Abstract
Transcriptional elongation is critical for gene expression regulation during embryogenesis. The super elongation complex (SEC) governs this process by mobilizing paused RNA polymerase II (RNAP2). Using exome sequencing, we discovered missense mutations in AFF4, a core component of the SEC in three unrelated probands with a novel syndrome that phenotypically overlaps Cornelia de Lange syndrome (CdLS), that we have named CHOPS syndrome (C for Cognitive impairment and Coarse facies, H for Heart defects, O for Obesity, P for Pulmonary involvement and S for Short stature and Skeletal dysplasia). Transcriptome and chromatin immunoprecipitation sequencing (ChIP-seq) analyses demonstrated similar alterations of genome-wide binding of AFF4, cohesin and RNAP2 between CdLS and CHOPS syndrome. Direct molecular interaction between SEC, cohesin and RNAP2 was demonstrated. This data supports a common molecular pathogenesis for CHOPS syndrome and CdLS caused by disturbance of transcriptional elongation due to alterations in genome-wide binding of AFF4 and cohesin.
Introduction
Transcriptional regulation during embryogenesis is of paramount importance in determining the precise temporospacial gene expression pattern. Recent evidence suggests that such transcriptional control is mainly achieved by the regulation of transcriptional elongation1. Approximately 50 base pairs after transcriptional initiation, RNA polymerase II (RNAP2) pauses in genes whose expression is mainly controlled by elongation phase. For this transcriptional pausing, NELF(negative elongation factor) and DSIF(DRB Sensitivity Inducing Factor)(a heterodimer of SPT4 and SPT5), are required 2. Serine residues located within the C-terminus heptapeptide repeats of RNAP2 undergo phosphorylation modifications during gene transcription. Ser5 phosphorylated RNAP2 (RNAP2 Ser5ph) is enriched at the promoter proximal region and Ser2 phosphorylation occurrs after transcriptional elongation starts. Therefore, Ser2-phosphorylated RNAP2 (RNAP2 Ser2ph) is regarded as actively elongating RNAP2 3. Mobilization of the paused RNAP2 machinery is governed by the super elongation complex (SEC) which is comprised of multiple proteins including AFF4, ELL2 and the positive transcription elongation factor b (P-TEFb) which is a heterodimer of CDK9 and cyclin T14. CDK9 phosphorylates the Ser2 residue of RNAP2 in order for initiation of transcriptional elongation 3.
Since many multiple congenital anomaly genetic conditions are due to the disruption of proper transcriptional processes during embryogenesis, disruption of the transcriptional elongation process would be a likely underlying pathogenic mechanism for developmental disorders however to date no causal links have been identified. Through the characterization of a novel genetic disorder caused by gain-of-function mutations in the AFF4 gene, encoding a critical component of the SEC, we have established a link between the dysregulated transcription observed amongst disorders of cohesin function (collectively termed “cohesinopathies”) and disruption of RNAP2 elongation, caused by altered genome-wide binding of AFF4 and cohesin.
Results
A novel genetic disorder caused by AFF4 missense mutations
Germline mutations of cohesin complex structural and regulatory components cause Cornelia de Lange syndrome (CdLS) (OMIM 122740, 300590 and 610759)5. CdLS is a multisystem developmental disorder characterized by craniofacial dysmorphisms, intellectual disabilities, growth retardation, limb anomalies and several other systemic abnormalities5. Mutations in NIPBL have been identified in nearly 60% of CdLS probands, and HDAC8, SMC1A, SMC3 and RAD21 mutations account for an additional small portion of patients with CdLS6–9. The probands (CHOPS T254S, CHOPS T254A and CHOPS R258W) found to have AFF4 mutations were originally suspected of having CdLS, due to intellectual disability, short stature and craniofacial dysmorphisms (Fig.1a). However, clinically, their physical features are distinctively different from typical CdLS probands and allowed us to classify this as a novel clinical entity (Supplementary Table 1). We propose “CHOPS syndrome” as an acronym to describe this novel genetic disorder: ‘C’ for Cognitive impairment and Coarse facies, ‘H’ for Heart defects, ‘O’ for Obesity, ‘P’ for Pulmonary involvement and ‘S’ for Short stature and Skeletal dysplasia.
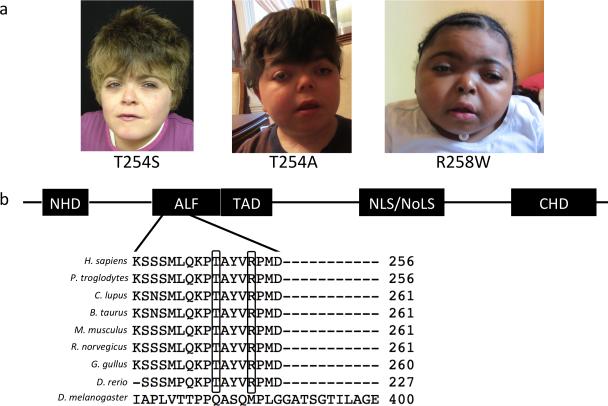
Figure 1.
Identification of novel genetic disorder and its causative gene. (a) CHOPS syndrome probands: CHOPS T254S (female with short stature, intellectual disability, chronic lung disease, obesity, brachydactyly, vertebral abnormalities, patent ductus aretriosus, horseshoe kidney and dysmorphic facial features), CHOPS T254A (male with short stature, intellectual disability, tracheomalacia, subglottic and tracheal stenosis, obesity, brachydactyly, cervical vertebrae abnormalities, ventricular septal defect and patent ductus arteriosus, cryptorchidism, hearing loss and dysmorphic facial features) and CHOPS R258W (female with short stature, intellectual disability, laryngomalacia, narrow oropharynx, brachydactyly, kyphoscoliosis, patent ductus arteriosus, ventricular septal defect, cataracts and dysmorphic facial features). Written permission to publish photograph was obtained from the parents of CHOPS syndrome probands. (b) AFF4 protein structure demonstrating the location of de novo missense mutations identified in 3 probands. Missense mutations altered highly conserved amino acid residues. NHD: N-terminal homology domain, TAD: transactivation domain, NLS: nuclear localization signal, NoLS: nucleolar localization signals, CHD: C-terminal homology domain.
The striking phenotypic similarities between the three unrelated probands, all born to unaffected non-consanguineous parents, led us to hypothesize that their clinical features were likely the result of a detrimental germline de novo mutation in the same gene. To test this hypothesis, we performed exome sequencing of these three probands. Each proband had 414 (CHOPS T254A), 720 (CHOPS R258W), 725 (CHOPS T254S) rare deleterious mutations (non-synonymous, protein truncating, deletion/duplication or splice site mutations) respectively. Exome statistics are listed in the Supplementary Table 2. Among these, variants in 16 genes were identified in common to all probands. However, the variants in 14 genes were found to also be present in in-house control samples, arguing against causality. Variants in the remaining two genes, AFF4 and ZFHX3, were scrutinized further. Genetic variants of AFF4 and ZFHX3 were confirmed by Sanger sequencing. All of the missense mutations found in AFF4 (c.760A>G (p.Thr254Ala), c.761C>G (p.Thr254Ser) and c.772C>T (p.Arg258Trp)) were de novo (not present in the 6 biological parents of the 3 probands) (Fig.1b and Supplementary Fig.1). All 3 missense mutations were located within the ALF (AF4/LAF4/FMR2) homology domain of AFF4, and these missense mutations altered highly evolutionarily conserved amino acids (Fig.1b). All of the identified ZFHX3 variants were inherited from one of the parents of each proband (all of whom were unaffected). Paternity was confirmed by STS markers in all three probands. Screening of an additional 25 probands with atypical features of CdLS for mutations in the AFF4 gene failed to identify mutations. However none of these probands exactly fit the CHOPS syndrome phenotype indicating that mutations in this gene are highly correlated with the specific phenotype as characterized by the 3 patients described here. Collectively this data supports that the missense mutations in the ALF homology domain of AFF4 cause CHOPS syndrome.
Mechanism of AFF4 mutations leading to CHOPS syndrome
A missense mutation in the ALF homology domain of Aff1 (Af4) was reported in the robotic mouse, an ataxia mouse model created by ENU mutagenesis10. The pathogenetic mechanism of this missense mutation is a gain-of-function effect due to decreased clearance of the protein by SIAH1 ubiquitin E3 ligase11. Given the vicinity of the location of the missense mutations in our probands to that described in the robotic mouse, we hypothesized that the missense mutations found in the three probands would likewise disrupt the ubiquitinylation-dependent proteasomal degradation of the AFF4 protein. To test this hypothesis, we created an AFF4 and SIAH1 overexpression model using HEK293T cells. When a wild type (WT) AFF4 expression vector was transfected with an SIAH1 expression vector, the amount of AFF4 protein was significantly decreased; however the AFF4 constructs containing the missense mutations found in the three CHOPS syndrome probands hindered the degradation of AFF4 with SIAH1 overexpression (Fig. 2a). The addition of MG132, which is a proteosomal degradation inhibitor, resulted in the recovery of AFF4 bands.
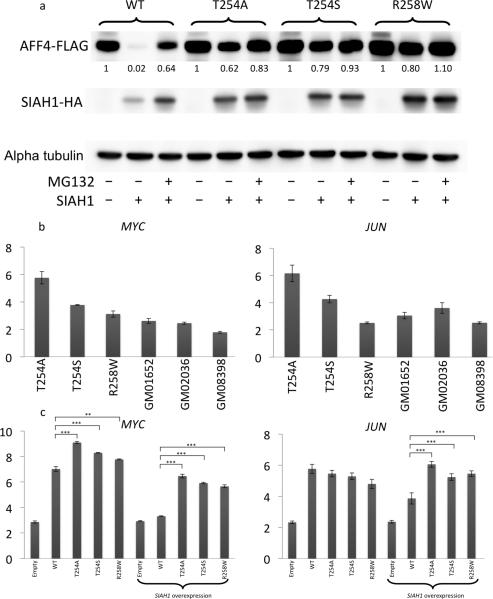
Figure 2.
Disease mechanism of CHOPS syndrome. (a) Decreased proteosomal degradation of mutant AFF4 in 293T cells. Western blot demonstrates disappearance of WT AFF4 bands with the addition of SIAH1 vector. However, such disappearance was not observed in 293T cells overexpressing mutant AFF4 vectors. The numbers beneath AFF4 bands indicate the signal intensities normalized to the band intensity of AFF4 only transfection condition of each category and alpha tubulin. (b, c) Expression level of MYC/JUN gene in patient-derived skin fibroblasts and 293T cell line with AFF4 overexpression. GM01652, GM02036 and GM08398 are control fibroblast cell lines. Elevation of MYC and JUN expression were observed in CHOPS syndrome skin fibroblast (2b) and 293T AFF4 overexpression model (2c). MYC and JUN expression was normalized against TBP. Error bars demonstrate mean ± 2 standard deviations. **P<0.01, ***P<0.001, two-tailed t-test, n= 3 per group.
Several genes including MYC and JUN have been identified as direct transcriptional targets of AFF412. Upregulation of MYC and JUN expression was confirmed in patient derived skin fibroblast samples by quantitative RT-PCR, although one proband with an AFF4 p.Arg258Trp alteration had a normal expression level of JUN (Fig. 2b). The expression levels of MYC and JUN were also evaluated in HEK293T cells with AFF4 overexpression vectors. With WT AFF4 overexpression, the expression level of MYC and JUN increased, confirming that MYC and JUN are downstream targets of the AFF4 protein. Overexpression of the mutant AFF4 construct resulted in a stronger activation of MYC expression. Without co-expression of SIAH1, the difference between WT AFF4 and mutant AFF4 overexpression was minimal, however, with the addition of an SIAH1 overexpression vector, the expression difference between WT and mutant constructs increased both for MYC and JUN (Fig. 2c).
Since the SEC plays an important role in the precise regulation of the immediate gene responses to various stimuli such as heat shock, retinoic acid or serum1,4 as well as developmental regulation, we evaluated the effects of the AFF4 missense mutations on the immediate gene response using patient-derived skin fibroblast cell lines. Patient skin fibroblast cell lines demonstrated up-regulation of the FOS and EGR1 genes, which was comparable to control cell lines, upon serum stimulation (Supplementary Fig.2). Therefore, the probands’ cell lines do not demonstrate an impaired response to serum stimulation in regards to the gene expression level of FOS and EGR1, and the mutant cells still maintain the capability of mounting a response to various stimuli.
Transcriptome analysis of CHOPS syndrome
Due to the critical role of the SEC in transcriptional regulation we evaluated for global gene expression abnormalities. We performed genome-wide expression profiling using Affymetrix U133plus2 chips on patient derived skin fibroblast cell lines. Two patient samples (CHOPS T254A and CHOPS T254S) and three age gender-matched control samples were used, with all samples run in duplicates. CHOPS R258W cell line was not used due to the lack of an appropriate ethnicity-matched control cell line. Using an FDR cutoff of 0.2, 288 genes were downregulated in patient skin fibroblasts, and 445 genes were upregulated in patient samples (Supplementary Table 3 and Supplementary Fig. 3). Gene ontogeny term analysis using DAVID revealed that upregulated genes are enriched for Homeobox proteins, skeletal system development/morphogenesis and anterior/posterior pattern formation as well as embryonic organ development/morphogenesis, and downregulated genes are enriched for actin binding genes and extracellular matrix genes (Supplementary Table 4). Previously, Luo et al. reported the results of AFF4 ChIP-seq and RNA-seq after siRNA knockdown of AFF412. Combining the data reported by Luo et al., we selected 7 genes as probable direct targets of AFF4, due to the presence of an AFF4 peak near their promoter regions and downregulation of their expression levels upon AFF4 knockdown. All 7 target genes were up-regulated by 9% to 127% (mean = 48.9%) in patients with AFF4 mutations compared to control samples (Table 1 and Supplementary Fig. 4). The up-regulation of 5 of these genes was statistically significant: MYC, JUN, TMEM100, ZNF711 and FAM13C. These observations further support the notion that AFF4 mutations render gain-of-function effects.
Table 1.
Expression Levels of AFF4 Target Genes.
| Gene | Gene Name | Mean-Control | Mean-Patient | Change In CHOPS | P value |
|---|---|---|---|---|---|
| ADAMTS1 | ADAM metallopeptidase with thrombospondin type 1 motif, 1 | 8.822 | 8.946 | +9.0% | 0.778 |
| FAM13C | family with sequence similarity 13, member C | 2.720 | 3.063 | +26.8% | 0.013 |
| IMMP2L | IMP2 inner mitochondrial membrane peptidase-like (S. cerevisiae) | 6.850 | 7.008 | +11.6% | 0.095 |
| JUN | jun proto-oncogene | 8.794 | 9.257 | +37.9% | 0.030 |
| MYC | v-myc myelocytomatosis viral oncogene homolog (avian) | 7.667 | 8.571 | +87.2% | 0.030 |
| TMEM100 | transmembrane protein 100 | 3.179 | 4.364 | +27.4% | 0.001 |
| ZNF711 | zinc finger protein 711 | 2.723 | 3.565 | +79.3% | 0.001 |
In this table, the group means are average expression levels of genes measured by microarrays, after normalization and log2-transformation. The processed measurements were compared between groups by the SAM method to generate the p values, and the un-log transformed measurements were used to calculate the percentage of expression change between groups. See details in Methods.
Transcriptome Similarities between CHOPS syndrome and CdLS
Given the phenotypic similarities between the three CHOPS syndrome probands with AFF4 mutations and individuals with CdLS, we compared the gene expression pattern of skin fibroblast samples with AFF4 mutations to those from CdLS probands with NIPBL frameshift mutations using the Affymetrics Human Gene 2.0 ST array. A general positive correlation (Pearson's correlation coefficient = 0.34, p < e-300) of gene expression pattern between CHOPS syndrome and CdLS was seen (Fig. 3a). We identified the top 250 dysregulated (over-expressed and under-expressed) genes in these two diagnoses. Expression levels of up- and down-regulated genes were similar between the CdLS probands and the probands with AFF4 mutations (Fig. 3b and 3c). Likewise, the expression levels of CdLS down-regulated genes were similarly under-expressed in the probands with AFF4 mutations. Dysregulated genes in CHOPS syndrome demonstrated a similar expression pattern as in the CdLS probands’ samples (Fig. 3b and 3c). There were 29 common genes within the top 250 most up-regulated genes in CdLS and CHOPS (odds ratio = 13.05, p = 4.9e-21, Fisher's test). On the other hand, the top 250 most down-regulated genes in CdLS and CHOPS syndrome included 27 common genes (odds ratio = 11.04, p = 4.4e-18, Fisher's test). For further analyses, we established hTERT immortalized cell lines from CHOPS syndrome as well as CdLS patients’ fibroblasts and performed total RNA-seq analyses (Supplementary Table 5). We observed significant positive correlations between transcriptional changes in CHOPS and CdLS (Supplementary Fig. 5) as we did for primary cultured cells. As summarized in Supplementary Fig.5b and 5c, we scored 302 up-regulated genes and 216 down-regulated genes in CHOPS syndrome (Supplementary Table 6). Among 78 up-regulated genes and 220 down-regulated genes in CdLS, 30 and 72 genes showed significantly similar tendencies to that observed in CHOPS syndrome, respectively. Although CHOPS R258W sample did not show overexpression of JUN in quantitative RTPCR (Fig. 2b), the profile of dysregulated genes are very similar in CHOPS R258W, compared to CHOPS T254A and CHOPS T254A (Supplementary Fig. 5d)
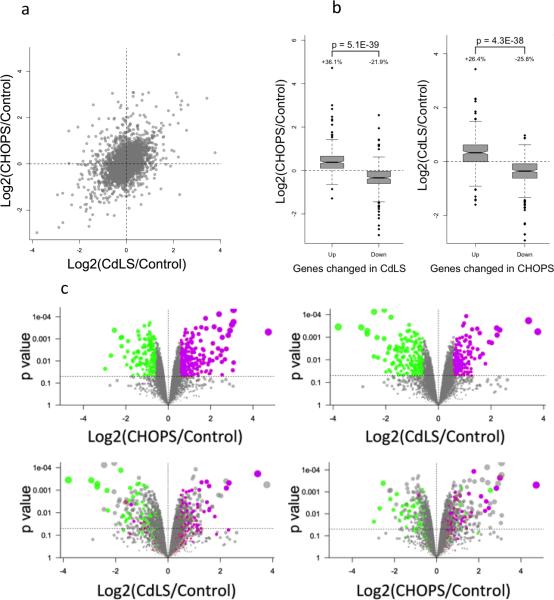
Figure 3.
Similar transcriptional profile between CHOPS syndrome and CdLS. (a) General comparison between CHOPS syndrome and CdLS demonstrated positive correlation between these two syndromes. (b) Top 250 dysregulated genes comparison. Left: Upregulated genes and downregulated genes in CdLS showed similar changes to CHOPS syndrome samples. Right: Dysregulated genes in CHOPS syndrome demonstrated similar changes to CdLS samples. The bottom of the box represents the 1st quartile, and the top of the box represents the 3rd quartile. The line in the middle of the box represents the median. The notches/whiskers demonstrate the confidence interval. (c) Volcano plots: Top left: volcano plots defining top 250 genes whose expression levels are higher (purple dots) and lower (green dots) in the CHOPs syndrome samples. These plots represent the top 250 genes that are either up- or down-regulated in the probands with the highest magnitude of difference and p < 0.05. Bottom left: Distribution of these 250 genes, whose expression is up- and down-regulated in CHOPS syndrome, in the CdLS samples. The same genes were labeled by the same colors in the bottom plot. Top right: Distribution of the top 250 genes whose expression was up- (purple dots) or down-regulated (green dots) in the CdLS syndrome samples. Bottom right: Distribution of these 250 genes, whose expression is up- or down-regulated in CdLS, in the CHOPS syndrome samples.
Chromatin accumulation of AFF4 in CHOPS syndrome
With the confirmation that the missense mutations found in the three CHOPS syndrome probands caused a decreased clearance of mutated AFF4 protein due to the acquired resistance to SIAH1-mediated proteasomal degradation, we then analyzed the amount of chromatin-associated SEC component in the CHOPS syndrome cell lines. The amount of AFF4 protein in patient derived skin fibroblasts was elevated, supporting the suggestion that the missense mutations found in these probands results in a more stable AFF4 protein (Fig.4a). The mRNA levels of AFF4 were comparable between probands’ and control samples (Supplementary Fig. 6a). In the probands’ samples, the wild type and mutant alleles were expressed in almost equal amounts (Supplementary Fig. 6b). We further analyzed the amount of chromatin associated AFF4, ELL2, and CDK9 in CHOPS syndrome cell lines and found that accumulation of AFF4 mainly occurred on the chromatin fraction in the CHOPS syndrome cell lines (Fig.4b). The increase in AFF4 accumulation in CHOPS syndrome T254A was due to the mutant AFF4 allele, since, in cell lines where the mutated AFF4 allele was deleted, the amount of AFF4 protein was clearly decreased (Fig.4b and Supplementary Fig. 7). The amount of other SEC components such as ELL2 and CDK9 did not show major alterations in the chromatin fraction of the CHOPS syndrome cell lines. The CdLS cell line with an NIPBL frameshift mutation (CDL006) did not show the elevation of any of these proteins (Fig. 4b).
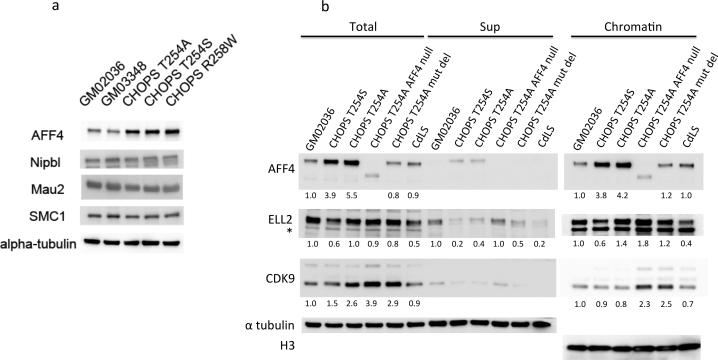
Figure 4.
Western blot analysis of the SEC components in CHOPS syndrome skin fibroblast cell lines. (a) CHOPS syndrome sample cell lysates demonstrate a stronger AFF4 band, however, the amounts of NIPBL, MAU2 and SMC1 were unchanged compared to the control samples. GM02036 and GM03348 are control samples. (b) Accumulation of AFF4 in the chromatin fraction was seen in the CHOPS syndrome samples, however, the amount of ELL2 and CDK9 remained unchanged. Numbers beneath the bands represents the signal intensities normalized against the band intensity of the control GM02036 sample, and also normalized against alpha tubulin in total cellular lysates and histone H3 in chromatin fraction samples. *The asterisk indicates a nonspecific band. CdLS sample used was CDL006 with a frameshift mutation of NIPBL.
Genome-wide chromatin binding of AFF4, RNAP2 and cohesin
To better understand the molecular mechanism leading to the similar transcriptional profiles between CHOPS syndrome and CdLS, we evaluated the genome-wide binding patterns of AFF4, elongating RNAP2, paused RNAP2, SPT5, and the cohesin subunit, RAD21, by performing chromatin immunoprecipitation sequencing (ChIP-seq) (Supplementary Table 7). To understand the shared characteristics of CdLS and CHOPS syndrome, we focused on differentially expressed genes in these two syndromes compared with a healthy control cell line.
Overall down-regulation and up-regulation of genes at the level of RNA correlated well with increasing or decreasing levels of RNAP2, elongating RNAP2 (Ser2ph), paused RNAP2 (Ser5ph) and SPT5 in both CdLS and CHOPS syndrome (Supplementary Fig. 8a and 8b). AFF4 binding around the TSS (transcription start site) was significantly elevated both in the CHOPS syndrome and CdLS samples among the transcriptionally up-regulated genes (Supplementary Fig. 8c and 8d). On the other hand, among transcriptionally down-regulated genes, such enhanced TSS binding of AFF4 was not observed (Supplementary Fig. 8c and 8d). However in both syndromes, we could not find a correlation between changes in AFF4 binding and transcription (Supplementary Fig. 8e). Interestingly, we observed an increase of RAD21 binding compared to the control at the TSSs of up-regulated genes and the opposite effect at TSSs of down-regulated genes (Supplementary Fig. 8c and 8d).
Molecular Interactions among SEC, cohesin and RNAP2
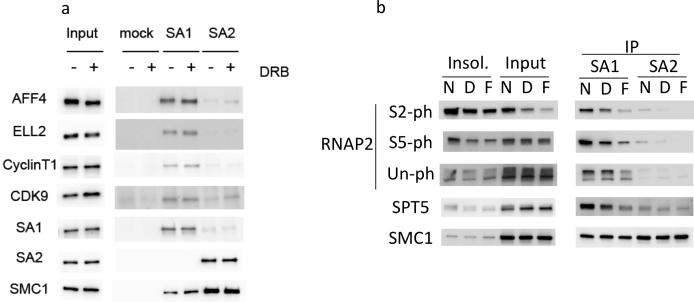
These findings prompted us to examine the physical interaction between SEC, cohesin and RNAP2 (Fig.5). In HeLa cell lysate, immunoprecipitation with a STAG1 (SA1) antibody yielded positive Western blot bands for various SEC components including AFF4, ELL2, CyclinT1 and CDK9, however, with a STAG2 (SA2) antibody, such interactions were not observed (Fig. 5a). Furthermore, we found that RNAP2 forms a complex with SMC1 and STAG1 (SA1) but not with STAG2 (SA2) in HeLa cell lysate (Fig. 5b). Phosphorylated forms of RNAP2 were preferentially enriched in the co-immunoprecipitated fraction with STAG1, but not with STAG2. Addition of elongation inhibitors such as DRB (5,6-Dichlorobenzimidazole 1-β-D-ribofuranoside) and flavopiridol, which is a CDK9 kinase inhibitor, decreased the amount of both RNAP2 Ser2ph and Ser5ph, precipitated with STAG1. Therefore, RNAP2 interacting with STAG1 is primarily the elongating form of RNAP2.
Figure 5.
Molecular interaction among SEC, cohesin and RNAP2. Results of immunoprecipitation-Western blot. (a) Protein interaction between SEC and cohesin. STAG1 (SA1) interacts with various SEC components including AFF4, ELL2, Cyclin T1 and CDK9. Immunoprecipitation with STAG2 (SA2) did not show such interaction with SEC. Addition of DRB slightly increased the amount of SEC components interacting with SA1. (b) Protein interaction between cohesin and RNAP2. STAG1 (SA1) interacts with various forms of RNAP2. Addition of DRB (D) and flavopiridol (F) decreased the amount of RNAP2 Ser2ph and Ser5ph precipitated with STAG1. RNAP2 was not immunoprecipitated with STAG2 (SA2) antibody.
Discussion
Through the identification of gain-of-function mutations in AFF4 as the cause of a novel syndrome consisting of a highly conserved pattern of features that we have termed CHOPS syndrome, we demonstrate that an altered RNAP2 distribution and resultant transcriptional elongation abnormalities underly the transcriptional dysregulation of CHOPS syndrome as well as CdLS.
CHOPS syndrome represents the first human developmental disorder caused by germline mutations in an SEC component, although synonymous mutations were previously suggested to correlate with autism in two individuals13,14. AFF4 is a scaffold protein comprising the core component of the SEC. In the probands reported here, the missense mutations create a resistance to ubiquitination-dependent proteasomal degradation, resulting in excessive amounts of mutant AFF4 protein accumulation. This persistence of AFF4 protein in turn results in an altered genomic distribution of AFF4 and leads to the unique transcriptome pattern of CHOPS syndrome. The transcriptome effects of AFF4 gain-of function mutations seen in CHOPS syndrome were opposite to that observed in AFF4 knock-down experiments, confirming that the disease mechanism of these mutant AFF4 proteins is a gain-of-function rather than haploinsufficiency or a dominant negative effect. In addition the phenotype of the CHOPS syndrome probands is different from that of the Aff4 knock-out mouse as well as in human subjects with genomic deletions encompassing the AFF4 gene lending further support to a gain-of-function effect15,16. Comparing the AFF4 binding patterns among the up-regulated genes, and down-regulated genes, AFF4 alteration was seen only in the up-regulated genes, further supporting the notion that the primary effect of AFF4 mutations found in CHOPS syndrome is a gain-of-function, leading to transcriptional activation (Supplementary Fig. 9). The fundamental role of transcriptional elongation in gene expression control during embryogenesis has been better understood recently1. Therefore, CHOPS syndrome can be regarded as a multiple congenital anomaly syndrome caused by disturbance of the transcriptional elongation process.
Although accumulation of AFF4 was observed in the transcriptionally activated genes in CHOPS syndrome and CdLS, there was no correlation between AFF4 ChIP-seq and RNA-seq/RNAP2 Ser2ph ChIP-seq in either syndrome (Supplementary Fig. 8). This finding suggests that AFF4 controls gene expression for only a subset of AFF4-bound genes, which is consistent with previously reported data by Luo et al., demonstrating that the knock down of AFF4 leads to down-regulation of expression of only a very small subset of genes (7 genes) among the genes that harbor an AFF4 ChIP-seq peak around TSS in 293T cell line 12. In CHOPS syndrome and CdLS, transcriptional alterations are more likely seen in the genes where expression is mainly regulated by the transcriptional elongation step. Similar findings were reported for other transcription elongation controller molecules such as DSIF and NELF 2,17.
In the CHOPS syndrome fibroblast cell lines, even with the massive accumulation of AFF4 in the chromatin fraction, the amount of other SEC components (i.e. ELL2 and CDK9) remained unchanged. However, the amount of these proteins forming SEC is likely to be increased, given that the primary effect seen in CHOPS syndrome is activation of AFF4 target genes. CDK9 is known to form a complex with BRD4 or 7SK RNA and HEXIM outside of the SEC as well as existing as a free CDK9 molecule 18–21. Therefore, the amount of CDK9 forming SEC is likely to be increased with the concurrent decrease of CDK9 forming complexes with BRD4 or 7SK RNA and HEXIM or free CDK9 in the nucleus. This speculation is supported by the fact that transfection of AFF4 alone exerted transcriptional activation effects on SEC target genes such as MYC or JUN, even without cooverexpression of other SEC components (Fig. 2c).
AFF4 is expressed in the fetal brain, lung and heart, and adult brain, heart, skeletal muscle and pancreatic tissues 22. It is very intriguing that all CHOPS syndrome probands manifested with chronic lung disease, given the strong expression of AFF4 in fetal lung. In addition, AFF4 is implicated in the regulation of the appetite, because hypothalamic AFF4 expression is up-regulated upon fasting 23. Therefore, it is possible that gain-of-function mutations of AFF4 have led to increased appetite, resulting the obesity seen in CHOPS syndrome probands.
While CHOPS syndrome and CdLS are clinically recognizable distinct entities there is some phenotypic overlap between these two diagnoses suggesting a common underlying pathogenesis. Given the apparently divergent molecular etiologies of each we were somewhat surprised to discover striking similarities in the transcriptome profile between CHOPS syndrome and CdLS.
Effects on gene transcription by mutated cohesin components/regulators has been proposed as the pathogenic mechanism of CdLS establishing a critical role for cohesin in transcriptional regulation 24. One mechanism by which cohesin controls transcription is through the creation of looping DNA structures that allow for physical interactions between distal regulatory elements and promoter regions25,26. However, the exact mechanism by which cohesin regulates transcription remains poorly understood.
We discovered that both CHOPS and CdLS syndromes display a similar disruption in behavior of AFF4 and RAD21. Interestingly, the genes significantly increased in CHOPS syndrome and CdLS demonstrate elevated AFF4 binding. In combination with the finding that similar sets of genes were up-regulated and down-regulated in both CdLS and CHOPS syndrome, we can conclude that the altered genome-wide distribution of AFF4 binding affects cohesin binding in CHOPS syndrome with the inverse relationship being true in CdLS (altered genome-wide distribution of cohesin affects AFF4 binding).
Furthermore, as a molecular link between CHOPS syndrome and CdLS, we demonstrated a physical interaction between SEC, STAG1-cohesin complex and RNAP2. This represents the first demonstration of such a physical interaction not only between SEC and cohesin but also between cohesin and RNAP2. Recently, functional differences between the STAG1 and STAG2 components of cohesin have started to emerge. Previously, the Stag1 knock-out mouse model demonstrated the importance of STAG1 in transcriptional regulation, as well as a critical function in chromosomal segregation27,28. Our results point towards a more significant role of STAG1 in the pathogenesis of CdLS.
Previously, a role for cohesin in transcription elongation has been proposed. Cohesin selectively binds genes in which RNAP2 pauses just downstream of the transcription start site, and it was hypothesized that cohesin facilitates transition of paused RNAP2 to elongation24,29. This role of cohesin in transcriptional elongation is likely facilitated by NIPBL/MAU2, with AFF4 and the SEC mediating its effect. Recently, the cohesin loaders (NIPBL/MAU2) were shown to be important in maintaining nucleosome-free regions in yeast models, and it was hypothesized that the derangements of nucleosome positioning cause transcriptional disturbance in the cohesinopathies30. Our observation suggests that NIPBL/MAU2 also plays a role in the transcriptional elongation process, which requires the presence of paused RNAP2 at gene promoters.
In summary, we report the identification of a novel genetic disorder, CHOPS syndrome, caused by mutations in AFF4, and demonstrate that altered genome-wide biding of AFF4 leading to RNAP2 gene body accumulation underlies the transcriptional abnormalities. We also demonstrate a molecular link between CHOPS syndrome and CdLS, and implicate a role for cohesin in transcriptional elongation. These observations underscore the importance of proper proximal pausing of RNAP2 as a crucial regulator of gene expression during human embryogenesis.
Methods
Human Subjects
All individuals enrolled in the study were evaluated by clinical geneticists experienced in the diagnosis of CdLS. All patients and family members were enrolled in the study under an Institutional Review Board-approved protocol of informed consent at The Children's Hospital of Philadelphia.
Exome sequencing and Sanger sequencing
Genomic DNA was extracted from peripheral blood cell. Exome capture was performed with Agilent SureSelect v 4 (Agilent). Captured DNA was sequenced using the Illumina HiSeq2000 with paired-end reads of 100 bp. The sequencing reads obtained in fastq formats were subject to the whole-exome sequence analysis pipeline. The reads were mapped to human genome GRCh37 (1000 genomes version) using Novoalign (www.novocraft.com) version 2.08. Base quality recalibration was also done as part of the mapping in Novoalign. Duplicate reads were marked using Picard (picard.sourceforge.net) version 1.79. The rest of the workflow was based on GATK package 31 best practices through indel realignment and base recalibration steps. Variant calling was done using UnifiedGenotyper with default parameters. The resulting vcf file were subject to variant effect prediction using SnpEff tool v3.1 32 using RefSeq transcripts. For the confirmation of genetic variants identified by exome sequence, Sanger sequence was performed. PCR primer sequences are available on request.
Reagents
MG132, DRB, mouse anti-FLAG monoclonal antibody andα-tubulin antibody was from Sigma Aldrich. Flavioridol was from Enzo Life Sciences. Anti-HA antibody was from Roche. AFF4 (ab57077)(used for Western blot), CDK9 (ab75848), MAU2 (ab46906), SA133, SA233, SMC133, RAD219 and Histone H333 antibodies were from Abcam. ELL2 antibody (A302-505A) and AFF4 antibody (A302-538A)(used for ChIP-seq analysis) were purchased from Bethyl Lab. NIPBL (sc-374625) and Cyclin T1 (sc-10750) antibodies are from Santa Cruz Biotechnology. SPT5 antibody was provided by Dr. Yuki Yamaguch, Department of Biological Information, Tokyo Institute of Technology, Japan. RNAP2 antibodies were used in a previous study, and were provided by Dr. Hiroshi Kimura, Graduate School of Frontier Biosciences, Osaka University, Japan34. Plasmids encoding Myc-DDK-tagged AFF4 (RC217692) and Myc-DDK-tagged SIAH1a (RC206576) were obtained from OriGene Technologies. To generate HA-His-tagged SIAH1a, the SIAH1a cDNA was subcloned into pCMV6-AC-HA-His, using the OriGene RapidShuttling kit (SfgI/MluI, PS200008) according to manufacturer directions. Mutagenesis of the AFF4 expression plasmid to introduce each of the nonsynonymous variants was performed using the QuikChange Site-Directed Mutagenesis Kit by Agilent Technologies following manufacturer's protocol. Plasmids were sequenced after site-directed mutagenesis to confirm the change and to rule out additional, nonspecific changes.
Cell culture
HEK293T and HeLa cells were cultured in DMEM containing 10% FBS and 1% Penicillin/Streptomycin. Cells were grown to 70% confluency and plasmid DNA was transiently transfected using Lipofectamine 2000 (Life Technologies), according to the manufacturer's instructions. At 24 hours after the transfection, media were changed to DMEM containing 10 μM MG132 for 7 hours where indicated. Cells were lysed with SDS sample buffer for Western blot. Skin fibroblast cell lines from 3 CHOPS syndrome and 2 CdLS probands were used in this study. The control fibroblast cell lines were ordered from the Coriell Cell Repository (coriell.org). Fibroblasts cell lines were uniformly cultured in RPMI 1640 or DMEM supplemented with 20% FBS, and antibiotics (100 units/ml of penicillin and 100ug/ml of streptomycin), and 1% L-glutamine. Skin fibroblast cell lines used for total RNA-seq and ChIP-seq were immortalized by exogeneous expression of hTERT.
Protein analyses
To obtain total cell extracts, cells were lysed with lysis buffer (20mM HEPES, 10mM KCl, 100mM NaCl, 1.5mM MgCl2, 0.34 M sucurose, 10% glycerol, 10mM NaF, 10mM beta-glycerophosphate, 10mM sodium butyrate, 1M DTT, 20% triton-X and cOmplete protease inhibitor cocktail (Roche). To obtain soluble cell extracts, the cells were lysed with lysis buffer and centrifuged at 1,500g. DNA in the chromatin fraction was digested by the treatment of benzonase (Novagen, Merck Millipore). Western blot band quantification was performed by ImageQuant TL (GE Healthcare Life Sciences).
Immunoprecipitation
HeLa cells were treated with none, 50μM DRB, 1 μM Flavioridol for 1.5 hours. The cells were harvested by trypsin and washed with PBS. Chromatin containing factions were prepared from cell pellets by the treatment of Buffer A (10mM Tris-HCl (pH7.5), 10mM NaCl, 3mM MgCl2, 0.2% NP-40, CompleteTM-EDTA free, PhosSTOP). The lysates were treated with Buffer B (40mM Tris-HCl(pH7.5), 200mM NaCl, 3mM MgCl2, 10% Glycerol, 0.2% NP-40, CompleteTM-EDTA, PhosSTOP) containing Benzonase. Chromain-bound protein(Input) and Insoluble fraction (Insol.) were separated by the centrifugation at 20000g for 15min. Immunoprecipitation from input fraction was performed using protein G magnetic beads conjugated with anti-SA1 antibody and SA2 antibody.
Expression array
Skin fibroblasts were plated onto T75 flask with cell number approximately 700,000 cells in U133P2, and 200,000 to 350,000 cells in Human Gene 2.0 arrays. Cells were harvested for RNA extraction at 70-80% confluency for U133P2, and 40-70% confluency for Human Gene 2.0 array. Samples used for expression array include two CHOPs syndrome samples (CHOPS T254S (CDL160): 6 year-old Caucasian female with T254S mut and CHOPS T254A (CDL444): 12 year-old Caucasian male with T254A mut) and three age gender matched controls (GM01652: 11 year-old Caucasian female, GM02036: 11 year-old Caucasian female and GM08398: 8 year-old Caucasian male) for U133P2 array. The samples used for Human Gene 2.0 arrays are two CHOPs syndrome samples (CHOPS T254S(CDL160) and CHOPS T254A(CDL444)), two CdLS samples (CDL006: 7 year-old Caucasian female with NIPBL mutation (NIPBL 742_743delCT;L248TfsX6) and CDL015: 10 year-old Caucasian male with NIPBL mutation (NIPBL 2969delG;G990DfsX2)), and four age gender matched control samples (GM01652, GM01864, GM02036 and GM03348) (GM03348: 8 year-old Caucasian female).
RNA was extracted using RNeasy mini (Qiagen). cRNA was purified, fragmented and lebelled using 3’ IVT Express Kit (Affymetrix). 100ng of RNA was used from each sample. First-strand cDNA was synthesized from the extracted RNA, then second-strand cDNA was synthesized from the first-strand cDNA. Subsequently, in vitro transcription and labeling of aRNA was performed. Labelled aRNA was hybridized to Affymetrix GeneChip Human Genome U133 Plus 2.0 Arrays (Affymetrix).
Purified sense-strand cDNA was made from extracted RNA using Ambion WT Expression Kit (Ambion). 100ng of RNA was used from each sample. Purified sense-strand cDNA samples were fragmented and labeled using Affymetrix GeneChip WT Terminal Labeling and Controls Kit (Affymetrix). Labeled cDNA was hybridized to Affymetrix GeneChip® Human Gene 2.0 ST Arrays (Affymetrix) for transcriptome analysis. Arrays were washed and stained according to standard Affymetrix protocols using Affymetrix GeneChip Fluidics Workstation 450 and scanned using an Affymetrix GeneChip Scanner 3000 7G. The DAT files were converted into cell intensities (CEL files) by Affymetrix Command Console software and these were used for subsequent analyses. The sequences from which these probes were derived were selected from GeneBank, dbEST, and Refseq.
Data were processed and analyzed within the R/Bioconductor environment (http://www.bioconductor.org). Original Affymetrix probes on both platforms were mapped to the current version of Entrez genes using the library files provided by the BRAINARRAY database (http://brainarray.mbni.med.umich.edu). Raw data were normalized and summarized by the RMA (Robust Multichip Averaging) method to obtain the gene expression levels. Pairwise comparison of sample groups was performed by the SAM (significance analysis of microarrays) method, which evaluated differential gene expression with fold change, p value, and FDR (false discovery rate). The fold changes and p values were used to create the volcano plots. The top 250 genes with the biggest fold change and p < 0.05 were imported into the DAVID (http://david.abcc.ncifcrf.gov) online tool for functional categorization. DAVID reported pre-defined gene sets or pathways whose members were significantly over-represented in the 250-gene list.
Quantitative RT-PCR
Extracted RNA samples were also used for quantitative RT-PCR. RNA was reverse-transcribed by TaqMan® Reverse Transcription Reagents (Life Technologies), and synthesized cDNA was used for quantitative PCR analysis by droplet digital PCR system (QX100, BioRad Laboratories)35,36. Gene expression was analyzed using TaqMan gene expression assay probes including AFF4 Hs00232683_m1, MYC Hs00153408_m1, JUN Hs01103582_s1, FOS Hs04194186_s1, and EGR1 Hs00152928_m1 (Life Technologies). Internal control probes used were TBP 4326322E and CYC 4326316E (Life Technologies). For allele specific mRNA detection of AFF4, Custom TaqMan® SNP Genotyping Assays (Life Technologies) were used, and TaqMan probes were designed for three AFF4 mutations found in CHOPS syndrome probands. Probe sequences are available on request. For serum stimulation, cells were cultured in the serum free media for two days, and then, cells were either left untreated or treated with serum for 30 min before RNA extraction. Gene expression levels of FOS and EGR1 genes, all of that were induced by serum stimulation, were measured by quantitative RT-PCR 1.
RNA-seq analyses
For total RNA-seq, we used immortalized skin fibroblast cell lines of CHOPS T254A(CDL444), CHOPS T254S(CDL160), CHOPS R258W, CDL006, GM01652, GM02036 and GM03348 (CHOPS R258W(CDL559): 8 year-old African-American female with R258W mut). All samples were sequenced on Illumina HiSeq2500 to single-end 50-bp reads, and mapped to the human genome (UCSC build hg19). Sequencing and mapping statistics are summarized in Supplementary Table 5. Total RNA was extracted using TRIzol (Life Technologies) and Nucleospin RNA (Macherey-Nagel) following the manufacturer's instructions. The RNA-seq analysis was performed using TopHat version 2.0.8b 37 and Cufflinks version 2.1.1 38 with the default parameter set. We applied Ensembl gene annotation (GRCh37) for both TopHat and Cufflinks. We considered protein-coding genes only and the gene-level expression values were estimated by FPKM score. We obtained a total of 519 and 299 deferentially expressed genes for CHOPS and CdLS, respectively (Supplementary Table 6).
ChIP-seq analyses
For ChIP-seq, we used immortalized skin fibroblast cell lines of CHOPS T254A (CDL444), CDL006 and GM2036 samples. ChIP was performed as previously described 39. Briefly cells were crosslinked by 1% formaldehyde for 10 min. Soluble cell lysates were prepared by sonication or both MNase treatment and sonication. And then the lysates were incubated with protein A or protein G Dynabeads (Life Technologies) conjugated with antibodies. After this, the beads were washed and eluted. After decrosslinking of elutes and Input, DNA was purified using PCR purification kit (Qiagen). DNA from Input and ChIP fractions was further sheared to an average size of approximately150 base pairs (bp) by ultrasonication (Covaris), end-repaired, ligated to sequencing adapters and amplified according to manufacturer's instructions (NEBNext ChIP-seq Library Prep Master Mix Set for Illumina; New England Biolabs). Sequenced reads were mapped to the human genome using Bowtie version 1.1.0 40, allowing two mismatches in the first 28 bases per read and outputting only uniquely mapped reads (-n2 –m1 option). Mapping statistics are summarized in Supplementary Table 7. The ChIP-seq analysis and visualization was performed using DROMPA version 2.5.3 41. To facilitate comparison of detected peaks between different ChIP experiments, the number of mapped reads was normalized with the total number of mapped reads. To eliminate uncertain sites, we ignored the low-mappable regions (mappability < 0.3 for the 1-kbp window). Scatter plot for RNAP2, AFF4 and Spt5 (Supplementary Figure 8a, 8b and 8e), we applied CG command to normalize and count mapped reads within each gene-body region. Averaged reads plot around transcription start sites (TSS) (Supplementary Figure 8c and 8d) was performed using PROFILE command. The RefSeq gene annotation was obtained from the UCSC genome browser (http://genome.ucsc.edu/).
Genome editing
For the introduction of AFF4 deletion, CRISPR/cas9 system was used 42. gRNA and hCas9 vectors were purchased from Addgene. AFF4 exon3 target sequence was introduced into an empty gRNA vector. gRNA target sequence is ATGGGCCGCACATAGGCAG. AFF4 ex3 targeting gRNA vector and hCas9 plasmids were transfected into skin fibroblast cell lines using Neon electroporation system (Life Technologies). After the electroporation, single cell cloning was performed. For each clone, the presence or absence of genome editing was confirmed by the Sanger sequencing.
Supplementary Material
Acknowledgements
We thank K. Nakagawa, S. Watanabe and Dr. Eric Rappaport for their technical assistance. This work was supported by Grant-in-Aid for Scientific Research (S) and for innovative science from MEXT (K.S.), and by research grants from the CdLS Foundation, development funds from the Children's Hospital of Philadelphia and PO1 HD052860 from the NHGRI (IDK).
Footnotes
Contributions
K.I. and I.D.K. initiated the human studies; K.I., S.N., K.G., J.S., E.H.Z., M.A.D., D.C., J.L.A., and I.D.K. characterized clinical data; K.I., M.C.D., R.R., C.P.V., M.K., and I.D.K. performed exome analysis and Sanger sequencing confirmation of the mutation; K.I., A.C.E., D.D., Z.M., M.B. and K.S. designed and performed the biochemical analyses; K.I. performed skin fibroblast expression studies and genome-editing experiments. Z.Z. analyzed the skin fibroblast expression array data; R.N., M.K., M.B., Y.K. and K.S. performed ChIP-seq and RNA-seq and its bioinformatics analysis; K.I., K.S. and I.D.K. drafted the manuscript. All authors analyzed the data, discussed the results and commented on the manuscript.
Accession codes
Expression array data was deposited into GEO website (Accession numbers: GSE64031 and GSE64034). ChIP-seq data and RNA-seq data were deposited into SRA database (accession number SRP050576).
Competing financial interests
None declared.
REFERENCES
- 1.Lin C, et al. Dynamic transcriptional events in embryonic stem cells mediated by the super elongation complex (SEC). Genes Dev. 2011;25:1486–1498. doi: 10.1101/gad.2059211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yamaguchi Y, Shibata H, Handa H. Transcription elongation factors DSIF and NELF: promoter-proximal pausing and beyond. Biochim. Biophys. Acta. 2013;1829:98–104. doi: 10.1016/j.bbagrm.2012.11.007. [DOI] [PubMed] [Google Scholar]
- 3.Heidemann M, Hintermair C, Voß K, Eick D. Dynamic phosphorylation patterns of RNA polymerase II CTD during transcription. Biochim. Biophys. Acta. 2013;1829:55–62. doi: 10.1016/j.bbagrm.2012.08.013. [DOI] [PubMed] [Google Scholar]
- 4.Lin C, et al. AFF4, a component of the ELL/P-TEFb elongation complex and a shared subunit of MLL chimeras, can link transcription elongation to leukemia. Mol. Cell. 2010;37:429–437. doi: 10.1016/j.molcel.2010.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu J, Krantz ID. Cornelia de Lange syndrome, cohesin, and beyond. Clin. Genet. 2009;76:303–314. doi: 10.1111/j.1399-0004.2009.01271.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gillis LA, et al. NIPBL mutational analysis in 120 individuals with Cornelia de Lange syndrome and evaluation of genotype-phenotype correlations. Am. J. Hum. Genet. 2004;75:610–623. doi: 10.1086/424698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deardorff MA, et al. Mutations in cohesin complex members SMC3 and SMC1A cause a mild variant of cornelia de Lange syndrome with predominant mental retardation. Am. J. Hum. Genet. 2007;80:485–494. doi: 10.1086/511888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deardorff MA, et al. RAD21 mutations cause a human cohesinopathy. Am. J. Hum. Genet. 2012;90:1014–1027. doi: 10.1016/j.ajhg.2012.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deardorff MA, et al. HDAC8 mutations in Cornelia de Lange syndrome affect the cohesin acetylation cycle. Nature. 2012;489:313–317. doi: 10.1038/nature11316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Isaacs AM, et al. A mutation in Af4 is predicted to cause cerebellar ataxia and cataracts in the robotic mouse. J. Neurosci. 2003;23:1631–1637. doi: 10.1523/JNEUROSCI.23-05-01631.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oliver PL, Bitoun E, Clark J, Jones EL, Davies KE. Mediation of Af4 protein function in the cerebellum by Siah proteins. Proc. Natl. Acad. Sci. U.S.A. 2004;101:14901–14906. doi: 10.1073/pnas.0406196101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luo Z, et al. The super elongation complex family of RNA polymerase II elongation factors: gene target specificity and transcriptional output. Mol. Cell. Biol. 2012;32:2608–2617. doi: 10.1128/MCB.00182-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sanders SJ, et al. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature. 2012;485:237–241. doi: 10.1038/nature10945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O'Roak BJ, et al. Exome sequencing in sporadic autism spectrum disorders identifies severe de novo mutations. Nat. Genet. 2011;43:585–589. doi: 10.1038/ng.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Urano A, et al. Infertility with defective spermiogenesis in mice lacking AF5q31, the target of chromosomal translocation in human infant leukemia. Mol. Cell. Biol. 2005;25:6834–6845. doi: 10.1128/MCB.25.15.6834-6845.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tzschach A, et al. Molecular cytogenetic analysis of a de novo interstitial deletion of 5q23.3q31.2 and its phenotypic consequences. Am. J. Med. Genet. A. 2006;140:496–502. doi: 10.1002/ajmg.a.31105. [DOI] [PubMed] [Google Scholar]
- 17.Sun J, et al. Genetic and genomic analyses of RNA polymerase II-pausing factor in regulation of mammalian transcription and cell growth. J. Biol. Chem. 2011;286:36248–36257. doi: 10.1074/jbc.M111.269167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang Z, Zhu Q, Luo K, Zhou Q. The 7SK small nuclear RNA inhibits the CDK9/cyclin T1 kinase to control transcription. Nature. 2001;414:317–322. doi: 10.1038/35104575. [DOI] [PubMed] [Google Scholar]
- 19.Nguyen VT, Kiss T, Michels AA, Bensaude O. 7SK small nuclear RNA binds to and inhibits the activity of CDK9/cyclin T complexes. Nature. 2001;414:322–325. doi: 10.1038/35104581. [DOI] [PubMed] [Google Scholar]
- 20.Jang MK, et al. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol. Cell. 2005;19:523–534. doi: 10.1016/j.molcel.2005.06.027. [DOI] [PubMed] [Google Scholar]
- 21.Yang Z, et al. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol. Cell. 2005;19:535–545. doi: 10.1016/j.molcel.2005.06.029. [DOI] [PubMed] [Google Scholar]
- 22.Taki T, et al. AF5q31, a newly identified AF4-related gene, is fused to MLL in infant acute lymphoblastic leukemia with ins(5;11)(q31;q13q23). Proc. Natl. Acad. Sci. U.S.A. 1999;96:14535–14540. doi: 10.1073/pnas.96.25.14535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Komori T, et al. Regulation of AMP-activated protein kinase signaling by AFF4 protein, member of AF4 (ALL1-fused gene from chromosome 4) family of transcription factors, in hypothalamic neurons. J. Biol. Chem. 2012;287:19985–19996. doi: 10.1074/jbc.M112.367854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu J, et al. Transcriptional dysregulation in NIPBL and cohesin mutant human cells. PLoS Biol. 2009;7:e1000119. doi: 10.1371/journal.pbio.1000119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wendt KS, et al. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature. 2008;451:796–801. doi: 10.1038/nature06634. [DOI] [PubMed] [Google Scholar]
- 26.Kagey MH, et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Remeseiro S, et al. Cohesin-SA1 deficiency drives aneuploidy and tumourigenesis in mice due to impaired replication of telomeres. EMBO J. 2012;31:2076–2089. doi: 10.1038/emboj.2012.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Remeseiro S, Cuadrado A, Gómez-López G, Pisano DG, Losada A. A unique role of cohesin-SA1 in gene regulation and development. EMBO J. 2012;31:2090–2102. doi: 10.1038/emboj.2012.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schaaf CA, et al. Genome-wide control of RNA polymerase II activity by cohesin. PLoS Genet. 2013;9:e1003382. doi: 10.1371/journal.pgen.1003382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lopez-Serra L, Kelly G, Patel H, Stewart A, Uhlmann F. The Scc2-Scc4 complex acts in sister chromatid cohesion and transcriptional regulation by maintaining nucleosome-free regions. Nat. Genet. 2014;46:1147–51. doi: 10.1038/ng.3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.DePristo MA, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cingolani P, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kon A, et al. Recurrent mutations in multiple components of the cohesin complex in myeloid neoplasms. Nat. Genet. 2013;45:1232–1237. doi: 10.1038/ng.2731. [DOI] [PubMed] [Google Scholar]
- 34.Stasevich TJ, et al. Regulation of RNA polymerase II activation by histone acetylation in single living cells. Nature. 2014 doi: 10.1038/nature13714. doi:10.1038/nature13714. [DOI] [PubMed] [Google Scholar]
- 35.Porensky PN, et al. A single administration of morpholino antisense oligomer rescues spinal muscular atrophy in mouse. Hum. Mol. Genet. 2012;21:1625–1638. doi: 10.1093/hmg/ddr600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Heredia NJ, et al. Droplet Digital™ PCR quantitation of HER2 expression in FFPE breast cancer samples. Methods. 2013;59:S20–23. doi: 10.1016/j.ymeth.2012.09.012. [DOI] [PubMed] [Google Scholar]
- 37.Kim D, et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Trapnell C, et al. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 2013;31:46–53. doi: 10.1038/nbt.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Komata M, et al. Chromatin immunoprecipitation protocol for mammalian cells. Methods Mol. Biol. 2014;1164:33–38. doi: 10.1007/978-1-4939-0805-9_4. [DOI] [PubMed] [Google Scholar]
- 40.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nakato R, Itoh T, Shirahige K. DROMPA: easy-to-handle peak calling and visualization software for the computational analysis and validation of ChIP-seq data. Genes Cells. 2013;18:589–601. doi: 10.1111/gtc.12058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mali P, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.