Abstract
Sumoylation plays important roles during DNA damage repair and responses. Recent broad-scope and substrate-based studies have shed light on the regulation and significance of sumoylation during these processes. An emerging paradigm is that sumoylation of many DNA metabolism proteins is controlled by DNA engagement. Such “on-site modification” can explain low substrate modification levels and has important implications in sumoylation mechanisms and effects. New studies also suggest that sumoylation can regulate a process through an ensemble effect or via major substrates. Additionally, we describe new trends in the functional effects of sumoylation, such as bi-directional changes in biomolecule binding and multi-level coordination with other modifications. These emerging themes and models will stimulate our thinking and research in sumoylation and genome maintenance.
Overview: the expanding universe of SUMO
SUMO (Small Ubiquitin-like Modifier) is a protein modifier that plays key roles in a wide range of cellular processes, making it essential for the viability of most eukaryotes. Like ubiquitin, SUMO is covalently linked to its substrates by a series of dedicated enzymes (Figure 1 and Box 1). Through dynamic alterations of a substrate’s biochemical properties, sumoylation and its reverse reaction, desumoylation, can elicit rapid and reversible biological changes. Research in the late 1990s and 2000s elucidated the enzymology of SUMO conjugation and de-conjugation, as well as the regulatory mechanisms for a few well-characterized substrates [1–4]. The broad-scope biochemical studies that followed, particularly those in recent years, have uncovered thousands of additional substrates in fungi, plants, invertebrates, and vertebrates [5–17]. In the meantime, focused, substrate-based studies have begun to describe in detail the various effects that sumoylation can have on the function of individual proteins. These parallel and complementary lines of work have provided new insights into the biological significance of sumoylation on many levels, from the global to the specific, from its pathological consequences for human disease to its molecular influence on a single reaction.
Figure 1. The dynamic SUMO cycle.
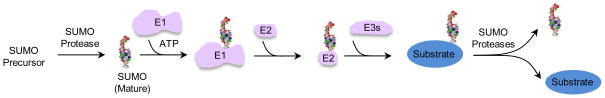
The maturation, activation, conjugation and deconjugation of the SUMO moiety (shown in 3D structure rendering) are depicted. SUMO enzymes are colored pink, and the substrate is in blue.
Box 1.
The principle of the sumoylation and desumoylation cycle. Sumoylation is a highly conserved process mechanistically similar to ubiquitylation but with SUMO (Small Ubiquitin-like Modifier)-specific enzymes. The maturation of SUMO is catalyzed by cysteine proteases called desumoylation enzymes (also referred to as desumolyases or SUMO-specific proteases). This cleavage reveals the terminal di-glycine motif required for conjugation to lysine residues on substrates. Mature SUMO is activated in an ATP-dependent manner by the dimeric E1 activating enzyme, and is then transferred to the E2 conjugating enzyme via a thioester transfer step. The E2 can conjugate SUMO to the target lysine through an isopeptide linkage either by direct recognition of the substrate or with the assistance of E3s (or ligases) that serve as substrate adapters. The reaction is reversed by the action of desumoylases, releasing the substrate and free SUMO for further rounds of modification. Most organisms have only one E1 and one E2, but several E3s and desumoylases. Plant and metazoan cells have higher numbers of enzyme isoforms of E3s and desumoylases compared with lower eukaryotic cells. The number of SUMO isoforms also varies among organisms. While yeast and lower eukaryotes encode only one form of SUMO, higher eukaryotes express several. In humans, SUMO-1 is different from SUMO-2/3 in sequence, expression and chain formation ability, and the three forms can target either the same or different substrates.
An arena that has seen particularly rapid progress pertains to the multiple processes that govern genome maintenance. Consistent with early findings that dysregulated sumoylation confers genome instability [18, 19], recent screens have found that SUMO substrates are enriched for enzymes and regulators of DNA metabolism [10–15]. These proteins as a group are also tightly regulated by other post-translational modifications (PTMs) [14–16, 20–26], highlighting the fact that preserving genome integrity requires the complex coordination of dynamic events by multiple PTMs. Here, we discuss several paradigms emerging from recent studies on SUMO-based regulation of DNA metabolism. Our topics include the tight regulation of substrate sumoylation and its implications, new features of the biological effects of sumoylation at both global and individual substrate levels, as well as the crosstalk and comparison with other PTMs. For additional topics, we refer readers to some excellent recent reviews [27–29].
Low level of substrate sumoylation can be linked to on-site modification
Amongst different PTMs, sumoylation is particularly known for its low level of modification as typically only a small percentage of a protein is sumoylated [3, 4]. While several hypotheses have been proposed to explain this phenomenon, based on recent work on DNA metabolism proteins, we suggest that this phenomenon could be explained by sumoylation only occurring when proteins are engaged with DNA substrates. We refer to this concept as “on-site sumoylation”, and summarize below the supporting evidence and its implications.
An early study demonstrated that sumoylation of the polymerase sliding clamp PCNA (Proliferating Cell Nuclear Antigen) depends on DNA association [30]. Recent characterization of additional substrates shows that this is not an isolated example; rather, the prerequisite of DNA association for sumoylation is a general trend. For instance, the non-homologous end joining (NHEJ) repair protein Yku70, the base excision repair protein Thymine DNA Glycosylase (TDG), the Rad1 nuclease, the viral polymerase processivity factor UL44, and the Fanconi anemia pathway proteins FANCI and FANCD2 all require DNA association for sumoylation [31–35]. In the case of the yeast Yku70 and human TDG proteins, which function upstream in their respective repair pathways, mutations abolishing their DNA binding prevent their sumoylation [34, 35]. Similarly, a DNA binding mutation in the PCNA-like UL44 protein also impairs its sumoylation [31]. In some cases, DNA repair engagement, rather than simply DNA association, is required for sumoylation, as seen for the yeast Rad1 nuclease and human FANCI and FANCD2 proteins [32, 33]. Taken together, these new findings in different organisms suggest that DNA association and repair engagement are conserved requirements for the sumoylation of multiple DNA metabolism proteins. Because typically only a small proportion of proteins is actively engaged with DNA substrates or in repair at a given time, on-site sumoylation provides an explanation for the low abundance of the sumoylated forms of these proteins.
We envision that on-site sumoylation is applicable to many additional substrates involved in DNA transactions based on further correlative evidence. First, sumoylation levels of some of these proteins increase in situations when they are more active. For example, the sumoylation of several DNA repair proteins is induced when cells are treated with the DNA damaging agents that elicit repair activities from that protein, with higher genotoxin dosage having a stronger effect [6, 33, 36]. Along the same lines, sumoylation levels of some substrates peak in the cell cycle stage at which they are most active [15, 37, 38]. Second, the absence of upstream DNA repair factors reduces the sumoylation of downstream proteins in homologous recombination (HR), nucleotide excision repair (NER), and inter-strand crosslink repair [5, 6, 32, 33, 39]. Third, substrates are hyper-sumoylated when their DNA association is prolonged, either by mutation of their catalytic sites or inhibition of downstream reactions. Topoisomerases and Rad1 specifically illustrate the former scenario [33, 40, 41], while the recombination protein Rad52 and the NER factor Rad4 provide examples of the latter [36, 42, 43]. Collectively, these findings show that sumoylation of many DNA metabolism proteins is tightly controlled in a manner correlated with their function. Based on these, we suggest that on-site sumoylation is likely generally applicable for many DNA metabolism proteins.
Possible mechanisms and implications of on-site modification
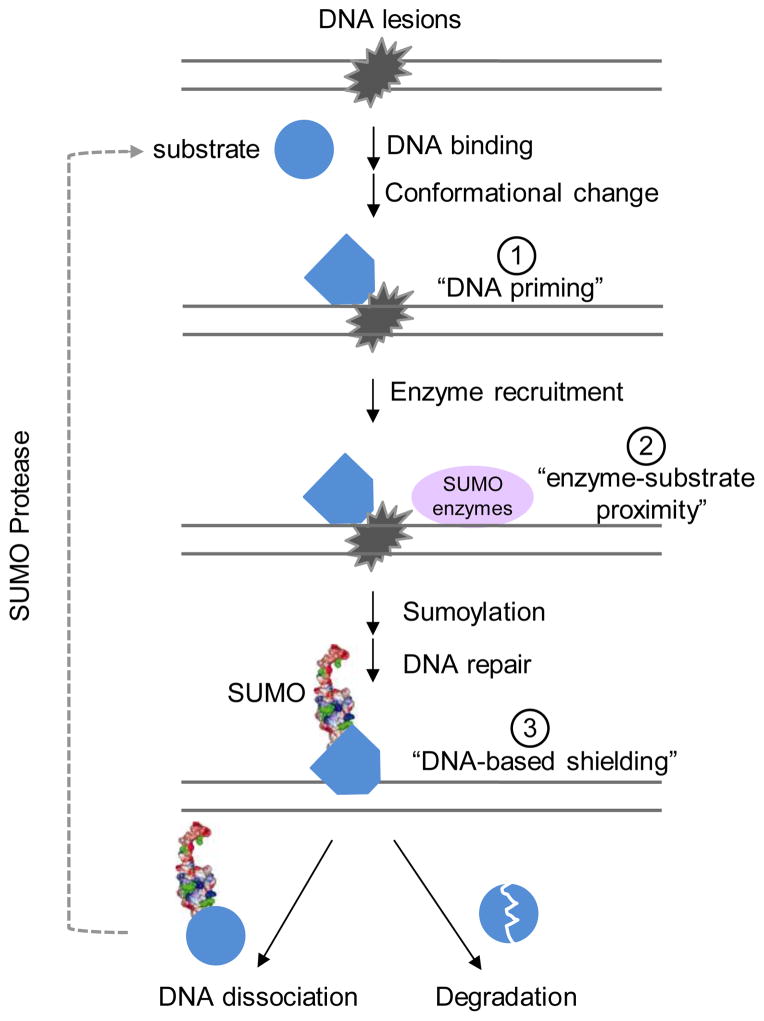
In principle, on-site sumoylation can be achieved by the following non-mutually exclusive possibilities (Figure 2); 1) conformation or property changes upon DNA association make substrates amenable to modification (referred to as “DNA priming”), 2) induced proximity between substrates and SUMO enzymes on DNA favors sumoylation (referred to as “enzyme-substrate proximity”), and 3) DNA association shields substrates from desumoylation (referred to as “DNA-based shielding”). Among these possibilities, the first two have already received substantial experimental support, as detailed below.
Figure 2. Possible mechanisms of on-site sumoylation.
Three models are drawn. Note that they can occur independently or sequentially, and only the latter is depicted for simplicity. Substrates (blue, circle) can undergo conformational changes upon binding to DNA lesions, as reflected by the change in the shape of the blue symbol, which may promote sumoylation via the proposed “DNA priming” model ➀. Alternatively or additionally, DNA lesion binding may bring substrates in close proximity with sumoylation enzymes (pink oval). This mechanism, namely “enzyme-substrate proximity” ➁, can function together with the DNA priming mechanism to promote substrate sumoylation at lesion sites, as indicated by the blue symbol with the miniature of SUMO structure on it. Sumoylation of proteins can lead to various effects, two of which are drawn here, namely DNA dissociation (left) or ubiquitin-dependent degradation (right). In the former case, the dissociated protein can be quickly desumoylated to allow recycling of the protein. By associating with DNA or repair substrates, the protein may be protected from desumoylation in a model called “DNA-based shielding” ➂.
The “DNA priming” model is bolstered by the observation that most of the aforementioned substrates undergo conformational changes upon DNA binding [44–48]. In addition, in vitro sumoylation of Rad52 is stimulated by single stranded DNA (ssDNA), which induces a conformational change in Rad52 [44]. Furthermore, sumoylation of PCNA by the SUMO ligase Siz1 is enhanced by double stranded DNA (dsDNA) even when the Siz1 DNA binding site is mutated [30]. Both these studies suggest a direct role for DNA in priming the substrate for modification. In these cases, we envision that DNA can serve as an allosteric factor for the sumoylation of DNA metabolism proteins through priming the substrates. An important implication of this idea is that it in turn provides an explanation for the conundrum that a few SUMO enzymes can specifically modify large numbers of substrates, as selectivity may be achieved by changes in the substrate, rather than by changes on the enzyme side. Future work will be needed to test this model more rigorously using additional substrates and with higher resolution techniques, including biophysical tools.
The second possibility of “enzyme-substrate proximity” is also supported by several lines of evidence [6, 27, 49, 50]. For example, the PIAS SUMO ligases were shown to colocalize with DNA double strand breaks (DSBs) and break repair substrates in mammalian cells [49, 50], and the Ubc9 SUMO E2 and Siz2 ligase were shown to interact with the DNA break resection protein Mre11 in yeast [6]. It is likely that the two mechanisms, namely “DNA priming” and “enzyme-substrate proximity”, operate simultaneously to achieve maximal on-site sumoylation levels. To our knowledge, no direct evidence has yet been reported for “DNA-based shielding” from desumoylation, but this will be an interesting possibility to test in the future. Additional work will also be needed to clarify the step-wise requirements for substrate sumoylation during on-site sumoylation. We note that DNA-independent control of sumoylation of DNA metabolism proteins does occur, an example of which is the tumor suppressor protein p53 [51]. Further exploration of both on-site and DNA-independent sumoylation for proteins involved in diverse DNA metabolism processes, such as DNA replication and transcription, will provide additional understanding of the control of sumoylation in genome maintenance.
From small to big: small changes collectively lead to large biological effects
The highly specific sumoylation events described above disfavor the idea that low level sumoylation is attributable to random bystander events and thus phenotypically irrelevant. Detailed examination of over a dozen DNA metabolism proteins has revealed that specific biological defects arise from loss of sumoylation in the case of every substrate (Table 1). In some cases, loss of sumoylation of a single substrate generates strong defects, but in most other cases, this causes much milder phenotypes than does loss of global sumoylation. These different effects can be explained by two models. We refer to the first as the “ensemble effect” model, whereby SUMO exerts a strong influence by inducing small changes in multiple proteins, and the second as the “star effect” model in which SUMO exerts a strong influence by eliciting large changes in the function of one or a few key factors (Figure 3). In this section, we discuss the main evidence for both models and their implications.
Table 1.
Effects of sumoylation on DNA repair proteins
| Substratea | Function(s)b | Molecular effect(s) of sumoylation | Modified lysine(s) | E3(s)c | Ref(s) |
|---|---|---|---|---|---|
| DNA association | |||||
| Yku70 | DNA end binding | Enhance DNA association | K588,591,592,596,597 | Mms21, Siz1,2 | [34] |
| Rad1 | DNA endonuclease | Lower DNA binding | K32 | Siz1, 2 | [33] |
|
| |||||
| Protein interactions | |||||
| RPA | ssDNA binding | Promote RAD51 interaction | K449, 577 | N/D | [110] |
| ATRIP | ATR kinase partner | Promote checkpoint protein interaction | K234, 289 | N/D | [72] |
| Srs2 | DNA translocase | Inhibit SUMO-PCNA interaction | K1081, 1089, 1142 | Siz1,2 | [80] |
| PCNA | Polymerase clamp | Favor Srs2 interaction, may disfavor Eco1interaction | K127, 164 | Siz1,2 | [76, 77, 84] |
| Saw1 | Nuclease scaffold | Favor Slx4 interaction, disfavor Rad1 interaction | K221 | Siz1,2 | [83] |
|
| |||||
| Protein level and stability | |||||
| FEN1 | Flap endonuclease | Promote its degradation | K168 | N/D | [109] |
| MDC1 | DNA checkpoint | Promote its degradation | K1840 | PIAS4 | [93] |
| FANCI | Damage recognition | Promote its degradation | K4,638,646,715,1248, 1288 | PIAS1, 4 | [32] |
| XPC | Damage recognition | Stabilize the protein | K655 | N/D | [39] |
|
| |||||
| Protein solubilty | |||||
| Sae2 | DSB end clipping | Increase soluble Sae2 protein levels | K97 | Siz1,2 | [52] |
|
| |||||
| Activity | |||||
| BRCA1 | Ubiquitin E3 | Stimulate its E3 activity in vitro | N/D | PIAS1, 4 | [50] |
|
| |||||
| Localization/recruitment | |||||
| XRCC4 | DNA ligase scaffold | Promote nuclear localization | K210 | PIAS1, xβ | [111] |
| TDP1 | Phosphodiesterase | Promote DNA damage site localization | K111 | N/D | [112] |
| Top2 | Topoisomerase | Promote centromeric localization | K1220, 1246, 1277 | Siz1,2 | [97, 113, 114] |
|
| |||||
| Multiple effects | |||||
| Lif1 | DNA ligase scaffold | Reduce ssDNA binding and self-association | K301 | Siz1,2 | [89] |
| BLM | DNA translocase | Promote RAD51 interaction, PML localization | K317, 331, 334, 347 | N/D | [115, 116] |
| Rad52 | Recombination mediator | Lower ssDNA binding/annealing, promote stability, nucleolar exclusion, Rad51/Ufd1 interaction | K10, 11, 220 | Siz2 | [36, 44, 70, 117] |
| TDG | DNA glycosylase | Decrease DNA binding, promote intermolecular SIM binding | K330 | N/D | [88, 91, 118- 120] |
Yeast protein names are with capitalized first letters followed by small letters, whereas mammalian protein names are all capitalized. The table lists only those substrates whose sumoylation effects have been studied in detail. Substrates are grouped by the effects of sumoylation.
Only the major function is indicated for each substrate.
The SUMO E3s in budding yeast include Siz1, Siz2 and Mms21. The mammalian SUMO E3s shown in the table include PIAS1, 4, and xβ. N/D denotes not determined.
Figure 3. Two models for SUMO-based regulation of DNA metabolism functions.
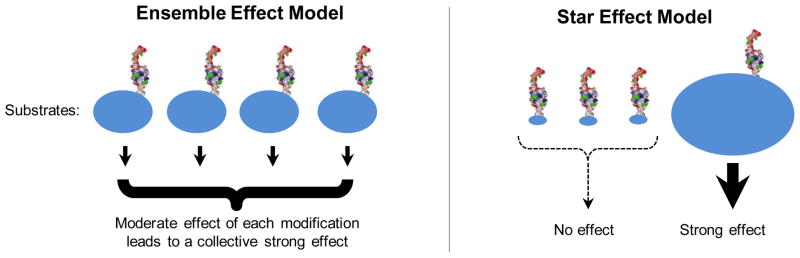
SUMO is depicted as its 3D structure rendering. Blue ovals represent substrates. The magnitude of the effects of sumoylation is reflected by the sizes of ovals and the thickness of the arrow underneath them. The ensemble effect model (left) suggests that sumoylation of each substrate has moderate effects, which collectively lead to strong biological consequences. The star effect model (right) suggests that sumoylation of key substrate(s) has strong biological effects, while that of others does not produce any effect. We note that these models are depicted to reflect extreme scenarios for the ease of discussion; intermediate situations can occur as well (not shown).
Two recent studies in yeast showed that loss of sumoylation of several proteins involved in HR confers a stronger phenotype than eliminating the sumoylation of only one substrate. In the first study, only mutating the sumoylation sites on five HR proteins involved in DNA strand exchange or annealing, namely Rfa1, Rfa2, Rfa3, Rad52 and Rad59, produced detectable defects in recombination and DNA damage resistance, whereas mutating those on a single protein did not [6]. In the second study, limiting the sumoylation of the DNA end resection factors Sae2 (Sporulation in the Absence of Spo Eleven) and the Mre11-Rad50-Xrs2 (MRX) complex resulted in a stronger defect in processing DNA ends at DSBs than compromising that of either [52]. The ensemble effect also appears to occur at telomeres [37, 53–56]. For example, sumoylation of the telomerase regulators, namely Cdc13 and Tpz1 in budding and fission yeast, respectively, restrains telomerase function by facilitating the action of the telomerase inhibitor complex Stn1-Ten1 [37, 53, 54]. However, eliminating Cdc13 sumoylation causes less severe telomere defects than those arising from impaired global sumoylation in a SUMO E2 mutant [37]. As several other telomere proteins are also sumoylated, it is possible that their sumoylation can compensate for the lack of Cdc13 sumoylation [37, 56]. Validation of this notion awaits the identification and simultaneous mutation of sumoylation sites in additional telomere proteins.
The effects of sumoylation of fission yeast Tpz1 are much stronger than that of budding yeast Cdc13 [37, 53, 54], suggesting an organism-based difference and providing an example for the star effect model. Another example for this model is that of the FANCI and FANCD2 proteins whose sumoylation appears to be critical for their function in DNA repair [32]. We note that both models represent extreme case scenarios, and an intermediate model wherein sumoylation of some substrates outweighs that of others in terms of biological significance likely applies in certain situations.
From a fitness perspective, the ensemble effect confers robustness, buffering the whole system against effects engendered by the loss of modification of a few substrates. The star effect hinges upon a single or few modifications, the lack of which would compromise function. Though less robust, this may be useful in situations when SUMO serves as a pivotal molecular switch. As many sumoylated proteins are found in the six main DNA repair pathways (Table 2), and in other DNA metabolism processes, such as DNA replication and chromatin regulation [5, 6, 11, 42, 57], expanding the research on additional substrates in the future will further evaluate the applicability of both models.
Table 2.
DNA repair pathways and examples of sumoylated proteins in each pathway
| DNA repair pathway | DNA lesionsa | Examples of sumoylated proteinsb |
|---|---|---|
| Base excision repair | Base lesions, single strand breaks | Apn1, Mag1, Ogg1, Ntg1 |
| Mismatch repair | Small insertions, deletions or mismatches | Mlh1, Msh3, Msh6, Pms1 |
| Nucleotide excision repair | Bulky helix-distorting lesions | Saw1, Rad7, Rad25, Rad16 |
| Post-replicative repair | Replication gaps, collapsed forks | PCNA, Rad5 |
| Homologous recombination | Double-strand breaks, gaps | Sae2, Rad52, Rad59, Srs2, Sgs1 |
| Non-homologous end joining | Double-strand breaks | Lif1, Yku70, Yku80 |
Examples fitting both models have also been shown for other PTMs in the DNA damage response (DDR). For example, checkpoint-mediated phosphorylation targets multiple DNA repair factors, and this division of labor ensures the ability to mount an efficient and flexible response to a large spectrum of insults, albeit within it, modification of some substrates elicits larger effects than that of others [58–64]. Similarly, multiple ubiquitylation events at DNA repair sites in mammalian cells coordinately promote the DDR such that elimination of single ubiquitylation events results in different extents of defects in DNA damage signaling [65–68]. These findings highlight the general usefulness and biological relevance of both the star effect and ensemble effect models in PTM-mediated regulation.
Several emerging trends in the varied functional effects of SUMO
Studies of sumoylation substrates in different cellular processes have demonstrated that SUMO influences protein function in multiple ways [69]. A wide spectrum of functional effects of SUMO is also seen for DNA metabolism proteins (Table 1 and Figure 4). Examples include effects on protein stability, solubility, localization, activity, and association with proteins and DNA. Below, we highlight some emerging trends on the biological effects of sumoylation in the DDR based on the most recent work.
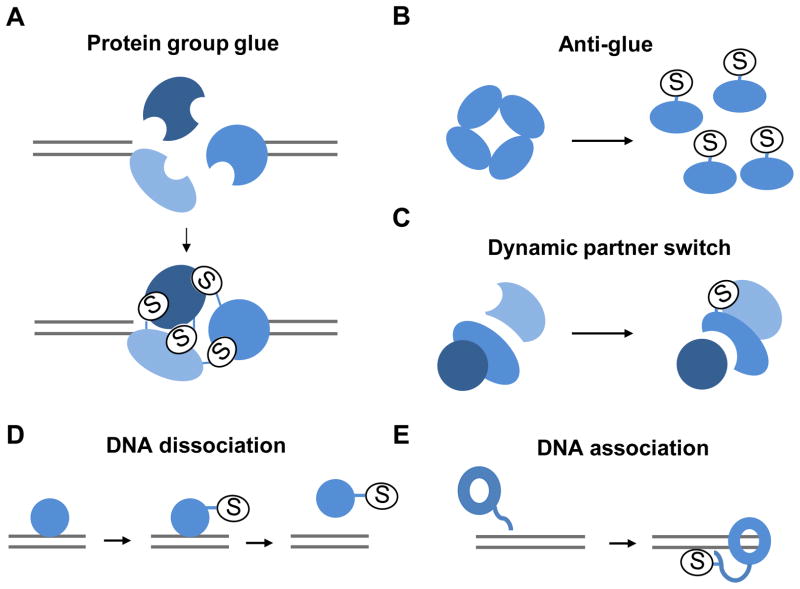
Figure 4. A few examples of the diverse functional effects of SUMO on DNA metabolism proteins.
SUMO-SIM (SUMO-interacting motif) binding promotes group interactions amongst proteins acting in the same pathway by multivalent interaction (protein group glue, A). Sumoylation can also promote dispersal from multimers (anti-glue, B), or a dynamic switch in protein interactions (C). In addition, SUMO can enhance either dissociation from DNA (D) or protein association with DNA (E, adapted from the study on Yku70 [34]). SUMO is depicted as an oval marked S.
One newly observed trend is that SUMO can exert multiple effects on a single substrate. For example, sumoylation of Rad52 reduces its ssDNA binding and annealing activities, and promotes its interaction with the Ufd1 segregase subunit for its removal from DNA [44, 70]. Both effects limit recombination. In contrast, sumoylation of Rad52 was also shown to prevent its degradation and foster its interaction with Rad51, a key recombination factor [36, 70], thus having pro-recombination effects. A challenge for the future will be to determine whether these different regulatory effects occur in a temporal sequence at different stages of recombination, or if they occur in different repair contexts based on cell cycle stage, genomic locus, or recombination subpathway. This information will help us grasp more completely the range of effects that sumoylation can elicit from a single protein.
Several recent studies also highlight the possibility that SUMO acts as a molecular glue to promote multiple interactions within a group of interacting proteins (Figure 4A). This idea was originally suggested for the PML (Pro-Myelocytic Leukemia) body nuclear structure [71], and more recently for the HR, DNA damage checkpoint, and SLX4 proteins [6, 72–74]. In these cases, several sumoylated proteins also possess SUMO-interacting motifs (SIMs), and thus can support multivalent interactions within the group of proteins through multiple SUMO-SIM interactions. In the case of PML, the “SUMO glue” supports the formation of a membrane-free compartment [71, 75]. It will be interesting to see how generally applicable this effect is. While SUMO can underlie group interactions in several DDR processes, it is important to note that the effects of SUMO on protein-protein interactions are diverse and not restricted to the above multivalent glue mode. For example, SUMO-SIM interactions can support simple binary protein interactions [76–79], and SUMO can even disfavor protein interactions in some cases, a so-called “anti-glue” effect [52, 80–82] (Figure 4B). In addition, when the pro- and anti- protein interaction effects of sumoylation occur on the same substrate toward different binding partners, sumoylation may lead to a binding partner switch [83, 84] (Figure 4C). As protein ubiquitylation and phosphorylation also modulate protein-protein interactions by mediating group- and pair-wise interactions during the DDR [85–87], a future question is how the different modifiers foster specific protein interactions in the DDR particularly when the same sets of proteins are subject to multiple kinds of modifications.
While the role of SUMO in mediating protein-protein interactions has been long recognized and well acknowledged, recent work has revealed widespread effects of sumoylation on association with DNA or chromatin. Early work had shown that sumoylation of TDG impairs its DNA binding [88]. Similar effects have recently been reported for several other substrates, such as Rad52, Rad1, the NHEJ factor Lif1, the helicase PICH (Plk1-Interacting Checkpoint Helicase) and p53 [33, 44, 51, 89, 90] (Figure 4D). In all cases, the sumoylated protein exhibits reduced DNA binding in vitro. The mechanism of this effect is best understood for TDG: conjugation of SUMO at the C-terminal domain of TDG causes a conformational change that affects its N-terminal DNA binding domain [91]. In the case of Rad1, SUMO may pose a steric hindrance to DNA binding, as sumoylation occurs in a domain implicated in DNA binding in the mammalian homolog [92]. In addition to these in vitro studies, in vivo chromatin association assays have provided additional examples wherein sumoylation disfavors DNA association directly or indirectly. For example, sumoylation of FANCI, FANCD2 and the DNA damage sensor MDC1 leads to their removal from chromatin by the SUMO-targeted ubiquitin ligase (STUbL) RNF4 [32, 93]. In addition, deficiency in sumoylation correlates with higher chromatin association for TopoII [38]. As some transcription factors also show this trend [94–96], it is likely that SUMO-mediated chromatin dissociation, either via direct alteration of DNA interaction or through stripping enzymes such as segregase or STUbL, is a general regulatory mode shared by several classes of DNA-binding proteins. The biological significance of such effects may be to remove an enzyme after function, as suggested for TDG and Rad1 [33, 88] (Figure 2), or to prevent non-productive DNA binding, as suggested for p53 [51], among other possibilities.
There are fewer reports for a positive role of SUMO in protein association with DNA or chromatin (Figure 4E). An early example is that of the Top2-SUMO fusion promoting its localization at centromeric chromatin [97]. A more recent example is that of the yeast Yku70 protein whose sumoylation at its C-terminal tail favors its DNA association [34] (Figure 4). As the Yku70 tail regulates its DNA binding [46, 47], sumoylation at this region may provide additional DNA binding interfaces through SUMO [98, 99], or impede the Ku ring from sliding inward on DNA [34].
It is noteworthy that the effects of SUMO on substrate DNA association in several cases are moderate [33, 34, 97], suggesting that sumoylation is suitable for fine-tuning this attribute of DNA repair. This could be important for proteins subject to intricate regulation such as those with multiple functions, or for specific situations, such as high lesion burdens as was shown for Rad1, where the effect of sumoylation is detectable only when lesion levels are high [33]. Understanding how SUMO can promote or weaken substrate-DNA interaction will require both mechanistic studies using purified sumoylated proteins, and in vivo approaches such as FRAP (fluorescence recovery after photo bleaching) for insight into mobility and chromatin association times. While these studies and others determining the range of substrates whose DNA binding is affected by SUMO are still awaited, the emerging trend from recent studies is that sumoylation provides an important tool to sculpt the landscape of proteins on chromatin.
The diverse regulatory effects of sumoylation discussed above (Table 1) are shared by other PTMs such as phosphorylation and ubiquitylation [27, 85, 87]. Such regulatory versatility is perfectly apt to control DNA metabolism processes that are both static at cytological levels by occurring within certain nuclear domains, and at the same time also highly dynamic through rapid alterations of protein and DNA interactions. It is possible that multivalent interactions, such as those mediated by multiple SUMO-SIM interactions, are more useful for the static aspects of these processes, while others, such as binary SUMO-SIM interactions and SUMO-dependent changes in DNA association, are effective in supporting the dynamic ones. The seamless integration of these different levels of control ultimately leads to a robust DDR.
Crosstalk with other PTMs in DNA repair and DDR
The DDR elicits waves of multiple PTMs and extensive crosstalk among them. The best-characterized example of this is the response at DSBs, in which ubiquitin ligases and DNA damage checkpoint kinases serve as recruitment and retention signals for modification enzymes and DNA repair factors in higher eukaryotes [29, 85, 100]. More recently, important roles for SUMO in activating both ubiquitin- and checkpoint kinase-mediated signaling pathways were revealed. Initially, two simultaneous reports showed that SUMO ligases are required for the formation of ubiquitin conjugates and for DNA repair [49, 50]. Since then, additional studies have revealed a variety of ways in which SUMO can regulate ubiquitin and checkpoint signaling at DSBs. We highlight a few examples of these effects.
Several recent studies show that SUMO influences the localization of ubiquitin ligases to DNA lesion sites. Sumoylation of three ubiquitin ligases, namely BRCA1 (breast cancer 1, early onset), HERC2 (HECT and RLD domain containing E3 ubiquitin protein ligase 2), and BMI1 (B lymphoma Mo-MLV insertion region 1 homolog), promotes their accrual at damage sites [50, 101, 102]. In the case of BRCA1, hybrid chains formed by ubiquitin and SUMO are bound more strongly by the SIMs and ubiquitin-interacting motifs (UIMs) of the repair scaffold protein RAP80 to facilitate BRCA1 accumulation [103, 104]. In addition, two studies suggest that a new SUMO ligase SLX4 can use its SIMs and UIMs for its targeting to different sites of DNA metabolism [73, 74]. The SIMs of SLX4 are important for targeting to fragile replication sites and DSB zones, while UIMs are required for its localization to interstrand crosslink sites [73, 74, 105, 106]. Thus, the same protein can bind to ubiquitin or SUMO to influence different types of genome stress responses.
Crosstalk between sumoylation and DNA damage checkpoint kinase pathways has also been recently revealed. Sumoylation promotes the full activation of checkpoint phosphorylation in yeast [5], whereas this requirement becomes much greater in mammals, as sumoylation of ATRIP (ATR-interacting protein), the binding partner of the main DNA damage checkpoint kinase ATR (ATM and Rad3-related), is required for checkpoint activation [72]. Sumoylation of ATRIP facilitates the recruitment of ATRIP and downstream proteins to DNA damage sites, likely via SUMO chain-mediated group interactions amongst the ATR pathway proteins [72]. Another paradigm shared between lower and higher eukaryotic cells is that compromised checkpoint signaling leads to increased sumoylation [5, 6, 107]. In human cells, this is followed by a decline in the levels of chromatin-bound sumoylated proteins due to RNF4-mediated degradation [107]. Interestingly removal of RNF4 rescues the replication fork collapse seen in ATR-deficient cells, suggesting an intricate interplay among sumoylation, ubiquitylation and checkpoint [107].
Besides crosstalk at the level of PTM enzymes as exemplified above, combinatorial modifications are seen at the level of substrates. Examples of independent modifications are Sae2 and the NER factor Rpb1 [52, 108]. In the case of Sae2, sumoylation and phosphorylation act concertedly to promote its solubility and function [52]. A very nice example of sequential cascades of modification is seen in the FEN1 nuclease: phosphorylation primes sumoylation, and sumoylation in turn leads to ubiquitylation and proteolysis [109]. These examples of intricate relationships among modification enzymes and modifications on shared substrates highlight the crosstalk between PTMs. It is likely that the combinatorial power of PTMs in the regulation of cellular functions is far greater than our current knowledge, and much more is yet to be uncovered about the additional means by which they support the spatial and temporal regulation of critical functions.
Concluding remarks
Large-scale screens for sumoylated proteins and targeted studies of individual substrates have shed much light on seminal questions regarding the regulation of sumoylation and its roles in the DDR and genome maintenance. Much of the evidence points to a far-reaching model wherein SUMO fine-tunes different functions of numerous proteins in situ to achieve a large biological effect. Key technical hurdles in the field have thus far been the preservation and detection of low abundance sumoylated forms and the unambiguous identification of sumoylation sites. There has been technical progress in these areas, such as the use of modified SUMO to aid purification and mass spectrometry analysis and the development of antibodies recognizing specific forms of SUMO or SUMO remnants [10, 11, 14–17]. Additionally, as SUMO often elicits small functional changes, uncovering the phenotype of a non-sumoylatable mutant is frequently challenging. Combining sumoylation mutants of multiple proteins functioning in the same step or pathway can be informative. Furthermore, better in vitro purification and characterization of sumoylated proteins are required to complement in vivo genetic and cell biological analyses of non-sumoylatable mutants. Lastly, while many insights continue to be gained from simple model systems such as yeast, further analysis of substrates and regulation in higher eukaryotic cells are necessary to flesh out key similarities and differences among species. Applying these approaches will continue to reveal unifying themes in SUMO-based regulation of genome maintenance across organisms in the future.
Highlights.
Sumoylation of many DNA metabolism proteins is tightly controlled by DNA engagement.
SUMO can influence a process through an ensemble effect or via major substrates.
SUMO causes diverse effects, such as bi-directional changes in biomolecule binding.
Crosstalk between SUMO and other modifiers occurs at multiple levels during DDR.
Acknowledgments
We thank Xiao Peng for her helpful suggestions during writing and other Zhao lab members for their comments. Due to space limitations, we apologize to colleagues whose work is not cited here. We acknowledge the support of NIH grant GM080670, American Cancer Society grant RSG-12-013-01-CCG, and a Leukemia and Lymphoma Society Scholar Award.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kerscher O, et al. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu Rev Cell Dev Biol. 2006;22:159–180. doi: 10.1146/annurev.cellbio.22.010605.093503. [DOI] [PubMed] [Google Scholar]
- 2.Geiss-Friedlander R, et al. Concepts in sumoylation: a decade on. Nat Rev Mol Cell Biol. 2007;8:947–956. doi: 10.1038/nrm2293. [DOI] [PubMed] [Google Scholar]
- 3.Hay RT. SUMO: a history of modification. Mol Cell. 2005;18:1–12. doi: 10.1016/j.molcel.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 4.Johnson ES. Protein modification by SUMO. Annu Rev Biochem. 2004;73:355–382. doi: 10.1146/annurev.biochem.73.011303.074118. [DOI] [PubMed] [Google Scholar]
- 5.Cremona CA, et al. Extensive DNA damage-induced sumoylation contributes to replication and repair and acts in addition to the Mec1 checkpoint. Mol Cell. 2012;45:422–432. doi: 10.1016/j.molcel.2011.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Psakhye I, et al. Protein group modification and synergy in the SUMO pathway as exemplified in DNA repair. Cell. 2012;151:807–820. doi: 10.1016/j.cell.2012.10.021. [DOI] [PubMed] [Google Scholar]
- 7.Miller MJ, et al. Proteomic analyses identify a diverse array of nuclear processes affected by small ubiquitin-like modifier conjugation in Arabidopsis. Proc Natl Acad Sci U S A. 2010;107:16512–16517. doi: 10.1073/pnas.1004181107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elrouby N, et al. Proteome-wide screens for small ubiquitin-like modifier (SUMO) substrates identify Arabidopsis proteins implicated in diverse biological processes. Proc Natl Acad Sci U S A. 2010;107:17415–17420. doi: 10.1073/pnas.1005452107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ma L, et al. Identification of small ubiquitin-like modifier substrates with diverse functions using the Xenopus egg extract system. Mol Cell Proteomics. 2014;13:1659–1675. doi: 10.1074/mcp.M113.035626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lamoliatte F, et al. Large-scale analysis of lysine SUMOylation by SUMO remnant immunoaffinity profiling. Nat Commun. 2014;5:5409. doi: 10.1038/ncomms6409. [DOI] [PubMed] [Google Scholar]
- 11.Tammsalu T, et al. Proteome-wide identification of SUMO2 modification sites. Sci Signal. 2014;7:rs2. doi: 10.1126/scisignal.2005146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tatham MH, et al. Comparative proteomic analysis identifies a role for SUMO in protein quality control. Sci Signal. 2011;4:rs4. doi: 10.1126/scisignal.2001484. [DOI] [PubMed] [Google Scholar]
- 13.Cubenas-Potts C, et al. Identification of SUMO-2/3 modified proteins associated with mitotic chromosomes. Proteomics. 2014;15:763–772. doi: 10.1002/pmic.201400400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hendriks IA, et al. Uncovering global SUMOylation signaling networks in a site-specific manner. Nat Struct Mol Biol. 2014;21:927–936. doi: 10.1038/nsmb.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schimmel J, et al. Uncovering SUMOylation dynamics during cell-cycle progression reveals FoxM1 as a key mitotic SUMO target protein. Mol Cell. 2014;53:1053–1066. doi: 10.1016/j.molcel.2014.02.001. [DOI] [PubMed] [Google Scholar]
- 16.Matic I, et al. Site-specific identification of SUMO-2 targets in cells reveals an inverted SUMOylation motif and a hydrophobic cluster SUMOylation motif. Mol Cell. 2010;39:641–652. doi: 10.1016/j.molcel.2010.07.026. [DOI] [PubMed] [Google Scholar]
- 17.Becker J, et al. Detecting endogenous SUMO targets in mammalian cells and tissues. Nat Struct Mol Biol. 2013;20:525–531. doi: 10.1038/nsmb.2526. [DOI] [PubMed] [Google Scholar]
- 18.Dou H, et al. SUMOylation and de-SUMOylation in response to DNA damage. FEBS Lett. 2011;585:2891–2896. doi: 10.1016/j.febslet.2011.04.002. [DOI] [PubMed] [Google Scholar]
- 19.Bergink S, et al. Principles of ubiquitin and SUMO modifications in DNA repair. Nature. 2009;458:461–467. doi: 10.1038/nature07963. [DOI] [PubMed] [Google Scholar]
- 20.Smolka MB, et al. Proteome-wide identification of in vivo targets of DNA damage checkpoint kinases. Proc Natl Acad Sci USA. 2007;104:10364–10369. doi: 10.1073/pnas.0701622104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matsuoka S, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 22.Chen SH, et al. A proteome-wide analysis of kinase-substrate network in the DNA damage response. J Biol Chem. 2010;285:12803–12812. doi: 10.1074/jbc.M110.106989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim W, et al. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol Cell. 2011;44:325–340. doi: 10.1016/j.molcel.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Wijk SJ, et al. Shared and unique properties of ubiquitin and SUMO interaction networks in DNA repair. Genes Dev. 2011;25:1763–1769. doi: 10.1101/gad.17593511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Merbl Y, et al. Profiling of ubiquitin-like modifications reveals features of mitotic control. Cell. 2013;152:1160–1172. doi: 10.1016/j.cell.2013.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu H, et al. A method for systematic mapping of protein lysine methylation identifies functions for HP1beta in DNA damage response. Mol Cell. 2013;50:723–735. doi: 10.1016/j.molcel.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ulrich HD. Two-way communications between ubiquitin-like modifiers and DNA. Nat Struct Mol Biol. 2014;21:317–324. doi: 10.1038/nsmb.2805. [DOI] [PubMed] [Google Scholar]
- 28.Jentsch S, et al. Control of nuclear activities by substrate-selective and protein-group SUMOylation. Annu Rev Genet. 2013;47:167–186. doi: 10.1146/annurev-genet-111212-133453. [DOI] [PubMed] [Google Scholar]
- 29.Jackson SP, et al. Regulation of DNA damage responses by ubiquitin and SUMO. Mol Cell. 2013;49:795–807. doi: 10.1016/j.molcel.2013.01.017. [DOI] [PubMed] [Google Scholar]
- 30.Parker JL, et al. SUMO modification of PCNA is controlled by DNA. EMBO J. 2008;27:2422–2431. doi: 10.1038/emboj.2008.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sinigalia E, et al. The human cytomegalovirus DNA polymerase processivity factor UL44 is modified by SUMO in a DNA-dependent manner. PLoS One. 2012;7:e49630. doi: 10.1371/journal.pone.0049630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gibbs-Seymour I, et al. Ubiquitin-SUMO circuitry controls activated Fanconi Anemia ID complex dosage in response to DNA damage. Mol Cell. 2015;57:150–164. doi: 10.1016/j.molcel.2014.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sarangi P, et al. Sumoylation of the Rad1 nuclease promotes DNA repair and regulates its DNA association. Nucleic Acids Res. 2014;42:6393–6404. doi: 10.1093/nar/gku300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hang LE, et al. Regulation of Ku-DNA association by Yku70 C-terminal tail and SUMO modification. J Biol Chem. 2014;289:10308–10317. doi: 10.1074/jbc.M113.526178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moriyama T, et al. SUMO-modification and elimination of the active DNA demethylation enzyme TDG in cultured human cells. Biochem Biophys Res Commun. 2014;447:419–424. doi: 10.1016/j.bbrc.2014.04.004. [DOI] [PubMed] [Google Scholar]
- 36.Sacher M, et al. Control of Rad52 recombination activity by double-strand break-induced SUMO modification. Nat Cell Biol. 2006;8:1284–1290. doi: 10.1038/ncb1488. [DOI] [PubMed] [Google Scholar]
- 37.Hang LE, et al. SUMOylation regulates telomere length homeostasis by targeting Cdc13. Nat Struct Mol Biol. 2011;18:920–926. doi: 10.1038/nsmb.2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Azuma Y, et al. SUMO-2/3 regulates Topoisomerase II in mitosis. J Cell Biol. 2003;163:477–487. doi: 10.1083/jcb.200304088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang QE, et al. Ubiquitylation-independent sumoylation of Xeroderma pigmentosum group C protein is required for efficient nucleotide excision repair. Nucleic Acids Res. 2007;35:5338–5350. doi: 10.1093/nar/gkm550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Horie K, et al. SUMO-1 conjugation to intact DNA Topoisomerase I amplifies cleavable complex formation induced by camptothecin. Oncogene. 2002;21:7913–7922. doi: 10.1038/sj.onc.1205917. [DOI] [PubMed] [Google Scholar]
- 41.Chen XL, et al. Topoisomerase I-dependent viability loss in Saccharomyces cerevisiae mutants defective in both SUMO conjugation and DNA repair. Genetics. 2007;177:17–30. doi: 10.1534/genetics.107.074708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Silver HR, et al. A role for SUMO in nucleotide excision repair. DNA Repair. 2011;10:1243–1251. doi: 10.1016/j.dnarep.2011.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ohuchi T, et al. Rad52 sumoylation and its involvement in the efficient induction of homologous recombination. DNA Repair. 2008;7:879–889. doi: 10.1016/j.dnarep.2008.02.005. [DOI] [PubMed] [Google Scholar]
- 44.Altmannova V, et al. Rad52 SUMOylation affects the efficiency of the DNA repair. Nucleic Acids Res. 2010;38:4708–4721. doi: 10.1093/nar/gkq195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Newman M, et al. Structure of an XPF endonuclease with and without DNA suggests a model for substrate recognition. EMBO J. 2005;24:895–905. doi: 10.1038/sj.emboj.7600581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Walker JR, et al. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature. 2001;412:607–614. doi: 10.1038/35088000. [DOI] [PubMed] [Google Scholar]
- 47.Rivera-Calzada A, et al. Structural model of full-length human Ku70-Ku80 heterodimer and its recognition of DNA and DNA-PKcs. EMBO Rep. 2007;8:56–62. doi: 10.1038/sj.embor.7400847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Joo W, et al. Structure of the FANCI-FANCD2 complex: insights into the Fanconi anemia DNA repair pathway. Science. 2011;333:312–316. doi: 10.1126/science.1205805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Galanty Y, et al. Mammalian SUMO E3-ligases PIAS1 and PIAS4 promote responses to DNA double-strand breaks. Nature. 2009;462:935–939. doi: 10.1038/nature08657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Morris JR, et al. The SUMO modification pathway is involved in the BRCA1 response to genotoxic stress. Nature. 2009;462:886–890. doi: 10.1038/nature08593. [DOI] [PubMed] [Google Scholar]
- 51.Wu SY, et al. Crosstalk between sumoylation and acetylation regulates p53-dependent chromatin transcription and DNA binding. EMBO J. 2009;28:1246–1259. doi: 10.1038/emboj.2009.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sarangi P, et al. Sumoylation influences DNA break repair partly by increasing the solubility of a conserved end resection protein. PLOS Genet. 2014;11(1):e1004899. doi: 10.1371/journal.pgen.1004899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Garg M, et al. Tpz1TPP1 SUMOylation reveals evolutionary conservation of SUMO-dependent Stn1 telomere association. EMBO Rep. 2014;15:871–877. doi: 10.15252/embr.201438919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Miyagawa K, et al. SUMOylation regulates telomere length by targeting the shelterin subunit Tpz1(Tpp1) to modulate shelterin-Stn1 interaction in fission yeast. Proc Natl Acad Sci U S A. 2014;111:5950–5955. doi: 10.1073/pnas.1401359111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Potts PR, et al. The SMC5/6 complex maintains telomere length in ALT cancer cells through SUMOylation of telomere-binding proteins. Nat Struct Mol Biol. 2007;14:581–590. doi: 10.1038/nsmb1259. [DOI] [PubMed] [Google Scholar]
- 56.Ferreira HC, et al. The PIAS homologue Siz2 regulates perinuclear telomere position and telomerase activity in budding yeast. Nat Cell Biol. 2011;13:867–874. doi: 10.1038/ncb2263. [DOI] [PubMed] [Google Scholar]
- 57.Pelisch F, et al. Dynamic SUMO modification regulates mitotic chromosome assembly and cell cycle progression in Caenorhabditis elegans. Nat Commun. 2014;5:5485. doi: 10.1038/ncomms6485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Binz SK, et al. The phosphorylation domain of the 32-kDa subunit of replication protein A (RPA) modulates RPA-DNA interactions. Evidence for an intersubunit interaction. J Biol Chem. 2003;278:35584–35591. doi: 10.1074/jbc.M305388200. [DOI] [PubMed] [Google Scholar]
- 59.Unal E, et al. DNA damage response pathway uses histone modification to assemble a double-strand break-specific cohesin domain. Mol Cell. 2004;16:991–1002. doi: 10.1016/j.molcel.2004.11.027. [DOI] [PubMed] [Google Scholar]
- 60.Flott S, et al. Slx4 becomes phosphorylated after DNA damage in a Mec1/Tel1-dependent manner and is required for repair of DNA alkylation damage. Biochem J. 2005;391:325–333. doi: 10.1042/BJ20050768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Herzberg K, et al. Phosphorylation of Rad55 on serines 2, 8, and 14 is required for efficient homologous recombination in the recovery of stalled replication forks. Mol Cell Biol. 2006;26:8396–8409. doi: 10.1128/MCB.01317-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen X, et al. Cell cycle regulation of DNA double-strand break end resection by Cdk1-dependent Dna2 phosphorylation. Nat Struct Mol Biol. 2011;18:1015–1019. doi: 10.1038/nsmb.2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Saugar I, et al. Temporal regulation of the Mus81-Mms4 endonuclease ensures cell survival under conditions of DNA damage. Nucleic Acids Res. 2013;41:8943–8958. doi: 10.1093/nar/gkt645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Szakal B, et al. Premature Cdk1/Cdc5/Mus81 pathway activation induces aberrant replication and deleterious crossover. EMBO J. 2013;32:1155–1167. doi: 10.1038/emboj.2013.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lu CS, et al. The RING finger protein RNF8 ubiquitinates Nbs1 to promote DNA double-strand break repair by homologous recombination. J Biol Chem. 2012;287:43984–43994. doi: 10.1074/jbc.M112.421545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tikoo S, et al. Ubiquitin-dependent recruitment of the Bloom syndrome helicase upon replication stress is required to suppress homologous recombination. EMBO J. 2013;32:1778–1792. doi: 10.1038/emboj.2013.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Feng L, et al. The E3 ligase RNF8 regulates KU80 removal and NHEJ repair. Nat Struct Mol Biol. 2012;19:201–206. doi: 10.1038/nsmb.2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Smogorzewska A, et al. Identification of the FANCI protein, a monoubiquitinated FANCD2 paralog required for DNA repair. Cell. 2007;129:289–301. doi: 10.1016/j.cell.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Flotho A, et al. Sumoylation: a regulatory protein modification in health and disease. Annu Rev Biochem. 2013;82:357–385. doi: 10.1146/annurev-biochem-061909-093311. [DOI] [PubMed] [Google Scholar]
- 70.Bergink S, et al. Role of Cdc48/p97 as a SUMO-targeted segregase curbing Rad51-Rad52 interaction. Nat Cell Biol. 2013;15:526–532. doi: 10.1038/ncb2729. [DOI] [PubMed] [Google Scholar]
- 71.Shen TH, et al. The mechanisms of PML-nuclear body formation. Mol Cell. 2006;24:331–339. doi: 10.1016/j.molcel.2006.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wu CS, et al. SUMOylation of ATRIP potentiates DNA damage signaling by boosting multiple protein interactions in the ATR pathway. Genes Dev. 2014;28:1472–1484. doi: 10.1101/gad.238535.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Guervilly JH, et al. The SLX4 Complex Is a SUMO E3 Ligase that Impacts on Replication Stress Outcome and Genome Stability. Mol Cell. 2015;57:123–137. doi: 10.1016/j.molcel.2014.11.014. [DOI] [PubMed] [Google Scholar]
- 74.Ouyang J, et al. Noncovalent Interactions with SUMO and Ubiquitin Orchestrate Distinct Functions of the SLX4 Complex in Genome Maintenance. Mol Cell. 2015;57:108–122. doi: 10.1016/j.molcel.2014.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bernardi R, et al. Structure, dynamics and functions of promyelocytic leukaemia nuclear bodies. Nat Rev Mol Cell Biol. 2007;8:1006–1016. doi: 10.1038/nrm2277. [DOI] [PubMed] [Google Scholar]
- 76.Pfander B, et al. SUMO-modified PCNA recruits Srs2 to prevent recombination during S phase. Nature. 2005;436:428–433. doi: 10.1038/nature03665. [DOI] [PubMed] [Google Scholar]
- 77.Papouli E, et al. Crosstalk between SUMO and ubiquitin on PCNA is mediated by recruitment of the helicase Srs2p. Mol Cell. 2005;19:123–133. doi: 10.1016/j.molcel.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 78.Xie Y, et al. The yeast Hex3.Slx8 heterodimer is a ubiquitin ligase stimulated by substrate sumoylation. J Biol Chem. 2007;282:34176–34184. doi: 10.1074/jbc.M706025200. [DOI] [PubMed] [Google Scholar]
- 79.Song J, et al. Identification of a SUMO-binding motif that recognizes SUMO-modified proteins. Proc Natl Acad Sci U S A. 2004;101:14373–14378. doi: 10.1073/pnas.0403498101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kolesar P, et al. Dual roles of the SUMO-interacting motif in the regulation of Srs2 sumoylation. Nucleic Acids Res. 2012;40:7831–7843. doi: 10.1093/nar/gks484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Krumova P, et al. Sumoylation inhibits alpha-synuclein aggregation and toxicity. J Cell Biol. 2011;194:49–60. doi: 10.1083/jcb.201010117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Janer A, et al. SUMOylation attenuates the aggregation propensity and cellular toxicity of the polyglutamine expanded Ataxin-7. Hum Mol Genet. 2010;19:181–195. doi: 10.1093/hmg/ddp478. [DOI] [PubMed] [Google Scholar]
- 83.Sarangi P, et al. A versatile scaffold contributes to damage survival via sumoylation and nuclease interactions. Cell Rep. 2014;9:143–152. doi: 10.1016/j.celrep.2014.08.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Moldovan GL, et al. PCNA controls establishment of sister chromatid cohesion during S phase. Mol Cell. 2006;23:723–732. doi: 10.1016/j.molcel.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 85.Ciccia A, et al. The DNA damage response: making it safe to play with knives. Mol Cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ulrich HD, et al. Ubiquitin signalling in DNA replication and repair. Nat Rev Mol Cell Biol. 2010;11:479–489. doi: 10.1038/nrm2921. [DOI] [PubMed] [Google Scholar]
- 87.Polo SE, et al. Dynamics of DNA damage response proteins at DNA breaks: a focus on protein modifications. Genes Dev. 2011;25:409–433. doi: 10.1101/gad.2021311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hardeland U, et al. Modification of the human Thymine-DNA Glycosylase by ubiquitin-like proteins facilitates enzymatic turnover. EMBO J. 2002;21:1456–1464. doi: 10.1093/emboj/21.6.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Vigasova D, et al. Lif1 SUMOylation and its role in non-homologous end-joining. Nucleic Acids Res. 2013;41:5341–5353. doi: 10.1093/nar/gkt236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sridharan V, et al. SUMOylation regulates Polo-like Kinase 1-interacting Checkpoint Helicase (PICH) during mitosis. J Biol Chem. 2015;290:3269–3276. doi: 10.1074/jbc.C114.601906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Steinacher R, et al. Functionality of human Thymine DNA Glycosylase requires SUMO-regulated changes in protein conformation. Curr Biol. 2005;15:616–623. doi: 10.1016/j.cub.2005.02.054. [DOI] [PubMed] [Google Scholar]
- 92.Bowles M, et al. Fluorescence-based incision assay for human XPF-ERCC1 activity identifies important elements of DNA junction recognition. Nucleic Acids Res. 2012;40:e101. doi: 10.1093/nar/gks284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Luo K, et al. Sumoylation of MDC1 is important for proper DNA damage response. EMBO J. 2012;31:3008–3019. doi: 10.1038/emboj.2012.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gronholm J, et al. Structure-function analysis indicates that sumoylation modulates DNA-binding activity of STAT1. BMC Biochem. 2012;13:20. doi: 10.1186/1471-2091-13-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sutinen P, et al. Nuclear mobility and activity of FOXA1 with Androgen Receptor are regulated by SUMOylation. Mol Endocrinol. 2014;28:1719–1728. doi: 10.1210/me.2014-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Rosonina E, et al. Sumoylation of transcription factor Gcn4 facilitates its Srb10-mediated clearance from promoters in yeast. Genes Dev. 2012;26:350–355. doi: 10.1101/gad.184689.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Takahashi Y, et al. SIZ1/SIZ2 control of chromosome transmission fidelity is mediated by the sumoylation of Topoisomerase II. Genetics. 2006;172:783–794. doi: 10.1534/genetics.105.047167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bayer P, et al. Structure determination of the small ubiquitin-related modifier SUMO-1. J Mol Biol. 1998;280:275–286. doi: 10.1006/jmbi.1998.1839. [DOI] [PubMed] [Google Scholar]
- 99.Eilebrecht S, et al. SUMO-1 possesses DNA binding activity. BMC Res Notes. 2010;3:146. doi: 10.1186/1756-0500-3-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bekker-Jensen S, et al. The ubiquitin- and SUMO-dependent signaling response to DNA double-strand breaks. FEBS Lett. 2011;585:2914–2919. doi: 10.1016/j.febslet.2011.05.056. [DOI] [PubMed] [Google Scholar]
- 101.Danielsen JR, et al. DNA damage-inducible SUMOylation of HERC2 promotes RNF8 binding via a novel SUMO-binding Zinc finger. J Cell Biol. 2012;197:179–187. doi: 10.1083/jcb.201106152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ismail IH, et al. CBX4-mediated SUMO modification regulates BMI1 recruitment at sites of DNA damage. Nucleic Acids Res. 2012;40:5497–5510. doi: 10.1093/nar/gks222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Guzzo CM, et al. RNF4-dependent hybrid SUMO-ubiquitin chains are signals for RAP80 and thereby mediate the recruitment of BRCA1 to sites of DNA damage. Sci Signal. 2012;5:ra88. doi: 10.1126/scisignal.2003485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hu X, et al. Rap80 protein recruitment to DNA double-strand breaks requires binding to both small ubiquitin-like modifier (SUMO) and ubiquitin conjugates. J Biol Chem. 2012;287:25510–25519. doi: 10.1074/jbc.M112.374116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kim Y, et al. Mutations of the SLX4 gene in Fanconi anemia. Nat Genet. 2011;43:142–146. doi: 10.1038/ng.750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lachaud C, et al. Distinct functional roles for the two SLX4 ubiquitin-binding UBZ domains mutated in Fanconi anemia. J Cell Sci. 2014;127:2811–2817. doi: 10.1242/jcs.146167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ragland RL, et al. RNF4 and PLK1 are required for replication fork collapse in ATR-deficient cells. Genes Dev. 2013;27:2259–2273. doi: 10.1101/gad.223180.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Chen X, et al. Rpb1 sumoylation in response to UV radiation or transcriptional impairment in yeast. PLoS One. 2009;4:e5267. doi: 10.1371/journal.pone.0005267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Guo Z, et al. Sequential posttranslational modifications program FEN1 degradation during cell-cycle progression. Mol Cell. 2012;47:444–456. doi: 10.1016/j.molcel.2012.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Dou H, et al. Regulation of DNA repair through deSUMOylation and SUMOylation of Replication Protein A complex. Mol Cell. 2010;39:333–345. doi: 10.1016/j.molcel.2010.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Yurchenko V, et al. SUMO modification of human XRCC4 regulates its localization and function in DNA double-strand break repair. Mol Cell Biol. 2006;26:1786–1794. doi: 10.1128/MCB.26.5.1786-1794.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hudson JJ, et al. SUMO modification of the neuroprotective protein TDP1 facilitates chromosomal single-strand break repair. Nat Commun. 2012;3:733. doi: 10.1038/ncomms1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Bachant J, et al. The SUMO-1 isopeptidase Smt4 is linked to centromeric cohesion through SUMO-1 modification of DNA Topoisomerase II. Mol Cell. 2002;9:1169–1182. doi: 10.1016/s1097-2765(02)00543-9. [DOI] [PubMed] [Google Scholar]
- 114.Takahashi Y, et al. In vivo modeling of polysumoylation uncovers targeting of Topoisomerase II to the nucleolus via optimal level of SUMO modification. Chromosoma. 2008;117:189–198. doi: 10.1007/s00412-007-0137-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Eladad S, et al. Intra-nuclear trafficking of the BLM helicase to DNA damage-induced foci is regulated by SUMO modification. Hum Mol Genet. 2005;14:1351–1365. doi: 10.1093/hmg/ddi145. [DOI] [PubMed] [Google Scholar]
- 116.Ouyang KJ, et al. SUMO modification regulates BLM and RAD51 interaction at damaged replication forks. PLoS Biol. 2009;7:e1000252. doi: 10.1371/journal.pbio.1000252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Torres-Rosell, et al. The Smc5-Smc6 complex and SUMO modification of Rad52 regulates recombinational repair at the ribosomal gene locus. Nat Cell Biol. 2007;9:923–931. doi: 10.1038/ncb1619. [DOI] [PubMed] [Google Scholar]
- 118.Baba D, et al. Crystal structure of Thymine DNA Glycosylase conjugated to SUMO-1. Nature. 2005;435:979–982. doi: 10.1038/nature03634. [DOI] [PubMed] [Google Scholar]
- 119.Baba D, et al. Crystal structure of SUMO-3-modified Thymine-DNA Glycosylase. J Mol Biol. 2006;359:137–147. doi: 10.1016/j.jmb.2006.03.036. [DOI] [PubMed] [Google Scholar]
- 120.Smet-Nocca C, et al. SUMO-1 regulates the conformational dynamics of Thymine-DNA Glycosylase regulatory domain and competes with its DNA binding activity. BMC Biochem. 2011:12. doi: 10.1186/1471-2091-12-4. [DOI] [PMC free article] [PubMed] [Google Scholar]