Abstract
The Hippo pathway was discovered as a conserved tumour suppressor pathway restricting cell proliferation and apoptosis. However, the upstream signals that regulate the Hippo pathway in the context of organ size control and cancer prevention are largely unknown. Here, we report that glucose, the ubiquitous energy source utilised for ATP generation, regulates the Hippo pathway downstream effector YAP. We show that both the Hippo pathway and AMP-activated protein kinase (AMPK) were activated during glucose starvation, resulting in phosphorylation of YAP and contributing to its inactivation. We also identified glucose-transporter 3 (GLUT3) as a YAP-regulated gene involved in glucose metabolism. Together, these results demonstrate that glucose-mediated energy homeostasis is an upstream event involved in regulation of the Hippo pathway and, potentially, an oncogenic function of YAP in promoting glycolysis, thereby providing an exciting link between glucose metabolism and the Hippo pathway in tissue maintenance and cancer prevention.
INTRODUCTION
The Hippo pathway was initially identified through genetic screening in Drosophila as having crucial roles in restricting tissue growth1–3. The evolutionarily conserved functions of this pathway in control of tissue and organ size were further demonstrated through genetically engineered mouse models. In mammalian systems, the Hippo pathway is composed of core kinase complexes (MST1/2 and LATS1/2), adaptor proteins (SAV1 for MST1/2 and MOB1 for LATS1/2), downstream effectors (YAP and TAZ) and nuclear transcription factors (TEAD1/2/3/4). MST1/2 kinase phosphorylates and activates LATS1/2 kinases. Active LATS1/2 phosphorylates YAP at Serine 127 (S127) and provides the docking site for the 14-3-3 protein, which sequesters YAP in cytoplasm. Moreover, LATS1/2 phosphorylates YAP at S381, which leads to YAP degradation through the β-TRCP E3 ligase complex4. Un-phosphorylated YAP translocates into the nucleus and functions as a transcriptional co-activator by binding to the TEAD family of transcription factors. The YAP-TEAD complex regulates transcription of genes that promote proliferation and inhibit apoptosis, two key events for organ size control. Nuclear protein VGLL4 directly competes with YAP for binding to TEAD transcription factors and consequently inhibits YAP’s transcriptional functions5,6.
Notably, ablation of Hippo pathway components leads to tumour formation7–9, which suggested that the Hippo pathway is a tumour suppressor pathway. As the major target of the Hippo pathway, YAP has been identified as an oncogene. Transgenic expression of YAP in mouse liver reversibly enlarged livers and eventually led to tumour formation10,11. Moreover, downregulation of Hippo pathway components and elevated activation of YAP/TAZ have been observed in various human cancers11,12, which further demonstrates the critical roles of the Hippo pathway in human cancer prevention.
Many studies in recent years have been devoted to identification of upstream regulators of the Hippo pathway in order to elucidate the mechanisms underlying organ size control. These studies uncovered many components of cell adhesion junction and tight junction as Hippo pathway regulators, findings that agree with the known cell density–dependent regulation of the Hippo pathway13. Cytoskeleton-mediated mechanical force also plays a key role in YAP regulation14–16. Moreover, G-protein-coupled receptors function upstream of the Hippo pathway through Rho GTPase and cytoskeleton remodeling17. However, the upstream signals that regulate the Hippo pathway in the context of organ size control and cancer prevention are still largely unknown.
In this study, we identified crosstalk between glucose metabolism and the Hippo pathway. The energy stress generated by a defect in glucose metabolism activated LATS kinase and AMPK kinase leading to phosphorylation of YAP and inhibition of its cellular functions. On the other hand, YAP promoted glucose metabolism through upregulation of glucose transporter 3 (GLUT3) expression at the transcriptional level. These findings revealed a critical crosstalk between energy homeostasis and the Hippo pathway underlining metabolic control of the Hippo pathway and a previously unknown function of the Hippo pathway in glucose metabolism.
RESULTS
Glucose homeostasis regulates YAP phosphorylation and localisation
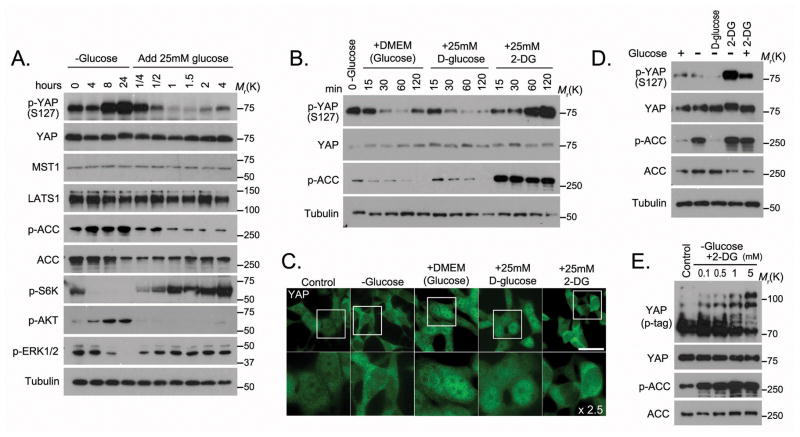
We explored whether any growth condition might control activation of the Hippo pathway and found that glucose starvation increased phosphorylation of YAP at S127 (Figure 1A), the major phosphorylation site regulated by the Hippo pathway. When glucose was added back to glucose-deprived cells, the phosphorylation of YAP at S127 dramatically decreased (Figure 1A). This glucose-stimulated effect was transient, since the phosphorylation of YAP gradually recovered after 2 hours or longer (Figure 1A). The levels of YAP upstream kinases LATS1 and MST1 were not affected by glucose (Figure 1A), while this glucose switch, as expected, regulated phosphorylation of ACC and activation of S6K, AKT and ERK (Figure 1A).
Figure 1. Glucose homeostasis controls YAP phosphorylation and localization.
(A) The phosphorylation of YAP was regulated by glucose deprivation and stimulation. HEK293A cells were glucose starved for the indicated intervals and then stimulated with glucose (25 mM) for the indicated intervals. Cell lysates from each time point were subjected to Western blotting. (B) Glucose but not 2-DG stimulated the dephosphorylation of YAP. Glucose starved HEK293A cells were released into 25mM glucose-containing Dulbecco modified essential medium (DMEM), glucose-free DMEM containing 25mM D-glucose or glucose-free DMEM containing 25mM 2-DG. Cell lysates from each time point were subjected to Western blotting. (C) The localisation of YAP was detected by immunostaining with YAP monoclonal antibody in cells released from energy stress (as described for Figure 1B) for 1 hour. Scale bar=20 μm. The region inside each box was enlarged 2.5 times below. (D) 2-DG induced YAP phosphorylation. HEK293A cells were treated with each indicated medium for 4 hours. The D-glucose and 2-DG concentrations were 25 mM in each medium. (E) 2-DG-induced YAP phosphorylation was dose dependent. HEK293A cells were treated with glucose-free medium supplemented with 2-DG at indicated concentrations for 4 hours, and cell lysates representing each dose were subjected to Western blotting. Phospho-tag-containing gel (p-tag) was used to detect YAP phosphorylation. Uncropped images of Western blots are shown in Supplementary Figure 7.
To further validate these findings, glucose-starved cells were given different types of glucose-containing medium: regular glucose-rich medium (25 mM glucose), or glucose-free medium containing either D-glucose (25 mM) or 2-deoxy-D-glucose (2-DG, 25 mM). 2-DG is a glucose molecule that has the 2-hydroxyl group replaced by hydrogen, so that it fails to undergo further glycolysis and therefore leads to energy stress due to reduced ATP production. The phosphorylation of YAP decreased in cells stimulated by the regular glucose-rich medium or the D-glucose–containing medium, but not in cells stimulated by 2-DG–containing medium (Figure 1B). The subcellular localisation of YAP corresponded to its phosphorylation status: YAP localised mostly in cytoplasm in glucose-starved cells (Figure 1C), and while glucose-rich medium and D-glucose–containing medium induced nuclear translocation of YAP, YAP was retained in the cytoplasm in cells cultured with 2-DG–containing medium (Figure 1C). These data suggest that glucose status regulates YAP phosphorylation and subcellular localisation.
To determine the role of 2-DG in YAP regulation, cells were treated transiently with glucose starvation, 2-DG or a combination. The combination increased YAP phosphorylation to a greater extent than 2-DG treatment alone (Figure 1D). Moreover, the effect of 2-DG treatment on YAP was dose dependent (Figure 1E). These data indicate that a 2-DG-induced defect in glucose metabolism could inhibit YAP function.
Release of energy stress activates YAP
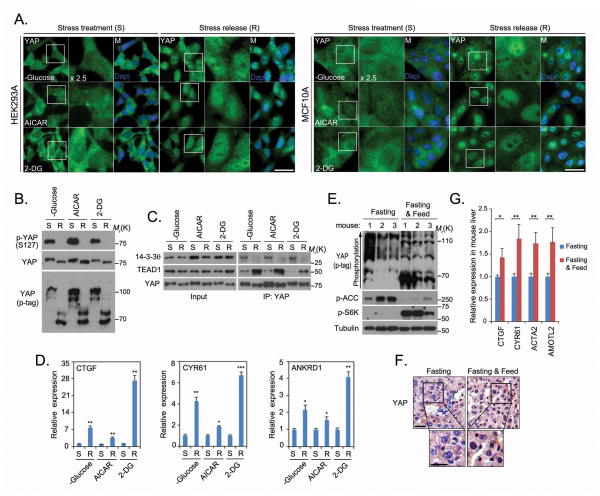
Glucose is widely used as an energy source in various organisms, and defects in glucose metabolism create energy stress in the cell18,19. To further address the role of energy stress in YAP function, we employed three different methods to induce energy stress in cells: glucose starvation, treatment with 2-DG, and treatment with 5-amino-1-β-D-ribofuranosyl-imidazole-4-carboxamide (AICAR). AICAR is phosphorylated by adenosine kinase to form ZMP, an analog of AMP, thereby mimicking energy stress conditions with a high AMP level. Under all three of these conditions, YAP localised mostly in the cytoplasm in both HEK293A and MCF10A cells (Figure 2A). Notably, however, YAP translocated into the nucleus when cells were released from energy stress (Figure 2A). The decrease of YAP phosphorylation upon release from energy stress was observed by both Western blotting with YAP phospho-specific antibody (S127) and the migration shift of YAP in phospho-tag gel (Figure 2B). Consistent with these findings were the observations that release from energy stress diminished YAP’s association with the 14-3-3 protein, while increasing its interaction with TEAD1 (Figure 2C). Accordingly, release from energy stress also increased the transcription of YAP downstream target genes CTGF, CYR61 and ANKRD1 (Figure 2D), indicating that release from energy stress activates YAP.
Figure 2. Release from energy stress activates YAP.
(A) Energy stress was induced in HEK293A and MCF10A cells by glucose starvation or treatment with AICAR (0.5 mM) or 2-DG (25 mM) for 16 hours. Cells were released from energy stress for 1 hour and endogenous YAP localisation was detected by immunostaining with the YAP monoclonal antibody. Nuclei were visualized by Dapi staining. Scale bar=20 μm. 2.5x enlargement of the boxed region is shown to the right of each image. (B) YAP phosphorylation decreased in cells released from energy stress. Lysates of HEK293A cells under one of the energy stresses (S) described in (a) or released from energy stress (R) for 1 hour were subjected to Western blotting. YAP phosphorylation was detected by phospho-tag-containing gel (p-tag). (C) Release from energy stress for 1 hour induced YAP-TEAD1 association but suppressed YAP-14-3-3 interaction. (D) Release of cells from energy stress increased YAP transcriptional activity. YAP-regulated gene transcripts (CTGF, CYR61 and ANKRD1) were detected by quantitative PCR in cells under energy stress or released from energy stress for 2 hours and normalised (mean±s.d, n=3 biological replicates). * p<0.05, ** p<0.01 and *** p<0.001 (Student t-test). (E) YAP phosphorylation was higher in livers of fasting mice than in livers of mice fed after fasting. Mice that had fasted for 16 hours were then fed (n=3) or fasted (n=3) for another 5 hours. Livers were collected and subjected to lysis and Western blotting. The arrow indicates increasing YAP phosphorylation. (F) YAP translocated into nucleus in the liver of a fasting mouse after feeding. A liver from a fasting mouse and a liver from a mouse fed after fasting from (E) were randomly chosen for immunohistochemical staining with YAP antibody, followed by hematoxylin staining to visualise the nucleus. Scale bar=30 μm. The boxed region is enlarged below. (G) YAP transcriptional activity increased in fasting mouse liver after feeding. Transcription of YAP-regulated genes (Ctgf, Cyr61, Acta2 and Amotl2) was detected by quantitative PCR in livers randomly chosen from mice in (E) (mean±s.d, n=3 biological replicates). * p<0.05 and ** p<0.01 (Student t-test). Statistics source data are shown in Supplementary Table 2. Uncropped images of Western blots are shown in Supplementary Figure 7.
We also investigated YAP activity in liver samples obtained from mice that had fasted for 16 hours and then were fed or fasted for another 5 hours. YAP was less phosphorylated in the liver samples from fed mice than in those from fasting mice (Figure 2E). Moreover, in the livers of fed mice, YAP translocated into nucleus (Figure 2F) and promoted transcription of its downstream target genes (Figure 2G). These results indicate the physiological relevance of energy homeostasis in the regulation of YAP activity.
AMPK associates with and phosphorylates YAP
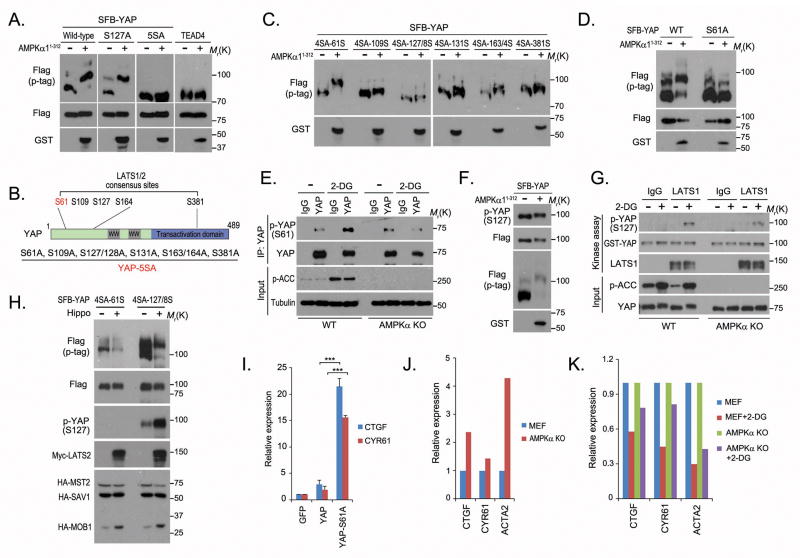
To identify the mechanisms underlying the regulation of YAP by energy stress, we isolated YAP-associated protein complexes in glucose-starved HEK293A cells through tandem affinity purification followed by mass spectrometry analysis. Surprisingly, the AMPK α and β subunits were identified on the prey list (Figure 3A). The specific interaction between AMPK and YAP was confirmed by pulldown assays (Figures 3B and 3C). Co-expression of YAP with an AMPKα1 C terminal–truncated mutant (amino acids 1–312), which has been demonstrated to be a constitutively active form of AMPK20,21, but not with its inactive counterpart (T172A), induced YAP migration shift in phospho-tag gel (Figure 3D), suggesting that YAP is phosphorylated by AMPK in vivo. Since AMPK is a master regulator of energy homeostasis and is activated by energy stress, our results indicate that AMPK may directly phosphorylate YAP in response to energy stress.
Figure 3. AMPK associates with and phosphorylates YAP.
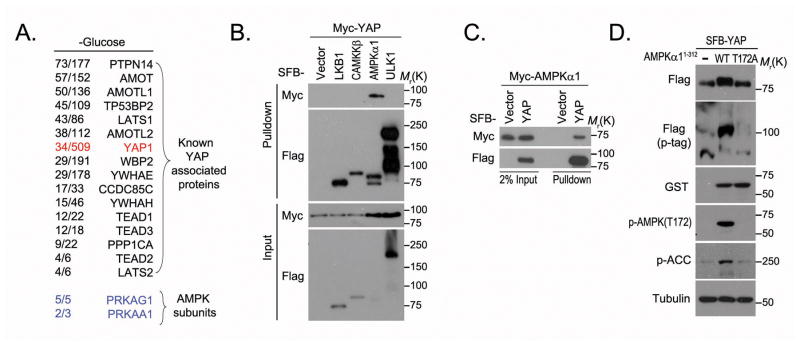
(A) AMPK kinase subunits were identified as YAP-associated proteins by tandem affinity purification–mass spectrometry in glucose-starved HEK293A cells. Bait protein is marked in red. Identified AMPKα1 and β1 subunits are marked in blue. The left column of numbers represents unique peptide number/total peptide number. The known YAP-associated proteins and the identified AMPK subunits are indicated. (B–C) YAP associated with AMPK. Indicated constructs were expressed in 293T cells for 24 hours, and cell lysates were subjected to pulldown assays with S protein beads. (D) AMPK phosphorylated YAP in vivo. GST-tagged rat AMPK (amino acids 1~312) or its kinase-dead mutant (T172A) was co-expressed with SFB-tagged YAP in 293T cells. The phosphorylation of YAP was assessed by phospho-tag gel (p-tag). Uncropped images of Western blots are shown in Supplementary Figure 7.
AMPK phosphorylates YAP on several residues including the S61 site
In searching for AMPK phosphorylation sites on YAP, we again used mobility shift in phospho-tag gel to assess YAP phosphorylation. We found that the YAP-5SA mutant13 could not be phosphorylated as dramatically as wild-type YAP by the active AMPK truncation mutant (Figure 4A), which indicated that one or more of these mutated serine residues is the AMPK phosphorylation site(s). The YAP-5SA mutant was originally generated to mutate all of the putative LATS kinase phosphorylation sites on YAP4,13,22. We sequenced this mutant and confirmed that it comprises eight serine sites mutated to alanine (Figure 4B). Mutation of each alanine back to serine generated six different YAP-4SA mutants, and of these only YAP-4SA-61S could be intensely phosphorylated by AMPK as evaluated by phospho-tag gel analysis (Figure 4C). The YAP-S61A mutant could not be strongly phosphorylated by AMPK in the same experiments (Figure 4D), indicating that the YAP S61 site is an AMPK phosphorylation site.
Figure 4. AMPK phosphorylates YAP at S61 and suppresses its transcriptional activity.
(A) AMPK active truncation mutant was co-expressed with indicated plasmids in 293T cells. The protein phosphorylation was assessed by phospho-tag gel (p-tag). (B) Schematic illustration of YAP protein domains and reported LATS kinase phosphorylation sites. The mutated serine sites in the YAP-5SA mutant are listed. (C) AMPK phosphorylated the YAP-4SA-61S mutant. AMPK active truncation mutant was co-expressed with indicated YAP-4SA mutants in 293T cells. (D) S61 was the AMPK phosphorylation site on YAP. AMPK active truncation mutant was co-expressed with YAP or YAP S61A mutant in 293T cells. The phosphorylation of YAP was assessed by phospho-tag gel (p-tag). (E) 2-DG induced AMPK-dependent YAP S61 phosphorylation. Wild-type MEFs (WT) and AMPKα-knockout MEFs (AMPKα KO) were treated with 25mM 2-DG in glucose-free medium for 4 hours. Endogenous YAP was immunoprecipitated (IP) and immunoblotted with indicated antibodies. (F) AMPK did not noticeably affect YAP phosphorylation at S127 site. (G) 2-DG–induced LATS1 kinase activation was observed in both WT and AMPKα KO MEFs. MEF cells were treated with 25 mM 2-DG in glucose-free medium for 4 hours. Endogenous LATS1 was immunoprecipitated and subjected to in vitro kinase assay with GST-YAP (2 μg) as substrate. (H) S61 was not the major phosphorylation site for LATS kinase. The Hippo pathway kinase complex (Myc-LATS2, HA-MOB1, HA-MST2 and HA-SAV1) was co-expressed with YAP-4SA-61S or YAP-4SA-127S mutant in 293T cells. (I) Phosphorylation of S61 suppressed YAP transcriptional activity. The transcripts of CTGF and CYR61 were detected in indicated YAP-stable cells by quantitative PCR (mean±s.d, n=3 biological replicates). *** p<0.001 (Student t-test). (J) AMPK suppressed YAP transcriptional activity. The transcription of YAP downstream genes was examined in WT and AMPKα KO MEF cells by quantitative PCR (mean, n=2 biological replicates). (K) Loss of AMPK partially rescued YAP activity in 2-DG–treated MEF cells. WT and AMPKα KO MEF cells were mock-treated or treated with 25 mM 2-DG in glucose-free medium for 4 hours. YAP downstream genes were examined by quantitative PCR (mean, n=2 biological replicates) and normalised to untreated cells. Statistics source data are shown in Supplementary Table 2. Uncropped images of Western blots are shown in Supplementary Figure 7.
We also performed in vitro kinase assays and analysed YAP phosphorylation site(s) by mass spectrometry as an independent method to identify AMPK phosphorylation sites. We again uncovered S61, as well as two other sites (S94 and T119), as AMPK phosphorylation sites (Supplementary Figure 1 and Supplementary Table 1), suggesting that AMPK may phosphorylate S61 as well as other sites on YAP. These data agree with the observation that energy stress was capable of inducing a mild shift of the YAP-5SA mutant in phospho-tag gel (Supplementary Figure 2A).
To further confirm that S61 is an AMPK phosphorylation site, we generated YAP S61 phospho-specific antibody and validated its specificity in vitro and in vivo (Supplementary Figures 2B–2D). Using this antibody, we showed that YAP S61 phosphorylation increased following 2-DG treatment in wild-type mouse embryonic fibroblasts (MEFs) but not in AMPK-knockout MEFs (Figure 4E), suggesting that AMPK phosphorylates YAP at the S61 site in response to energy stress. Notably, the S61 site is evolutionarily conserved (but not in Drosophila) and shows some similarity to the consensus AMPK phosphorylation site (Supplementary Figure 2E).
AMPK phosphorylates YAP in parallel with the Hippo pathway
Although YAP S127 phosphorylation increased following energy stress (Figure 1), AMPK did not appear to be the major contributor to the phosphorylation of YAP at the S127 site (Figure 4F). LATS1 phosphorylation and MST1 phosphorylation were not significantly affected by the active AMPK truncation mutant (Supplementary Figures 3A and 3B). Moreover, LATS1 kinase activation induced by 2-DG treatment was not dramatically different in wild-type or AMPK knockout MEFs (Figure 4G and Supplementary Figure 3C). These results indicate that energy stress activates the Hippo pathway in a manner that is largely independent of AMPK. The S61 site was previously predicted to be a LATS phosphorylation site4,13,22. However, the Hippo core kinase complex could phosphorylate only the YAP-4SA-127S mutant but not the YAP-4SA-61S mutant (Figure 4H). These results indicate that energy stress may activate two parallel cellular signaling pathways, AMPK and the Hippo pathway, which phosphorylate YAP at least at the S61 and S127 sites, respectively.
AMPK-mediated YAP phosphorylation inhibits YAP transcriptional activity
Previous studies suggested that phosphorylation of YAP increased its interaction with the 14-3-3 protein, which sequesters YAP in the cytoplasm11,13,22. The 5SA mutant of YAP blocked YAP’s association with the 14-3-3 protein, translocated YAP into the nucleus and increased YAP transcriptional functions11,13,22. Since the YAP S61 site is among the putative phosphorylation sites mutated in the 5SA mutant, we first examined whether AMPK would affect the interaction between YAP and the 14-3-3 protein. AMPK-mediated YAP phosphorylation did not increase the association of YAP with 14-3-3 (Supplementary Figure 4A). The S127 site, but not the S61 site, was the binding site for the 14-3-3 protein (Supplementary Figures 4B and 4C). The active AMPK truncation mutant did not change YAP localisation (Supplementary Figure 4D), and the YAP S61 mutant did not regulate YAP nuclear localisation (Supplementary Figure 4E). However, we noticed that the S61A single mutation increased the transcription of YAP downstream target genes (CTGF and CYR61) (Figure 4I). These data suggest that YAP transcriptional activity was suppressed by S61 phosphorylation, although the binding between YAP and TEAD was not noticeably affected by the phospho-mimetic mutant of the S61 site (S61D) (Supplementary Figure 4F). Exactly how AMPK-dependent S61 phosphorylation inhibits YAP transcriptional activity remains unknown. It is possible that this AMPK-dependent phosphorylation at the YAP S61 site may affect the binding of YAP with some yet unknown proteins required for YAP activity, and we are now investigating this hypothesis.
On the other hand, the YAP S94 site was also identified as an AMPK phosphorylation site (Supplementary Figure 1 and Supplementary Table 1), and the phospho-mimetic mutant of this site (S94D) greatly diminished YAP-TEAD interaction (Supplementary Figure 4F). Moreover, a single phospho-mimetic mutation of either S61 or S94 suppressed YAP transcriptional activity, and the combined phospho-mimetic mutation of both sites further suppressed it (Supplementary Figure 4G). These data indicate that AMPK may inhibit YAP transcriptional activities by phosphorylating multiple sites on YAP.
To further confirm the suppressive role of AMPK in YAP regulation, we examined YAP transcriptional activity in wild-type or AMPK-deficient MEFs. The transcription of YAP downstream genes was elevated in AMPK-knockout MEFs (Figure 4J). Moreover, loss of AMPK attenuated the relative suppressive effect on YAP induced by 2-DG compared to that in control MEFs (Figure 4K). Together, these results indicate that AMPK is a negative regulator of YAP in response to energy stress.
Energy stress activates LATS kinase but not MST kinase in the Hippo pathway
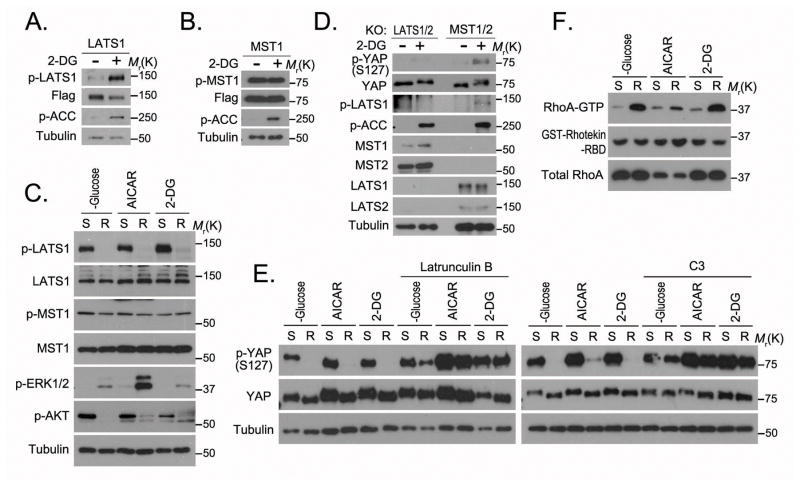
The observation that energy stress regulated YAP S127 phosphorylation (Figure 1) indicates that the Hippo pathway was also activated by energy stress. Interestingly, 2-DG treatment increased LATS1 kinase phosphorylation (T1079) (Figure 5A) but not MST1 kinase phosphorylation (T183) (Figure 5B). Release from energy stress rapidly decreased the LATS1 phosphorylation but not MST1 phosphorylation (Figure 5C). Moreover, YAP S127 phosphorylation was increased by 2-DG in MST1/2 double-knockout cells (Figure 5D). These data indicate that energy stress induces YAP S127 phosphorylation through activation of LATS kinases but not MST kinases in the Hippo pathway.
Figure 5. Energy stress increases LATS kinase activation independent of MST kinases.
(A–B) 2-DG treatment activated LATS1 but not MST1. HEK293A cells stably expressing SFB-LATS1 (A) or MST1 (B) were treated with 2-DG (25 mM) in glucose-free medium for 4 hours, and cell lysates were subjected to Western blotting. (C) LATS1 activation decreased in cells released from various energy stresses (glucose starvation, AICAR, 2-DG as described for Figure 2B). Lysates of cells under energy stress (S) or released from energy stress for 1 hour (R) were subjected to Western blotting. (D) LATS kinases but not MST kinases were required for 2-DG–induced YAP S127 phosphorylation. LATS1/2-double knockout cells (KO) and MST1/2-double knockout cells were treated with 2-DG (25 mM) in glucose-free medium for 4 hours. Cell lysates were subjected to Western blotting. (E) Actin cytoskeleton and Rho GTPase were necessary for the decrease of YAP phosphorylation in cells released from energy stress. Cells under energy stress (glucose starvation, AICAR, 2-DG) were pretreated with actin depolymeriser latrunculin B (1 μg/mL) for 30 minutes or Rho protein inhibitor C3 (2 μg/mL) for 2 hours and released from energy stress for 1 hour. Cell lysates were subjected to Western blotting. (F) Rho GTPase activation increased in cells released from energy stress. Indicated cell lysates were subjected to GST pulldown assay with GST-Rhotekin-RBD (Rho binding domain; 2 μg). GTP-formed RhoA in the pulldown samples, as well as the input controls, was detected by RhoA antibody. Uncropped images of Western blots are shown in Supplementary Figure 7.
Energy stress induces LATS kinase activation mainly through Rho GTPase and cytoskeleton
Previous studies suggested that Rho GTPase–mediated remolding of actin cytoskeleton could inhibit LATS kinase and activate YAP independent of MST kinase14,17. We therefore examined the roles of actin cytoskeleton and Rho GTPase in energy stress–regulated YAP phosphorylation. In cells released from energy stress, latrunculin B–mediated actin depolymerisation reversed the decrease of YAP S127 phosphorylation (Figure 5E). Rho GTPase inhibitor C3 also rescued the decrease of YAP S127 phosphorylation (Figure 5E). Furthermore, release from energy stress increased the Rho GTPase activity (GTP-formed Rho) (Figure 5F). Although detailed mechanisms are still unclear, several studies have shown that the activity of Rho GTPase is regulated by glucose treatment23–25, which is consistent with our findings presented in Figure 5F. Thus, energy stress may activate LATS kinases by modulating Rho GTPase activity. It remains to be determined whether actin cytoskeleton plays a direct and specific role in this process.
Active YAP promotes glycolysis
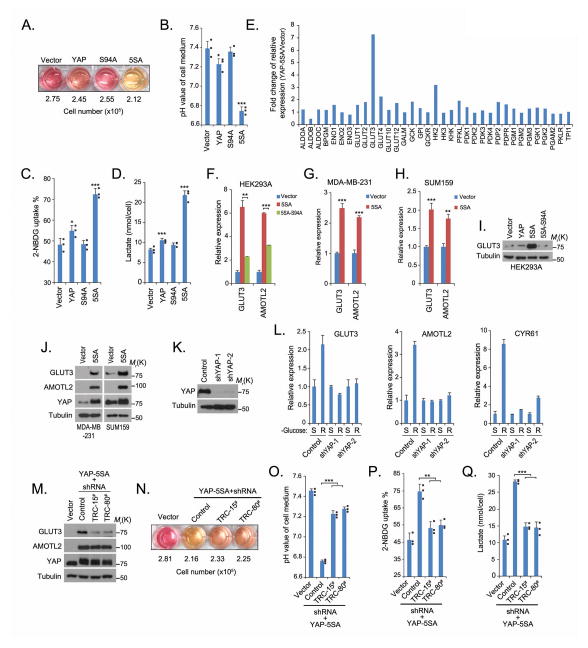
The results presented so far established a connection between glucose homeostasis and the regulation of YAP. The next question is the functional significance of this regulation. We noticed, interestingly, that the culture medium for YAP-5SA cells quickly turned yellow compared with the culture media for the vector control cells, the wild-type YAP cells or the TEAD non-binding mutant YAP-S94A26 cells (Figure 6A), although the numbers of cells in each culture were similar. The yellow colour of the medium indicated that it had been acidified and the level of glycolysis increased in these cells27. Indeed, the YAP-5SA medium showed a lower pH value but higher glucose uptake and lactate production than media from other cell lines (Figures 6B–6D). These data suggest that YAP is involved in the glucose metabolism pathway and that this involvement is likely mediated by its target genes.
Figure 6. YAP regulates GLUT3 transcription in the glucose metabolic pathway.
(A–B) YAP activity increased the acidification of cell culture medium. The colour of the medium (A) varied in the same-time cultures of the indicated YAP-stable cell lines. The pH was measured (B) in cultures of the indicated YAP-stable cell lines (mean±s.d, n=3 biological replicates). * p<0.05 and *** p<0.001 (Student t-test). (C–D) YAP-5SA mutation promotes glycolysis. 2-NBDG uptake (C) and lactate production (D) were examined in indicated YAP stable cells (mean±s.d, n=3 biological replicate) as described in Methods. * p<0.05 and *** p<0.001 (Student t-test). (E) GLUT3 was identified as a YAP-regulated gene. The transcripts of glucose metabolism–related genes were detected by quantitative PCR in YAP-5SA cells and control vector–transfected cells. Fold increases are shown (mean, n=2 biological replicates). (F–H) The control of GLUT3 transcription by YAP was confirmed in various cell lines by quantitative PCR (mean±s.d, n=3 biological replicates). ** p<0.01 and *** p<0.001 (Student t-test). (I and J) YAP increased the level of GLUT3 protein in various cell lines. (K–L) Loss of YAP attenuated the increased GLUT3 expression in cells released from 16 hours of glucose starvation. The downregulation of YAP protein was confirmed by Western blotting (K). The transcription of GLUT3, AMOTL2 and CYR61 was detected by quantitative PCR in cells under glucose starvation or released from glucose starvation for 2 hours and normalised (mean±s.d, n=3 biological replicates). (M) GLUT3 was knockdown in YAP-5SA HEK293A cells by two different GLUT3 shRNAs. (N–O) GLUT3 knockdown suppressed the acidification of YAP-5SA cell medium. The colour of the medium varied in the cultures of the indicated cell lines (N). The pH was measured in the indicated cells (mean±s.d, n=3 biological replicates) (O). *** p<0.001 (Student t-test). (P–Q) Knockdown of GLUT3 suppressed YAP-5SA–induced glycolysis. 2-NBDG uptake (P) and lactate production (Q) were examined (mean±s.d, n=3 biological replicates) as described in Methods. ** p<0.01 and *** p<0.001 (Student t-test). Statistics source data are shown in Supplementary Table 2. Individual data are plotted for Figures 6B, 6C, 6D, 6O, 6P and 6Q. Uncropped images of Western blots are shown in Supplementary Figure 7.
YAP up-regulates GLUT3 in glucose metabolism pathway
To identify YAP regulated genes involved in glucose metabolism, we examined the transcription of a panel of glucose metabolism–related genes in cells expressing the YAP-5SA mutant and in vector-control cells. The analysis showed that GLUT3 transcription was much higher in YAP-5SA cells than in vector-control cells (Figure 6E). The dependence on YAP transcriptional activity was confirmed in YAP-5SA-S94A cells, in which GLUT3 transcription decreased (Figure 6F). Similar findings were obtained from other cell lines (Figures 6G and 6H). Furthermore, a higher level of GLUT3 protein was detected in YAP-5SA cells than in other cells by Western blotting (Figures 6I and 6J). Down-regulation of YAP (Figure 6K) suppressed the increase of GLUT3 transcription in cells released from energy stress (Figure 6L). We identified a conserved TEAD-binding site in the GLUT3 promoter, and ENCODE data showed that this site was bound by TEAD (Supplementary Figure 5). These results indicate that YAP-TEAD may directly regulate the transcription of GLUT3. Moreover, results from two other studies26,28 suggested that the expression of GLUT3 was up-regulated by active YAP, which support our findings that YAP regulates GLUT3 expression.
To determine the role of GLUT3 in YAP-mediated glycolysis, we knocked down GLUT3 in YAP-5SA cells (Figure 6M) and found that this down-regulation of GLUT3 partially reversed medium acidification, glucose uptake and lactate production in cells expressing the YAP-5SA mutant (Figures 6N–6Q). These data suggest that GLUT3 is at least one of the downstream effectors involved in YAP-regulated glycolysis.
GLUT3 expression correlates with YAP protein level in human liver and colon cancers
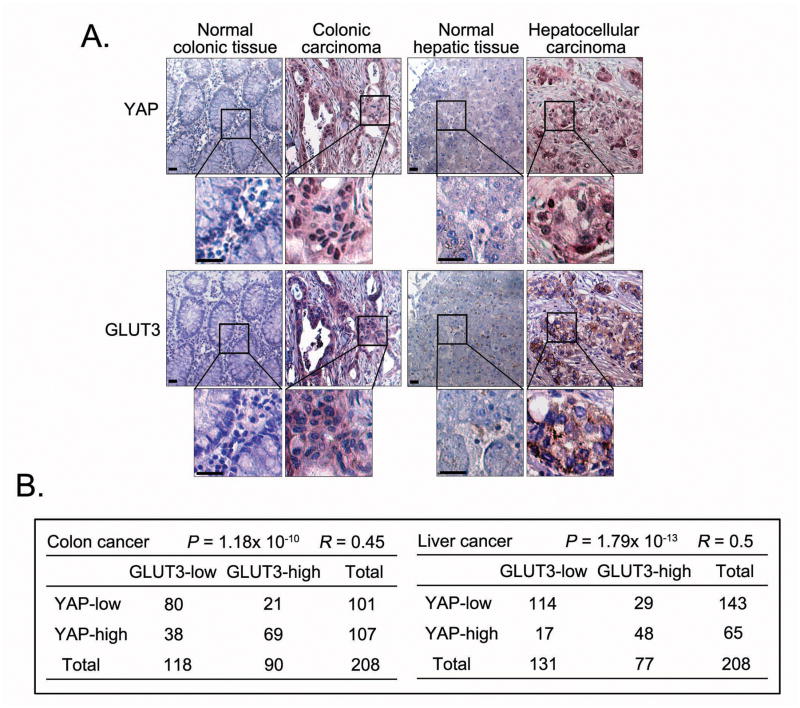
Since high expression of GLUT3 has been identified in various types of cancers29–34, we examined expression of YAP and GLUT3 in samples of human cancer, specifically colon and liver cancers. Indeed, the expression of YAP and GLUT3 was higher in tumour samples than in normal tissues (Figure 7A), and their expressions were positively correlated in tumour samples (Figure 7B). These data indicate a potential role for YAP in regulating GLUT3 expression in human cancers.
Figure 7. YAP and GLUT3 expression positively correlate with each other in human colon and liver cancers.
(A) Representative normal colon and colon carcinoma specimens and normal liver and liver carcinoma specimens in tissue arrays were subjected to immunohistochemical staining of YAP and GLUT3. Brown staining indicates positive immunoreactivity. The region in each box is enlarged below. Scale bar=30 μm. (B) Correlations between YAP and GLUT3 protein levels in human colon and liver tumours were analysed. Statistical significance was determined by the chi-square test; R: correlation coefficient.
DISCUSSION
In this study, we unexpectedly discovered that AMPK, the master kinase that senses the cellular AMP/ATP ratio, directly phosphorylates YAP and inhibits YAP transcriptional activity. We demonstrated that AMPK directly phosphorylates YAP at multiple sites and thus suppresses YAP transcriptional activity. We primarily investigated phosphorylation at S61 by AMPK, since this phosphorylation is the major event contributing to YAP migration shift on phospho-tag gel. We also noticed a minor shift of the YAP-5SA mutant on phospho-tag gel, indicating that there are additional phosphorylation sites. Indeed, our mass spectrometry analysis supports these findings and revealed two more AMPK phosphorylation sites on YAP, S94 and T119. The estimated phosphorylation on these three sites indicated that S61 (11.27%) and S94 (2.39%) are relatively dominant phosphorylation sites compared to T119 (0.53%). The S94 site is known to be required for YAP’s binding to TEAD and mutation of this site (S94A) abolishes TEAD-dependent YAP transcriptional activity26. The phospho-mimetic mutant of S94 (S94D) also inhibited the interaction between YAP and TEAD as well as transcription of YAP downstream target genes. In this issue, Guan and colleagues report similar findings about AMPK-dependent YAP phosphorylation on the S94 site, complementing our results (#35 Guan paper reference). Together, with their findings, our results suggest that AMPK may phosphorylate multiple sites on YAP and inhibit YAP transcriptional activity.
We also revealed that energy stress increases S127 phosphorylation and cytoplasmic localisation of YAP by activating the Hippo pathway. We explored the crosstalk between AMPK and the Hippo pathway, since both pathways are activated following energy stress. In LATS- or MST- deficient cells, AMPK activity, as indicated by ACC S79 phosphorylation, was still increased by 2-DG treatment (Figure 5D), suggesting that AMPK is activated by energy stress independent of the Hippo pathway. On the other hand, 2-DG-induced LATS kinase activation was slightly decreased in AMPK-knockout MEF cells (Figure 4G and Supplementary Figure 3C), indicating the involvement of both AMPK-dependent and -independent pathways in the regulation of LATS kinase activity in response to energy stress. The AMPK-dependent regulation of the Hippo pathway is in agreement with recent finding that AMPK may activate the Hippo pathway by increasing the levels of angiomotin family proteins under energy stress36. Our findings showed that the AMPK-independent pathway may be regulated by Rho GTPase, although the detailed mechanisms remain to be elucidated. Similar findings are reported by Guan and colleagues in this issue (#35 reference). Therefore, both AMPK and the Hippo pathway are activated following energy stress, which act together to suppress YAP activity (Supplementary Figure 6).
Our results demonstrated that the Hippo pathway is intimately associated with glucose homeostasis. This is highly significant since it establishes a connection between a pathway involved in organ size control and nutrient availability. The discovery of AMPK as a kinase for YAP, reported here, extends our understanding of YAP regulation outside of the classic Hippo pathway. The identification of GLUT3 as a YAP-regulated gene offers a potential link between the Hippo pathway and cancer metabolism. As hallmarks of cancer19, tumour cells are known to reprogram their signaling pathways and metabolism to support their uncontrolled proliferation and survival. Our study proposes yet another oncogenic function of YAP, via promotion of glucose uptake and glycolysis, which warrants further investigation.
METHODS
Antibodies
Anti-YAP antibody (1:1000 dilution) was raised by immunising rabbits with bacterially expressed and purified GST-fused human full-length YAP protein. Anti-AMOTL2 antibody (1:1000 dilution) was raised by immunising rabbits with bacterially expressed and purified GST-fused human AMOTL2 (amino acids 1~675). Antisera were affinity-purified by using the AminoLink Plus immobilisation and purification kit (Pierce). Anti-YAP S61 phospho-specific antibody (1:1000 dilution) was raised against keyhole limpet hemocyanin conjugated- phospho-peptide Biotin-QIVHVRGD(phospho-S)ETDLEALC, and anti-serum was affinity purified through SulfoLink peptide coupling gel. Additional anti-YAP (sc101199, 1:200 dilution for immunostaining) and anti-phospho-YAP (S127) (4911S, 1:1000 dilution) antibodies were purchased from Santa Cruz Biotechnology and Cell Signaling Technology respectively. Anti-α-tubulin (T6199-200UL, 1:5000 dilution) and anti-flag (M2) (F3165-5MG, 1:5000 dilution) monoclonal antibodies were obtained from Sigma-Aldrich. Anti-Myc (sc-40, 1:1000 dilution), anti-GST (sc-138, 1:1000 dilution) and anti-14-3-3θ (sc-732, 1:1000 dilution) monoclonal antibodies were purchased from Santa Cruz Biotechnology. Anti-TEAD1 (610922, 1:1000 dilution) monoclonal antibody was obtained from Millipore. Anti-phosho-AKT1 (Ser473) (9271S, 1:1000 dilution), anti-phospho-ERK1/2 (Thr202/Tyr204) (9100S, 1:1000 dilution), anti-phospho-ACC (Ser79) (3661S, 1:1000 dilution), anti-ACC (3662S, 1:1000 dilution), anti-phospho-p70 S6K (Thr389) (9205S, 1:1000 dilution), anti-phospho-AMPKα (Thr172) (2531S, 1:1000 dilution), anti-AMPKα (2532S, 1:1000 dilution), anti-phospho-LATS1 (Thr1079) (8654S, 1:1000 dilution), anti-LATS1(9153S, 1:1000 dilution and 3477S, 1:100 dilution for immunoprecipitation), anti-phospho-MST1 (Thr183)/MST2 (Ser180) (3681S, 1:1000 dilution) and anti-MST1(3682S, 1:1000 dilution) polyclonal antibodies were purchased from Cell Signaling Technology. Anti-GLUT3 (sc-30107, 1:1000 dilution) polyclonal antibody was purchased from Santa Cruz Biotechnology. Anti-RhoA (ARH03-A, 1:1000 dilution) monoclonal antibody and GST-Rhotekin-RBD protein (RT01-A) were obtained from Cytoskeleton, Inc.
Constructs and viruses
All constructs were generated by polymerase chain reaction (PCR) and subcloned into pDONOR201 vector using Gateway Technology (Invitrogen) as the entry clones. As needed, the entry clones were subsequently recombined into gateway-compatible destination vectors for the expression of N- or C-terminal-tagged fusion proteins. pEBG-AMPKα1 (amino acids 1–312) was purchased from Addgene (Plasmid #27632). pCMV-Flag YAP2 5SA (Plasmid #27371), HA-MST2 (Plasmid #33098), HA-SAV1 (Plasmid #32834) and HA-MOB1 (Plasmid #32835) were all obtained from Addgene. PCR-mediated site-directed mutagenesis was used to generate serial point mutations as indicated. The GST-ACC (amino acids 1~130) construct was kindly provided by Dr. Zhijun Luo (Boston University).
HA-Flag–tagged retroviruses for various YAP mutants were generated by co-transfecting with pCL-Ampho packaging vector in the BOSC23 cell line. Cells were infected twice by 0.45 μm–filtered retrovirus with the addition of polybrene (8 μg/mL) and selected with puromycin (2 μg/mL). Pooled stable cells were used for all the experiments after validation by immunostaining and Western blotting.
The SFB-YAP lentiviral expression vector was generated by inserting the gateway response fragment (attR1-ccdB-attR2)–fused SFB tag into the XbaI and SwaI multi-clonal sites of the pCDH-CMV-EF1-GFP vector (kindly provided by Dr. M. James You, MD Anderson Cancer Center). YAP was cloned into this vector through a gateway-based LR reaction. YAP shRNA1 (Plasmid# 27368) and shRNA2 (Plasmid# 27369) were obtained from Addgene. GLUT3 shRNAs (TRCN0000043615 and TRCN0000042880) were purchased from OpenBiosystem. All lentiviral supernatants were generated by transient transfection of 293T cells with helper plasmids pSPAX2 and pMD2G (kindly provided by Dr. Zhou Songyang, Baylor College of Medicine) and harvested 48 hours after transfection. Supernatants were passed through the 0.45μm filter and used to infect cells with the addition of polybrene (8 μg/mL).
Cell culture and transfection
HEK293T and HeLa cells were purchased from American Type Culture Collection (ATCC) and maintained in Dulbecco modified essential medium (DMEM) supplemented with 10% fetal bovine serum (FBS) at 37°C in 5% CO2 (v/v). HEK293A cells were kindly provided by Dr. Jae-Il Park (MD Anderson Cancer Center). MCF10A cells, purchased from ATCC, were kindly provided from Dr. Dihua Yu (MD Anderson Cancer Center). MCF10A cells were maintained in DMEM/F12 medium supplemented with 5 % horse serum, 200 ng/mL epidermal growth factor, 500 ng/mL hydrocortisone, 100 ng/mL cholera toxin and 10 μg/ml insulin at 37°C in 5% CO2 (v/v). SUM159 cells, purchased from ATCC, were kindly provided by Dr. Li Ma (MD Anderson Cancer Center) and maintained in F12 medium containing 5% FBS, insulin (5 μg/mL), hydrocortisone (1 μg/mL) and HEPES (10 mM). The MDA-MB-231 cells, purchased from ATCC, were kindly provided by Dr. Mien-Chie Hung (MD Anderson Cancer Center) and cultured in DMEM with 10% FBS. AMPKα-knockout MEFs and wild-type MEFs were kindly provided by Dr. Benoit Viollet (University Paris Descartes, Institut Cochin) and cultured in DMEM with 10% FBS. All culture media contained 1% penicillin and streptomycin antibiotics. Plasmid transfection was performed with the polyethylenimine reagent.
Tandem affinity purification of SFB-tagged protein complexes
HEK293A cells were infected twice with SFB-YAP lentivirus. Pooled cells stably expressing the tagged YAP were selected by culturing in medium containing puromycin (2 μg/mL), and the protein expression was confirmed by immunostaining and Western blotting. Cells were washed once and cultured in glucose-free DMEM (Invitrogen) with 10% dialysed FBS (Gemini Bio-Products) for 18 hours.
For affinity purification, glucose-starved HEK293A cells were subjected to lysis in NETN buffer (100 mM NaCl, 20 mM Tris-Cl, 0.5 mM EDTA, 0.5 % Nonidet P-40) with protease and phosphatase inhibitors at 4°C for 20 minutes. Crude lysates were subjected to centrifugation (14,000 rpm) at 4°C for 15 minutes. Supernatants were incubated with streptavidin-conjugated beads (Amersham) for 4 hours at 4°C. The beads were washed three times with NETN buffer, and bound proteins were eluted with NETN buffer containing biotin (2 mg/mL; Sigma) overnight at 4°C. The elutes were incubated with S protein beads (Novagen) for 4 hours. The beads were washed three times with NETN buffer and subjected to sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). Protein bands were excised and subjected to mass spectrometry analysis (performed by the Taplin Mass Spectrometry Facility, Harvard Medical School).
Mouse fasting experiment
Five-week old C57/B6 female mice were randomly assigned to fasting and fasting-feed groups. The animal experiments were performed in accordance with a protocol approved by the Institutional Animal Care and Use Committee (IACUC) of MD Anderson Cancer Center. Mice that had fasted for 16 hours were then fed (n=3) or fasted (n=3) for another 5 hours. Livers from these animals were collected and subjected to indicated experiments.
Immunofluorescent staining
Cells cultured on coverslips were fixed by 4% paraformaldehyde for 10 minutes at room temperature and then extracted with 0.5% Triton X-100 solution for 5 minutes. After blocking with Tris-buffered saline and Tween 20 solution containing 1% bovine serum albumin, cells were incubated with indicated primary antibodies for 1 hour at room temperature. After that, cells were washed and incubated with fluorescein isothiocyanate or rhodamine-conjugated second primary antibodies (1:3000 dilution, Jackson ImmunoResearch) for 1 hour. Cells were counterstained with 100 ng/mL 4′,6-diamidino-2-phenylindole (DAPI) for 2 minutes to visualise nuclear DNA. The cover slips were mounted onto glass slides with anti-fade solution and visualised on a Nikon ECLIPSE E800 fluorescence microscope with a Nikon Plan Fluor 60× oil objective lens (NA 1.30).
RNA extraction, reverse transcription and real-time PCR
RNA samples were extracted with TRIZOL reagent (Invitrogen). Reverse transcription assay was performed by using the ProtoScript M-MuLV Taq RT-PCR Kit (New England Biolabs) according to the manufacturer’s instructions. Real-time PCR was performed by using Power SYBR Green PCR master mix (Applied Biosystems). For quantification of gene expression, the 2−ΔΔCt method was used. GAPDH expression was used for normalisation. The sequence information for each primer used for gene expression analysis is as follows:
For human cell lines:
CTGF-Forward: 5′-CCAATGACAACGCCTCCTG-3′
CTGF-Reverse: 5′-GAGCTTTCTGGCTGCACCA-3′
CYR61-Forward: 5′-AGCCTCGCATCCTATACAACC-3′
CYR61-Reverse: 5′-GAGTGCCGCCTTGTGAAAGAA-3′
ANKRD1-Forward: 5′-CACTTCTAGCCCACCCTGTGA-3′
ANKRD1-Reverse: 5′-CCACAGGTTCCGTAATGATTT-3′
GAPDH-Forward: 5′-ATGGGGAAGGTGAAGGTCG-3′
GAPDH-Reverse: 5′-GGGGTCATTGATGGCAACAATA-3′
AMOTL2-Forward: 5′-AGCTTCAATGAGGGTCTGCT-3′
AMOTL2-Reverse: 5′-TGAAGGACCTTGATCACTGC-3′
GLUT3-Forward: 5′-TTGAACACCTGCATCCTTGA-3′
GLUT3-Reverse: 5′-GACAGCCCATCATCATTTCC-3′
For mouse cell lines or tissues:
Amotl2-Forward: 5′-AGAGATTGGAATCGGCAAAC-3′
Amotl2-Reverse: 5′-TTCTCCTGTTCCTGTTGCTG-3′
Cyr61-Forward: 5′-ATGATGATCCAGTCCTGCAA-3′
Cyr61-Reverse: 5′-TAGGCTGTACAGTCGGAACG-3′
Gapdh-Forward: 5′-TGTTCCTACCCCCAATGTGT-3′
Gapdh-Reverse: 5′-TGTGAGGGAGATGCTCAGTG-3′
Acta2-Forward: 5′-AGGGCTGTTTTCCCATCCATCG-3′
Acta2-Reverse: 5′-TCTCTTGCTCTGGGCTTCATCC-3′
Ctgf-Forward: 5′-CAAGGACCGCACAGCAGTT-3′
Ctgf-Reverse: 5′-AGAACAGGCGCTCCACTCTG-3′
Glucose uptake assay
Cells were seeded in 10-cm dishes. Twenty-four hours later, cells were washed and refreshed with glucose-free DMEM with 10% dialysed FBS. Eighteen hours later, cells were treated with 2-NBDG (50 μM; Invitrogen) for 1 hour and washed with ice-cold phosphate-buffered saline solution twice. Glucose uptake was quantified in the propidium iodide–negative population of cells using fluorescence-activated cell sorting analysis.
Lactate production assay
Cells were seeded in 6-cm dishes. Forty-eight hours later, cell medium was removed and lactate concentration was determined by using Lactate Plus test strips and a Lactate Plus meter (Nova Biomedical). The cells remaining in the dish were harvested and counted by using a hemocytometer on a microscope. The rate of lactate production was calculated (lactate production = total lactate mole number/cell number).
Immunohistochemical analysis
Colon and liver tissue arrays were purchased from US Biomax. Samples were deparaffinised and rehydrated, and antigens were retrieved by applying Unmask Solution (Vector Laboratories) in a steamer for 30 minutes. To block endogenous peroxidase activity, the sections were treated with 1% hydrogen peroxide in methanol for 30 minutes. After 1 hour pre-incubation in 10% goat serum to prevent non-specific staining, the samples were incubated with an antibody to YAP (4912S, Cell Signaling Technology, 1:20 dilution) or GLUT3 (HPA006539, Sigma, 1:500 dilution) at 4°C overnight. The sections were incubated with SignalStain Boost Detection Reagent for 30 minutes at room temperature. Colour was developed with the SignalStain DAB Chromogen diluted solution (all reagents were obtained from Cell Signaling Technology). Sections were counterstained with Mayer hematoxylin. The correlation between YAP and GLUT3 and the correlation of YAP or GLUT3 with tissue type (normal versus cancer) were determined by the chi-square test.
Statistical analysis
Each experiment was repeated twice or more, unless otherwise noted. No samples or animals were excluded from the analysis. For the mouse fasting experiment, no statistical method was used to predetermine sample size. The samples or animals were randomly assigned to different groups. A laboratory technician who provided animal care and collected the livers was blinded to the group allocation during all animal experiments and outcome assessment. Differences between groups were analysed by the Student t-test and Pearson chi-square analysis. A p value < 0.05 was considered statistically significant.
Supplementary Material
Acknowledgments
We thank all of our colleagues in Dr. Chen’s laboratory for insightful discussion and technical assistance, especially Drs. Jingsong Yuan, Gargi Ghosal and Binoj C. Nair. We also thank Drs. Jae-Il Park, Li Ma, Yutong Sun, Jinsong Zhang, Aifu Lin and Hyemin Lee for reagents, insightful suggestions and comments on this work. We thank Dr. Fa-Xing Yu (University of California, San Diego) for technical help. We thank the shRNA-ORFeome Core Facility at MD Anderson Cancer Center for the ORFs and shRNAs. We thank Dr. Benoit Viollet for providing AMPK wild-type and knockout MEFs. We thank Kathryn Hale for proof-reading the manuscript. We thank Ross Tomaino for assistance with the mass spectrometry analysis. This work was supported in part by the U.S. Department of Defense Era of Hope research scholar award to J.C. (W81XWH-09-1-0409 and W81XWH-05-1-0470). This work was partly supported by the U.S. National Cancer Institute through the MD Anderson Cancer Center Support Grant (CA016672).
Footnotes
AUTHOR CONTRIBUTIONS
W.W. performed all of the experiments with assistance from Z-D.X., X.L., K.E.Z., B.G, R.L.J and J.C. W.W. and J.C. designed the experiments. J.C. supervised the study. W.W and J.C. wrote the manuscript. All authors commented on the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Pan D. The hippo signaling pathway in development and cancer. Dev Cell. 2010;19:491–505. doi: 10.1016/j.devcel.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhao B, Li L, Lei Q, Guan KL. The Hippo-YAP pathway in organ size control and tumorigenesis: an updated version. Genes Dev. 2010;24:862–874. doi: 10.1101/gad.1909210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Halder G, Johnson RL. Hippo signaling: growth control and beyond. Development. 2011;138:9–22. doi: 10.1242/dev.045500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhao B, Li L, Tumaneng K, Wang CY, Guan KL. A coordinated phosphorylation by Lats and CK1 regulates YAP stability through SCF(beta-TRCP) Genes Dev. 2010;24:72–85. doi: 10.1101/gad.1843810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jiao S, et al. A peptide mimicking VGLL4 function acts as a YAP antagonist therapy against gastric cancer. Cancer Cell. 2014;25:166–180. doi: 10.1016/j.ccr.2014.01.010. [DOI] [PubMed] [Google Scholar]
- 6.Zhang W, et al. VGLL4 functions as a new tumor suppressor in lung cancer by negatively regulating the YAP-TEAD transcriptional complex. Cell Res. 2014;24:331–343. doi: 10.1038/cr.2014.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu L, et al. Hippo signaling is a potent in vivo growth and tumor suppressor pathway in the mammalian liver. Proc Natl Acad Sci U S A. 2010;107:1437–1442. doi: 10.1073/pnas.0911427107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cai J, et al. The Hippo signaling pathway restricts the oncogenic potential of an intestinal regeneration program. Genes Dev. 2010;24:2383–2388. doi: 10.1101/gad.1978810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee KP, et al. The Hippo-Salvador pathway restrains hepatic oval cell proliferation, liver size, and liver tumorigenesis. Proc Natl Acad Sci U S A. 2010;107:8248–8253. doi: 10.1073/pnas.0912203107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Camargo FD, et al. YAP1 increases organ size and expands undifferentiated progenitor cells. Curr Biol. 2007;17:2054–2060. doi: 10.1016/j.cub.2007.10.039. [DOI] [PubMed] [Google Scholar]
- 11.Dong J, et al. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell. 2007;130:1120–1133. doi: 10.1016/j.cell.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mo JS, Park HW, Guan KL. The Hippo signaling pathway in stem cell biology and cancer. EMBO Rep. 2014 doi: 10.15252/embr.201438638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao B, et al. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 2007;21:2747–2761. doi: 10.1101/gad.1602907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao B, et al. Cell detachment activates the Hippo pathway via cytoskeleton reorganization to induce anoikis. Genes Dev. 2012;26:54–68. doi: 10.1101/gad.173435.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Halder G, Dupont S, Piccolo S. Transduction of mechanical and cytoskeletal cues by YAP and TAZ. Nat Rev Mol Cell Biol. 2012;13:591–600. doi: 10.1038/nrm3416. [DOI] [PubMed] [Google Scholar]
- 16.Dupont S, et al. Role of YAP/TAZ in mechanotransduction. Nature. 2011;474:179–183. doi: 10.1038/nature10137. [DOI] [PubMed] [Google Scholar]
- 17.Yu FX, et al. Regulation of the Hippo-YAP pathway by G-protein-coupled receptor signaling. Cell. 2012;150:780–791. doi: 10.1016/j.cell.2012.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jones RG, Thompson CB. Tumor suppressors and cell metabolism: a recipe for cancer growth. Genes Dev. 2009;23:537–548. doi: 10.1101/gad.1756509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism. Nat Rev Cancer. 2011;11:85–95. doi: 10.1038/nrc2981. [DOI] [PubMed] [Google Scholar]
- 20.Egan DF, et al. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science. 2011;331:456–461. doi: 10.1126/science.1196371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Crute BE, Seefeld K, Gamble J, Kemp BE, Witters LA. Functional domains of the alpha1 catalytic subunit of the AMP-activated protein kinase. J Biol Chem. 1998;273:35347–35354. doi: 10.1074/jbc.273.52.35347. [DOI] [PubMed] [Google Scholar]
- 22.Hao Y, Chun A, Cheung K, Rashidi B, Yang X. Tumor suppressor LATS1 is a negative regulator of oncogene YAP. J Biol Chem. 2008;283:5496–5509. doi: 10.1074/jbc.M709037200. [DOI] [PubMed] [Google Scholar]
- 23.Fujishiro SH, et al. ERK1/2 phosphorylate GEF-H1 to enhance its guanine nucleotide exchange activity toward RhoA. Biochem Biophys Res Commun. 2008;368:162–167. doi: 10.1016/j.bbrc.2008.01.066. [DOI] [PubMed] [Google Scholar]
- 24.Wu SZ, et al. Akt and RhoA activation in response to high glucose require caveolin-1 phosphorylation in mesangial cells. Am J Physiol Renal Physiol. 2014;306:F1308–1317. doi: 10.1152/ajprenal.00447.2013. [DOI] [PubMed] [Google Scholar]
- 25.Zhang Y, Peng F, Gao B, Ingram AJ, Krepinsky JC. High glucose-induced RhoA activation requires caveolae and PKCbeta1-mediated ROS generation. Am J Physiol Renal Physiol. 2012;302:F159–172. doi: 10.1152/ajprenal.00749.2010. [DOI] [PubMed] [Google Scholar]
- 26.Zhao B, et al. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 2008;22:1962–1971. doi: 10.1101/gad.1664408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stubbs M, McSheehy PM, Griffiths JR, Bashford CL. Causes and consequences of tumour acidity and implications for treatment. Mol Med Today. 2000;6:15–19. doi: 10.1016/s1357-4310(99)01615-9. [DOI] [PubMed] [Google Scholar]
- 28.Zhang H, Pasolli HA, Fuchs E. Yes-associated protein (YAP) transcriptional coactivator functions in balancing growth and differentiation in skin. Proc Natl Acad Sci U S A. 2011;108:2270–2275. doi: 10.1073/pnas.1019603108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamamoto T, et al. Over-expression of facilitative glucose transporter genes in human cancer. Biochem Biophys Res Commun. 1990;170:223–230. doi: 10.1016/0006-291x(90)91263-r. [DOI] [PubMed] [Google Scholar]
- 30.Mellanen P, Minn H, Grenman R, Harkonen P. Expression of glucose transporters in head-and-neck tumors. Int J Cancer. 1994;56:622–629. doi: 10.1002/ijc.2910560503. [DOI] [PubMed] [Google Scholar]
- 31.Boado RJ, Black KL, Pardridge WM. Gene expression of GLUT3 and GLUT1 glucose transporters in human brain tumors. Brain Res Mol Brain Res. 1994;27:51–57. doi: 10.1016/0169-328x(94)90183-x. [DOI] [PubMed] [Google Scholar]
- 32.Younes M, Brown RW, Stephenson M, Gondo M, Cagle PT. Overexpression of Glut1 and Glut3 in stage I nonsmall cell lung carcinoma is associated with poor survival. Cancer. 1997;80:1046–1051. doi: 10.1002/(sici)1097-0142(19970915)80:6<1046::aid-cncr6>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 33.Ayala FR, et al. GLUT1 and GLUT3 as potential prognostic markers for Oral Squamous Cell Carcinoma. Molecules. 2010;15:2374–2387. doi: 10.3390/molecules15042374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ha TK, Chi SG. CAV1/caveolin 1 enhances aerobic glycolysis in colon cancer cells via activation of SLC2A3/GLUT3 transcription. Autophagy. 2012;8:1684–1685. doi: 10.4161/auto.21487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guan’s paper in this issue
- 36.DeRan M, et al. Energy stress regulates hippo-YAP signaling involving AMPK-mediated regulation of angiomotin-like 1 protein. Cell Rep. 2014;9:495–503. doi: 10.1016/j.celrep.2014.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.