Summary
The pursuit of quantitative biological information with imaging requires robust labeling approaches that can be used in multiple applications and with a variety of detectable colors and properties. In addition to conventional fluorescent proteins, chemists and biologists have come together to provide a range of approaches that combine dye chemistry with the convenience of genetic targeting. This hybrid-tagging approach combines the rational design of properties available through synthetic dye chemistry with the robust biological targeting available with genetic encoding. In this review, we discuss the current range of approaches that have been exploited for dye targeting, or targeting and activation, and some of the recent applications that are uniquely enabled by these hybrid-tagging approaches.
Introduction
Fluorescent proteins have revolutionized researchers’ ability to visualize subcellular structures and study biochemical activities with unprecedented spatial and temporal resolution. Since the first use of the green fluorescent protein (GFP) outside its original host Aequorea Victoria in 1994 (Chalfie et al., 1994), considerable efforts have been devoted to improving its spectral and biochemical properties and optimizing its behavior in live cells and organisms. Nowadays GFP and its color variants are routinely used by cell biologists to provide a wealth of information on a wide variety of biological processes. The facts that fluorescent proteins (FPs) do not require any substrates or cofactors and they are genetically encoded make them an ideal tool for specific protein labeling. Despite these advantages, FPs have a few shortcomings (Giepmans et al., 2006; Marks and Nolan, 2006). First, with a molecular weight of 30 kD, FPs can interfere with the folding, structure, localization and function of the tagged protein. Second, as the chromophore of FPs is buried in the β-barrel structure, FPs are less accessible for modifications without loss in fluorescence. Third, FPs are much less bright and photostable than the best organic fluorophores and therefore are not optimal for applications requiring a high photon-output, such as single molecule super-resolution imaging. Fourth, FPs generally show a narrower dynamic range than organic flurophores in response to environmental changes. These drawbacks have motivated researchers to develop alternative chemical labeling strategies.
In chemical labeling, instead of using a FP, a dye-free protein domain or polypeptide sequence is genetically fused to the protein of interest. This protein domain or polypeptide sequence is subsequently modified with an organic fluorophore or other targeting molecule through covalent conjugation or high-affinity binding (Figure 1). Thus, chemical labeling presents the combined advantages of specific labeling through the genetic fusion to the protein of interest and the superior sensitivity and versatility of synthesized and designed organic probes. In addition, the absence of an intrinsic fluorophore allows a single genetic fusion construct to be used for multiple distinct applications, simply by choosing different organic probes to combine with the tagged constructs, whether for in-vitro or in-vivo applications.
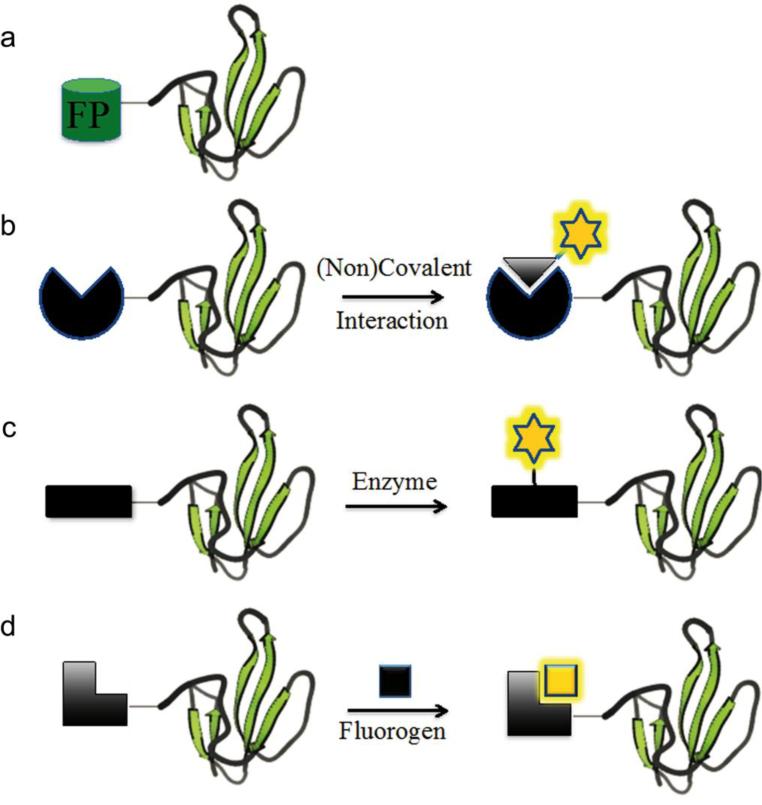
Figure 1. Chemical labeling of proteins of interest.
(a) The protein of interest is fused to a fluorescent protein with intrinsic fluorescence. (b) The protein of interest is fused to a protein domain tag, which can bind to a fluorophore derivative through covalent or non-covalent interactions. (c) The protein of interest is fused to a peptide tag, which then acts as the enzyme substrate for covalent fluorophore attachment. (d) The protein of interest is fused to a protein or peptide tag, which then binds and activates a fluorogen through covalent or non-covalent interactions.
In 2014 the Nobel Prize in Chemistry recognized super-resolution imaging, a breakthrough in optical microscopy that enabled researchers to visualize and investigate biological processes at the individual molecule level inside living cells. Such achievements would be impossible without the innovation of chemical labeling in the past two decades, delivering dyes with high brightness and photostability to targeted sites in living cells and organisms. In this review, we focus on the development of various chemical labeling strategies and technologies and highlight the unique properties and advantages of chemical tags in recent applications (Table 1). With the unforeseeable potential of chemical labeling techniques, we expect it will not be long before more innovative methods will be applied to solve sophisticated biological problems.
Table 1.
Properties and major applications of chemical tags discussed in this review.
| Tag name | M.W.(KDa) | Probe Linkage Mechanism | Application | Restriction |
|---|---|---|---|---|
| SNAP/CLIP | 20 | Covalent Fluorogenic |
Multicolor Intracellular labeling Super-resolution imaging Protein-protein interaction FRET Enviroment sensitive substrate (calcium/membrane) In vivo QDs targeting |
|
| Halo | 33 | Covalent | Multicolor Intracellular labeling in vivo Protein degradation QDs targeting |
|
| TMP-eDHFR | 18 | (Non)covalent Fluorogenic |
Super-resolution imaging LRET CALI Protein degradation Intracellular labeling |
|
| PYP | 14 | Covalent Fluorogenic |
Enviroment sensitive probe (polar/non-polar) Intracellular |
Coumarin fluorogenic probes |
| FKBP12/SLF | 12 | Noncovalent | CALI FRET |
|
| Poly-his Tag | <1 | (Non)covalent | FRET | Cell surface only |
| AP/Biotin Ligase | <2 | Covalent | Orthogonality Protein-protein interaction QDs targeting |
Cell surface only |
| LAP1/Lipoic acid ligase LAP2/Lipoic acid ligase |
<3 <2 |
Covalent Covalent |
Protein-protein interaction Intracellular labeling Two steps for mutiple fluorophore QDs targeting |
one step for coumarin only Slow kinetics, potential toxicity |
| Tetracysteine tag | <1 | Covalent Fluorogenic |
Pulse-Chase Protein-protein interaction FRET CALI EM Protein folding/conformation change |
FlAsH and ReAsH only Reducing enviroment only in vitro only |
| FAP | 26 | Non-covalent Fluorogenic |
Multicolor Intracellular labeling Super-resolution imaging Protein trafficking Signal amplification pH sensing QDs targeting |
Limited orthogonal choices inside cells |
Tag abbreviated as follows: Trimethoprim-e. coli dihydrofolate reductase (TMP-eDHFR), Photoactive Yellow Protein (PYP), Acceptor Peptide (AP), Fluorogen Activating Protein (FAP).
Applications abbreviated as follows: Forster Resonant Energy Transfer (FRET), Quantum Dots (QDs), Luminescence Resonant Energy Transfer (LRET), Chromophore Assisted Light Inactivation (CALI).
Protein domain based chemical tags
In this labeling strategy, a protein domain is fused to the target protein to subsequently incorporate the fluorescent label. One of the most widely used chemical tags in this class is the SNAP-tag developed by Johnsson's group (Keppler et al., 2003). SNAP-tag is based on a DNA repair protein human O6-alkylgaunine-DNA-alkyltransferase (hAGT), a 20 KDa protein that transfers the alkyl group on the O6 position of benzyl-gaunine to a reactive cysteine residue. By employing random mutagenesis followed by phage display and a yeast three hybrid assay, an AGT mutant has been identified that shows a 17 fold and 52 fold faster reactivity than the starting mutant and the wild type protein, respectively (Gronemeyer et al., 2006). In addition, the mutant AGT accepts a large number of derivatized substrates with different fluorescent labels, shows a low affinity towards DNA and is stable under oxidizing environment, such as the cell surface. To date, a variety of SNAP tag fusion proteins have been generated both extracellularly and in the cytoplasm and have been applied in recently developed super-resolution imaging and single particle tracking (Benke et al., 2012). Moreover, an AGT variant that recognizes benzyl-cytosine as the substrate has also been established and is named the CLIP tag (Gautier et al., 2008). SNAP-tag and CLIP-tag act orthogonally and have been used to detect protein-protein interactions by selective cross-linking: the interaction between two partners, one fused to the SNAP tag and the other to the CLIP tag, was visualized by a bifunctional molecule composed of the two tag substrates connected by a fluorophore. This molecule therefore bridges the interacting partners with high selectivity. (Gautier et al., 2009). When linked together, SNAP-tag and CLIP tag can act as a FRET pair and be used to detect changes in the concentration of a metabolite (Brun et al., 2011; Brun et al., 2009). SNAP-tag has also been engineered to sense the local calcium changes when a BODIPY based calcium sensor BOCA is conjugated to the SNAP substrate BG, leading to a 180-fold increase in fluorescence signal in response to calcium binding (Kamiya and Johnsson, 2010). More recently, the SNAP-tag labeling technology has been applied to living organisms, for example, the zebrafish embryo (Campos et al., 2011). After injecting the mRNA encoding SNAP-fusion proteins at an early stage of embryo development, subcellular structures can be visualized by fluorescence microscopy later at different developmental stages. Furthermore, SNAP tag labeled embryos can tolerate acid-based fixations, which was shown to be incompatible with fluorescent proteins. Like any chemical labeling strategies, multiple wash steps are required in SNAP-tagging to remove the unreacted fluorophore to minimize the background signal. To overcome such limitations, researchers were motivated to develop fluorogenic SNAP probes for wash-free applications. In 2011 Sun et al. reported the design of a fluorogenic SNAP-tag where a quencher is linked to the guanine group (Sun et al., 2011). As a result, after reacting with AGT the quencher bound guanine acts as the leaving group while the fluorophore is now covalently linked to the protein tag AGT. Another fluorogenic SNAP tag was generated by taking advantage of a polarity sensitive, solvatochromic dye: Nile Red (Prifti et al., 2014). Nile Red shows a strong increase in fluorescence when exposed to a low polarity environment, such as the plasma membrane (Greenspan and Fowler, 1985). When conjugated to a BG derivative, Nile Red can be targeted to the SNAP-receptor protein on the plasma membrane and become fluorescent (Prifti et al., 2014). These fluorogenic labeling systems are therefore well-suited for tracking dynamic cellular events in real time.
SNAP tag mediated genetic targeting has been recently applied to animals (Yang et al., 2014). The SNAP tag was fused to a membrane associated CaaX sequence and then targeted to the Rosa26 locus in mice. When such RosaSnapcaax line was crossed with lines expressing Cre in different neuronal tissues, specific fluorescence signal was detected on neuronal endings in the skin or cornea from live tissue confocal imaging after the tissues were subjected to BG substrate treatment. Moreover, when used with a photosensitizing substrate BG-fluorescein in chromophore-assisted light inactivation (CALI) experiments, axons with SNAPCaax expression showed complete degeneration and loss of mechanical sensory functions upon illumination. Such light mediated cell ablation is highly specific and selective due to genetic targeting of the SNAP tag in Cre expressing tissues and local administration of the substrates. Therefore, these SNAP tag expressing animals provide an invaluable tool for in vivo functional studies at the tissue level.
Another protein based self-labeling tag is the Halo tag. The Halo tag protein (33 KDa) is based on the bacterial enzyme haloalkane dehalogenase which functions to remove halides from haloalkanes in a two-step process (Los et al., 2008). It was found that mutation at his272 in the wild type dehalogenase leads to a trapped reaction intermediate with the alkyl group covalently linked to an aspartate residue in the enzyme (Pries et al., 1995). A modified haloalkane dehalogenase has been developed, which specifically recognizes chloroalkanes linked to fluorophores, biotin and agarose beads (Los and Wood, 2007). Because haloalkane dehalogenase is of prokaryote origin, the background in mammalian cells is low. The Halo tag technology has been applied to label β1-integrin with the substrate linked to Atto655 in STED (Stimulated emission depletion) microscopy (Schroder et al., 2009), to investigate peroxisome dynamics (Huybrechts et al., 2009), to image tumors in various colors in live animals (Kosaka et al., 2009) and to label proteins in bacteria and mammalian cells for super-resolution imaging (Lee et al., 2010). In addition, Halo tag has proven useful for CALI applications using an eosin ligand and pulse-chase experiments to examine the stability of receptor proteins (He et al., 2011). As a functional extension of this tagging approach, Neklesa et al. designed a small molecule induced degradation system in which a hydrophobic moiety is linked to the Halo tag ligand (Neklesa et al., 2011). As a result, the hydrophobic group is anchored on the surface of the Halo tag fusion protein, leading to its degradation in the proteasome through the cellular quality control mechanism. This hydrophobic tagging induced protein degradation was demonstrated in cultured cells, in zebrafish embryo and can be used to suppress tumor progression by selectively removing tagged oncogenes in mouse models (Neklesa et al., 2011).
In addition to covalent interactions between the ligand and the protein tag, non-covalent interactions have also been exploited for protein labeling, given the high affinity and selectivity of the interaction. Cornish and colleagues took advantage of a well characterized interaction pair: methotrexate and the E. coli dihydrofolate reductase (eDHFR) (Miller et al., 2004). eDHFR is a relatively small protein tag with a molecular weight of 18 KDa. Successful labeling was achieved when a protein of interest was fused with eDHFR and stained with Texas Red-methotrexate. To overcome the potential toxicity and background signal from labeling endogenous DHFR in mammalian cells, trimethoprim (TMP) was later used to replace methotrexate (Miller et al., 2005). Specifically, TMP has a high affinity for eDHFR (1 nM), but a significantly lower affinity (> 1 uM) for the mammalian counterpart. In addition to protein labeling, TMP has been conjugated to the photoswitchable organic fluorophore ATTO655 for dSTORM super-resolution imaging and ~20 nm resolution of the human histone H2B protein was achieved in live cells (Wombacher et al., 2010). Furthermore, the TMP tag has been combined with the SNAP tag to label numerous spliceosomal proteins in yeast cell extract, which allowed the elucidation of the kinetics and order of the spliceosome assembly pathway (Hoskins et al., 2011). Such studies would be impossible to carry out with low photon output, blinking fluorescent proteins. The TMP tag has also been successfully used in other fluorescence related applications. When TMP was linked to a terbium complex, luminescence resonance energy transfer from eDHFR fusion proteins to GFP was detected and can be used to visualize protein-protein interactions (Rajapakse et al., 2010). TMP-fluorescein has proven to be an effective fluorophore for CALI and has been used to examine the function of non-muscle myosin II in maintaining a coherent cytoskeleton network (Cai et al., 2010). Long et al. used the TMP-eDHFR pair to improve the biological compatibility of iron oxide nanoparticles for studying focal adhesions in response to magnetic force (Long et al., 2011). Magnetic nanoparticles were coated with TMP through dopamine anchors and then introduced into cells expressing eDHFR. It was found that cells detach from the plate after being under the magnetic attraction for 4 hours, suggesting that TMP-eDHFR nanoparticles are a useful tool to manipulate cells in a noncontact way. In another application, a general strategy for inhibitor mediated protein degradation was developed and TMP-Boc3Arg (tert-butyl carbamate-proteceted arginine) induced rapid and robust degradation of eDHFR in the cell lysate, probably caused by tagging eDHFR with the hydrophobic Boc3Arg moiety (Long et al., 2012). Recently, a covalent TMP tag was developed by installing a latent acrylamide electrophile on TMP that reacts with a Cys residue just outside the binding pocket in eDHFR through proximity-induced reactivity (Gallagher et al., 2009). The positions of the Cys residue and the acrylamide electrophile were further optimized to give rise to the second generation covalent TMP tag, which showed a much faster labeling half-life (t1/2 = 8 min) and targeted a number of intracellular proteins robustly (Chen et al., 2012). The covalent TMP tag therefore will benefit applications that require long-term or permanent protein labeling.
Jing et al. further extended the eDHFR-TMP system to develop a fluorogenic TMP-tag with significantly improved signal to noise ratio for live cell imaging (Jing and Cornish, 2013). This fluorogenic tag (TMP-BHQ1-Atto520) contains TMP, Atto520 as the fluorophore and BHQ1 as the quencher. In the presence of eDHFR, a proximity induced SN2 reaction takes place between a Cys (L28C) as the nucleophile on eDHFR and a tosylate linker in the tag, leading to the cleavage of the quencher and a 20 fold fluorescence enhancement. When compared with the nonfluorogenic tag TMP-Atto520, TMP-BHQ1-Atto520 showed reduced non-specific background on the cells. With the modular design of the fluorogenic TMP tag, it is anticipated that a wide variety of fluorophores with diverse properties and functions can be readily incorporated.
Hori et al. developed another fluorogenic system utilizing the photoactive yellow protein (PYP) as the protein tag (Hori et al., 2013; Hori et al., 2009). PYP is a small protein (14 kDa) discovered in purple bacteria (Kumauchi et al., 2008). It was found that PYP binds to its natural substrate CoA thioester of 4-hydroxycinnamic acid and derivatives of coumarin through transthioesterification with Cys69 (Kyndt et al., 2002). A rational probe design led to FCTP, a fluorogenic probe composed of 7-hydroxycoumarin- 3-carboxylic acid thioester and fluorescein with ethylene glycol as a flexible linker in between (Hori et al., 2009). In the absence of PYP, FCTP is not fluorescent due to intramolecular quenching of the two dyes. However, upon binding to PYP, the coumarin moiety is covalently attached to PYP and separated from fluorescein, leading to markedly enhanced fluorescence of fluorescein. The next generation PYP probes are based on environment-sensitive dialkylaminocoumarin derivatives (Hori et al., 2013). Such probes are not fluorescent in a polar environment but become intensely fluorescent in a low-polarity environment, for example, when buried in the binding pocket of the PYP-tag. Two of these probes with increased water solubility (TMBDMA and CMBDMA) showed specific and quick labeling of intracellular proteins without washing (Hori et al., 2013). TMBDMA and CMBDMA were also used to image DNA methylation in live cells (Hori et al., 2013).
Nolan and colleagues created another chemical tag by taking advantage of the interaction between the receptor-ligand pair FKBP12(F36V) and SLF’ (Marks et al., 2004a). FKBP12 is a 12 KDa immunophilin. It was hypothesized that when bulky groups, “bumps” are introduced to synthetic ligands of the wild type FKBP12, the binding affinity decreases while compensating “holes” in the wild type FKBP12 will restore the interaction with the bumped synthetic ligands. Based on molecular design and remodeling, Clackson et al. identified a synthetic ligand SLF’ exhibiting a 0.094 nM Kd to the mutant FKBP12(F36V), 1000-fold increased selectivity over the wild type protein (Clackson et al., 1998). A fluorescein SLF’ derivative was used to label FKBP12(F36V) fusion proteins expressed at varied levels and CALI was performed to abolish the enzyme activity of β-galactosidase (Marks et al., 2004a). Robers et al. optimized the labeling condition of the FKBP12(F36V) tag and demonstrated its feasibility as a partner with fluorescent proteins in FRET based assays (Robers et al., 2009).
As an alternative to molecular modeling, Marks et al. set out to screen a phage display library to select for peptides that display a high affinity to the fluorophore Texas red (Marks et al., 2004b). The screening yielded two peptides (a 9 mer and a 13 mer) that bind to Texas red with subnanomolar affinity. Because X-rhod dyes have the Texas red chromophore structure and a BAPTA calcium-sensing moiety, the authors used the identified peptide to create fusion proteins to sense the local calcium change in cultured cells. This approach is another example that provides genetic targeting of small molecules in the cellular milieu.
Metal-chelation based peptide tags
The selective oligo-histidine (His6 and His10) and the Ni(II)-nitrilotriacetic acid (NTA) interaction has become an indispensable tool for recombinant protein purification and surface immobilization. The his-tag was recently extended to labeling proteins on the cell surface (Goldsmith et al., 2006; Guignet et al., 2004) and single molecule FRET in vitro (DeRocco et al., 2010) by using chromophore conjugated Ni-NTA. To improve the binding affinity of Ni-NTA to poly-his, Lata et al. synthesized multivalent chelator headgroups containing two, three and four NTA moieties (Lata et al., 2005). Tris-NTA showed ~20 nM KD, an enhancement in affinity by 3 orders of magnitude compared to mono-NTA. Multivalent interactions of Tris-NTA conjugates and polyhis are highly stable, leading to > 1 hour of complex lifetimes (Lata et al., 2005). However, all of the reported NTA chromophores showed a loss of fluorescence upon complexing with Ni(II), due to its paramagnetic nature. Peneva et al. reported a perylene (dicarboximide) dye connected to NTA, which retained its fluorescence upon binding to Ni(II) (Peneva et al., 2008). The noncovalent interaction between poly-his and Ni-NTA has been rendered covalent recently through a proximity induced nucleophilic reaction: the electrophilic tosyl group in the NTA moiety forms a covalent bond with a his residue in the tag, releasing Ni-NTA (Uchinomiya et al., 2009). To overcome the drawbacks associated with Ni(II), namely fluorescence quenching and cytotoxicity, Tsien and colleagues developed the dye HisZiFiT based on a fluorescein scaffold that utilizes Zn(II) instead of Ni(II) to bind to His6 (Hauser and Tsien, 2007). Zn(II) is diamagnetic and present in biological systems, thus better-suited for in vivo protein labeling than Ni(II). HisZiFiT shows a ~40 nM KD to His6 and was used to address a controversy concerning stromal interaction molecule 1 (Hauser and Tsien, 2007). These metal chelating chemical tags have so far been limited to labeling proteins on the cell surface, due to the impermeability of the probes and potential toxicity of the metal.
Peptide tags mediated by enzymes
Peptide tags are generally less invasive due to their small size than protein tags. However it can be difficult to obtain desired specificity with a peptide tag. For example, in the FlAsH labeling approach extensive wash steps are required to remove the dye from binding to intracellular targets that share a similar sequence as the tetracysteine recognition motif (Hoffmann et al., 2010). To overcome the shortcomings of peptide tags, alternative labeling methodologies were developed utilizing enzymes. The specificity of such labeling is conferred by the peptide sequence as the substrates of the enzyme. The Ting lab established the use of the E. Coli biotin ligase (BirA) as one of the most advanced enzyme mediated chemical tags (Chen et al., 2005). BirA recognizes and ligates biotin to the lysine side chain in a 15 amino-acid acceptor peptide (AP) sequence, which allows the subsequent modification with a functional group or fluorophore. It was shown that ketone1, a biotin analogue can also be used as a BirA substrate (Chen et al., 2005). Ketone 1 is not present on natural proteins and endogenous proteins were not labeled under the same conditions. Although biotinylation of a target protein has been demonstrated in different cellular compartments (Howarth and Ting, 2008), protein labeling with BirA has been restricted to the cell surface for mainly two reasons: endogenous biotin and small molecules containing ketone in the cell would compete with the functionalized substrate; because BirA has a high specificity towards its limited substrate, labeling of the AP needs to be completed in two steps and the second step is slow and inefficient in the cellular context. Chen et al. later identified an orthogonal system to BirA and AP, namely the yeast biotin ligase (yBL) and yeast acceptor peptide (yAP) through screening a phage display library (Chen et al., 2007). Combining the two systems allowed simultaneous labeling of two independent receptor proteins in the same cell with quantum dots. However, yAP has a low catalytic value and the catalytic efficiency of the yAP-yBL pair is about 800 fold lower than the BirA-AP pair. To expand the substrate range of biotin ligase, Slavoff et al. examined the biotinylation reaction with biotin ligases from nine species and eight biotin isosters, using p67, a domain from one of the human biotin acceptor proteins (Slavoff et al., 2008). It was found that biotin ligases from S. cerevisiae and P. horikoshii accept alkyne and azide biotin analogues. These newly identified enzymes and substrates open up new avenues for protein labeling.
Another chemical tagging approach mediated by enzymes utilizes lipoic acid ligase. E.coli lipoic acid ligase (LplA) naturally couples lipoic acid to a lysine side chain in E2p, E2o and H-protein involved in oxidative metabolism (Green et al., 1995). Ting and colleagues reengineered LplA to accept an alkyl azide that can then be selectively functionalized through a strain-promoted [3+2] cycloaddtion (Fernandez-Suarez et al., 2007). The LplA acceptor peptide (LAP1), a 22 amino-acid-sequence, was also developed to replace the original large protein substrates. Although LplA is mechanistically similar to BirA, it was demonstrated that there is adequate bioorthogonality between the two labeling approaches and two receptors can be tagged simultaneously in the same cell (Fernandez-Suarez et al., 2007). LplA has also been employed to ligate a fluorinated aryl azide photo-cross-linker to detect protein-protein interactions (Baruah et al., 2008). The peptide sequence for LplA was further decreased to 13 amino acid through screening a yeast cell surface LAP library (Puthenveetil et al., 2009). A novel peptide sequence LAP2 was identified by FACS and showed improved kinetics over LAP1. Uttamapinant et al. extended LplA labeling from the cell surface to the interior of living cells (Uttamapinant et al., 2010). A blue fluorophore, 7-hydroxycoumarin was chosen for intracellular labeling because of its small size and hydrophobicity. The authors then created several LplA mutants based on the crystal structure of the LplA-lipoic acid complex. Two of the mutants, LplA(W37V) and LplA(W37I) showed robust labeling of LAP2 fusion proteins both from the cell lysate and when targeted to a number of cellular structures. This approach, called PRIME (PRobe Incorporation Mediated by Enzymes) for the first time proved that it is possible to ligate a fluorophore to the substrate in one step by a ligase. Although PRIME is an attractive methodology for specific intracellular labeling based on LplA, it suffers from the fact that coumarin is the only choice as the substrate. Coumarin is not an ideal fluorophore for live cell microscopy due to its spectral properties. To incorporate a diverse set of fluorophores, Yao et al. exploited 10-azidodecanoic acid as the substrate for LplA(W37I) and the functional group handle to react with fluorescent dyes ranging from fluorescein to ATTO 647N in a two-step scheme (Yao et al., 2012). This labeling approach is limited by the kinetics of the second step, the strain-promoted cycloaddition reaction and may not be compatible with the fast intracellular dynamics. Copper-catalyzed azide-alkyne cycloaddition (CuAAC) is at least ten times faster than strain-promoted cycloaddition (Uttamapinant et al., 2012). However, CuAAC has been limited to in vitro applications because of the toxicity of copper(I) (Wolbers et al., 2006). To address the toxicity issue of copper(I) and maintain the kinetics of CuAAC, Uttamapinant et al. explored the possibility of using a copper chelating azide (Uttamapinant et al., 2012). When picolyl azide 8 was coupled to the PRIME labeling, fast and site-specific protein labeling and RNA labeling was achieved. The newest-generation copper(I) ligand, BTTAA further accelerated the reaction (Soriano Del Amo et al., 2010). Thus, chelation assisted CuAAC enables biocompatible lableling with low concentrations of copper(I), which opens the door to labeling highly sensitive cells and tissues, such as neuron cultures. PRIME labeing has also been extended to resorufin, a small, bright, red phenoxazine dye (Liu et al., 2014). Computational design was employed to search for resorufin ligases using the original LplA sequence as a template. A triple LplA mutant (E20A/F147A/H149G) emerged from in silico analysis and proved to carry out covalent attachment of resorufin to LAP specifically and efficiently. In addition to conventional protein labeling and fluorescence imaging in living cells, this resorufin ligase was used to perform STED based super-resolution imaging and electron microscopy (Liu et al., 2014).
Phosphopantetheine transferases (PPTases) from E.coli (AcpS) and B.subtilis (Sfp) have also been used to selectively label cell surface proteins. The natural function of PPTase is to covalently attach a phosphopantetheine (Ppant) from coenzymeA (CoA) to a serine residue on acyl-and peptidyl-carrier proteins (ACP and PCP). Carrier proteins play an important role in fatty acid synthesis, nonribosomal peptide synthesis, polyketide synthesis and lysine biosynthesis (Ehmann et al., 1999). Ample biochemical and structural evidence suggests that the β-mercaptoethylamine group of CoA is not located in the binding pocket of PPTase and is not responsible for the transfer of Ppant to carrier proteins (Gehring et al., 1997; Parris et al., 2000; Reuter et al., 1999). Therefore, a number of functionalities, including haptens, fluorophores, affinity labels and quantum dots have been introduced to the substrate CoA through a maleimide thiol reaction (George et al., 2004; Johnsson et al., 2005). AcpS and Sfp have been employed for multi-color labeling to selectively tag two different proteins (Vivero-Pol et al., 2005). ACP and PCP domains are 80-100 residues in size. An 11-residue DSLEFIASKLA sequence (ybbR) was found to be sufficient to be modified by Sfp, which dramatically reduced the tag size (Yin et al., 2005). Zhou et al. performed phage display screening and identified two 12 residue clones S6 and A1 that can be selectively recognized by Sfp and AcpS, respectively (Zhou et al., 2007). S6 and A1 are orthogonal to each other, permitting sequential labeling of two proteins with little cross-reactivity. The A1 peptide has been used to label the membrane pool of Smoothened which enabled the study of its trafficking pattern in response to hedgehog (Wang et al., 2009). Furthermore, based on NMR studies of the A1 peptide, an 8-residue peptide was generated that undergoes efficient phosphopantethein conjugation with AcpS (Zhou et al., 2008).
Other enzyme mediated chemical tags include the transglutaminase tag (Lin and Ting, 2006), the sortase tag (Popp et al., 2007) and the formylglycine-generating enzyme (FGE) tag (Wu et al., 2009). These labeling approaches have so far been only applicable to cell surface proteins and in vitro tagging.
Fluorogen based chemical tags
The fact that these fluorescent dye-labeled chemical tags require washing to remove unbound dye led to the development of fluorogenic labeling agents, those that convert from a weakly fluorescent form to a highly fluorescent form upon binding to the genetically encoded protein or peptide domain. The very first fluorogenic chemical tag is based on the tetracysteine sequence (CCXXCC), the TC tag, reported by Tsien and colleagues in 1998 (Griffin et al., 1998). The TC tag is recognized by a membrane permeable, bis-arsenical ligand FlAsH (fluorescein arsenical hairpin binder) and its red-shifted analogue ReAsH (resorufin arsenical hairpin binder). FlAsH and ReAsH are quenched when complexed with the ligand ethanedithiol but exhibit strong fluorescence and high affinity upon binding to the TC tag (Luedtke et al., 2007). The original design of the tetracysteine sequence features the four cysteines placed on one side of the α-helix assuming that cysteines at i, i+1, i+4 and i+5 positions will accommodate bis-arsenical binding (Griffin et al., 1998). However it was later found that substituting the two central amino acids to Pro-Gly led to an increase in quantum yield, association rate and affinity (apparent KD = 4pM) (Adams et al., 2002). This result was surprising because Pro-Gly is found in β-turns, rather than α-helices, suggesting that the preferred conformation for the tetracysteine motif is a hairpin turn, not an α-helix. Notably reversing Gly and Pro resulted in forming a complex with decreased brightness and stability (Adams et al., 2002). Because FlAsH and ReAsH bind to intracellular proteins with cysteine pairs nonspecifically, it is necessary to destain the sample with dithiols such as EDT or dimercaptopropanol (BAL), which limits the use of TC tag in animals (Hoffmann et al., 2010). In order to decrease the nonspecific background and increase the detection sensitivity for proteins expressed at low levels, a retroviral library containing different sequences flanking the tetracysteine motif was constructed (Martin et al., 2005). The library was then expressed in mammalian cells and a high concentration of dithiol was applied to screen for clones exhibiting improved dithiol resistance by FACS. Two sequences FLNCCPGCCMEP and HRWCCPGCCKTF were identified that showed higher signal to noise ratio when directly fused to β-actin under stringent dithiol wash conditions (Martin et al., 2005). NMR studies focusing on FLNCCPGCCMEP and ReAsH revealed that the central CPGC backbone forms a structure similar to a β-turn type II and one arsenic binds to i and i+1 thiols and another to i+4 and i+5 thiols (Madani et al., 2009). The structural requirements for arsenic display and fluorescence have mainly limited the tetracysteine ligands to FlAsH and ReAsH. For example, several rhodamine analogs of FlAsH were found to be non-fluorescent, even in the presence of tetracysteine peptide (Adams et al., 2002). Bhunia et al. designed a general platform that separates the bis-arsenical targeting moiety from the fluorescent label, permitting the synthesis of a wide variety of SplAsH dyes (Spiralactam Arsenical Hairpin binder) (Bhunia and Miller, 2007). Unlike FlAsH and ReAsH, free SplAsH dyes are still fluorescent. Ying et al. later adopted this strategy and synthesized SplAsH-biotin for affinity purification of tetracysteine tagged proteins (Ying and Branchaud, 2011).
Because the thiol group on cysteine is required to react with FlAsH and ReAsH, TC labeling has been mainly used for intracellular proteins in a reducing environment. To label proteins in the ER, Golgi and on the cell surface, strong reducing reagents such as phosphines need to be added to cells acutely (Hoffmann et al., 2010). The TC tag has been widely used for protein labeling, including examples where protein function is less disrupted by the small TC tag than fluorescent proteins (Andresen et al., 2004; Das et al., 2009; Dyachok et al., 2006; Enninga et al., 2005). In addition, the TC tag has been used to address a number of cell biology questions. For example, FlAsH and ReAsH were used in pulse chase experiments to differentiate young and old connexin43 and its assembly pattern in gap junctions (Gaietta et al., 2002). FlAsH and ReAsH have also been paired with fluorescent proteins, for example, CFP and GFP, respectively to study conformation changes of individual proteins and protein-protein interactions (Bhattacharya et al., 2012; Hoffmann et al., 2005; Inobe et al., 2011; Roberti et al., 2007). FlAsH and ReAsH have also been paired with each other for FRET experiments by exploiting the different binding affinities between two TC motifs to simultaneously label two proteins in the cell (Zurn et al., 2010). FlAsH and ReAsH have been used as efficient photosensitizers to inactivate TC tagged proteins in CALI experiments (Kasprowicz et al., 2008; Marek and Davis, 2002; Tour et al., 2003). Moreover, ReAsH shows a high output of singlet oxygen and has been successfully used to photoconvert diaminobenzidine tetrahydrochloride (DAB) in correlated light and electron microscopy (Gaietta et al., 2002; Gaietta et al., 2006), (Lichtenstein et al., 2009). In a recent study, two FlAsH molecules were linked together to create xCrAsH (a dimeric biarsenical derivative of carboxyfluorescein) for crossing linking protein pairs and monitoring protein-protein interactions (Rutkowska et al., 2011).
The fact that any mutations at the two central positions of the TC sequence cause a decrease in binding affinity and brightness suggests that it may be possible to replace PG with a large amino acid sequence between the two cysteine pairs such that the conformation of the cysteine pairs in 3D can recapitulate the required orientation and geometry in the linear TC motif for fluorescent labeling. This hypothesis led to the development of bipartite tetracysteine display by Schepartz and colleagues (Luedtke et al., 2007). The two C-C pairs can be split and put in separate regions of one protein to report conformational change or on two different proteins to report protein-protein interactions. In a proof of principle study by Luedtke et al. it was shown that model proteins containing the bipartite tetracysteine motif form stable complexes with FlAsH and ReAsH and exhibit strong fluorescence, only when these proteins are folded properly (Luedtke et al., 2007). Proteins with single point mutations result in a misfolded state or assembly and lose both affinity and brightness. To test the feasibility of bipartite tetracysteine display in non-alpha helical structures, C-C motifs were introduced into loops of p53 and C-X-C motifs into adjacent beta-strands of EmGFP (Goodman et al., 2009). Limited binding with ReAsH was observed with EmGFP variants while p53 variants favored ReAsH binding significantly, suggesting that flexible regions, such as loops that are close in 3D may be good binding sites for bipartite tetracysteine display. The cysteine split display strategy has been employed in a number of applications, for example, to report on the folding state of retinoic-acid binding protein (CRABP) (Krishnan and Gierasch, 2008) to sense the Src kinase phosphorylation (Ray-Saha and Schepartz, 2010), to detect amyloid-β aggregation and fibril formation in Alzheimer's disease (Lee et al., 2011), to quantify protein dimerization on the plasma membrane (Pace et al., 2011) and to study the conformational change of EGFR in response to different ligands (Scheck et al., 2012). Similar to the linear TC tagging, high resolution electron microscopy can be applied to bipartite tetracysteine display on interacting proteins (Dexter and Schepartz, 2010)
In analogy to the arsenic-tetracysteine interaction, another short peptide tag was developed utilizing the bis-boronic acid rhodamine dye (RhoBo) and the tetraserine sequence SSPGSS (Halo et al., 2009). Although originally designed as a monosaccharide sensor (Kim et al., 2003), RhoBo has an affinity to the tetraserine sequence at least 105 higher than a number of monosaccharides and shows limited fluorescence on the cell surface, a saccharide-rich environment (Halo et al., 2009). Since RhoBo does not use the toxic element arsenic and is inert to redox conditions, it is an attractive fluorogenic probe for protein labeling. However, because there are > 100 human proteins containing the SSPGSS motif, further work is required to identify optimized tetraserine motifs for highly specific, targeted intracellular labeling.
Szent-Gyorgyi et al. recently developed a new dye-protein system based on the fluorogen concept (Szent-Gyorgyi et al., 2008). Fluorogen Activating Proteins (FAPs) (~ 25 KDa) are based on specific molecular recognition and activation between a single chain antibody fragment and small, otherwise non-fluorescent dye molecules such as thiazole orange (TO) and malachite green (MG) (Szent-Gyorgyi et al., 2008). TO and MG are known fluorogens that interact with DNA (Nygren et al., 1998) or RNA aptamers (Babendure et al., 2003). When free in solution, these fluorogens have extremely low quantum yields, due to free rotation of aromatic moieties within the chromophore leading to excited state de-activation. Upon binding to cognate FAPs, the fluorogens are constrained in the binding pocket of FAP, reducing these excited state rotations and stabilizing the fluorescent conformation, resulting in thousands-fold increases in fluorescence emission.
The FAPs were initially isolated by screening a yeast cell surface display library containing ~109 synthetically recombined scFv clones by FACS for fluorescence resulting from protein-dye interactions. Clones displaying high affinity and high quantum yield against cognate dyes were enriched through multiple rounds of sorting. Several FAP-dye pairs were characterized; with some showing single digit nanomolar affinity from both purified and yeast cell surface-displayed FAP. Remarkably, no cross-reactivity was observed between TO-1 and MG-2p FAPs, thus permitting multi-color labeling and imaging. Two classes of MG based fluorogens were synthesized: cell surface receptors tagged with FAP are readily detected with an impermeable dye MG-2p or MG-11p while proteins within the secretory compartments in the cell can be stained with a membrane permeable dye MG-ester. Recently, another MG-based, cell impermeable fluorogen MG-B-Tau was designed to improve cell exclusion and nonspecific binding and showed fast binding kinetics on the cell surface (kon =5 × 105 M−1 s−1) (Yan et al., 2014a). TO and MG fluorogens are well suited for flow cytometry and microscopy assays with commonly used laser lines: 488 nm or 514 nm excitation for TO and 633 nm for MG. To expand the fluorescence emission range with FAPs, Ozhalici-Unal et al. took advantage of the yeast cell surface display library and isolated FAP clones for Dimethylindole Red (DIR), another unsymmetrical cyanine dye (Ozhalici-Unal et al., 2008). One of the DIR clones, K7 showed low binding affinity (14 nM) and relatively high quantum yield (0.33). Surprisingly, in addition to binding to the bulky fluorogen DIR, K7 can also accommodate a few smaller TO-1 related fluorogens, suggesting considerable plasticity of the binding site. This promiscuity of K7 allows for fluorescence readout from 450 nm to 750nm using one single FAP, spanning most of the visible and near-IR spectrum. Further efforts were made to select FAPs that can activate an oxazole thiazole blue derived blue-fluorescing cyanine dye (Zanotti et al., 2011). Moreover, DIR has been modified with an electron-withdrawing cyano group and showed significantly improved photostability while retaining a high binding affinity for the FAP (Shank et al., 2009). Saunders et al. took advantage of the fact that MG fluorogens emit around 670 nm where there is little cellular autofluorescence and developed a bifunctional fusion protein composed of a fluorescein binding and quenching scFv and a high-affinity MG-binding FAP (Saunders et al., 2014). When bound to fluorescein-conjugated antibodies, this reagent effectively quenches fluorescein and converts the conjugate to a far-red excitation/emission probe, effectively replacing the fluorescein with a high brightness, high stability MG-FAP complex. Because fluorescein conjugated antibodies are among the most commonly used, this reagent offers a straightforward and convenient method to reduce cellular background and improve the signal to noise ratio for both live and fixed-cell labeling.
The non-covalent interaction between the fluorogen and FAP is similar to a ligand and its receptor. Therefore, the fraction of the FAP-fluorogen complex can be tuned by adjusting the fluorogen concentration. Fractional occupancy increases with the increased fluorogen concentration and can be modeled as a hyperbolic binding function. Low fractional occupancy or low density labeling can then be readily achieved with controllable low concentrations of the free fluorogen. Based on this principle, FAP based localization super-resolution microscopy was developed for both fixed and live cell imaging (Yan et al., 2014b). This system is advantageous over traditional approaches (STORM (Rust et al., 2006) and PALM (Betzig et al., 2006)) because it does not require special imaging buffers or photoactivation or photoswitching using a second laser line. Similarly, Schwartz et al. demonstrated the use of the FAP-fluorogen system for single particle tracking experiments. FAP was fused to the γ subunit of the high affinity IgE receptor (FcεRI) (Schwartz et al., 2014). This labeling scheme, for the first time, enabled the tracking of FcεRI independent of a separately labeled IgE molecule and provided new insights into FcεRI signaling and dynamics, both free and when bound to an activating (cytokinergic) IgE molecule. This work demonstrates that the FAP-based tunable labeling system is a viable tool for studying the biology of many receptor proteins at the single molecule level.
The cell-impermeable fluorogens have enabled selective labeling of receptor proteins, including the β2 adrenergic receptor (β2-AR), the insulin-regulated glucose transporter (GLUT4), and the cystic fibrosis transmembrane conductance regulator (CFTR) and quantitative analysis of β2-AR endocytosis by flow cytometry and microscopy (Fisher et al., 2010; Holleran et al., 2010). On the other hand, intracellular protein labeling with FAP is less straightforward. This is because antibodies are naturally secreted into the extracellular space or membrane bound and do not fold efficiently in the cytoplasm (Farinas and Verkman, 1999). Nonetheless, an engineered FAP clone H6 that does not contain any disulfide bond has been successfully expressed in the cytoplasm by making an N-terminal fusion to β-actin and used in STED nanoscopy (Fitzpatrick et al., 2009). Recently, crystallography studies revealed that two L5*VL domains form homodimers when complexed with MG (Szent-Gyorgyi et al., 2013). This led to the construction of dL5**, a covalent tandem dimer of L5 with point mutations (E52D L91S) to confer tighter binding and increased quantum yield (Szent-Gyorgyi et al., 2013). dL5** has been successfully used to target proteins both on the plasma membrane and inside the cells (Saurabh et al., 2014; Wang et al., 2014; Yan et al., 2014a).
The FAP-fluorogen complex forms a modular probe in two ways. First, the same FAP clone can be paired with different fluorogens or fluorogen derivatives for different biological applications, such as pH measurement during protein trafficking (Grover et al., 2012) and fluorescence detection of low abundance proteins on the cell surface (Szent-Gyorgyi et al., 2010). Second, the FAP-fluorogen complex is generally transferrable, from one protein of interest to another, when fused to protein affinity reagents, such as affibodies. Wang et al. expressed FAP fused to an EGFR Affibody ZEGFR:1907 as one single recombinant protein (Wang et al., 2014). In this case, the affibody serves as an affinity probe with high specificity for EGFR while FAP provides the signal readout when bound to various fluorogens. A new MG-based signal amplifying dendrimer Hexa-Cy3-MG was used to label the endogenous EGFR on A431 cells, leading to a significantly improved detection sensitivity (Wang et al., 2014). By altering the order of adding MG-B-Tau (Yan et al., 2014a), an improved cell impermeable dye and EGF, subpopulations of EGFR undergoing endocytosis can be tracked and quantified (Wang et al., 2014).
The highly specific interaction between FAP and fluorogen suggests that fluorogens can serve as an affinity handle to incorporate other functional groups to the FAP-fluorogen complex and provide targeted labeling and fluorescence readout. This “localization” concept has been experimentally demonstrated before, for example, the calcium sensor Indo-1, Fura -2 or BOCA-1 was linked to O6-benzylguanine (BG), the substrate for the SNAP tag to report the local calcium changes in the cell (Bannwarth et al., 2009; Kamiya and Johnsson, 2010; Ruggiu et al., 2010). In this case, BG acts as a structural moiety to bring the fluorophore/functional group to the tagged protein and it is not fluorescent. In addition to targeting functional groups to cells, such “hybrid” systems have been used to target quantum dots to cells. Quantum dots (QD) are semiconductor nanocrystals (Bruchez et al., 1998). Due to their superior brightness and photostability, QDs are well suited for single molecule, long term tracking experiments. However, QDs are generally targeted to proteins via antibodies, which is limited by the availability and low affinity of many antibodies. Saurabh et al. developed a FAP/MG-biotin based approach to target streptavidin-QD conjugates to both membrane and intracellular proteins through genetical fusion with FAP (Saurabh et al., 2014). Previously, QDs have been targeted to proteins uing E.coli biotin ligase (BirA) (Howarth et al., 2005) and lipoic acid ligase (LplA) (Liu et al., 2012). However, for both approaches, the enzymes need to be provided exogenously, either by expression and purification and added to the cells, or by coexpression in the ER (BirA-ER) to biotinylate AP in the secretory compartments. In addition, LplA mediated QD targeting is achieved using a two step protocol to sequentially conjugate 10-bromodecanoic acid and then HaloTag derivatized QD, which is not commercially available.
On the other hand, in the scenario of fluorogen-based biosensors, when an environmentally sensitive probe is conjugated to the fluorogen, a FRET pair is formed once the fluorogen binds to FAP and becomes activated. This rational design has led to the development of fluorogen based tandem dyes as a platform for a wide variety of biosensors.
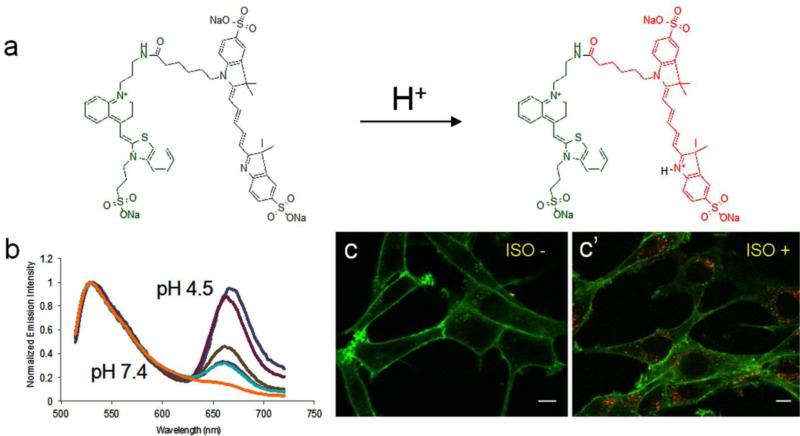
One example of such tandem dye is TO1-Cypher5. Cypher5 is a water-soluble pH sensitive cyanine dye. With a pKa of 6.1, Cypher 5 is only fluorescent under acidic conditions. Traditionally, Cypher5 has been conjugated to antibodies to study receptor internalization and the pH change in the endocytic compartments (Adie et al., 2002; Nordberg et al., 2007; Wenzel et al., 2012). However, antibody conjugation is not practical and readily available. To selectively label receptors present on the cell surface, Grover et al. utilized TO1-Cypher5 to label FAP-tagged β2 adrenergic receptor (β2-AR) (Grover et al., 2012). Because of the spectral overlap between TO-1 and Cypher5, intramolecular energy transfer takes place from the donor TO1 to the acceptor Cypher5. From neutral pH to acidic pH, the extinction coefficient of Cypher5 increases 8 fold, leading to a significant change in the ratio of Cypher5 emission over TO1 emission. This change therefore correlates with a well-defined ratiometric signature against the pH. As the receptor internalizes from the plasma membrane (pH7.4) to endosomes (pH5-6), pH changes are easily detected with TO1-Cypher5 (Figure 2).
Figure 2.
Structure and spectral properties of TO1-CypHer5 (Grover et al., 2012). (a) Structure of TO1-CypHer5. TO1 is linked to pH dependent CypHer5, forming a FRET pair. The pH sensitivity is conferred by the protonation of the free indole nitrogen atom. (b) The emission spectra of TO1-CypHer5 when bound to FAP at various pH, normalized to the TO1 emission peak. Excitation wavelength: 488 nm. (c – c’) Color change of TO1-CypHer5 during FAP tagged β2-AR endocytosis. Upon addition of the agonist, isoproterenol (10 μM) to the cells, FAP-β2-AR undergoes endocytosis, leading to a significant increase of red emission from the acidic vesicles while the plasma membrane (pH 7.4) still shows exclusively green emission. Scalebar 10 μm.
Another application of the tandem dye is the development of fluorogenic dyedrons (Szent-Gyorgyi et al., 2010). Dyedrons are comprised of multiple Cy3 donors linked to a single malachite green (MG) acceptor. When free in aqueous solution, although there is efficient energy transfer from Cy3 to MG, the energy is dissipated nonradiatively due to free rotation of the MG chromophore, which acts as a quencher. Upon binding to a cognate FAP, MG is held rigidly in the binding site of FAP, leading to fluorescence emission, with enhanced excitation at the donor wavelength proportional to the number of donor molecules attached. The properties of dyedrons were characterized with L5, a FAP clone affinity matured to show increased binding affinity and quantum yield against MG. Tight binding was again observed with dyedrons and the highest Kd is 15 nM, suggesting that addition of multiple Cy3 molecules does not abolish the binding between MG and FAP, although it is significantly reduced relative to free MG binding (<1 nM). The extinction coefficient of dyedrons increases in proportion to the number of Cy3 molecules, with the highest measured at 530, 000 M−1 cm−1, about 10 times higher than a typical fluorescent protein (Newman et al., 2011). Like MG, free dyedrons in solution are essentially dark (quantum yield: <0.0005). The intrinsic high brightness of dyedrons coupled with the extremely low background makes them an ideal tool to detect low copy number proteins with a high signal to noise ratio. Currently dyedrons are limited to tagging cell surface proteins due to their negative charges.
Similarly, MG has been linked to tetramethylrhodamine (TMR) to give TMR-MG (Yushchenko et al., 2012). Like TO1-Cypher5 and dyedrons, TMR-MG was designed for intramolecular energy transfer. Surprisingly, instead of being quenched, one of the isomers TMR-para-MG showed pronounced fluorescence at TMR emission in the absence of FAP. It was discovered that this property of TMR-para-MG is caused by the pH dependence of MG. At pH 7.4, 65% of MG-NH2 is in the dye form while the rest is in the colorless carbinol form. As a result, TMR can only be quenched to ~70% by the dye form of MG through FRET and the unquenched TMR (30%) still remains fluorescent. Similar to other cationic dyes, TMR-para-MG localizes in mitochondria (pH 8.0) after crossing the cell membrane and can be easily detected with TMR emission. This unique color-switching behavior of TMR-para-MG provides a distinct advantage over other tandem dyes: TMR-para-MG labels FAP-tagged structures with MG emission and cells not expressing FAP can be visualized by mitochondrial staining in the TMR channel. Moreover, it was demonstrated that TMR-para-MG can be applied to two-photon imaging because of the high two-photon absorption cross section of TMR.
Clearly, the FAP-fluorogen system possesses a few distinct advantages for a range of biological applications. As a genetically encoded tag, FAP allows for exclusive cell surface labeling with an impermeable fluorogen. FAP labeling has also been extended to the interior of the cell by employing a disulfide-bond free FAP and a cell-permeable fluorogen. These labeling schemes provide unique spatial control by chemically separating the cell surface target from the intracellular pool. In addition, because fluorescence signal is turned on only after the addition of the fluorogen, temporal control is readily achieved and this system is well suited for pulse-chase and order of addition experiments, which can be designed to detect various components of the cellular trafficking pool. Free fluorogens exhibit an extremely low background in a number of physiological buffers. Therefore no washing steps are needed before detection. In this two component system, both the FAP and the fluorogen can be tuned to obtain desired properties. For example, fluorogens can be designed to display improved spectral properties, brightness and photostability. On the other hand, through random mutagenesis and directed evolution, FAPs can be selected to accommodate a large repertoire of fluorogens. With further effort, we expect that the FAP-fluorogen system will be useful for addressing exciting unexplored biological questions.
Conclusion
Since the development of the first chemical tag by Tsien and colleagues in 1998, chemical tagging has become an invaluable tool for cell biologists. Although the chemical tagging approaches described here have already provided a breakthrough to study the activity and dynamics of proteins in their native environment, each of the methods still has some shortcomings and the choice of the labeling technique should match the biological question to be answered. Ideally, the chemical tag and labeling approach should meet the following criteria. The tag size should be small enough such that it does not perturb the structure and function of the protein of interest. The fluorophore conjugates and the byproduct from the labeling reaction should not exhibit any cytotoxicity and be biocompatible. The fluorophore conjugates should be inert to the oxidative/reducing environment and be able to function both on the cell surface and in the cytoplasm. The labeling reaction should be fast, highly specific and orthogonal to the mammalian system. Lastly, the fluorophores should have a high photon output and generate minimum background to ensure a high signal to noise ratio. With continuous effort to developing novel chemical labeling strategies, we foresee that chemical tagging will be widely adopted to study sophisticated biological questions and greatly advance our understanding of human disease and normal cell function.
References
- Adams SR, Campbell RE, Gross LA, Martin BR, Walkup GK, Yao Y, Llopis J, Tsien RY. New biarsenical ligands and tetracysteine motifs for protein labeling in vitro and in vivo: synthesis and biological applications. Journal of the American Chemical Society. 2002;124:6063–6076. doi: 10.1021/ja017687n. [DOI] [PubMed] [Google Scholar]
- Adie EJ, Kalinka S, Smith L, Francis MJ, Marenghi A, Cooper ME, Briggs M, Michael NP, Milligan G, Game S. A pH-sensitive fluor, CypHer 5, used to monitor agonist-induced G protein-coupled receptor internalization in live cells. Biotechniques. 2002;33:1152–1154. 1156–1157. doi: 10.2144/02335dd10. [DOI] [PubMed] [Google Scholar]
- Andresen M, Schmitz-Salue R, Jakobs S. Short tetracysteine tags to beta-tubulin demonstrate the significance of small labels for live cell imaging. Mol Biol Cell. 2004;15:5616–5622. doi: 10.1091/mbc.E04-06-0454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babendure JR, Adams SR, Tsien RY. Aptamers switch on fluorescence of triphenylmethane dyes. Journal of the American Chemical Society. 2003;125:14716–14717. doi: 10.1021/ja037994o. [DOI] [PubMed] [Google Scholar]
- Bannwarth M, Correa IR, Sztretye M, Pouvreau S, Fellay C, Aebischer A, Royer L, Rois E, Johnsson K. Indo-1 derivatives for local calcium sensing. ACS chemical biology. 2009;4:179–190. doi: 10.1021/cb800258g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baruah H, Puthenveetil S, Choi YA, Shah S, Ting AY. An engineered aryl azide ligase for site-specific mapping of protein-protein interactions through photo-cross-linking. Angewandte Chemie. 2008;47:7018–7021. doi: 10.1002/anie.200802088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benke A, Olivier N, Gunzenhauser J, Manley S. Multicolor single molecule tracking of stochastically active synthetic dyes. Nano Lett. 2012;12:2619–2624. doi: 10.1021/nl301018r. [DOI] [PubMed] [Google Scholar]
- Betzig E, Patterson GH, Sougrat R, Lindwasser OW, Olenych S, Bonifacino JS, Davidson MW, Lippincott-Schwartz J, Hess HF. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313:1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- Bhattacharya D, Ansari IH, Mehle A, Striker R. Fluorescence Resonance Energy Transfer-Based Intracellular Assay for the Conformation of Hepatitis C Virus Drug Target NS5A. J Virol. 2012;86:8277–8286. doi: 10.1128/JVI.00645-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhunia AK, Miller SC. Labeling tetracysteine-tagged proteins with a SplAsH of color: a modular approach to bis-arsenical fluorophores. Chembiochem. 2007;8:1642–1645. doi: 10.1002/cbic.200700192. [DOI] [PubMed] [Google Scholar]
- Bruchez M, Jr., Moronne M, Gin P, Weiss S, Alivisatos AP. Semiconductor nanocrystals as fluorescent biological labels. Science. 1998;281:2013–2016. doi: 10.1126/science.281.5385.2013. [DOI] [PubMed] [Google Scholar]
- Brun MA, Griss R, Reymond L, Tan KT, Piguet J, Peters RJ, Vogel H, Johnsson K. Semisynthesis of fluorescent metabolite sensors on cell surfaces. Journal of the American Chemical Society. 2011;133:16235–16242. doi: 10.1021/ja206915m. [DOI] [PubMed] [Google Scholar]
- Brun MA, Tan KT, Nakata E, Hinner MJ, Johnsson K. Semisynthetic fluorescent sensor proteins based on self-labeling protein tags. Journal of the American Chemical Society. 2009;131:5873–5884. doi: 10.1021/ja900149e. [DOI] [PubMed] [Google Scholar]
- Cai Y, Rossier O, Gauthier NC, Biais N, Fardin MA, Zhang X, Miller LW, Ladoux B, Cornish VW, Sheetz MP. Cytoskeletal coherence requires myosin-IIA contractility. J Cell Sci. 2010;123:413–423. doi: 10.1242/jcs.058297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos C, Kamiya M, Banala S, Johnsson K, Gonzalez-Gaitan M. Labelling cell structures and tracking cell lineage in zebrafish using SNAP-tag. Dev Dyn. 2011;240:820–827. doi: 10.1002/dvdy.22574. [DOI] [PubMed] [Google Scholar]
- Chalfie M, Tu Y, Euskirchen G, Ward WW, Prasher DC. Green fluorescent protein as a marker for gene expression. Science. 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- Chen I, Choi YA, Ting AY. Phage display evolution of a peptide substrate for yeast biotin ligase and application to two-color quantum dot labeling of cell surface proteins. Journal of the American Chemical Society. 2007;129:6619–6625. doi: 10.1021/ja071013g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen I, Howarth M, Lin W, Ting AY. Site-specific labeling of cell surface proteins with biophysical probes using biotin ligase. Nature methods. 2005;2:99–104. doi: 10.1038/nmeth735. [DOI] [PubMed] [Google Scholar]
- Chen Z, Jing C, Gallagher SS, Sheetz MP, Cornish VW. Second-Generation Covalent TMP-Tag for Live Cell Imaging. Journal of the American Chemical Society. 2012 doi: 10.1021/ja303374p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clackson T, Yang W, Rozamus LW, Hatada M, Amara JF, Rollins CT, Stevenson LF, Magari SR, Wood SA, Courage NL, Lu X, Cerasoli F, Jr., Gilman M, Holt DA. Redesigning an FKBP-ligand interface to generate chemical dimerizers with novel specificity. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:10437–10442. doi: 10.1073/pnas.95.18.10437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das SC, Panda D, Nayak D, Pattnaik AK. Biarsenical labeling of vesicular stomatitis virus encoding tetracysteine-tagged m protein allows dynamic imaging of m protein and virus uncoating in infected cells. J Virol. 2009;83:2611–2622. doi: 10.1128/JVI.01668-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeRocco V, Anderson T, Piehler J, Erie DA, Weninger K. Four-color single-molecule fluorescence with noncovalent dye labeling to monitor dynamic multimolecular complexes. Biotechniques. 2010;49:807–816. doi: 10.2144/000113551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dexter RJ, Schepartz A. Direct visualization of protein association in living cells with complex-edited electron microscopy. Angewandte Chemie. 2010;49:7952–7954. doi: 10.1002/anie.201003217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyachok O, Isakov Y, Sagetorp J, Tengholm A. Oscillations of cyclic AMP in hormone-stimulated insulin-secreting beta-cells. Nature. 2006;439:349–352. doi: 10.1038/nature04410. [DOI] [PubMed] [Google Scholar]
- Ehmann DE, Gehring AM, Walsh CT. Lysine biosynthesis in Saccharomyces cerevisiae: mechanism of alpha-aminoadipate reductase (Lys2) involves posttranslational phosphopantetheinylation by Lys5. Biochemistry. 1999;38:6171–6177. doi: 10.1021/bi9829940. [DOI] [PubMed] [Google Scholar]
- Enninga J, Mounier J, Sansonetti P, Tran Van Nhieu G. Secretion of type III effectors into host cells in real time. Nature methods. 2005;2:959–965. doi: 10.1038/nmeth804. [DOI] [PubMed] [Google Scholar]
- Farinas J, Verkman AS. Receptor-mediated targeting of fluorescent probes in living cells. J Biol Chem. 1999;274:7603–7606. doi: 10.1074/jbc.274.12.7603. [DOI] [PubMed] [Google Scholar]
- Fernandez-Suarez M, Baruah H, Martinez-Hernandez L, Xie KT, Baskin JM, Bertozzi CR, Ting AY. Redirecting lipoic acid ligase for cell surface protein labeling with small-molecule probes. Nature biotechnology. 2007;25:1483–1487. doi: 10.1038/nbt1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher GW, Adler SA, Fuhrman MH, Waggoner AS, Bruchez MP, Jarvik JW. Detection and quantification of beta2AR internalization in living cells using FAP-based biosensor technology. J Biomol Screen. 2010;15:703–709. doi: 10.1177/1087057110370892. [DOI] [PubMed] [Google Scholar]
- Fitzpatrick JA, Yan Q, Sieber JJ, Dyba M, Schwarz U, Szent-Gyorgyi C, Woolford CA, Berget PB, Waggoner AS, Bruchez MP. STED nanoscopy in living cells using Fluorogen Activating Proteins. Bioconjugate chemistry. 2009;20:1843–1847. doi: 10.1021/bc900249e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaietta G, Deerinck TJ, Adams SR, Bouwer J, Tour O, Laird DW, Sosinsky GE, Tsien RY, Ellisman MH. Multicolor and electron microscopic imaging of connexin trafficking. Science. 2002;296:503–507. doi: 10.1126/science.1068793. [DOI] [PubMed] [Google Scholar]
- Gaietta GM, Giepmans BN, Deerinck TJ, Smith WB, Ngan L, Llopis J, Adams SR, Tsien RY, Ellisman MH. Golgi twins in late mitosis revealed by genetically encoded tags for live cell imaging and correlated electron microscopy. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:17777–17782. doi: 10.1073/pnas.0608509103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher SS, Sable JE, Sheetz MP, Cornish VW. An in vivo covalent TMP-tag based on proximity-induced reactivity. ACS chemical biology. 2009;4:547–556. doi: 10.1021/cb900062k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier A, Juillerat A, Heinis C, Correa IR, Jr., Kindermann M, Beaufils F, Johnsson K. An engineered protein tag for multiprotein labeling in living cells. Chem Biol. 2008;15:128–136. doi: 10.1016/j.chembiol.2008.01.007. [DOI] [PubMed] [Google Scholar]
- Gautier A, Nakata E, Lukinavicius G, Tan KT, Johnsson K. Selective cross-linking of interacting proteins using self-labeling tags. Journal of the American Chemical Society. 2009;131:17954–17962. doi: 10.1021/ja907818q. [DOI] [PubMed] [Google Scholar]
- Gehring AM, Lambalot RH, Vogel KW, Drueckhammer DG, Walsh CT. Ability of Streptomyces spp. acyl carrier proteins and coenzyme A analogs to serve as substrates in vitro for E. coli holo-ACP synthase. Chem Biol. 1997;4:17–24. doi: 10.1016/s1074-5521(97)90233-7. [DOI] [PubMed] [Google Scholar]
- George N, Pick H, Vogel H, Johnsson N, Johnsson K. Specific labeling of cell surface proteins with chemically diverse compounds. Journal of the American Chemical Society. 2004;126:8896–8897. doi: 10.1021/ja048396s. [DOI] [PubMed] [Google Scholar]
- Giepmans BN, Adams SR, Ellisman MH, Tsien RY. The fluorescent toolbox for assessing protein location and function. Science. 2006;312:217–224. doi: 10.1126/science.1124618. [DOI] [PubMed] [Google Scholar]
- Goldsmith CR, Jaworski J, Sheng M, Lippard SJ. Selective labeling of extracellular proteins containing polyhistidine sequences by a fluorescein-nitrilotriacetic acid conjugate. Journal of the American Chemical Society. 2006;128:418–419. doi: 10.1021/ja0559754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman JL, Fried DB, Schepartz A. Bipartite tetracysteine display requires site flexibility for ReAsH coordination. Chembiochem. 2009;10:1644–1647. doi: 10.1002/cbic.200900207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green DE, Morris TW, Green J, Cronan JE, Jr., Guest JR. Purification and properties of the lipoate protein ligase of Escherichia coli. Biochem J. 1995;309(Pt 3):853–862. doi: 10.1042/bj3090853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenspan P, Fowler SD. Spectrofluorometric studies of the lipid probe, nile red. Journal of lipid research. 1985;26:781–789. [PubMed] [Google Scholar]
- Griffin BA, Adams SR, Tsien RY. Specific covalent labeling of recombinant protein molecules inside live cells. Science. 1998;281:269–272. doi: 10.1126/science.281.5374.269. [DOI] [PubMed] [Google Scholar]
- Gronemeyer T, Chidley C, Juillerat A, Heinis C, Johnsson K. Directed evolution of O6-alkylguanine-DNA alkyltransferase for applications in protein labeling. Protein Eng Des Sel. 2006;19:309–316. doi: 10.1093/protein/gzl014. [DOI] [PubMed] [Google Scholar]
- Grover A, Schmidt BF, Salter RD, Watkins SC, Waggoner AS, Bruchez MP. Genetically encoded pH sensor for tracking surface proteins through endocytosis. Angewandte Chemie. 2012;51:4838–4842. doi: 10.1002/anie.201108107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guignet EG, Hovius R, Vogel H. Reversible site-selective labeling of membrane proteins in live cells. Nature biotechnology. 2004;22:440–444. doi: 10.1038/nbt954. [DOI] [PubMed] [Google Scholar]
- Halo TL, Appelbaum J, Hobert EM, Balkin DM, Schepartz A. Selective recognition of protein tetraserine motifs with a cell-permeable, pro-fluorescent bisboronic acid. Journal of the American Chemical Society. 2009;131:438–439. doi: 10.1021/ja807872s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser CT, Tsien RY. A hexahistidine-Zn2+-dye label reveals STIM1 surface exposure. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:3693–3697. doi: 10.1073/pnas.0611713104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Xu Y, Zhang C, Gao X, Dykema KJ, Martin KR, Ke J, Hudson EA, Khoo SK, Resau JH, Alberts AS, MacKeigan JP, Furge KA, Xu HE. Identification of a lysosomal pathway that modulates glucocorticoid signaling and the inflammatory response. Sci Signal. 2011;4:ra44. doi: 10.1126/scisignal.2001450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann C, Gaietta G, Bunemann M, Adams SR, Oberdorff-Maass S, Behr B, Vilardaga JP, Tsien RY, Ellisman MH, Lohse MJ. A FlAsH-based FRET approach to determine G protein-coupled receptor activation in living cells. Nature methods. 2005;2:171–176. doi: 10.1038/nmeth742. [DOI] [PubMed] [Google Scholar]
- Hoffmann C, Gaietta G, Zurn A, Adams SR, Terrillon S, Ellisman MH, Tsien RY, Lohse MJ. Fluorescent labeling of tetracysteine-tagged proteins in intact cells. Nat Protoc. 2010;5:1666–1677. doi: 10.1038/nprot.2010.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holleran J, Brown D, Fuhrman MH, Adler SA, Fisher GW, Jarvik JW. Fluorogen-activating proteins as biosensors of cell-surface proteins in living cells. Cytometry A. 2010;77:776–782. doi: 10.1002/cyto.a.20925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori Y, Norinobu T, Sato M, Arita K, Shirakawa M, Kikuchi K. Development of fluorogenic probes for quick no-wash live-cell imaging of intracellular proteins. Journal of the American Chemical Society. 2013;135:12360–12365. doi: 10.1021/ja405745v. [DOI] [PubMed] [Google Scholar]
- Hori Y, Ueno H, Mizukami S, Kikuchi K. Photoactive yellow protein-based protein labeling system with turn-on fluorescence intensity. Journal of the American Chemical Society. 2009;131:16610–16611. doi: 10.1021/ja904800k. [DOI] [PubMed] [Google Scholar]
- Hoskins AA, Friedman LJ, Gallagher SS, Crawford DJ, Anderson EG, Wombacher R, Ramirez N, Cornish VW, Gelles J, Moore MJ. Ordered and dynamic assembly of single spliceosomes. Science. 2011;331:1289–1295. doi: 10.1126/science.1198830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howarth M, Takao K, Hayashi Y, Ting AY. Targeting quantum dots to surface proteins in living cells with biotin ligase. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:7583–7588. doi: 10.1073/pnas.0503125102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howarth M, Ting AY. Imaging proteins in live mammalian cells with biotin ligase and monovalent streptavidin. Nat Protoc. 2008;3:534–545. doi: 10.1038/nprot.2008.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huybrechts SJ, Van Veldhoven PP, Brees C, Mannaerts GP, Los GV, Fransen M. Peroxisome dynamics in cultured mammalian cells. Traffic. 2009;10:1722–1733. doi: 10.1111/j.1600-0854.2009.00970.x. [DOI] [PubMed] [Google Scholar]
- Inobe T, Fishbain S, Prakash S, Matouschek A. Defining the geometry of the two-component proteasome degron. Nat Chem Biol. 2011;7:161–167. doi: 10.1038/nchembio.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing C, Cornish VW. A fluorogenic TMP-tag for high signal-to-background intracellular live cell imaging. ACS chemical biology. 2013;8:1704–1712. doi: 10.1021/cb300657r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnsson N, George N, Johnsson K. Protein chemistry on the surface of living cells. Chembiochem. 2005;6:47–52. doi: 10.1002/cbic.200400290. [DOI] [PubMed] [Google Scholar]
- Kamiya M, Johnsson K. Localizable and highly sensitive calcium indicator based on a BODIPY fluorophore. Analytical chemistry. 2010;82:6472–6479. doi: 10.1021/ac100741t. [DOI] [PubMed] [Google Scholar]
- Kasprowicz J, Kuenen S, Miskiewicz K, Habets RL, Smitz L, Verstreken P. Inactivation of clathrin heavy chain inhibits synaptic recycling but allows bulk membrane uptake. J Cell Biol. 2008;182:1007–1016. doi: 10.1083/jcb.200804162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keppler A, Gendreizig S, Gronemeyer T, Pick H, Vogel H, Johnsson K. A general method for the covalent labeling of fusion proteins with small molecules in vivo. Nature biotechnology. 2003;21:86–89. doi: 10.1038/nbt765. [DOI] [PubMed] [Google Scholar]
- Kim KK, Escobedo JO, St Luce NN, Rusin O, Wong D, Strongin RM. Postcolumn HPLC detection of mono- and oligosaccharides with a chemosensor. Org Lett. 2003;5:5007–5010. doi: 10.1021/ol035978q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosaka N, Ogawa M, Choyke PL, Karassina N, Corona C, McDougall M, Lynch DT, Hoyt CC, Levenson RM, Los GV, Kobayashi H. In vivo stable tumor-specific painting in various colors using dehalogenase-based protein-tag fluorescent ligands. Bioconjugate chemistry. 2009;20:1367–1374. doi: 10.1021/bc9001344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan B, Gierasch LM. Cross-strand split tetra-Cys motifs as structure sensors in a beta-sheet protein. Chem Biol. 2008;15:1104–1115. doi: 10.1016/j.chembiol.2008.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumauchi M, Hara MT, Stalcup P, Xie A, Hoff WD. Identification of six new photoactive yellow proteins--diversity and structure-function relationships in a bacterial blue light photoreceptor. Photochemistry and photobiology. 2008;84:956–969. doi: 10.1111/j.1751-1097.2008.00335.x. [DOI] [PubMed] [Google Scholar]
- Kyndt JA, Meyer TE, Cusanovich MA, Van Beeumen JJ. Characterization of a bacterial tyrosine ammonia lyase, a biosynthetic enzyme for the photoactive yellow protein. FEBS letters. 2002;512:240–244. doi: 10.1016/s0014-5793(02)02272-x. [DOI] [PubMed] [Google Scholar]
- Lata S, Reichel A, Brock R, Tampe R, Piehler J. High-affinity adaptors for switchable recognition of histidine-tagged proteins. Journal of the American Chemical Society. 2005;127:10205–10215. doi: 10.1021/ja050690c. [DOI] [PubMed] [Google Scholar]
- Lee HL, Lord SJ, Iwanaga S, Zhan K, Xie H, Williams JC, Wang H, Bowman GR, Goley ED, Shapiro L, Twieg RJ, Rao J, Moerner WE. Superresolution imaging of targeted proteins in fixed and living cells using photoactivatable organic fluorophores. Journal of the American Chemical Society. 2010;132:15099–15101. doi: 10.1021/ja1044192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Culyba EK, Powers ET, Kelly JW. Amyloid-beta forms fibrils by nucleated conformational conversion of oligomers. Nat Chem Biol. 2011;7:602–609. doi: 10.1038/nchembio.624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenstein A, Gaietta GM, Deerinck TJ, Crum J, Sosinsky GE, Beyer EC, Berthoud VM. The cytoplasmic accumulations of the cataract-associated mutant, Connexin50P88S, are long-lived and form in the endoplasmic reticulum. Exp Eye Res. 2009;88:600–609. doi: 10.1016/j.exer.2008.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CW, Ting AY. Transglutaminase-catalyzed site-specific conjugation of small-molecule probes to proteins in vitro and on the surface of living cells. Journal of the American Chemical Society. 2006;128:4542–4543. doi: 10.1021/ja0604111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu DS, Nivon LG, Richter F, Goldman PJ, Deerinck TJ, Yao JZ, Richardson D, Phipps WS, Ye AZ, Ellisman MH, Drennan CL, Baker D, Ting AY. Computational design of a red fluorophore ligase for site-specific protein labeling in living cells. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:E4551–4559. doi: 10.1073/pnas.1404736111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu DS, Phipps WS, Loh KH, Howarth M, Ting AY. Quantum dot targeting with lipoic acid ligase and HaloTag for single-molecule imaging on living cells. ACS nano. 2012;6:11080–11087. doi: 10.1021/nn304793z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long MJ, Gollapalli DR, Hedstrom L. Inhibitor mediated protein degradation. Chem Biol. 2012;19:629–637. doi: 10.1016/j.chembiol.2012.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long MJ, Pan Y, Lin HC, Hedstrom L, Xu B. Cell compatible trimethoprim-decorated iron oxide nanoparticles bind dihydrofolate reductase for magnetically modulating focal adhesion of mammalian cells. Journal of the American Chemical Society. 2011;133:10006–10009. doi: 10.1021/ja202767g. [DOI] [PubMed] [Google Scholar]
- Los GV, Encell LP, McDougall MG, Hartzell DD, Karassina N, Zimprich C, Wood MG, Learish R, Ohana RF, Urh M, Simpson D, Mendez J, Zimmerman K, Otto P, Vidugiris G, Zhu J, Darzins A, Klaubert DH, Bulleit RF, Wood KV. HaloTag: a novel protein labeling technology for cell imaging and protein analysis. ACS chemical biology. 2008;3:373–382. doi: 10.1021/cb800025k. [DOI] [PubMed] [Google Scholar]
- Los GV, Wood K. The HaloTag: a novel technology for cell imaging and protein analysis. Methods Mol Biol. 2007;356:195–208. doi: 10.1385/1-59745-217-3:195. [DOI] [PubMed] [Google Scholar]
- Luedtke NW, Dexter RJ, Fried DB, Schepartz A. Surveying polypeptide and protein domain conformation and association with FlAsH and ReAsH. Nat Chem Biol. 2007;3:779–784. doi: 10.1038/nchembio.2007.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madani F, Lind J, Damberg P, Adams SR, Tsien RY, Graslund AO. Hairpin structure of a biarsenical-tetracysteine motif determined by NMR spectroscopy. Journal of the American Chemical Society. 2009;131:4613–4615. doi: 10.1021/ja809315x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marek KW, Davis GW. Transgenically encoded protein photoinactivation (FlAsH-FALI): acute inactivation of synaptotagmin I. Neuron. 2002;36:805–813. doi: 10.1016/s0896-6273(02)01068-1. [DOI] [PubMed] [Google Scholar]
- Marks KM, Braun PD, Nolan GP. A general approach for chemical labeling and rapid, spatially controlled protein inactivation. Proceedings of the National Academy of Sciences of the United States of America. 2004a;101:9982–9987. doi: 10.1073/pnas.0401609101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks KM, Nolan GP. Chemical labeling strategies for cell biology. Nature methods. 2006;3:591–596. doi: 10.1038/nmeth906. [DOI] [PubMed] [Google Scholar]
- Marks KM, Rosinov M, Nolan GP. In vivo targeting of organic calcium sensors via genetically selected peptides. Chem Biol. 2004b;11:347–356. doi: 10.1016/j.chembiol.2004.03.004. [DOI] [PubMed] [Google Scholar]
- Martin BR, Giepmans BN, Adams SR, Tsien RY. Mammalian cell-based optimization of the biarsenical-binding tetracysteine motif for improved fluorescence and affinity. Nature biotechnology. 2005;23:1308–1314. doi: 10.1038/nbt1136. [DOI] [PubMed] [Google Scholar]
- Miller LW, Cai Y, Sheetz MP, Cornish VW. In vivo protein labeling with trimethoprim conjugates: a flexible chemical tag. Nature methods. 2005;2:255–257. doi: 10.1038/nmeth749. [DOI] [PubMed] [Google Scholar]
- Miller LW, Sable J, Goelet P, Sheetz MP, Cornish VW. Methotrexate conjugates: a molecular in vivo protein tag. Angewandte Chemie. 2004;43:1672–1675. doi: 10.1002/anie.200352852. [DOI] [PubMed] [Google Scholar]
- Neklesa TK, Tae HS, Schneekloth AR, Stulberg MJ, Corson TW, Sundberg TB, Raina K, Holley SA, Crews CM. Small-molecule hydrophobic tagging-induced degradation of HaloTag fusion proteins. Nat Chem Biol. 2011;7:538–543. doi: 10.1038/nchembio.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman RH, Fosbrink MD, Zhang J. Genetically encodable fluorescent biosensors for tracking signaling dynamics in living cells. Chem Rev. 2011;111:3614–3666. doi: 10.1021/cr100002u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordberg E, Friedman M, Gostring L, Adams GP, Brismar H, Nilsson FY, Stahl S, Glimelius B, Carlsson J. Cellular studies of binding, internalization and retention of a radiolabeled EGFR-binding affibody molecule. Nucl Med Biol. 2007;34:609–618. doi: 10.1016/j.nucmedbio.2007.05.010. [DOI] [PubMed] [Google Scholar]
- Nygren J, Svanvik N, Kubista M. The interactions between the fluorescent dye thiazole orange and DNA. Biopolymers. 1998;46:39–51. doi: 10.1002/(SICI)1097-0282(199807)46:1<39::AID-BIP4>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- Ozhalici-Unal H, Pow CL, Marks SA, Jesper LD, Silva GL, Shank NI, Jones EW, Burnette JM, 3rd, Berget PB, Armitage BA. A rainbow of fluoromodules: a promiscuous scFv protein binds to and activates a diverse set of fluorogenic cyanine dyes. Journal of the American Chemical Society. 2008;130:12620–12621. doi: 10.1021/ja805042p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pace CJ, Huang Q, Wang F, Palla KS, Fuller AA, Gao J. A FlAsH-tetracysteine assay for quantifying the association and orientation of transmembrane alpha-helices. Chembiochem. 2011;12:1018–1022. doi: 10.1002/cbic.201000736. [DOI] [PubMed] [Google Scholar]
- Parris KD, Lin L, Tam A, Mathew R, Hixon J, Stahl M, Fritz CC, Seehra J, Somers WS. Crystal structures of substrate binding to Bacillus subtilis holo-(acyl carrier protein) synthase reveal a novel trimeric arrangement of molecules resulting in three active sites. Structure. 2000;8:883–895. doi: 10.1016/s0969-2126(00)00178-7. [DOI] [PubMed] [Google Scholar]
- Peneva K, Mihov G, Herrmann A, Zarrabi N, Borsch M, Duncan TM, Mullen K. Exploiting the nitrilotriacetic acid moiety for biolabeling with ultrastable perylene dyes. Journal of the American Chemical Society. 2008;130:5398–5399. doi: 10.1021/ja711322g. [DOI] [PubMed] [Google Scholar]
- Popp MW, Antos JM, Grotenbreg GM, Spooner E, Ploegh HL. Sortagging: a versatile method for protein labeling. Nat Chem Biol. 2007;3:707–708. doi: 10.1038/nchembio.2007.31. [DOI] [PubMed] [Google Scholar]
- Pries F, Kingma J, Krooshof GH, Jeronimus-Stratingh CM, Bruins AP, Janssen DB. Histidine 289 is essential for hydrolysis of the alkyl-enzyme intermediate of haloalkane dehalogenase. J Biol Chem. 1995;270:10405–10411. doi: 10.1074/jbc.270.18.10405. [DOI] [PubMed] [Google Scholar]
- Prifti E, Reymond L, Umebayashi M, Hovius R, Riezman H, Johnsson K. A fluorogenic probe for SNAP-tagged plasma membrane proteins based on the solvatochromic molecule Nile Red. ACS chemical biology. 2014;9:606–612. doi: 10.1021/cb400819c. [DOI] [PubMed] [Google Scholar]
- Puthenveetil S, Liu DS, White KA, Thompson S, Ting AY. Yeast display evolution of a kinetically efficient 13-amino acid substrate for lipoic acid ligase. Journal of the American Chemical Society. 2009;131:16430–16438. doi: 10.1021/ja904596f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajapakse HE, Gahlaut N, Mohandessi S, Yu D, Turner JR, Miller LW. Time-resolved luminescence resonance energy transfer imaging of protein-protein interactions in living cells. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:13582–13587. doi: 10.1073/pnas.1002025107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray-Saha S, Schepartz A. Visualizing tyrosine kinase activity with bipartite tetracysteine display. Chembiochem. 2010;11:2089–2091. doi: 10.1002/cbic.201000234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter K, Mofid MR, Marahiel MA, Ficner R. Crystal structure of the surfactin synthetase-activating enzyme sfp: a prototype of the 4'-phosphopantetheinyl transferase superfamily. Embo J. 1999;18:6823–6831. doi: 10.1093/emboj/18.23.6823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robers M, Pinson P, Leong L, Batchelor RH, Gee KR, Machleidt T. Fluorescent labeling of proteins in living cells using the FKBP12 (F36V) tag. Cytometry A. 2009;75:207–224. doi: 10.1002/cyto.a.20649. [DOI] [PubMed] [Google Scholar]
- Roberti MJ, Bertoncini CW, Klement R, Jares-Erijman EA, Jovin TM. Fluorescence imaging of amyloid formation in living cells by a functional, tetracysteine-tagged alpha-synuclein. Nature methods. 2007;4:345–351. doi: 10.1038/nmeth1026. [DOI] [PubMed] [Google Scholar]
- Ruggiu AA, Bannwarth M, Johnsson K. Fura-2FF-based calcium indicator for protein labeling. Organic & biomolecular chemistry. 2010;8:3398–3401. doi: 10.1039/c000158a. [DOI] [PubMed] [Google Scholar]
- Rust MJ, Bates M, Zhuang X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nature methods. 2006;3:793–795. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutkowska A, Haering CH, Schultz C. A FlAsH-based cross-linker to study protein interactions in living cells. Angewandte Chemie. 2011;50:12655–12658. doi: 10.1002/anie.201106404. [DOI] [PubMed] [Google Scholar]
- Saunders MJ, Block E, Sorkin A, Waggoner AS, Bruchez MP. A bifunctional converter: fluorescein quenching scFv/fluorogen activating protein for photostability and improved signal to noise in fluorescence experiments. Bioconjugate chemistry. 2014;25:1556–1564. doi: 10.1021/bc500273n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saurabh S, Beck LE, Maji S, Baty CJ, Wang Y, Yan Q, Watkins SC, Bruchez MP. Multiplexed modular genetic targeting of quantum dots. ACS nano. 2014;8:11138–11146. doi: 10.1021/nn5044367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheck RA, Lowder MA, Appelbaum JS, Schepartz A. Bipartite Tetracysteine Display Reveals Allosteric Control of Ligand-Specific EGFR Activation. ACS chemical biology. 2012 doi: 10.1021/cb300216f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder J, Benink H, Dyba M, Los GV. In vivo labeling method using a genetic construct for nanoscale resolution microscopy. Biophys J. 2009;96:L01–03. doi: 10.1016/j.bpj.2008.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz SL, Yan Q, Telmer CA, Lidke KA, Bruchez MP, Lidke DS. Fluorogen-Activating Proteins Provide Tunable Labeling Densities for Tracking FcepsilonRI Independent of IgE. ACS chemical biology. 2014 doi: 10.1021/cb5005146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shank NI, Zanotti KJ, Lanni F, Berget PB, Armitage BA. Enhanced photostability of genetically encodable fluoromodules based on fluorogenic cyanine dyes and a promiscuous protein partner. Journal of the American Chemical Society. 2009;131:12960–12969. doi: 10.1021/ja9016864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slavoff SA, Chen I, Choi YA, Ting AY. Expanding the substrate tolerance of biotin ligase through exploration of enzymes from diverse species. Journal of the American Chemical Society. 2008;130:1160–1162. doi: 10.1021/ja076655i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano Del Amo D, Wang W, Jiang H, Besanceney C, Yan AC, Levy M, Liu Y, Marlow FL, Wu P. Biocompatible copper(I) catalysts for in vivo imaging of glycans. Journal of the American Chemical Society. 2010;132:16893–16899. doi: 10.1021/ja106553e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Zhang A, Baker B, Sun L, Howard A, Buswell J, Maurel D, Masharina A, Johnsson K, Noren CJ, Xu MQ, Correa IR., Jr Development of SNAP-tag fluorogenic probes for wash-free fluorescence imaging. Chembiochem. 2011;12:2217–2226. doi: 10.1002/cbic.201100173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szent-Gyorgyi C, Schmidt BF, Creeger Y, Fisher GW, Zakel KL, Adler S, Fitzpatrick JA, Woolford CA, Yan Q, Vasilev KV, Berget PB, Bruchez MP, Jarvik JW, Waggoner A. Fluorogen-activating single-chain antibodies for imaging cell surface proteins. Nature biotechnology. 2008;26:235–240. doi: 10.1038/nbt1368. [DOI] [PubMed] [Google Scholar]
- Szent-Gyorgyi C, Schmidt BF, Fitzpatrick JA, Bruchez MP. Fluorogenic dendrons with multiple donor chromophores as bright genetically targeted and activated probes. Journal of the American Chemical Society. 2010;132:11103–11109. doi: 10.1021/ja9099328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szent-Gyorgyi C, Stanfield RL, Andreko S, Dempsey A, Ahmed M, Capek S, Waggoner AS, Wilson IA, Bruchez MP. Malachite green mediates homodimerization of antibody VL domains to form a fluorescent ternary complex with singular symmetric interfaces. Journal of molecular biology. 2013;425:4595–4613. doi: 10.1016/j.jmb.2013.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tour O, Meijer RM, Zacharias DA, Adams SR, Tsien RY. Genetically targeted chromophore-assisted light inactivation. Nature biotechnology. 2003;21:1505–1508. doi: 10.1038/nbt914. [DOI] [PubMed] [Google Scholar]
- Uchinomiya SH, Nonaka H, Fujishima SH, Tsukiji S, Ojida A, Hamachi I. Site-specific covalent labeling of His-tag fused proteins with a reactive Ni(II)-NTA probe. Chem Commun (Camb) 2009:5880–5882. doi: 10.1039/b912025d. [DOI] [PubMed] [Google Scholar]
- Uttamapinant C, Tangpeerachaikul A, Grecian S, Clarke S, Singh U, Slade P, Gee KR, Ting AY. Fast, cell-compatible click chemistry with copper-chelating azides for biomolecular labeling. Angewandte Chemie. 2012;51:5852–5856. doi: 10.1002/anie.201108181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uttamapinant C, White KA, Baruah H, Thompson S, Fernandez-Suarez M, Puthenveetil S, Ting AY. A fluorophore ligase for site-specific protein labeling inside living cells. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:10914–10919. doi: 10.1073/pnas.0914067107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivero-Pol L, George N, Krumm H, Johnsson K, Johnsson N. Multicolor imaging of cell surface proteins. Journal of the American Chemical Society. 2005;127:12770–12771. doi: 10.1021/ja0533850. [DOI] [PubMed] [Google Scholar]
- Wang Y, Telmer CA, Schmidt BF, Franke JD, Ort S, Arndt-Jovin DJ, Bruchez MP. Fluorogen Activating Protein-Affibody Probes: Modular, No-Wash Measurement of Epidermal Growth Factor Receptors. Bioconjugate chemistry. 2014 doi: 10.1021/bc500525b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Zhou Z, Walsh CT, McMahon AP. Selective translocation of intracellular Smoothened to the primary cilium in response to Hedgehog pathway modulation. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:2623–2628. doi: 10.1073/pnas.0812110106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenzel EM, Morton A, Ebert K, Welzel O, Kornhuber J, Cousin MA, Groemer TW. Key physiological parameters dictate triggering of activity-dependent bulk endocytosis in hippocampal synapses. PloS one. 2012;7:e38188. doi: 10.1371/journal.pone.0038188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolbers F, ter Braak P, Le Gac S, Luttge R, Andersson H, Vermes I, van den Berg A. Viability study of HL60 cells in contact with commonly used microchip materials. Electrophoresis. 2006;27:5073–5080. doi: 10.1002/elps.200600203. [DOI] [PubMed] [Google Scholar]
- Wombacher R, Heidbreder M, van de Linde S, Sheetz MP, Heilemann M, Cornish VW, Sauer M. Live-cell super-resolution imaging with trimethoprim conjugates. Nature methods. 2010;7:717–719. doi: 10.1038/nmeth.1489. [DOI] [PubMed] [Google Scholar]
- Wu P, Shui W, Carlson BL, Hu N, Rabuka D, Lee J, Bertozzi CR. Site-specific chemical modification of recombinant proteins produced in mammalian cells by using the genetically encoded aldehyde tag. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:3000–3005. doi: 10.1073/pnas.0807820106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Q, Schmidt BF, Perkins LA, Naganbabu M, Saurabh S, Andreko SK, Bruchez MP. Near-instant surface-selective fluorogenic protein quantification using sulfonated triarylmethane dyes and fluorogen activating proteins. Organic & biomolecular chemistry. 2014a doi: 10.1039/c4ob02309a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Q, Schwartz SL, Maji S, Huang F, Szent-Gyorgyi C, Lidke DS, Lidke KA, Bruchez MP. Localization microscopy using noncovalent fluorogen activation by genetically encoded fluorogen-activating proteins. Chemphyschem : a European journal of chemical physics and physical chemistry. 2014b;15:687–695. doi: 10.1002/cphc.201300757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang G, de Castro Reis F, Sundukova M, Pimpinella S, Asaro A, Castaldi L, Batti L, Bilbao D, Reymond L, Johnsson K, Heppenstall PA. Genetic targeting of chemical indicators in vivo. Nature methods. 2014 doi: 10.1038/nmeth.3207. [DOI] [PubMed] [Google Scholar]
- Yao JZ, Uttamapinant C, Poloukhtine A, Baskin JM, Codelli JA, Sletten EM, Bertozzi CR, Popik VV, Ting AY. Fluorophore targeting to cellular proteins via enzyme-mediated azide ligation and strain-promoted cycloaddition. Journal of the American Chemical Society. 2012;134:3720–3728. doi: 10.1021/ja208090p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin J, Straight PD, McLoughlin SM, Zhou Z, Lin AJ, Golan DE, Kelleher NL, Kolter R, Walsh CT. Genetically encoded short peptide tag for versatile protein labeling by Sfp phosphopantetheinyl transferase. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:15815–15820. doi: 10.1073/pnas.0507705102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying LQ, Branchaud BP. Purification of tetracysteine-tagged proteins by affinity chromatography using a non-fluorescent, photochemically stable bisarsenical affinity ligand. Bioconjugate chemistry. 2011;22:987–992. doi: 10.1021/bc200038t. [DOI] [PubMed] [Google Scholar]
- Yushchenko DA, Zhang M, Yan Q, Waggoner AS, Bruchez MP. Genetically targetable and color-switching fluorescent probe. Chembiochem. 2012;13:1564–1568. doi: 10.1002/cbic.201200334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanotti KJ, Silva GL, Creeger Y, Robertson KL, Waggoner AS, Berget PB, Armitage BA. Blue fluorescent dye-protein complexes based on fluorogenic cyanine dyes and single chain antibody fragments. Organic & biomolecular chemistry. 2011;9:1012–1020. doi: 10.1039/c0ob00444h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Cironi P, Lin AJ, Xu Y, Hrvatin S, Golan DE, Silver PA, Walsh CT, Yin J. Genetically encoded short peptide tags for orthogonal protein labeling by Sfp and AcpS phosphopantetheinyl transferases. ACS chemical biology. 2007;2:337–346. doi: 10.1021/cb700054k. [DOI] [PubMed] [Google Scholar]
- Zhou Z, Koglin A, Wang Y, McMahon AP, Walsh CT. An eight residue fragment of an acyl carrier protein suffices for post-translational introduction of fluorescent pantetheinyl arms in protein modification in vitro and in vivo. Journal of the American Chemical Society. 2008;130:9925–9930. doi: 10.1021/ja802657n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurn A, Klenk C, Zabel U, Reiner S, Lohse MJ, Hoffmann C. Site-specific, orthogonal labeling of proteins in intact cells with two small biarsenical fluorophores. Bioconjugate chemistry. 2010;21:853–859. doi: 10.1021/bc900394j. [DOI] [PubMed] [Google Scholar]