/AMPK signaling axis, including response to glucose and nutrients.
2. Materials and methods
2.1. Mice
Wild type C57BL/6J mice and B6.129S4-Park2tm1Shn/J deficient in Park2 Ub-ligase E3 (PARK2−/−) were purchased from The Jackson Laboratory (Bar Harbor, ME). Male mice, 8–10 weeks of age, were used for experiments. The mice were maintained on a 12-h light-dark cycle with free access to food and water. All experiments were conducted in accordance with protocols approved by the University of Alabama at Birmingham Animal Care and Use Committee.
2.2. Reagents and Antibodies
MG132 (carbobenzoxy-Leu-Leu-leucinal) was purchased from EMD Millipore (Billerica, MA). PYR41, PYDZ4409, Chloroquine, Triphenylmethylphosphonium (TPMP), and N-acetyl-L-cysteine (NAC), glucose and mannitol were obtained from Sigma-Aldrich (Saint Louis, MO). Mito-TEMPO and 5-Aminoimidazole-4-carboxamide-1-β-D-ribofuranoside (AICAR) were purchased from Enzo Life Sciences (Farmingdale, NY). MitoSox was purchased from Invitrogen (Carlsbad, CA). Antibodies specific for Phospho-Thr172-AMPKα, total AMPKα, Phospho-Ser79-ACC, total-ACC and Ubiquitin were obtained from Cell signaling Technology (Danvers, MA). β-actin and GRP75 antibody were from Santa Cruz Biotechnology (Santa Cruz, CA). Ubiquitin expressing DNA construct was obtained from Addgene (Cambridge, MA).Anti-LC3B and anti-VDAC1 antibodies were from Abcam (Cambridge, MA). Emulsion oil solution containing 4’,6-diamidino-2-phenylindole (DAPI) was from Vector Laboratories (Burlingame, CA).
2.3. Cell culture
Macrophage cell line Raw 264.7 cells (ATCC) were cultured in DMEM medium with 8% FBS + 4.5g/l Glucose + L-glutamine. Thioglycollate elicited peritoneal macrophages were purified and cultured as previously described [33]. Bovine Aortic Endothelial Cells (BAEC) were cultured in DMEM medium containing 8% FBS whereas Human Epithelial Kidney Cells (HEK293) were cultured in RPMI1640 medium containing 8% FBS. Mouse embryonic fibroblasts (MEFs) were cultured in DMEM medium with 8% FBS. Wild type (AMPKα1/2+/+) and AMPK deficient MEFs (AMPKα1/2−/−) were provided by Dr. Benoit Viollet (University of Paris, France). Cells were maintained at 37°C, 5% CO2 in humidified conditions.
2.4. Western Blot analysis
Western blot analysis was performed as described previously [42–44]. Each experiment was carried out three or more times.
2.5. Measurement of cellular bioenergetics
The bioenergetics of macrophages was determined using the XF24 analyzer from Seahorse Bioscience which measures O2 consumption and proton production (pH) in intact cells, which can be ascribed to oxidative phosphorylation and glycolysis using inhibitors of metabolism as previously described [45]. Measurements were performed using macrophages (7.5 × 104) that were plated on XF24 plates after which they were treated with the compounds of interest. The plate was then washed with XF assay buffer (DMEM, 5% FBS supplemented with 5.5 mM, D-glucose, 4 mM L-glutamine, and 1 mM pyruvate, pH 7.4) and incubated in XF buffer for 30–60 minutes before the assay. After the assay, the cells were lysed with RIPA buffer and protein concentration was determined by DC Lowry assay. All results are corrected to protein levels.
2.6. Detection of Mitochondrial ROS formation
The intracellular level of superoxide generation by macrophages was measured using superoxide sensitive, mitochondria-targeted probe mitoSOX [46]. Briefly, Raw 264.7 cells loaded with mitoSOX (5µM) for 60 minutes were treated with MG132 (10µM) for 45 minutes and then fluorescence was determined using FACS analysis [47].
2.7. Mitochondria isolation
Mitochondria were isolated from cultured cells using the nitrogen cavitation method as previously described with modification [48]. Briefly, the cells were washed once with ice-cold PBS and then scraped in 1ml of cavitation buffer (5mM HEPES, 3mM MgCl2, 1mM EGTA, 250 mM sucrose, containing protease and phosphatase inhibitors) and collected into a pre-cooled cavitation chamber (Parr Instrument Co., Moline, IL, USA). The cell suspension was subjected to 2000 p.s.i. for 5 minutes at 4°C and then released through outflow tubing attached to the valve localized at the bottom of the cavitation chamber. The crude mitochondrial fraction was collected after centrifuged at 500 g for 5 minutes at 4°C. The crude mitochondria were layered over a 1M/1.5 discontinuous sucrose gradients and centrifuged at 28,000 g for 60 minutes at 4°C. Mitochondria in a diffuse white band between the 1 M and 1.5 M sucrose layers were transferred to a 1.5-mL microcentrifuge tube and diluted 1 : 2 (v/v) in dilution buffer (5 mM HEPES, pH 7.4, 3 mM MgCl2, 1 mM EGTA, containing protease and phosphatase inhibitors). After gentle mixing the mitochondria were centrifuged at 20,800 g for 20 minutes at 4°C and pellet re-suspended in in RIPA lysis buffer.
2.8. Imaging mitochondria and Ub-protein conjugates
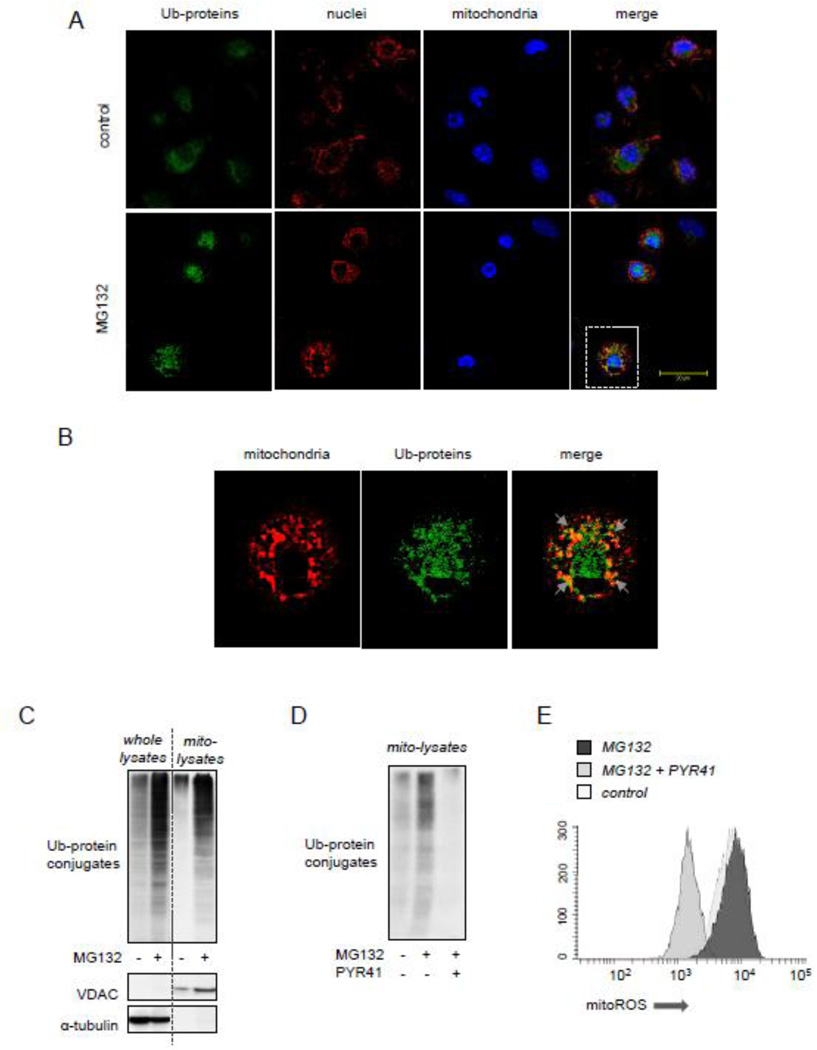
Peritoneal macrophages were incubated with 4% paraformaldehyde in PBS for 20 min at room temperature, then washed with PBS and permeabilized with 0.1% TritonX-100/PBS for 4 minutes. The cells were then washed and incubated with 3% BSA in PBS for 45 min, followed by the addition of anti-Ubiquitin mouse monoclonal and anti-GRP75 rabbit polyclonal IgG overnight at 4°C. The cells were then washed and incubated with fluorescent anti-mouse or anti-rabbit antibodies (Alexa-488 or Alexa-555) for 90 minutes at room temperature. After the cells were washed with PBS, they were mounted with emulsion oil solution containing DAPI to visualize nuclei. Confocal microscopy was performed as described previously, using a Leica DMIRBE inverted epifluorescence/Nomarski microscope (Leica Microsystems, Wetzlar, Germany) outfitted with Leica TCS NT laser confocal optics [49].
2.9. Statistical analysis
Multigroup comparisons were performed using one-way ANOVA with Tukey's post hoc test. Student's t test for comparisons between two groups. A value of P less than 0.05 was considered significant. Analyses were performed on SPSS version 16.0 (IBM, Armonk, NY) for Windows (Microsoft Corp., Redmond, WA).
RESULTS
2.10. Inhibition of 26S proteasome is associated with rapid activation of AMPK
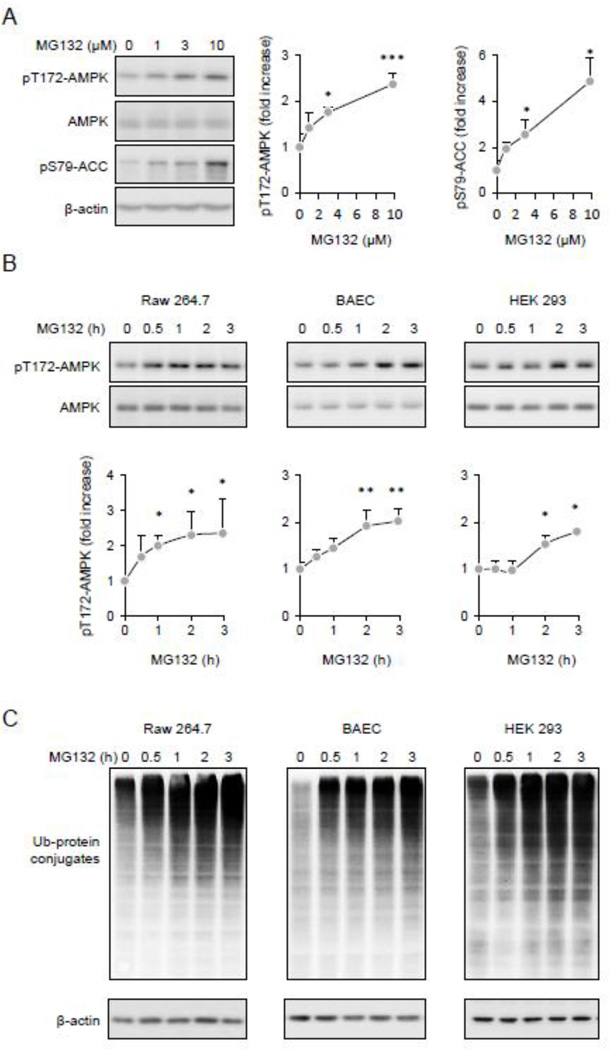
Although both cellular metabolism and protein turnover are involved in regulating cellular homeostasis, little is known about the influence of the ubiquitin/proteasome degradative pathway on AMPK activity. To examine this issue, Thr172-AMPK phosphorylation status was determined in Raw 264.7 macrophages before and after exposure to the cell-permeable 26S proteasome inhibitor MG132. As shown in Figure 1A, inclusion of MG132 in macrophage cultures resulted in dose-dependent activation of AMPK, as shown by increased levels of phospho-Thr172-AMPK and phospho-Ser79-ACC, a downstream target of AMPK. The MG132-dependent activation of AMPK was also found in cell populations other than macrophages, including endothelial and epithelial cells. As shown in Figures 1B and C, AMPK activation was accompanied by the accumulation of non-degraded ubiquitinated proteins. Of note, while inhibition of the 26 proteasome resulted in significant phosphorylation of Thr172AMPK, total amounts of the AMPKα subunit were not altered.
Figure 1.
Inhibition 26S proteasome and accumulation of Ub-protein conjugates is associated with AMPK activation. (A). Representative Western blots show the amount of pThr172-AMPK or pSer79ACC, total AMPK and β-actin in Raw 264.7 cells treated with MG132 (0, 1, 3, or 10 µM) for 60 minutes. Quantitative data of optical bend densitometry are shown. Mean ± SD, n = 3, * P < 0.05, ** P < 0.01. (B and C). Raw 264.7 cells, BAEC or HEK 293 cells were treated with MG132 (10 µM) for indicated time. Representative Western blots (B) and quantitative data (C) show the extent of pThr172-AMPK, total AMPK, Ub-protein conjugates and β-actin Mean ± SD, n = 3, * P < 0.05, ** P < 0.01.
2.11. Accumulation of non-degraded Ub-protein conjugates is involved in AMPK activation
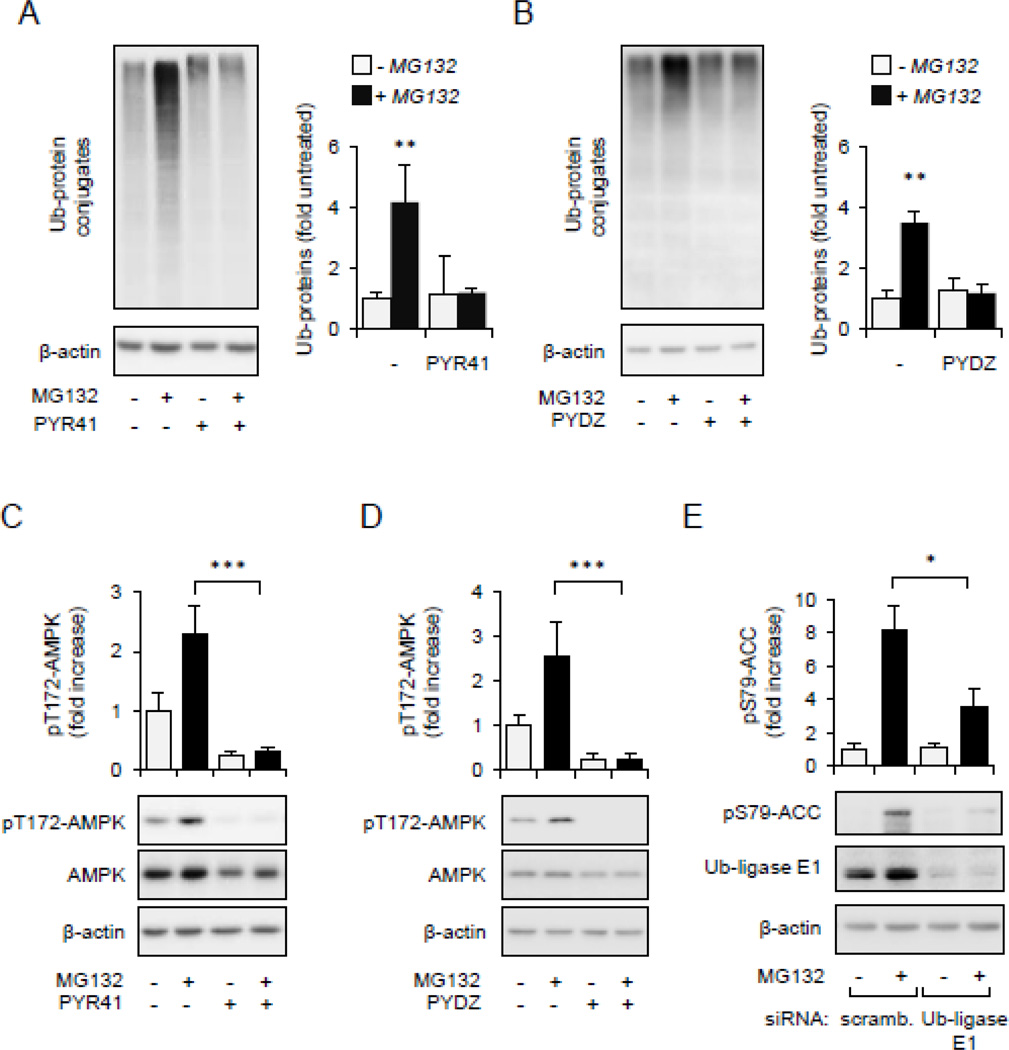
To examine if accumulation of non-degraded Ub-protein conjugates after 26S proteasome inhibition is responsible for AMPK activation, Raw 264.7 macrophages were treated with MG132 in the presence or absence of PYR41 or PYDZ4409, inhibitors of Ubiquitin-activating enzyme E1. As shown in Figures 2A–D, exposure to PYR41 or PYDZ4409 effectively diminished the accumulation of Ub-protein conjugates in MG132-treated cells and prevented AMPK activation. In confirmation of these results, we found that siRNA-dependent knockdown of Ub-activating enzyme E1 also diminished activation of AMPK in MG132-treated cells, as shown by significant decrease in phosphorylation of pSer79-ACC (Figure 2E). Collectively, these findings suggest that accumulation of non-degraded ubiquitinated proteins are implicated in AMPK activation.
Figure 2.
AMPK activity is dependent on accumulation of ubiquitinated proteins. Raw 264.7 cells were pre-treated with Ub-ligase E1 inhibitor PYR41 (0 or 50 µM) or PYZD4409 (0 or 50 µM) for 30 minutes followed by incubation with MG132 (0 or 10 µM) for additional 60 minutes. (A). Representative Western blots show the amount of Ub-protein conjugates and β–actin or (B) the extent of AMPK phosphorylation. Mean ± SD, n = 3, *** P < 0.001.
2.12. Mitochondrial bioenergetics and ROS are involved in AMPK activation after 26S proteasome inhibition
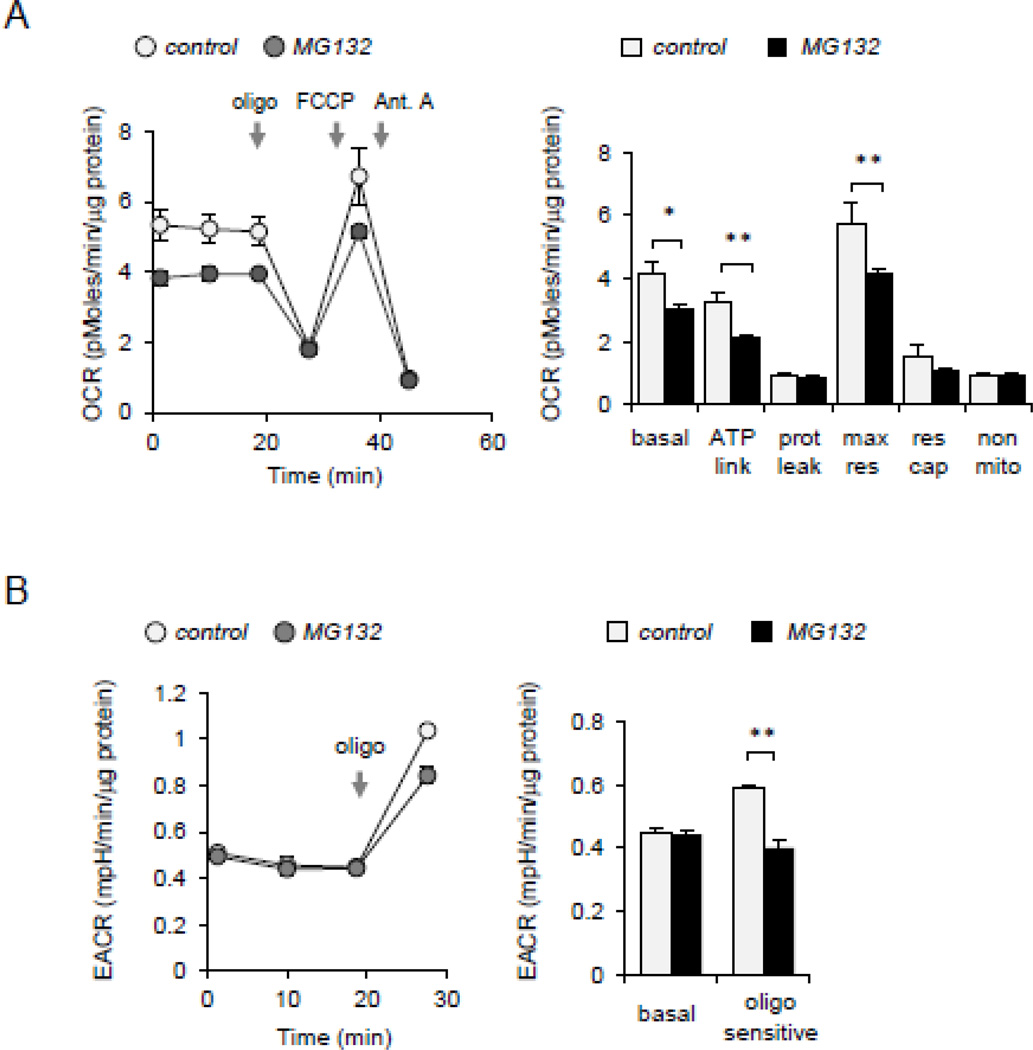
Previous studies have shown that dissipation of mitochondrial membrane potential (Δψm) and increase in mitochondrial ROS formation are important for AMPK activation in many cell populations [32, 33, 50, 51]. To examine whether alterations in cellular bioenergetics occurred after 26S proteasome inhibition, oxygen consumption and glycolysis rate was determined in MG132 treated macrophages. As shown in Figure 3A, culture of macrophages with MG132 resulted in significant decrease in basal, maximal oxygen consumption rate but with no significant change in bioenergetic reserve capacity. Basal ECAR (extracellular acidification rate) was not changed, which taken with the OCR measurements, suggests both a decreased bioenergetic demand and decreased mitochondrial capacity in the MG132 cells. Significant decrease in the oligomycin-stimulated ECAR was observed in MG132 treated cells, consistent with a suppressed glycolytic maximal capacity (Figure 3B).
Figure 3.
Cellular bioenergetics is implicated in AMPK activation. (A). Raw 264.7 cells were treated with MG132 (0 or 10 µM) for 60 minutes and then bioenergetic status determined using Seahorse Extracellular Flux Analyzer. The OCR was monitored over time and after subsequent injection of oligomycin (1 µg/ml), carbonyl cyanide 4-(trifluoromethoxy) phenylhydrazone (0.5 µM), and antimycin A (ant. A) (10 µM). Bar graph shows the indices of mitochondrial respiratory function, including basal OCR, ATP-linked OCR (ATP link), protein leak (prot leak), maximal respiration (max res), and non-mitochondrial respiration (non-mito) in control or MG132-treated Raw 264.7 macrophages. (B) ECAR was determined in control and MG132-treated cells. Measurements were performed before and after inclusion of oligomycin. Mean ± SEM, n = 3 – 8, * P < 0.05; ** P < 0.01.
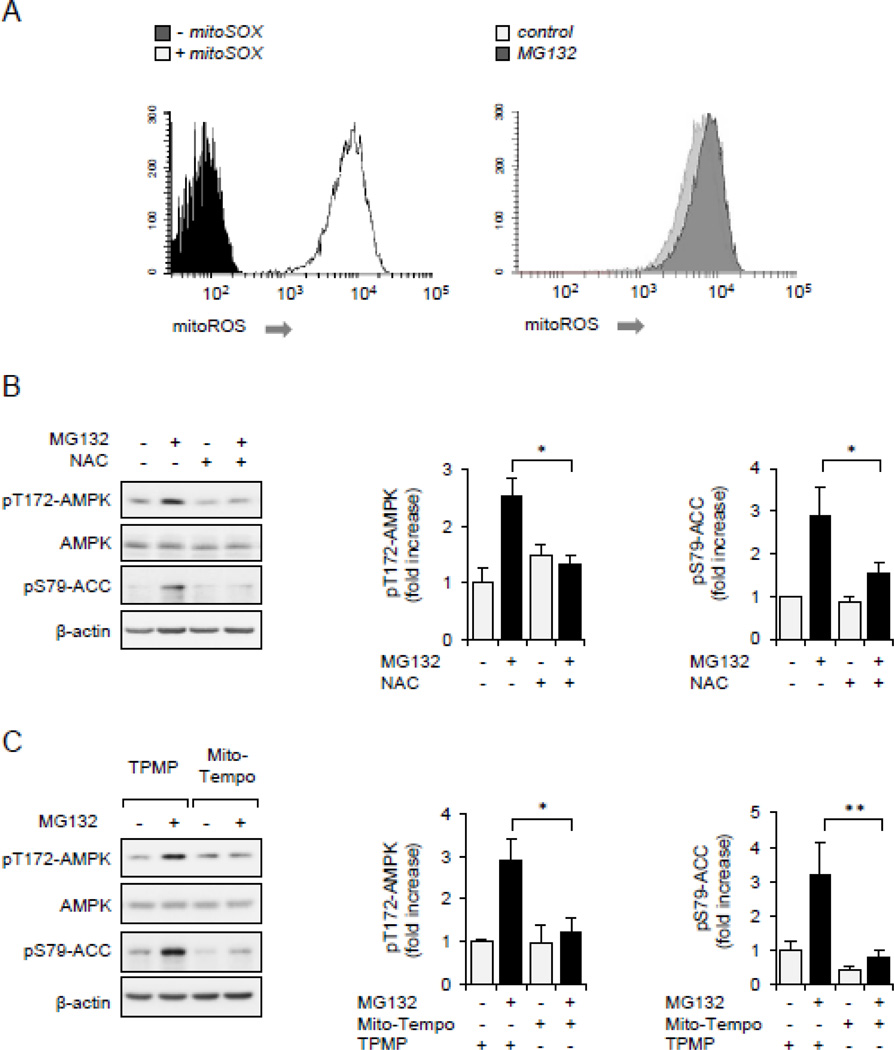
As shown in Figure 4A, exposure of Raw 264.7 macrophages to MG132 for 60 minutes resulted in marked increase in superoxide formation, as compared to levels present in untreated cells. To examine a role for ROS in activating AMPK, Raw 264.7 cells were pretreated with the ROS scavenger NAC for 30 minutes, or incubated with the mitochondria-targeted antioxidant MitoTEMPO or control compound TPMP for 15 minutes, followed by culture with MG132 for an additional 60 minutes. As shown in Figures 4B and C, both NAC and MitoTEMPO, but not the control TPMP, diminished AMPK activation in MG132 treated cells (Figures 4B and C). These results indicate that alterations in mitochondrial redox signaling are involved in 26S proteasome associated modulation of AMPK activation state.
Figure 4.
ROS is associated with AMPK activation. (A).Raw 264.7 cells were incubated with fluorogenic probe mitoSOX (0 or 5 µM) for 60 minutes (left panel), or mitoSOX loaded cells were treated with MG132 (0, 10 µM) for 45 minutes and fluorescence determined using flow cytometry (right panel). (B and C).Raw 264.7 cells were pretreated with NAC (0 or 20 mM) for 30 minutes, TPMP or MitoTempo (0 or 1 µM) 1 for 15 minutes followed by inclusion of MG132 (0 or 10 µM) for an additional 60 minutes. Representative Western blots and quantitative data of pThr172-AMPK and pSer79-ACC are shown. Mean ± SEM, n = 3 – 8, * P < 0.05; ** P < 0.01.
2.13. Inhibition of Ub-ligase E1 diminishes FCCP-dependent activation of AMPK
Recent studies have suggested that accumulation of mitochondria-associated Ub-proteins, particularly in the area of the mitochondrial other membrane (MOM), are involved in regulating mitochondrial structure and function [52, 53]. As shown in Figures 5A and B, confocal microscopy revealed that Ub-protein conjugates are partially overlapped with mitochondrial staining in MG312-treated peritoneal macrophages. Moreover, Western Blot analysis evidenced that MG132-dependent activation of AMPK was associated with significant accumulation of Ub-protein conjugates in mitochondrial fractions (Figure 5C), while diminished amounts of Ub-proteins were found after culture of macrophages with the Ub-ligase E1 inhibitor PYR41 (Figure 5D). Of note, PYR41 also prevented MG132-dependent increases of both ROS formation and AMPK activation (Figures 5E and 2C).
Figure 5.
Accumulation of Ub-protein conjugates is associated with mitoROS-dependent activation of AMPK. Peritoneal macrophages were incubated with or without MG312 (10 µM) for 2 hours followed by staining cells for Ub-protein and GRP75, a mitochondrial marker. (A and B) Representative images show mitochondria (red), Ub-protein conjugates (green) and nuclei (blue). Area of interest (dotted lines in A) is magnified and shown in panel (B). Arrows indicate overlap between mitochondria and Ub-protein conjugates. (C) The amount of Ub-protein conjugates, VDAC, and α-tubulin was determined using Western blot analysis of whole cell or mitochondrial extracts obtained from Raw 264.7 that were treated with MG132 (0 or 10 µM) for 60 minutes. (D). Raw 264.7 cells were pretreated with PYR41 (0 or 50 µM) for 30 minutes followed by exposure to MG132 (0 or 10 µM) for additional 60 minutes. Ub-protein conjugates obtained from mitochondrial fractions are shown. (E) The extent of ROS production was determined in Raw 264.7 macrophages pre-treated with PYR41 (0 or 50 µM) for 30 minutes followed by inclusion of MG132 (0 or 10 µM) for additional 60 minutes. MitoSOX fluorescence intensity was determined using flow cytometry.
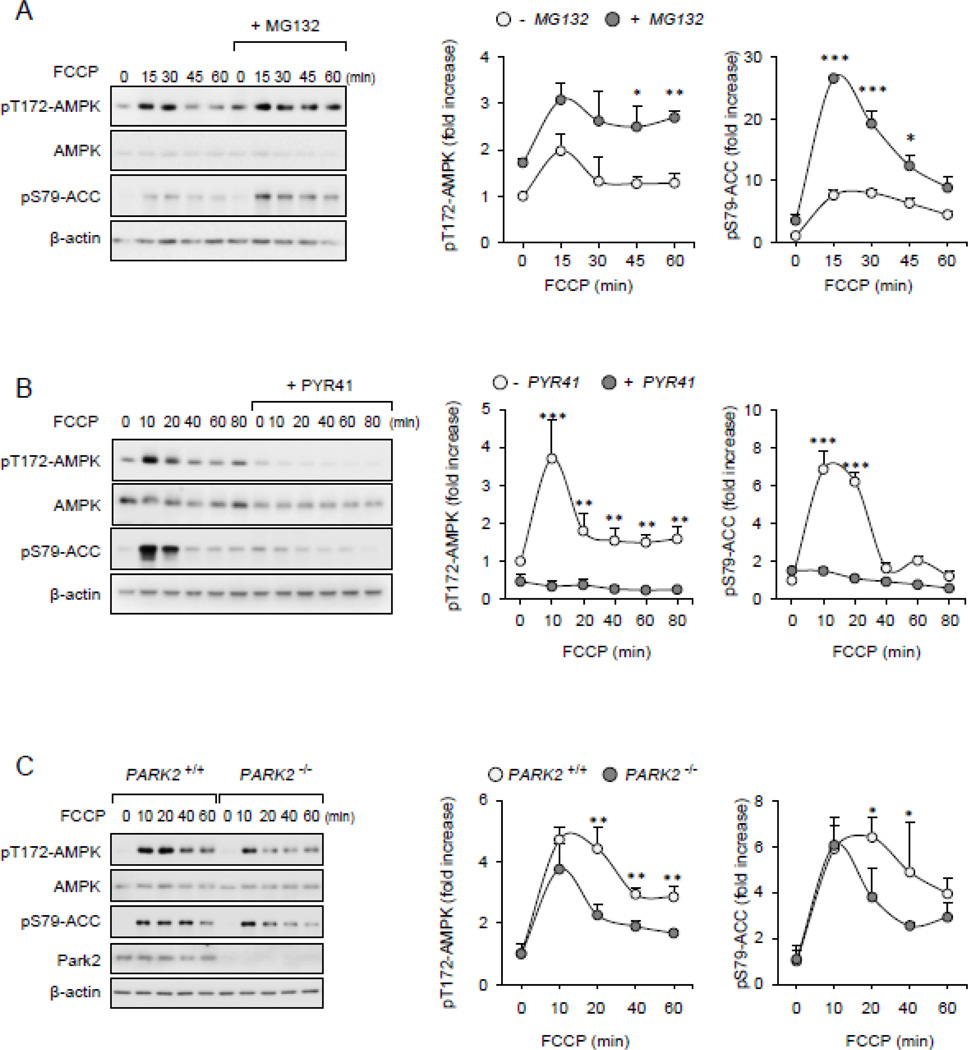
FCCP-induced dissipation of mitochondrial membrane potential (Δψm) has been previously demonstrated to be associated with the appearance of ubiquitinated proteins in MOM [54]. Therefore, we examined if stabilization or inhibition in the formation of mitochondria-associated Ub-protein conjugates affects AMPK activity. As shown in Figures 6A and B, exposure of FCCP-treated cells to MG132 resulted in more robust and prolonged activation of AMPK, whereas pre-treatment with the Ub-ligase E1 inhibitor PYR-41 had opposite effects. These results suggest that accumulation of Ub-protein conjugates affect the mitochondria-AMPK signaling axis following depolarization of mitochondrial membrane potential.
Figure 6.
FCCP-dependent activation of AMPK is diminished upon inclusion of E1 inhibitors. (A and B) Raw 264.7 were subsequently (A) MG132 (0 or 10 µM) or (B) PYR41 (0 or 50 µM) for 30 minutes followed by inclusion of FCCP (500 nM) for indicated time points. Representative Western blots and quantitative data show the amounts of pThr172-AMPK, pSer79-ACC (mean ± SD, n = 3, * P < 0.05; ** P < 0.01 or *** P < 0.001. (C). Wild type (PARK+/+) and Park2 deficient (PARK2−/−) peritoneal macrophages were incubated with FCCP (500 nM) for indicate time followed by Western Blot analysis of phospho- or total AMPK and ACC. Mean ± SD, n = 3, * P < 0.05; ** P < 0.01.
2.14. Park2 Ub-ligase E3 modulates AMPK activity
Although the experiments described above indicate that accumulation of mitochondria-associated Ub-protein conjugates is likely involved in AMPK activation, the role of the specific Ub-ligases E3 in regulating AMPK action was not explored. Among the group of recently described mitochondria-associated Ub-ligases E3, Park2 ligase has been found to process ubiquitination of proteins associated with MOM, particularly during depolarization of the mitochondrial membrane potential [54]. To determine a possible role for Park2 on AMPK activity, we used peritoneal macrophages isolated form control (wild type) and Park2 deficient mice (PARK2−/−). As shown in Figure 6C, FCCP-induced AMPK activation was reduced in Park2 deficient as compared to wild type macrophages. These results suggest that enrichment of mitochondria-associated Ub-proteins, including proteins ubiquitinated by Park2 Ub-ligase E3, is involved in AMPK activation.
2.15. Ub-protein conjugates affect glucose associated modulation of AMPK activation
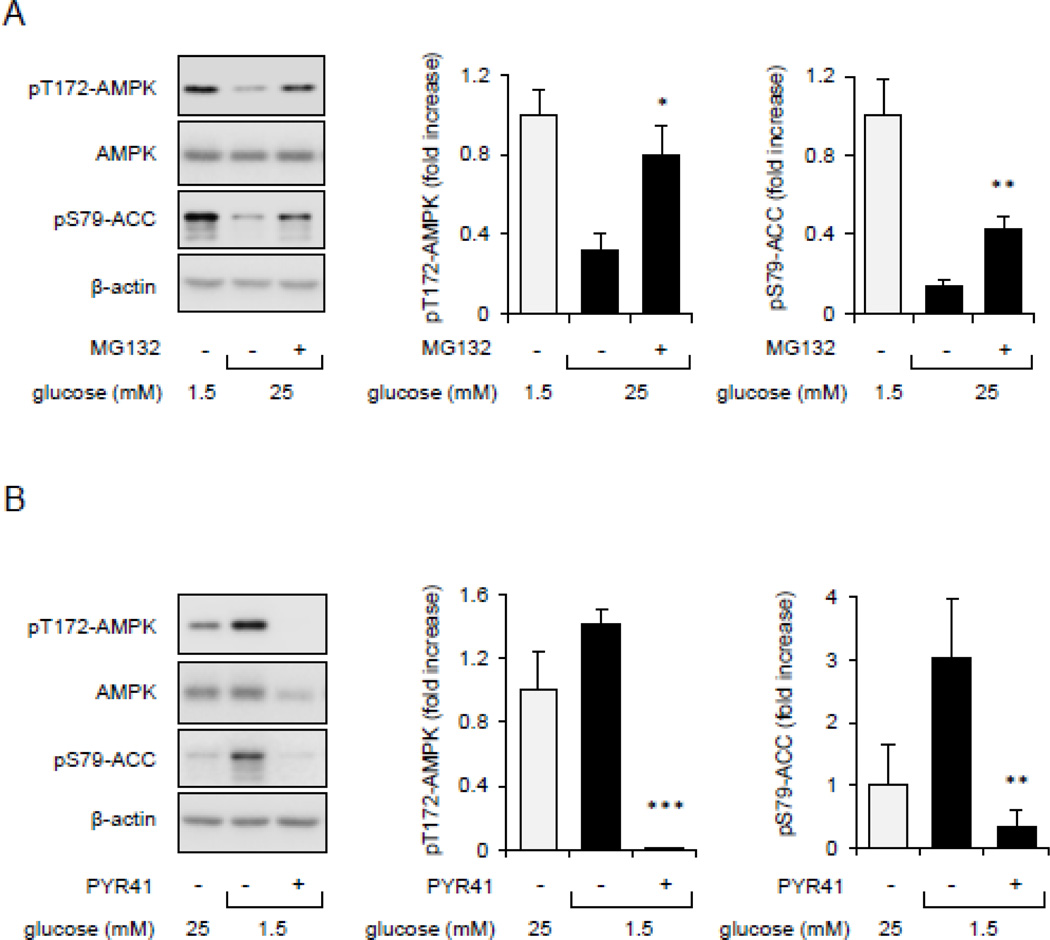
Previous studies have shown that bioavailability of glucose and other nutrients can affect AMPK function [26]. Therefore, we examined whether Ub-protein conjugates may influence glucose-dependent regulation of AMPK activity. To explore this possibility, Raw 264.7 macrophages were cultured in low glucose (1.5 mM) and low serum (FBS; 0.5%) medium for 60 minutes and then exposed to higher concentrations of glucose (25 mM) or mannitol (25 mM) as a control. As shown in Figure 7A, increasing glucose concentrations in cultures effectively diminished phosphorylation of Thr172-AMPK as well as of the AMPK downstream target pSer79-ACC. Of note, proteasome inhibition through inclusion of MG132 in the cultures prevented the inhibitory effects of glucose on AMPK activation. We also found that while glucose starvation effectively increased AMPK activity, such activation diminished upon cellular exposure to the Ub-ligase E1 inhibitor PYR-41 (Figure 7B). These results suggest that the accumulation of Ub-protein conjugates influence glucose-dependent stimulation of AMPK activity.
Figure 7.
The effects of protein ubiquitination on glucose and nutrients-dependent regulation of AMPK activity. (A). Raw 264.7 cells were cultured in low glucose/serum medium for 60 minutes followed by inclusion of glucose (25 mM) and MG132 (0 or 10 µM) for additional 60 minutes. (B). Cells cultured in 25 mM glucose medium were exposed to low glucose (1.5 mM) in the presence or absence of PYR-41 (50 µM) for 60 minutes. Western blot and quantitative data in panels A and B show the extent of Thr172-AMPK and Ser79-ACC phosphorylation. Mean ± SEM, n = 3 – 7, * P < 0.05 or ** P < 0.01.
2.16. Ub-protein conjugates affect AMPK-dependent autophagy/mitophagy
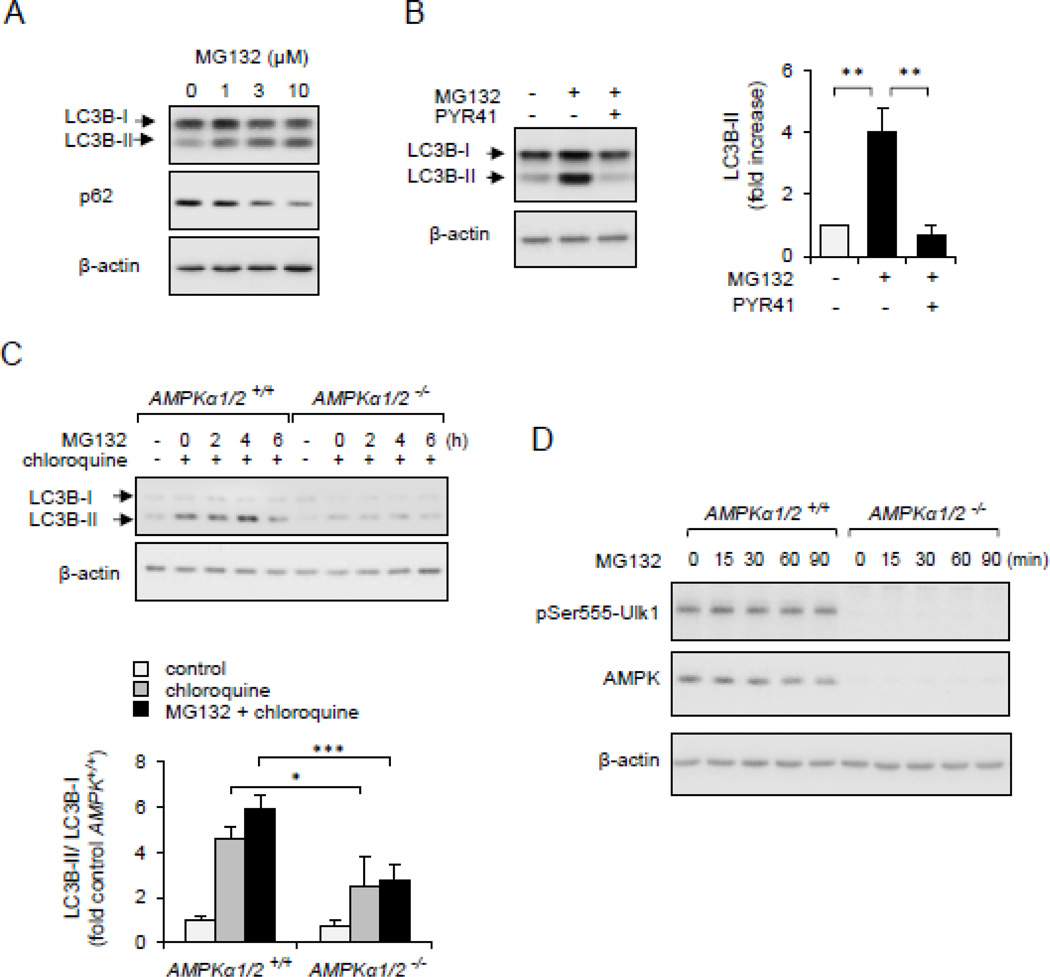
Autophagy is an important route of intracellular protein and organelles degradation/recycling [13]. Recent studies have suggested that cross-talk exists between autophagy and the ubiquitin-proteasome system [55, 56]. As shown on Figure 8A, activation of autophagy was found in MG132-treated HEK 293 epithelial cells, as shown by increased expression of the autophagy marker LC3B-II, a lipidated form of LC3B that is recruited to autophagosomal membranes during autophagy. Accumulation of GFP-LC3B-II aggregates was completely prevented by the Ub-ligase E1 inhibitor PYR41 (Figure 8B). These findings are consistent with previous studies that showed involvement of Ub-protein conjugates in regulating autophagy/mitophagy [57].
Figure 8.
Non-degraded Ub-protein conjugates are involved in AMPK-dependent autophagy/mitophagy. Panel (A). HEK 293 cells were dose-dependently treated with MG132 for 4 hours. Western blots of LC3B-I, LC3B-II, p62 β-actin is shown. (B). HEK293 cells were first incubated with PYR41 (0 or 50 µM) followed by exposure to MG132 (0 or 10 µM) for 4 hours. Western blots and quantitative data show the extent of LC3B-I, LC3B-II and β-actin. Mean ± SEM, n = 3, * P < 0.05 or ** P < 0.01. (C). MEFs, wild type (AMPKα1/2+/+) or AMPK deficient fibroblasts (AMPKα1/2−/−), were time dependently treated with MG132 (10 µM) followed by Western Blot analysis of LC3B-I, LC3B-II and β-actin. Chloroquine was applied for 30 minutes, as indicated. (D).Western blot analysis quantitative data show the extent of pSer555-Ulk1, AMPKα and β-actin in MG132-treated MEF AMPKα1/2+/+ or AMPKα1/2−/−. Mean ± SEM, n = 3, * P < 0.05 or *** P < 0.001.
The relationship between AMPK activation and autophagy/mitophagy was determined in wild type (AMPKα1/2+/+) and AMPK deficient MEF cells (AMPKα1/2−/−). Relatively short exposure of fibroblasts with MG132 without chloroquine produced little of no accumulation of LC3-BII. However, inclusion of MG132 and chloroquine in cell cultures resulted in robust increase in the amounts of LC3-BII in wild type, but not in AMPK deficient cells (Figure 8C). As shown in Figure 6D, AMPKα1/2−/− MEF cells showed lack of AMPK-dependent phosphorylation of Ser555Ulk1, an essential component of pathways associated with mitophagy (Figure 8D) [9].
3. DISCUSSION
In the present studies, we found that dysregulation of the ubiquitin/proteasome degradative pathway resulted in rapid activation of AMPK. In particular, AMPK activation was associated with accumulation of non-degraded Ub–protein conjugates, including enrichment of ubiquitinated proteins in mitochondrial fractions. While many studies have explored the relationship between cellular bioenergetics and AMPK activation, there is only limited information showing that ubiquitination may affect AMPK function. The 26S proteasome inhibitor bortezomib has recently been shown to induce protective autophagy through an AMPK-dependent pathway [58]. In contrast, ubiquitination of the AMPKβ subunit was implicated in diminished AMPK function in adipose tissue, while ubiquitination of the AMPKα subunit had only negligible effects on AMPK activation [59, 60].
While our results do not preclude cell type-dependent and Ub-ligase E3 specific regulation of AMPK, they do suggest that a major regulatory mechanism of AMPK activity is related to the accumulation of Ub-protein conjugates in mitochondria, followed by alterations in mitochondrial bioenergetics and ROS formation [61–63]. The importance of mitochondrial bioenergetics in ubiquitination associated pathways is supported by studies showing that inhibitors of oxidative phosphorylation or dissipation in mitochondrial membrane potential (Δψm) stimulate ubiquitination of many proteins associated with the outer mitochondrial membrane (OMM) [54, 64]. We found that diminished protein ubiquitination or the accumulation of Ub-protein conjugates, due to inhibition of the Ub-ligase E1 or 26S proteasome, had modulatory effects on AMPK activation in FCCP-treated macrophages (Figures 6A and B). In additional experiments, we found that events occurring downstream of non-degraded Ub-protein conjugates, such as alterations in mitochondrial bioenergetics and ROS formation, were involved in AMPK activation. Such findings are consistent with previous studies that demonstrated the ability of mitochondrial bioenergetics and mitochondrially derived ROS to affect AMPK activity [30, 33, 50].
Recent studies indicate that a specific subset of E3 Ub-ligases, including Park2, Mitol/March5, Mullan, Mdm30, and Mfb1, are involved in ubiquitination and degradation of mitochondria-associated proteins [54, 65–67]. Although the processes involved in extraction of ubiquitinated proteins from the mitochondria and degradation of mitochondrial proteins are not completely delineated, knockdown of mitochondria-associated Ub-ligase E3 showed their involvement in mitochondrial function and morphology [52, 53].
Although protein turnover is implicated in regulating the mitochondrial/AMPK signaling axis, identification of the specific Ubiquitin-ligases and substrates involved in mitochondrial bioenergetics and associated redox signaling may further support this emerging concept. Among mitochondria-associated Ub-ligases, Park2 was implicated in ubiquitination of many MOM proteins, particularly after Δψm dissipation [54]. Our results indicate that deficiency of Park2 produced at least a partial decrease in AMPK activation in FCCP-treated macrophages (Figure 6C). Although Park2 is involved in regulating mitochondrial function [54], the precise mechanism through which ubiquitinated proteins affect mitochondrial redox pathways has not been delineated.
Bioavailability of glucose and nutrients has a major impact on AMPK activity and downstream effects on cellular metabolism [26]. We found that glucose -dependent regulation of AMPK activity can be influenced by either accumulation or absence of Ub-protein conjugates. Such findings suggest that modulation in the formation of Ub-protein conjugates or the activity of specific Ub-ligases may be a therapeutic approach to target AMPK activity and effects in diabetes and obesity. Recent study has shown that hyperglycemia enhances 26S proteasome activity and also that accelerated degradation of Ub-protein conjugates contributed to vascular inflammation in murine models of diabetes [68]. Moreover, mice that received the 26S proteasome inhibitor MG132 showed improvement of diabetic nephropathy [69]. AMPK activation is also known to diminish tissue injury associated with neutrophil and macrophage pro-inflammatory activation, such as acute lung injury [42].
AMPK is a central metabolic switch between anabolic and catabolic processes, preserving energy homeostasis and cellular viability [26]. AMPK modulates protein turnover at several levels, including mTOR-dependent protein synthesis, transcriptional regulation, and affecting 26S proteasome function [26, 70]. AMPK can also activate alternative protein degradative pathways, including autophagy and mitophagy [9, 41]. Despite differences in mechanisms of protein degradation as well as in client proteins, cross-talk between the ubiquitin-proteasome and autophagy-lysosome system is implicated in many cellular functions [55, 56]. Our results indicate that under injurious inflammatory responses, such as those that occur in diabetes and aging in which the degradative ubiquitination/proteasome system is altered, AMPK-dependent autophagy should improve cellular bioenergetics and normalize redox status.
4. Conclusion
Our results indicate that proteasome/ubiquitin protein degradation pathways play a key role in regulating mitochondrial/AMPK signaling axis. In particular, these results delineate a new mechanism through which protein turnover links mitochondria bioenergetics, redox signaling and AMPK function. Our findings suggest that protein turnover affects activation of AMPK by glucose and the ability of AMPK to influence autophagy and mitophagy.
Supplementary Material
Figure 9.
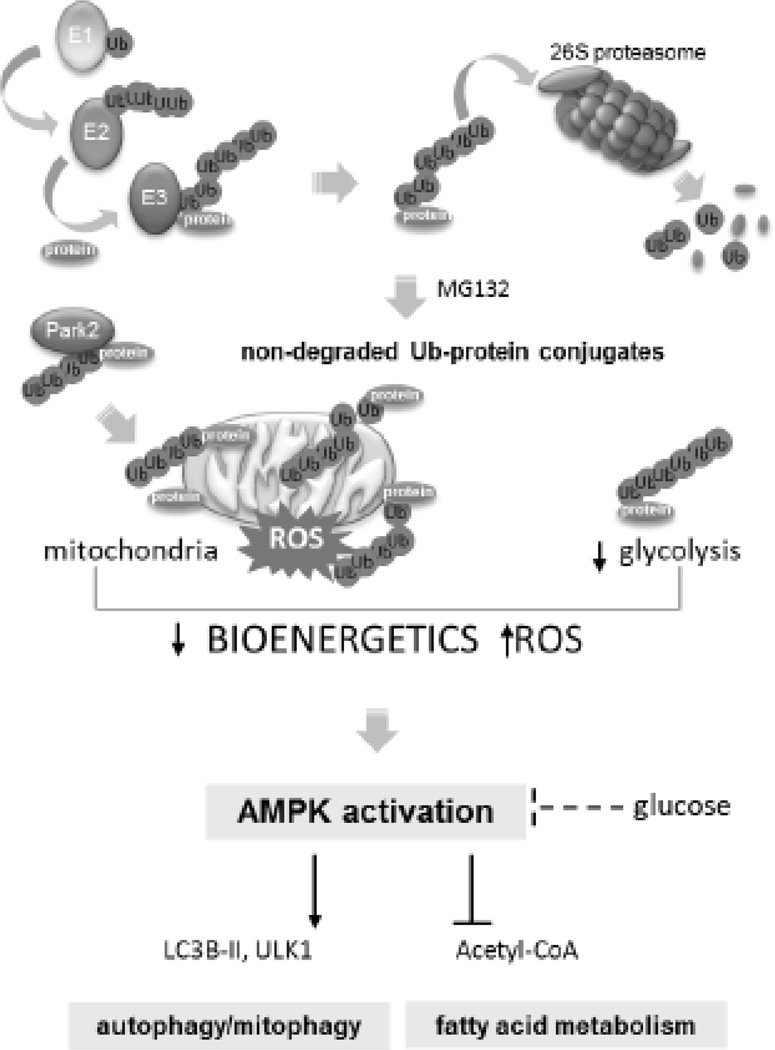
Proteasome/ubiquitin protein degradative pathway controls AMPK function. Proteasome inhibition and accumulation of non-degraded Ub-protein conjugates is associated with alterations in mitochondrial bioenergetics and ROS formation followed by activation of AMPK. Protein ubiquitination can also affect glucose-dependent regulation of AMPK activity. Once activated AMPK can influence bioenergetic and degradative pathways, including autophagy, mitochondrial quality control mitophagy and promotes fatty acids oxidation.
Highlights.
Proteasome inhibition and accumulation of Ub-proteins increased AMPK activity
Activation of AMPK is mediated through alterations in mitochondrial bioenergetics and ROS formation
Ub-proteins influenced AMPK activation by diminished mitochondrial membrane potential, glucose bioavailability and autophagy/mitophagy
Ubiquitin/proteasomal degradative pathways play a central role in regulating mitochondrial/AMPK signaling axis
Acknowledgement
We thank Dr. Zhongyu Liu (University of Alabama at Birmingham) for excellent technical support and Dr. Dantuma (Karolinska Institutet, Stockholm, Sweden) for Ubiquitin expressing plasmid (Dantuma et al, J Cell Biol. 2006).
Grants
This work was supported in part by National Institutes of Health Grants GM87748 and HL107585 to Jaroslaw W. Zmijewski.
Abbreviations
- AMPK
AMP-activated protein kinase AICAR
- ROS
reactive oxygen species
- Δψm
mitochondrial membrane potential
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest
All authors declare no conflict of interest
Contributors
S.J., D.W.P., Y.G. and S. R. performed experiments. S.J., S.R., V.D.U., E.A. and J.W.Z. interpreted results and revised the manuscript. S.J. and JZ designed research, S.J. EA and J.Z. wrote the manuscript.
REFERENCES
- 1.Glickman MH, Ciechanover A. Physiol Rev. 2002;82:373–428. doi: 10.1152/physrev.00027.2001. [DOI] [PubMed] [Google Scholar]
- 2.Hershko A. Cell Death Differ. 2005;12:1191–1197. doi: 10.1038/sj.cdd.4401702. [DOI] [PubMed] [Google Scholar]
- 3.Ravid T, Hochstrasser M. Nat Rev Mol Cell Biol. 2008;9:679–690. doi: 10.1038/nrm2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gallastegui N, Groll M. Trends Biochem Sci. 2010;35:634–642. doi: 10.1016/j.tibs.2010.05.005. [DOI] [PubMed] [Google Scholar]
- 5.Hershko A, Heller H, Elias S, Ciechanover A. J Biol Chem. 1983;258:8206–8214. [PubMed] [Google Scholar]
- 6.O'Neill LA. J Biol Chem. 2009;284:8209. doi: 10.1074/jbc.R800070200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mattiroli F, Sixma TK. Nat Struct Mol Biol. 2014;21:308–316. doi: 10.1038/nsmb.2792. [DOI] [PubMed] [Google Scholar]
- 8.Wilkinson KD. Semin Cell Dev Biol. 2000;11:141–148. doi: 10.1006/scdb.2000.0164. [DOI] [PubMed] [Google Scholar]
- 9.Egan DF, Shackelford DB, Mihaylova MM, Gelino S, Kohnz RA, Mair W, Vasquez DS, Joshi A, Gwinn DM, Taylor R, Asara JM, Fitzpatrick J, Dillin A, Viollet B, Kundu M, Hansen M, Shaw RJ. Science. 2011;331:456–461. doi: 10.1126/science.1196371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Metzger MB, Pruneda JN, Klevit RE, Weissman AM. Biochim Biophys Acta. 2014;1843:47–60. doi: 10.1016/j.bbamcr.2013.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Finley D. Annu Rev Biochem. 2009;78:477–513. doi: 10.1146/annurev.biochem.78.081507.101607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lander GC, Estrin E, Matyskiela ME, Bashore C, Nogales E, Martin A. Nature. 2012;482:186–191. doi: 10.1038/nature10774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shaid S, Brandts CH, Serve H, Dikic I. Cell Death Differ. 2013;20:21–30. doi: 10.1038/cdd.2012.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gomes LC, Scorrano L. Biochim Biophys Acta. 2013;1833:205–212. doi: 10.1016/j.bbamcr.2012.02.012. [DOI] [PubMed] [Google Scholar]
- 15.Ashrafi G, Schwarz TL. Cell Death Differ. 2013;20:31–42. doi: 10.1038/cdd.2012.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paul S. BioEssays : news and reviews in molecular, cellular and developmental biology. 2008;30:1172–1184. doi: 10.1002/bies.20852. [DOI] [PubMed] [Google Scholar]
- 17.Kevei E, Hoppe T. Nat Struct Mol Biol. 2014;21:290–292. doi: 10.1038/nsmb.2806. [DOI] [PubMed] [Google Scholar]
- 18.Corn JE, Vucic D. Nat Struct Mol Biol. 2014;21:297–300. doi: 10.1038/nsmb.2808. [DOI] [PubMed] [Google Scholar]
- 19.Ciechanover A, Brundin P. Neuron. 2003;40:427–446. doi: 10.1016/s0896-6273(03)00606-8. [DOI] [PubMed] [Google Scholar]
- 20.Song R, Peng W, Zhang Y, Lv F, Wu HK, Guo J, Cao Y, Pi Y, Zhang X, Jin L, Zhang M, Jiang P, Liu F, Meng S, Zhang X, Jiang P, Cao CM, Xiao RP. Nature. 2013;494:375–379. doi: 10.1038/nature11834. [DOI] [PubMed] [Google Scholar]
- 21.Lipkowitz S, Weissman AM. Nat Rev Cancer. 2011;11:629–643. doi: 10.1038/nrc3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fakharzadeh SS, Trusko SP, George DL. EMBO J. 1991;10:1565–1569. doi: 10.1002/j.1460-2075.1991.tb07676.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Welcsh PL, King MC. Human molecular genetics. 2001;10:705–713. doi: 10.1093/hmg/10.7.705. [DOI] [PubMed] [Google Scholar]
- 24.Moldovan GL, D'Andrea AD. Annu Rev Genet. 2009;43:223–249. doi: 10.1146/annurev-genet-102108-134222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kahn BB, Alquier T, Carling D, Hardie DG. Cell metabolism. 2005;1:15–25. doi: 10.1016/j.cmet.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 26.Hardie DG, Ross FA, Hawley SA. Nat Rev Mol Cell Biol. 2012;13:251–262. doi: 10.1038/nrm3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xiao B, Heath R, Saiu P, Leiper FC, Leone P, Jing C, Walker PA, Haire L, Eccleston JF, Davis CT, Martin SR, Carling D, Gamblin SJ. Nature. 2007;449:496–500. doi: 10.1038/nature06161. [DOI] [PubMed] [Google Scholar]
- 28.Hawley SA, Pan DA, Mustard KJ, Ross L, Bain J, Edelman AM, Frenguelli BG, Hardie DG. Cell Metab. 2005;2:9–19. doi: 10.1016/j.cmet.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 29.Woods A, Johnstone SR, Dickerson K, Leiper FC, Fryer LG, Neumann D, Schlattner U, Wallimann T, Carlson M, Carling D. Current biology : CB. 2003;13:2004–2008. doi: 10.1016/j.cub.2003.10.031. [DOI] [PubMed] [Google Scholar]
- 30.Zmijewski JW, Banerjee S, Bae H, Friggeri A, Lazarowski ER, Abraham E. J Biol Chem. 2010;285:33154–33164. doi: 10.1074/jbc.M110.143685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klaus A, Zorman S, Berthier A, Polge C, Ramirez S, Michelland S, Seve M, Vertommen D, Rider M, Lentze N, Auerbach D, Schlattner U. PLoS ONE. 2013;8:e62497. doi: 10.1371/journal.pone.0062497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zou MH, Kirkpatrick SS, Davis BJ, Nelson JS, Wiles WGt, Schlattner U, Neumann D, Brownlee M, Freeman MB, Goldman MH. J Biol Chem. 2004;279:43940–43951. doi: 10.1074/jbc.M404421200. [DOI] [PubMed] [Google Scholar]
- 33.Jiang S, Park DW, Stigler WS, Creighton J, Ravi S, Darley-Usmar V, Zmijewski JW. J Biol Chem. 2013 doi: 10.1074/jbc.M113.489468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kukidome D, Nishikawa T, Sonoda K, Imoto K, Fujisawa K, Yano M, Motoshima H, Taguchi T, Matsumura T, Araki E. Diabetes. 2006;55:120–127. [PubMed] [Google Scholar]
- 35.Towler MC, Hardie DG. Circ Res. 2007;100:328–341. doi: 10.1161/01.RES.0000256090.42690.05. [DOI] [PubMed] [Google Scholar]
- 36.Liu TF, Brown CM, El Gazzar M, McPhail L, Millet P, Rao A, Vachharajani VT, Yoza BK, McCall CE. J Leukoc Biol. 2012;92:499–507. doi: 10.1189/jlb.0212078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.O'Neill LA, Hardie DG. Nature. 2013;493:346–355. doi: 10.1038/nature11862. [DOI] [PubMed] [Google Scholar]
- 38.Xing J, Wang Q, Coughlan K, Viollet B, Moriasi C, Zou MH. Am J Pathol. 2013;182:1021–1030. doi: 10.1016/j.ajpath.2012.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Park CS, Bang BR, Kwon HS, Moon KA, Kim TB, Lee KY, Moon HB, Cho YS. Biochem Pharmacol. 2012;84:1660–1670. doi: 10.1016/j.bcp.2012.09.025. [DOI] [PubMed] [Google Scholar]
- 40.Eid AA, Lee DY, Roman LJ, Khazim K, Gorin Y. Mol Cell Biol. 2013;33:3439–3460. doi: 10.1128/MCB.00217-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim J, Kundu M, Viollet B, Guan KL. Nat Cell Biol. 2011;13:132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao X, Zmijewski JW, Lorne E, Liu G, Park YJ, Tsuruta Y, Abraham E. Am J Physiol Lung Cell Mol Physiol. 2008;295:L497–L504. doi: 10.1152/ajplung.90210.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zmijewski JW, Banerjee S, Bae H, Friggeri A, Lazarowski ER, Abraham E. J Biol Chem. 285:33154–33164. doi: 10.1074/jbc.M110.143685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zmijewski JW, Banerjee S, Abraham E. J Biol Chem. 2009;284:22213–22221. doi: 10.1074/jbc.M109.028902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dranka BP, Benavides GA, Diers AR, Giordano S, Zelickson BR, Reily C, Zou L, Chatham JC, Hill BG, Zhang J, Landar A, Darley-Usmar VM. Free Radic Biol Med. 2011;51:1621–1635. doi: 10.1016/j.freeradbiomed.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liang HL, Sedlic F, Bosnjak Z, Nilakantan V. Free Radic Biol Med. 2010;49:1550–1560. doi: 10.1016/j.freeradbiomed.2010.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mukhopadhyay P, Rajesh M, Hasko G, Hawkins BJ, Madesh M, Pacher P. Nat Protoc. 2007;2:2295–2301. doi: 10.1038/nprot.2007.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Frezza C, Cipolat S, Scorrano L. Nat Protoc. 2007;2:287–295. doi: 10.1038/nprot.2006.478. [DOI] [PubMed] [Google Scholar]
- 49.Bae HB, Zmijewski JW, Deshane JS, Tadie JM, Chaplin DD, Takashima S, Abraham E. Faseb J. 2011;25:4358–4368. doi: 10.1096/fj.11-190587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Choi HC, Song P, Xie Z, Wu Y, Xu J, Zhang M, Dong Y, Wang S, Lau K, Zou MH. J Biol Chem. 2008;283:20186–20197. doi: 10.1074/jbc.M803020200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 51.Weisova P, Anilkumar U, Ryan C, Concannon CG, Prehn JH, Ward MW. Biochim Biophys Acta. 2012;1817:744–753. doi: 10.1016/j.bbabio.2012.01.016. [DOI] [PubMed] [Google Scholar]
- 52.Nagashima S, Tokuyama T, Yonashiro R, Inatome R, Yanagi S. Journal of biochemistry. 2014;155:273–279. doi: 10.1093/jb/mvu016. [DOI] [PubMed] [Google Scholar]
- 53.Yonashiro R, Ishido S, Kyo S, Fukuda T, Goto E, Matsuki Y, Ohmura-Hoshino M, Sada K, Hotta H, Yamamura H, Inatome R, Yanagi S. EMBO J. 2006;25:3618–3626. doi: 10.1038/sj.emboj.7601249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sarraf SA, Raman M, Guarani-Pereira V, Sowa ME, Huttlin EL, Gygi SP, Harper JW. Nature. 2013;496:372–376. doi: 10.1038/nature12043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Korolchuk VI, Menzies FM, Rubinsztein DC. Autophagy. 2009;5:862–863. doi: 10.4161/auto.8840. [DOI] [PubMed] [Google Scholar]
- 56.Korolchuk VI, Menzies FM, Rubinsztein DC. FEBS Lett. 2010;584:1393–1398. doi: 10.1016/j.febslet.2009.12.047. [DOI] [PubMed] [Google Scholar]
- 57.Kraft C, Peter M, Hofmann K. Nat Cell Biol. 2010;12:836–841. doi: 10.1038/ncb0910-836. [DOI] [PubMed] [Google Scholar]
- 58.Min H, Xu M, Chen ZR, Zhou JD, Huang M, Zheng K, Zou XP. Cancer chemotherapy and pharmacology. 2014;74:167–176. doi: 10.1007/s00280-014-2451-7. [DOI] [PubMed] [Google Scholar]
- 59.Qi J, Gong J, Zhao T, Zhao J, Lam P, Ye J, Li JZ, Wu J, Zhou HM, Li P. Embo J. 2008;27:1537–1548. doi: 10.1038/emboj.2008.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zungu M, Schisler JC, Essop MF, McCudden C, Patterson C, Willis MS. Am J Pathol. 2011;178:4–11. doi: 10.1016/j.ajpath.2010.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Taylor EB, Rutter J. Biochem Soc Trans. 2011;39:1509–1513. doi: 10.1042/BST0391509. [DOI] [PubMed] [Google Scholar]
- 62.Campello S, Strappazzon F, Cecconi F. Biochim Biophys Acta. 2014;1837:451–460. doi: 10.1016/j.bbabio.2013.11.010. [DOI] [PubMed] [Google Scholar]
- 63.Xu S, Peng G, Wang Y, Fang S, Karbowski M. Mol Biol Cell. 2011;22:291–300. doi: 10.1091/mbc.E10-09-0748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Azzu V, Brand MD. J Cell Sci. 2010;123:578–585. doi: 10.1242/jcs.060004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zemirli N, Pourcelot M, Ambroise G, Hatchi E, Vazquez A, Arnoult D. The FEBS journal. 2014 doi: 10.1111/febs.12846. [DOI] [PubMed] [Google Scholar]
- 66.Cohen MM, Leboucher GP, Livnat-Levanon N, Glickman MH, Weissman AM. Mol Biol Cell. 2008;19:2457–2464. doi: 10.1091/mbc.E08-02-0227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Durr M, Escobar-Henriques M, Merz S, Geimer S, Langer T, Westermann B. Mol Biol Cell. 2006;17:3745–3755. doi: 10.1091/mbc.E06-01-0053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liu H, Yu S, Xu W, Xu J. Arterioscler Thromb Vasc Biol. 2012;32:2131–2140. doi: 10.1161/ATVBAHA.112.253385. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 69.Huang W, Yang C, Nan Q, Gao C, Feng H, Gou F, Chen G, Zhang Z, Yan P, Peng J, Xu Y. BioMed research international. 2014;2014:684765. doi: 10.1155/2014/684765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xu J, Wang S, Viollet B, Zou MH. PLoS One. 2012;7:e36717. doi: 10.1371/journal.pone.0036717. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.