Abstract
Cell division in bacteria requires the construction of two new polar caps for the daughter cells. To constrict the cell membrane and build these new surface layers, bacteria employ a multiprotein machine called the divisome. Over the years, most of the essential division proteins have been identified and localized to the ring-like divisome apparatus. The challenge now is to determine the molecular function of these factors, how they cooperate to bring about the dramatic transformation of the mother cell envelope, and what coordinates their activity with other major cell cycle events. In this review, we discuss recent progress in these areas with an emphasis on results from the model organisms Escherichia coli and Bacillus subtilis.
Introduction
The bacterial cell cycle culminates with the onset of cell division. The process initiates with the polymerization of the tubulin-like FtsZ protein into a ring structure (the Z-ring) just underneath the cytoplasmic membrane [1,2**]. Following Z-ring assembly, numerous essential and non-essential division proteins are recruited to midcell to form the mature division apparatus called the divisome or the septal ring [3]. Over the years, most, if not all, of the core proteins required for divisome activity have likely been identified [3,4]. A great deal has also been learned about the regulators that control Z-ring positioning to ensure that division takes place at the appropriate location. Despite this progress, major questions remain unanswered. Not all of the factors controlling Z-ring formation are known, including those that coordinate its assembly with the replication and segregation of the chromosome. Also, the precise functions of many core division proteins remain to be determined. Finally, although the steps of divisome assembly have been well characterized, the factors controlling the switch from an assembly phase to active cell constriction remain largely mysterious. This review focuses on recent work that has shed light on these outstanding questions. For a more in-depth overview of cell division, the reader is referred to several excellent reviews [3–6].
Connecting Z-ring formation to the chromosome
In the model bacteria Escherichia coli and Bacillus subtilis, the regulation of Z-ring placement is mediated by two negative regulators: the Min system and the nucleoid [7–11] (Figure 1). The output of the Min system is the FtsZ antagonist MinC, which together with its partner protein MinD, interferes with Z-ring formation [12–17*]. In E. coli the MinCD complex oscillates from pole-to-pole [13,14,18], whereas in B. subtils it is targeted to both cell poles [19]. However, the end result is the same in both cases; polar Z-ring formation is inhibited, and midcell Z-ring assembly is favored.
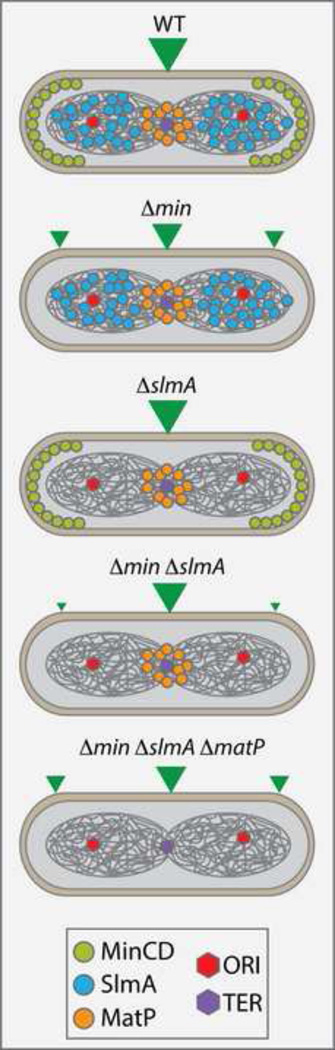
Figure 1. Determinants of division site positioning in E. coli.
Shown is an illustration summarizing the results of Bailey et al. 2014 [29**] showing that MatP and the Ter macrodomain of the chromosome can serve as a determinant of division site positioning in addition to Min and SlmA. Green triangles indicate possible division sites with their size reflecting preference for a particular site. See text for details.
The phenomenon of nucleoid occlusion reflects the negative effect of the chromosome on division [9,10]. Division inhibitors that associate with the nucleoid to mediate nucleoid occlusion were identified several years ago: Noc in B. subtilis and SlmA in E. coli [20,21]. The target of Noc regulation remains unknown. SlmA, on the other hand, directly antagonizes FtsZ assembly [22,23*,24,25*,26,27**]. Irrespective of their precise molecular target, Noc and SlmA share a surprising number of features considering that they belong to different protein families. Both proteins bind to distinct, yet specific, DNA sequences that are broadly distributed around the origin proximal two-thirds of their respective chromosomes, but absent near the replication terminus (Ter region) [22,24,28]. Coupled with the known dynamics of chromosome regions during the replication cycle, this binding site distribution is thought to be one of the possible mechanisms for coordinating chromosome replication and segregation with division [22,24,28] (Figure 1). Mutants defective for the nucleoid occlusion proteins also share the property of being synthetically lethal with Min system inactivation [20,21]. Cells lacking both systems fail to divide in rich medium and form long filamentous cells [20,21]. Interestingly, Z-ring formation is not completely random in these cells. Robust structures were still primarily observed between segregated nucleoids in the cell filaments. It was thus suggested that additional positional queues exist to guide Z-ring formation and position it relative to the chromosome [20,21].
A breakthrough in this area was recently reported by Bailey and co-workers [29**]. Their quantitative study of cell division positioning in Min− SlmA− E. coli cells grown in minimal medium, a condition previously shown to suppress the synthetic lethal phenotype [21], revealed that ΔslmA ΔminC cells divided more accurately at midcell than a singe ΔminC mutant [29**]. Surprisingly, they also observed a dramatic drop in the number of polar (minicell) divisions displayed by the ΔslmA ΔminC mutant relative to cells lacking MinC alone, which showed the classic minicell phenotype [29**]. These findings t hus suggested that a new positional marker at midcell becomes a dominant feature guiding Z-ring assembly when SlmA is inactivated in ΔminC cells. Further investigation implicated the chromosomal terminus organization protein MatP [30,31*] as the potential marker [29**]. This possibility was intriguing because MatP interacts with the ZapB protein, which together with ZapA associates with FtsZ and helps to coalesce the Z-ring structure [32**–35*]. Espeli and co-workers [32**] showed that this network of interactions is important for “anchoring” the Ter chromosomal domain to midcell after it localizes to this region during replication. Bailey and colleagues show that in ΔslmA ΔminC cells these interactions can also stimulate Z-ring formation at midcell [29**]. It currently remains to be determined whether the Ter region provides an important guide for Z-ring positioning in wild-type cells or if the connection between the Z-ring and the Ter domain simply functions to maintain and/or stabilize the midcell localization of these macromolecular structures. In either case, these two reports [29**,32**] highlight the potential for distinct domains in the chromosome and their associated binding proteins to function as landmarks for the proper organization of cellular processes.
Using outgrowing B. subtilis spores as a model, Rodrigues and Harry also recently observed precise midcell Z-ring formation in the absence of Min and nucleoid occlusion [36**]. This finding has led to the proposal that midcell is identified independently of these factors and that Min and Noc may primarily function to ensure the efficient utilization of this site. Although the identity of the factor(s) that determine(s) this positioning is not clear, several previous studies from the Harry laboratory implicate the early stages of chromosome replication in Z-ring formation and positioning [37–39]. Further support for a link between DNA replication and cell division in B. subtilis was also recently reported by Arjes and colleagues [40**]. They find that after several mass doublings following division inhibition, the resulting cell filaments are unable to initiate new rounds of replication. Intriguingly, Arjes and colleagues a lso find that cell division is inhibited after several generations following a block in the initiation of DNA replication [40**]. It therefore appears that, contrary to the widely held view in the field, there is an obligatory link between cell division and DNA replication, at least in B. subtilis. Although the mechanism of this coupling remains unclear, an exciting possibility is that the factors involved here [40**] are also responsible for the phenomena observed by Harry and co-workers [36**–39] connecting early stages of replication with Z-ring formation.
Controlling divisome activity
In E. coli, recruitment of essential divisome components to midcell proceeds via a mostly linear dependency pathway starting with the FtsZ-interacting proteins FtsA and ZipA that anchor the Z-ring to the membrane and ending with the bitopic membrane protein FtsN [41–52]. Because it is the last divisome protein in the recruitment pathway, FtsN has long been though to play a role in the switch from divisome assembly to the constriction phase of division [53]. This idea was reinforced with the demonstration that FtsN joins the divisome in a self-enhancing process involving its small, membrane-proximal, essential domain (EFtsN) and its C-terminal, peptidoglycan (PG)-binding SPOR domain (SFtsN) [54,55]. Based on this observation, it was proposed that cell constriction is driven by a positive feedback loop in which EFtsN stimulates the synthesis and remodeling of cell wall material by other divisome components to create the recruitment signal for SFtsN, which brings more EFtsN to the division site to stimulate more cell wall synthesis, and so on [54] (Figure 2).
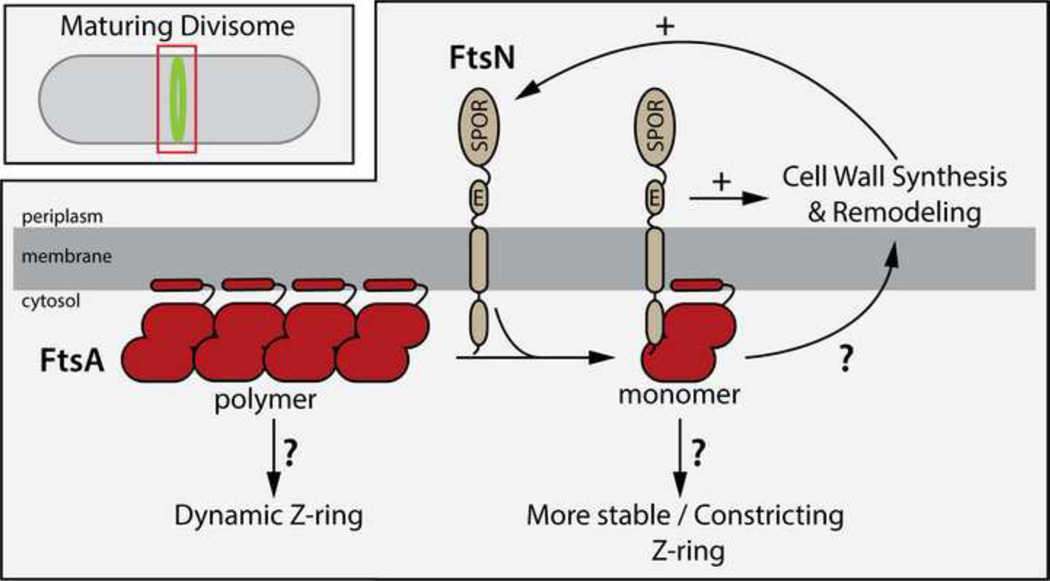
Figure 2. Potential role of FtsA-FtsN interactions in triggering cell constriction.
Shown is a diagram depicting potential events occurring at a maturing divisome focusing on the FtsA-FtsN interaction. As described in the text, a growing number of studies suggest the attractive possibility that FtsA serves as a “sensor” of divisome assembly. As late components of the divisome like FtsN are recruited to the structure, they promote the formation of a reduced polymeric form of FtsA. Once a threshold level of this altered FtsA form accumulates at midcell, it may trigger changes in FtsZ polymer dynamics to initiate contraction of the ring in conjunction with the activation of divisome components associated with cell wall synthesis by both FtsA and FtsN. These activities of FtsA and its partner FtsN may coordinate transitions in the Z-ring pattern with cell wall remodeling processes on the other side of the membrane. Although not shown, it has also been proposed that ZipA may promote the formation of FtsA monomers to stimulate the recruitment of downstream divisome proteins like FtsN [3,62,64]. See text for details.
In addition to binding FtsZ, FtsA was recently demonstrated to interact directly with the cytoplasmic N-terminus of FtsN (NFtsN) [56**]. This interaction was shown to be important for the initial localization of FtsN to the divisome [57*], suggesting that the FtsA-NFtsN interaction may be responsible for initiating the proposed positive feedback loop that promotes constriction (Figure 2). Clues as to how this process may work have come from the isolation and analysis of ftsA mutants in E. coli that bypass the normal requirement for other essential division proteins [58–60]. The first “bypass mutant” identified was ftsA*(R286W) [59]. It was isolated as a suppressor that allowed cells to survive in the absence of the other membrane anchor of FtsZ, ZipA. Subsequent studies indicated that this allele and/or other alleles of ftsA were also able to bypass the essential functions of divisome proteins FtsK and FtsN, and also suppress the division defects of certain temperature-sensitive ftsQ mutants [58,60,61]. Pichoff and co-workers [62**] recently found that the activity of FtsA bypass variants is likely related to defects in FtsA-FtsA interactions. They used a genetic screen to identify FtsA derivatives with a reduced ability to self-interact as assessed by in vivo assays. Strikingly, this collection of variants included the original FtsA* variant. They subsequently showed that all self-interaction defective variants of FtsA could function as ZipA bypass suppressors [62**]. Furthermore, in addition to bypassing the function of essential division proteins, many of the poorly self-interacting FtsA variants were also shown to promote early cell division [62**], suggesting that a reduction in FtsA-FtsA interactions stimulates division. Importantly, the FtsA-NFtsN interaction described above involves the 1c domain of FtsA [53,56**]. Based on a structural analysis of FtsA polymers [63**], this interaction is likely to interfere with FtsA-FtsA interactions. Thus, when all of the genetic and biochemical studies are taken together, the results point to a competition between FtsA-FtsA and FtsA-NFtsN interactions in the control of constriction initiation with both EFtsN and monomeric FtsA stimulating the process [3,56**,57*,62**,64*] (Figure 2). One possible scenario suggested previously [3,62**,64*] is that ZipA disrupts FtsA-FtsA interactions at the Z-ring to generate free FtsA interfaces for the recruitment of downstream divisome proteins like FtsN and the eventual activation of constriction. Thus, FtsA* variants with a reduced capacity to interact bypass ZipA function. Alternatively, or in addition to serving as a recruitment factor for other division proteins, the polymeric status of FtsA and its interaction with FtsN may also serve as a sensor or signal [56**,57*,65] used to monitor the status of divisome assembly and promote constriction only after the machinery is deemed stable enough to successfully complete division. In this case, FtsA may be progressively converted to a reduced polymeric form as the machine assembles, at first spontaneously, but then stimulated by the self-enhanced recruitment of FtsN. This conversion of FtsA would proceed until a threshold level of monomers or small oligomers is achieved, which, together with FtsN, would then somehow trigger a change in other components of the machinery to to stimulate ring closure. How this activation might occur is not clear, but recent results indicate that the FtsQLB subcomplex of the divisome is involved and may receive signals from both FtsA and FtsN in order to activate cell wall synthesis and remodeling at the division site to stimulate constriction [66*,67*].
In addition to its potential role in sensing divisome assembly, the direct connection of FtsA with FtsZ also puts it in position to play a key role in modulating Z-ring activity and/or dynamics during the division cycle. This possible function was highlighted by a recent study of FtsZ polymer dynamics on supported lipid bilayers [68**] that showed dramatic changes in the FtsZ patterns formed depending on whether FtsZ was recruited to the membrane by FtsA or ZipA. Polymer bundles that associated with the membrane surface via an interaction with ZipA were found to form relatively stable patterns [68**]. Polymers brought to the membrane by FtsA, on the other hand, formed rapidly rotating swirls [68**]. These dynamic patterns were shown to result from the ability of FtsA to destabilize the FtsZ polymer network [68**]. This observation connects with in vivo results indicating that the FtsA/FtsZ ratio is important for proper division and that too much FtsA can inhibit Z-ring formation [69,70]. Thus, FtsA may, at least initially, promote the formation of a more dynamic, less stable pattern of FtsZ polymers at midcell. An attractive possibility is that changes in FtsA polymerization status brought about through the recruitment of downstream divisome proteins like FtsN may change the effect of FtsA on FtsZ polymer dynamics such that it now stabilizes the Z-ring pattern at midcell and promotes constriction (Figure 2). In support of this possibility, the self-interaction defective FtsA(R286W) derivative has been shown alter the Z-ring to reduce its sensitivity to negative-regulators [59]. However, this variant of FtsA was not found to alter the FtsZ swirl patterns formed in the supported lipid bilayer experiments [68**], suggesting that factors in addition to changes in FtsA self-association status are likely needed to more faithfully reconstitute FtsZ pattern formation at the membrane in vivo. Nevertheless, the possibility that FtsA polymer status controls Z-ring and/or divisome activity remains highly attractive and warrants further investigation.
Conclusions
Most of the straightforward aspects of divisome assembly, such as determining what proteins localize to the structure and when they get there, have been extensively characterized over the last two decades. We now face questions that are much more difficult to solve. What are all of these proteins doing at the division site? How do they work together to transform the cell envelope? What couples their activity to other major cellular processes like DNA replication? As described in this review and the highlighted references, progress in these areas is being made thanks to multidisciplinary efforts that encompass everything from genetic analysis and in vivo imaging to biochemical reconstitutions and structural biology. We look forward to seeing how the mechanistic picture of bacterial cell division evolves as we continue to apply these and other emerging technologies to understand this fundamental process.
Highlights.
-
-
The chromosome-binding protein MatP plays a role in division site positioning
-
-
DNA replication and cell division are obligatorily coupled in Bacillus subtilis
-
-
FtsA-FtsA interactions play a critical role in divisome maturation and activation
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Bi EF, Lutkenhaus J. FtsZ ring structure associated with division in Escherichia coli. Nature. 1991;354:161–164. doi: 10.1038/354161a0. [DOI] [PubMed] [Google Scholar]
- 2. Szwedziak P, Wang Q, Bharat TAM, Tsim M, Löwe J. Architecture of the ring formed by the tubulin homologue FtsZ in bacterial cell division. eLife. 2014 doi: 10.7554/eLife.04601. (in press). ** High resolution imaging reveals the molecular architecture of the Z-ring.
- 3.Lutkenhaus J, Pichoff S, Du S. Bacterial cytokinesis: From Z ring to divisome. Cytoskeleton. 2012;69:778–790. doi: 10.1002/cm.21054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Typas A, Banzhaf M, Gross CA, Vollmer W. From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat Rev Microbiol. 2012;10:123–136. doi: 10.1038/nrmicro2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Boer PAJ. Advances in understanding E. coli cell fission. Curr Opin Microbiol. 2010;13:730–737. doi: 10.1016/j.mib.2010.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Adams DW, Errington J. Bacterial cell division: assembly, maintenance and disassembly of the Z ring. Nat Rev Microbiol. 2009;7:642–653. doi: 10.1038/nrmicro2198. [DOI] [PubMed] [Google Scholar]
- 7.de Boer PA, Crossley RE, Rothfield LI. A division inhibitor and a topological specificity factor coded for by the minicell locus determine proper placement of the division septum in E. coli. Cell. 1989;56:641–649. doi: 10.1016/0092-8674(89)90586-2. [DOI] [PubMed] [Google Scholar]
- 8.Levin PA, Shim JJ, Grossman AD. Effect of minCD on FtsZ ring position and polar septation in Bacillus subtilis. J Bacteriol. 1998;180:6048–6051. doi: 10.1128/jb.180.22.6048-6051.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Woldringh CL, Mulder E, Huls PG, Vischer N. Toporegulation of bacterial division according to the nucleoid occlusion model. Res Microbiol. 1991;142:309–320. doi: 10.1016/0923-2508(91)90046-d. [DOI] [PubMed] [Google Scholar]
- 10.Woldringh CL, Mulder E, Valkenburg JA, Wientjes FB, Zaritsky A, Nanninga N. Role of the nucleoid in the toporegulation of division. Res Microbiol. 1990;141:39–49. doi: 10.1016/0923-2508(90)90096-9. [DOI] [PubMed] [Google Scholar]
- 11.Yu XC, Margolin W. FtsZ ring clusters in min and partition mutants: role of both the Min system and the nucleoid in regulating FtsZ ring localization. Molecular Microbiology. 1999;32:315–326. doi: 10.1046/j.1365-2958.1999.01351.x. [DOI] [PubMed] [Google Scholar]
- 12.Bi E, Lutkenhaus J. Cell division inhibitors SulA and MinCD prevent formation of the FtsZ ring. J Bacteriol. 1993;175:1118–1125. doi: 10.1128/jb.175.4.1118-1125.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Raskin DM, de Boer PA. Rapid pole-to-pole oscillation of a protein required for directing division to the middle of Escherichia coli. Proc Natl Acad Sci USA. 1999;96:4971–4976. doi: 10.1073/pnas.96.9.4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu Z, Lutkenhaus J. Topological regulation of cell division in Escherichia coli involves rapid pole to pole oscillation of the division inhibitor MinC under the control of MinD and MinE. Molecular Microbiology. 1999;34:82–90. doi: 10.1046/j.1365-2958.1999.01575.x. [DOI] [PubMed] [Google Scholar]
- 15.Lackner LL, Raskin DM, de Boer PAJ. ATP-dependent interactions between Escherichia coli Min proteins and the phospholipid membrane in vitro. J Bacteriol. 2003;185:735–749. doi: 10.1128/JB.185.3.735-749.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu Z, Saez C, Lutkenhaus J. Recruitment of MinC, an inhibitor of Z-ring formation, to the membrane in Escherichia coli: role of MinD and MinE. J Bacteriol. 2003;185:196–203. doi: 10.1128/JB.185.1.196-203.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ghosal D, Trambaiolo D, Amos LA, Löwe J. MinCD cell division proteins form alternating copolymeric cytomotive filaments. Nat Commun. 2014;5:5341. doi: 10.1038/ncomms6341. * Structural analysis shows that MinC and MinD associate to form copolymeric protein filaments.
- 18.Raskin DM, de Boer PA. MinDE-dependent pole-to-pole oscillation of division inhibitor MinC in Escherichia coli. J Bacteriol. 1999;181:6419–6424. doi: 10.1128/jb.181.20.6419-6424.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marston AL, Errington J. Selection of the midcell division site in Bacillus subtilis through MinD-dependent polar localization and activation of MinC. Molecular Microbiology. 1999;33:84–96. doi: 10.1046/j.1365-2958.1999.01450.x. [DOI] [PubMed] [Google Scholar]
- 20.Wu LJ, Errington J. Coordination of cell division and chromosome segregation by a nucleoid occlusion protein in Bacillus subtilis. Cell. 2004;117:915–925. doi: 10.1016/j.cell.2004.06.002. [DOI] [PubMed] [Google Scholar]
- 21.Bernhardt TG, de Boer PAJ. SlmA, a nucleoid-associated, FtsZ binding protein required for blocking septal ring assembly over Chromosomes in E. coli. Mol Cell. 2005;18:555–564. doi: 10.1016/j.molcel.2005.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tonthat NK, Arold ST, Pickering BF, Van Dyke MW, Liang S, Lu Y, Beuria TK, Margolin W, Schumacher MA. Molecular mechanism by which the nucleoid occlusion factor, SlmA, keeps cytokinesis in check. EMBO J. 2011;30:154–164. doi: 10.1038/emboj.2010.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cho H, Bernhardt TG. Identification of the SlmA active site responsible for blocking bacterial cytokinetic ring assembly over the chromosome. PLoS Genet. 2013;9:e1003304. doi: 10.1371/journal.pgen.1003304. * Genetic and biochemical analysis identifies the FtsZ-binding site of SlmA, the location of which provides a possible mechanism for the activation of SlmA upon DNA binding.
- 24.Cho H, McManus HR, Dove SL, Bernhardt TG. Nucleoid occlusion factor SlmA is a DNA-activated FtsZ polymerization antagonist. Proc Natl Acad Sci USA. 2011;108:3773–3778. doi: 10.1073/pnas.1018674108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tonthat NK, Milam SL, Chinnam N, Whitfill T, Margolin W, Schumacher MA. SlmA forms a higher-order structure on DNA that inhibits cytokinetic Z-ring formation over the nucleoid. Proc Natl Acad Sci USA. 2013;110:10586–10591. doi: 10.1073/pnas.1221036110. * Structural analysis shows that SlmA binds as a dimer of dimers to DNA and significantly distorts DNA structure. The SlmA binding sites were also found to be located near genes coding for membrane proteins, suggesting that these sites are localized near the membrane to help SlmA antagonize the formation of membrane-associated FtsZ assemblies.
- 26.Du S, Park K-T, Lutkenhaus J. Oligomerization of FtsZ converts the FtsZ tail motif (conserved carboxy-terminal peptide) into a multivalent ligand with high avidity for partners ZipA and SlmA. Molecular Microbiology. 2014 doi: 10.1111/mmi.12854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Du S, Lutkenhaus J. SlmA Antagonism of FtsZ Assembly Employs a Two-pronged Mechanism like MinCD. PLoS Genet. 2014;10:e1004460. doi: 10.1371/journal.pgen.1004460. ** Genetic and biochemical experiments reveal that SlmA antagonizes Ft sZ polymerization via a two-step mechanism. First, activated SlmA bound to its DNA site associates with the conserved C-terminal tail of FtsZ. The isolation of FtsZ variants that still associate with SlmA but remain resistant to depolymerization suggests that additional interactions between SlmA and FtsZ then result in the breakdown of FtsZ polymers. This mechanism is strikingly similar to current models for MinC action.
- 28.Wu LJ, Ishikawa S, Kawai Y, Oshima T, Ogasawara N, Errington J. Noc protein binds to specific DNA sequences to coordinate cell division with chromosome segregation. EMBO J. 2009;28:1940–1952. doi: 10.1038/emboj.2009.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bailey MW, Bisicchia P, Warren BT, Sherratt DJ, Männik J. Evidence for Divisome Localization Mechanisms Independent of the Min System and SlmA in Escherichia coli. PLoS Genet. 2014;10:e1004504. doi: 10.1371/journal.pgen.1004504. ** As discussed in the main text, this paper identifies MatP and the Ter macrodomain of the chromosome as a positional marker for Z-ring assembly.
- 30.Mercier R, Petit M-A, Schbath S, Robin S, Karoui ElM, Boccard F, Espéli O. The MatP/matS site-specific system organizes the terminus region of the E. coli chromosome into a macrodomain. Cell. 2008;135:475–485. doi: 10.1016/j.cell.2008.08.031. [DOI] [PubMed] [Google Scholar]
- 31. Dupaigne P, Tonthat NK, Espéli O, Whitfill T, Boccard F, Schumacher MA. Molecular Basis for a Protein-Mediated DNA-Bridging Mechanism that Functions in Condensation of the E. coli Chromosome. Mol Cell. 2012;48:560–571. doi: 10.1016/j.molcel.2012.09.009. * Structure of MatP suggests a mechanism by which the protein organizes the Ter macrodomain of the chromosome.
- 32. Espéli O, Borne R, Dupaigne P, Thiel A, Gigant E, Mercier R, Boccard F. A MatP-divisome interaction coordinates chromosome segregation with cell division in E. coli. EMBO J. 2012;31:3198–3211. doi: 10.1038/emboj.2012.128. ** An interaction between MatP and the Z-ring protein ZapB is identified and found to be important for anchoring the Ter macrodomain of the chromosome to midcell.
- 33.Galli E, Gerdes K. Spatial resolution of two bacterial cell division proteins: ZapA recruits ZapB to the inner face of the Z-ring. Molecular Microbiology. 2010;76:1514–1526. doi: 10.1111/j.1365-2958.2010.07183.x. [DOI] [PubMed] [Google Scholar]
- 34.Gueiros-Filho FJ, Losick R. A widely conserved bacterial cell division protein that promotes assembly of the tubulin-like protein FtsZ. Genes Dev. 2002;16:2544–2556. doi: 10.1101/gad.1014102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Buss J, Coltharp C, Huang T, Pohlmeyer C, Wang S-C, Hatem C, Xiao J. In vivo organization of the FtsZ-ring by ZapA and ZapB revealed by quantitative super-resolution microscopy. Molecular Microbiology. 2013;89:1099–1120. doi: 10.1111/mmi.12331. * Super-resolution microscopy reveals a role for ZapA and ZapB in bringing together clusters of FtsZ polymers to efficiently form the canonical Z-ring structure.
- 36. Rodrigues CDA, Harry EJ. The Min system and nucleoid occlusion are not required for identifying the division site in Bacillus subtilis but ensure its efficient utilization. PLoS Genet. 2012;8:e1002561. doi: 10.1371/journal.pgen.1002561. ** Analysis of division site positioning in outgrowing spores of B. subtilis indicates that the Min system and Noc protein are not required for precise midcell division. Thus, additional positional markers for division are likely to exist.
- 37.Moriya S, Rashid RA, Rodrigues CDA, Harry EJ. Influence of the nucleoid and the early stages of DNA replication on positioning the division site in Bacillus subtilis. Molecular Microbiology. 2010;76:634–647. doi: 10.1111/j.1365-2958.2010.07102.x. [DOI] [PubMed] [Google Scholar]
- 38.Harry EJ, Rodwell J, Wake RG. Co-ordinating DNA replication with cell division in bacteria: a link between the early stages of a round of replication and mid-cell Z ring assembly. Molecular Microbiology. 1999;33:33–40. doi: 10.1046/j.1365-2958.1999.01439.x. [DOI] [PubMed] [Google Scholar]
- 39.Regamey A, Harry EJ, Wake RG. Mid-cell Z ring assembly in the absence of entry into the elongation phase of the round of replication in bacteria: co-ordinating chromosome replication with cell division. Molecular Microbiology. 2000;38:423–434. doi: 10.1046/j.1365-2958.2000.02130.x. [DOI] [PubMed] [Google Scholar]
- 40. Arjes HA, Kriel A, Sorto NA, Shaw JT, Wang JD, Levin PA. Failsafe mechanisms couple division and DNA replication in bacteria. Curr Biol. 2014;24:2149–2155. doi: 10.1016/j.cub.2014.07.055. ** As discussed in the main text, and contrary to a common assumption in the field, the results described in this report indicate an obligatory coupling between the initiation of DNA replication and cell division in B. subtilis.
- 41.Pichoff S, Lutkenhaus J. Unique and overlapping roles for ZipA and FtsA in septal ring assembly in Escherichia coli. EMBO J. 2002;21:685–693. doi: 10.1093/emboj/21.4.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hale CA, de Boer PA. Direct binding of FtsZ to ZipA, an essential component of the septal ring structure that mediates cell division in E. coli. Cell. 1997;88:175–185. doi: 10.1016/s0092-8674(00)81838-3. [DOI] [PubMed] [Google Scholar]
- 43.Goehring NW, Beckwith J. Diverse paths to midcell: assembly of the bacterial cell division machinery. Curr Biol. 2005;15:R514–R526. doi: 10.1016/j.cub.2005.06.038. [DOI] [PubMed] [Google Scholar]
- 44.Chen JC, Beckwith J. FtsQ, FtsL and FtsI require FtsK, but not FtsN, for co-localization with FtsZ during Escherichia coli cell division. Molecular Microbiology. 2001;42:395–413. doi: 10.1046/j.1365-2958.2001.02640.x. [DOI] [PubMed] [Google Scholar]
- 45.Buddelmeijer N, Judson N, Boyd D, Mekalanos JJ, Beckwith J. YgbQ, a cell division protein in Escherichia coli and Vibrio cholerae, localizes in codependent fashion with FtsL to the division site. Proc Natl Acad Sci USA. 2002;99:6316–6321. doi: 10.1073/pnas.092128499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hale CA, de Boer PA. Recruitment of ZipA to the septal ring of Escherichia coli is dependent on FtsZ and independent of FtsA. J Bacteriol. 1999;181:167–176. doi: 10.1128/jb.181.1.167-176.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schmidt KL, Peterson ND, Kustusch RJ, Wissel MC, Graham B, Phillips GJ, Weiss DS. A predicted ABC transporter, FtsEX, is needed for cell division in Escherichia coli. J Bacteriol. 2004;186:785–793. doi: 10.1128/JB.186.3.785-793.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang L, Khattar MK, Donachie WD, Lutkenhaus J. FtsI and FtsW are localized to the septum in Escherichia coli. J Bacteriol. 1998;180:2810–2816. doi: 10.1128/jb.180.11.2810-2816.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weiss DS, Chen JC, Ghigo JM, Boyd D, Beckwith J. Localization of FtsI (PBP3) to the septal ring requires its membrane anchor, the Z ring, FtsA, FtsQ, and FtsL. J Bacteriol. 1999;181:508–520. doi: 10.1128/jb.181.2.508-520.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mercer KLN, Weiss DS. The Escherichia coli cell division protein FtsW is required to recruit its cognate transpeptidase, FtsI (PBP3), to the division site. J Bacteriol. 2002;184:904–912. doi: 10.1128/jb.184.4.904-912.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hale CA, de Boer PAJ. ZipA is required for recruitment of FtsK, FtsQ, FtsL, and FtsN to the septal ring in Escherichia coli. J Bacteriol. 2002;184:2552–2556. doi: 10.1128/JB.184.9.2552-2556.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Addinall SG, Cao C, Lutkenhaus J. FtsN, a late recruit to the septum in Escherichia coli. Molecular Microbiology. 1997;25:303–309. doi: 10.1046/j.1365-2958.1997.4641833.x. [DOI] [PubMed] [Google Scholar]
- 53.Corbin BD, Geissler B, Sadasivam M, Margolin W. Z-ring-independent interaction between a subdomain of FtsA and late septation proteins as revealed by a polar recruitment assay. J Bacteriol. 2004;186:7736–7744. doi: 10.1128/JB.186.22.7736-7744.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gerding MA, Liu B, Bendezú FO, Hale CA, Bernhardt TG, de Boer PAJ. Self-enhanced accumulation of FtsN at Division Sites and Roles for Other Proteins with a SPOR domain (DamX, DedD, and RlpA) in Escherichia coli cell constriction. J Bacteriol. 2009;191:7383–7401. doi: 10.1128/JB.00811-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lutkenhaus J. FtsN--trigger for septation. J Bacteriol. 2009;191:7381–7382. doi: 10.1128/JB.01100-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Busiek KK, Eraso JM, Wang Y, Margolin W. The early divisome protein FtsA interacts directly through its 1c subdomain with the cytoplasmic domain of the late divisome protein FtsN. J Bacteriol. 2012;194:1989–2000. doi: 10.1128/JB.06683-11. ** In vitro assays reveal a direct interaction between the N-terminal domain of FtsN and a domain of FtsA important for FtsA-FtsA interactions. The results suggest that the interaction may be involved a signal transduction between the cell wall synthetic machinery and the Z-ring to promote envelope constriction.
- 57. Busiek KK, Margolin W. A role for FtsA in SPOR-independent localization of the essential Escherichia colicell division protein FtsN. Molecular Microbiology. 2014;92:1212–1226. doi: 10.1111/mmi.12623. * This paper adds to the growing connection between FtsA, FtsN, and the initiation of cell constriction by the divisome.
- 58.Geissler B, Margolin W. Evidence for functional overlap among multiple bacterial cell division proteins: compensating for the loss of FtsK. Molecular Microbiology. 2005;58:596–612. doi: 10.1111/j.1365-2958.2005.04858.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Geissler B, Elraheb D, Margolin W. A gain-of-function mutation in ftsA bypasses the requirement for the essential cell division gene zipA in Escherichia coli. Proc Natl Acad Sci USA. 2003;100:4197–4202. doi: 10.1073/pnas.0635003100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bernard CS, Sadasivam M, Shiomi D, Margolin W. An altered FtsA can compensate for the loss of essential cell division protein FtsN in Escherichia coli. Molecular Microbiology. 2007;64:1289–1305. doi: 10.1111/j.1365-2958.2007.05738.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Goehring NW, Petrovska I, Boyd D, Beckwith J. Mutants, suppressors, and wrinkled colonies: mutant alleles of the cell division gene ftsQ point to functional domains in FtsQ and a role for domain 1C of FtsA in divisome assembly. J Bacteriol. 2007;189:633–645. doi: 10.1128/JB.00991-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Pichoff S, Shen B, Sullivan B, Lutkenhaus J. FtsA mutants impaired for self-interaction bypass ZipA suggesting a model in which FtsA's self-interaction competes with its ability to recruit downstream division proteins. Molecular Microbiology. 2012;83:151–167. doi: 10.1111/j.1365-2958.2011.07923.x. ** The reported genetic analysis of FtsA reveals that defects in FtsA self interaction can stimulate cell constriction and bypass the essential function of other divisome components.
- 63. Szwedziak P, Wang Q, Freund SMV, Löwe J. FtsA forms actin-like protofilaments. EMBO J. 2012;31:2249–2260. doi: 10.1038/emboj.2012.76. ** This paper shows that FtsA forms actin-like polymers even though it has a unique domain structure relative to other proteins in the actin family.
- 64. Pichoff S, Du S, Lutkenhaus J. The bypass of ZipA by overexpression of FtsN requires a previously unknown conserved FtsN motif essential for FtsA-FtsN interaction supporting a model in which FtsA monomers recruit late cell division proteins to the Z ring. Molecular Microbiology. 2014 doi: 10.1111/mmi.12907. (in press). * Reports the unique ability of FtsN overproduction to bypass t he essential function of ZipA and supports a role for the FtsA-FtsN interaction in stimulating cell division.
- 65.Herricks JR, Nguyen D, Margolin W. A thermosensitive defect in the ATP binding pocket of FtsA can be suppressed by allosteric changes in the dimer interface. Molecular Microbiology. 2014;94:713–727. doi: 10.1111/mmi.12790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Tsang M-J, Bernhardt TG. A role for the FtsQLB complex in cytokinetic ring activation revealed by an ftsL allele that accelerates division. Molecular Microbiology. 2014 doi: 10.1111/mmi.12905. (in press). *Genetic analysis of ftsL reveals a role for the FtsQLB complex in the initiation of constriction. See also reference 67.
- 67. Liu B, Persons L, Lee L, de Boer PAJ. Roles for both FtsA and the FtsQLB subcomplex in FtsN-stimulated cell constriction in Escherichia coli. Molecular Microbiology. 2014 doi: 10.1111/mmi.12906. (in press). * Mutants in ftsL, ftsB, and ftsA are identified as suppressors that bypass the requirement for the essential domain of FtsN. The results indicate that the FtsQLB complex is likely to receive signals from FtsA and FtsN to promote the onset of cell constriction.
- 68. Loose M, Mitchison TJ. The bacterial cell division proteins FtsA and FtsZ self-organize into dynamic cytoskeletal patterns. Nat Cell Biol. 2013;16:38–46. doi: 10.1038/ncb2885. ** This paper shows that FtsZ polymers recruited to the membrane surface display dramatically different dynamics depending on whether they are recruited to the membrane by FtsA or ZipA. Interestingly, FtsA appears to promote the formation of highly dynamic FtsZ polymer structures that may mimic the early stages of Z-ring formation prior to the recruitment of downstream components.
- 69.Dai K, Lutkenhaus J. The proper ratio of FtsZ to FtsA is required for cell division to occur in Escherichia coli. J Bacteriol. 1992;174:6145–6151. doi: 10.1128/jb.174.19.6145-6151.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dewar SJ, Begg KJ, Donachie WD. Inhibition of cell division initiation by an imbalance in the ratio of FtsA to FtsZ. J Bacteriol. 1992;174:6314–6316. doi: 10.1128/jb.174.19.6314-6316.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]