Abstract
A series of 17-cyclopropylmethyl-3,14β-dihydroxy-4,5α-epoxy-6α-(isoquinoline-3′-carboxamido)morphinan (NAQ) analogues were synthesized and pharmacologically characterized to study their structure-activity relationship at the mu opioid receptor (MOR). The competition binding assay showed two-atom spacer and aromatic side chain were optimal for MOR selectivity. Meanwhile, substitutions at the 1′- and/or 4′-position of the isoquinoline ring retained or improved MOR selectivity over the kappa opioid receptor while still possessing above 20-fold MOR selectivity over the delta opioid receptor. In contrast, substitutions at the 6′-and/or 7′-position of the isoquinoline ring reduced MOR selectivity as well as MOR efficacy. Among this series of ligands, compound 11 acted as an antagonist when challenged with morphine in warm-water tail immersion assay and produced less significant withdrawal symptoms compared to naltrexone in morphine-pelleted mice. Compound 11 also antagonized the intracellular Ca2+ increase induced by DAMGO. Molecular dynamics simulation studies of 11 in three opioid receptors indicated orientation of the 6’-nitro group varied significantly in the different “address” domains of the receptors and played a crucial role in the observed binding affinities and selectivity. Collectively, the current findings provide valuable insights for future development of NAQ-based MOR selective ligands.
Keywords: MOR, antagonist, SAR, selectivity
1. Introduction
There are three main types of opioid receptors: mu (MOR), kappa (KOR) and delta (DOR), all of which belong to the Class A rhodopsin-like G protein-coupled receptor (GPCR) superfamily.1 The MOR interacts mostly with Gai/o.2 Following MOR activation, G protein-gated inwardly rectifying K+ (GIRK) channels3 open up while voltage-gated Ca2+ channels (VGCC)4 and intracellular adenylate cyclase-mediated cyclic adenosine monophosphate (cAMP)5 production are inhibited. All of these cascades lead to membrane potential decrease, neuronal excitability, neurotransmitter release, and downstream signaling through their second messenger systems which ultimately affect gene expression.6 Behavioral effects manifested through MOR activation include antinociception as well as reward-related behaviors such as substance (e.g. opioid and alcohol) abuse and addiction.6–8
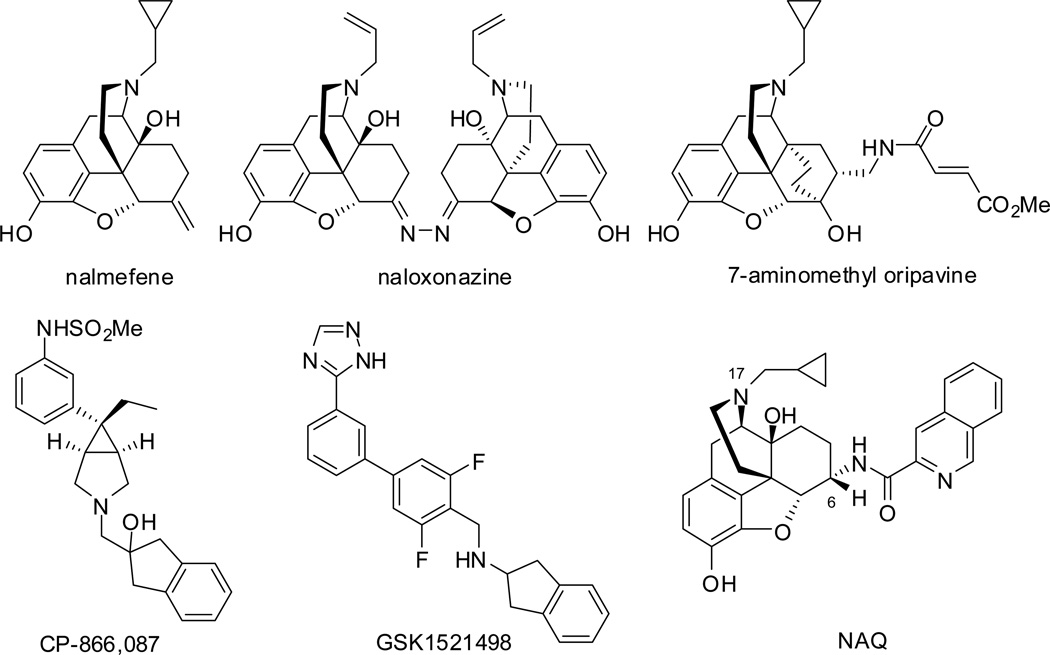
As highlighted in the 2013 World Drug Report, “Opioids remained the most commonly reported group of substances involved in drug-related deaths”.9 Opioid overdose was a major cause of mortality. Currently, the available treatment for opioid overdose is administration of opioid antagonists, such as naloxone (short action) and naltrexone (long action).10 Opioid abuse and addiction is a risk factor for opioid overdose. The MOR full agonist methadone, partial agonist buprenorphine, and antagonist naltrexone are used to treat opioid addiction presently.11–13 These three medications serve as “proof-of-concept” that targeting MOR would deliver therapeutic effect for opioid addiction while their associated shortcomings, such as overdose risk or hepatotoxicity,14–16 call for new molecules with improved pharmacological profiles. Under such a context, scientists and researchers have identified a number of MOR selective ligands. Some of the representative antagonists are illustrated in Figure 1.17–22 Among them, several newly developed ligands showed promising therapeutic benefit for alcohol (CP-866,08720) and food (GSK152149821) addiction, while limited success has been achieved for opioid addiction.
Figure 1.
Representative MOR antagonists used as therapeutic agents or under clinical/preclinical investigation.
Our research interest in developing MOR selective antagonists for treatment of neurological disorders led to the identification of a potent and highly selective MOR ligand NAQ, a C(6)-isoquinoline substituted naltrexone derivative based on a MOR homology model and the “message-address” concept (Figure 1).22 NAQ acted as a low-efficacy MOR partial agonist in the [35S]GTPγS binding assay, but antagonized the effects of MOR full agonist [d-Ala2-MePhe4-Gly(ol)5]enkephalin (DAMGO) in the [35S]GTPγS binding assay and MOR agonist morphine in the warm-water tail immersion assay.22, 23 NAQ produced weak intracranial self-stimulation (ICSS) facilitation in opioid-naïve rats but more robust ICSS facilitation in opioid-experienced rats. It also significantly reversed morphine withdrawal-associated depression of ICSS. These effects of NAQ were similar to that of nalbuphine.24 The ICSS results thus agree with the in vitro characterization of NAQ as a low-efficacy MOR partial agonist and indicate that NAQ may serve as a relatively safe option for treatment of opioid withdrawal or dependence.
NAQ, however, also showed relatively high efficacy and moderate potency in the DOR [35S]GTPγS binding assay.23 It has been proposed that DOR activation is also involved in the development of morphine dependence.25 Thus a NAQ analogue showing less DOR activation while retaining the selective MOR low-efficacy partial agonistic and antagonistic properties would possess more promising therapeutic potentials. We herein report the structure-activity relationship (SAR) studies of the first generation of NAQ analogues during such a pursuit. All the synthesized compounds were first evaluated for binding affinities in opioid receptor radioligand competition binding assays, MOR functions in the MOR [35S]GTPγS binding assay, and acute antinociceptive agonistic and antagonistic effects in the warm-water tail immersion assay. Compound 11 (NNQ), which demonstrated in vivo opioid antagonism activity, was further tested for its potential opioid-withdrawal symptoms in chronic morphine-dependent mice. This new ligand was also docked into the recently determined crystal structures of three opioid receptors followed by molecular dynamics simulations to gain insights into its selectivity profile to guide future molecular design.
2. Results and discussion
2.1. Molecular design
Previous modeling studies in which NAQ was docked into the recently determined crystal structures of the MOR26, KOR27 and DOR28 yielded two different binding sites as “address” domains that interacted with the side chain of NAQ (i.e. the spacer plus the isoquinoline ring): one located at the top of transmembrane helices 6 and 7, and the other at the interface of helix 5 and extracellular loop 2 (ECL2).29 While several residues (such as Trp318 and Lys303) within these MOR “address” domains were proposed to form favorable hydrogen bonding or aromatic stacking interactions with the side chain of NAQ, others (such as Glu229) may also provide favorable interactions upon modification of the NAQ side chain. To further enrich our understanding of the postulated MOR “address” domains and more importantly, to study the SAR of NAQ, a series of NAQ analogues were designed based on our modeling studies and the Craig plots30. The following features were varied: the distance between the isoquinoline ring and the epoxymorphinan skeleton, the aromatic character of the C(6) side chain, and the substitutions on the isoquinoline ring. To have a wide range of the partition constant π, the Hammett substituent constant σ, and the resonance constant ER values for appropriate SAR studies, different functional groups, such as −Cl and −NO2, −CN, −OMe and −OH, −NMe2 and −Me were chosen as the substituents on the isoquinoline ring to cover all the quadrants of the Craig plots.
2.2. Chemistry
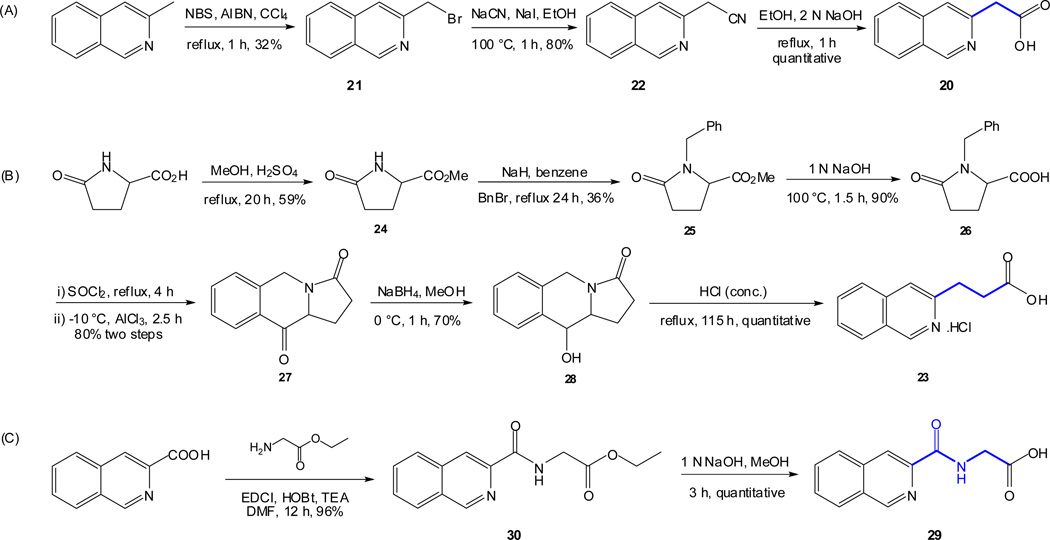
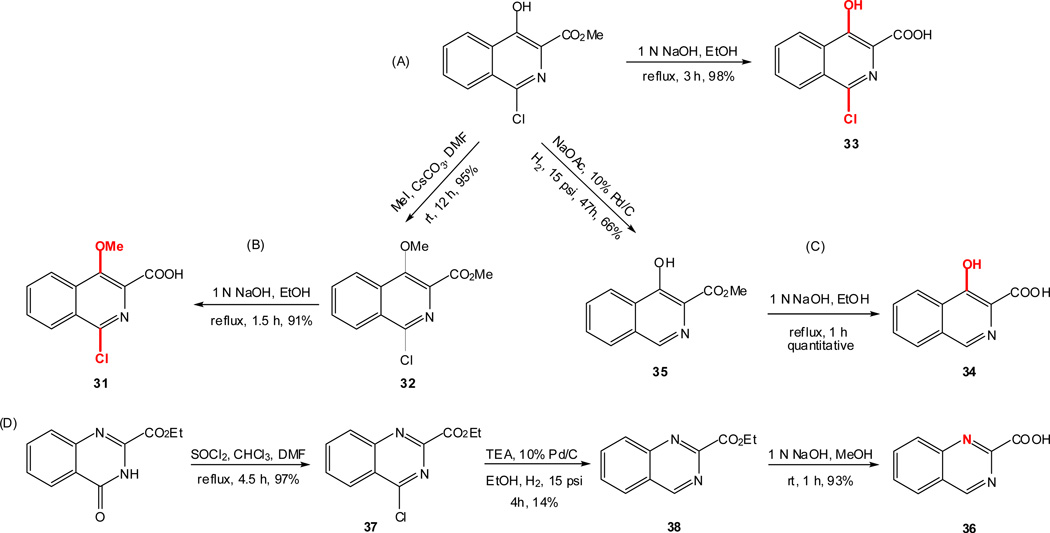
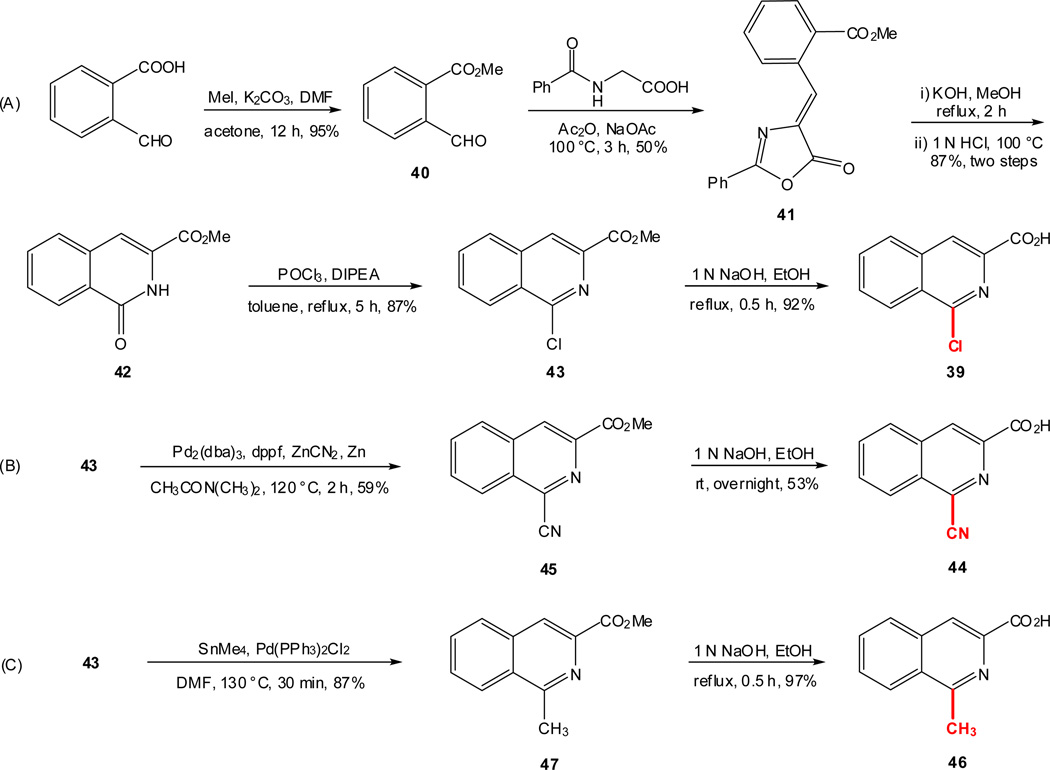
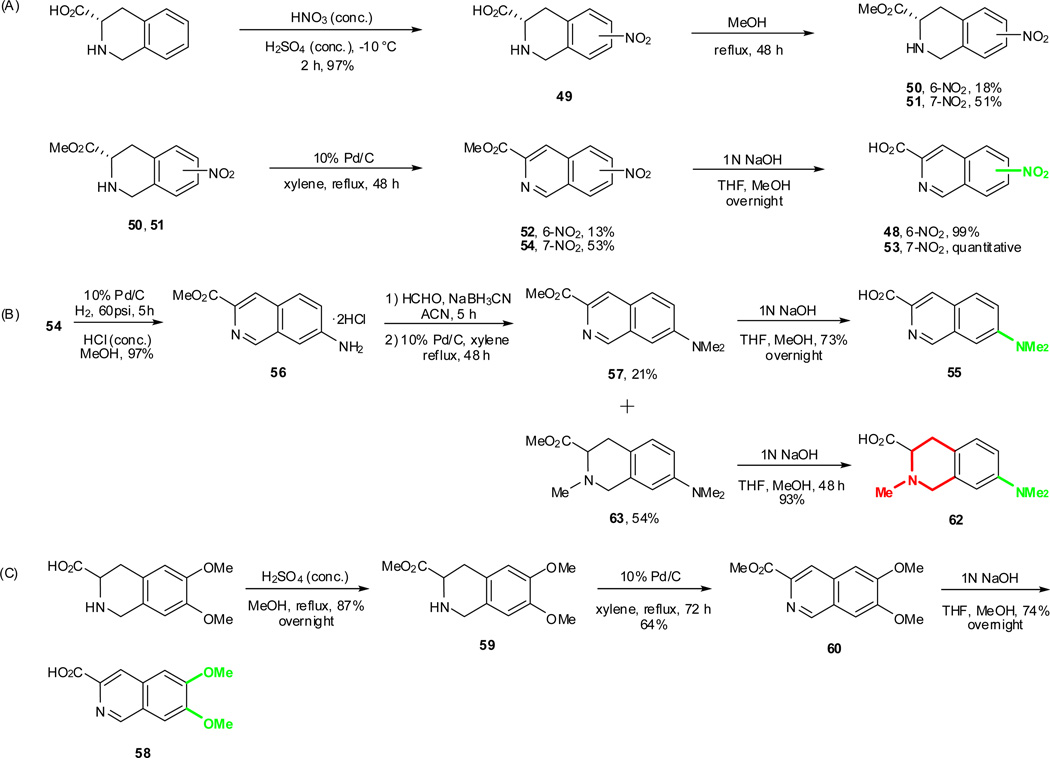
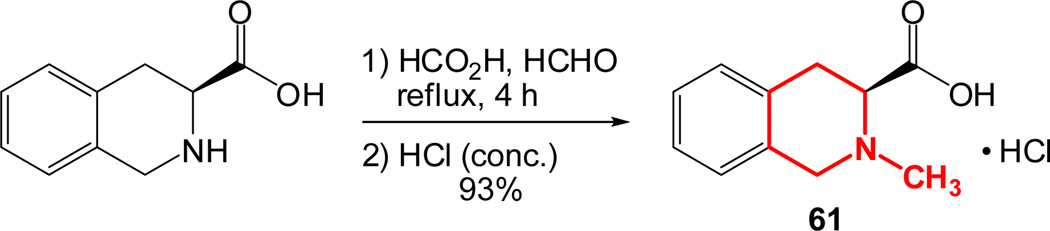
As outlined in the previous section, the proposed SAR study of NAQ would mainly focus on its side chain while keeping the epoxymorphinan core intact. Thus, the syntheses of substituted isoquinoline-3-carboxylic acids and their saturated analogues became the first major task. Interestingly, there are very limited reports on the synthesis of isoquinoline-3-carboxylic acids or their esters. Based on these literature reports and others describing similar functional group transformation as well as commercial availability of starting materials and synthetic challenges, five groups of desirable substituted isoquinoline-3-carboxylic acids or saturated analogues, including several new compounds, were synthesized via multistep synthesis (Scheme 1–Scheme 5, see details in supplementary data). The majority of the reactions went smoothly with good to excellent yields. To be noted, the reductive amination of intermediate 56 with NaBH3CN also reduced the aromatic ring containing the nitrogen atom, which further yielded two products, 57 and 63, after being treated with 10% Pd/C in boiling xylene (Scheme 4). (S)-2-Methyl-1,2,3,4-tetrahydroisoquinoline-3-carboxylic acid hydrochloride (61) was prepared by Eschweiler-Clarke reaction adopting the reported procedure31 (Scheme 5).
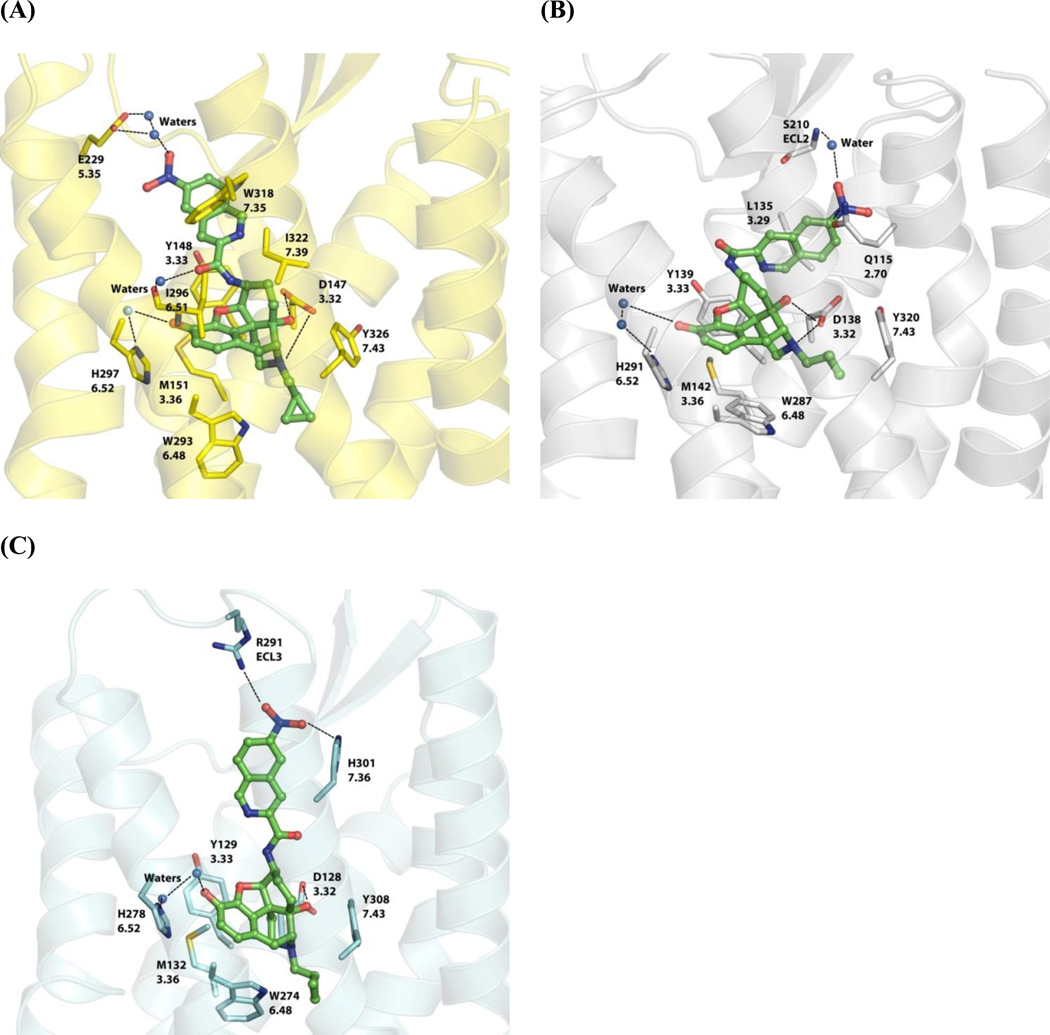
Figure 5.
Lowest interaction energy-associated NNQ poses after a 15-ns molecular dynamic simulation with three opioid receptors: (A) MOR; (B) KOR; (C) DOR. NNQ is represented by balls and sticks (green carbon atoms) and the interacting opioid receptor residues are shown as capped sticks (MOR = yellow; KOR = white; DOR = cyan). Ionic/dipole-dipole interactions and hydrogen bonds are shown with black dashed lines. Opioid receptor amino acid residues are labeled with their sequence number and Ballesteros-Weinstein index53.
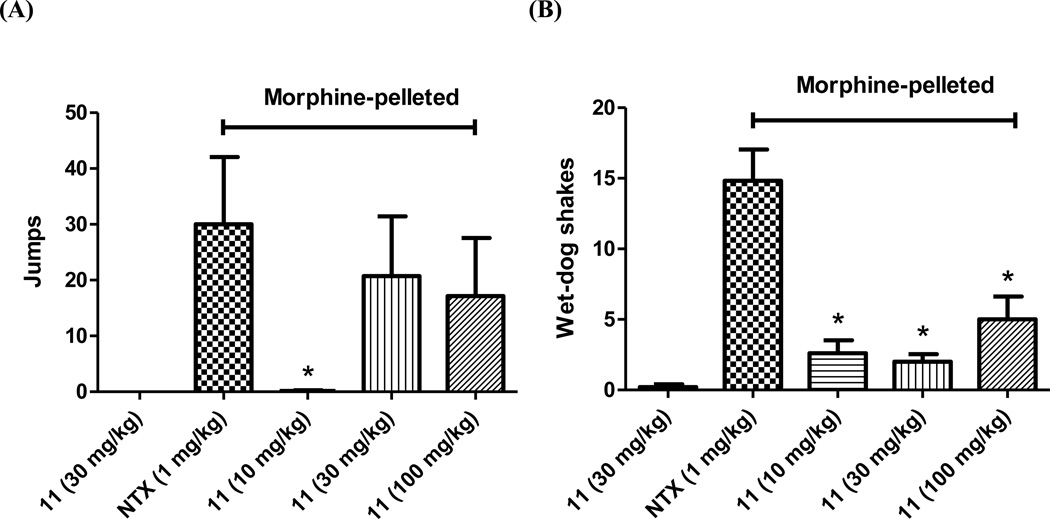
Figure 4.
Compound 11 (s.c.) in opioid-withdrawal assays in chronic morphine-exposed mice (n ≥ 6): (A) Escape jumps; (B) Wet-dog shakes. The first column in each figure represents placebo-pelleted mice while the second to the fifth represent morphine-pelleted mice. * indicates P < 0.05, compared to naltrexone (NTX, s.c).
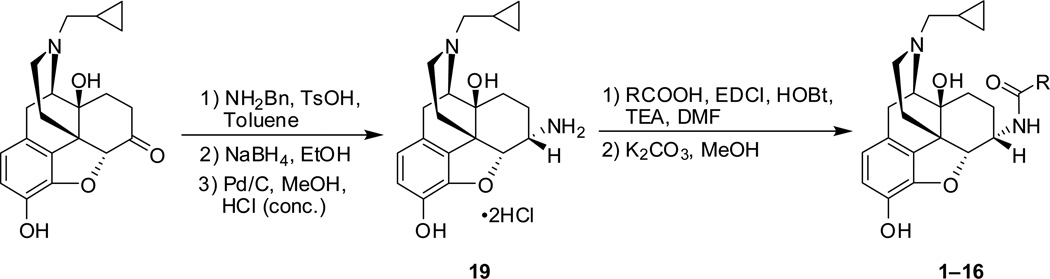
To prepare the epoxymorphinan core part in the designed NAQ analogues, naltrexone was reacted with benzylamine and sodium borohydride, followed by catalytic hydrogenation in the presence of concentrated hydrochloric acid to yield 6α-naltrexamine dihydrocholoride (6α-NTA·2HCl)32 in a total yield of 79%. Then the prepared substituted isoquinoline-3-carboxylic acids or their saturated counterparts were coupled to 6α-NTA·2HCl using EDCI/HOBT method33–35. After treating the coupling mixture with K2CO3 in methanol, the products were then purified by silica gel column chromatography with reasonable yields (Scheme 6). A total of sixteen compounds were successfully synthesized as the first generation of NAQ analogues.
Scheme 1.
Synthetic routes of isoquinoline-3-carboxylic acids 20, 23, and 29 with different spacer lengths.
2.3. In vitro and in vivo pharmacological studies
Both in vitro and in vivo assays were carried out to characterize the first generation of NAQ analogues. The known MOR antagonist naltrexone (NTX) was tested along as a control in all the studies.
2.3.1. In vitro radioligand binding assay and MOR [35S]GTPγS functional assay
The synthesized NAQ analogues were first evaluated in radioligand competition binding assays and MOR [35S]GTPγS functional assay on mouse opioid receptor transfected Chinese hamster ovarian (CHO) cell membranes for their binding affinity, selectivity and MOR agonism/antagonism in vitro. The competitive radioligand binding assays were performed as described previously.36, 37 To compare with previous results as well as for consistency purpose, [3H]naloxone (NLX), [3H]naltrindole (NTI), and [3H]diprenorphine (DPN) were used to label the MOR, DOR and KOR, respectively. The MOR [35S]GTPγS binding assay was conducted to determine the efficacy of each ligand at the MOR as reported earlier.23 The results were interpreted as the potency (EC50) and the relative efficacy (% Emax, the maximum response of each compound to the response of the MOR full agonist DAMGO at 3 µM) for MOR activation. A portion of the results have been briefly reported earlier.35
As seen in Table 1, all sixteen NAQ analogues retained sub-nanomolar to single digit nanomolar binding affinities at the MOR except the one carrying methoxy groups at the 6′- and 7′-positions of the isoquinoline ring (14). Therefore these two positions may not be suitable to introduce electron-donating groups simultaneously when a potent MOR ligand is desirable.
Table 1.
Binding Affinity, Selectivity and Stimulation of MOR [35S]GTPγS Binding for NAQ Analogues.a
| Compd. | R | Ki(nM) |
Selectivity |
MOR [35S]GTPγS
Binding |
ClogP | ||||
|---|---|---|---|---|---|---|---|---|---|
| µ | κ | δ | µ/κ | δ/µ | EC50 (nM) | %Emax of DAMGO | |||
| NTX | NA | 0.33 ± 0.02 | 1.44 ± 0.11 | 143.5 ± 13.7 | 4.4 | 435 | 0.16 ± 0.04 | 5.4 ± 0.8 | |
| NAQ |  |
1.11 ± 0.07 | 13.3 ± 1.1 | 161.9 ± 15.0 | 12 | 146 | 3.3 ± 0.4 | 20.8 ± 1.2 | |
| 1 | 1.20 ± 0.04 | 1.10 ± 0.15 | 12.5 ± 0.7 | 0.9 | 10 | 4.6 ± 0.6 | 18.6 ± 1.1 | ||
| 2 |  |
0.68 ± 0.05 | 1.61 ± 0.04 | 8.4 ± 0.7 | 2.4 | 12 | 1.14 ± 0.11 | 27.38 ± 0.35 | |
| 3 |  |
2.7 ± 1.4 | 0.61 ± 0.04 | 9.2 ± 0.4 | 0.2 | 3.4 | 14.1 ± 4.1 | 13.9 ± 1.9 | |
|
4 (NCQ) |
 |
0.55 ± 0.01 | 22.2 ± 2.1 | 33.9 ± 0.5 | 40 | 62 | 1.74 ± 0.13 | 51.0 ± 0.4 | |
| 5 |  |
0.73 ± 0.07 | 18.3 ± 1.9 | 17.4 ± 1.8 | 25 | 24 | 1.23 ± 0.09 | 19.8 ± 0.8 | |
| 6 |  |
0.45 ± 0.02 | 4.0 ± 0.4 | 32.8 ± 1.5 | 8.9 | 73 | 1.06 ± 0.03 | 20.5 ± 0.9 | |
| 7 |  |
1.11 ± 0.06 | 5.1 ± 0.3 | 78.8 ± 0.7 | 4.6 | 71 | 6.0 ± 1.5 | 21.6 ± 0.8 | |
| 8 |  |
1.26 ± 0.04 | 10.8 ± 1.2 | 79.8 ± 2.4 | 8.6 | 63 | 2.62 ± 0.38 | 26.4 ± 0.9 | |
| 9 |  |
0.99 ± 0.07 | 10.1 ± 0.5 | 129.9 ± 9.6 | 10 | 131 | 3.32 ± 0.24 | 37.5 ± 0.7 | |
| 10 |  |
2.1 ± 0.2 | 29.1 ± 0.7 | 117.5 ± 7.3 | 14 | 56 | 7.2 ± 0.5 | 22.2 ± 0.4 | |
|
11 (NNQ) |
 |
5.7 ± 1.7 | 27.9 ± 2.0 | 94.7 ± 1.1 | 4.9 | 16 | 31.5 ± 18.7 | 12.5 ± 1.4 | 3.10 |
| 12 |  |
3.6 ± 1.1 | 16.0 ± 1.4 | 55.3 ± 1.8 | 4.5 | 16 | 42.7 ± 23.7 | 13.4 ± 0.6 | 3.10 |
| 13 |  |
4.4 ± 0.4 | 88.0 ± 6.9 | 68.6 ± 2.5 | 20 | 16 | 4.70 ± 0.62 | 6.32 ± 0.30 | 3.83 |
| 14 |  |
13.4 ± 0.4 | 61.0 ± 2.8 | 69.2 ± 4.8 | 4.6 | 5.2 | 85.2 ± 8.6 | 14.0 ± 0.4 | |
| 15 |  |
2.9 ± 1.3 | 6.76 ± 0.54 | 9.3 ± 0.3 | 2.3 | 3.2 | 19.4 ± 10.3 | 24.8 ± 1.3 | |
| 16 |  |
1.79 ± 0.06 | 4.91 ± 0.23 | 4.5 ± 0.1 | 2.7 | 2.5 | 11.4 ± 0.9 | 26.2 ± 0.6 | |
The values are the means ± S.E.M. of four independent experiments. Membranes were prepared from mMOR-, mKOR-, and mDOR-CHO cells. The Bmax values for the MOR, KOR, and DOR are 3.20 ± 0.41, 1.500 ± 0.139, and 3.667 ± 0.002 pmol/mg. [3H]Naloxone, [3H]naltrindole and [3H]diprenorphine were used to label the MOR, DOR and KOR, respectively. The percentage maximal stimulation to DAMGO (% Emax of DAMGO) is the maximal response of the compound compared to that of 3 µM DAMGO (normalized to 100%).
Comparing the binding data at both MOR and KOR, 1′,4′-disubstitution (4 and 5), 1′-substitution (8–10), or an electron-donating group at the 4′- or 7′-position (6 and 13) yielded comparable or improved MOR/KOR selectivity relative to NAQ, while the other structural modifications moderately decreased MOR/KOR selectivity. Among this series of NAQ analogues, 4 showed the highest MOR/KOR selectivity, nearly 10-fold higher than that of naltrexone.
With respect to the MOR and the DOR binding affinities, ligands containing different electronic characteristics substituents at the 1′- and/or 4′-position of the isoquinoline ring (4–10) showed >50-fold MOR/DOR selectivity, except for 5. The remaining analogues showed little to medium MOR/DOR selectivity. Compound 9 with a 1′-cyano group was the most MOR/DOR selective ligand in this series of NAQ analogues and displayed similar MOR/DOR selectivity as NAQ.
A majority of the first generation of NAQ analogues showed less than 30% of the maximal stimulation observed with 3 µM DAMGO in the MOR [35S]GTPγS binding assay, except for 4 and 9 (Table 1). Among them, 3 with a glycine-unit spacer and 11–14 carrying substitutions with different electronic characteristics at the 6′- and/or 7′-position of the isoquinoline ring had less than 15% MOR stimulation relative to DAMGO. Interestingly, NAQ and each of its analogues examined here were 1 to 2 orders of magnitude less potent than naltrexone in the MOR [35S]GTPγS binding assay, especially for 11, 12 and 14 (≥ 200-fold). The EC50 and Emax for morphine (M) and buprenorphine (B) in the mMOR [35S]GTPγS binding assay were previously tested in our labs: M, 120 ± 24 nM and 106 ± 3.0 %; B, 3.6 ± 1.2 nM and 43 ± 2.4 %.38 It thus seems that 4, the most efficacious MOR ligand in this series of NAQ analogues, is more potent but less efficacious than morphine, whereas it possesses similar potency and efficacy as buprenorphine.
Collectively, the in vitro competition binding studies of NAQ revealed that a two-atom spacer and an aromatic ring system is favorable for MOR selectivity whereas different positions and electronic characteristics of substituents on the isoquinoline ring affect MOR selectivity in different patterns. Nonetheless, the in vitro radioligand binding studies of the first generation of NAQ analogues identified five ligands (4, 5, 9, 10, and 13) with over 10-fold MOR/KOR and 15-fold MOR/DOR selectivity, which is comparable to that of the reported MOR antagonist cyprodime39. As for the MOR efficacy study, the most significant observation was that substitutions at the 6′- and/or 7′-position of the isoquinoline ring and the glycine-unit spacer reduced MOR efficacy. The studies also provide us valuable information for future molecular design. For example, combining the encouraging MOR selectivity feature of 1′,4′-disubstitutions with the low MOR efficacy property of 6′/7′-substitution might yield more selective MOR ligands with relative low efficacy.
2.3.2. Warm-water tail immersion assay
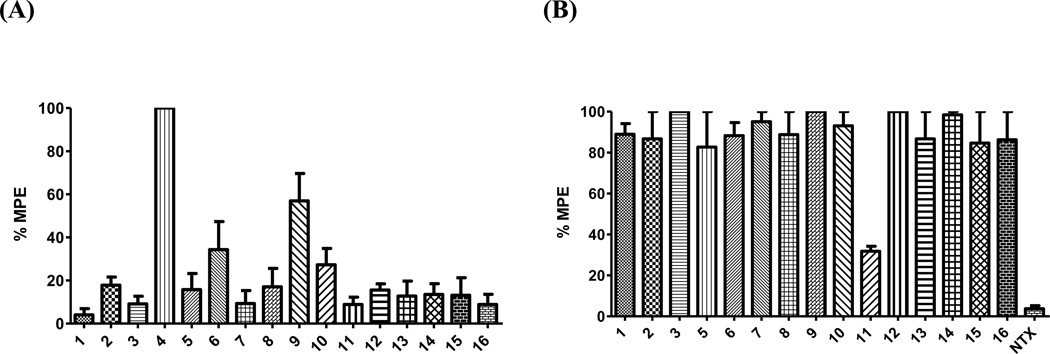
The warm-water tail immersion assay is one of the tests used to measure the efficacy of analgesics.40 The advantage of this assay is its relative reproducibility41 and was thus employed in the current study. Each NAQ analogue was tested for its ability to produce antinociception and/or to antagonize the antinociceptive effects of morphine in mice. The results are interpreted as the percentage of maximum possible effect (% MPE). A higher % MPE indicates a stronger antinociception effect by the ligand. Figure 2A depicts the intrinsic antinociceptive effects of the sixteen NAQ analogues at 10 mg/kg (This dose was chosen so as to compare with 10 mg/kg morphine, which provides 90–100% MPE in the tail immersion test). Not surprisingly, 4 (NCQ), which showed the highest MOR maximal stimulation in the in vitro [35S]GTPγS binding assay, acted as a full agonist in the in vivo warm-water tail immersion assay, resembling the MOR agonist morphine. The ED50 values of 4 and morphine were calculated to be 0.73 (0.55–0.97), and 3.24 (2.44–4.31) mg/kg (95% CL), respectively. The much smaller difference between the potencies of 4 and morphine in the tail immersion assay (∼9-fold) compared to that in the MOR [35S]GTPγS binding assay (∼70 fold) suggests that pharmacokinetics may play significant roles in the in vivo study. Compound 9 produced a % MPE of 56.9 ± 12.7, which was also consistent with its second highest MOR maximal stimulation in the [35S]GTPγS binding assay. The % MPEs of the remaining NAQ analogues, except for 6 and 10, were less than 20%, indicating either weak opioid receptor agonism (as shown in the in vitro MOR [35S]GTPγS binding assay) or unfavorable pharmacokinetic properties (such as low CNS permeability) since tail immersion response is mainly mediated at the spinal levels of CNS42.
Figure 2.
Warm-water tail immersion assay in mice (n ≥ 6) at 56 ± 0.1 °C. All tested compounds were administered subcutaneously (s.c). (A) Antinociceptive effects of NAQ analogues 1–16. Compounds (10 mg/kg) were injected at Time 0. Twenty minutes after injection, tail flick was assessed with hot water. (B) Blockage of the antinociceptive effect of morphine by NAQ analogues 1–3, 5–16, and naltrexone (NTX). Tested compounds (1 mg/kg) were injected at Time 0. Five minutes later, morphine (10 mg/kg) were administered. Twenty minutes after morphine injection, tail flick was tested using hot water.
Figure 2B portrays the antinociceptive effect of morphine at 10 mg/kg in the presence of each NAQ analogue at 1 mg/kg. Due to the full agonism of 4 as seen in Figure 2A, it was not tested in this antagonism study. Compound 11 (NNQ) effectively blocked morphine-mediated antinociception. Its calculated AD50 value is 0.92 (0.51–1.67) mg/kg (95% CL), which is less potent than naltrexone (Table 2). This may be partially due to their different binding affinities at the MOR: naltrexone is nearly 20-fold more potent than 11 (Table 1). The other fourteen NAQ analogues had marginal impact on the antinociceptic effect of morphine. For example, compound 13 showed similar affinity to the MOR compared to 11 with lower MOR efficacy than 11 in the in vitro [35S]GTPγS binding assay but showed no significant antinociception effect. This could possibly due to more rapid metabolic excretion rate of the dimethylamino moiety43 compared to a nitro group44 at the in vivo level so that 13 could not block the antinoceiception of morphine in the in vivo warm-water tail withdrawal assay.
Table 2.
AD50 Values of 11 to Antagonize Morphine-mediated (10 mg/kg) Antinociceptive Effect in Warm-Water Tail Immersion Assay in Mice (n ≥ 6)a
| Compd. | AD50 values (mg/kg (95% CL)) |
|---|---|
| Naltrexone | 0.006 (0.003–0.014) |
| NAQb | 0.45 (0.27–0.78) |
| 11 (NNQ) | 0.92 (0.51–1.67) |
All drugs and test compounds were administered subcutaneously (s.c.) to a group of at least six mice.
Data taken from Li, et al.22
Of note, 11 and 12 are regionisomers. They shared a similar pharmacological profile including MOR binding affinity, selectivity, [35S]GTPγS binding, and intrinsic agonism in the tail immersion test. On the other hand, they acted differently when challenged with morphine in the tail immersion assay. Compound 11 was able to antagonize morphine while compound 12 showed no effect. Distinct pharmacological profiles of regioisomers have been reported for other opioid ligands, such as 5’-guanidinonaltrindole (5’-GNTI) and 6’-GNTI. In that case, both ligands have comparable binding affinities at the MOR and the DOR, with 5’-GNTI is 8-fold more potent than 6’-GNTI at the KOR. However, 5’-GNTI acts as a KOR antagonist while 6’-GNTI behaves like a KOR full agonist when tested on the electrically stimulated guinea pig ileal longitudinal muscle.45 It was suggested that a conformational change of transmembrane helix 6 that was induced by the interaction between 6’-guanidinium and Glu297 led to KOR activation. Further investigation is certainly warranted to see if similar scenario might happen to compound 12.
To be noticed, the binding affinity of morphine at the mMOR-CHO membrane under the same assay conditions is 1.00 ± 0.03 nM ([3H]DAMGO as the radioligand), which is comparable to most of the NAQ analogues, except for 14 (Table 1). Because the majority of these NAQ analogues (1 mg/kg) didn’t antagonize the antinociceptive effects of 10 mg/kg morphine, we decided not to pursue further studies on them except 4 and 11.
2.3.3. The KOR and DOR [35S]GTPγS binding assays for 4 and 11
To further understand the underlying mechanisms of 4 (a full agonist) and 11 (low-efficacy partial agonist and moderate-potency antagonist) in the warm-water tail immersion assay, KOR and DOR [35S]GTPγS functional assays were performed for these two compounds to see if KOR and DOR play any roles for the observed activities. The KOR full agonist (±)-trans-2-(3,4-dichlorophenyl)-N-methyl-N-(−2-pyrrolidin-1-ylcyclohexyl)acetamide (U50,488)46 and DOR full agonist 4-[(R)-[(2S,5R)-4-allyl-2,5-dimethylpiperazin-1-yl](3-methoxyphenyl)methyl]-N,N-diethylbenzamide (SNC80)47 were included in the assays as reference compounds for a maximal effect at the KOR and the DOR, respectively.
As seen in Table 3, compared to naltrexone and NAQ, 4 acted as a low-potency KOR partial agonist with low efficacy (7.1 ±0.5 %). Compound 11 displayed a comparable KOR efficacy to naltrexone, while nearly 80-fold less potent than naltrexone. The potency of 11 was nearly 6-fold lower than that of NAQ though it had a relatively higher KOR efficacy than NAQ. The reduced potency of 11 at KOR [35S]GTPγS binding compared to both naltrexone and NAQ is particularly encouraging since KOR activation has been reported to be considerably involved in dysphoria and anhedonia associated with drug withdrawal, stress-induced aversion states, and stress-induced relapse-like behavior.48 NAQ, 4 and 11 all induced ∼10-fold higher maximal stimulation than naltrexone at the DOR. However, intriguingly, they were 7- to 25-fold less potent compared to naltrexone.
Table 3.
Stimulation of KOR and DOR [35S]GTPγS Binding for 4 and 11a
| Compd. | KOR [35S]GTPγS
Binding |
DOR [35S]GTPγS
Binding |
Receptor Function Selectivityb |
|||
|---|---|---|---|---|---|---|
| EC50 (nM) | % Emax
of U50,488 |
EC50 (nM) | % Emax
of SNC80 |
κ/µ | δ/µ | |
| NTX | 0.81 ± 0.08 | 20.8 ± 0.9 | 4.4 ± 1.6 | 5.6 ± 0.6 | 5.1 | 28 |
| NAQc | 10.9 ± 7.9 | 13.1 ± 2.0 | 98.6 ± 23.7 | 53.5 ± 5.4 | 3.3 | 30 |
| 4 (NCQ) | 26.4 ± 3.9 | 7.1 ± 0.5 | 32.6 ± 5.1 | 54.8 ± 0.1 | 15 | 19 |
| 11 (NNQ) | 62.6 ± 10.4 | 21.1 ± 0.4 | 108.1 ± 0.8 | 67.7 ± 2.0 | 2.0 | 3.4 |
The values are the means ± S.E.M. of three independent experiments. The percentage maximal stimulation (% Emax) to (±)-trans-U50,488 or SNC80 is the maximal response of the compound compared to that of (±)-trans-U50,488 (3 µM) or SNC80 (5 µM).
Calculated as the ratio of the EC50 values of a ligand at given two opioid receptors in the [35S]GTPγS binding assay.
Data taken from Yuan, et al.23
With respect to their relative potencies in the opioid receptor [35S]GTPγS binding assays, 4 retained moderate MOR/DOR function selectivity compared to naltrexone and NAQ, while displayed a significantly higher MOR/KOR function selectivity than both naltrexone and NAQ. Compound 11, however, exhibited less MOR/DOR function selectivity than either naltrexone or NAQ and showed similar MOR/KOR function selectivity to naltrexone and NAQ. Thus the full agonism of 4 in the warm-water tail immersion assay could be primarily mediated by the MOR. Hough and coworkers have shown that the KOR full agonist U50,488 (s.c) produced 25% MPE at 55 °C in the tail immersion assay while the DOR full agonist [D-Pen2, D-Pen5]enkephalin (intracerebroventricular) exerted a DOR-independent antinociceptive effect at this temperature.49 Based on their report and the function selectivity profile of 11, the weak partial agonism of 11 (8.9 ±3.4 % MPE) as seen in the tail immersion agonism study (Figure 2A) might be mediated through both the MOR and the KOR. Since the analgesic effect of morphine has been mainly attributed to MOR activation, the antagonism of 11 as seen in the tail immersion antagonism study (Figure 2B) would be mainly due to its action at the MOR.
2.3.4. Ca2+ flux assays for 4 and 11
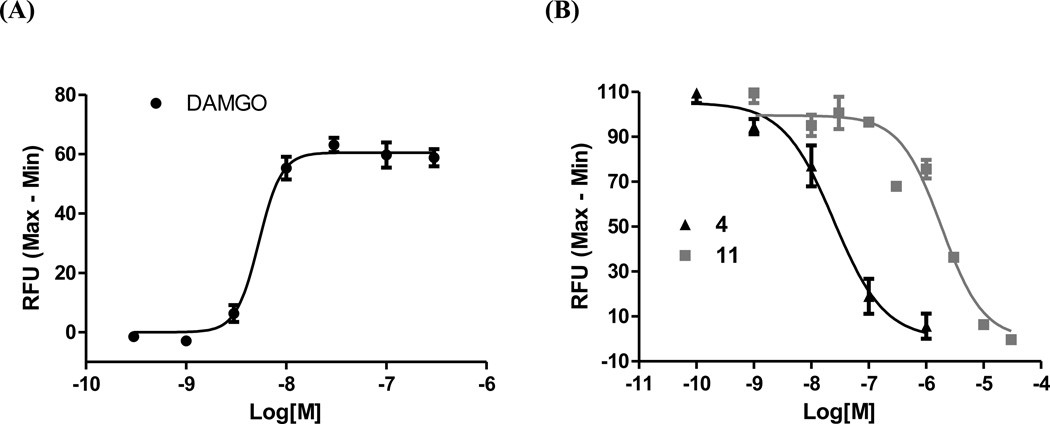
Cytosolic Ca2+ level undergoes rapid and transient increase upon activation of Gq-signaling pathways in CHO cells.50 The intracellular Ca2+ measurement has thus become one of the primary assays to assess GPCR function. To further profile 4 and 11 at the MOR, Ca2+ flux functional assays were carried out in Gαqi5 transfected hMOR-CHO cells. Compound 11 displayed no apparent agonism to augment intracellular Ca2+ level. Interestingly, 4 did not show any significant agonist activity either when compared to the MOR full agonist DAMGO. As reported in the literature recently, a number of GPCR ligands have been identified to display “biased signaling”, i.e., the ability of a ligand to activate only some subset(s) of its receptor’s signaling repertoire. Different ligand-induced receptor conformational states, diversified G proteins and scaffolding/signaling partners, as well as different receptor oligomerization states can all lead to distinctive function and signaling.51 In light of this phenomenon, we hypothesize that the characteristic agonism of 4 in [35S]GTPγS binding and warm-water tail immersion assays compared to its insignificant agonism in Ca2+ flux assay might be due to its biased signaling towards different signaling pathways, which would be interesting to further verify. The determined EC50 of DAMGO is 7.5 ± 1.6 nM (Figure 3A), which is comparable to the data obtained from Cytosensor micrometer (9.61 ± 2.78 nM)50. Both 4 and 11 dose-dependently blocked the Ca2+ increase trigged by DAMGO (Figure 3B). The calculated IC50 values for 4 and 11 are 23.7 ± 3.9 nM and 2728 ± 452 nM, respectively. Compound 4 was about 10-fold more potent than 11 in the cell membrane-based radioligand competition binding assay (Table 1). Therefore, the observed 100-fold higher potency of 4 to 11 in Ca2+ flux antagonism study indicates intrinsic signal amplification occurs in the whole cell-based assay. The IC50 value for naltrexone under same testing conditions is 15.5 ± 0.1 nM.
Figure 3.
Ca2+ flux assays in Gqi5 transfected hMOR-CHO cells. (A) DAMGO dose-dependently increased intracellular Ca2+ levels. (B) Compounds 4 and 11 antagonized the intracellular Ca2+ increase triggered by activation of the MOR with DAMGO. The results shown are representative of at least three independent experiments.
2.3.5. Opioid withdrawal assays
Since 11 antagonized morphine-mediated antinociception in the warm-water tail immersion assay, it was further tested in the chronic opioid withdrawal assays in morphine-pelleted mice. First, 11 itself did not induce any significant withdrawal symptoms in placebo-pelleted mice at 30 mg/kg (Figure 4A and 4B, first columns) while naltrexone (1 mg/kg) induced significant withdrawal signs almost immediately after injection in morphine-pelleted mice and gave 30.0 ± 12.1 times escape jumps and 14.8 ± 2.2 times wet-dog shakes (Figure 4A and 4B, second columns). Second, 11 did not precipitate significant jumps in morphine-pelleted mice until 30 mg/kg and reached a plateau at 100 mg/kg (Figure 4A, columns 3–5). Compared to naltrexone, 11 also produced significantly less wet-dog shakes, even at dose as high as 100 mg/kg (Figure 4B, columns 3–5). Thus, 11 had marginal opioid withdrawal potential at a dose 10-fold of its AD50 value and caused less significant withdrawal symptoms than naltrexone overall. This might be due to its weak partial agonism as seen in the tail immersion agonism study (Figure 2A). Such observations render 11, an opioid low-efficacy partial agonist/antagonist lacking severe opioid withdrawal capacity, as a promising lead compound towards developing novel ligands for substance addiction treatment.
2.4. Molecular modeling studies
To provide insights into the opioid receptor selectivity profile of 11 (NNQ) compared with NAQ and guide future molecular design, molecular modeling studies of NNQ in three opioid receptors were conducted. NNQ was first docked into the optimized crystal structures of three opioid receptors (see details in experimental part). Molecular dynamics (MD) simulations on the best scored solutions were then carried out for all three receptors embedded in a lipid bilayer and solvated with water to allow the receptor-ligand complexes to equilibrate in this simulated biological environment. Energy analyses were then performed on the resulting NNQ-receptor complexes. The most favorable non-bonded interaction energy, calculated using NAMD (a molecular dynamic program) Energy, between NNQ and its surrounding environment (including protein and water molecules) within different cutoff distances of the ligand were determined and summarized in Table 4.
Table 4.
NNQ–Opioid Receptor Interaction Energies (kcal/mol).
| Radiusa(Å) | MOR-NNQ | KOR-NNQ | DOR-NNQ | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Eb | VDWc | Total | Eb | VDWc | Total | Eb | VDWc | Total | |
| 10 | –13.76 | –71.64 | –85.40 | –22.52 | –66.35 | –88.87 | –8.37 | –63.49 | –71.86 |
| 8 | –14.69 | –69.89 | –84.58 | –21.43 | –66.04 | –87.47 | –8.18 | –63.24 | –71.42 |
| 6 | –17.44 | –67.83 | –85.27 | –23.00 | –62.90 | –85.90 | –6.71 | –63.82 | –70.53 |
| 5 | –16.45 | –63.89 | –80.34 | –19.27 | –58.52 | –77.79 | –11.70 | –60.49 | –72.19 |
Distance from the docked ligand NNQ
E: Electrostatic interaction
VDW: Van der Waals’ interaction.
As seen in Table 4, the choice of cutoff distance from the ligand in general did not significantly affect the non-bonded interaction energy between the receptor and the ligand NNQ, except for those of MOR and KOR at 5 Å, in which moderate increases of total energy were observed compared to the corresponding values at larger cutoff radii. The total interaction energies at 5 Å for the NNQ–receptor complexes followed the trend: MOR < KOR < DOR, that is NNQ bound most favorably to the MOR followed by the KOR and then to the DOR, which is supported by a similar trend observed in the experimental binding affinities of NNQ at the three receptors (Table 1).
The binding poses associated with the lowest total energy for NNQ in the three opioid receptors are shown in Figure 5. The epoxymorphinan core remained in its crystallographically-determined binding site and maintained the same interactions as previously observed.26–28 However, orientation of the 6′-nitro group, located in the extracellular loop ‘address’ regions of the opioid receptors,52 was more varied. For the MOR-NNQ complex, the 6′-nitro group was directed toward the top of helix 5, interacting with Glu2295.35 through two bridging water molecules (Figure 5A, 5.35 is the Ballesteros-Weinstein index53). In contrast, the isoquinoline ring was placed between helixes 2 and 3 in the KOR-NNQ complex, with the 6′-nitro group interacting with Gln1152.70 and the backbone NH group of Ser210ECL2 via a water molecule (Figure 5B). However, in the DOR-NNQ complex (Figure 5C), the isoquinoline ring was oriented toward helix 7 with the nitro group interacting with Arg291ECL3 and His3017.36 via hydrogen bonds. This resulted in the loss of the ionic interaction between the carboxylate group of Asp128 and the protonated N(17) atom of the epoxymorphinan skeleton, leaving only one hydrogen bond formed between Asp128 residue and the 14-hydroxyl group. This loss of favorable hydrogen bonding interaction was also reflected by the relatively high electrostatic interaction energy for the DOR–NNQ complex, compared to the MOR–NNQ and KOR–NNQ complexes (Table 4).
Taken together, the molecular modeling study of the interactions between 11 (NNQ) and three opioid receptors well matched with the experimental binding data. Thus it could be utilized as a reliable tool to guide future molecular design. The modeling study also implicated that the nitrogen atom in the nitro group was involved in a dipole-dipole interaction which only occurred in the KOR–NNQ complex. Thus replacing the nitro group with another hydrogen bond acceptor lacking dipole-dipole interaction potential might improve its MOR selectivity over the KOR.
3. Conclusion
In conclusion, the first generation of NAQ analogues were synthesized and pharmacologically characterized to study their structure–activity relationship (SAR) at three opioid receptors, particularly the mu opioid receptor (MOR). All ligands bound to the MOR with sub-nanomolar to nanomolar affinity. Substitutions at the 1′- and/or 4′-position of the isoquinoline ring retained or increased MOR selectivity over the KOR while still possessing more than 20-fold MOR selectivity over the DOR. In contrast, substitution at the 6′- and/or 7′-position of the isoquinoline ring decreased MOR selectivity. A two-atom spacer and an aromatic side chain were preferred for optimal MOR selectivity. Meanwhile, substitutions at the 6′- and/or 7′-position of the isoquinoline ring also reduced MOR efficacy while other structural modifications either retained or increased MOR efficacy in the [35S]GTPγS binding assay. Among this series of NAQ analogues, 4 behaved as a full agonist in the warm-water tail immersion assay, while 11 acted as a low-efficacy partial agonist or a moderate-potency antagonist in the absence or presence of morphine, respectively. The full agonism of 4 in the tail immersion study was mediated primarily by the MOR whereas the weak partial agonism of 11 may be mediated by both the MOR and the KOR. Nevertheless, 11 exerted its antagonism mainly through the MOR. Both 4 and 11 antagonized the intracellular Ca2+ increase induced by DAMGO in Gqi5 transfected hMOR-CHO cells. Furthermore, 11 produced less significant withdrawal symptoms than naltrexone in the chronic opioid withdrawal assay. In addition, molecular dynamics simulation studies of 11 in the crystal structures of three opioid receptors revealed that orientation of the 6′-nitro group varied considerably in the structurally different ‘address’ regions of the receptors and played an important role in the observed binding affinities. All together, the current study identified a new compound (11, NNQ) carrying interesting pharmacological profiles which warrants further investigation on the SAR of NAQ, particularly at the 6’-position of the isoquinoline ring. Meanwhile, the potential application of 11 in treatment of different neurological disorders, such as substance addiction remains to be explored.
4. Experimental
4.1. Chemical synthesis
General methods
Reagents were purchased from either Sigma-Aldrich or Alfa Aesar. TLC analyses were carried out on the Analtech Uniplate F254 plates. Chromatographic purification was carried out on silica gel (230∼400 mesh, Merck) columns. Melting points were obtained with a Fisher scientific micro melting point apparatus without correction. All IR spectra were acquired on a Nicolet iS10 instrument. 1H (400 MHz) and 13C (100 MHz) nuclear magnetic resonance (NMR) spectra were recorded at ambient temperature with tetramethylsilane as the internal standard on Varian Mercury 400 MHz NMR spectrometer. LC-MS analysis was performed on a Waters Micromass high resolution (HR) QTOF-II instrument (ESI source), or an Applied Bio Systems 3200 Q trap with a turbo V source for TurbolonSpray. HPLC analysis was done with a Varian ProStar 210 system on Microsorb-MV 100–5 C8/C18 column (250 mm × 4.6 mm) eluting with acetonitrile (0.1% TFA)/water at 1 mL/min over 15 to 45 min. Elemental analysis was conducted by Atlantic Microlab, Inc. All the analytical methods listed above were used to determine the purity of the newly synthesized compounds and analyses indicated by the symbols of the elements or HRMS were within ± 0.4 % of the theoretical values. Yields were not maximized.
General procedure for amide coupling/saponification reaction
On an ice-water bath, to the solution of acid (3 eq, prepared in house, see Supplementary data for details) in anhydrous DMF (7 mL), was added 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide hydrochloride (EDCI, 3 eq), hydrobenzotriazole (HOBt, 3 eq), 4 Å molecular sieves, and TEA (8.0 eq) under N2 protection. Fifteen minutes later, a solution of 6α-naltrexamine hydrochloride (19, 1.0 eq) in DMF (5 mL) was added dropwise. The resulted mixture was allowed to warm up to ambient temperature gradually. Upon completion of the reaction, the mixture was then filtered through celite. The filtrate was concentrated to remove DMF. Methanol (10 mL), and K2CO3 (3 eq) were then added to the residue and stirred at ambient temperature overnight. The mixture was then filtered through celite again. The filtrate was concentrated to remove methanol. The residue was partitioned between CH2Cl2 (50 mL) and brine (50 mL). The organic layer was separated and dried over anhydrous MgSO4, concentrated under reduced pressure. The residue was then purified by column chromatography, eluting with CH2Cl2/MeOH (1% NH3·H2O) to afford the corresponding compound as free base. Upon confirmation of the structure by 1H NMR and 13C NMR, the compound was then transformed into hydrochloride salt by dissolving the free base in MeOH (0.1 mL) and DCM (2 mL), adding HCl methanol solution (1.25 M, 4 eq) under an ice-water bath, and stirred for 5 min. Diethyl ether (10 mL) was then added. Two hours later, the precipitate was collected by filtration, dried in vacuum to give the target compound as hydrochloride salt, which was then used in the HPLC, LC-MS, elemental analysis, and biological screenings.
17-Cyclopropylmethyl-3,14β-dihydroxy-4,5α-epoxy-6α-[2-(isoquinolin-3-yl)acetamido)morphinan (1)
The title compound was prepared in 70% yield. Hydrochloride salt: 1H NMR (400 MHz, DMSO-d6) δ 9.67 (s, 1H), 9.31 (brs, 1H, exchangeable), 8.84 (brs, 1H, exchangeable), 8.37 (d, J = 8.56 Hz, 1H), 8.33 (d, J = 8.04 Hz, 1H), 8.17–8.15 (m, 2H), 8.04 (t, J = 7.58 Hz, 1H), 7.87 (t, J = 7.46 Hz, 1H), 6.75 (d, J = 8.12 Hz, 1H), 6.58 (d, J = 8.16 Hz, 1H), 6.29 (brs, 1H), 4.64 (d, J = 3.88 Hz, 1H), 4.45 (m, 1H), 4.06 (s, 2H), 3.90 (d, J = 6.64 Hz, 1H), 3.34 (d, J = 19.96 Hz, 1H), 3.26 (m, 1H), 3.09–2.97 (m, 2H), 2.94 (m, 1H), 2.70 (m, 1H), 2.43 (m, 1H), 1.87 (m, 1H), 1.62 (m, 1H), 1.50–1.35 (m, 2H), 1.08–0.95 (m, 2H), 0.67 (m, 1H), 0.61 (m, 1H), 0.47 (m, 1H), 0.39 (m, 1H); 13C NMR (100 MHz, DMSO-d6) δ 169.19, 148.48, 147.37, 141.37, 140.74, 140.50, 138.04, 131.87, 131.35, 130.11, 128.49, 127.87, 126.71, 123.43, 121.08, 119.65, 89.12, 71.00, 63.70, 59.21, 47.52, 47.17, 47.11, 40.01, 31.67, 30.62, 25.02, 21.02, 6.86, 6.24, 3.42. HRMS m/z found 512.2699, calculated 512.2544 (M + H)+. IR (Diamond, cm−1) vmax 3232.8, 1651.9, 1615.6, 1236.4, 1117.6, 765.6, 747.5. mp 203 °C dec.
17-Cyclopropylmethyl-3,14β-dihydroxy-4,5α-epoxy-6α-[3-(isoquinolin-3-yl)propanamido)morphinan (2)
The title compound was prepared in 89% yield. 1H NMR (400 MHz, CDCl3) δ9.19 (s, 1H), 7.92 (d, J = 8.2 Hz, 1H), 7.74 (d, J = 8.2 Hz, 1H), 7.66 (dt, J = 1.04 Hz, 6.95 Hz, 1H), 7.54 (m, 2H), 6.73 (d, J = 8.04 Hz, 1H), 6.49 (d, J = 8.12 Hz, 1H), 6.41 (d, J = 8.44 Hz, 1H, exchangeable), 4.49 (m, 2H), 3.27 (m, 2H), 3.06 (d, J = 5.12 Hz, 1H), 2.99 (d, J = 18.52 Hz, 1H), 2.69 (dt, J = 3.28 Hz, 7.32 Hz, 2H), 2.63 (m, 1H), 2.54 (dd, J = 6.48 Hz, 18.56 Hz, 1H), 2.33 (m, 2H), 2.18 (m, 2H), 1.65 (m, 1H), 1.55–1.43 (m, 2H), 1.28 (m, 1H), 0.83 (m, 2H), 0.52 (m, 2H), 0.11 (m, 2H); 13C NMR (100 MHz, CDCl3) δ 171.63, 153.19, 151.90, 145.73, 137.74, 136.68, 131.10, 130.81, 127.63, 127.22, 126.86, 126.29, 125.74, 119.39, 119.13, 117.66, 89.77, 69.45, 62.25, 59.64, 47.00, 45.89, 43.28, 36.56, 33.44, 33.15, 28.84, 22.84, 21.25, 9.33, 3.92, 3.84. HRMS m/z found 526.2710, calculated 526.2700 (M + H)+. IR (Diamond, cm−1) vmax 1628.3, 1502.9, 1117.4, 859.0, 748.0. mp 106–108 °C.
17-Cyclopropylmethyl-3,14β-dihydroxy-4,5α-epoxy-6α-[2-(isoquinoline-3-carboxamido)acetamido}morphinan (3)
The title compound was prepared in 72% yield. 1H NMR (400 MHz, CD3OD) δ9.24 (s, 1H), 8.49 (s, 1H), 8.12 (d, J = 8.12 Hz, 1H), 8.02 (d, J = 8.12 Hz, 1H), 7.81 (dt, J = 1.28 Hz, 7.54 Hz, 1H), 7.75 (dt, J = 1.16 Hz, 7.50 Hz, 1H), 6.63 (d, J = 8.08 Hz, 1H), 6.49 (d, J = 8.12 Hz, 1H), 4.58 (d, J = 3.36 Hz, 1H), 4.51 (dt, J = 4.19 Hz, 12.95 Hz, 1H), 4.20 (s, 2H), 3.12 (d, J = 6.68 Hz, 1H), 3.03 (d, J = 18.6 Hz, 1H), 2.63 (d, J = 7.00 Hz, 1H), 2.58 (dd, J = 6.90 Hz, 18.70 Hz, 1H), 2.35 (m, 1H), 2.33 (m, 1H), 2.26 (d, J = 11.72 Hz, 2H), 1.71 (dt, J = 9.24 Hz, 14.80 Hz, 1H), 1.55 (m, 1H), 1.48 (d, J = 9.60 Hz, 1H), 1.42 (dd, J = 8.80 Hz, 14.80 Hz, 1H), 1.03 (m, 1H), 0.85 (m, 1H), 0.51 (m, 2H), 0.13 (m, 2H); 13C NMR (100 MHz, CD3OD) δ 170.44, 167.48, 152.91, 147.11, 144.33, 139.39, 137.12, 132.46, 132.13, 131.24, 130.40, 129.08, 128.89, 126.78, 121.35, 120.21, 118.40, 90.52, 71.10, 63.33, 60.63, 48.36, 47.73, 44.42, 43.69, 34.83, 30.43, 23.73, 21.72, 10.15, 4.59, 4.07. HRMS m/z found 555.2635, calculated 555.2602 (M + H)+. IR (Diamond, cm−1) vmax 3295.8, 2928.6, 1651.7, 1505.9, 1231.8. mp 138–140 °C. Anal. (C32H34N4O5·2HCl·H2O) C, H, N (see details in Supplementary data).
17-Cyclopropylmethyl-3,14β-dihydroxy-4,5α-epoxy-6α-(1-chloro-4-methoxyisoquinoline-3-carboxamido)morphinan (4)
The title compound was prepared in 70% yield. 1H NMR (400 MHz, CDCl3) δ 8.33–8.30 (m, 2H), 8.10 (d, J = 8.44 Hz, 1H, exchangeable), 7.86–7.76 (m, 2H), 6.71 (d, J = 8.08 Hz, 1H), 6.56 (d, J = 8.12 Hz, 1H), 4.84–4.78 (m, 2H), 4.15 (s, 3H), 3.14 (d, J = 6.04 Hz, 1H), 3.06 (d, J = 18.48 Hz, 1H), 2.69–2.60 (m, 2H), 2.42–2.23 (m, 4H), 1.90–1.79 (m, 2H), 1.58 (d, J = 10.44 Hz, 1H), 1.47 (m, 1H), 1.18 (m, 1H), 0.87 (m, 1H), 0.54 (m, 2H), 0.14 (m, 2H); 13C NMR (100 MHz, CDCl3) δ 162.45, 152.84, 145.24, 143.86, 137.28, 134.72, 133.27, 131.48, 130.96, 130.13, 129.15, 126.66, 126.08, 123.52, 119.27, 117.06, 90.50, 69.59, 63.81, 62.28, 59.73, 47.37, 46.20, 43.23, 33.57, 29.24, 22.92, 21.09, 9.40, 3.94, 3.87. HRMS m/z found 562.2128, calculated 562.2103 (M + H)+. IR (Diamond, cm−1) vmax 3256.8, 1652.1, 1503.9, 1313.2, 1117.0, 945.5, 768.8, 726.8. mp 202–204 °C.
17-Cyclopropylmethyl-3,14β-dihydroxy-4,5α-epoxy-6α-(1-chloro-4-hydroxyisoquinoline-3-carboxamido)morphinan (5)
The title compound was prepared in 44% yield. Hydrochloride salt: 1H NMR (400 MHz, DMSO-d6) δ13.61 (s, 1H, exchangeable), 9.42 (s, 1H, exchangeable), 8.88 (brs, 1H, exchangeable), 8.37 (m, 1H), 8.32 (m, 1H), 8.25 (d, J = 8.72 Hz, 1H, exchangeable), 8.03–8.01 (m, 2H), 6.75 (d, J = 8.08 Hz, 1H), 6.63 (d, J = 8.12 Hz, 1H), 6.36 (s, 1H, exchangeable), 4.85 (d, J = 3.96 Hz, 1H), 4.71 (m, 1H), 3.93 (d, J = 6.96 Hz, 1H), 3.41–3.27 (m, 2H), 3.15–3.05 (m, 2H), 2.96 (m, 1H), 2.72 (m, 1H), 2.50 (m, 1H), 1.94 (dt, J = 8.97 Hz, 15.36 Hz, 1H), 1.69–1.66 (m, 2H), 1.50 (dd, J = 8.92 Hz, 15.32 Hz, 1H), 1.19–1.08 (m, 2H), 0.70 (m, 1H), 0.63 (m, 1H), 0.49 (m, 1H), 0.42 (m, 1H); 13C NMR (100 MHz, CD3OD) δ 169.81, 155.29, 147.22, 140.78, 140.56, 132.36 (× 2), 131.39, 130.59, 129.87, 127.42, 124.28, 122.98, 121.92, 121.12, 120.04, 89.14, 71.05, 63.73, 59.10, 47.23, 46.83, 31.62, 30.32, 24.92, 21.19, 6.85, 6.28, 3.34 (× 2). HRMS m/z found 548.1970, calculated 548.1947 (M + H)+. IR (Diamond, cm−1) vmax 3076.8, 2952.6, 1620.6, 1532.1, 1319.0, 1118.3, 1032.3, 948.3, 768.0. mp 207–210 °C.
17-Cyclopropylmethyl-3,14β-dihydroxy-4,5α-epoxy-6α-(4-hydroxyisoquinoline-3-carboxamido)morphinan (6)
The title compound was prepared in 60% yield. 1H NMR (0400 MHz, CDCl3) δ 13.36 (s, 1H, exchangeable), 8.58 (s, 1H), 8.34 (d, J = 8.04 Hz, 1H), 8.29 (d, J = 8.44 Hz, 1H, exchangeable), 7.90 (d, J = 7.64 Hz, 1H), 7.75–7.67 (m, 2H), 6.74 (d, J = 8.08 Hz, 1H), 6.56 (d, J = 8.12 Hz, 1H), 4.79 (m, 2H), 3.14 (d, J = 6.52 Hz, 1H), 3.06 (d, J = 18.48 Hz, 1H), 2.69–2.61 (m, 2H), 2.42–2.24 (m, 4H), 1.87 (dt, J = 9.01 Hz, 14.54 Hz, 1H), 1.77 (m, 1H), 1.60 (m, 1H), 1.47 (dd, J = 9.22 Hz, 14.14 Hz, 1H), 1.19 (m, 1H), 0.87 (m, 1H), 0.57–0.53 (m, 2H), 0.14 (m, 2H); 13C NMR (100 MHz, CDCl3) δ 169.20, 155.12, 145.36, 141.77, 137.42, 131.39, 130.87, 129.94, 129.58, 128.31, 127.14, 126.04, 122.79, 122.13, 119.42, 117.29, 90.41, 69.62, 62.25, 59.76, 47.45, 45.93, 43.17, 33.72, 29.33, 22.99, 21.04, 9.40, 3.95, 3.87. HRMS m/z found 514.2514, calculated 514.2339 (M + H)+. IR (Diamond, cm−1) vmax 3211.9, 1624.9, 1529.8, 1456.4, 1118.2, 952.0, 764.1, 745.5. mp 128–131 °C.
17-Cyclopropylmethyl-3,14β-dihydroxy-4,5α-epoxy-6α-(4-quinazoline-2-carboxamido)morphinan (7)
The title compound was prepared in 63% yield. 1H NMR (400 MHz, CDCl3) δ9.45 (s, 1H), 8.45 (d, J = 8.96 Hz, 1H), 8.17 (d, J = 8.44 Hz, 1H), 7.99-7.93 (m, 2H), 7.72 (dt, J = 0.76 Hz, 7.52 Hz, 1H), 6.73 (d, J = 8.08 Hz, 1H), 6.56 (d, J = 8.12 Hz, 1H), 4.88 (m, 1H), 4.82 (d, J = 4.4 Hz, 1H), 3.14 (d, J = 6.16 Hz, 1H), 3.06 (d, J = 18.48 Hz, 1H), 2.69–2.60 (m, 2H), 2.42-2.22 (m, 4H), 1.89–1.80 (m, 2H), 1.58 (m, 1H), 1.48 (m, 1H), 1.25 (m, 1H), 0.88 (m, 1H), 0.55 (m, 2H), 0.14 (m, 2H); 13C NMR (100 MHz, CDCl3) δ 162.07, 160.68, 153.97, 149.87, 145.26, 137.59, 134.77, 130.92, 129.38, 129.22, 127.15, 125.79, 124.87, 119.25, 117.33, 90.08, 69.65, 62.32, 59.68, 47.32, 46.70, 43.28, 33.44, 28.95, 22.96, 21.13, 9.35, 3.91, 3.84. HRMS m/z found 499.2515, calculated 499.2340 (M + H)+. IR (Diamond, cm−1) vmax 3369.5, 1675.0, 1506.4, 1117.3, 777.8. mp 151–153 °C.
17-Cyclopropylmethyl-3,14β-dihydroxy-4,5α-epoxy-6α-(1-chloroisoquinoline-3-carboxamido)morphinan (8)
The title compound was prepared in 29% yield. 1H NMR (400 MHz, CDCl3) δ 8.55 (s, 1H), 8.36 (d, J = 8.24 Hz, 1H), 8.18 (d, J = 8.76 Hz, 1H, exchangeable), 7.99 (d, J = 7.52 Hz, 1H), 7.83-7.75 (m, 2H), 6.74 (d, J = 8.12 Hz, 1H), 6.57 (d, J = 8.12 Hz, 1H), 4.86-4.81 (m, 2H), 3.13 (d, J = 6.52 Hz, 1H), 3.07 (d, J = 18.48 Hz, 1H), 2.69-2.61 (m, 2H), 2.42–2.24 (m, 4H), 1.89–1.73 (m, 2H), 1.62 (d, J = 8.96 Hz, 1H), 1.48 (m, 1H), 1.21 (m, 1H), 0.88 (m, 1H), 0.57–0.53 (m, 2H), 0.15–0.12 (m, 2H); 13C NMR (100 MHz, CDCl3) δ 162.82, 150.33, 145.21, 142.79, 137.88, 137.42, 131.80, 130.86, 130.00, 128.68, 127.93, 126.58, 125.94, 120.68, 119.33, 117.22, 90.35, 69.63, 62.31, 59.72, 47.39, 46.46, 43.24, 33.59, 29.15, 22.95, 21.13, 9.40, 3.90, 3.82. HRMS m/z found 532.2002, calculated 532.1998 (M + H)+. IR (diamond, cm−1) vmax 3380.5, 2926.0, 1656.5, 1506.2, 1259.8, 1116.5, 1033.8, 988.7, 786.4. mp 210–212 °C.
17-Cyclopropylmethyl-3,14β-dihydroxy-4,5α-epoxy-6α-(1-cyanoisoquinoline-3-carboxamido)morphinan (9)
The title compound was prepared in 40% yield. 1H NMR (400 MHz, CDCl3) δ 8.82 (s, 1H), 8.39 (m, 1H), 8.16 (d, J = 8.68 Hz, 1H, exchangeable), 8.09 (m, 1H), 7.92–7.87 (m, 2H), 6.75 (d, J = 8.08 Hz, 1H), 6.61 (d, J = 8.08 Hz, 1H), 4.84 (m, 2H), 3.15 (d, J = 6.36 Hz, 1H), 3.08 (d, J = 18.48 Hz, 1H), 2.69–2.63 (m, 2H), 2.42–2.27 (m, 4H), 1.89–1.79 (m, 2H), 1.60 (m, 1H), 1.50 (m, 1H), 1.30 (m, 1H), 0.87 (m, 1H), 0.55 (m, 2H), 0.15 (m, 2H); 13C NMR (100 MHz, CDCl3) δ 162.39, 144.98, 144.18, 137.62, 136.51, 133.16, 132.48, 131.38, 130.70, 130.08, 128.97, 125.63, 125.52, 124.35, 119.56, 117.44, 115.46, 89.90, 69.72, 62.31, 59.69, 47.35, 46.61, 43.36, 33.42, 28.78, 22.96, 21.21, 9.43, 3.95, 3.87. HRMS m/z found 523.2339, calculated 523.23398 (M + H)+. Salt: IR (diamond, cm−1) vmax 3076.2, 2161.7, 1507.3, 1318.1, 1116.8, 1033.1, 793.8, 748.9. mp 251 °C dec.
17-Cyclopropylmethyl-3,14β-dihydroxy-4,5α-epoxy-6α-(1-methylisoquinolin-3-carboxamido)morphinan (10)
The title compound was prepared in 65% yield. 1H NMR (400 MHz, CDCl3) δ 8.54 (d, J = 8.44 Hz, 1H, exchangeable), 8.45 (s, 1H), 8.11 (d, J = 8.20 Hz, 1H), 7.93 (d, J = 7.92 Hz, 1H), 7.73–7.63 (m, 2H), 6.72 (d, J = 8.08 Hz, 1H), 6.56 (d, J = 8.08 Hz, 1H), 4.87–4.84 (m, 2H), 3.13 (d, J = 6.40 Hz, 1H), 3.06 (d, J = 18.48 Hz, 1H), 2.93 (s, 3H), 2.68–2.60 (m, 2H), 2.41–2.22 (m, 4H), 1.88–1.80 (m, 2H), 1.58 (m, 1H), 1.48 (dd, J = 8.94 Hz, 11.70 Hz, 1H), 1.23 (m, 1H), 0.87 (m, 1H), 0.55 (m, 2H), 0.14 (m, 2H); 13C NMR (100 MHz, CDCl3) δ 164.29, 157.57, 145.29, 142.41, 137.30, 136.10, 131.04, 130.41, 128.86, 128.61, 128.47, 126.12, 125.67, 119.15, 119.08, 116.96, 90.57, 69.63, 62.34, 59.71, 47.38, 46.17, 43.27, 33.53, 29.11, 22.94, 22.48, 21.31, 9.41, 3.92, 3.83. HRMS m/z found 512.2569, calculated 512.2544 (M + H)+. Salt: IR (diamond, cm−1) vmax 3207.9, 1660.7, 1506.7, 1457.6, 1320.3, 1117.4, 1032.6, 782.8. mp 224 °C dec.
17-Cyclopropylmethyl-3,14β-dihydroxy-4,5α-epoxy-6α-(6-nitroisoquinoline-3-carboxamido)morphinan (11)
The title compound was prepared in 62% yield. [α]26D −308.89° (c = 0.09, MeOH). 1H NMR (400 MHz, DMSO-d6) δ9.63 (s, 1H), 9.27 (d, J = 1.96 Hz, 1H), 9.16 (brs, 1H), 8.94 (s, 1H), 8.53 (d, J = 8.96 Hz, 1H), 8.51 (d, J = 8.32 Hz, 1H), 8.49 (dd, J = 2.18 Hz, 9.02 Hz, 1H), 6.63 (d, J = 8.08 Hz, 1H), 6.51 (d, J = 8.08 Hz, 1H), 4.67–4.62 (m, 2H), 3.12 (d, J = 6.4 Hz, 1H), 3.01 (d, J = 18.56 Hz, 1H), 2.51–2.50 (m, 2H), 2.38–2.32 (m, 2H), 2.29–2.14 (m, 2H), 1.71 (dt, J = 9.22 Hz, 14.47 Hz, 1H), 1.60 (t, J = 11.24 Hz, 1H), 1.46–1.37 (m, 2H), 1.05 (m, 1H), 0.90 (m, 1H), 0.50 (m, 2H), 0.14 (m, 2H); 13C NMR (100 MHz, DMSO-d6) δ 162.58, 152.04, 148.56, 145.39, 144.61, 138.01, 135.04, 130.76, 130.64, 130.36, 124.81, 124.45, 122.33, 121.31, 118.89, 117.22, 88.73, 69.12, 61.23, 58.69, 46.56, 45.86, 42.80, 33.49, 29.23, 22.36, 20.64, 8.98, 3.84, 3.37. HRMS m/z found 543.2243, calculated 543.2238 (M + H)+. IR (Diamond, cm−1) vmax 3368.3, 1663.2, 1630.6, 1531.8, 1507.1, 1457.3, 1341.8, 1116.3. mp 141–143 °C.
17-Cyclopropylmethyl-3,14β-dihydroxy-4,5α-epoxy-6α-(7-nitroisoquinoline-3-carboxamido)morphinan (12)
The title compound was prepared in 37% yield. [α]26D −320.00° (c = 0.1, MeOH). 1H NMR (400 MHz, CDCl3) δ9.37 (s, 1H), 9.00 (m, 1H), 8.73 (s, 1H), 8.53 (dd, J = 2.06 Hz, 8.94 Hz, 1H), 8.46 (d, J = 8.64 Hz, 1H), 8.16 (d, J = 8.96 Hz, 1H), 6.73 (d, J = 8.04 Hz, 1H), 6.58 (d, J = 8.04 Hz, 1H), 4.89–4.82 (m, 2H), 3.15 (d, J = 6.36 Hz, 1H), 3.08 (d, J = 18.4 Hz, 1H), 2.70–2.61 (m, 2H), 2.42–2.24 (m, 4H), 1.89–1.82 (m, 2H), 1.62–1.59 (m, 1H), 1.49 (dd, J = 9.28 Hz, 12.4 Hz, 1H), 1.23–1.19 (m, 1H), 0.88 (m, 1H), 0.56 (m, 2H), 0.14 (m, 2H); 13C NMR (100 MHz, CDCl3) δ 163.03, 152.80, 147.13, 146.82, 145.21, 138.68, 137.28, 130.91, 130.14, 128.33, 126.12, 124.42, 124.22, 119.98, 119.41, 117.08, 90.45, 69.61, 62.21, 59.71, 47.41, 46.46, 43.19, 33.62, 29.16, 22.94, 21.09, 9.39, 3.96, 3.86. HRMS m/z found 543.2218, calculated 543.2238 (M + H)+. IR (Diamond, cm−1) Vmax 3367.7, 1664.1, 1631.3, 1522.7, 1508.7, 1487.5, 1459.2, 1342.9, 1117.0. mp 230 °C dec.
17-Cyclopropylmethyl-3,14β-dihydroxy-4,5α-epoxy-6α-(7-dimethylaminoisoquinoline-3-carboxamido)morphinan (13)
The title compound was prepared in 75% yield. [α]26D −224.62° (c = 0.1, MeOH). 1H NMR (400 MHz, CDCl3) δ8.91 (s, 1H), 8.45 (s, 1H), 8.32 (d, J = 8.72 Hz, 1H, exchangeable), 7.80 (d, J = 9.08 Hz, 1H), 7.34 (dd, J = 9.12 Hz, 2.48 Hz, 1H), 6.94 (d, J = 2.16 Hz, 1H), 6.73 (d, J = 8.08 Hz, 1H), 6.55 (d, J = 8.12 Hz, 1H), 4.86–4.79 (m, 2H), 3.20-3.00 (m, 8H), 2.68–2.59 (m, 2H), 2.38–2.27 (m, 4H), 1.90–1.76 (m, 2H), 1.59 (m, 1H), 1.46 (m, 1H), 1.16 (m, 1H), 0.87 (m, 1H), 0.57–0.52 (m, 2H), 0.15–0.11 (m, 2H); 13C NMR (100 MHz, CDCl3) δ 164.63, 150.28, 149.12, 145.46, 140.09, 137.49, 131.73, 130.99, 129.02, 128.27, 125.88, 120.54, 120.22, 119.19, 117.21, 104.52, 90.72, 69.62, 62.31, 59.75, 47.33, 46.07, 43.22, 40.42 (×2), 33.62, 29.38, 22.95, 21.21, 9.39, 3.94, 3.86. HRMS m/z found 541.3001, calculated 541.2809 (M + H)+. IR (Diamond, cm−1) Vmax 3375.0, 1616.2, 1518.4, 1504.5, 1156.4, 810.0, 726.1. mp 268 °C dec.
17-Cyclopropylmethyl-3,14β-dihydroxy-4,5α-epoxy-6α-(6,7-dimethoxyisoquinoline-3-carboxamido)morphinan (14)
The title compound was prepared in 72% yield. [α]26D −262.59° (c = 0.25, MeOH). 1H NMR (400 MHz, DMSO-d6) δ9.16 (s, 1H, exchangeable), 9.13 (s, 1H), 8.42 (m, 2H), 7.625 (s, 1H), 7.619 (s, 1H), 6.63 (d, J = 8.0 Hz, 1H), 6.50 (d, J = 8.04 Hz, 1H), 4.92 (brs, 1H, exchangeable), 4.61 (m, 2H), 3.97 (s, 3H), 3.95 (s, 3H), 3.09 (d, J = 6.48 Hz, 1H), 3.00 (d, J = 18.52 Hz, 1H), 2.65–2.56 (m, 2H), 2.40–2.27 (m, 2H), 2.26–2.13 (m, 2H), 1.69 (dt, J = 9.32 Hz, J = 14.49 Hz, 1H), 1.56 (m, 1H), 1.45–1.36 (m, 2H), 0.99 (m, 1H), 0.88 (m, 1H), 0.50 (m, 2H), 0.13 (m, 2H); 13C NMR (100 MHz, DMSO-d6) δ 163.46, 153.21, 151.30, 148.62, 145.38, 141.84, 137.89, 132.08, 130.70, 125.69, 124.88, 118.75, 118.45, 117.09, 106.20, 105.80, 89.00, 69.06, 61.19, 58.71, 55.86, 55.74, 46.55, 45.52, 42.65, 33.62, 29.27, 22.25, 20.69, 9.06, 3.74, 3.36. HRMS m/z found 558.2615, calculated 558.2599 (M + H)+. IR (Diamond, cm−1) vmax 3486.3, 3382.4, 1669.1, 1506.4, 1457.0, 1250.1, 1220.7, 1177.2, 1004.9, 830.7. mp 250 °C dec.
17-Cyclopropylmethyl-3,14β-dihydroxy-4,5α-epoxy-6α-[(S)-(2-methyl-1,2,3,4-tetrahydroisoquinoline-3-carboxamido)]morphinan (15)
The title compound was prepared in 87% yield. 1H NMR (400 MHz, CDCl3) δ 7.15–7.09 (m, 3H), 7.01 (m, 1H), 6.69 (d, J = 8.08 Hz, 1H), 6.53 (d, J = 8.16 Hz, 1H), 4.93 (brs, 1H), 4.66 (d, J = 4.64 Hz, 1H), 4.52 (m, 1H), 3.78 (d, J = 14.96 Hz, 1H), 3.64 (d, J = 14.92 Hz, 1H), 3.29 (t, J = 6.92 Hz, 1H), 2.99–3.05 (m, 4H), 2.64 (d, J = 5.84 Hz, 1H), 2.55 (dd, J = 6.50 Hz, 18.46 Hz, 1H), 2.43 (s, 3H), 2.33 (dd, J = 3.6 Hz, 6.40 Hz, 2H), 2.22 (m, 2H), 1.48–1.66 (m, 3H), 1.19 (m, 1H), 0.81–0.95 (m, 2H), 0.53 (m, 2H), 0.11 (m, 2H); 13C NMR (100 MHz, CDCl3) δ 172.03, 145.06, 137.57, 133.94, 133.17, 130.81, 128.23, 126.91, 126.26, 126.18, 125.61, 119.29, 117.43, 89.73, 69.51, 64.56, 62.25, 59.67, 55.88, 47.13, 45.74, 43.29, 42.07, 33.28, 29.49, 28.62, 22.86, 21.29, 9.40, 3.90, 3.85. HRMS m/z found 516.2854, calculated 516.2857 (M + H)+. IR (Diamond, cm−1) vmax 3200.0, 2930.5, 1641.3, 1504.4, 1253.9, 1118.8. mp 137–139 °C. Anal. (C31H37N3O4·2HCl·H2O) C, H, N (see details in Supplementary data).
17-Cyclopropylmethyl-3,14β-dihydroxy-4,5α-epoxy-6α-(7-dimethylamino-2-methyl-1,2,3,4-tetrahydroisoquinoline-3-carboxamido)morphinan (16)
The title compound was prepared in 76% yield as a mixture of diastereoisomers with a ratio of 3/2 based on the proton NMR spectrum analysis. [α]26D −308.96° (c = 0.25, MeOH). Hydrochloride salt of one isomer: 1H NMR (400 MHz, DMSO-d6) δ 10.75 (brs, 1H, exchangeable), 9.27 (brs, 1H, exchangeable), 8.97–8.95 (m, 2H, exchangeable), 7.15 (d, J = 7.60 Hz, 1H), 6.86 (m, 1H), 6.75 (d, J = 8.08 Hz, 1H), 6.64 (m, 1H), 6.58 (d, J = 8.08 Hz, 1H), 6.45 (brs, 1H, exchangeable), 4.65 (d, J = 3.8 Hz, 1H), 4.60–4.25 (m, 4H), 3.98 (m, 1H), 3.35 (d, J = 20.16 Hz, 1H), 3.30-2.80 (m, 15H), 2.71 (m, 1H), 2.45 (m, 1H), 1.93 (m, 1H), 1.70–1.35 (m, 3H), 1.09 (m, 1H), 1.01 (m, 1H), 0.69 (m, 1H), 0.62 (m, 1H), 0.48 (m, 1H), 0.40 (m, 1H); 13C NMR (100 MHz, DMSO-d6) δ 166.32, 147.40, 145.83, 138.65, 129.04, 128.66 (×2), 122.25, 121.21, 119.36, 118.59, 114.76, 111.47, 86.88, 69.23, 64.85, 62.91, 60.80, 57.00, 54.80, 45.59, 45.18, 41.51, 40.71, 29.92, 29.45, 28.96, 23.51, 19.71, 19.54, 5.65, 5.17, 2.57. Hydrochloride salt of another isomer: 1H NMR (400 MHz, DMSO-d6) δ 10.75 (brs, 1H, exchangeable), 9.27 (brs, 1H, exchangeable), 8.97–8.95 (m, 2H, exchangeable), 7.09 (d, J = 7.32 Hz, 1H), 6.86 (m, 1H), 6.75 (d, J = 8.08 Hz, 1H), 6.64 (m, 1H), 6.58 (d, J = 8.08 Hz, 1H), 6.45 (brs, 1H, exchangeable), 4.70 (d, J = 3.8 Hz, 1H), 4.60–4.25 (m, 4H), 3.98 (m, 1H), 3.35 (d, J = 20.16 Hz, 1H), 3.30–2.80 (m, 15H), 2.71 (m, 1H), 2.45 (m, 1H), 1.93 (m, 1H), 1.70–1.35 (m, 3H), 1.09 (m, 1H), 1.01 (m, 1H), 0.69 (m, 1H), 0.62 (m, 1H), 0.48 (m, 1H), 0.40 (m, 1H); 13C NMR (100 MHz, DMSO-d6) δ 166.35, 147.40, 145.93, 138.65, 128.81, 128.66 (× 2), 122.25, 121.21, 119.36, 118.59, 114.76, 111.47, 86.73, 69.23, 64.85, 62.91, 60.80, 57.00, 54.80, 45.67, 45.22, 41.51, 40.87, 29.98, 29.67, 28.92, 23.51, 19.73, 19.54, 5.65, 5.17, 2.57. Hydrochloride salts of the mixture: HRMS m/z found 559.3279, calculated 559.32788 (M + H)+; IR (Diamond, cm−1) vmax 3348, 3233, 1671, 1618, 1459, 1236, 1118, 1034, 811; mp >250 °C.
4.2. Biological evaluation
4.2.1. Drugs
Morphine sulfate was purchased from Mallinckrodt (St. Louis, MO). Naloxone and naltrexone were purchased from Sigma-Aldrich (St. Louis, MO). All drugs and test compounds were dissolved in pyrogen-free isotonic saline (Baxter Healthcare, Deerfield, IL).
4.2.2. Animals
Male Swiss-Webster mice (Harlan, Indianapolis, IN) weighing 25 to 30 g were housed six per cage in animal care quarters at 22 ± 2 °C on a 12 h light/dark cycle. Food and water were available ad libitum. The mice were brought to a test room (22 ± 2 °C, 12 h light/dark cycle), marked for identification, and allowed 18 h to recover from transport and handling. Protocols and procedures were approved by the Institutional Animal Care and Use Committee at Virginia Commonwealth University Medical Center and complied with the recommendations of the International Association for the Study of Pain.
4.2.3. In vitro competitive radioligand binding and functional assay
The radioligand binding assay and the [35S]GTPγS binding assay were conducted using monocloned mice opioid receptor-expressed Chinese hamster ovarian (CHO) cell lines as described previously.36,37 Briefly, for the competition binding assay, [3H]NLX, [3H]DPN, and [3H]NTI were used to label the MOR, the KOR, and the DOR, respectively. Aliquots of a membrane protein (30 µg) were incubated with the corresponding radioligand in the presence of different concentrations of the ligand under investigation in TME buffer (50 mM Tris, 3 mM MgCl2, 0.2 mM EGTA, pH 7.7) at 30 °C for 1.5 h. The bound radioactive ligand was separated from the free radioligand by filtration using the Brandel harvester (Biomedical Research & Development Laboratories, MD). Specific (i.e., opioid receptor-related) binding was determined as the difference in binding obtained in the absence and presence of 5 µM naltrexone, 5 µM (±)-trans-U50,488, and 5 µM SNC80 for the MOR, the KOR, and the DOR, respectively. The potency of the drugs in displacing the specific binding of the radioligand was determined by linear regression analysis of Hill plots. The IC50 values were determined and converted to Ki values using the Cheng-Prusoff equation. The [35S]GTPγS functional assays were conducted in the same cell membranes used for the receptor binding assays. Membrane proteins (10 µg) were incubated with varying concentrations of compounds, GDP (10 µM) and 0.1 nM [35S]GTPγS in assay buffer (50 mM Tris, 3 mM MgCl2, 100 mM NaCl, 0.2 mM EGTA, pH 7.7) for 1.5 h at 30 °C. Nonspecific binding was determined with 20 µM unlabeled GTPγS while basal binding was assessed in the presence of GDP and absence of drug. DAMGO (3 µM), (±)-trans-U50,488 17 (5 µM), and SNC80 18 (5 µM) were included as the internal standard in the assay for a maximal effect of a full agonist for the MOR, KOR, and DOR, respectively. The bound radioactive ligand was separated from the free radioligand by rapid filtration and three washes and then subjected to liquid scintillation spectrophotometry. Percent maximal stimulation is defined as (net stimulated binding by test compound/by internal standard) × 100%. The normalized data were then subjected to nonlinear regression analysis to determine EC50 and efficacy (Emax) values with Prism 3.0 software (GraphPad Software, San Diego, CA).
4.2.4. Ca2+ flux assay
The Ca2+ flux assay was performed similarly to the procedure described earlier29,54 except as indicated below. The hMOR-CHO cells were maintained in DMEM/F12 supplemented with 10% fetal bovine serum, 100 units/mL penicillin, 100 µg/mL streptomycin, and 0.25 mg/mL hygromycin B at 37 °C and 5% CO2. Four hours after transfection, cells were plated at 30,000 cells per well into a clear bottom, black 96-well plate (Greiner Bio-one) and incubated for twenty four hours. The growth media was then decanted and the wells were washed with 100 µL of 50:1 HBSS:HEPES assay buffer. Cells were then incubated with either 80 µL (agonism study) or 55 µL (antagonism study) of Fluo4 loading buffer (40 µL 2 µM Fluo4-AM (Invitrogen), 84 µL 2.5 mM probenacid, in 8 mL or 5.5 mL assay buffer) for 30 min. For antagonism study, 25 µL of varying concentrations of testing compounds were added in triplicates and the plate was incubated for an additional 15 min. Plates were then read on a FlexStation3 microplate reader (Molecular Devices) at 494/516 ex/em for a total of 120 s. After 15 s of reading, 20 µL of varying concentrations of testing compounds in triplicates (agonism study) or 500 nM of DAMGO (NIDA, antagonism study) in assay buffer, or assay buffer alone, was added to the wells to bring the total volume up to 100 µL. The changes in Ca2+ flux were monitored and peak height values were recorded. The obtained values were then subjected to nonlinear regression analysis to determine EC50 or IC50 values using Prism 3.0 (GraphPad Software, San Diego, CA). All experiments were repeated at least three times.
4.2.5. Tail immersion test
The warm-water tail immersion assay was performed according to Coderre and Rollman55 using a water bath with the temperature maintained at 56 ± 0.1 °C. Before injecting, the baseline latency (control) of the mice was determined. Only mice with a reaction time from 2 to 4 s were used. The average baseline latency for the experiment was 3.0 ± 0.1 s. For the agonism study, the test latency after drug treatment was assessed at 20 minutes (time that morphine effect starting to peak) following drug administration. For the antagonism study, NAQ analogues were administered five minutes before morphine injection, and 20 minutes after morphine treatment the test latency was assessed. A 10 s maximum cutoff time was imposed to prevent tissue damage. Antinociception was quantified according to the method of Harris and Pierson56 as the percentage of maximum possible effect (% MPE), which was calculated as: % MPE = [(test latency – control latency)/(10 – control latency)] × 100. Percent MPE was calculated for each mouse using at least six mice per drug.
4.2.6. Opioid withdrawal assays
A 75 mg morphine pellet was implanted into the base of the neck of male Swiss-Webster mice following the reported procedure.23 The animals were allowed to recover in their home cages before testing. Mice were then allowed for 30 minutes habituation to an open-topped, square, clear Plexiglas observation chamber (26 × 26 × 26 cm3) with lines partitioning the bottom into quadrants before given antagonist. Withdrawal was precipitated at 72 hours from pellet implantation with naltrexone (1.0 mg/kg, s.c), and the testing compound (s.c.) at indicated doses. Withdrawal commenced within 3 minutes after antagonist administration. Escape jumps and wet dog shakes were quantified by counting their occurrences over 20 minutes for each mouse using at least six mice per drug.
4.2.7. Statistical analysis
One-way ANOVA followed by the post hoc Dunnett test were performed to assess significance using the Prism 3.0 software (GraphPad Software, San Diego, CA).
4.3. Molecular modeling studies
The molecular structure of the ligand (11, NNQ) was sketched in SYBYL-X 2.0, and its Gasteiger-Hückel charges were assigned before energy minimization (10,000 iterations) with the Tripos force field (TFF). The X-ray crystal structures for MOR (4DKL)26, KOR (4DJH)27 and DOR (4EJ4)28 were retrieved from PDB Data Bank (http://www.pdb.org). SYBYL-X 2.0 was also used to prepare the obtained protein coordinates for ligand docking by extracting the crystallized ligand and the fusion protein at intracellular loop 3, followed by addition of hydrogen atoms and subsequent energy minimization of only the added hydrogen atoms. Automated docking on these “cleaned” receptor structures was performed utilizing a genetic algorithm-based docking program GOLD 5.2.57 The binding site was defined to include all atoms within 10 Å of the γ-carbon atom of Asp3.32 for the three opioid crystal structures along with a hydrogen bond constraint between the N(17) nitrogen atom and the carboxylate group oxygen atoms of Asp3.32. The best CHEM-PLP-scored solutions were chosen for molecular dynamics (MD) studies. Gaps in the protein sequence including those due to extraction of the fusion proteins were modeled and refined employing MODELLER9v10.58,59 Force field parameter and topology files for NNQ were generated utilizing SwissParam.60 Density functional theory (DFT) calculations at the 6–31G* level were employed to calculate partial atomic charges of the NNQ atoms using NWChem 6.0.61 Coordinates for the spatial arrangement of the receptors within the lipid bilayer were retrieved from the Orientations of Proteins in Membranes (OPM) database.62 The simulation system, consisting of the receptor–ligand complex embedded in a lipid (POPC) bilayer surrounded with saline solution (0.15 M NaCl) was created in VMD 1.9.163 using the CHARMM force field topology file64,65. All simulations were performed under hybrid CHARMM force field parameters that included protein, lipids and ligand with a time-step of 2 femtoseconds (fs). Periodic boundary conditions were employed, and Particle Mesh Ewald (PME) summation was used to calculate long-range electrostatic interactions. Non-bonded interactions were calculated with a smooth cutoff between 10 to 12 Å with a frequency of 1 fs. The temperature was maintained at 310 K via Langevin dynamics. All molecular modeling simulations were performed using NAMD 2.866. MD simulations were carried out in four stages. In the first stage, equilibration of the fluid-like lipid bilayer was performed via minimization (1000 iterations) followed by NPT equilibration (pressure equilibration, 0.5 fs) of the lipid tails only. In the second stage, an NPT equilibration of the system was run for a period of 1 ns with harmonic constraints placed on protein and NNQ atoms (5 kcal/(mol-Å)). The harmonic restraint was released in stage 3 and the entire system was equilibrated using the NVT canonical ensemble for a further 1 ns. The final production run was conducted for 15 ns using an NVT ensemble. Energy landscape analysis was performed using the NAMD Energy 1.4 plug-in; non-bonded interaction analyses were performed at various distances with a dielectric constant of 6.5.67f The best-scored poses based on the NAMD non-bonded interactions were selected for further analysis. Figures were generated using the PyMOL Molecular Graphics System, Version 1.5.0.4.
Supplementary Material
Scheme 2.
Synthetic routes of isoquinoline-3-carboxylic acids 31, 33, 34, and 36 with 4 ’ or 1’,4’-substitutions.
Scheme 3.
Synthetic routes of isoquinoline-3-carboxylic acids 39, 44, and 46 with 1’-substitution.
Scheme 4.
Synthetic routes of isoquinoline-3-carboxylic acids 48, 53, 55, 58, and 1,2,3,4-tetrahydroisoquinoline-3-carboxylic acid 62 with 6 ’ and/or 7’-substitutions.
Scheme 5.
Synthetic route of (S)-2-methyl-1,2,3,4-tetrahydroisoquinoline-3-carboxylic acid hydrochloride 61.
Scheme 6.
Synthetic route used to generate the first generation of NAQ analogues 1–16.
Acknowledgements
We are grateful to Drs. Lee-Yuan Liu-Chen (Temple University) and Ping-Yee Law (University of Minnesota) for the generous gift of mice opioid receptor-expressed CHO cell lines. Y.Y. thanks Orgil Elbegdorj, Irma B. Adams, Joanna C. Jacob, Christopher K. Arnatt, and Thomas Raborg for their technical guidance on the radioligand binding and Ca2+ flux assays. Y.Y. would also like to thank Yunfei Mao and Brianna Mackie for providing HRMS data. The work was funded by PHS grants from NIH/NIDA DA024022 (YZ), NIH Grant R01 DA017204 (PDM), and National Center for Research Resources Award S10RR027411 (VCU). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute on Drug Abuse or the National Institutes of Health.
Glossary
Abbreviations
- cAMP
cyclic adenosine monophosphate
- CHO
Chinese hamster ovary
- DAMGO
[d-Ala2-MePhe4-Gly(ol)5]enkephalin
- DFT
density functional theory
- DOR
delta opioid receptor
- DPN
diprenorphine
- EDCI
1-ethyl-3-(3-dimethylaminopropyl)carbodiimide
- GIRK
G protein-gated inwardly rectifying K+
- GPCR
G protein-coupled receptor
- HOBt
hydrobenzotriazole
- KOR
kappa opioid receptor
- MD
molecular dynamics
- MOR
mu opioid receptor
- NTX
naltrexone
- NTI
naltrindole
- NAQ
17-cyclopropylmethyl-3,14β-dihydroxy-4,5α-epoxy-6α-(isoquinoline-3′-carboxamido)morphinan
- NAMD
NAnoscale Molecular Dynamics program
- NCQ
17-cyclopropylmethyl-3,14β-dihydroxy-4,5α-epoxy-6α-(1-chloro-4-methoxyisoquinoline-3- carboxamido)morphinan
- NNQ
17-cyclopropylmethyl-3,14β-dihydroxy-4,5α-epoxy-6α-(6-nitroisoquinoline-3-carboxamido)morphinan
- PME
Particle Mesh Ewald
- OPM
Orientations of Proteins in Membranes
- SAR
structure-activity relationship
- TFF
Tripos Force Field
- VGCC
voltage-gated Ca2+ channels
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplementary data
Acid side chain synthesis and characterization; HPLC, elemental analysis, and NMR spectra of the final compounds. This material can be found at.
References
- 1.Dhawan BN, Cesselin F, Raghubir R, Reisine T, Bradley PB, Portoghese PS, Hamon M. Pharmacol. Rev. 1996;48:567. [PubMed] [Google Scholar]
- 2.Chakrabarti S, Prather PL, Yu L, Law PY, Loh HH. J. Neurochem. 1995;64:2534. doi: 10.1046/j.1471-4159.1995.64062534.x. [DOI] [PubMed] [Google Scholar]
- 3.Miyake M, Christie MJ, North RA. Proc. Natl. Acad. Sci. U. S. A. 1989;86:3419. doi: 10.1073/pnas.86.9.3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ortiz-Miranda SI, Dayanithi G, Coccia V, Custer EE, Alphandery S, Mazuc E, Treistman S, Lemos JR. J. Neuroendocrinol. 2003;15:888. doi: 10.1046/j.1365-2826.2003.01076.x. [DOI] [PubMed] [Google Scholar]
- 5.Barchfeld CC, Maasen ZF, Medzihradsky F. Life Sci. 1982;31:1661. doi: 10.1016/0024-3205(82)90180-1. [DOI] [PubMed] [Google Scholar]
- 6.Thorsell A. Alcohol Alcohol. 2013;48:402. doi: 10.1093/alcalc/agt030. [DOI] [PubMed] [Google Scholar]
- 7.Herz A. J.Neural Transm Suppl. 1983;18:227. [PubMed] [Google Scholar]
- 8.Sadée W, Wang Z. Adv. Exp. Med. Biol. 1995;373:85. doi: 10.1007/978-1-4615-1951-5_12. [DOI] [PubMed] [Google Scholar]
- 9.World Drug Report. Report from the United Nations Office on Drugs and Crime. 2013 http://www.unodc.org/unodc/secured/wdr/wdr2013/World Drug Report 2013.pdf. [Google Scholar]
- 10.White JM, Irvine RJ. Addiction. 1999;94:961. [PubMed] [Google Scholar]
- 11.Bart G. J. Addict. Dis. 2012;31:207. doi: 10.1080/10550887.2012.694598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stotts AL, Dodrill CL, Kosten TR. Expert Opin. Pharmacother. 2009;10:1727. doi: 10.1517/14656560903037168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Veilleux JC, Colvin PJ, Anderson J, York C, Heinz AJ. Clin. Psychol. Rev. 2010;30:155. doi: 10.1016/j.cpr.2009.10.006. [DOI] [PubMed] [Google Scholar]
- 14.Isbell H, Vogel VH. Am. J. Psychiatry. 1949;105:909. doi: 10.1176/ajp.105.12.909. [DOI] [PubMed] [Google Scholar]
- 15.Minozzi S, Amato L, Vecci S, Davoli M, Kirchmayer U, Verster A. Cochrane Database Syst. Rev. 2011:CD001333. doi: 10.1002/14651858.CD001333.pub2. [DOI] [PubMed] [Google Scholar]
- 16.Walsh SL, Preston KL, Stitzer ML, Cone EJ, Bigelow GE. Clin. Pharmacol. Ther. 1994;55:569. doi: 10.1038/clpt.1994.71. [DOI] [PubMed] [Google Scholar]
- 17.Niciu MJ, Arias AJ. CNS Drugs. 2013;27:777. doi: 10.1007/s40263-013-0096-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chien C-C, Lee Y-J, Fan L-W, Ho I-K, Tien L-T. Toxicol. Lett. 2012;212:61. doi: 10.1016/j.toxlet.2012.04.022. [DOI] [PubMed] [Google Scholar]
- 19.Moynihan HA, Derrick I, Broadbear JH, Greedy BM, Aceto MD, Harris LS, Purington LCS, Thomas MP, Woods JH, Traynor JR, Husbands SM, Lewis JW. J. Med. Chem. 2012;55:9868. doi: 10.1021/jm301096s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McHardy SF, Heck SD, Guediche S, Kalman M, Allen MP, Tu M, Bryce DK, Schmidt AW, Vanase-Frawley M, Callegari E, Doran S, Grahame NJ, McLean S, Liras S. MedChemComm. 2011;2:1001. [Google Scholar]
- 21.Ziauddeen H, Chamberlain SR, Nathan PJ, Koch A, Maltby K, Bush M, Tao WX, Napolitano A, Skeggs AL, Brooke AC, Cheke L, Clayton NS, Sadaf Farooqi I, O’Rahilly S, Waterworth D, Song K, Hosking L, Richards DB, Fletcher PC, Bullmore ET. Mol. Psychiatry. 2013;18:1287. doi: 10.1038/mp.2012.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li G, Aschenbach LC, Chen J, Cassidy MP, Stevens DL, Gabra BH, Selley DE, Dewey WL, Westkaemper RB, Zhang Y. J. Med. Chem. 2009;52:1416. doi: 10.1021/jm801272c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yuan Y, Li G, He H, Stevens DL, Kozak P, Scoggins KL, Mitra P, Gerk PM, Selley DE, Dewey WL, Zhang Y. ACS Chem. Neurosci. 2011;2:346. doi: 10.1021/cn2000348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Altarifi AA, Yuan Y, Zhang Y, Selley DE, Negus SS. Psychopharmacology. 2015;232:815. doi: 10.1007/s00213-014-3719-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abdelhamid EE, Sultana M, Portoghese PS, Takemori AE. J. Pharmacol. Exp. Ther. 1991;258:299. [PubMed] [Google Scholar]
- 26.Manglik A, Kruse AC, Kobilka TS, Thian FS, Mathiesen JM, Sunahara RK, Pardo L, Weis WI, Kobilka BK, Granier S. Nature. 2012;485:321. doi: 10.1038/nature10954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu H, Wacker D, Mileni M, Katritch V, Han GW, Vardy E, Liu W, Thompson AA, Huang X-P, Carroll FI, Mascarella SW, Westkaemper RB, Mosier PD, Roth BL, Cherezov V, Stevens RC. Nature. 2012;485:327. doi: 10.1038/nature10939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Granier S, Manglik A, Kruse AC, Kobilka TS, Thian FS, Weis WI, Kobilka BK. Nature. 2012;485:400. doi: 10.1038/nature11111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zaidi SA, Arnatt CK, He H, Selley DE, Mosier PD, Kellogg GE, Zhang Y. Bioorg. Med. Chem. 2013;21:6405. doi: 10.1016/j.bmc.2013.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Craig PN. J. Med. Chem. 1971;14:680. doi: 10.1021/jm00290a004. [DOI] [PubMed] [Google Scholar]
- 31.Gray AP, Platz RD, Henderson TR, Chang TC, Takahashi K, Dretchen KL. J. Med. Chem. 1988;31:807. doi: 10.1021/jm00399a022. [DOI] [PubMed] [Google Scholar]
- 32.Sayre LM, Portoghese PS. J. Org. Chem. 1980;45:3366. [Google Scholar]
- 33.Yuan Y, Elbegdorj O, Chen J, Akubathini SK, Beletskaya IO, Selley DE, Zhang Y. Bioorg. Med. Chem. Lett. 2011;21:5625. doi: 10.1016/j.bmcl.2011.06.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Y, Elbegdorj O, Yuan Y, Beletskaya IO, Selley DE. Bioorg. Med. Chem. Lett. 2013;23:3719. doi: 10.1016/j.bmcl.2013.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yuan Y, Elbegdorj O, Beletskaya IO, Selley DE, Zhang Y. Bioorg. Med. Chem. Lett. 2013;23:5045. doi: 10.1016/j.bmcl.2013.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yuan Y, Elbegdorj O, Chen J, Akubathini SK, Zhang F, Stevens DL, Beletskaya IO, Scoggins KL, Zhang Z, Gerk PM, Selley DE, Akbarali HI, Dewey WL, Zhang Y. J. Med. Chem. 2012;55:10118. doi: 10.1021/jm301247n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yuan Y, Zaidi SA, Elbegdorj O, Aschenbach LCK, Li G, Stevens DL, Scoggins KL, Dewey WL, Selley DE, Zhang Y. J. Med. Chem. 2013;56:9156. doi: 10.1021/jm4012214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Selley DE, Sim LJ, Xiao R, Liu Q, Childers SR. Mol. Pharmacol. 1997;51:87. doi: 10.1124/mol.51.1.87. [DOI] [PubMed] [Google Scholar]
- 39.Spetea M, Schuellner F, Moisa RC, Berzetei-Gurske IP, Schraml B, Doerfler C, Aceto MD, Harris LS, Coop A, Schmidhammer H. J. Med. Chem. 2004;47:3242. doi: 10.1021/jm031126k. [DOI] [PubMed] [Google Scholar]
- 40.Janssen PAJ, Niemegeers CJE, Dony JGH. Arzneim. Forsch. 1963;13:502. [PubMed] [Google Scholar]
- 41.Wilson SG, Mogil JS. Behav. Brain Res. 2001;125:65. doi: 10.1016/s0166-4328(01)00281-9. [DOI] [PubMed] [Google Scholar]
- 42.Caggiula AR, Epstein LH, Perkins KA, Saylor S. Psychopharmacology. 1995;122:301. doi: 10.1007/BF02246552. [DOI] [PubMed] [Google Scholar]
- 43.Logan CJ, Hutson DH, Hesk D. Xenobiotica. 1985;15:391. doi: 10.3109/00498258509045009. [DOI] [PubMed] [Google Scholar]
- 44.Kestell P, Pruijn FB, Siim BG, Palmer BD, Wilson WR. Cancer Chemother. Pharmacol. 2000;46:365. doi: 10.1007/s002800000165. [DOI] [PubMed] [Google Scholar]
- 45.Sharma SK, Jones RM, Metzger TG, Ferguson DM, Portoghese PS. J. Med. Chem. 2001;44:2073. doi: 10.1021/jm010095v. [DOI] [PubMed] [Google Scholar]
- 46.Von Voightlander PF, Lewis RA. Prog. Neuro-Psychopharmacol. Biol. Psychiatry. 1982;6:467. [Google Scholar]
- 47.Calderon SN, Rothman RB, Porreca F, Flippen-Anderson JL, McNutt RW, Xu H, Smith LE, Bilsky EJ, Davis P, Rice KC. J. Med. Chem. 1994;37:2125. doi: 10.1021/jm00040a002. [DOI] [PubMed] [Google Scholar]
- 48.Butelman ER, Yuferov V, Kreek MJ. Trends Neurosci. 2012;35:587. doi: 10.1016/j.tins.2012.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hough LB, Nalwalk JW, Chen Y, Schuller A, Zhu Y, Zhang J, Menge WM, Leurs R, Timmerman H, Pintar JE. Brain Res. 2000;880:102. doi: 10.1016/s0006-8993(00)02776-1. [DOI] [PubMed] [Google Scholar]
- 50.Coward P, Chan SD, Wada HG, Humphries GM, Conklin BR. Anal. Biochem. 1999;270:242. doi: 10.1006/abio.1999.4061. [DOI] [PubMed] [Google Scholar]
- 51.Urban JD, Clarke WP, von Zastrow M, Nichols DE, Kobilka B, Weinstein H, Javitch JA, Roth BL, Christopoulos A, Sexton PM, Miller KJ, Spedding M, Mailman RB. J. Pharmacol. Exp. Ther. 2007;320:1. doi: 10.1124/jpet.106.104463. [DOI] [PubMed] [Google Scholar]
- 52.Filizola M, Devi LA. Nature. 2012;485:314. doi: 10.1038/485314a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ballesteros JA, Weinstein H. Methods Neurosci. 1995;25:366. [Google Scholar]
- 54.Yuan Y, Arnatt CK, El-Hage N, Dever SM, Jacob JC, Selley DE, Hauser KF, Zhang Y. Medchemcomm. 2013;4:847. doi: 10.1039/C3MD00080J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Coderre TJ, Rollman GB. Life Sci. 1983;32:2139. doi: 10.1016/0024-3205(83)90103-0. [DOI] [PubMed] [Google Scholar]
- 56.Harris LS, Pierson AK. J. Pharmacol. Exp. Ther. 1964;143:141. [PubMed] [Google Scholar]
- 57.Jones G, Willett P, Glen RC, Leach AR, Taylor R. J. Mol. Biol. 1997;267:727. doi: 10.1006/jmbi.1996.0897. [DOI] [PubMed] [Google Scholar]
- 58.Eswar N, Eramian D, Webb B, Shen M-Y, Sali A. Methods Mol. Biol. 2008;426:145. doi: 10.1007/978-1-60327-058-8_8. [DOI] [PubMed] [Google Scholar]
- 59.Fiser A, Do RKG, Šali A. Protein Sci. 2000;9:1753. doi: 10.1110/ps.9.9.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zoete V, Cuendet MA, Grosdidier A, Michielin O. J. Comput. Chem. 2011;32:2359. doi: 10.1002/jcc.21816. [DOI] [PubMed] [Google Scholar]
- 61.Valiev M, Bylaska EJ, Govind N, Kowalski K, Straatsma TP, Van Dam HJJ, Wang D, Nieplocha J, Apra E, Windus TL, de Jong WA. Comput. Phys. Commun. 2010;181:1477. [Google Scholar]
- 62.Lomize MA, Lomize AL, Pogozheva ID, Mosberg HI. Bioinformatics. 2006;22:623. doi: 10.1093/bioinformatics/btk023. [DOI] [PubMed] [Google Scholar]
- 63.Humphrey W, Dalke A, Schulten K. J. Mol. Graph. 1996;14:33. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 64.Feller SE, Gawrisch K, MacKerell AD., Jr J. Am. Chem. Soc. 2002;124:318. doi: 10.1021/ja0118340. [DOI] [PubMed] [Google Scholar]
- 65.MacKerell AD, Jr, Feig M, Brooks CLIII. J. Comput. Chem. 2004;25:1400. doi: 10.1002/jcc.20065. [DOI] [PubMed] [Google Scholar]
- 66.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorsh id E, Villa E, Chipot C, Skeel RD, Kalé L, Schulten K. J. Comput. Chem. 2005;26:1781. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kukic P, Farrell D, McIntosh LP, García-Moreno EB, Jensen KS, Toleikis Z, Teilum K, Nielsen JE. J. Am. Chem. Soc. 2013;135:16968. doi: 10.1021/ja406995j. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.