Abstract
The structural manipulation of small molecule metabolites occurs in all organisms and plays a fundamental role in essentially all biological processes. Despite an increasing interest in developing new, non-enzymatic chemical reactions capable of functioning in the presence of living organisms, the ability of such transformations to interface with cellular metabolism and influence biological function is a comparatively underexplored area of research. This review will discuss efforts to combine non-enzymatic chemistry with microbial metabolism. We will highlight recent and historical uses of non-biological reactions to study microbial growth and function, the use of non-enzymatic transformations to rescue auxotrophic microorganisms, and the combination of engineered microbial metabolism and biocompatible chemical reactions for organic synthesis.
1.1 Introduction
Microorganisms are the most abundant and diverse organisms on Earth. Through billions of years of evolution they have acquired the ability to inhabit an astonishing array of environments [1]. These include the extreme temperatures found in deep-sea hydrothermal vents (hyperthermophiles) [2] and in subterranean ice sheets (psychrophiles) [3]; the high pressure of the Mariana Trench (piezophiles) [4]; and various sites in and on the human body (the human microbiota) [5]. To support growth in these habitats, many microbes have coevolved strategies to support their metabolism that involve chemistry not encoded by their genomes. In many cases survival is therefore dependent on a microbe’s ability to interface its metabolism with the surrounding chemical environment. Conceptually this parallels the way synthetic chemists make molecules in a laboratory setting, using non-enzymatic reagents and catalysts to synthesize molecules essential to our everyday lives.
Over the last several decades there has been a steadily growing interest in developing non-biological chemical transformations for use in a cellular setting [6–12]. Although such reactions have been demonstrated to occur in both cells and in living organisms and are powerful tools for studying natural biological processes (bioorthogonal chemistry), the question of whether or not their reactivity can interface with cellular metabolism and alter biological function remains comparatively underexplored. One approach to achieving this goal is the development of biocompatible chemistry: non-enzymatic reactions that alter the structures of metabolites as they are produced by living organisms. There are multiple ways in which biocompatible transformations could be integrated with cellular metabolism (Figure 1). They could modify the end products of metabolic pathways, support metabolic functions by generating key substrates or nutrients in vivo, or be fully integrated into cellular metabolism.
Figure 1.
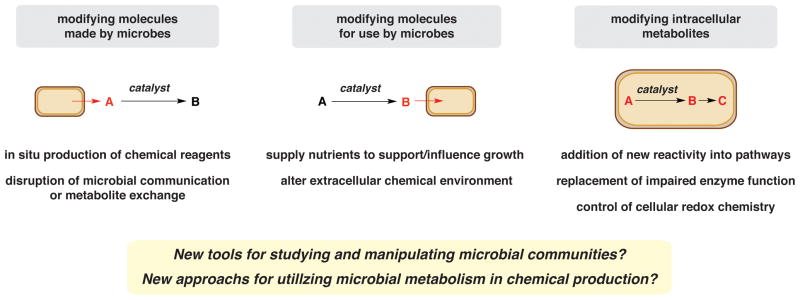
Approaches to interfacing biocompatible chemistry with microbial metabolism and potential applications of this technology.
One of the largest challenges encountered in merging non-enzymatic chemistry with metabolism is the apparent mismatch between the approaches used in synthetic organic chemistry (non-aqueous solvents, rare earth and transition metals, reactive intermediates, extreme temperatures and pH) and the growth conditions required to support a living organism (aqueous media, ambient temperature, neutral pH). The complexity of the cellular and extracellular environments and the typically low concentrations of cellular metabolites are also potential concerns. As has been the case with bioorthogonal reactions, we feel that these obstacles may be overcome through reaction screening optimization and that the potential benefits associated with the realization of this approach justify the challenges associated with developing such transformations.
Biocompatible chemistry would provide scientists with a unique toolkit for manipulating and augmenting biological function in vivo that would not require genetic manipulation and could be applied to both cultured and uncultured organisms. This review will discuss recent achievements in combining non-enzymatic chemistry with metabolism, with a particular emphasis on studies involving microorganisms. We will focus on non-enzymatic reactions that have a direct influence on metabolic function. We will not include examples that use solely genetic approaches to manipulate metabolism or studies that use non-enzymatic de-caging strategies to control protein function in vivo. We will highlight key experiments that have seeded interest in this topic and provide our perspective on the future challenges and opportunities for this area of research.
2.1 The role of non-enzymatic chemistry in natural microbial habitats
Before discussing examples of non-biological transformations that have been designed by chemists and chemical biologists to function in the presence of living systems, we will briefly discuss several examples of how microbes have evolved to utilize non-enzymatic reactions to facilitate growth in natural settings. The influence of non-enzymatic chemistry on microbial metabolism is a phenomenon often observed in natural microbial populations. Non-enzymatic transformations have also frequently been proposed to play important roles in the origin of life and metabolic pathways. In 1924 biochemist Aleksandr Oparin first hypothesized that carbon-based metabolism could have originated from non-enzymatic chemistry enabled by the highly reducing geochemical environment of the early Earth [13–16]. In support of this hypothesis, ferrous iron has been shown to catalyze transformations that in modern organisms constitute glycolysis and pentose phosphate pathway chemistry, suggesting that the origins of cellular metabolism could date back to non-enzymatic chemistry operating in the iron-rich oceans of the prebiotic world [17].
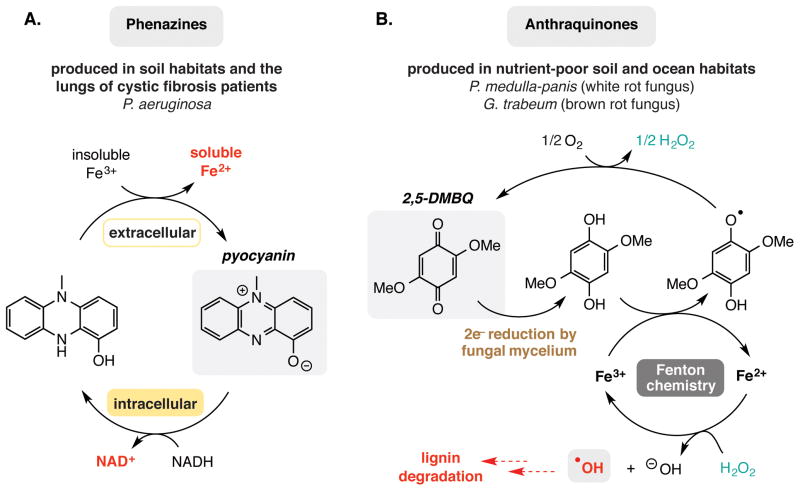
The chemistry of environmental iron impacts microbial survival in limiting habitats [18–22]. Under oxygen-limiting conditions many facultative anaerobic microbes use extracellular iron and manganese ions as terminal electron acceptors [23]. This use of redox-active environmental metals is especially remarkable due to the chemical challenges associated with their acquisition. For example, geological manganese and iron exist in metal oxides/oxyhydroxides that are inaccessible to reduction due to their insolubility. Microbes overcome this issue by excreting small molecules capable of solubilizing these metal ions (siderophores), as well as redox active small molecules that can directly reduce metal oxides (extracellular electron shuttles) [24]. An example of a microbe that uses redox-active small molecules to facilitate respiration of metals is the opportunistic pathogen Pseudomonas aeruginosa, which secretes redox-active pigments collectively known as phenazines [25]. Secreted, reduced phenazines support respiration under anaerobic conditions by transferring electrons to nearby extracellular oxidants (frequently Fe2+/3+). This generates ATP through the Entner-Doudoroff pathway, which involves the non-enzymatic regeneration of NAD+ from accumulating NADH (Figure 2A) [26]. Phenazines and Fe2+ have been detected in the sputum of cystic fibrosis patients at levels consistent with this process supporting the anaerobic survival of P. aeruginosa in this environment and contributing to chronic infection [27,28]. This observation has led to research into iron-chelation therapy as a treatment for persistent P. aeruginosa infections [29–31].
Figure 2.
Non-enzymatic transformations that influence the survival of environmental microbes. A. Extracellular reduction of iron(II) by secreted phenazines enables the survival of Pseudomonas aeruginosa. B. Extracellular iron reduction by secreted quinones facilitates lignin degradation by fungi.
Microbes in soil and aquatic ecosystems also use iron to support their metabolism by harnessing its reactivity to promote the non-enzymatic degradation of lignin [32]. Lignin is complex polymer containing aromatic functionality and is a critical component of plant cell walls. It represents one of the largest sources of fixed carbon on Earth and its biodegradation therefore occupies a significant role in global carbon cycling [33]. The white and brown rot fungi Perenniporia medulla-panis and Gloeophyllum trabeum degrade lignin to humic acids using an extracellular hydroquinone-quinone redox couple to environmental Fe3+ to produce hydrogen peroxide from oxygen [34]. This enables the non-enzymatic degradation of lignin via the generation of highly reactive hydroxyl radicals using Fenton chemistry (Figure 2B). In this context both G. trabeum and P. medulla-panis use non-enzymatic redox chemistry as a strategy to replace the function of lignin peroxidase, the enzyme used by other microbes to degrade lignin. The humic acids released in this process serve as a primary source of carbon and as terminal electron acceptors for a variety of organisms in nutrient and oxygen-poor soil and aquatic environments.
3.1 Developing non-enzymatic chemistry to control biological function
The ability of chemists to develop chemical reactions that extend beyond what is available to living organisms in natural environments opens up the possibility of developing new ways of linking microbial metabolism to non-enzymatic chemistry. Specifically, biocompatible non-enzymatic chemistry could enable the chemical manipulation of small organic molecules such as cofactors and metabolites in ways that could impact cellular function. Indeed, chemists have achieved this goal by harnessing the reactivity of both stoichiometric small organic reagents and transition metal catalysts.
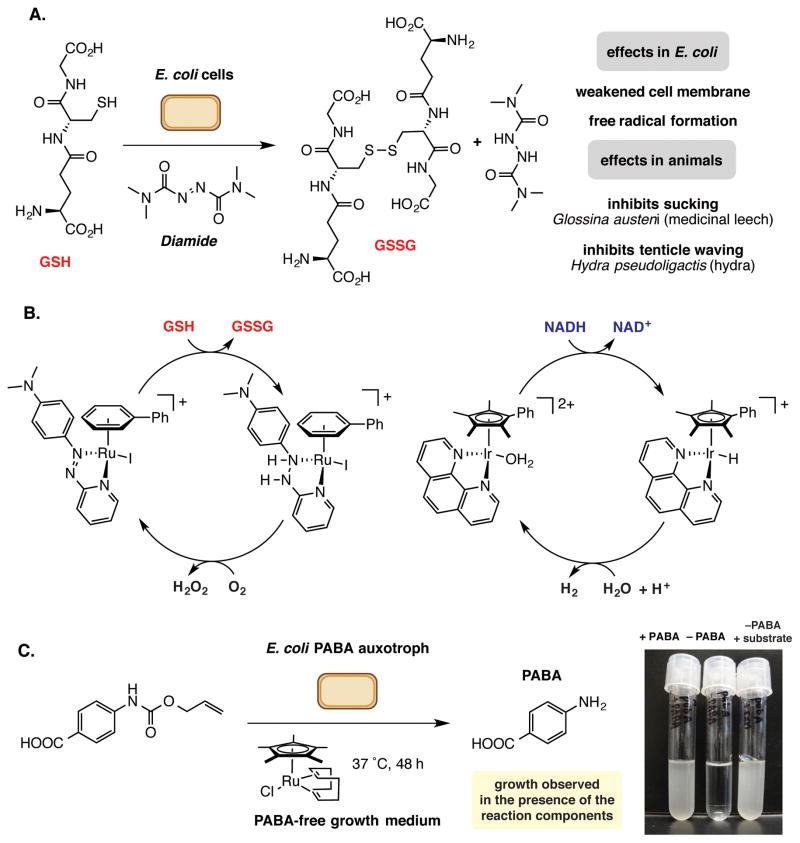
Much of the seminal work in this area has focused on using non-enzymatic redox chemistry to manipulate the redox state of cellular components. In 1969, Kosower reported the stoichiometric use of the organic reagent diamide to oxidize the endogenous antioxidant glutathione (GSH) to glutathione disulfide (GSSG) in vivo in E. coli. (Figure 3A) [35]. In addition to detoxifying reactive oxygen species and free radicals in the cell, GSH also serves as a cofactor for enzymes that mediate redox chemistry. Under normal cellular conditions over 90% of GSH is in its reduced form. In vivo manipulation of GSH and GSSG levels impacts a variety of cellular functions, especially in cells recovering from oxidative damage. In 2014, Ward and coworkers also employed stoichiometric diamide to oxidize the GSH present in E. coli cell lysates, improving the efficiency of transition metal mediated reactions in this environment [36]. Oxidation of GSH in vivo has also been accomplished using transitional metal complexes (Figure 3B). Sadler and coworkers have shown the ruthenium piano-stool complexes efficiently oxidize GSH to GSSG without issues of poisoning for living human ovarian and lung cancer cell lines [37]. The same group has also reported an iridium-catalyzed transfer-hydrogenation that can operate in vivo to modulate redox balance in cells by converting the co-factor NADH to NAD+ and hydrogen [38]. The non-enzymatic oxidation of NADH has the potential to impact the functions of enzymes that use this redox cofactor and may enhance or reduce growth depending on an organism’s mode of metabolism. Such catalysts could also potentially alter redox balance in ways that could be beneficial in the context of engineered metabolic pathways.
Figure 3.
Examples of non-enzymatic chemistry with the potential to control biological functions. A. Diamide can be used to oxidize glutathione in living organisms, altering intracellular redox chemistry and compromising cell membrane integrity. B. Ru-catalyzed oxidation of glutathione and Ir-catalyzed generation of oxidized nicotinamide via transfer hydrogenation. C. Generation of the essential metabolite (PABA) using a Ru-catalyzed alloc-deprotection reaction enables the growth of auxotrophic strains of E. coli.
While the chemistry mentioned above affects the homeostasis of organisms by oxidizing cellular reductants, researchers have also been interested in developing catalytic anti-oxidants to reduce the strain placed on organisms and cells by excess reactive oxygen species [39]. The most prevalent catalytic antioxidants are manganese(II) porphyrin complexes that mimic the activity of superoxide dismutase (SOD), reducing superoxide, peroxynitrite, carbonate anion radical, hypochlorite, nitric oxide, lipid peroxyl radicals, and alkoxyl radicals, all of which can cause oxidative stress to cells [40]. There has been substantial interest in developing SOD mimics as pharmaceuticals, prompting many efforts to understand the in vivo activities of metalloporphyrins. SOD-deficient E. coli mutants have proven to be useful tools for investigating the minimal requirements of a functional SOD mimic [41]. Mutants lacking SOD enzymes exhibit growth defects in certain minimal media and restoration of growth is indicative of antioxidant activity. This system has enabled the efficient identification of catalytic antioxidants with high activity, low toxicity, and efficient cellular uptake.
Biocompatible chemistry can also be used to generate metabolites that play essential roles in metabolism. A recent study from our own laboratory has demonstrated the possibility of controlling microbial growth using non-enzymatic reactions (Figure 3C) [42]. In this report we were able to support the growth of multiple p-aminobenzoic acid auxotrophs via the [Cp*Ru(cod)Cl]-catalyzed deprotection of N-alloc-PABA in situ. Similarly, we used an iron-mediated hydroxylation reaction to rescue the growth of a p-hydroxybenzoic acid auxotroph. In both cases, the growth of the auxotrophic organism was dependent upon the success of the non-enzymatic reaction. Rescued cultures retained their auxotrophies upon re-inoculation into media lacking key nutrients, indicating that rescue was not the result of genetic changes in the organism over the course of the experiment. The product yields and/or isomer distributions were low for both reactions, highlighting how minor modulations of metabolite levels in a cell can have a dramatic impact on growth and function. Both of these transformations rescue auxotrophy by providing alternate routes to essential compounds and not by directly mimicking the missing biochemical function. The development of biocompatible catalysts that can replace the function of an essential metabolic enzyme is an important future goal that could also represent a new strategy for treating metabolic disorders.
4.1 Combining microbial metabolism and non-enzymatic chemistry for small molecule production
In addition to influencing core biological functions, the merging of biocompatible chemistry with microbial metabolism provides an opportunity to modify metabolites in ways that benefit efforts to utilize these organisms for chemical production. Advances in the field of synthetic biology have enabled the use of engineered microbes for the production of small molecules such as pharmaceuticals and commodity chemicals. Despite these achievements, many important chemical scaffolds cannot be accessed using this approach due to the differences in the scope of reactivity available to enzymes in comparison with that available to non-enzymatic catalysts and reagents [43]. A possible solution to this problem could involve combining biocompatible reactions with engineered metabolism for small molecule synthesis, a strategy that has been relatively underexplored [44]. To the best of our knowledge, the cooperative use of non-enzymatic chemistry from organic synthesis and microbial metabolism originated with Neuberg’s bisulfite-steered glycerol fermentations during the First World War [45,46]. Neuberg observed substantial overproduction of glycerol from glucose by Saccharomyces cerevisiae when sodium bisulfite was added to the fermentation. It was later discovered that the bisulfite anion formed a stable adduct with acetaldehyde, a metabolite generated en route to ethanol, the typical fermentation end product. Bisulfite adducts are used in organic chemistry as a method for protecting aldehydes [47]. This non-enzymatic reaction interfered with the reduction of acetaldehyde to ethanol, forcing accumulating NADH to be reoxidized to NAD+ via an alternate pathway, the reduction of dihydroxyacetone phosphate, which yielded glycerol as the primary product. This process contributed significantly to the German war effort by allowing the manufacture of glycerol from sugar for use in the production of explosives.
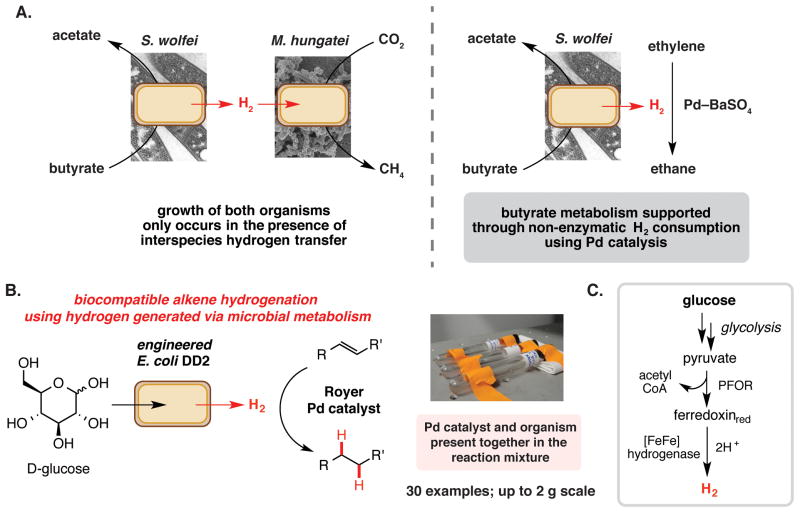
Our lab is developing new biocompatible chemical transformations that can be combined with microbial metabolism to enable chemical production. We have begun this work by identifying transition metal mediated reactions that can use metabolites generated in situ by living bacteria. Our initial studies focused on transformations that could engage the diffusible microbial metabolite hydrogen gas. Bacterially-produced hydrogen had previously been utilized for non-enzymatic chemistry on an analytical scale. Mountfort and co-workers demonstrated the use of hydrogenation chemistry to decouple the metabolism of the proton-reducing syntrophic bacterium from a partner methanogen (Figure 4A) [48]. The syntrophic bacterium Syntrophomonas wolfei and the methanogen Metanospirillum hungatei co-exist in an obligate metabolic partnership that couples the thermodynamically unfavorable oxidation of butyrate to acetate (acetogenesis) to the thermodynamically favorable production of methane from CO2 (methanogenesis) via the interspecies exchange of hydrogen. Culturing these organisms in the presence of ethylene gas and super-stoichiometric amounts of Pd-BaSO4 resulted in the formation of ethane. This observation indicated that in the absence of a partner methanogen, S. wolfei was able to couple butyrate oxidation and hydrogen production to the reduction of ethylene by the palladium catalyst. This study demonstrated for the first time that it is possible to decouple the metabolic functions of two obligate syntrophs using a non-enzymatic transition metal catalyzed hydrogenation.
Figure 4.
Chemical synthesis using biocompatible chemistry and microbial metabolism. A. A non-enzymatic hydrogenation reaction can decouple microbial syntrophy. Acetogenesis by S. wolfei in the absence of interspecies hydrogen transfer is observed with the addition of ethylene and a heterogeneous palladium catalyst. B. Hydrogen produced by engineered E. coli can engage in non-enzymatic alkene reduction reactions using the Royer palladium catalyst. C. The engineered metabolic pathway in E. coli DD2 responsible for H2 production from D-glucose. Royer palladiums catalyst = 2.44% palladium on polyethyleneimine (PEI)/silica gel.
Inspired by this observation, our laboratory recently reported a biocompatible alkene hydrogenation reaction that utilizes hydrogen generated by living bacteria (Figure 4B) [49]. We used a strain of E. coli engineered to produce hydrogen via a single inducible pathway consisting of a pyruvate ferredoxin oxidoreductase, a ferredoxin, and a [Fe–Fe] hydrogenase (Figure 4C). After demonstrating that heterogeneous transition metal catalysts could promote alkene hydrogenation in growth media using bacterially-generated hydrogen, screening identified the Royer palladium catalyst (palladium on polyethyleneimine/SiO2) as an optimal hydrogenation catalyst. After further optimization, we identified conditions that could be applied to a range of alkene substrates and could be utilized for gram scale reactions. Growth of the hydrogen-producing E. coli cells was unaffected by the reaction components. To the best of our knowledge, this is the first time that the metabolic output of a living organism and a non-enzymatic chemical transformation have been directly combined for preparative scale chemical synthesis. Intended to compliment synthetic biology efforts, this study represents a crucial first step towards integrating biocompatible reactions with cellular metabolism for small molecule synthesis.
5.1 Outlook
The specific examples summarized here demonstrate the ability of non-enzymatic, biocompatible transformations to influence the metabolism of microorganisms. We believe that the timing is right for an expansion of this approach due to recent advances in organic chemistry and microbiology. Although discovering and optimizing biocompatible reactions poses technical challenges, many of these are conceptually identical to the obstacles faced by synthetic chemists in developing new reactions. Biocompatible chemistry may therefore take advantage of new, high-throughput approaches to reaction discovery that have been implemented by organic chemists [50–53]. Additionally, organic chemists are continuously developing mild bond-forming methods that operate under ambient and aqueous reaction conditions [54]. Access to increasingly powerful tools for synthesizing and assembling DNA has also made it possible to manipulate microbial metabolic pathways in new ways with unprecedented efficiency [55]. Microbial genome sequencing efforts are also rapidly expanding the ‘toolkit’ of enzymes available for metabolic engineering [56–59]. These advances will continue to expand the range of metabolites that can be accessed using microbial metabolism and interfaced with non-enzymatic chemistry, as well as enhance the speed with which such processes are developed.
What are the most exciting potential future applications of interfaced chemistry and metabolism? In the context of chemical production, interfacing biocompatible reactions with the products of engineered microbial metabolism could rapidly diversify the structures of metabolites. The ability to access a range of potentially new chemical scaffolds directly from fermentations could change the way engineered organisms are used for chemical production, making microbial metabolism a useful source of new molecular scaffolds that could be evaluated for desired properties in addition to providing a route to chemicals of established importance. Engineered microbial metabolism could also be used to generate unstable, toxic, or inefficiently manufactured reagents directly in reaction mixtures, obviating the need to transport and store large quantities of these materials. Before this goal can be realized, biocompatible reactions must be optimized for process scale applications with regards to cost and efficiency.
Non-enzymatic reactions that can influence cellular metabolism may also have applications in microbiology and medicine. One can envision developing biocompatible reactions that modify the structures of metabolites exchanged in microbial communities. Transformations of this type could potentially alter community structure by disrupting cell-cell communication (quorum sensing) or manipulating metabolite exchange. This technology could be applied to reduce the susceptibility of communities like the human gut microbiota to dysbiosis or to invasion by pathogens. Reactions that interface with microbial metabolites could also perhaps aid in efforts to grow previously unculturable microbes by replacing essential metabolic interactions. Thinking beyond microorganisms, the development of biocompatible transformations that can directly replace the functions of metabolic enzymes could provide a new approach to treating human disease, particularly metabolic disorders that arise from deficiencies in enzyme function. Exploring both of these applications will require the identification of new catalysts that are not only functional and biocompatible, but also possess improved efficiency and selectivity.
6.1 Conclusion
The use of non-enzymatic, biocompatible reactions represents a new way to utilize, manipulate, and complement microbial metabolism. Small molecules synthesized by living organisms participate in and control an array of biological processes, making biocompatible chemistry a potentially powerful approach to influencing cellular function. Recent work has shown that reaction discovery and optimization efforts similar to those used in synthetic organic chemistry can access non-enzymatic transformations that function cooperatively with the metabolism of a living organism. We hope that these studies inspire future efforts to develop further transformations the can be utilized in this way, as well as new applications of natural and engineered metabolism in chemical manufacturing.
Highlights.
Microorganisms have evolved to utilize non-enzymatic reactions to facilitate their survival in many environments.
Non-enzymatic reactions developed by chemists can be interfaced with microbial metabolism and can have an influence on biological function.
The products of engineered metabolism can be combined with non-enzymatic reactions to enable chemical synthesis.
Acknowledgments
This work was supported by the National Institutes of Health (DP2 GM105434). S. Wallace acknowledges receipt of a Marie Curie International Outgoing Fellowship from the European Commission.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
* of special interest
** of outstanding interest
- 1.Curtis TP, Sloan WT. Prokaryotic diversity and its limits: Microbial community structure in nature and implications for microbial ecology. Curr Opin Microbiol. 2004;7:221–226. doi: 10.1016/j.mib.2004.04.010. [DOI] [PubMed] [Google Scholar]
- 2.Dick GJ, Anantharaman K, Baker BJ, Li M, Reed DC, Sheik CS. The microbiology of deep-sea hydrothermal vent plumes: ecological and biogeographic linkages to seafloor and water column habitats. Front Microbiol: Extreme Microbiol. 2013;4:1–16. doi: 10.3389/fmicb.2013.00124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Christner BC, Priscu JC, Achberger AM, Barbante C, Carter SP, Christianson K, Michaud AB, Mikucki JA, Mitchell AC, Skidmore ML, et al. A microbial ecosystem beneath the West Antarctic ice sheet. Nature. 2014;512:310–313. doi: 10.1038/nature13667. [DOI] [PubMed] [Google Scholar]
- 4.Glud RN, Wenzhöfer F, Middelboe M, Oguri K, Turnewitsch R, Canfield DE, Kitazato H. High rates of microbial carbon turnover in sediments in the deepest oceanic trench on Earth. Nat Geosci. 2013;6:284–288. [Google Scholar]
- 5.The Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McKay CS, Finn MG. Click Chemistry in Complex Mixtures: Bioorthogonal Bioconjugation. Chem Biol. 2014;21:1075–1101. doi: 10.1016/j.chembiol.2014.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shih H-W, Kamber DN, Prescher JA. Building better bioorthogonal reactions. Curr Opin Chem Biol. 2014;21:103–111. doi: 10.1016/j.cbpa.2014.07.002. [DOI] [PubMed] [Google Scholar]
- 8.Lang K, Chin JW. Cellular incorporation of unnatural amino acids and bioorthogonal labeling of proteins. Chem Rev. 2014;114:4764–4806. doi: 10.1021/cr400355w. [DOI] [PubMed] [Google Scholar]
- 9.Lang K, Chin JW. Bioorthogonal reactions for labeling proteins. ACS Chem Biol. 2014;9:16–20. doi: 10.1021/cb4009292. [DOI] [PubMed] [Google Scholar]
- 10.Patterson DM, Nazarova LA, Prescher JA. Finding the right (bioorthogonal) chemistry. ACS Chem Biol. 2014;9:592–605. doi: 10.1021/cb400828a. [DOI] [PubMed] [Google Scholar]
- 11.Bertozzi CR. A decade of bioorthogonal chemistry. Acc Chem Res. 2011;44:651–653. doi: 10.1021/ar200193f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sletten EM, Bertozzi CR. Bioorthogonal chemistry: fishing for selectivity in a sea of functionality. Angew Chem Int Ed. 2009;48:6974–6998. doi: 10.1002/anie.200900942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oparin AI. Proiskhozhdenie zhinzny. Idz. Moskovski Rabochii; Moscow: 1924. [Google Scholar]
- 14.Oparin AI. Vozniknovenie zhinzny na zemle. Izd. AN SSSR; Moscow: 1936. [Google Scholar]
- 15.Oparin AI. The origin of Life. Moscow 1938 and New York, Dover: 1952. [Google Scholar]
- 16.Miller SL, Schapf JW, Lazcano A. Oparin’s “origin of life”: sixty years later. J Mol Evol. 1997;44:351–353. [PubMed] [Google Scholar]
- 17*.Keller MA, Turchyn AV, Ralser M. Non-enzymatic glycolysis and pentose phosphate pathway-like reactions in a plausible Archean ocean. Mol Syst Biol. 2014;10:1–12. doi: 10.1002/msb.20145228. Ferrous iron was demonstrated to catalyze a range of metabolically relevant transformations in an artificial Archean ocean, providing support for the hypothesis that the origins of cellular metabolism could stem from non-enzymatic chemistry operating in the iron-rich oceans of the prebiotic world. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Emerson D, Fleming EJ, McBeth JM. Iron-oxidising bacteria: an environmental and genomic perspective. Annu Rev Microbiol. 2010;64:561–583. doi: 10.1146/annurev.micro.112408.134208. [DOI] [PubMed] [Google Scholar]
- 19.Fleming EJ, Cetinić I, Chan CS, Whitney King D, Emerson D. Ecological succession among iron-oxidizing bacteria. ISME J. 2014;8:804–815. doi: 10.1038/ismej.2013.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bird LJ, Bonnefoy V, Newman DK. Bioenergetic challenges of microbial iron metabolisms. Trends Microbiol. 2011;19:330–340. doi: 10.1016/j.tim.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 21.Cornelis P, Andrews SC. Iron uptake and homeostasis in microorganisms. Caister Academic Press; UK: Jun, 2010. [Google Scholar]
- 22.Kopf SH, Henny C, Newman DK. Ligand-enhanced abiotic iron oxidation and the effects of chemical versus biological iron cycling in anoxic environments. Environ Sci Technol. 2013;47:2602–2611. doi: 10.1021/es3049459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lovely DR. Dissimilatory metal reduction. Annu Rev Microbiol. 1993;47:362–290. doi: 10.1146/annurev.mi.47.100193.001403. [DOI] [PubMed] [Google Scholar]
- 24.Richter K, Schicklberger M, Gescher J. Dissimilatory reduction of extracellular electron acceptors in anaerobic respiration. Appl Environ Microb. 2011;78:913–921. doi: 10.1128/AEM.06803-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Y, Kern SE, Newman DK. Endogenous phenazine antibiotics promote anaerobic survival of Pseudomonas aeruginosa via extracellular electron transport. J Bacteriol. 2010;192:365–369. doi: 10.1128/JB.01188-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thomashow LS. Phenazines in the environment: microbes, habitats, and ecological relevance. In: Chincholkar S, Thomashow LS, editors. Microbial Phenazines. Chapter 10. Heidelberg, Germany: Springer-Verlag; 2013. [Google Scholar]
- 27.Hunter RC, Klepac-Ceraj V, Lorenzi MM, Grotzinger H, Martin TR, Newman DK. Phenazine content in the cystic fibrosis respiratory tract negatively correlates with lung function and microbial complexity. Am J Respir Cell Mol Biol. 2012;47:738–745. doi: 10.1165/rcmb.2012-0088OC. [DOI] [PubMed] [Google Scholar]
- 28*.Hunter RC, Asfour F, Dingemans J, Osuna BL, Samad T, Malfroot, Cornelis P, Newman DK. Ferrous iron is a significant component of bioavailable iron in cystic firbrosis airways. mBio. 2013;4:1–8. doi: 10.1128/mBio.00557-13. Iron acquisition is important for the growth of microbial community in cystic fibrosis patients, and limiting iron acquisition can disrupt microbial communities, perhaps leading to new therapeutic strategies. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O’May CY, Sanderson K, Roddam LF, Kirov LF, Reid DW. Iron-binding compounds impair Pseudomonas aeruginosa biofilm formation, especially under anaerobic conditions. J Med Microbiol. 2009;58:765–773. doi: 10.1099/jmm.0.004416-0. [DOI] [PubMed] [Google Scholar]
- 30.Moreau-Marquis S, O’Toole GA, Stanton BA. Tobramycin and FDA-approved iron chelators eliminate Pseudomonas aeruginosa biofilms on cystic fibrosis cells. Am J Respir Cell Mol Biol. 2009;41:305–313. doi: 10.1165/rcmb.2008-0299OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Musk DJ, Jr, Hergenrother PJ. Chelated iron sources are inhibitors of Pseudomonas aeruginosa biofilms and distribute efficiently in an in vitro model of drug delivery to the human lung. J Appl Microbiol. 2008;105:380–388. doi: 10.1111/j.1365-2672.2008.03751.x. [DOI] [PubMed] [Google Scholar]
- 32.Dashtban M, Schraft H, Syed TA, Qin W. Fungal biodegradation and enzymatic modification of lignin. Int J Biochem Mol Biol. 2010;1:36–50. [PMC free article] [PubMed] [Google Scholar]
- 33.Jensen KA, Jr, Houtman CJ, Ryan ZC, Hammel KE. Pathways for extracellular fenton chemistry in the brown rot basidiomycete Gloeophyllum trabeum. Appl Environ Microb. 2001;67:2705–2711. doi: 10.1128/AEM.67.6.2705-2711.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gómez-Toribio V, García-Martín AB, Martínez MJ, Martínez AT, Guillén F. Induction of extracellular hydroxyl radical production by white-rot fungi through quinone redox cycling. Appl Environ Microb. 2009;75:3944–3953. doi: 10.1128/AEM.02137-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kosower EM, Kowoser NS. Lest I forget thee, glutathione…. Nature. 1969;224:117–120. doi: 10.1038/224117a0. [DOI] [PubMed] [Google Scholar]
- 36*.Wilson YM, Duerrenberger M, Nogueira ES, Ward TR. Neutralizing the detrimental effect of glutathione on precious metal catalysis. J Am Chem Soc. 2014;136:8928–8932. doi: 10.1021/ja500613n. Various non-enzymatic oxidants were assessed for their ability to oxidize glutathione. Diamide was identified as an efficient and biocompatible non-enzymatic oxidant, permitting the Ir-catalyzed transfer hydrogenation of imines using a streptavidin-based artificial imine reductase in E. coli cell lysates. [DOI] [PubMed] [Google Scholar]
- 37.Dougan SJ, Habtermariam A, McHale SE, Parsons S, Sadler PJ. Catalytic organometallic anticancer complexes. Proc Natl Acad Sci U S A. 2008;105:11628–11633. doi: 10.1073/pnas.0800076105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Betanzos-Lara S, Liu Z, Habtermariam A, Pizarro AM, Qamar B, Sadler PJ. Organometallic ruthenium and iridium transfer-hydrogenation catalysts using coenzyme NADH as a cofactor. Angew Chem Int Ed. 2012;51:3897–3900. doi: 10.1002/anie.201108175. [DOI] [PubMed] [Google Scholar]
- 39.Batinic-Haberle I, Rajic Z, Tovmasyan A, Reboucas JS, Ye X, Leong KW, Dewhirst MW, Vujaskovic Z, Benov L, Spasojevic I. Diverse functions of cationic Mn(III) N-substituted pyridylporphyrins, recognized as SOD mimics. Free Radic Biol Med. 2011;51:1035–1053. doi: 10.1016/j.freeradbiomed.2011.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miriyala S, Spasojevic I, Tovmasyan A, Salvemini D, Vujaskovic Z, St Clair D, Batinic-Haberle I. Manganese superoxide dismutase, MnSOD and its mimics. Biochim Biophys Acta. 2012;1822:794–814. doi: 10.1016/j.bbadis.2011.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tovmasyan A, Reboucas JS, Benov L. Simple biological systems for assessing the activity of superoxide dismutase mimics. Antioxid Redox Signal. 2014;20:2416–2436. doi: 10.1089/ars.2013.5576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42**.Lee Y, Umeano A, Balskus EP. Rescuing auxotrophic microorganisms with nonenzymatic chemistry. Angew Chem Int Ed. 2013;52:11800–11803. doi: 10.1002/anie.201307033. Two transition metal catalyzed transformations were shown to support the growth of auxotrophic strains of E. coli via the generation of essential nutrients in vivo. Growth of the organisms was shown to be dependent on the success of the non-enzymatic reaction. This study demonstrates the use of a non-enzymatic reaction to control biological function. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Keasling JD, Mendoza A, Baran PS. Synthesis: a constructive debate. Nature. 2012;492:188–189. doi: 10.1038/492188a. [DOI] [PubMed] [Google Scholar]
- 44.Wallace S, Balskus EP. Opportunities for merging chemical and biological synthesis. Curr Opin Biotechnol. 2014;30:1–8. doi: 10.1016/j.copbio.2014.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Neuberg C, Hirsch J. Uber den Verlauf der alkoholischen-Garung bei alkalischer Reaktion. II. Garung mit lebender Hefe in alkalischen Losungen. Biochem Z. 1919;96:175–202. [Google Scholar]
- 46.Neuberg C, Hirsch J. Die dritte Vergarungform des Zurkers. Biochem Z. 1919;100:304–322. [Google Scholar]
- 47.Wuts PGM, Greene TW. Sodium bisulfite adducts: RCH(OH)SO3Na. Greene’s Protective Groups in Organic Synthesis. 4. Hoboken, New Jersey, USA: John Wiley & Sons, Inc; 2006. [Google Scholar]
- 48.Kaspar HF, Holland AJ, Mountfort DO. Simultaneous butyrate oxidation by Syntrophomonas wolfei and catalytic olefin reduction in absence of interspecies hydrogen transfer. Arch Microbiol. 1987;147:334–339. [Google Scholar]
- 49**.Sirasani G, Tong L, Balskus EP. A biocompatible alkene hydrogenation merges organic synthesis with microbial metabolism. Angew Chem Int Ed. 2014;53:7785–7788. doi: 10.1002/anie.201403148. Hydrogen generated via engineered microbial metabolism was utilized in a non-enzymatic alkene hydrogenation reaction using a biocompatible palladium catalyst (Royer catalyst). This was demonstrated on a range of alkene substrates and on a preparative scale. This study represents the first use of microbial metabolism as a source of reagents for preparative chemical synthesis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Robbins DW, Hartiwig JF. A simple, multidimensional approach to high-throughput discovery of catalytic reactions. Science. 2011;333:1423–1427. doi: 10.1126/science.1207922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McNally A, Prier CK, MacMillan DW. Discovery of an α-amino C–H arylation reaction using the strategy of accelerated serendipity. Science. 2011;334:1114–1117. doi: 10.1126/science.1213920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Collins KD, Gensch T, Glorius F. Contemporary screening approaches to reaction discovery and development. Nat Chem. 2014;6:859–871. doi: 10.1038/nchem.2062. [DOI] [PubMed] [Google Scholar]
- 53.Collins KD, Rühling A, Glorius F. Application of a robustness screen for the evaluation of synthetic organic methodology. Nat Prot. 2014;9:1348–1353. doi: 10.1038/nprot.2014.076. [DOI] [PubMed] [Google Scholar]
- 54.Kolodych S, Rasolofonjatovo E, Chaumontet M, Nevers M-C, Créminon, Taran F. Discovery of chemoselective and biocompatible reactions using a high-throughput immunoassay screening. Angew Chem Int Ed. 2013;52:12056–12060. doi: 10.1002/anie.201305645. [DOI] [PubMed] [Google Scholar]
- 55.Lee JW, Na D, Park JM, Lee J, Choi S, Lee SY. Systems metabolic engineering of microorganisms for natural and non-natural chemicals. Nat Chem Bio. 2012;8:536–546. doi: 10.1038/nchembio.970. [DOI] [PubMed] [Google Scholar]
- 56.Woolston BM, Edgar S, Stephanopoulos G. Metabolic engineering: past and future. Annu Rev Chem Biomol Eng. 2013;4:259–288. doi: 10.1146/annurev-chembioeng-061312-103312. [DOI] [PubMed] [Google Scholar]
- 57.Chen Y, Nielsen J. Advances in metabolic pathway and strain engineering paving the way for sustainable production of chemical building blocks. Curr Opin Biotechnol. 2013;24:965–972. doi: 10.1016/j.copbio.2013.03.008. [DOI] [PubMed] [Google Scholar]
- 58.Keasling J. Manufacturing molecules through metabolic engineering. Science. 2010;330:1355–1358. doi: 10.1126/science.1193990. [DOI] [PubMed] [Google Scholar]
- 59.Carothers JM, Goler JA, Keasling JD. Chemical synthesis using synthetic biology. Curr Opin Biotechnol. 2009;20:498–503. doi: 10.1016/j.copbio.2009.08.001. [DOI] [PubMed] [Google Scholar]