MicroRNAs are short (20–40 nucleotides) non-coding RNA molecules that are responsible for the post-transcriptional regulation of gene expression. Aberrant expression of microRNAs has been associated with various malignancies.1,2 Specifically, downregulation of microRNA-142 (miR-142) has been shown to occur in acute myeloid leukemia (AML).3 Interestingly, also gene mutations in miR-142, miR-632 and miR-891 have been recently described in de novo AML.4 So far, little is known about mutations in miR-142 and other related microRNAs in myeloid malignancies. The aim of this study was to analyze mutations in the miR-142 and related microRNAs, such as miR-632, and miR-891, in a large cohort of 935 patients with AML or myelodysplastic syndrome (MDS).
The patient group consisted of 416 de novo AML patients (excluding AML M3) who entered the multicenter treatment trials AMLSHG-0199 or AMLSHG-0295, 326 patients undergoing allogeneic hematopoietic stem cell transplantation (HSCT) for secondary AML after a prior diagnosis of MDS (sAML) (n=170) or primary MDS (n=156), and 193 primary MDS patients not undergoing intensive therapy or allogeneic HSCT. Informed consent was obtained in accordance with the Declaration of Helsinki. This study was approved by the institutional review board of Hannover Medical School.
The genomic region of the miR-142 gene (containing miR-142-5p and miR-142-3p) (all patients), miR-632 (CN-AML and MDS patients) and miR-891 (CN-AML and MDS patients) were sequenced by Sanger sequencing as previously described5 and with primers listed in Online Supplementary Table S1. Patients’ samples were also assessed for other frequently mutated genes in AML and MDS, as previously described (Online Supplementary Appendix).6–9 All mutations were confirmed in an independent experiment. Sequence analysis was performed with the Mutation Surveyor software (Softgenetics, State College, PA, USA). The somatic or germ-line status of mutations in miR-142 was established by evaluating T cells (CD3+CD11b‒CD14‒CD33‒) purified from diagnostic samples by flow cytometry when available (n=2).
Gene expression levels of miR-142-3p and miR-142-5p were assessed by real-time reverse transcriptase polymerase chain reaction (RT-PCR) as detailed in the Online Supplementary Appendix. Overall survival (OS), complete remission (CR) and relapse-free survival (RFS) were calculated as previously described.10,11 The statistical analyses were performed with the statistical software package SPSS 21.0 (IBM Corporation, Armonk, NY, USA).
In the whole cohort, we identified 7 patients with mutations in miRNAs (0.74%) (Table 1). All mutations were heterozygous (Online Supplementary Figure S1). MiR-142-3p was the most frequently affected miRNA with 5 patients showing a mutation (0.53%) (Figure 1A and Online Supplementary Figure S1). The somatic origin could be confirmed for miR-142-3p by analyzing T cells which did not show the mutation (n=2). All of these mutations affected the seed region of miR-142-3p, thereby potentially changing the target specificity of miR-142. In line with this, target prediction for miR-142-3p with targetscan (release 5.2: June 2011; available from: www.targetscan.org/vert_50/) showed 250 conserved targets, while three of four mutations had much fewer predicted targets [pos54U>C: 245 conserved targets, pos55A>G: 76, pos56G>U: 95, and pos57U>C: 4 conserved targets (Online Supplementary Table S2)]. The overlap of the predicted target genes was 0 to 7.9% between wild-type and mutated miR-142-3p in these four positions, suggesting that mutations in miR-142 are loss-of-function mutations. Functional validation of the mutated miRNAs confirmed that mutations of miR-142-3p pos54U>C, pos56G>U, and pos57U>C had little or no effect on protein expression of miR-142-3p targets CCNT2 and MAP3K7IP2 (TAB2)12 in leukemic and fibroblast cells in contrast to wild-type miR-142-3p, which knocked down protein expression (Figure 1B and Online Supplementary Figure S2). The partial knockdown of target genes by the mutation at pos55A>G can be explained by the wobble effect of guanosine.
Table 1.
Characteristics of miR-142 and miR-632 mutated patients.
Figure 1.
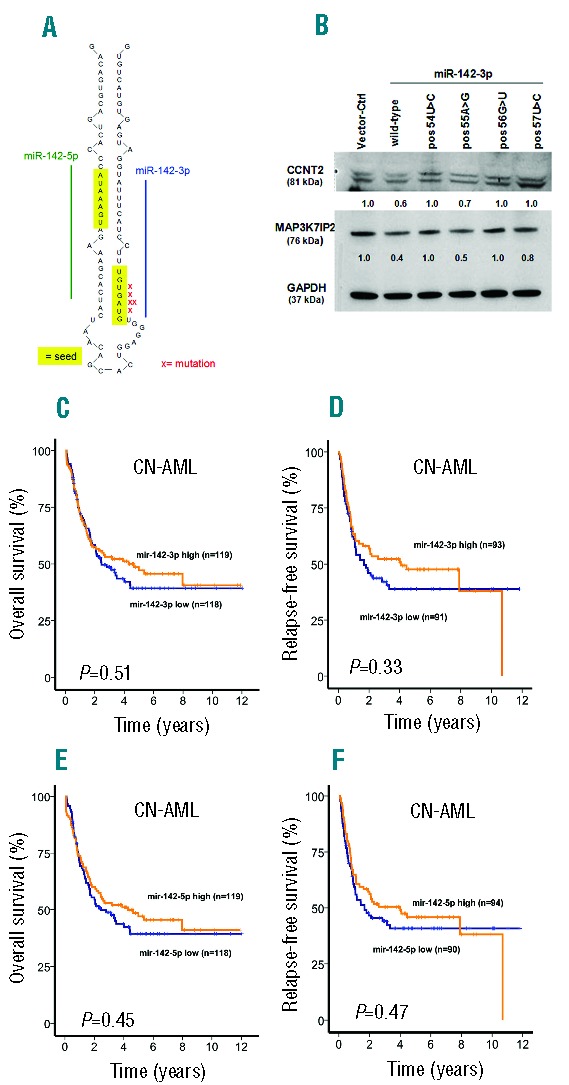
(A) Location of mutations in miR-142 in patients with AML and MDS. (B) Western Blot of miR-142-3p target genes CCNT2 and MAP3K7IP2 in THP1 cells transduced with CTL or miR-142-3p wild-type or mutated vectors as indicated. (C) Overall and (D) relapse-free survival of patients with CN-AML according to miR-142-3p expression levels above or below the median expression (log rank test). (E) Overall and (F) relapse-free survival of patients with CN-AML according to miR-142-5p expression levels above or below the median expression (log rank test).
Mutations in miR-142 occurred in male and female patients (Table 1). Of the 5 patients with mutations in miR-142, only one patient carried the diagnosis of de novo AML (0.2% in de novo AML), while 2 patients were diagnosed with sAML (1.2% in sAML) and 2 patients had MDS (0.56% in MDS, corresponding to 0.77% in MDS/sAML from MDS). Apart from one patient who underwent allogeneic transplantation for sAML and was alive 2.73 years later, all other patients with follow-up data died of the disease in less than one year. Four patients had normal cytogenetics; while for one patient cytogenetic information was not available. Myelodysplasia-related gene mutations, such as mutations in the splicing genes or chromatin remodeling genes, were found in 2 patients (one patient with mutated ASXL1 and SRSF2, one patient with mutated U2AF1). One patient had a concomitant mutation in NRAS and IDH1 (Table 1). The associated mutational profile could possibly suggest that miR-142 mutations play a role in the pathogenesis of MDS rather than de novo AML. Two patients were identified with heterozygous missense mutations in miR-632 (Table 1). One of these patients was a female AML patient with a normal karyotype but with concomitant mutations in IDH2 and DNMT3A; the patient died six months after diagnosis. The other miR-632 mutated patient carried the diagnosis of MDS and was classified as refractory anemia (RA) (Table 1). While the 2 patients had different nucleotides of miR-632 affected by the mutations, neither variant affected the seed region. Interestingly, miR-632 expression was found to be lower in MDS patients as compared to normal controls in a prior study13 suggesting that a functional loss or reduction of miR-632 could be involved in the pathogenesis of MDS. None of the evaluated patients carried a mutation in miRNA-891A suggesting that this miRNA is only very rarely mutated in myeloid malignancies.
Because of data suggesting that aberrant miR-142-3p expression is associated with myeloid malignancies,14,15 we also evaluated the expression of miR-142-3p and miR-142-5p in 237 patients with CN-AML. Relative miR-142-3p and miR-142-5p transcript levels ranged from 0.0014 to 15.80 and from 0 to 15.14 of miRNA/RNU48 copies in CN-AML patients, respectively. The relative transcript levels of miR-142-3p and miR-142-5p in the miR-142 mutated CN-AML patients were 1.79 and 1.16, respectively. Relative miR-142-3p and miR-142-5p transcript levels were highly correlated (R=0.89, P<0.001) (Online Supplementary Figure S3).
We found that patients with levels above or below the median miR-142-3p expression showed no difference in OS (HR 0.89, 95%CI: 0.63–1.26; P=0.51), RFS (HR 0.82, 95%CI 0.56–1.22; P=0.033) and CR (78% vs. 77%; P=0.85) (Figure 1C and D). Similar results were obtained for miR-142-5p with expression above or below the median not affecting OS (HR 0.88, 95%CI: 0.62–1.24; P=0.45), RFS (HR 0.87, 95%CI: 0.59–1.28; P=0.47) and CR (79% vs. 76%; P=0.62) (Figure 1E and F). CN-AML patients with high versus low expression of miR-142-3p and miR-142-5p showed no significant difference in clinical parameters such as age, sex, FAB subtype, blast count, type of AML, ECOG performance status as well as in mutation frequencies of FLT3-ITD, NPM1, CEBPA, DNMT3A, WT1, NRAS, IDH1 and IDH2 (Online Supplementary Tables S3 and S4). Only a trend towards a higher NRAS mutation rate could be observed in the miR-142-3p high versus low expressing group (17 vs. 8.5%; P=0.057).
In summary, miR-142 is recurrently but infrequently mutated in MDS, de novo and secondary AML evolving from MDS. Mutations in miR-632 and miR-891 are even rarer. MicroRNA mutations co-occur with MDS-related gene aberrations like IDH mutations and are associated with normal cytogenetics and poor survival. We show that miR-142-3p mutations affecting the seed region of the miRNA reduce target specificity. These data suggest that loss of target control by miR-142 may be a novel mechanism in the pathogenesis of leukemia.
Acknowledgments
We thank all patients and contributing doctors for participation in this study. We thank Iris Dallmann for virus production.
Footnotes
Funding: this work was supported by grants 109003, 110284, 110292 and 111267 from Deutsche Krebshilfe; grant DJCLS R13/14 from the Deutsche-José-Carreras Leukämie-Stiftung e.V; DFG grants: HE 5240/5-1, SCHE550/6-1 ED34/4-1; the Dieter-Schlag-Stiftung grant from 2013; the German Federal Ministry of Education and Research grant 01EO0802; HiLF grant from Hannover Medical School and a grant from the MHH Tumorstiftung.
Information on authorship, contributions, and financial & other disclosures was provided by the authors and is available with the online version of this article at www.haematologica.org.
References
- 1.Ji J, Shi J, Budhu A, et al. MicroRNA expression, survival, and response to interferon in liver cancer. N Engl J Med. 2009;361(15):1437–1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Calin GA, Ferracin M, Cimmino A, et al. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med. 2005;353(17):1793–1801. [DOI] [PubMed] [Google Scholar]
- 3.Wang F, Wang XS, Yang GH, et al. miR-29a and miR-142-3p downregulation and diagnostic implication in human acute myeloid leukemia. Mol Biol Rep. 2012;39(3):2713–2722. [DOI] [PubMed] [Google Scholar]
- 4.Network TCGAR Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368(22):2059–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Damm F, Heuser M, Morgan M, et al. Single nucleotide polymorphism in the mutational hotspot of WT1 predicts a favorable outcome in patients with cytogenetically normal acute myeloid leukemia. J Clin Oncol. 2010;28(4):578–585. [DOI] [PubMed] [Google Scholar]
- 6.Thol F, Kade S, Schlarmann C, et al. Frequency and prognostic impact of mutations in SRSF2, U2AF1, and ZRSR2 in patients with myelodysplastic syndromes. Blood. 2012;119(15):3578–3584. [DOI] [PubMed] [Google Scholar]
- 7.Thol F, Suchanek KJ, Koenecke C, et al. SETBP1 mutation analysis in 944 patients with MDS and AML. Leukemia. 2013;27(10):2072–2075. [DOI] [PubMed] [Google Scholar]
- 8.Thol F, Damm F, Lüdeking A, et al. Incidence and prognostic influence of DNMT3A mutations in acute myeloid leukemia. J Clin Oncol. 2011;29(21):2889–2896. [DOI] [PubMed] [Google Scholar]
- 9.Thol F, Bollin R, Gehlhaar M, et al. Mutations in the cohesin complex in acute myeloid leukemia: clinical and prognostic implications. Blood. 2014;123(6):914–920. [DOI] [PubMed] [Google Scholar]
- 10.Thol F, Winschel C, Sonntag AK, et al. Prognostic significance of expression levels of stem cell regulators MSI2 and NUMB in acute myeloid leukemia. Ann Hematol. 2013;92(3):315–323. [DOI] [PubMed] [Google Scholar]
- 11.Thol F, Yun H, Sonntag AK, et al. Prognostic significance of combined MN1, ERG, BAALC, and EVI1 (MEBE) expression in patients with myelodysplastic syndromes. Ann Hematol. 2012. April 10 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 12.Wang XS, Gong JN, Yu J, et al. MicroRNA-29a and microRNA-142-3p are regulators of myeloid differentiation and acute myeloid leukemia. Blood. 2012;119(21):4992–5004. [DOI] [PubMed] [Google Scholar]
- 13.Erdogan B, Facey C, Qualtieri J, et al. Diagnostic microRNAs in myelodysplastic syndrome. Exp Hematol. 2011;39(9):915–926 e912. [DOI] [PubMed] [Google Scholar]
- 14.Bellon M, Lepelletier Y, Hermine O, Nicot C. Deregulation of microRNA involved in hematopoiesis and the immune response in HTLV-I adult T-cell leukemia. Blood. 2009;113(20):4914–4917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lv M, Zhang X, Jia H, et al. An oncogenic role of miR-142-3p in human T-cell acute lymphoblastic leukemia (T-ALL) by targeting glucocorticoid receptor-alpha and cAMP/PKA pathways. Leukemia. 2012;26(4):769–777. [DOI] [PubMed] [Google Scholar]


