Abstract
Isothermal titration calorimetry (ITC) has become a standard and widely available tool to measure the thermodynamic parameters of macromolecular associations. Modern applications of the method, including global analysis and drug screening, require the acquisition of multiple sets of data; sometimes these data sets number in the hundreds. Therefore, there is a need for quick, precise, and automated means to process the data, particularly at the first step of data analysis, which is commonly the integration of the raw data to yield an interpretable isotherm. Herein, we describe enhancements to an algorithm that previously has been shown to provide an automated, unbiased, and high-precision means to integrate ITC data. These improvements allow for the speedy and precise serial integration of an unlimited number of ITC data sets, and they have been implemented in the freeware program NITPIC, version 1.1.0. We present a comprehensive comparison of the performance of this software against an older version of NITPIC and a current version of Origin, which is commonly used for integration. The new methods recapitulate the excellent performance of the previous versions of NITPIC while speeding it up substantially, and their precision is significantly better than that of Origin. This new version of NITPIC is therefore well suited to the serial integration of many ITC data sets.
Keywords: Isothermal titration calorimetry, serial integration, automated processing, singular value decomposition
1. Introduction
Isothermal titration calorimetry (ITC) is commonly used to examine the thermodynamics of biomolecular interactions. For example, protein-protein [1], protein-DNA [2], and protein-ligand [3] binding can be thermodynamically characterized with this method. Thermodynamic parameters for detergent demicellization [4], the insertion of proteins into membranes [5], and other aspects of membrane biology, like protein or peptide uptake/release [6] and membrane solubilization and reconstitution [7] may also be studied with ITC. The rich information content and the label-free nature of the experiment make it the method of choice for many researchers.
The ITC experiment is conceptually simple [1,3,8]. For an interacting system, a solution of one interacting partner (M) is deposited into a heat-sensing cell, while a solution of the other partner (L) is placed into a motorized syringe. In a typical heat-compensating calorimeter [9], the cell is maintained at the same temperature as a reference cell; both have associated heating elements, and a constant reference power is applied to the reference cell’s heater. Thus, the sample-cell heater must be constantly engaged to maintain isothermicity between the two cells; it is this feedback signal to the sample-cell heater that is monitored as a function of time during the experiment (i.e. the “thermogram”). If no heat-evolving processes occur, only this baseline heat would be monitored. The syringe is inserted into the cell, and, after an equilibration period, L is injected stepwise into M. As a result of the interaction of L and M, heat is either taken up or evolved, and this is reflected in the thermogram as either more or less power applied to the sample-cell heater (i.e. positive or negative deflections from the baseline power), respectively. These deflections return to the baseline value, and thus each injection is marked by a “peak” in the thermogram. Ideally, the injections continue until M is saturated with L.
The most common method of analyzing the thermogram is to integrate each injection peak with respect to time. This operation yields the heat evolved for each injection; when graphed as a function of total concentration of L (or commonly as the molar ratio of L to M), an “isotherm” results. These transformed data are analyzed to derive parameter estimates, typically KA, ΔH, and a term compensating for concentration errors or incompetent material not able to form specific complexes (sometimes termed N). However, before integration is performed, the baseline power must be established and effectively subtracted from the thermogram. This step is fraught with difficulty: the baseline is never identical in repeated experiments, it may feature stochastic high- and low-frequency elements, and it is only truly sampled in the time periods between injections. This last point underscores the central difficulty of determining the baseline: at the points where it is most important to know it, i.e. during the power deflections that constitute the signal in the experiment (termed the “injection period” herein), the baseline is obscured by this signal.
The software programs distributed by calorimeter manufacturers treat this baseline problem in various ways. In general, they attempt to establish straight or smoothly varying baselines in the injection period that connect the periods in which the baseline is accurately sampled (herein termed the “baseline periods”). However, in recognition of the inaccuracies introduced by this approach, they allow for the manual adjustment of the baseline. Although these adjustments often yield significant improvements in the baseline, they also are time-consuming and are subject to the biases of the researcher.
These difficulties are amplified by the frequent need to perform multiple titrations if one is to embrace the mechanistic insights available from modern calorimetry. For even relatively simple bi-molecular interactions, many titrations collected under different solution conditions are necessary to derive parameters related to proton-linked binding [10–12], salt-ion-linked binding [13], and the change in heat capacity upon interaction (ΔCp) [13,14]. If more than one binding partner is indicated, then phenomena such as cooperativity are best studied using multiple titrations and global analysis [15,16]. Finally, many medium-throughput drug-discovery [17,18] efforts involve the calorimetric screening of tens to hundreds of potential candidate small molecules [19,20]. Such applications may involve the manual adjustment of several to several hundred baselines; the potential for operator fatigue and error is therefore substantial.
An automated, unbiased means of baseline determination was recently introduced by Schuck and coworkers [21]. This advanced approach was integrated into the freeware program NITPIC. In brief, the program A) reads thermograms acquired by isothermal titration calorimeters, B) uses signal-decomposition techniques to provide an automatic, precise, and unbiased estimate of the baseline, C) calculates integrated heats evolved during each injection, including estimated errors in these heats, and D) exports a data table that is suitable for subsequent analysis, particularly in the freeware program SEDPHAT [15]. NITPIC consistently and significantly outperforms the manufacturers’ software, providing isotherms that have substantially less noise [21].
Here we describe several enhancements into the most recent versions of NITPIC that better facilitate applications involving large numbers of titrations. As a result of an extensive code-remodeling effort, the current algorithm is at least four times faster than that introduced in version 1.0.0, first made available in 2012. Further, the program now features convenient methods to enqueue and automatically integrate an unlimited number of thermograms. The current version also contains methods to detect and identify problems that can occur during integration. Finally, it includes an intuitive graphical system to summarize the results and flag experiments requiring additional user attention, allowing the user to interact with the results. Our findings also represent the first comprehensive comparison of the performance of NITPIC and Origin software (the latter is distributed with MicroCal’s calorimetric hardware), wherein we find that NITPIC almost always provides a more precisely defined isotherm to analysis software.
2. Methods
2.1 Selection of interacting systems
The interacting systems investigated in this report as illustrations of the power of serial integration in NITPIC were not selected with any particular goal in mind besides broad diversity. They include protein-protein and protein-small molecule interactions. The other main criterion for selection was that the data had not been published heretofore.
2.1 Protein purification
2.1.1 α-Chymotrypsin and soybean trypsin inhibitor
Soybean trypsin inhibitor (SBTI) and α-chymotrypsin (α-CT) were purchased from Worthington Biochemical. The proteins, supplied as lyophilized powders, were dissolved in phosphate-buffered saline (PBS), pH 7.4. They were then purified by size exclusion chromatography using an S75 (GE Healthcare) column that had been equilibrated in the same buffer. Fractions containing the proteins were pooled, and the proteins were concentrated to 100 μM and 20 μM for SBTI and α-CT, respectively. The proteins were stored at 4° C until needed for the titrations.
2.1.2 Tp0655
A recombinant version of Treponema pallidum Tp0655 was prepared as described [22], as were its three polyamine ligands.
2.1.3 PDK2
Recombinant human pyruvate dehydrogenase kinase isoform 2 (PDK2) and its inhibitors (PS10 and PS8) were prepared as described [23].
2.1.4 HIF2α
The human hypoxia-inducible factor 2α (HIF2α) Per/Arnt/Sim (PAS)-B domain (residues 239 – 350) was heterologously expressed and purified as previously described [20]. The purified protein was flash frozen and stored at −80° C prior to experimentation. The small molecule “HIF2α benzoxadiazole antagonist” (HBA; N-(3-Chloro-5-fluorophenyl)-4-nitrobenzo[c][1,2,5]oxadiazol-5-amine) was prepared as described [20].
2.1.5 SynIIIC
The expression plasmid for the C domain of synapsin isotherm III (SynIIIC) from rat was obtained from Dr. Thomas Sudhof. The expressed SynIIIC protein comprised glutathione-S-transferase (GST), a linker peptide (GSPGISGGGGGI), and residues 89-403 of rat synapsin III. BL21(DE3) cells transformed with the plasmid were grown in 1 L of terrific broth [24] at 37° C until the cell density reached an OD600 of 0.6. The temperature of the culture was lowered to 25° C, and protein production was induced by the addition of isopropyl-β-D-thiogalactopyranoside to a final concentration of 0.4 mM. After 17 hours, the cells were harvested by centrifugation and frozen in liquid nitrogen. For purification of SynIIIC, the cells were thawed on ice, then suspended in 40 mL of lysis buffer (20 mM Tris pH 8.0, 50 mM NaCl, 2 mM ethylenediaminetetraacetic acid (EDTA), and 1 mM dithiothreitol (DTT)). Once suspended, 4 mg of lysozyme and one protease inhibitor tablet (Roche Pharmaceuticals) were added, followed by incubation on ice for 45 minutes. Triton X-100 was added and the slurry was sonicated. Cell debris was pelleted by ultracentrifugation at 45,000 rpm in a Beckman Ti45 rotor. The resulting supernatant was incubated with 1g of glutathione-agarose beads (Sigma Chemical Corp.) that had been hydrated. The incubation was for 4 hours at 4° C. The beads were packed into a column and washed by two 25-mL additions of Wash Buffer I (20 mM Tris pH 8.0, 100 mM NaCl, 2 mM EDTA, 1 mM DTT, 0.1% Triton X-100), followed by one 100-mL addition of Wash Buffer II (20 mM Tris pH 8.0, 150 mM NaCl). The beads were suspended in 5 mL of Thrombin Buffer (20 mM Tris pH 8.0, 150 mM NaCl, 2.5 mM CaCl2) and treated with 100 units of bovine thrombin (Sigma Chemical Corp.) for 45 minutes at room temperature. The column was repacked under flow, and the eluent, which contained SynIIIC, was collected. The thrombin treatment was repeated four more times. The pooled eluents were dialyzed against MonoS buffer (50 mM MES pH 6.5, 100 mM NaCl, 1 mM EDTA, and 1 mM DTT). The dialysate was applied to a MonoS (GE Healthcare) column and eluted using a linear gradient of NaCl. Fractions containing SynIIIC were pooled, concentrated, and applied to a Superdex 200 (16/60) column (GE Healthcare) that had been equilibrated with a buffer containing 50 mM Tris pH 7.5, 150 mM NaCl, and 1 mM DTT. Fractions that contained SynIIIC were pooled and concentrated to approximately 15 mg/mL. The protein was exhaustively dialyzed against the SynIIIC ITC buffer: 50 mM HEPES pH 7.4, 25 mM NaCl, 1 mM ethylene glycol tetraacetic acid (EGTA), and 10 mM 2-mercaptoethanol. Disodium ATP was dissolved in the identical buffer and diluted as needed in this buffer.
2.2 Isothermal titration calorimetry
Titrations were performed in either a VP-ITC (Malvern Instruments, Malvern, UK) or an iTC200 (Malvern). The buffer compositions of the cell contents and the injectant were equilibrated by dialysis, where possible. In some cases, gel-filtration or dialysis buffer was used to suspend soluble small-molecule injectants. When dimethyl sulfoxide (DMSO) was present in the ligand solution, this solvent was added to the protein solution to achieve the same final concentration of DMSO. Generally, the interaction partners M at concentrations between 2 and 20 μM were placed into the sample cell, and solutions of L at roughly 10–15-fold higher concentrations were introduced into the syringe. Small-molecule solubility considerations required that, in the case of the HIF2α/HBA interaction, M be defined as the small molecule and L as the protein HIF2α. The titrations consisted of 15–32 injections of the syringe contents into the cell contents after equilibrating the cell while stirring at temperatures between 15 and 25° C. Conditions for all of the titrations performed in this study are shown in Table 1.
Table 1.
Experimental conditions for the 53 thermograms
| Filename | #Inj | Inj. Vol. (μL) | calorimeter | M | [M] (μM) | L | [L] (μM) | Temperature (°C) |
|---|---|---|---|---|---|---|---|---|
| 0655dap040307.itc | 32 | 8 | VP-ITC | Tp0655 | 20.8 | DAPa | 400 | 20 |
| 0655dap040307b.itc | 32 | 8 | VP-ITC | Tp0655 | 20.8 | DAP | 400 | 20 |
| 0655dap040307c.itc | 32 | 8 | VP-ITC | Tp0655 | 20.8 | DAP | 400 | 20 |
| 0655put040307a.itc | 32 | 8 | VP-ITC | Tp0655 | 20.4 | Putrescine | 200 | 20 |
| 0655put040307b.itc | 32 | 8 | VP-ITC | Tp0655 | 20.4 | Putrescine | 200 | 20 |
| 0655put040307c.itc | 32 | 8 | VP-ITC | Tp0655 | 20.4 | Putrescine | 200 | 20 |
| 0655spmd032407a.itc | 32 | 8 | VP-ITC | Tp0655 | 19.5 | Spermidine | 420 | 20 |
| 0655spmd032407b.itc | 32 | 8 | VP-ITC | Tp0655 | 19.5 | Spermidine | 420 | 20 |
| 0655spmd032407c.itc | 32 | 8 | VP-ITC | Tp0655 | 19.5 | Spermidine | 420 | 20 |
| 20131030r1.itc | 16 | 2.5 | iTC200 | EDTA | 400 | Ca2+ | 5,000 | 25 |
| 20131030r2.itc | 16 | 2.5 | iTC200 | EDTA | 400 | Ca2+ | 5,000 | 25 |
| 20131030r3.itc | 16 | 2.5 | iTC200 | EDTA | 100 | Ca2+ | 1,250 | 25 |
| 20131030r4.itc | 16 | 2.5 | iTC200 | EDTA | 50 | Ca2+ | 625 | 25 |
| 20131030r5.itc | 16 | 2.5 | iTC200 | EDTA | 25 | Ca2+ | 312.5 | 25 |
| 20131030r6.itc | 16 | 2.5 | iTC200 | EDTA | 12.5 | Ca2+ | 156.25 | 25 |
| 20131030r7.itc | 16 | 2.5 | iTC200 | EDTA | 6.25 | Ca2+ | 78.125 | 25 |
| 20140625r4.itc | 38 | 1 | iTC200 | α-CT | 20 | SBTI | 100 | 25 |
| 20140625r5.itc | 38 | 1 | iTC200 | α-CT | 20 | SBTI | 100 | 25 |
| 20140625r6.itc | 38 | 1 | iTC200 | α-CT | 20 | SBTI | 200 | 25 |
| EDTACaCl22.itc | 19 | 2 | iTC200 | EDTA | 400 | Ca2+ | 5000 | 25 |
| H70ATp34hLF051209a.itc | 32 | 8 | VP-ITC | hLF | 18.6 | Tp34 | 332 | 20 |
| H70ATp34hLF051209b.itc | 32 | 8 | VP-ITC | hLF | 18.6 | Tp34 | 332 | 20 |
| H70ATp34hLF051209c.itc | 32 | 8 | VP-ITC | hLF | 18.6 | Tp34 | 332 | 20 |
| h70aTp34hLFa.itc | 32 | 8 | VP-ITC | hLF | 15 | Tp34 | 200 | 20 |
| h70aTp34hLFb.it | 32 | 8 | VP-ITC | hLF | 15 | Tp34 | 200 | 20 |
| S3C0730A.ITC | 30 | 8 | VP-ITC | SynIIIC | 25 | ATP | 339 | 20 |
| S3C0730B.ITC | 30 | 8 | VP-ITC | SynIIIC | 25 | ATP | 345 | 20 |
| S3C0730C.ITC | 30 | 8 | VP-ITC | SynIIIC | 25 | ATP | 346 | 20 |
| s3c071603.itc | 30 | 8 | VP-ITC | SynIIIC | 21.1 | ATP | 300 | 20 |
| S3CNOCA4.ITC | 30 | 8 | VP-ITC | SynIIIC | 19 | ATP | 274 | 20 |
| s3cplusca22.itc | 30 | 8 | VP-ITC | SynIIIC | 19.3 | ATP | 400 | 20 |
| s3cplusca23.itc | 30 | 8 | VP-ITC | SynIIIC | 19.2 | ATP | 380 | 20 |
| sam0716a.itc | 18 | 2 | iTC200 | PDK2 | 40 | PS10 | 400 | 15 |
| sam0716b.itc | 19 | 2 | iTC200 | PDK2 | 40 | PS10 | 400 | 15 |
| sam0716d.itc | 19 | 2 | iTC200 | PDK2 | 20 | PS10 | 200 | 15 |
| sam0716e.itc | 19 | 2 | iTC200 | PDK2 | 10 | PS10 | 100 | 15 |
| sam0717a.itc | 19 | 2 | iTC200 | PDK2 | 5 | PS10 | 50 | 15 |
| sam0717b.itc | 19 | 2 | iTC200 | PDK2 | 20 | PS10 | 200 | 15 |
| sam0717c.itc | 19 | 2 | iTC200 | PDK2 | 10 | PS10 | 100 | 15 |
| sam0717d.itc | 19 | 2 | iTC200 | PDK2 | 5 | PS10 | 50 | 15 |
| sam0717e.itc | 19 | 2 | iTC200 | PDK2 | 2.5 | PS10 | 25 | 15 |
| sam0717f.itc | 19 | 2 | iTC200 | PDK2 | 2.5 | PS10 | 25 | 15 |
| sam0718a.itc | 19 | 2 | iTC200 | HBA | 5 | HIF2α | 200 | 25 |
| sam0718b.itc | 19 | 2 | iTC200 | HBA | 5 | HIF2α | 100 | 25 |
| sam0718d.itc | 19 | 2 | iTC200 | HBA | 5 | HIF2α | 50 | 25 |
| sam0719a.itc | 18 | 2 | iTC200 | HBA | 5 | HIF2α | 50 | 25 |
| sam0719c.itc | 19 | 2 | iTC200 | HBA | 5 | HIF2α | 100 | 25 |
| sam0722b.itc | 15 | 2 | iTC200 | HBA | 5 | HIF2α | 100 | 25 |
| sam0722c.itc | 19 | 2 | iTC200 | HBA | 5 | HIF2α | 100 | 25 |
| ScottTso1.itc | 18 | 2 | iTC200 | PDK2 | 40 | PS8 | 500 | 25 |
| ScottTso2.itc | 25 | 1.5 | iTC200 | PDK2 | 40 | PS8 | 400 | 25 |
| Tp34hLFHep.itc | 32 | 8 | VP-ITC | hLF | 30 | Tp34 | 620 | 20 |
| Tp34hLFPi.itc | 32 | 8 | VP-ITC | hLF | 30 | Tp34 | 620 | 20 |
1,3-diaminopropane
2.3 Analysis
For the tests using manufacturer-supplied software, VP-ITC and iTC200 thermograms were integrated with OriginPro 7, version 7.0552. The integrations were performed in the calorimeter model-specific modules supplied with this software, i.e. data collected from a VP-ITC were processed with the “VP-ITC” mode, and data collected from an iTC200 were processed in “iTC200” mode. The integrated isotherms were exported as ASCII text files after integration, and then imported into SEDPHAT version 12.01. Immediately afterward, experimental parameters such as the concentrations of M and L, cell volume, and experimental temperature were inputted. The “A + B <-> AB Hetero-Association” model was chosen in SEDPHAT; in all cases “B” was defined as the species in the syringe. This model provides log(KA), ΔH, and incompetent fractions (see below) as adjustable parameters. Notably, this model is incorrect for the α-CT/SBTI interaction, which is known to associate in a 2:1 complex [25]. For the purposes of this report, it can be treated as a 1:1 interaction with a large incompetent fraction of α-CT. Appropriate initial -parameter values for log(KA) and ΔH were manually entered. In contrast to other analysis programs, such as that commonly packaged with calorimeters, SEDPHAT does not define an “N” value for isotherms, which is frequently referred to as “stoichiometry” [9]. Instead, SEDPHAT assumes that all concentrations and association models are correct; in a given isotherm, a defect that causes apparently early or late saturation of the cell component is treated as an “incompetent fraction” of one of the binding partners; this is a refinable parameter [15]. Thus, in this initiation step, it was necessary to decide which binding partner, A or B, was to have its incompetent fraction refined. This choice is relevant, because the parameter cannot refine to values below zero or above 1. Operationally, if N would refine to a value greater than 1 in Origin, the incompetent fraction of B should be refined, and vice versa. After the parameters were initialized, they were refined by fitting to the isotherm alternately using the “Marquardt-Levenberg” and “Simplex” algorithms available in SEDPHAT. This refinement procedure was repeated until convergence had been achieved, as judged by the lack of changes in the reduced chi-square of the fit upon multiple fitting actuations.
Because NITPIC is specifically designed to work with SEDPHAT, analysis of isotherms provided by NITPIC is considerably streamlined compared to the procedure outlined above. In the serial mode used for these analyses, NITPIC writes files in the .sedphat format, which, after imported into SEDPHAT, sets the model and appropriate experimental and adjustable parameters automatically. The same fitting algorithms described above for isotherms derived from Origin were used for the NITPIC isotherms.
A difference in the outputs from NITPIC and Origin necessitated a deviation in the normal NITPIC/SEDPHAT analysis procedure. NITPIC provides error estimates for each data point in the isotherm [21] (see Section 3.1.3), allowing SEDPHAT to weight individual data points in the isotherm respectively in the fitting algorithm. However, Origin does not calculate such errors, and thus, all data points are necessarily weighted equally. The weights calculated by NITPIC should allow SEDPHAT to arrive at the best parameter estimates. Nonetheless, to perform the most apt comparison between the two programs, we disabled the weighted-fitting option for all isotherms presented in this study, regardless of the program used to integrate them. All analyses were performed on an Apple MacBook Pro running 64-bit Windows 7 Ultimate Service Pack 1 natively under BootCamp. The computer had an Intel i7 M620 CPU running at 2.67 GHz.
3. Theory
3.1 The NITPIC algorithm
The mathematical details of the NITPIC algorithm are given by Keller et al. [21]. Here, we briefly recapitulate the general strategy. In essence, the algorithm has three parts: (1) determine which parts of the thermogram are in the injection period and which are in the baseline period; (2) adjust the injection period and other baseline parameters by examining their effects on an unbiased measure of isotherm noise, wrmsd; and (3) finalize the integrations and output the result.
3.1.1 Injection/baseline border
Perhaps the most essential decision to be made in the algorithm is which parts of the thermogram represent the injection periods and which represent the baseline periods. The thermogram may naturally be divided into n subsections, where n is the number of injections. Each of these subsections will have a period where the reading is deflected from the baseline (the injection period), which will occur immediately after the syringe contents are introduced into the sample cell. The beginning of the injection period is easily identified from the file formats outputted by the manufacturers’ data-acquisition/analysis software packages. If the experiment was performed properly, each subsection will also have a time period after the injection period when the signal returned to the baseline value (the baseline period). Integration of the baseline period is likely to contaminate the data with noise features. The algorithm must therefore decide the location of the boundary between the two. To accomplish this, the data subsections are arranged as a rectangular matrix T, with the n injections serving as columns, and the time points after injection (τ) occupying the rows. Then, singular value decomposition is performed on the matrix such that
| (Eq. 1) |
The column vectors in the U matrix may be thought of as “shape components” that contribute to the overall appearance of all injections. The NITPIC algorithm examines the two most prominent shape components and estimates an end for the injection period.
3.1.2 Adjusting Parameters
At this point, the baseline is defined either by connecting the points just pre- and post-injection with a straight line or with a smoothly varying function. Which type of connection is used, adjustments to the injection period, and other parameters are assessed based on a model-free measure of the noise in the isotherm that results from the integration of the thermogram after subtraction of the putative baseline. This integration results in a set of heats of injection Q, and these are treated locally by omitting Qi and fitting a straight line or polynomial function to a subset of the data composed of Qi−ν, …, Qi−1, Qi+1, …, Qi+ν, where ν is set by default to 5, but is user-adjustable and sometimes adjusted by the program in suitable cases. The distance of Qi from this fitted line, δ(Qi), is determined. This operation is performed iteratively for all possible injections, and then the isotherm noise, wrmsd, is calculated as
| (Eq. 2) |
where wi is a weighting term related to the goodness of fit of the respective line. Decisions that result in a lower wrmsd are generally accepted. Notably, at this point, if the injection period encompasses more than 75% of the interinjection period, the user is warned via a “pop-up” window.
3.1.3 Finalizing the integrations
An individual injection Ii is defined by the power signal pi and the baseline bi: Ii = pi – bi. By definition, Ii may be reconstructed from the elements of the SVD-derived matrices as:
| (Eq. 3) |
with s, v, and u representing elements of their respective matrices, and m representing the total number of shape components calculated in the matrix decomposition. As with many applications that employ SVD, we find that only a subset of the shape-component vectors in U contribute significantly to the shapes of the injections; the remaining components may be considered to be noise elements. Thus, there exists a number z < m that fulfills
| (Eq. 4) |
The remaining components, uz+1, …, um, are treated as noise elements in the baseline such that a new baseline during the injection period βi may be defined as
| (Eq. 5) |
Now, we may state that . The number z is adjusted such that the heats integrated from , QSVD, do not differ significantly from the native heats and such that the choice of z does not negatively impact wrmsd. QSVD is outputted in the .DAT format popularized by MicroCal’s implementation of Origin, and also in a format (.nitpic) that can be read and analyzed by SEDPHAT. Additionally, straight or quadratic lines are fitted to the before- and after-injection baselines of Ii. By extrapolating the error estimates from these lines into the injection period, the integration errors are estimated and outputted for use in SEDPHAT. All of these features were released with NITPIC version 1.0.0 in 2012.
4. Calculation
4.1 Improvements to the Algorithm
4.1.1 Speed
To provide a foundation for later serial integrations (see Section 4.2), it was desirable to minimize the time taken by the algorithm to achieve integration while simultaneously retaining its excellent performance. A code-remodeling project was undertaken to eliminate inefficiencies in the algorithm. In particular, unnecessary “for loops” and unnecessary iterations therein were targeted for elimination. This strategy has resulted in significant improvements in the speed of the algorithm (see Section 5.2.1). Collectively, these changes are referred to as the “Jackrabbit” code changes below.
4.1.2 Warnings
Although the algorithm has performed well through the integration of thousands of thermograms, difficulties sometimes arise. Two classes of problems are treated here: SVD components and baseline systematicity.
It can occur that the number z (Section 3.1.3) is chosen to be too large. Thus, too many components of matrix U are included in the reconstructed injections ISVD (and conversely, too few are included in the calculation of the SVD-influenced baseline β). The overly large choice of z can lead to unwanted noise components being included in the reconstructed injections. This problem manifests as a very close correspondence between b and β (see Section 3.1.3). An algorithm for detecting this issue has been developed. It is based on the notion that there is no inherent reason that the noise in the injection period should be less than the noise in the baseline period. Hence, a test for this problem naturally presents itself: is the calculated noise in the injection baseline significantly lower than that of the baseline periods defined by the program?
To estimate the noise in the defined baseline periods, we define an overall measure of baseline noise, δb. It is:
| (Eq. 6) |
where Nb is the number of data points in the baseline period, is the after-injection data for injection i and data point j, and the line fitted to the data in the after-injection period is . Similarly, we define δi to be:
| (Eq. 7) |
where NI is the number of data points in the injection period, is the jth data point in the injection period of the ith injection, and βi (τj) are the data points in the SVD-modified baseline. If δi < 0.5δb, an iconified warning (Fig. 1A) to the user is produced, and a notation in the run log is made. Clicking on the warning icon provides the user an explanation of the potential problem and a potential remedy (i.e. a suggestion to limit the number of SVD components included in the reconstruction of the injection).
Figure 1. NITPIC Warning Icons.
(A) The “S” nit. The nit with an “S” on its abdomen denotes that the “too many SVD-shape components in the injection reconstruction” warning is active. (B) The “E” nit. This icon warns the user of significant systematicity in the early-injection period of the baseline.
The other problem that sometimes occurs is that the algorithm produces a baseline having a systematic “sawtooth” appearance in the early injections (see Section 5.1.2). This can be caused by the injection period being too short. An algorithm was added to NITPIC to try to detect this phenomenon. A proxy for the slope of the estimated baseline of injection i, is calculated as
| (Eq. 8) |
and a corresponding slope proxy for the after-injection period of injection i, , is also calculated:
| (Eq. 9) |
where the notation τb3 represents the third data point in the baseline period. If the signs of and are opposite for more than 70% of the early injections (usually defined as about the first half of the injections), another iconified warning (Fig. 1B) and log entry are initiated. As above, clicking on the warning icon presents the user with information on why the warning has occurred.
4.2 Serial Integration
4.2.1 Enqueuing the Data
In NITPIC 1.1.0, the user first must place all data sets in a common file system directory. These files are enqueued by using the NITPIC GUI (graphical user interface) to select the location of the data. Any number of files may be included, and any mixture of calorimeter types may be included. Data collected from TA Instruments calorimeters, however, will need to be exported as an .xml file prior to loading into and analysis with NITPIC; instructions for doing so are included in the NITPIC documentation. No data from TA Instruments calorimeters were included in this study.
4.2.2 Additional Requirements
Attention to all user-provided parameters should be given prior to serial integration. In particular, all syringe concentrations provided for the cell and syringe contents must be nonzero and correctly stated in the data files (as entered in the data acquisition software at the time of data collection), as these cannot be entered or corrected during a serial integration session.
4.2.3 Starting Serial Integration
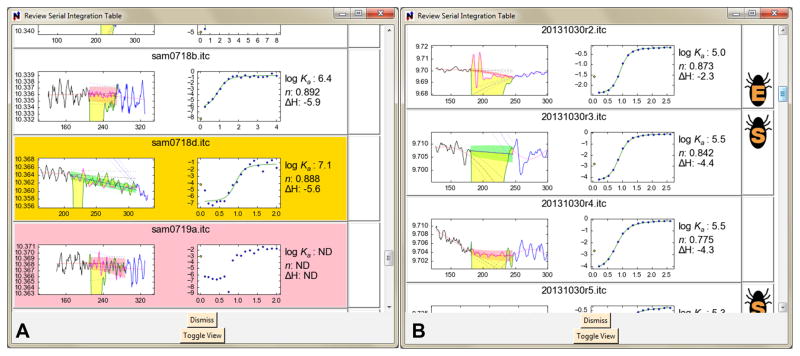
The user actuates serial integration by pressing the “Serial Execute” button in the software’s user-interface menu bar. Immediately, serial integration begins: a progress monitor appears, and a table of results is populated as they become available. NITPIC attempts to fit a simple 1:1 binding model to every resulting isotherm, and the results of this fit along with wrmsd are shown in the table. At the end of serial integration, the user may peruse the table of results or toggle the appearance of the table to a series of graphical buttons, one for each integration result. Each button has a close-up view of the second injection of the thermogram, the resulting isotherm (and fit if applicable), and the parameters derived from the fit to the isotherm (again, if applicable). This graphical view also flags four potential problems with the integration: the fact that too much of the interinjection period may have been identified as the injection period (gold background), the failure of the isotherm-fitting routine (pink background), and the two defects described in Section 4.1.2 (iconified warnings). Examples are shown in Fig. 2. Left-clicking on a button causes everything regarding the respective integration result to be loaded into memory and displayed in the program’s main graphing window. The user may interact with the integration result by examining individual injections and summary plots. Afterward, the user may load another integration result from the graphical isotherm buttons. The examination/loading cycle can continue indefinitely.
Figure 2. Two views of the graphical table after serial injection.
(A) A view showing colored-background warnings. The gold background signifies that the injection period was greater than or equal to 75% of the entire interinjection period. The pink background alerts the user that isotherm fitting failed.
4.3 Other improvements
4.3.1 Subtracting control titrations
Since the introduction of the initial version of NITPIC, the option of subtracting control titrations has become available and is fully implemented in version 1.1.0. There is a built-in assumption that the control titration was carried out very similarly to the subject titration, i.e. there were similar numbers of injections with an identical injection-volume scheme and interinjection spacings. Control-titration subtraction is implemented in two different ways. For single experiments, the user may choose “Subtract Control Titration from Current” from the File Menu after an integration has been performed. The program will use the parameters derived for the current titration to integrate the control titration and these control heats will be subtracted from the subject titrations heats. In the serial-fitting mode, another option is available. If the user is integrating a series of related isotherms from which a single control titration should be subtracted, this may be indicated in the “Prepare Files for Serial Integration” dialog. In this mode, the program will examine all experimental thermograms and choose the longest interinjection period that still will yield good data. This interinjection period is held constant as the experimental and control thermograms are integrated, with the control heats being subtracted from the experimental heats. In both modes, the errors in the integrated heats are propagated. That is, if σe,i is the error in the ith injection of the experimental titration, and σc,i is the error in the corresponding injection in the control titration, then
| (Eq. 10) |
where σs,i is the error in the resulting subtracted injection i.
4.3.2 Improved SEDPHAT interface
There are also new features in NITPIC that aid its interface with SEDPHAT. In the serial-integration mode, all titrations may optionally be placed into a single SEDPHAT-style configuration file. The user may then open this one file, which will automatically load all integrated experiments. This feature is useful when all subject titrations are to be fitted using a single global model. Another option is to add the current integration result into an existing SEDPHAT configuration file. Thus, an additional experiment may be easily added to a previously characterized global fitting session.
4.4 Software availability
The enhancements mentioned above have been implemented in NITPIC version 1.1.0. It is freely available at the web site http://biophysics.swmed.edu/MBR/software.html.
5. Results
5.1 Integration Notifications
Although NITPIC almost always produces excellent isotherms, suboptimal results are sometimes obtained. In this work, we have developed algorithms to detect two occasional problems that can occur when using NITPIC: the inclusion of too few SVD-derived shape components in the estimation of the injection-period baseline and errors in the choice of the boundary between the injection period and the baseline period. When integrating a large number of thermograms, the need to examine all of the resulting isotherms individually is cumbersome. We therefore consider the following means to identify problematic integrations to be essential to the ultimate utility of serial integration.
5.1.1 Erroneous SVD truncation
In ITC thermograms, injection-period baseline cannot be directly measured because it is subsumed by the injection signal. However, if the experiment was carried out correctly, the baseline just before and just after each injection is measured. For the purposes of integration-error estimation, NITPIC fits lines to these pre- and post-injection baseline periods, and the difference between these fitted lines and the actual signals can be thought of as the inherent stochastic noise of data acquisition. To estimate the baseline, NITPIC forms a smooth connection between these pre- and post-injection periods. To this noiseless line, it adds an estimate of the high-frequency noise using SVD truncation (Section 3.1.3). Presumably, the noise apparent from the fits to the pre- and post-injection periods should be similar in magnitude to that estimated from the SVD truncation. However, if too few shape components have been used to estimate the baseline in the injection period, there will be almost no noise in the estimate. This is not desirable, because it indicates that high-frequency noise components have been included in the integration of the peak.
An extreme example of this phenomenon occurs in NITPIC’s default behavior toward a thermogram called “20131030r3.itc” herein (middle panel, right half of Fig. 2B). Examining injection 9 (Fig. 3A), it is clear that the program has included too few shape components in estimating the injection-period baseline; the high-frequency noise on either side of the injection period is substantially higher than that estimated in the injection period. Indeed, there is essentially no noise in the injection-period baseline! In this injection, there is a small negative spike as the injection signal recovers to the baseline (circled feature in Fig. 3A). NITPIC included this spike in the integration of this peak, but it probably was a noise feature, i.e. there should have been a compensating spike in the baseline. An examination of the scree plot of singular values (Fig. 3B) that were included in the estimates of the injections and in the injection-period baselines shows that only one out of sixteen possible shape components were used for the baseline estimation (and, conversely, fifteen of these components were used for estimation of the shape of the injection).
Figure 3. Shape components and the estimated baseline in the injection period.
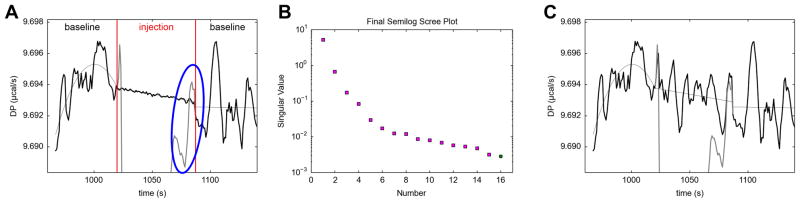
(A) A close-up of Injection 9 of “20131030r3.itc” before adjustments were made. The red lines show the divisions between the before-injection baseline period, the injection period, and the after-injection baseline period. The thin black lines depict fits to the baseline periods or the linear connection between the two (in the injection period) chosen by NITPIC. The thick black lines in the baseline periods are the actual power signals. The thick black line in the injection period is the estimated baseline with high-frequency noise included. The gray line in the injection period is the actual injection power signal. Circled in blue is a sharp negative feature in this signal. (B) The semilog scree plot for this integration. The markers are the values of the respective singular values from the SVD of the thermogram. The magenta squares are singular components that were included in the reconstruction of the injection, and the blue circle is the component that was used to estimate the noise in the injection-period baseline. (C) A close-up of Injection 9 after limiting the injection-period signal to be reconstructed with only 6 SVD-shape components. The coloration is the same as in part (A). The remaining 10 components have been added to the injection-period baseline estimate in this case.
By making use of the quantities δi and δb (Eqs. 6 & 7), NITPIC 1.1.0 successfully identified this problem and warned the user. Additionally, the program makes a suggestion of how many SVD components to include in the injection estimate; six, in this case, leaving nine for the estimate of the injection-period baseline. Including only this number of components in the injection estimate will cause δi to be greater than or equal to 0.5 · δb. This is a conservative estimate of the best number of shape components. In NITPIC, the user is allowed to provide the maximum and minimum number of SVD components included in the injection estimate. By setting the maximum to six and re-integrating the thermogram, the estimated noise in the injection-period baseline increases (Fig. 3C), and now the negative spike is accounted for in the baseline estimate and the excursions of the estimate from the smooth connector are commensurate with those in the flanking baselines. Experience has shown that sometimes it is desirable to restrict the number of shape components used for the reconstruction of the injection-period signal to fewer than that suggested by NITPIC. Additional tests have demonstrated that the δb and δi metrics reliably diagnose errors in SVD truncation (not shown).
5.1.2 Baseline systematicity
A fundamental aspect of the NITPIC algorithm is the definition of which time periods of the thermogram may be defined as injections and which may be deemed baseline. The mathematical basis of this choice is described in Sections 3.1.1 and 3.1.2, and more thoroughly by Keller et al. [21]. Despite the robustness of this approach, errors are sometimes made in the assignment and refinement of the boundary. We find that noisy thermograms with a low overall heat signal tend to include too much of the thermogram in the injection periods. This problem was addressed in the first version of NITPIC by warning the user when the injection periods encompass 75% or more of the thermogram. Strategies for alleviating this problem are detailed in the NITPIC manual (which is distributed with the software).
But the opposite problem can arise, i.e. that the injection periods are too short, causing a sawtooth-like appearance in the calculated baseline (Fig. 4A). This strong systematicity usually occurs in the injections with the most heat signal, which are almost always the initial injections. Rarely, these imperfections can cause significant errors in the integrated heats, and it was therefore desirable to alert the user to this potential pitfall.
Figure 4. Baseline systematicity in NITPIC.
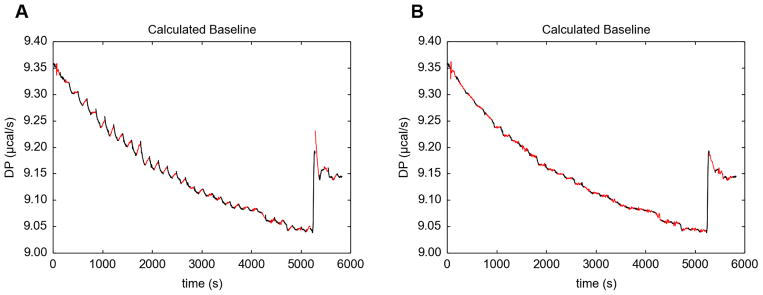
(A) The baseline before adjustment. Shown is the NITPIC-estimated baseline for the entire “H70ATp34hLF051209b.itc” thermogram. The black parts are the baseline periods, while the red lines depict the estimated baseline in the injection periods. In this plot, 45% of the interinjection time is attributed to the injections. (B) The baseline after adjustment. The coloration is the same as in part (A), but now the injection periods have been set at 65%.
NITPIC version 1.1.0 takes a very simple approach to detecting this problem (Section 4.1.2). It calculates two values, sI and sb, which serve as proxies for the slopes of the injection-period baseline and post-injection baseline, respectively. If the signs of these two values are opposite for more than 70% of the early injections, a warning is generated. The user can then examine the baseline and injections to verify the need for changes. NITPIC offers a way for the user to set a minimum injection-period time as a percentage of the interinjection time; a judicious choice of this value followed by reintegration should resolve the systematicity.
In the specific example shown in Fig. 4A (from the thermogram “H70ATp34hLF051209b.itc”), NITPIC’s default choice of injection period was 45% of the interinjection time. This appears to be too short. Therefore, NITPIC’s user interface was used to restrict this choice to a minimum of 65%. After reintegration, the overall baseline is much less systematic (Fig 4B). The increase in injection period causes a statistically insignificant 7% increase in the enthalpy of this interaction as refined by SEDPHAT, where our criterion of statistical insignificance is that the new value (4.9 kcal/mol) does not lie outside the 68.3% error interval for the previous value (4.3–4.9 kcal/mol).
5.2 Serial Integration
We executed three serial integrations on a set of 53 isotherms obtained over the past 11 years using MicroCal calorimeters (Table 2). The thermograms were not chosen using any particular experimental criteria. Indeed, the sole selection criterion was that the thermogram had not been published elsewhere. The resulting isotherms were all analyzed in SEDPHAT, and relevant parameters were recorded.
Table 2.
Results from the serial integration of 53 thermograms
| Parameters | NITPIC 1.1.0 | NITPIC 1.0.3 | OriginPro 7.0552 | ||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Name | #Inj | calorimeter | cell | ligand | seconds | log(Ka) | KD (M) | ΔH (kcal/mol) |
incA | incB | rmsd | seconds | logKa | KD (M) | ΔH (kcal/mol) |
incA | incB | rmsd | seconds | logKa | KD (M) | ΔH (kcal/mol) |
incA | incB | rmsd |
| 0655dap040307.itc | 32 | VP-ITC | Tp0655 | DAP | 3 | 4.717 | 1.92E-05 | −3.574 | 0.204 | 0 | 19.034 | 12 | 4.718 | 1.91E-05 | −3.57 | 0.204 | 0 | 19.034 | 1 | 4.569 | 2.70E-05 | −4.699 | 0.205 | 0 | 42.228 |
| 0655dap040307b.itc | 32 | VP-ITC | Tp0655 | DAP | 3 | 4.441 | 3.62E-05 | −6.455 | 0.361 | 0 | 24.298 | 12 | 4.439 | 3.64E-05 | −6.51 | 0.365 | 0 | 24.298 | 1 | 4.116 | 7.66E-05 | −159.184 | 0.953 | 0 | 64.588 |
| 0655dap040307c.itc | 32 | VP-ITC | Tp0655 | DAP | 4 | 4.358 | 4.38E-05 | −9.904 | 0.543 | 0 | 24.530 | 12 | 4.357 | 4.4E-05 | −9.96 | 0.545 | 0 | 24.530 | 1 | 4.377 | 4.20E-05 | −10.17 | 0.597 | 0 | 48.75 |
| 0655put040307a.itc | 32 | VP-ITC | Tp0655 | Putrescine | 3 | 7.341 | 4.56E-08 | −7.518 | 0 | 0.059 | 188.669 | 13 | 7.342 | 4.55E-08 | −7.51 | 0 | 0.059 | 186.641 | 1 | 7.385 | 4.12E-08 | −7.35 | 0 | 0.058 | 213.766 |
| 0655put040307b.itc | 32 | VP-ITC | Tp0655 | Putrescine | 3 | 6.783 | 1.65E-07 | −7.802 | 0 | 0.060 | 224.837 | 12 | 6.773 | 1.69E-07 | −7.800 | 0 | 0.061 | 222.491 | 1 | 6.798 | 1.59E-07 | −7.737 | 0 | 0.079 | 263.072 |
| 0655put040307c.itc | 32 | VP-ITC | Tp0655 | Putrescine | 4 | 6.932 | 1.17E-07 | −7.280 | 0 | 0.055 | 307.574 | 12 | 6.932 | 1.17E-07 | −7.280 | 0 | 0.055 | 307.574 | 1 | 6.954 | 1.11E-07 | −7.191 | 0 | 0.055 | 337.007 |
| 0655spmd032407a.itc | 32 | VP-ITC | Tp0655 | Spermidine | 3 | 5.637 | 2.31E-06 | −2.655 | 0.082 | 0 | 15.570 | 12 | 5.635 | 2.32E-06 | −2.660 | 0.082 | 0 | 15.431 | 1 | 5.666 | 2.16E-06 | −2.603 | 0.067 | 0 | 35.548 |
| 0655spmd032407b.itc | 32 | VP-ITC | Tp0655 | Spermidine | 3 | 5.625 | 2.37E-06 | −2.650 | 0.057 | 0 | 19.754 | 12 | 5.618 | 2.41E-06 | −2.670 | 0.059 | 0 | 20.042 | 1 | 5.708 | 1.96E-06 | −2.61 | 0.06 | 0 | 52.239 |
| 0655spmd032407c.itc | 32 | VP-ITC | Tp0655 | Spermidine | 3 | 5.578 | 2.64E-06 | −2.713 | 0.058 | 0 | 33.444 | 12 | 5.577 | 2.65E-06 | −2.720 | 0.057 | 0 | 34.54 | 1 | 5.592 | 2.56E-06 | −2.69 | 0.038 | 0 | 70.598 |
| 20131030r1.itc | 16 | iTC200 | EDTA | Ca2+ | 2 | 5.122 | 7.55E-06 | −4.254 | 0.072 | 0 | 13.145 | 8 | 5.122 | 7.55E-06 | −4.254 | 0.072 | 0 | 13.145 | 5 | 5.139 | 7.26E-06 | −4.246 | 0.065 | 0 | 12.642 |
| 20131030r2.itc | 16 | iTC200 | EDTA | Ca2+ | 2 | 5.371 | 4.26E-06 | −4.532 | 0.127 | 0 | 11.378 | 8 | 5.371 | 4.26E-06 | −4.354 | 0.127 | 0 | 11.378 | 5 | 5.387 | 4.10E-06 | −4.534 | 0.119 | 0 | 9.366 |
| 20131030r3.itc | 16 | iTC200 | EDTA | Ca2+ | 2 | 5.535 | 2.92E-06 | −4.354 | 0.158 | 0 | 6.407 | 8 | 5.537 | 2.9E-06 | −4.351 | 0.158 | 0 | 5.441 | 5 | 5.516 | 3.05E-06 | −4.392 | 0.155 | 0 | 8.102 |
| 20131030r4.itc | 16 | iTC200 | EDTA | Ca2+ | 2 | 5.567 | 2.71E-06 | −4.310 | 0.225 | 0 | 13.544 | 8 | 5.567 | 2.71E-06 | −4.310 | 0.225 | 0 | 13.544 | 5 | 5.594 | 2.55E-06 | −4.279 | 0.219 | 0 | 21.406 |
| 20131030r5.itc | 16 | iTC200 | EDTA | Ca2+ | 2 | 5.440 | 3.63E-06 | −5.234 | 0.279 | 0 | 31.916 | 8 | 5.44 | 3.63E-06 | −5.234 | 0.279 | 0 | 31.916 | 5 | 5.463 | 3.44E-06 | −5.226 | 0.267 | 0 | 42.288 |
| 20131030r6.itc | 16 | iTC200 | EDTA | Ca2+ | 2 | 5.662 | 2.18E-06 | −3.821 | 0.346 | 0 | 88.958 | 8 | 5.662 | 2.18E-06 | −3.821 | 0.346 | 0 | 88.958 | 4 | 5.57 | 2.69E-06 | −4.488 | 0.4 | 0 | 39.934 |
| 20131030r7.itc | 16 | iTC200 | EDTA | Ca2+ | 2 | 5.012 | 9.73E-06 | −19.139 | 0.755 | 0 | 86.271 | 9 | 5.058 | 8.75E-06 | −13.835 | 0.680 | 0 | 86.262 | 5 | 5.803 | 1.57E-06 | −2.51558 | 0.165 | 0 | 100.221 |
| 20140625r4.itc | 38 | iTC200 | α-CT | SBTI | 4 | 6.892 | 1.28E-07 | 10.354 | 0.586 | 0 | 484.261 | 17 | 6.899 | 1.26E-07 | 10.339 | 0.585 | 0 | 495.586 | 10 | 7.22 | 6.02E-08 | 9.697 | 0.552 | 0 | 942.101 |
| 20140625r5.itc | 38 | iTC200 | α-CT | SBTI | 4 | 6.983 | 1.04E-07 | 10.526 | 0.602 | 0 | 281.659 | 17 | 6.991 | 1.02E-07 | 10.541 | 0.601 | 0 | 292.562 | 10 | 7.266 | 5.42E-08 | 10.318 | 0.595 | 0 | 331.93 |
| 20140625r6.itc | 38 | iTC200 | α-CT | SBTI | 5 | 7.002 | 9.95E-08 | 10.511 | 0.592 | 0 | 276.222 | 17 | 7.001 | 9.98E-08 | 10.511 | 0.592 | 0 | 276.875 | 10 | 6.97 | 1.07E-07 | 10.875 | 0.586 | 0 | 468.162 |
| EDTACaCl22.itc | 19 | iTC200 | EDTA | Ca2+ | 2 | 4.993 | 1.02E-05 | −3.986 | 0.014 | 0 | 48.977 | 10 | 4.976 | 1.06E-05 | −4.051 | 0.016 | 0 | 43.171 | 6 | 4.943 | 1.14E-05 | −4.185 | 0.022 | 0 | 100.378 |
| H70ATp34hLF051209a.itc | 32 | VP-ITC | hLF | Tp34 | 3 | 5.967 | 1.08E-06 | 5.049 | 0 | 0.188 | 98.069 | 12 | 5.967 | 1.08E-06 | 5.049 | 0 | 0.188 | 98.069 | 1 | 6.253 | 5.58E-07 | 5.534 | 0 | 0.239 | 358.18 |
| H70ATp34hLF051209b.itc | 32 | VP-ITC | hLF | Tp34 | 3 | 5.896 | 1.27E-06 | 4.567 | 0 | 0.151 | 81.668 | 13 | 5.89 | 1.29E-06 | 4.622 | 0 | 0.156 | 85.138 | 1 | 5.8 | 1.58E-06 | 6.741 | 0 | 0.276 | 666.889 |
| H70ATp34hLF051209c.itc | 32 | VP-ITC | hLF | Tp34 | 2 | 5.806 | 1.56E-06 | 5.153 | 0 | 0.163 | 143.841 | 13 | 5.806 | 1.56E-06 | 5.150 | 0 | 0.163 | 143.841 | 1 | 6.384 | 4.13E-07 | 6.094 | 0 | 0.198 | 401.103 |
| h70aTp34hLFa.itc | 32 | VP-ITC | hLF | Tp34 | 3 | 5.403 | 3.95E-06 | 4.322 | 0 | 0.004 | 52.190 | 12 | 5.403 | 3.95E-06 | 4.322 | 0 | 0.004 | 52.190 | 1 | 5.444 | 3.60E-06 | 5.193 | 0 | 0.114 | 159.832 |
| h70aTp34hLFb.it | 32 | VP-ITC | hLF | Tp34 | 2 | 5.494 | 3.21E-06 | 3.790 | 0 | 0.026 | 125.212 | 13 | 5.494 | 3.21E-06 | 3.790 | 0 | 0.026 | 125.212 | 1 | 5.886 | 1.30E-06 | 3.546 | 0 | 0.163 | 597.007 |
| S3C0730A.ITC | 30 | VP-ITC | SynIIIC | ATP | 3 | 7.015 | 9.66E-08 | −6.988 | 0.181 | 0 | 141.710 | 12 | 7.015 | 9.66E-08 | −6.988 | 0.181 | 0 | 141.710 | 1 | 7.033 | 9.27E-08 | −7.18 | 0.156 | 0 | 207.125 |
| S3C0730B.ITC | 30 | VP-ITC | SynIIIC | ATP | 3 | 5.604 | 2.49E-06 | −7.000 | 0.077 | 0 | 93.240 | 11 | 5.604 | 2.49E-06 | −7.000 | 0.077 | 0 | 93.240 | 1 | 5.693 | 2.03E-06 | −7.058 | 0.035 | 0 | 225.172 |
| S3C0730C.ITC | 30 | VP-ITC | SynIIIC | ATP | 3 | 6.783 | 1.65E-07 | −7.103 | 0.202 | 0 | 255.676 | 12 | 6.783 | 1.65E-07 | −7.103 | 0.202 | 0 | 255.676 | 1 | 6.832 | 1.47E-07 | −7.104 | 0.183 | 0 | 271.069 |
| s3c071603.itc | 30 | VP-ITC | SynIIIC | ATP | 3 | 6.838 | 1.45E-07 | −7.027 | 0.172 | 0 | 148.414 | 12 | 6.838 | 1.45E-07 | −7.027 | 0.172 | 0 | 148.414 | 1 | 6.928 | 1.18E-07 | −6.987 | 0.161 | 0 | 332.376 |
| S3CNOCA4.ITC | 30 | VP-ITC | SynIIIC | ATP | 3 | 6.866 | 1.36E-07 | −6.618 | 0.178 | 0 | 162.566 | 12 | 6.866 | 1.36E-07 | −6.618 | 0.178 | 0 | 162.566 | 1 | 6.806 | 1.56E-07 | −7.195 | 0.112 | 0 | 432.289 |
| s3cplusca22.itc | 30 | VP-ITC | SynIIIC | ATP | 2 | 5.471 | 3.38E-06 | −7.185 | 0.052 | 0 | 43.658 | 11 | 5.467 | 3.41E-06 | −7.205 | 0.050 | 0 | 43.212 | 1 | 5.505 | 3.13E-06 | −7.087 | 0.011 | 0 | 68.922 |
| s3cplusca23.itc | 30 | VP-ITC | SynIIIC | ATP | 2 | 5.372 | 4.25E-06 | −7.738 | 0.119 | 0 | 91.118 | 11 | 5.372 | 4.25E-06 | −7.740 | 0.119 | 0 | 91.118 | 1 | 5.395 | 4.03E-06 | −7.407 | 0.042 | 0 | 92.884 |
| sam0716a.itc | 18 | iTC200 | PDK2 | PS10 | 1 | 6.652 | 2.23E-07 | −13.08 | 0.051 | 0 | 441.837 | 10 | 6.607 | 2.47E-07 | −12.961 | 0.052 | 0 | 277.425 | 5 | 6.741 | 1.81E-07 | −11.939 | 0.078 | 0 | 1126.34 |
| sam0716b.itc | 19 | iTC200 | PDK2 | PS10 | 1 | 6.671 | 2.13E-07 | −12.965 | 0.123 | 0 | 107.282 | 9 | 6.671 | 2.13E-07 | −12.965 | 0.123 | 0 | 107.282 | 5 | 6.78 | 1.66E-07 | −12.988 | 0.133 | 0 | 174.522 |
| sam0716d.itc | 19 | iTC200 | PDK2 | PS10 | 2 | 6.607 | 2.47E-07 | −13.915 | 0.456 | 0 | 132.631 | 10 | 6.573 | 2.67E-07 | −13.910 | 0.460 | 0 | 128.594 | 5 | 6.769 | 1.70E-07 | −13.600 | 0.428 | 0 | 158.77 |
| sam0716e.itc | 19 | iTC200 | PDK2 | PS10 | 2 | 6.616 | 2.42E-07 | −16.199 | 0.518 | 0 | 315.218 | 10 | 6.616 | 2.42E-07 | −16.199 | 0.518 | 0 | 315.218 | 5 | 6.462 | 3.45E-07 | −19.154 | 0.548 | 0 | 677.199 |
| sam0717a.itc | 19 | iTC200 | PDK2 | PS10 | 2 | 7.387 | 4.10E-08 | −14.681 | 0.273 | 0 | 3127.302 | 8 | 7.387 | 4.1E-08 | −14.680 | 0.273 | 0 | 3127.302 | 5 | 10 | 1E-10 | −15.788 | 0.19 | 0 | 3996.092 |
| sam0717b.itc | 19 | iTC200 | PDK2 | PS10 | 1 | 6.717 | 1.92E-07 | −13.251 | 0.349 | 0 | 52.36 | 10 | 6.717 | 1.92E-07 | −13.251 | 0.349 | 0 | 52.36 | 5 | 6.782 | 1.65E-07 | −13.008 | 0.326 | 0 | 142.795 |
| sam0717c.itc | 19 | iTC200 | PDK2 | PS10 | 2 | 6.944 | 1.14E-07 | −13.017 | 0.497 | 0 | 708.617 | 10 | 6.944 | 1.14E-07 | −13.017 | 0.497 | 0 | 708.617 | 5 | 7.166 | 6.82E-08 | −12.815 | 0.508 | 0 | 742.886 |
| sam0717d.itc | 19 | iTC200 | PDK2 | PS10 | 2 | 6.669 | 2.14E-07 | −14.393 | 0.382 | 0 | 508.933 | 9 | 6.669 | 2.14E-07 | −14.393 | 0.382 | 0 | 508.932 | 5 | 6.756 | 1.75E-07 | −14.139 | 0.353 | 0 | 637.264 |
| sam0717e.itca | 19 | iTC200 | PDK2 | PS10 | 2 | N/A | N/A | N/A | N/A | N/A | N/A | 7 | N/A | N/A | N/A | N/A | N/A | N/A | 5 | N/A | N/A | N/A | N/A | N/A | N/A |
| sam0717f.itca | 19 | iTC200 | PDK2 | PS10 | 1 | N/A | N/A | N/A | N/A | N/A | N/A | 8 | N/A | N/A | N/A | N/A | N/A | N/A | 5 | N/A | N/A | N/A | N/A | N/A | N/A |
| sam0718a.itca | 19 | iTC200 | HBA | HIF2α | 2 | N/A | N/A | N/A | N/A | N/A | N/A | 10 | N/A | N/A | N/A | N/A | N/A | N/A | 5 | N/A | N/A | N/A | N/A | N/A | N/A |
| sam0718b.itc | 19 | iTC200 | HBA | HIF2α | 2 | 6.435 | 3.67E-07 | −5.901 | 0.108 | 0 | 192.891 | 10 | 6.435 | 3.67E-07 | −5.902 | 0.108 | 0 | 192.891 | 5 | 6.348 | 4.49E-07 | −6.376 | 0.176 | 0 | 207.461 |
| sam0718d.itc | 19 | iTC200 | HBA | HIF2α | 1 | 7.173 | 6.71E-08 | −5.622 | 0.112 | 0 | 600.778 | 8 | 7.173 | 6.71E-08 | 5.622 | 0.112 | 0 | 600.778 | 5 | 7.225 | 5.96E-08 | −5.069 | 0.109 | 0 | 510.112 |
| sam0719a.itcb | 18 | iTC200 | HBA | HIF2α | 2 | 7.384 | 4.13E-08 | −4.777 | 0.221 | 0 | 285.804 | 8 | 7.384 | 4.13E-08 | −4.777 | 0.221 | 0 | 285.804 | 5 | 7.801 | 1.58E-08 | −4.756 | 0.142 | 0 | 324.357 |
| sam0719c.itc | 19 | iTC200 | HBA | HIF2α | 2 | 6.829 | 1.48E-07 | −5.305 | 0.204 | 0 | 241.43 | 10 | 6.835 | 1.46E-07 | −5.312 | 0.206 | 0 | 244.102 | 5 | 6.9 | 1.26E-07 | −5.201 | 0.183 | 0 | 272.67 |
| sam0722b.itc | 15 | iTC200 | HBA | HIF2α | 2 | 7.096 | 8.02E-08 | −4.193 | 0.140 | 0 | 281.838 | 9 | 7.096 | 8.02E-08 | −4.193 | 0.140 | 0 | 281.838 | 4 | 6.762 | 1.73E-07 | −4.697 | 0.051 | 0 | 248.599 |
| sam0722c.itc | 19 | iTC200 | HBA | HIF2α | 2 | 6.979 | 1.05E-07 | −4.896 | 0 | 0.022 | 195.112 | 11 | 6.979 | 1.05E-07 | −4.896 | 0 | 0.022 | 195.112 | 6 | 6.784 | 1.64E-07 | −5.590 | 0 | 0.06 | 247.682 |
| ScottTso1.itc | 18 | iTC200 | PDK2 | PS8 | 1 | 6.306 | 4.94E-07 | −9.656 | 0.187 | 0 | 144.536 | 9 | 6.306 | 4.94E-07 | −9.656 | 0.187 | 0 | 144.536 | 5 | 6.296 | 5.06E-07 | −9.780 | 0.178 | 0 | 269.089 |
| ScottTso2.itc | 25 | iTC200 | PDK2 | PS8 | 2 | 5.915 | 1.22E-06 | −11.366 | 0.410 | 0 | 615.077 | 12 | 5.915 | 1.22E-06 | −11.366 | 0.410 | 0 | 615.077 | 6 | 5.778 | 1.67E-06 | −12.482 | 0.404 | 0 | 1442.478 |
| Tp34hLFHep.itca | 32 | VP-ITC | hLF | Tp34 | 3 | N/A | N/A | N/A | N/A | N/A | N/A | 12 | N/A | N/A | N/A | N/A | N/A | N/A | 1 | N/A | N/A | N/A | N/A | N/A | N/A |
| Tp34hLFPi.itc | 32 | VP-ITC | hLF | Tp34 | 3 | 4.947 | 1.13E-05 | 5.88 | 0.838 | 0 | 35.402 | 11 | 4.958 | 1.1E-05 | 5.302 | 0.820 | 0 | 36.54 | 1 | 4.651 | 2.23E-05 | 517.590 | 0.997 | 0 | 66.453 |
The isotherm contained features that made it inappropriate for fitting to the 1:1 binding model; thermograms are included for time-comparison purposes only.
Injection 8 was omitted from the fitting for all three isotherms. It was a clear outlier that was strongly impacting the analyses.
The three integrations were undertaken using NITPIC 1.1.0, NITPIC 1.0.3, and Origin version 7.0552. NITPIC version 1.0.3 was chosen because it was the last version released before the streamlining Jackrabbit code changes (Section 4.1.1) were implemented. Thus, it should provide a good estimate of the efficacy of the changes and serve as a control to spot any degradation in the quality of the outputted isotherms. The version of Origin used in this study was supplied in conjunction with an iTC200 calorimeter purchased in October of 2013, and thus is considered current.
5.2.1 Time performance
A key component of the practicality of serial integration is the speed of the integrations. Slow integration performance would disincentivize the conduction of properly designed experiments that require numerous isotherms. Thus, to significantly reduce or eliminate the barrier to performing such experiments, the Jackrabbit code changes (Section 4.1.1) were implemented in NITPIC 1.1.0. The resulting speed gains can be discerned from Table 2. For all thermograms, NITPIC 1.1.0 was several-fold faster than version 1.0.3, with up to 6-fold speed increases measured under the conditions of this study. A 4.5-fold average speed performance improvement across all datasets was observed. Some thermograms (with small numbers of injections) were integrated in only 1 s in the new version. Overall, NITPIC version 1.1.0 took two minutes and thirty-eight seconds (2:38) to integrate the 53 thermograms; version 1.0.3 took 9:53.
The Origin algorithms, which are different for iTC200- and the VP-ITC- derived data, also demonstrated good speed performance. The iTC200 method adjusts the definition of injection and baseline on an individual-injection basis, and only integrates these injection regions. The VP-ITC algorithm simply integrates the entire interinjection region. The VP-ITC algorithm never exceeded 1 s for the integration of a 32-injection thermogram, but the iTC200 algorithm exhibited integration times that were slower than NITPIC 1.1.0. On average, NITPIC 1.1.0 was 2.7 times faster than the Origin iTC200 algorithm. Not shown are the times necessary to export the data from Origin and to import them into SEDPHAT, which can add 15–30 s to each Origin-based analysis. This time overhead is not present with NITPIC in the serial integration mode, because it writes its results in SEDPHAT’s native format.
5.2.2 Quality performance
The speed gains achieved by the Jackrabbit code changes would not be worthwhile if they degrade the quality of the resulting isotherms. It was thus necessary to examine all of the isotherms produced in the serial integration. The r.m.s.d.’s of the fits to the isotherms were used as the metric of isotherm quality. This is because, as demonstrated earlier [21], lower r.m.s.d.’s will result in more precisely determined parameters.
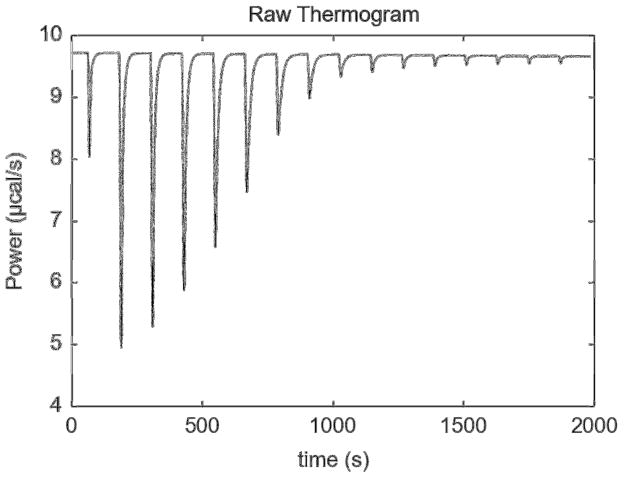
In almost all cases (Table 2), the NITPIC versions produced higher-quality isotherms than Origin. These differences were stark in some thermograms, e.g. “H70ATp34hLF051209b.itc”, wherein NITPIC’s resulting fitted r.m.s.d.’s were reduced by a divisor of ca. 8 compared to those derived from Origin. This was an outlier, however, and, on average, this divisor was about 1.6. Nonetheless, even this seemingly modest improvement should provide significantly more precise parameter estimates from a SEDPHAT fitting session. By this criterion, Origin outperformed both versions of NITPIC on two thermograms having unusually high signal-to-noise ratios: “20131030r1.itc” and “20131030r2.itc” (e.g., Fig. 5). These and similarly named thermograms were collected from a pair of common calorimeter testing solutions (5 mM CaCl2 and 400 μM EDTA), where the reagents were tested at very high concentrations (20131013r1) and were subsequently serially diluted to provide thermograms with progressively lower signal-to-noise ratios (20131013r2, -r3, etc.). Following the second two-fold serial dilution (20131030r3 through -r6), NITPIC significantly outperforms Origin. Although the NITPIC-derived r.m.s.d.’s from the analyses of 20131030r7.itc, the most-diluted sample, are lower, they feature aberrant KA and ΔH values, whereas those derived from Origin for this thermogram were more consistent with measurements from more concentrated samples. The origin of this phenomenon is unknown, but merits scrutiny in future work.
Figure 5. A high-signal thermogram.
The raw thermogram of titration “20131030r1.itc” is shown.
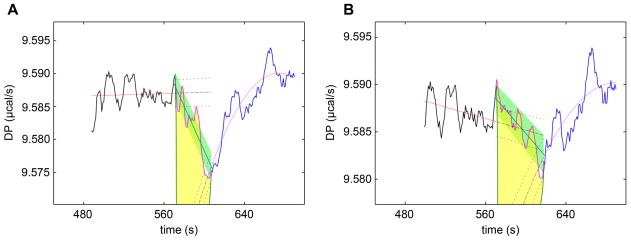
Having established that the NITPIC approach overall provides more accurate parameter estimates, it was then necessary to compare the outcomes of the two NITPIC versions to assess whether the small but significant changes in the algorithm between versions 1.0.3 and 1.1.0 compromised the quality of the integrations from the latter. The majority of the results are identical with the two versions. Where differences occur, they are small, and often the r.m.s.d.’s are smaller using version 1.1.0. One notable exception to these trends is found in thermogram “sam0716a.itc”: the r.m.s.d. derived from the use of version 1.1.0 was 1.6-fold higher than that resulting from version 1.0.3. This result appears to be due entirely to the choice of injection-period width, with version 1.1.0 selecting 40%, and version 1.0.3 choosing a 31% period width. Strong cases can be made for both choices; version 1.1.0 may have defined some of the baseline period as injection, increasing the noise, but there is evidence (particularly in injections 4 and 5; see Fig. 6) that the choice of 31% made by version 1.0.3 may have cut off the injection period too early. Thus, the choice made by version 1.1.0 may result in more isotherm noise, but this might be a more accurate reflection of the quality of the isotherm.
Figure 6. Injection 5 of the thermogram “sam0716a.itc”.
(A) As treated by NITPIC 1.0.3. (B) As treated by NITPIC 1.1.0. “DP” stands for “differential power”. Both figures have the same color scheme: The black line at the left is the pre-injection baseline; associated with it are the red line (a linear fit to the data) and the black long-dashed line (extending into the injection period), with accompanying error bars (black short dashes). The blue line at the right is the post-injection baseline; it has associated with it a purple line (a quadratic fit to the data), a blue, long-dashed line (the extrapolation into the injection period), and accompanying error bars for the extrapolation (blue with short dashes). The magenta line is the estimated baseline; the solid green line shows the linear baseline that NITPIC chose; the green-shaded area is the error estimate for the integration of the peak; and the yellow-shaded area is the area integrated to form the fifth point of the isotherm.
6. Discussion
6.1 Speed improvements in NITPIC
As mentioned in Section 1, the advent of calorimetric strategies to address complex biomolecular binding problems, such as proton-linked binding, derivation of heat-capacity changes, and refinement of cooperativity parameters require multiple isotherms to be analyzed. Ideally, these isotherms should be analyzed globally [12,15,16], and their errors should be scrutinized closely [15,26,27]. But before the analysis steps can be undertaken, the thermograms must be integrated. In many instances, the baselines of the thermograms will not have sufficient quality to allow for unattended use of the baseline derived by Origin. Thus, if this program is used, manual adjustment of the baseline is usually required, adding unwanted time and user biases to the integration step. This situation certainly does not improve when there are tens or hundreds of thermograms to integrate.
The NITPIC algorithm avoids these problems. The user has only a few choices that can be made, and manual adjustment of the baseline is not allowed; this philosophy removes user bias. The only expectation that NITPIC builds into its analysis is that of a smoothly varying isotherm, which should be fulfilled for most applications of the ITC. Even if it is not, NITPIC still performs well [7]. With the introduction of the Jackrabbit code changes (Section 4.1.1), speed is no longer an issue. Indeed, NITPIC on average was faster than the iTC200 algorithm of Origin (Table 2). While NITPIC was slower than the VP-ITC algorithm of Origin, this amounted to only a few seconds, and neglects the time-saving file formats that NITPIC provides that are used in subsequent analyses with SEDPHAT. NITPIC almost always provides a more precisely defined isotherm than Origin. However, without serial integration enabled, the NITPIC user would still be required to input the files into the program and actuate the generation of output files. Removing these obstacles to processing large numbers of thermogram files therefore was a main goal of the current work.
6.2 Serial Integration
6.2.1 The goals of serial integration
To achieve the maximum utility of serial integration, several criteria were laid out. First, the processing run should be fully automatic, i.e. no user intervention should be required at any time during the analysis. This required automated file loading, processing, and writing of output files. It also necessitated that NITPIC avoid displaying real-time warnings that would be useful and appropriate in a single-integration context. Next, because of the hands-off nature of the processing, the program should be able to identify some occasional shortcomings in the analysis and notify the user of data sets that exhibit them. Finally, there should be an optimized user interface that allows the user to easily and intuitively interact with the results of the serial integration.
6.2.2 Unattended integration
The first criterion was easily achieved. The user initiates serial fitting from the File Menu in the normal user interface. The program prompts for a list of files in a manner familiar to users of the operating system. Once the list of files has been indicated, the user initiates the integration session by pressing a single button in the user interface; the serial integration may be unattended for its duration. Importantly, the user has the ability to abort the session. A warning that may arise during a normal, non-serial run occurs when the chosen integration period is equal to or exceeds 75% of the interinjection period. This warning is suppressed in the serial-integration mode; it is noted internally, however, and the user is alerted to this potential problem in the user interface (Sections 4.2.3 and 6.2.4). During the run, the necessary files are written to a new file folder (called “NITPICserialInt”) for recalling the individual integration sessions and for loading the resulting isotherms into SEDPHAT.
6.2.3 New warnings
To relieve the user of the necessity of scrutinizing all of the integrations and isotherms, two new post-hoc integration warnings were developed in this work. They concern the number of shape components that were used to estimate the baseline and the appearance of systematic baselines in early, high-magnitude injections (Sections 4.1.2 and 5.1; Figs. 3 & 4). Experience has shown that these problems are faithfully detected by the algorithms developed herein. To alert the user of their presence in a serial integration, a set of iconified warnings (Fig. 1) were developed and implemented in the user interface to the serial-integration results (Section 6.2.4). These warnings are also present during single-file integrations in the new version of NITPIC, 1.1.0.
6.2.4 The post-integration user interface
As summarized in Section 4.2.3, the user interface is offered in tabular form. The first table is a text table that shows information about the fitting of the isotherm and the wrmsd. However, the more useful form is available by clicking on a button at the bottom of the table labeled “Toggle View”. After initiating the toggling for the first time, it may take several seconds for the view to be switched, because iconified versions of the integrations are being generated. This time overhead in switching could have been avoided by generating the icons during the serial integration. However, in the interest of the most stable and quick serial-integration session, the icon generation was postponed until after the serial integration is complete. This approach also recognizes that some users may not want to use the iconified table, and therefore they should not be forced to endure the delays introduced by icon generation. After the icons have been generated once, they never need to be made again; thus, the user may instantaneously switch between the text table and the iconified one.
The iconified table (Fig. 2) is less information dense, but offers intuitive summaries of the integrations at a glance. Beneath the name of the analyzed file, a close-up of the second injection is displayed (traditionally in ITC analysis, this is the first analyzed injection, as the first injection is discarded as likely bearing a spurious heat value). This view provides insight into whether NITPIC chose an appropriate injection period. The fitted isotherm is also shown on the icon, allowing a quick assessment of the quality of the result. Finally, the K, ΔH, and N values from the fitting session are displayed in text to the far right. Clicking on this part of the icon loads the integration result into NITPIC’s main graphing window, where the user may inspect it more closely.
Four possible warnings are available in this version of the table in the form of strong visual cues. Any suppressed warnings concerning the length of the injection period (Sections 5.1.2 & 6.2.2) are shown here as gold backgrounds to the icon (Fig. 2A). This phenomenon can occur when the signal-to-noise ratio of the thermogram is low. This problem can usually be ameliorated by changing a few parameters in NITPIC (see the NITPIC manual). Failure of the fitting routine to converge is signified by a pink background color in the icon. The cause of this problem is usually due to low heat signals or to isotherms that do not adopt the canonical sigmoid shape. Although this problem is not considered serious, sometimes adjustments to the initial parameters used for isotherm fitting relieve it. The other two warnings, for too few shape components in the SVD-reconstructed injection-period baseline signal and systematic baselines, take the form of the same icons that are documented in Fig. 1. As noted in Section 4.2.3, there is no limitation on the number of files that may be examined using the graphical table, nor are there limits on the number of examination cycles. Closure of the program does not eliminate the results table; it may be recalled by the user at any time after the serial integration.
6.3 Future directions
The advances made in this version of NITPIC should enable further improvements. For instance, as data are gathered regarding the efficacy of the new warning methods, it may be possible to correct the problems during the serial integration run, or automatically after the integration run. Three of the warnings (too long injection period, too many shape components, systematic baselines) are done post hoc; therefore, correcting them requires reinitializing the software with new parameter sets. This approach would necessarily add time to the automated integration session; however, eliminating the need for manual reintegration of problem thermograms may be desirable for some users. Although the new version is very fast, very large groups of thermograms still require significant amounts of time to process. For example, there is an automated version of the iTC200 available that can perform up to 384 titrations unattended. This large group of thermograms would take over ten minutes to integrate using the current software. Parallelization of the integration tasks could reduce the overall integration time substantially. “On-the-fly” analyses of thermograms as they appear during such an automated experiment may also be possible.
Algorithmic enhancements were made to NITPIC, used to integrate ITC data
The speed of the new algorithm is excellent
The new algorithm’s precision is comparable to the old one
The new method is generally better than other commonly used integration software
Acknowledgments
The authors wish to thank Yogarany Chelliah and Drs. Shih-Chia Tso, Ranjit Deka, Kevin Gardner, David Chuang, Michael Norgard, and Johann Deisenhofer for providing proteins and ligands used in this study. The Norgard Lab’s work on Tp0655 was supported by NIH grant AI56305 (to Norgard), and the Chuang Lab’s research on PDK2 was supported by NIH grant DK62306 (to Chuang). The HIF2α work in the Gardner Lab was supported by grants RP100846 and RP130513 from the Cancer Prevention Research Institute of Texas. The synapsin III-C studies were supported by the Howard Hughes Medical Institute, when Deisenhofer was an investigator there. We thank Drs. Verna Frasca and Ivana Simonovic for acquiring some thermograms used in this work.
Abbreviations
- α-CT
bovine α-chymotrypsin
- DAP
1,3-diaminopropane
- DP
differential power
- DMSO
dimethyl sulfoxide
- DTT
dithiothreitol
- EDTA
ethylenediaminetetraacetic acid
- EGTA
ethylene glycol tetraacetic acid
- GST
glutathione-S-transferase
- GUI
graphical user interface
- HBA
HIF2α benzoxadiazole antagonist
- HIF2α
hypoxia-inducible factor 2α
- ITC
isothermal titration calorimetry
- PAS
Per/Arnt/Sim
- PBS
phosphate-buffered saline
- r.m.s.d
root-mean-square deviation
- SynIIIC
Synapsin IIIC
- SBTI
Soybean trypsin inhibitor
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Pierce MM, Raman CS, Nall BT. Isothermal titration calorimetry of protein-protein interactions. Methods. 1999;19:213–21. doi: 10.1006/meth.1999.0852. [DOI] [PubMed] [Google Scholar]
- 2.Oda M, Nakamura H. Thermodynamic and kinetic analyses for understanding sequence-specific DNA recognition. Genes Cells. 2000;5:319–26. doi: 10.1046/j.1365-2443.2000.00335.x. [DOI] [PubMed] [Google Scholar]
- 3.Leavitt S, Freire E. Direct measurement of protein binding energetics by isothermal titration calorimetry. Curr Opin Struct Biol. 2001;11:560–6. doi: 10.1016/s0959-440x(00)00248-7. [DOI] [PubMed] [Google Scholar]
- 4.Majhi PR, Blume A. Thermodynamic Characterization of Temperature-Induced Micellization and Demicellization of Detergents Studied by Differential Scanning Calorimetry. Langmuir. 2001;17:3844–3851. [Google Scholar]
- 5.Jahnke N, Krylova OO, Hoomann T, Vargas C, Fiedler S, Pohl P, et al. Real-time monitoring of membrane-protein reconstitution by isothermal titration calorimetry. Anal Chem. 2014;86:920–927. doi: 10.1021/ac403723t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vargas C, Klingler J, Keller S. Membrane partitioning and translocation studied by isothermal titration calorimetry. Methods Mol Biol. 2013;1033:253–270. doi: 10.1007/978-1-62703-487-6_16. [DOI] [PubMed] [Google Scholar]
- 7.Heerklotz H, Tsamaloukas AD, Keller S. Monitoring detergent-mediated solubilization and reconstitution of lipid membranes by isothermal titration calorimetry. Nat Protoc. 2009;4:686–97. doi: 10.1038/nprot.2009.35. [DOI] [PubMed] [Google Scholar]
- 8.Freyer MW, Lewis EA. Isothermal titration calorimetry: experimental design, data analysis, and probing macromolecule/ligand binding and kinetic interactions. Methods Cell Biol. 2008;84:79–113. doi: 10.1016/S0091-679X(07)84004-0. [DOI] [PubMed] [Google Scholar]
- 9.Wiseman T, Williston S, Brandts JF, Lin LN. Rapid measurement of binding constants and heats of binding using a new titration calorimeter. Anal Biochem. 1989;179:131–137. doi: 10.1016/0003-2697(89)90213-3. [DOI] [PubMed] [Google Scholar]
- 10.Armstrong KM, Baker BM. A comprehensive calorimetric investigation of an entropically driven T cell receptor-peptide/major histocompatibility complex interaction. Biophys J. 2007;93:597–609. doi: 10.1529/biophysj.107.104570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baker BM, Murphy KP. Evaluation of linked protonation effects in protein binding reactions using isothermal titration calorimetry. Biophys J. 1996;71:2049–55. doi: 10.1016/S0006-3495(96)79403-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coussens NP, Schuck P, Zhao H. Strategies for assessing proton linkage to bimolecular interactions by global analysis of isothermal titration calorimetry data. J Chem Thermodyn. 2012;52:95–107. doi: 10.1016/j.jct.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ladbury JE, Williams MA. The extended interface: measuring non-local effects in biomolecular interactions. Curr Opin Struct Biol. 2004;14:562–569. doi: 10.1016/j.sbi.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 14.Tan A, Tanner JJ, Henzl MT. Energetics of OCP1-OCP2 complex formation. Biophys Chem. 2008;134:64–71. doi: 10.1016/j.bpc.2008.01.005. [DOI] [PubMed] [Google Scholar]
- 15.Houtman JCD, Brown PH, Bowden B, Yamaguchi H, Appella E, Samelson LE, et al. Studying multisite binary and ternary protein interactions by global analysis of isothermal titration calorimetry data in SEDPHAT: application to adaptor protein complexes in cell signaling. Protein Sci. 2007;16:30–42. doi: 10.1110/ps.062558507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Freiburger LA, Auclair K, Mittermaier AK. Elucidating protein binding mechanisms by variable- c ITC. Chembiochem. 2009;10:2871–2873. doi: 10.1002/cbic.200900614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Freire E. Do enthalpy and entropy distinguish first in class from best in class? Drug Discov Today. 2008;13:869–74. doi: 10.1016/j.drudis2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ladbury JE. Calorimetry as a tool for understanding biomolecular interactions and an aid to drug design. Biochem Soc Trans. 2010;38:888–93. doi: 10.1042/BST0380888. [DOI] [PubMed] [Google Scholar]
- 19.Sarver RW, Bills E, Bolton G, Bratton LD, Caspers NL, Dunbar JB, et al. Thermodynamic and structure guided design of statin based inhibitors of 3-hydroxy-3-methylglutaryl coenzyme A reductase. J Med Chem. 2008;51:3804–13. doi: 10.1021/jm7015057. [DOI] [PubMed] [Google Scholar]
- 20.Scheuermann TH, Li Q, Ma HW, Key J, Zhang L, Chen R, et al. Allosteric inhibition of hypoxia inducible factor-2 with small molecules. Nat Chem Biol. 2013;9:271–276. doi: 10.1038/nchembio.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keller S, Vargas C, Zhao H, Piszczek G, Brautigam CA, Schuck P. High-precision isothermal titration calorimetry with automated peak-shape analysis. Anal Chem. 2012;84:5066–73. doi: 10.1021/ac3007522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Machius M, Brautigam CA, Tomchick DR, Ward P, Otwinowski Z, Blevins JS, et al. Structural and biochemical basis for polyamine binding to the Tp0655 lipoprotein of Treponema pallidum: putative role for Tp0655 (TpPotD) as a polyamine receptor. J Mol Biol. 2007;373:681–694. doi: 10.1016/j.jmb.2007.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tso SC, Qi X, Gui WJ, Wu CY, Chuang JL, Wernstedt-Asterholm I, et al. Structure-guided development of specific pyruvate dehydrogenase kinase inhibitors targeting the ATP-binding pocket. J Biol Chem. 2014;289:4432–4443. doi: 10.1074/jbc.M113.533885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2. Cold Spring Harbor Publications; Cold Spring Harbor, NY: 1989. [Google Scholar]
- 25.Bösterling B, Quast U. Soybean trypsin inhibitor (Kunitz) is doubleheaded: kinetics of the interaction of α-chymotrypsin with each side. Biochim Biophys Acta. 1981;657:58–72. doi: 10.1016/0005-2744(81)90130-3. [DOI] [PubMed] [Google Scholar]
- 26.Brautigam CA. Fitting two- and three-site models to isothermal titration calorimetric data. Methods. 2015 doi: 10.1016/j.ymeth.2014.11.018. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bevington PR, Robinson DK. Data reduction and error analysis for the physical sciences. 2. WCB/McGraw-Hill; Boston, MA: 1992. [Google Scholar]