Abstract
Background
Intercellular communication by the hedgehog cell signaling pathway is necessary for tooth development throughout the vertebrates, but it remains unclear which specific developmental signals control cell behavior at different stages of odontogenesis. To address this issue, we have manipulated hedgehog activity during zebrafish tooth development and visualized the results using confocal microscopy.
Results
We first established that reporter lines for dlx2b, fli1, NF-κB, and prdm1a are markers for specific subsets of tooth germ tissues. We then blocked hedgehog signaling with cyclopamine and observed a reduction or elimination of the cranial neural crest derived dental papilla, which normally contains the cells that later give rise to dentin-producing odontoblasts. Upon further investigation we observed that the dental papilla begins to form and then regresses in the absence of hedgehog signaling, through a mechanism unrelated to cell proliferation or apoptosis. We also found evidence of an isometric reduction in tooth size that correlates with the time of earliest hedgehog inhibition.
Conclusions
We hypothesize that these results reveal a previously uncharacterized function of hedgehog signaling during tooth morphogenesis, regulating the number of cells in the dental papilla and thereby controlling tooth size.
Keywords: teeth, cell signaling, odontoblasts, cyclopamine, morphogenesis
Introduction
Soon after Sonic hedgehog (Shh) was identified as the key signaling molecule responsible for the zone of polarizing activity in the developing vertebrate limb bud (Riddle et al., 1993), much attention was given to understanding other important signaling roles the hedgehog pathway might be playing in vertebrate organogenesis, including in tooth development (Bitgood and McMahon, 1995; Chiang et al., 1996). Hedgehog signaling has since been found to be an essential part of odontogenesis in evolutionarily diverse vertebrate species (Fraser et al., 2008; Smith et al., 2009; Handrigan and Richman, 2010; Jackman et al., 2010), and defects in the pathway are responsible for human pathologies that include holoprosencephaly (Nanni et al., 1999), the related Solitary median maxillary central incisor syndrome (Nanni et al., 2001), and odontogenic keratocysts (Lench et al., 1997). Thus, learning more about how hedgehog signaling functions during tooth development has the potential to enhance both our understanding of vertebrate dental organogenesis and its evolution.
In both mammals and teleost fish, only the Shh ligand, or its teleost duplicate semi-ortholog shha, is expressed in developing dental tissues (Kronmiller et al., 1995; Cobourne et al., 2004; Jackman et al., 2010). In very early stages of tooth development, Shh expression is widespread in the epithelial layer that later gives rise to ameloblasts that secrete the enamel of the mature, mineralized tooth (Bitgood and McMahon, 1995; ten Berge et al., 1998) or enameloid in non-tetrapods (Huysseune and Sire, 1998; Kawasaki et al., 2005). At these early stages, Shh expression is absent from the adjacent cranial neural crest-derived mesenchymal cells that form an embryonic structure called the dental papilla (Rothova et al., 2012) and later differentiate into odontoblasts that make the dentin layer of the mature tooth. However, expression of Patched hedgehog receptors are widespread in developing dental tissues (Thomas et al., 1997; Motoyama et al., 1998; Jackman et al., 2010), indicating that many different types of cells within a tooth germ are likely capable of responding to a hedgehog signal. It is thus interesting to consider whether hedgehog cell signaling may control multiple functions during different stages of odontogenesis.
Several different kinds of overall effects on tooth development have been recorded when hedgehog signaling has been inhibited in different species and at different developmental stages. The initial mouse Shh knockout mutant had no teeth but also lacked the surrounding tissues, leaving the specificity of a tooth hedgehog requirement unresolved (Chiang et al., 1996). Later mouse experiments revealed that chemical hedgehog pathway inhibition just after tooth initiation resulted in early developmental arrest (Cobourne et al., 2001), and a conditional knockout disabling Shh activity during dental morphogenesis resulted in smaller than normal tooth germs that gave rise to severely malformed teeth (Dassule et al., 2000). Outside of mammals, chemical inhibition of hedgehog signaling with the antagonist cyclopamine (Chen et al., 2002) has been reported to cause tooth developmental malformations in species ranging from teleost fish (Fraser et al., 2008; Jackman et al., 2010) to snakes (Buchtova et al., 2008). However, the cellular mechanisms responsible for these dental effects of hedgehog inhibition remain largely uncharacterized.
While there is some evidence that hedgehog signaling prevents apoptosis during odontogenesis (Cobourne et al., 2001), research has focused more extensively on its role in controlling cell proliferation as a cellular developmental mechanism of its action. Several of the developmental arrest and morphogenesis defects resulting from hedgehog inhibition have been associated with a decrease in cell proliferation (Gritli-Linde et al., 2002; Nakatomi et al., 2006; Buchtova et al., 2008). A compelling model of tooth cusp formation in mammalian molars places Shh in a role of controlling morphogenesis by regulating epithelial proliferation surrounding its expression in a signaling center known as the enamel knot (Jernvall et al., 1994; Vaahtokari et al., 1996; Salazar-Ciudad, 2012). Similarly, cell signaling from the enamel knot to the underlying mesenchymal cells may be involved in the formation of the molar dental papilla via localized stimulation of cell proliferation (Rothova et al., 2012). However, more empirical data on how hedgehog signaling affects developmental cellular behaviors at all stages of odontogenesis is needed to test these models and expand their scope to account for other possible hedgehog signaling functions during tooth development.
It has previously been shown that hedgehog signaling via the shha ligand is required for tooth initiation in the easily visualized embryos of zebrafish and that hedgehog requirements in zebrafish teeth continue throughout odontogenesis (Jackman et al., 2010). However, the resolution of these experiments was insufficient to distinguish cellular mechanisms of hedgehog action. Here we visualize developing zebrafish tooth germs at high resolution with fluorescence confocal microscopy and report the effects of chemical hedgehog signaling inhibition on their morphogenesis. We find that hedgehog inhibition just after tooth initiation results in the later specific loss of the dental mesenchyme-derived dental papilla, and that this loss completely prevents mature tooth formation. Further characterizing this effect, we find that the dental papilla begins to form and then regresses after early hedgehog signaling inhibition, but that neither cell proliferation nor apoptosis appear to mediate this phenotype. We also report that later hedgehog inhibition results in the formation of a dental papilla with a lower-than-normal cell count, which correlates with an isometric reduction in tooth size. We propose that a normal function of hedgehog signaling after the initiation of odontogenesis is to maintain cells in the dental papilla, ultimately controlling tooth size.
Results
Fluorescence imaging of zebrafish tooth morphogenesis
To better understand how manipulating hedgehog signaling alters tooth development and thus learn more about how the pathway controls normal odontogenesis, we first developed a system to view gene expression and morphology at high resolution in developing zebrafish tooth germs (Fig. 1). We define stages of tooth development based on previous descriptive work in zebrafish (Huysseune and Sire, 1998; Van der heyden et al., 2000; Laurenti et al., 2004), modified slightly by our own observations. For simplicity, the first tooth to form on each side of the midline we refer to as tooth #1 (4V1 in Laurenti et al. (2004)), and the next two subsequently forming teeth as #2 (3V1) and #3 (5V1). The stages of development for tooth germ #1 are: initiation placode (36 hours post fertilization (hpf)), early morphogenesis (44-48 hpf), mid-morphogenesis (52-54 hpf), late morphogenesis (56-60 hpf), cell differentiation (72 hpf), and attachment/eruption (96 hpf).
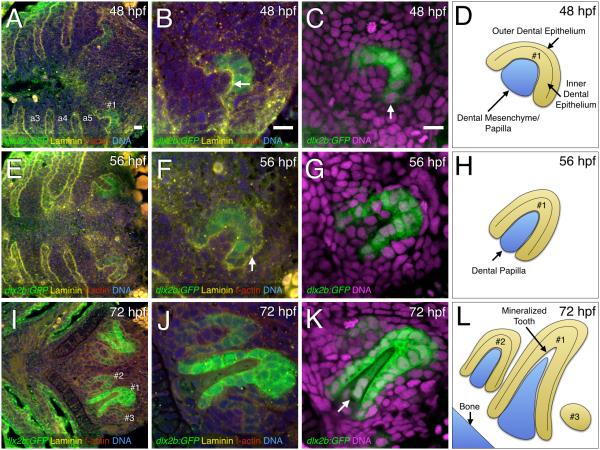
Figure 1. Fluorescence visualization of cellular details during tooth germ morphogenesis.
(A, E, I) Ventral views, anterior to the left, of the zebrafish pharyngeal region. (B-C, F-G, J-K) Closeups of right-side tooth germs. (A-B, E-F, I-J) Four-color stains of dlx2b:GFP reporter expression (green), laminin protein (yellow), f-actin (red), and DNA (blue). (C, G, K) Two-color stains with dlx2b:GFP (green) and DNA (magenta). (A-B) Laminin expression at 48 hpf and 56 hpf stages at the boundaries of the pharyngeal arches (a3-a5) as well as the interface between the dental epithelium and dental mesenchyme of tooth germ #1 (B, arrow). (C) dlx2b:GFP reporter expression mostly in the inner dental epithelium (arrow). (D) Schematic drawing of a early morphogenesis stage tooth germ #1 at 48 hpf. (E-F) Laminin expression highlighting the outer dental epithelium in a late morphogenesis stage tooth germ #1 at 56 hpf (F, arrow). (G-H) dlx2b:GFP expression in the inner dental epithelium and schematic drawing. (I-J) F-actin staining along with dlx2b:GFP expression in cell differentiation stage tooth germ #1 at 72 hpf. Tooth germs #2 and #3 are also present. (K) dlx2b:GFP expression in the inner dental epithelium and the dental papilla (arrow) at 72 hpf. Tooth germs #2 and #3 are mostly outside the focal plane in this specimen. (L) Schematic drawing of typical orientations of the tooth germs at 72 hpf. Scale bars = 10 μm (A-C).
From the analysis of a reporter line and fluorescent histological markers we have composed a summary of tooth morphogenesis for orientation (Fig. 1D, 1H, 1L). At the early morphogenesis stage at 48 hpf, the inner and outer layers of the dental epithelium of tooth germ #1 are broadly curved and surround a group of dental mesenchyme cells that are beginning to assume the shape of the dental papilla (Fig. 1D). By 56 hpf, the late morphogenesis stage tooth germ is elongated in both the dental epithelium and the dental papilla along the long axis of the incipient tooth (Fig. 1H). By the cellular differentiation stage of 72 hpf, tooth #1 has almost fully elongated and is starting to mineralize, growing close to the bone to which it will attach by 96 hpf (ceratobranchial #5; Fig. 1L). Also at 72 hpf, tooth #2 is in mid-morphogenesis and tooth #3, if visible, is still at an initiation placode or early morphogenesis stage. We could often distinguish between the inner and outer dental epithelial layers, but because we could not always do so, we sometimes refer to these two epithelial layers together as a single dental epithelium.
To provide a context in which to view reporter transgenic expression during tooth morphogenesis, we used a combination of fluorescent labels. An antibody against laminin was used to visualize basal lamina (Thesleff et al., 1981), and clearly indicated the pharyngeal arches and the interface between the dental epithelium and dental mesenchyme at relatively early stages of tooth formation (Fig. 1). An f-actin label was also used to visualize the cortical cytoplasm of cells in the tooth-forming pharyngeal region (Gaete and Tucker, 2013), and was particularly helpful in helping to highlight cellular morphology at later stages (e.g. Fig. 1I, 1J). Additionally, a DNA stain indicated the size and position of cell nuclei in tooth germs and surrounding tissues. These markers in combination with the dlx2b:GFP reporter expression provided a clear picture of the developing tooth and surrounding tissues. dlx2b:GFP reporter expression was mostly localized to the inner dental epithelium during morphogenesis stages (Fig. 1A-1G), but by 72 hpf was expressed both in the inner dental epithelium and in the dental papilla of tooth #1 (Fig. 1I-K).
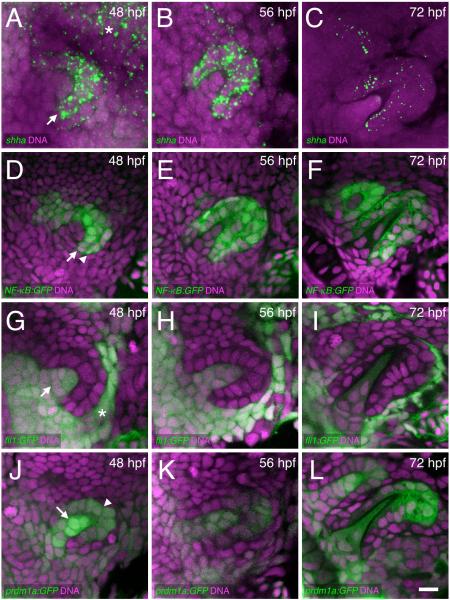
Because the shha gene was previously identified as the only hedgehog ligand expressed during zebrafish tooth development (Jackman et al., 2010), we wanted to visualize it at higher resolution in this system. During morphogenesis stages, shha mRNA expression was observed in the dental epithelium (primarily in the inner dental epithelium) and at relatively lower levels in nearby non-dental pharyngeal epithelium (Fig. 2A-B). Expression of shha appeared lower by the cellular differentiation stage but was still present in the dental epithelium (Fig. 2C). We additionally identified three other GFP transgenic reporter lines that help to demarcate the dental epithelium and mesenchyme at these stages of development (Fig. 2D-L). Similarly to shha mRNA, NF-κB:GFP reporter expression had an epithelium-restricted expression pattern, except that at 48 and 56 hpf it was expressed both in the inner and outer dental epithelium (Fig. 2D-E), and by 72 hpf a small amount of expression was seen in the dental papilla of tooth germ #1 (Fig. 2F). Conversely, fli1:GFP, a marker of cranial neural crest cells (Lawson and Weinstein, 2002), was expressed in both dental and nearby non-dental mesenchyme and the dental papilla at all stages examined, but was completely absent from the dental epithelium (Fig. 2G-I). A fourth reporter, prdm1a:GFP, was localized to developing tooth germs with an expression pattern resembling a partial composite of the previous two (Fig. 2J-L). prdm1a:GFP was expressed both in the dental mesenchyme and nearby cranial neural crest cells, but levels appeared relatively higher in the dental mesenchyme especially at the early morphogenesis stage (Fig. 2J). This reporter was also expressed in the dental epithelium, with expression concentrated in the distal portion of both the inner and outer dental epithelium surrounding the tooth tip at all stages examined.
Figure 2. Cellular details of hedgehog epithelial expression and tooth germ morphogenesis.
Ventral views, anterior to the left, of right-side tooth germs. DNA is stained in magenta. (A-C) shha mRNA FISH (green) with expression visible in the inner dental epithelium (arrow) and nearby pharyngeal epithelium (asterisk). (D-F) NF-κB:GFP reporter expression in both the inner (arrow) and outer dental epithelium (arrowhead). (G-I) fli1:GFP expression in the dental papilla (arrow) and nearby non-dental mesenchyme (asterisk). (J-L) prdm1a:GFP expression in both the dental papilla (arrow) and in distal parts of the dental epithelium (arrowhead), with papilla expression appearing relatively higher at 48 hpf. Scale bar = 10 μm (L).
Together these data reveal the specific location of shha signaling during odontogenesis and provide markers of tooth germ tissue subtypes to help interpret experimental manipulations.
Hedgehog signaling is required for dental papilla formation
It has been previously reported that inhibition of hedgehog signaling in zebrafish using 50 μM cyclopamine (CyA) results in the complete loss of a mineralized tooth if treatment begins at or before the initiation placode stage of 36 hpf (Jackman et al., 2010). However, this study provided only a gross view of mineralized tooth morphology and gene expression after such treatment, so we used the visualization methods described above to pinpoint more specific developmental phenotypes that result from hedgehog signaling disruption.
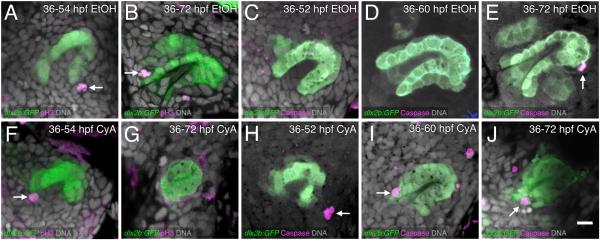
For these experiments, embryos were exposed to 50 μm CyA, or to 0.2% EtOH as a control, and grown in this medium until analysis. Control exposure either from 30-72 hpf (n = 6), 32-72 hpf (n = 6), 36-72 hpf (n = 19), or from 48-72 hpf (n = 16) did not appear to affect tooth development whatsoever, and the resulting tooth germs were morphologically normal as judged by dlx2b:GFP expression (Fig. 3A-C). In contrast, CyA treatment starting at 30 or 32 hpf resulted in a complete or almost complete elimination of the tooth germ by 72 hpf (n = 12, Fig. 3D-F). This result was consistent with the previously observed lack of mineralized tooth development and most tooth-related gene expression from similar CyA treatments (Jackman et al., 2010). However, slightly later, 36-72 hpf CyA exposure often resulted in a more striking phenotype, with a relatively normal dental epithelium but completely absent dental papilla (n = 20, Fig. 3G-H). Additionally, CyA treatment from 48-72 hpf resulted in an intermediate phenotype with an abnormally small dental papilla (n = 15, Fig. 3I).
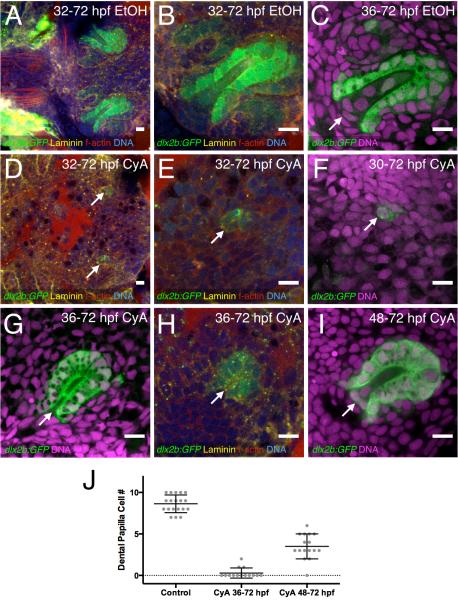
Figure 3. Hedgehog signaling is required for dental papilla formation.
(A-C) dlx2b:GFP expression in normal-appearing tooth germs exposed to 0.2% EtOH as a control from 32 or 36-72 hpf. The base of the dental papilla of tooth #1 is indicated (C, arrow). (D-F) Tooth forming region after 50μM CyA exposure from 30 or 32-72 hpf, showing a few disorganized-appearing GFP-positive cells (arrows). (G-H) Tooth germ after later CyA treatment from 36-72 hpf lacking a dental papilla (arrow). (I) Tooth germ after CyA exposure from 48-72 hpf, with a smaller than normal dental papilla (arrow). (J) Graph of dental papilla cell numbers at 72 hpf. Gray dots represent individual counts, central bars are the mean, and flanking error bars indicate standard error. All three pairwise comparisons are significantly different. Tooth germ orientations are as in Fig. 1. Scale bars = 10 μm.
Quantifying these dental papilla cell number observations revealed that control teeth contained a mean of 8.6 cells at 72 hpf, 36-72 hpf CyA treatments had only 0.3 cells, and 48-72 hpf treatments contained 3.5 cells (Fig. 3J), with each mean significantly different from the others (t-tests, P < 0.0001). Cell numbers in the dental epithelium often appeared only slightly reduced in 36-72 and 48-72 hpf CyA treatments relative to controls, but we were unable to precisely quantify this due to limitations in visualization of the more complex and larger 3D shape of the dental epithelium. However, for some sense of comparison, we were able to count cells in a few exceptionally clear control 72 hpf embryos, and these had on average 29 dental epithelial cells (n = 4).
We concluded from these experiments that hedgehog activity is particularly important for the formation of the dental papilla during early morphogenesis stages of tooth formation.
Cyclopamine exposure does not prevent early dental papilla formation
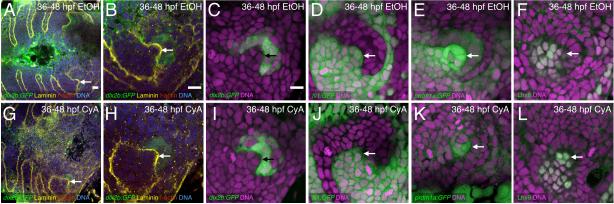
We initially hypothesized that the 36-72 hpf CyA treatment described above would inhibit dental papilla formation from its earliest stages in odontogenesis. However to test this idea, we treated embryos with CyA from 36-48 hpf and used several different markers to visualize the tooth germs at this early morphogenesis stage (Fig. 4). The markers examined included dlx2b:GFP (with DNA counterstain alone n = 7, or in combination with laminin and f-actin stains n = 6); fli1:GFP (n = 6); prdm1a:GFP (n = 6); and an antibody against zebrafish Lhx6 (n = 6), the mRNA of which has been reported to be expressed in the dental papilla and adjacent mesenchyme at this stage (Jackman et al., 2004). We found evidence of dental papilla formation in all CyA treated specimens examined, suggesting that the dental papilla begins to form even with hedgehog inhibition, and only later regresses to the state seen at 72 hpf. Dental papilla cell numbers were not quantified in these early morphogenesis stage tooth germs because dlx2b:GFP does not stain the mesenchyme at this earlier stage and we had no other dental papilla marker specific enough to distinguish the boundary between dental and adjacent non-dental mesenchyme (fli1:GFP, prdm1a:GFP, and Lhx6 all appear to stain both the dental papilla and adjacent non-dental mesenchyme). However, laminin staining outlining the interface between the dental epithelium and the mesenchyme of the dental papilla revealed that the dental papilla often appeared smaller overall and wider at the base in 36-48 hpf CyA treated tooth germs relative to controls (Fig. 4H). However, regardless of its exact morphology, the dental papilla does appear to begin to form and then later regress after hedgehog inhibition.
Figure 4. Hedgehog inhibition does not prevent early dental papilla formation.
Tooth germ #1 after 36-48 hpf EtOH control (A-F) or CyA exposure (G-L). Arrows indicate the dental papilla. Stains are as described in Figs. 1 and 2 with the addition of an Lhx6 antibody stain marking the dental papilla and adjacent non-dental mesenchyme (F, L). In all cases early dental papilla formation is visible in CyA treated tooth germs, but its shape does not always appear normal. Scale bars = 10 μm (A-C).
Cell proliferation and apoptosis after hedgehog inhibition
We examined cell proliferation and apoptosis associated with hedgehog inhibition in order to observe whether either of these two developmental cell behaviors might account for the regression of the dental papilla after 36-48 hpf CyA exposure and its loss by 72 hpf. To do so, we used antibody staining against phosphorylated Histone H3 (pH3) to mark cells in mitosis (Murphey et al., 2006) and against activated Caspase 3 to mark apoptotic cells (Sorrells et al., 2013). We examined the germs of tooth #1 exposed to CyA starting at the initiation placode stage (36 hpf), with fixation and antibody staining at mid-morphogenesis (52-54 hpf), late morphogenesis (60 hpf), or cell differentiation stages (72 hpf).
For proliferation, we visualized and quantified pH3-positive cell numbers in tooth germs from 36-54 and 36-72 hpf treatments (Fig. 5). Treatments from 36-60 hpf appeared similar to 36-72 hpf and were not quantified. The most striking observation from these experiments was that there was very little evidence of proliferation in the dental papilla at any of these stages, in either control or CyA treated individuals. For example, pooling controls from both quantified stages, we saw no pH3-positive cells in the dental papilla in 25 visualized tooth germs, whereas we counted 16 pH3-positive cells in the dental epithelium of these same developing teeth (mean of 0.64 mitotic cells per tooth germ). The pH3-positive cells we did observe were often at the base of the dental epithelium (Fig. 5A, B, F). Also of note was a lack of observed proliferation in the dental epithelium in CyA treated tooth germs at 72 hpf (Fig. 5G). The absence of dental papilla cell proliferation, especially in untreated individuals, suggests that a reduction in proliferation after hedgehog inhibition is unlikely to be the cause by which 36-72 hpf CyA treated tooth germs lose their dental papilla.
Figure 5. Hedgehog inhibition induces unusual late tooth germ apoptosis but proliferation appears relatively normal.
Antibody labels (magenta) for pH3 (proliferation; A-B, F-G) or Caspase 3 (apoptosis; C-E, H-J) in the dlx2b:GFP reporter line (green) with DNA label (gray). Treatments are EtOH control (A-E) or with CyA exposure starting at 36 hpf (F-J). pH3 proliferation staining at or near the base of the dental epithelium in control (A) and CyA treated (F) tooth germs at 54 hpf (arrows). Proliferation is visible in a 72 hpf control tooth germ (arrow, B), but only background staining is seen after CyA treatment (G). (C-E) Caspase 3 apoptosis staining is absent from control tooth germs, except near the tip of tooth #1 at 72 hpf (arrow, D). Apoptosis can be seen in the tooth forming region after 36-52 hpf CyA exposure but not localized to the tooth germ (arrow, H). Caspase 3-positive cells in the dental epithelium after 36-60 hpf CyA treatment (arrow, I). An apoptotic cell is visible in the base of the tooth germ after 36-72 hpf CyA exposure (arrow, J). Tooth germ orientations are as in Fig. 1. Scale bar = 10 μm (J).
Regarding apoptosis, we observed and counted Caspase 3-positive cells in tooth germs with treatment times of 36-52, 36-60, and 36-72 hpf (Fig. 5). In contrast to the proliferation described above, we observed no evidence of apoptosis in 36-52 hpf or 36-60 hpf tooth germs treated with the EtOH control (Fig. 5C-D, n = 24 tooth germs examined at each stage). However, in control tooth germs treated from 36-72 hpf we did see caspase-positive cells, but only in the distal part of the dental epithelium of tooth germ #1 and thus not likely involved with dental papilla formation (Fig. 5E, n = 7 positive cells in 24 tooth germs examined). In contrast, we saw many apoptotic cells after CyA treatment, especially at later stages, but never in the dental papilla. Caspase-positive cells were frequently observed in the vicinity of 36-52 hpf CyA treated tooth germs, but rarely in the germ itself (Fig. 5H, 1 cell in 24 tooth germs). In 36-60 hpf CyA treatments, apoptotic cells were occasionally seen in the dental epithelium (Fig. 5I, 8 cells in 26 tooth germs examined). However, after 36-72 hpf CyA exposure, caspase-positive cells were observed in the dental epithelium of almost every tooth germ examined (43 cells in 26 tooth germs). Many of these cells were located at the base of the dental epithelium (e.g. Fig. 5J), but no caspase-positive cells were seen in cells that could be positively identified as within the dental papilla at any stage examined.
Overall we found no apoptosis clearly associated with the dental papilla after hedgehog signaling inhibition, and proliferation during morphogenesis stages appeared relatively normal. Together, these data suggest that another mechanism is likely responsible for the regression of the papilla after hedgehog inhibition (see Discussion).
Hedgehog signaling is required for subsequent tooth initiation
While completing the experiments described above, we noted that subsequent tooth formation beyond tooth #1 appeared to be inhibited after CyA treatment and thus examined this issue more carefully. Because hedgehog signaling was already known to be required for the formation of all but the earliest initiation placode stages of tooth germ #1 (Jackman et al., 2010), it might follow that later-forming teeth would also have a similar requirement. However, this had not been specifically tested, and gene expression differences have been reported between tooth germ #1 and subsequently forming teeth (Laurenti et al., 2004), suggesting that later teeth may be induced by different mechanisms than tooth #1.
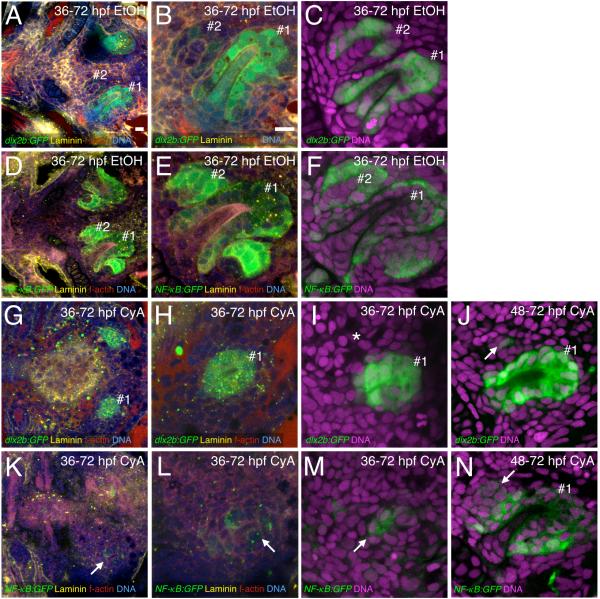
Thus, we examined GFP reporter expression in tooth germ #2 after CyA treatment, using the laminin, f-actin, nuclear stain combination to give a general sense of tissue morphology and the presence of tooth germs and the dlx2b:GFP and NF-κB:GFP lines to help mark the dental epithelium (Fig. 6). In control embryos treated from 36-72 hpf, we found evidence of tooth germ #2 in the vast majority of tooth-forming regions examined (Fig. 6A-F; GFP+: n = 26/28 dlx2b:GFP, 12/12 NF-κB:GFP). In contrast, in embryos treated with CyA from 36-72 hpf, we typically saw no GFP expression in the location of tooth #2, with the exception of occasional small patches of GFP+ cells that did not appear to be part of an organized tooth germ (Fig. 6G-I, K-M; GFP+: n = 2/28 dlx2b:GFP, 0/16 NF-κB:GFP).
Figure 6. Blocking hedgehog signaling restricts subsequent tooth initiation.
Tooth germ dental epithelium labeled with dlx2b:GFP (A-C, G-J) and NF-κB:GFP (D-F, K-N). (A-F) Tooth germs #1 and #2 in control transgenic individuals. (G-I) 36-72 hpf CyA treated individuals with dlx2b:GFP expression in the position of tooth germ #1 but no evidence of tooth germ #2 (asterisk I). (J) 48-72 hpf CyA treated individual with a single dlx2b:GFP+ cell in the position of tooth germ #2 (arrow). (K-M) NF-κB:GFP individuals after 36-72 hpf CyA treatment with very weak and disorganized GFP expression in the tooth-forming region (arrows). (N) Cluster of NF-κB:GFP expressing cells in the position of tooth germ #2 after 48-72 hpf CyA treatment (arrow). Additional stains and tooth germ orientations are as in Fig. 1. Scale bars = 10 μm (A, B).
Interestingly, reporter expression in the NF-κB:GFP line in tooth germ #1 also appeared greatly reduced (n = 6/16) or eliminated (n = 10/16) after 36-72 hpf CyA exposure (Fig. 6K-M). One explanation for this is that NF-κB activity has a more specific requirement for hedgehog signaling than does the expression of dlx2b. However, we also noticed that in tooth #1, NF-κB:GFP expression appeared to fade from the inner dental epithelium relative to the outer dental epithelium by 72 hpf (Fig. 6F). Therefore, the lack of NF-κB:GFP expression in tooth #1 after 36-72 hpf CyA treatment could indicate that CyA exposure reduces the outer dental epithelium, resulting in a tooth germ composed of mostly inner dental epithelium with weaker NF-κB:GFP expression.
Later hedgehog inhibition from 48-72 hpf produced similar results, except that tooth germ #1 was always visible in both reporter lines (Fig. 6J, N). Tooth germ GFP expression was typically absent from the region of tooth #2 after this later CyA treatment (GFP-: n = 20/22 dlx2b:GFP, 12/19 NF-κB:GFP), and when it was present, it consisted of only a few GFP+ cells with no obvious organization (Fig. 6J, N; GFP+: n = 2/22 dlx2b:GFP, 7/19 NF-κB:GFP).
These results are consistent with there being a similar hedgehog signaling requirement for the initiation of the first tooth and subsequently forming tooth germs.
Later stage hedgehog signaling is required for isometric tooth growth
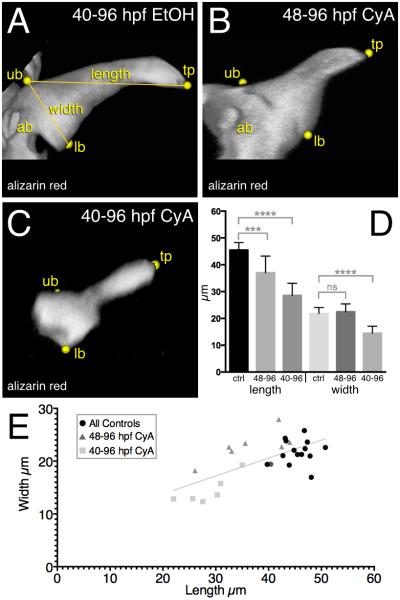
It was previously reported that CyA treatment starting at 36 hpf completely inhibited mineralized tooth formation but that later exposures resulted in small, possibly misshapen teeth (Jackman et al., 2010). To better understand the morphological changes to tooth mineralization associated with hedgehog inhibition, we collected 3D data of tooth #1 using confocal microscopy of alizarin red stained teeth and performed a simple morphometric analysis (Fig. 7). Three unambiguous landmarks identified in all teeth examined were the tip of the tooth, and the uppermost and lowermost aspects of the base of the tooth relative to the downward curvature of the tooth towards the tip (Fig. 7A). The base of the tooth was distinguishable from sometimes-present, adjacent attachment bone by discontinuities and differences in shape when viewed in 3D. We measured the distance from the tooth tip to the upper base as a proxy for tooth length, and between the upper and lower base to estimate the width. The degree of mineralization varied, especially in the earlier CyA treatment group (Fig. 7C), and we only included in the dataset measurements from renderings where all three landmarks were unambiguously identifiable.
Figure 7. Reduction in mineralized tooth size associated with hedgehog inhibition.
(A-C) 3D renderings of 96 hpf mineralized tooth #1 using alizarin red staining and confocal microscopy. (A) 40-96 hpf EtOH treated control tooth #1. (B) 48-96 hpf CyA exposed tooth. (C) 40-96 hpf CyA treated tooth. Labels: tp = tooth tip, ub = upper point of base, lb = lower base, ab = attachment bone. (D) Graph of measurement means. Error bars = standard deviation. (E) XY plot of all measurements. Best fit line is shown (R2 = 0.46, P < 0.0001). Significance tests: ***, P < 0.001; ****, P < 0.0001; ns = not significant.
We compared the shape of tooth #1 after CyA or control treatments starting at 40 or 48 hpf and with visualization at the attachment/eruption stage of 96 hpf. The shape of control teeth were all very similar (40-96 hpf n = 8, 48-96 hpf n = 6), with a mean length of 45.51 μm (n = 14, 2.797 SD) and width of 21.77 μm (2.298 SD). Teeth exposed to CyA from 48-96 hpf were significantly shorter in length than controls (mean 37.00 μm, n = 8, 6.239 SD, t-test P = 0.0003), but not significantly different in width at the base (mean 22.48 μm, 2.918 SD, t-test P = 0.5). Teeth exposed to CyA from 40-96 hpf were significantly shorter than controls in both length (mean 28.57 μm, n = 6, 4.537 SD, t-test P < 0.0001) and in width (mean 14.45 μm, 2.648 SD, t-test P < 0.0001; Fig. 6D).
Plotting the length vs. width measurements from this experiment (Fig. 7E), there is a linear correlation between length and width for all CyA treatment time points (linear regression R2 = 0.46, P < 0.0001), suggesting that hedgehog inhibition starting at earlier stages is causing teeth to become generally smaller in an isometric fashion. We did not observe severely deformed teeth after CyA exposure as reported previously (Jackman et al., 2010). We speculate that this is because at low resolution it is difficult to distinguish between the attachment bone and the mineralized tooth itself, which was quite apparent when visualized with these methods (e.g. Fig. 6A), and we suspect that this bone was previously mistaken for a deformed tooth.
Discussion
A hedgehog signaling requirement for dental papilla formation
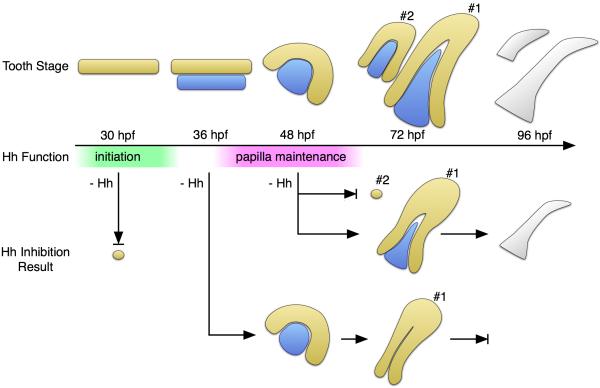
Based on the hedgehog inhibition experiments described above, we propose that there are at least two separate phases during tooth development where hedgehog signaling has a required function (Fig. 8). The first “initiation” phase is at the very start of odontogenesis and is involved in the establishment of the dental epithelium. The second “papilla-maintenance” phase takes place during tooth germ morphogenesis, where hedgehog signaling is required for maintaining the cells within the dental papilla. There are several potential alternatives for how hedgehog signaling may be acting on the cells of the developing tooth germ to promote the establishment of the dental papilla in this latter phase.
Figure 8. Summary of proposed hedgehog functions in zebrafish tooth development.
Top row: timeline of the development of the first two teeth. The dental epithelium (DE) is shown in yellow, and the dental mesenchyme (DM) and dental papilla (DP) in blue. Stages with respect to tooth #1: pre-initiation (30 hpf), initiation placode (36 hpf), early morphogenesis (48 hpf), cell differentiation (72 hpf), and attachment/eruption (96 hpf). Bottom rows: observed results after hedgehog inhibition with cyclopamine. Arrows ending in a bar represent arrest of tooth development. Time windows of hedgehog signaling requirements for initiation (green) and dental papilla maintenance (magenta) are indicated.
When we first observed that hedgehog inhibition at the early initiation placode stage resulted in the formation of a later tooth germ lacking a dental papilla (Fig. 3), we suspected that an elimination of mesenchymal cell proliferation might be responsible, especially given the long history of hedgehog signaling regulating cell division in tooth development and in other systems (Gritli-Linde et al., 2002; Nakatomi et al., 2006; Buchtova et al., 2008). Particularly relevant to this idea was a recent study examining cell fate in the development of the mouse molar dental papilla, where cell proliferation was invoked as a key mechanism for the formation of this structure, although cell movement was also observed (Rothova et al., 2012). We were thus surprised when we carefully examined intermediate stages after hedgehog inhibition and found that the early stages of dental papilla formation appeared relatively normal (Fig. 4). In addition, we detected very little cell proliferation in control tooth germs in stages between when the papilla has started to form during morphogenesis and when the cells begin differentiating into odontoblasts (Fig. 5). We have occasionally observed dividing cells in the dental papilla when we have visualized tooth germs in other contexts, but it is a rare occurrence. Together, these observations suggest that during morphogenesis stages there is relatively little cell proliferation in the zebrafish dental papilla, and thus a reduction in proliferation is not a likely mechanism for the papilla loss we see after initiation placode stage hedgehog inhibition.
Another explanation might be that hedgehog signaling normally inhibits apoptosis in the dental papilla during the papilla-maintenance phase, leading to papilla loss through programmed cell death when this signaling is blocked. In support of this idea, activation of apoptosis has been observed in mouse tooth germs when hedgehog signaling was inhibited (Cobourne et al., 2001), but this cell death was reported to be in epithelial tissues. We observed apoptosis after hedgehog inhibition, but primarily at later stages when the papilla had already completely regressed (Fig. 5). We also did not observe apoptosis localized to the dental papilla itself, except possibly at the base of the tooth germ at 72 hpf after most or all of the papilla was already gone (Fig. 5J). The apoptosis we did observe was almost exclusively seen in the dental epithelium or surrounding non-dental tissues. Thus, we conclude that preventing apoptosis is an unlikely normal function of hedgehog signaling during the papilla-maintenance phase.
However, a developmental cell behavior we have not yet been able to test in this system is cell movement. As mentioned above, when fate-mapping dental papilla cells in mouse, a certain amount of cell migration both into and out of the forming dental papilla has been observed, although cell proliferation was focused on as a likely more important mechanism of dental papilla formation (Rothova et al., 2012). These authors speculated that cell signals from the epithelium, including Shh, are controlling the formation of the dental papilla by regulating proliferation and eventual odontoblast differentiation. These thoughts are congruent with classic dental tissue transplantation experiments demonstrating that odontoblast differentiation requires a signal from the dental epithelium (Thesleff and Hurmerinta, 1981), and later speculation that this signal is Shh (Koyama et al., 1996). We suggest that our results are generally consistent with this model, except that in zebrafish tooth germs hedgehog signaling may be controlling cell movement instead of cell proliferation in the dental papilla. We hypothesize that shha signaling from the dental epithelium prevents cells from migrating out of the dental papilla before they differentiate into odontoblasts. Mammals may also use this mechanism during the formation of the dental papilla, in addition to extensive cell proliferation to enable larger teeth. The development of methods for live cell imaging and lineage tracing in zebrafish tooth germs will allow for this cell movement hypothesis to be tested in the future.
Establishment of subsequent tooth germs
In addition to the loss of the dental papilla from tooth germ #1, we also observed that subsequent teeth fail to form after hedgehog inhibition beginning at 36 hpf. We looked for but never observed tooth #2 or #3 in individuals used for the calcium stain morphometric analysis (Fig. 7), and we found no evidence for the presence of organized tooth germs #2 or #3 using reporter transgenics and histological analysis (Fig. 6). It has been suggested that tooth germ #1 might serve as a trigger for the initiation of subsequent tooth germs (Verstraeten et al., 2013). This notion comes partly from observations that some aspects of gene expression differ between tooth germ #1 and subsequently forming teeth (Laurenti et al., 2004), and it has been proposed that subsequent teeth form via Fgf signaling from the first tooth germ, mediated by N-cadherin proteins (Verstraeten et al., 2013). Given these ideas, it follows that only the first tooth germ may require hedgehog signaling, and that subsequent tooth germ initiation is hedgehog independent. However, we see very similar hedgehog dependance for at least the first three teeth that form and we suggest that hedgehog signaling is required for the early epithelial development of each of them, as well as later for the dental papilla maintenance of at least tooth #1.
Hedgehog regulation of tooth size
The decrease in tooth size that we observe after morphogenesis stage hedgehog inhibition is reminiscent of results from Dassule et al. (2000), in which a Shh conditional knockout mouse was observed to produce smaller than normal tooth germs after pathway inactivation. However, contrary to our findings, this study did not note any particular effect on the dental papilla and also reported severe morphological changes to mature teeth after inhibition. These discrepancies may result from the previous study's restriction of hedgehog inhibition to a single stage and/or from the growth of tooth germs in kidney capsules that may have influenced their normal development. Overall, the results of Dassule et al. (2000) are very similar to the early morphogenesis stage (48 hpf) hedgehog inhibition experiments we report here, which resulted in small tooth size. It is therefore possible that the same late hedgehog signaling mechanism has been evolutionarily conserved between teleost fish and mammalian tooth development.
We observed both a reduction in the number of dental papilla cells after hedgehog inhibition and an overall reduction in tooth size under identical treatments; however, there are several ways in which these two observations may be interrelated. It is possible that a reduction of cell number in the dental papilla limits the number of odontoblasts that eventually differentiate in the tooth germ. Unlike in mammalian teeth, teleost odontoblasts contribute extracellular matrix to both the inner dentin and outer enameloid layers of the mature tooth (Poole, 1967). Enameloid is similar to tetrapod enamel in that ameloblasts from the inner dental epithelium contribute to its hypermineralized matrix, but it differs substantially in that odontoblasts also make a collagen contribution to its initial matrix as it develops (Shellis and Miles, 1974; Sasagawa, 1995; Sire et al., 2009). However, it has also been shown in teleosts that ameloblasts themselves exhibit gene expression suggesting that they also make a large collagen contribution to the enameloid matrix (Huysseune et al., 2008). Despite this, because there is some connection between odontoblasts and both major layers of the mature teleost tooth, it is possible that changes to odontoblast numbers in a teleost fish tooth germ would have a more profound effect on overall tooth size than similar changes in a mammalian tooth. Thus if hedgehog inhibition is either directly or indirectly limiting odontoblast cell number, it could merely represent a side effect that the resulting mature tooth is smaller than normal.
However, an interesting extension of this idea is the possibility that hedgehog signaling controls dental papilla/odontoblast cell number for the express purpose of regulating tooth size. The validity and evolutionary breadth of this hypothesis could be investigated by performing hedgehog inhibition experiments similar to those described here in vertebrate species that normally make teeth of substantially differing sizes, such as Astyanax mexicanus (Seritrakul et al., 2012). The window of Shh expression in the normal tooth germs of these species could be carefully characterized to look for a correlation between the timing of hedgehog signaling activity and the size of the resulting tooth. From a broad perspective, the regulation of organ size has been one of the enduring puzzles of developmental biology (Bryant and Simpson, 1984; Stanger et al., 2007), and a late hedgehog signaling function during tooth morphogenesis may represent part of its solution.
Experimental Procedures
Animal husbandry and lines
Zebrafish (Danio rerio, Hamilton 1822) used in this study were from an in-house stock originally derived from the Tü inbred line (Zebrafish International Resource Center) outcrossed with fish from a commercial supplier (LiveAquaria.com, Rhinelander, WI, USA). Embryos were raised at 28.5°C in an embryo medium consisting of 30% Danieu’s medium with 0.002% methylene blue to inhibit fungal growth. Embryo and larval staging was performed as described previously (Kimmel et al., 1995) with developmental time reported as hours post-fertilization (hpf). Green fluorescent protein (GFP) reporter lines employed: dlx2b:GFP = Tg(dlx2b:EGFP)cs1 (Jackman and Stock, 2006), fli1:GFP = Tg(fli1:EGFP)y1 (Lawson and Weinstein, 2002), prdm1a:GFP = Tg(−60prdm1a:EGFP)i111 (Elworthy et al., 2008), and NF-κB:GFP = Tg(NF-κB:GFP)nc1 (Kanther et al., 2011). Animal care was in accordance with the Institutional Animal Care and Use Committee at Bowdoin College.
Immunohistochemistry, in situ hybridization, and histology
All staining was performed on intact, whole embryos or larvae. Primary antibodies used for immunohistochemistry were anti-GFP (A10260 or 332600, Life Technologies), phospho-Histone H3 (06-570, EMD Millipore), Lhx6 (GTX127337, GeneTex), Laminin (L9393, Sigma-Aldrich), and active Caspase 3 (559565, BD Biosciences). Secondary antibodies were HRP-anti-mouse (626520, Life Technologies), HRP-anti-rabbit (A10547, Life Technologies) and Alexa Fluor 633 anti-rabbit (A21071, Life Technologies). Fluorescence from HRP-conjugated antibodies was developed using the TSA Plus system for tyramide signal amplification (PerkinElmer), with either fluorescein or Cy5 as the fluorophore. F-actin staining was with Alexa Fluor 555 Phalloidin (A34055, Life Technologies). DNA staining was with Sytox Orange or Sytox Blue (Life Technologies). Combinations were as follows: 2-color: anti-GFP-HRP/fluorescein TSA, and Sytox Orange; 3-color: anti-GFP-HRP/fluorescein TSA, anti-Caspase 3/HRP-anti-rabbit/Cy5 TSA, and Sytox Orange; 4-color: anti-GFP/HRP-anti-mouse/fluorescein TSA, anti-Laminin/Alexa Fluor 633 anti-rabbit, Alexa Fluor 555 Phalloidin, and Sytox Blue.
For immunohistochemistry, embryos were fixed overnight at 4°C in 4% formaldehyde in phosphate buffered saline (PBS), blocked 1 hour in PBS + 10% blocking reagent (Roche Diagnostics) + 10% heat inactivated sheep serum + 1% DMSO + 0.1% Tween-20, and incubated overnight in 1:200 dilution of primary antibody. For double antibody labels, the primary antibodies were incubated simultaneously if derived from different species. Embryos were then washed 30 min PBS + 0.1% Tween-20 (PBST). If the anti-GFP-HRP direct conjugate primary antibody was used (A10260, Life Technologies), embryos were developed 30 min in TSA amplification diluent + substrate, and washed 30 min PBST. When doing a double-label with two HRP conjugated antibodies, HRP activity was quenched with a 1 hour incubation in PBST + 2% H2O2. Secondary antibodies were then applied at 1:200 dilution and incubated overnight in the same manner as for the primaries. We found that we could also add the DNA stain (1:50,000 Sytox Orange or Blue) as well as the f-actin stain (1:200 Alexa Fluor 555 Phalloidin) to this step and it did not seem to interfere with secondary antibody binding. Embryos were then washed 30 min PBST, and a second TSA reaction was performed as above if appropriate. Finally, embryos were washed briefly with PBST, equilibrated for 5 minutes in 50% glycerol 50% PBST, and then overnight in 100% glycerol to clear for visualization.
Fluorescence mRNA in situ hybridization (FISH) was performed as described in Talbot et al. (2010) with the addition of an overnight incubation in a 1:50,000 dilution of Sytox Green DNA stain (Life Technologies) to visualize cell nuclei. Cy3 TSA was used for developing.
Mineralized teeth were visualized by alizarin red staining. Embryos were fixed as described above, incubated in 0.5% KOH for 10 min, stained in 0.5% KOH + 0.0005% alizarin red for 2 hrs, washed in 0.5% KOH until background staining was eliminated (0-3 hrs), and then equilibrated into glycerol as above.
Mounting and microscopy
For confocal microscopy, whole embryos or larvae were first dissected with 1.5 mm insect pins to remove the yolk and heart, and to cut off the lower trunk and tail just posterior to the pectoral fins to facilitate positioning of the head. Heads were then placed ventral side up on a microscope slide in a very small drop of glycerol (~5-10 μl)and a 9 mm coverslip with modeling clay feet at each corner was placed on top. The coverslip was then pressed down slightly with fine forceps and moved from side to side if necessary to immobilize the head and position the tooth-forming region just beneath the coverslip in a level orientation.
Images were captured with a Zeiss 510 Meta laser scanning confocal microscope employing a Plan-Neofluar 40x 1.3 n.a. oil objective. All images presented were cropped to 225 × 225 μm or 75 × 75 μm, and represent either the entire posterior pharyngeal region or a closeup of the tooth-forming region on one side of the midline. 3D stacks for cell counts were taken at optimum z resolution as determined by the Zeiss software, typically ~0.5 μm. Brightness/contrast levels and color lookup tables were adjusted independently for each channel but evenly across the images with Fiji/ImageJ (Schindelin et al., 2012). Colors are presented in 2 and 3-color experiment figures as green (primary stain) and magenta (nuclear stain or second antibody) with double-label appearing white, as recommended to aid in visualization for color-blind readers (Wong, 2011). 4-color panels are presented with colors matching the fluorophores as accurately as possible, with the exception of the non-visible, far-red, 633 nm emission for laminin staining being represented as yellow.
Cyclopamine treatment, cell counts, and tooth measurements
Embryos were dechorionated and placed into 12-well plates in 1 ml of embryo medium just before chemical treatment. Cyclopamine (CyA) dissolved in EtOH, or EtOH alone as a control, was added to the medium to a final concentration of 50 μM CyA and/or 0.2% EtOH. Surrounding wells were filled with water to greatly reduce or eliminate evaporation. Treatment and control wells were made in randomized locations within the plates between experiments to control for possible position effects.
Dental papilla cells were quantified from confocal stacks, and tooth germ cell nuclei were counted from several different slice orientations using Volocity software (PerkinElmer). Cells of the dental papilla at 72 hpf in all treatments were found to have faint dlx2b:GFP expression which aided in their identification. This was especially helpful at the base of the dental papilla, where dlx2b:GFP-positive cells often bulged out slightly away from the dental epithelium and took on a flattened and slightly curved nuclear morphology that would have been difficult to distinguish as part of the dental papilla without the transgene marker.
To measure mineralized tooth morphology, confocal stacks were viewed with the Fiji/ImageJ 3D Viewer (Schmid et al., 2010; Schindelin et al., 2012). Landmarks were assigned with the points tool while freely rotating the 3D image, coordinates were recorded, and measurements were calculated separately by spreadsheet. Saved points files were later reloaded into the 3D images and the position of all landmarks double-checked.
Computation
Statistics and graphs were done with the assistance of Prism 6 (GraphPad Software, Inc.) and with spreadsheets from the Handbook of Biological Statistics (McDonald, 2009). Comparison of means were with unpaired, two-tailed Student’s t-tests. Schematic diagrams were composed in iDraw (Indeeo Inc.), and figures were assembled with Keynote (Apple Inc.) and Pixelmator (Pixelmator Team Ltd.).
Key Findings.
Hedgehog cell signaling is required for tooth dental papilla formation
Hedgehog pathway inhibition results in reduced tooth size
Subsequent tooth initiation also requires hedgehog signaling
Acknowledgements
We thank Kristin Artinger and John Rawls for sharing zebrafish transgenic reporter lines, Marko Melendy and Martha Mixon for animal husbandry and technical support, and David Rivers and an anonymous reviewer for insightful comments on the manuscript. The work was supported by an Institutional Development Award (IDeA) from the National Institute of General Medical Sciences of the National Institutes of Health under grant number P20GM0103423, and a Major Research Instrumentation award from the National Science Foundation.
References
- Bitgood MJ, McMahon AP. Hedgehog and Bmp genes are coexpressed at many diverse sites of cell-cell interaction in the mouse embryo. Dev Biol. 1995;172:126–138. doi: 10.1006/dbio.1995.0010. [DOI] [PubMed] [Google Scholar]
- Bryant PJ, Simpson P. Intrinsic and extrinsic control of growth in developing organs. Q Rev Biol. 1984;59:387–415. doi: 10.1086/414040. [DOI] [PubMed] [Google Scholar]
- Buchtova M, Handrigan GR, Tucker AS, Lozanoff S, Town L, Fu K, Diewert VM, Wicking C, Richman JM. Initiation and patterning of the snake dentition are dependent on Sonic hedgehog signaling. Dev Biol. 2008;319:132–145. doi: 10.1016/j.ydbio.2008.03.004. [DOI] [PubMed] [Google Scholar]
- Chen JK, Taipale J, Cooper MK, Beachy PA. Inhibition of Hedgehog signaling by direct binding of cyclopamine to Smoothened. Genes Dev. 2002;16:2743–2748. doi: 10.1101/gad.1025302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang C, Litingtung Y, Lee E, Young KE, Corden JL, Westphal H, Beachy PA. Cyclopia and defective axial patterning in mice lacking Sonic hedgehog gene function. Nature. 1996;383:407–413. doi: 10.1038/383407a0. [DOI] [PubMed] [Google Scholar]
- Cobourne MT, Hardcastle Z, Sharpe PT. Sonic hedgehog regulates epithelial proliferation and cell survival in the developing tooth germ. J Dent Res. 2001;80:1974–1979. doi: 10.1177/00220345010800110501. [DOI] [PubMed] [Google Scholar]
- Cobourne MT, Miletich I, Sharpe PT. Restriction of sonic hedgehog signalling during early tooth development. Development. 2004;131:2875–2885. doi: 10.1242/dev.01163. [DOI] [PubMed] [Google Scholar]
- Dassule HR, Lewis P, Bei M, Maas R, McMahon AP. Sonic hedgehog regulates growth and morphogenesis of the tooth. Development. 2000;127:4775–4785. doi: 10.1242/dev.127.22.4775. [DOI] [PubMed] [Google Scholar]
- Elworthy S, Hargrave M, Knight R, Mebus K, Ingham PW. Expression of multiple slow myosin heavy chain genes reveals a diversity of zebrafish slow twitch muscle fibres with differing requirements for Hedgehog and Prdm1 activity. Development. 2008;135:2115–2126. doi: 10.1242/dev.015719. [DOI] [PubMed] [Google Scholar]
- Fraser GJ, Bloomquist RF, Streelman JT. A periodic pattern generator for dental diversity. BMC Biol. 2008;6:32. doi: 10.1186/1741-7007-6-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaete M, Tucker AS. Organized emergence of multiple-generations of teeth in snakes is dysregulated by activation of Wnt/beta-catenin signalling. PLoS One. 2013;8:e74484. doi: 10.1371/journal.pone.0074484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gritli-Linde A, Bei M, Maas R, Zhang XM, Linde A, McMahon AP. Shh signaling within the dental epithelium is necessary for cell proliferation, growth and polarization. Development. 2002;129:5323–5337. doi: 10.1242/dev.00100. [DOI] [PubMed] [Google Scholar]
- Handrigan GR, Richman JM. Autocrine and paracrine Shh signaling are necessary for tooth morphogenesis, but not tooth replacement in snakes and lizards (Squamata) Dev Biol. 2010;337:171–186. doi: 10.1016/j.ydbio.2009.10.020. [DOI] [PubMed] [Google Scholar]
- Huysseune A, Sire J-Y. Early development of the zebrafish (Danio rerio) pharyngeal dentition (Teleostei, Cyprinidae) Anatomy and embryology. 1998;198:289–305. doi: 10.1007/s004290050185. [DOI] [PubMed] [Google Scholar]
- Huysseune A, Takle H, Soenens M, Taerwe K, Witten PE. Unique and shared gene expression patterns in Atlantic salmon (Salmo salar) tooth development. Dev Genes Evol. 2008;218:427–437. doi: 10.1007/s00427-008-0237-9. [DOI] [PubMed] [Google Scholar]
- Jackman WR, Draper BW, Stock DW. Fgf signaling is required for zebrafish tooth development. Dev Biol. 2004;274:139–157. doi: 10.1016/j.ydbio.2004.07.003. [DOI] [PubMed] [Google Scholar]
- Jackman WR, Stock DW. Transgenic analysis of Dlx regulation in fish tooth development reveals evolutionary retention of enhancer function despite organ loss. Proc Natl Acad Sci U S A. 2006;103:19390–19395. doi: 10.1073/pnas.0609575103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackman WR, Yoo JJ, Stock DW. Hedgehog signaling is required at multiple stages of zebrafish tooth development. BMC Dev Biol. 2010;10:119. doi: 10.1186/1471-213X-10-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jernvall J, Kettunen P, Karavanova I, Martin LB, Thesleff I. Evidence for the role of the enamel knot as a control center in mammalian tooth cusp formation: non-dividing cells express growth stimulating Fgf-4 gene. Int J Dev Biol. 1994;38:463–469. [PubMed] [Google Scholar]
- Kanther M, Sun X, Muhlbauer M, Mackey LC, Flynn EJ, 3rd, Bagnat M, Jobin C, Rawls JF. Microbial colonization induces dynamic temporal and spatial patterns of NF-kappaB activation in the zebrafish digestive tract. Gastroenterology. 2011;141:197–207. doi: 10.1053/j.gastro.2011.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki K, Suzuki T, Weiss KM. Phenogenetic drift in evolution: the changing genetic basis of vertebrate teeth. Proc Natl Acad Sci U S A. 2005;102:18063–18068. doi: 10.1073/pnas.0509263102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Koyama E, Yamaai T, Iseki S, Ohuchi H, Nohno T, Yoshioka H, Hayashi Y, Leatherman JL, Golden EB, Noji S, Pacifici M. Polarizing activity, Sonic hedgehog, and tooth development in embryonic and postnatal mouse. Dev Dyn. 1996;206:59–72. doi: 10.1002/(SICI)1097-0177(199605)206:1<59::AID-AJA6>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- Kronmiller JE, Nguyen T, Berndt W, Wickson A. Spatial and temporal distribution of sonic hedgehog mRNA in the embryonic mouse mandible by reverse transcription/polymerase chain reaction and in situ hybridization analysis. Arch Oral Biol. 1995;40:831–838. doi: 10.1016/0003-9969(95)00053-r. [DOI] [PubMed] [Google Scholar]
- Laurenti P, Thaeron C, Allizard F, Huysseune A, Sire JY. Cellular expression of eve1 suggests its requirement for the differentiation of the ameloblasts and for the initiation and morphogenesis of the first tooth in the zebrafish (Danio rerio) Dev Dyn. 2004;230:727–733. doi: 10.1002/dvdy.20080. [DOI] [PubMed] [Google Scholar]
- Lawson ND, Weinstein BM. In vivo imaging of embryonic vascular development using transgenic zebrafish. Dev Biol. 2002;248:307–318. doi: 10.1006/dbio.2002.0711. [DOI] [PubMed] [Google Scholar]
- Lench NJ, Telford EA, High AS, Markham AF, Wicking C, Wainwright BJ. Characterisation of human patched germ line mutations in naevoid basal cell carcinoma syndrome. Hum Genet. 1997;100:497–502. doi: 10.1007/s004390050541. [DOI] [PubMed] [Google Scholar]
- McDonald J. Handbook of Biological Statistics. Sparky House Publishing; Baltimore: 2009. [Google Scholar]
- Motoyama J, Takabatake T, Takeshima K, Hui C. Ptch2, a second mouse Patched gene is co-expressed with Sonic hedgehog. Nat Genet. 1998;18:104–106. doi: 10.1038/ng0298-104. [DOI] [PubMed] [Google Scholar]
- Murphey RD, Stern HM, Straub CT, Zon LI. A chemical genetic screen for cell cycle inhibitors in zebrafish embryos. Chem Biol Drug Des. 2006;68:213–219. doi: 10.1111/j.1747-0285.2006.00439.x. [DOI] [PubMed] [Google Scholar]
- Nakatomi M, Morita I, Eto K, Ota MS. Sonic hedgehog signaling is important in tooth root development. J Dent Res. 2006;85:427–431. doi: 10.1177/154405910608500506. [DOI] [PubMed] [Google Scholar]
- Nanni L, Ming JE, Bocian M, Steinhaus K, Bianchi DW, Die-Smulders C, Giannotti A, Imaizumi K, Jones KL, Campo MD, Martin RA, Meinecke P, Pierpont ME, Robin NH, Young ID, Roessler E, Muenke M. The mutational spectrum of the sonic hedgehog gene in holoprosencephaly: SHH mutations cause a significant proportion of autosomal dominant holoprosencephaly. Hum Mol Genet. 1999;8:2479–2488. doi: 10.1093/hmg/8.13.2479. [DOI] [PubMed] [Google Scholar]
- Nanni L, Ming JE, Du Y, Hall RK, Aldred M, Bankier A, Muenke M. SHH mutation is associated with solitary median maxillary central incisor: a study of 13 patients and review of the literature. Am J Med Genet. 2001;102:1–10. doi: 10.1002/1096-8628(20010722)102:1<1::aid-ajmg1336>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- Poole DFG. Phylogeny of tooth tissues: enameloid and enamel in recent vertebrates with a note in the history of cementum. In: Miles AEW, editor. Structural and chemical organization of teeth. Academic Press; New York: 1967. pp. 111–149. [Google Scholar]
- Riddle RD, Johnson RL, Laufer E, Tabin C. Sonic hedgehog mediates the polarizing activity of the ZPA. Cell. 1993;75:1401–1416. doi: 10.1016/0092-8674(93)90626-2. [DOI] [PubMed] [Google Scholar]
- Rothova M, Peterkova R, Tucker AS. Fate map of the dental mesenchyme: dynamic development of the dental papilla and follicle. Dev Biol. 2012;366:244–254. doi: 10.1016/j.ydbio.2012.03.018. [DOI] [PubMed] [Google Scholar]
- Salazar-Ciudad I. Tooth patterning and evolution. Curr Opin Genet Dev. 2012;22:585–592. doi: 10.1016/j.gde.2012.10.006. [DOI] [PubMed] [Google Scholar]
- Sasagawa I. Fine structure of tooth germs during the formation of enameloid matrix in Tilapia nilotica, a teleost fish. Arch Oral Biol. 1995;40:801–814. doi: 10.1016/0003-9969(95)00050-y. [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid B, Schindelin J, Cardona A, Longair M, Heisenberg M. A high-level 3D visualization API for Java and ImageJ. BMC Bioinformatics. 2010;11:274. doi: 10.1186/1471-2105-11-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seritrakul P, Samarut E, Lama TT, Gibert Y, Laudet V, Jackman WR. Retinoic acid expands the evolutionarily reduced dentition of zebrafish. FASEB J. 2012;26:5014–5024. doi: 10.1096/fj.12-209304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shellis R, Miles A. Autoradiographic study of the formation of enameloid and dentine matrices in teleost fishes using tritiated amino acids. Proceedings of the Royal Society of London. 1974:51–72. Series B, Biological Sciences. [Google Scholar]
- Sire JY, Donoghue PC, Vickaryous MK. Origin and evolution of the integumentary skeleton in non-tetrapod vertebrates. J Anat. 2009;214:409–440. doi: 10.1111/j.1469-7580.2009.01046.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MM, Fraser GJ, Chaplin N, Hobbs C, Graham A. Reiterative pattern of sonic hedgehog expression in the catshark dentition reveals a phylogenetic template for jawed vertebrates. Proc Biol Sci. 2009;276:1225–1233. doi: 10.1098/rspb.2008.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorrells S, Toruno C, Stewart RA, Jette C. Analysis of apoptosis in zebrafish embryos by whole-mount immunofluorescence to detect activated Caspase 3. J Vis Exp. 2013:e51060. doi: 10.3791/51060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanger BZ, Tanaka AJ, Melton DA. Organ size is limited by the number of embryonic progenitor cells in the pancreas but not the liver. Nature. 2007;445:886–891. doi: 10.1038/nature05537. [DOI] [PubMed] [Google Scholar]
- Talbot JC, Johnson SL, Kimmel CB. hand2 and Dlx genes specify dorsal, intermediate and ventral domains within zebrafish pharyngeal arches. Development. 2010;137:2507–2517. doi: 10.1242/dev.049700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ten Berge D, Brouwer A, Korving J, Martin JF, Meijlink F. Prx1 and Prx2 in skeletogenesis: roles in the craniofacial region, inner ear and limbs. Development. 1998;125:3831–3842. doi: 10.1242/dev.125.19.3831. [DOI] [PubMed] [Google Scholar]
- Thesleff I, Barrach HJ, Foidart JM, Vaheri A, Pratt RM, Martin GR. Changes in the distribution of type IV collagen, laminin, proteoglycan, and fibronectin during mouse tooth development. Dev Biol. 1981;81:182–192. doi: 10.1016/0012-1606(81)90361-4. [DOI] [PubMed] [Google Scholar]
- Thesleff I, Hurmerinta K. Tissue interactions in tooth development. Differentiation. 1981;18:75–88. doi: 10.1111/j.1432-0436.1981.tb01107.x. [DOI] [PubMed] [Google Scholar]
- Thomas BL, Tucker AS, Qui M, Ferguson CA, Hardcastle Z, Rubenstein JL, Sharpe PT. Role of Dlx-1 and Dlx-2 genes in patterning of the murine dentition. Development. 1997;124:4811–4818. doi: 10.1242/dev.124.23.4811. [DOI] [PubMed] [Google Scholar]
- Vaahtokari A, Aberg T, Jernvall J, Keranen S, Thesleff I. The enamel knot as a signaling center in the developing mouse tooth. Mech Dev. 1996;54:39–43. doi: 10.1016/0925-4773(95)00459-9. [DOI] [PubMed] [Google Scholar]
- Van der heyden C, Huysseune A, Sire JY. Development and fine structure of pharyngeal replacement teeth in juvenile zebrafish (Danio rerio) (Teleostei, Cyprinidae) Cell Tissue Res. 2000;302:205–219. doi: 10.1007/s004410000180. [DOI] [PubMed] [Google Scholar]
- Verstraeten B, van Hengel J, Sanders E, Van Roy F, Huysseune A. N-cadherin is required for cytodifferentiation during zebrafish odontogenesis. J Dent Res. 2013;92:365–370. doi: 10.1177/0022034513477424. [DOI] [PubMed] [Google Scholar]
- Wong B. Color blindness. Nat Methods. 2011;8:441. doi: 10.1038/nmeth.1618. [DOI] [PubMed] [Google Scholar]