Abstract
Methionine adenosyltransferase 2B (MAT2B) encodes for variant proteins V1 and V2 that interact with GIT1 to increase ERK activity and growth in human liver and colon cancer cells. MAT2B or GIT1 overexpression activates MEK. This study explores the mechanism for MEK activation. We examined protein-protein interactions by co-immunoprecipitation and verified by confocal microscopy and pull-down assay using recombinant or in vitro translated proteins. Results were confirmed in an orthotopic liver cancer model. We found that MAT2B and GIT1-mediated MEK1/2 activation was not mediated by PAK1 or Src in HepG2 or RKO cells. Instead, MAT2B and GIT1 interact with B-Raf and c-Raf and enhance recruitment of Raf proteins to MEK1/2. MAT2B-GIT1 activates c-Raf, which is the key mediator for MEK/12 activation, because this still occurred in RKO cells that express constitutively active B-Raf mutant. The mechanism lies with the ability of MAT2B-GIT1 to activate Ras and promote B-Raf/c-Raf heterodimerization. Interestingly, MAT2B but not GIT1 can directly interact with Ras, which increases protein stability. Finally, increased Ras-Raf-MEK signaling occurred in phenotypically more aggressive liver cancers overexpressing MAT2B variants and GIT1. In conclusion, interaction between MAT2B and GIT1 serves as a scaffold and facilitates signaling in multiple steps of the Ras/Raf/MEK/ERK pathway, further emphasizing the importance of MAT2B/GIT1 interaction in cancer growth.
Methionine adenosyltransferase (MAT) is an essential enzyme expressed in all mammalian cells that catalyzes the formation of S-adenosylmethionine (SAMe), the principal biological methyl donor.1 There are three mammalian MAT genes. MAT1A and MAT2A encode for the catalytic subunit (α1 and α2) of the different MAT isoforms, and MAT2B encodes for a regulatory subunit (β) that modulates the activity of the MAT2A-encoded isoenzyme (MATII).1 MAT1A is predominantly expressed in normal hepatocytes, whereas MAT2A is expressed in all extrahepatic tissues.1 MAT2B shares a similar expression pattern as MAT2A.2 There are two major variant proteins encoded by MAT2B, MAT2BV1 (or V1) and MAT2BV2 (or V2), and both variants can increase MATII efficiency by lowering the Michaelis constant of MATII for its substrates methionine and ATP.2,3 In addition to regulating the kinetic properties of MATII, MAT2B is overexpressed in hepatocellular carcinoma (HCC) and colon cancer and offers the cancer cell a growth advantage.2,4
A key mechanism for MAT2B to enhance growth is ERK1/2 activation.2,4 Our previous work found that increased ERK1/2 activation occurs only when both MAT2B variants are present in addition to GIT1, a scaffold protein that facilitates c-Src–dependent mitogen-activated protein kinase (MAPK) activation.4 We found that both MAT2B variants directly interact with GIT1, and when these proteins are overexpressed, there is enhanced recruitment of ERK2 to MEK1 and the activity of both ERK1/2 and MEK1 increased.4 This finding proved to be important in tumorigenesis because overexpression of either V1 or V2 with GIT1 enhanced growth and lung metastasis in an orthotopic HCC model.4 Conversely, knockdown of endogenous V1, V2, or GIT1 lowered MEK1 and ERK1/2 activity.4 Thus, our previous work established MAT2B-GIT1 as a scaffold that facilitates MEK-ERK signaling.4 However, we did not examine how MAT2B-GIT1 complex activates MEK. Our current work examined the signaling pathways that can lead to MEK activation and identified MAT2B-GIT1 as a scaffold that acts on multiple levels of the Ras-Raf-MEK-ERK signaling cascade to facilitate their activation in human liver and colon cancer cells.
Materials and Methods
Cell Culture
HepG2, Hep3B, SW480, and RKO cell lines were obtained from the Cell Separation and Culture Core facility at the University of Southern California Research Center for Liver Diseases. NCM460 normal colon epithelial cells were from INCELL Corporation (San Antonio, TX) and grown in M3:base cell culture medium supplemented with 10% fetal bovine serum at 37°C in a 5% CO2 humidified incubator. HepG2 cells were maintained in Dulbecco’s modified Eagle’s medium (Corning, Manassas, VA) and Hep3B and RKO cells in modified Eagle’s medium (Corning) each with 10% fetal bovine serum (Seradigm, Radnor, PA). SW480 cells were maintained in L15 medium (Corning) with 10% fetal bovine serum in a humidified incubator without CO2.
Transfection and Quantitative PCR
Human GIT1 and MAT2B V1 and V2 expression plasmids were described previously.4 siRNA against GIT1 was purchased from Santa Cruz Biotechnology (Santa Cruz, CA), and siRNA against V1 and V2 were described previously.4 For gene overexpression experiments, 1.5 × 105 HepG2, Hep3B, RKO, and SW480 cells in 12-well plates were transiently transfected with V1, V2, GIT1 expression plasmids, or empty vector using Superfect (Qiagen, Valencia, CA) according to the manufacturer's protocol. For gene knockdown studies, 10 nmol/L siRNA against V1 and V2, 8 nmol/L siRNA against GIT1 (Santa Cruz Biotechnology), or 10 nmol/L scramble control were delivered into HepG2 or RKO cells by Lipofectamine RNAiMAX (Life Technologies, Grand Island, NY) following the manufacturer's protocol. For combination overexpression and knockdown experiments, 1.5 × 105 RKO cells were co-transfected with 10 nmol/L siRNA against c-Raf (Santa Cruz Biotechnology) and 1 μg of V1, V2, or GIT1 expression vector. Equal amounts of scramble control siRNA plus empty vector were used as a control. Forty-eight hours after transfection, total RNA or whole cell lysates were prepared for real-time quantitative PCR and Western blotting, respectively. Total RNA isolation, cDNA synthesis, real-time quantitative, primers, and Taqman probes were described previously.4 Probes for c-Raf, H-Ras, and K-Ras were purchased from Life Technologies.
Western Blotting
For Western blotting, 20 μg of whole cell extract were separated by 10% SDS-PAGE. Blots were probed with antibodies against phospho (p)-MEK1/2 (S218/222), p-MEK1/2 (S298), total MEK/12, p-c-Raf (S338), total c-Raf, p-B-Raf (T598/S601), total B-Raf, pSrc (Y416), total Src, pan Ras, K-Ras (Santa Cruz Biotechnology), GIT1, MAT2B, β-actin (Sigma, St Louis, MO), or V5 (Life Technologies). Western blotting was performed as previously described.2,4
Immunoprecipitation Studies
Immunoprecipitation studies were performed as described previously.4 Briefly, cells were lyzed in a modified radioimmunoprecipitation assay buffer with protease and phosphatase inhibitors. A total of 500 μg of whole cell lysate was used to co-immunoprecipitate with antibodies against MAT2B, GIT1, MEK1/2, or B-Raf. Western blots were performed to detect MAT2B, GIT1, V5, MEK1/2, c-Raf, B-Raf, PAK1, Ras, and SOS1/2. Clean Blot IP Detector Reagent (Thermo Scientific, Rockford, IL) was used to reduce background. Normal IgG (Santa Cruz Biotechnology) was used as a control.
In Vitro Translation and Protein Binding Assay
Full-length human MEK1 from pEGFP-N1-MEK1 (Addgene plasmid 14746; Addgene, Cambridge, MA) was subcloned into p-CDNA3.1-V5-His vector (Life Technologies) to generate p-CDNA3.1-V5-His-MEK1. Human B-Raf expression plasmid p-cDNA3.1-Hygro-BRaf was obtained from Addgene (Addgene plasmid 40775). Human GIT1 expression plasmid Xpress-GIT1was a gift from Dr. Bradford C. Berk (University of Rochester, Rochester, NY). In vitro transcription translation of full-length GIT1 or B-Raf proteins was performed using the TNT Quick Coupled Transcription/Translation System (Promega, Madison, WI) as per the manufacturer's protocol.
Human c-Raf recombinant protein (rc-Raf) was purchased from GeneTex (Irvine, CA). Human Ras recombinant protein (rRas) was purchased from Cytoskeleton, Inc. (Denver, CO). Recombinant human MAT2B protein (rMAT2B) was from Novus. In vitro pull-down assay was performed as described previously.4 To determine the influence of MAT2B and GIT1 on recruitment of B-Raf or c-Raf to MEK1, MEK1 protein was immobilized to protein A/G plus agarose (Santa Cruz Biotechnology) by incubating the protein with anti-MEK1 antibody for 16 hours and then incubating the two with the beads for an additional 2 hours at 4°C. The complex was then washed and incubated with full-length human B-Raf protein or rc-Raf protein for 2 hours, then rMAT2B, in vitro transcription-translation GIT1 (IVTGIT1), or both were added for an additional 16 hours, after which the beads were washed six times with protein-binding buffer. The protein-binding complexes were resolved on SDS-PAGE gel and detected by Western blot. To determine whether MAT2B and GIT1 enhance recruitment of B-Raf to c-Raf, human rc-Raf protein immobilized to protein A/G plus agarose was incubated with IVTB-Raf for 2 hours, then IVTGIT1 and rMAT2B proteins were added and incubated for 16 hours, after which the beads were washed. To determine direct interactions between Ras and MAT2B or GIT1, human rRas protein immobilized to protein A/G plus agarose was incubated with rMAT2B or IVTGIT1 alone or together for 16 hours and beads were washed. To determine whether MAT2B and GIT1 enhance recruitment of c-Raf and B-Raf to Ras, immobilized rRas protein was incubated with IVTB-Raf plus rc-Raf proteins for 4 hours, then rMAT2B plus IVTGIT1 proteins were added for 16 hours, after which beads were washed.
Immunofluorescence and Confocal Microscopy
Immunolabeling of MAT2B and B-Raf or c-Raf proteins in the same cell was performed according to a standard protocol (Jackson ImmunoResearch Laboratories, West Grove, PA), with minor modifications. Antibodies used are from Jackson ImmunoResearch Laboratories unless indicated otherwise. Briefly, cells on the coverslips were incubated in 0.1% Triton X-100 in phosphate-buffered saline for 15 minutes at room temperature and then blocked in 5% donkey serum for 1 hour. The coverslips were then incubated with anti-MAT2B antibody (Sigma) at 1:100 dilution for 2 hours, followed by incubation with a fluorescein isothiocyanate–conjugated donkey anti-rabbit antibody for 1 hour at room temperature. After washing, cells were blocked in 5% goat serum, followed by incubation with anti–B-Raf or c-Raf antibodies (1:50 dilution; Santa Cruz Biotechnology) for 2 hours and then with Cy3-conjugated goat anti-mouse antibody for 1 hour, washed in phosphate-buffered saline for 30 minutes, and mounted with mounting medium that contained DAPI (Sigma). The images were visualized and captured by an Eclipse TE300 confocal microscope (Nikon Instruments Inc., Melville, NY).
Ras Activity Assay
To evaluate Ras activation status, a Ras pull-down activation assay (Cytoskeleton, Inc., Denver, CO) was performed according to the manufacturer's protocol. Briefly, cell lysates were incubated with glutathione S-transferase–tagged Raf1-RBD protein bound to colored glutathione agarose beads for 1 hour at 4°C with rotation. Beads that contained activated Ras were collected by centrifugation, washed with wash buffer provided by a kit, and prepared and separated by SDS-PAGE. Activated Ras (c-Raf-RBD bound) was analyzed by immunoblotting with a mouse monoclonal anti-Pan Ras antibody provided by the kit. Activated Ras levels were normalized to c-Raf-RBD level, and total Ras levels and total Ras expression levels were normalized to actin levels. Total cell lysates (500 μg) were loaded with GTPγS as a positive control and GDP as a negative control for the pull-down assay.
Orthotopic Liver Cancer Model
The orthotopic liver cancer model that allowed examination of the effect of MAT2B V1 and V2 plus GIT1 overexpression was previously described.4 Briefly, 6-week-old male BALB/c nude mice were injected in the left hepatic lobe with Huh-7 cells stably expressing MAT2BV1 and V2 or empty vector (1 × 106 cells/50 μL) and concurrently under the same anesthesia injected intrasplenically with lentiviral vector expressing GIT1 or empty vector (2 × 109 transducing units in 0.05 mL). Two weeks later mice received tail vein injection with the same lentiviral vector expressing GIT1 or empty vector. All mice were sacrificed on day 21, when we reported increased tumor volume and presence of lung metastasis in those overexpressing V1 or V2 with GIT1.4 Tumors overexpressing V1+GIT1, V2+GIT1, or Vec+Vec were processed for co-immunoprecipitation and Western blotting as above. All protocols, use, and the care of the animals were reviewed and approved by the Institutional Animal Care and Use Committee at the University of Southern California (Los Angeles, CA).
Statistical Analysis
Densitometric values were measured using Quantity One Software version 4.6.5 (BioRad Laboratories, Hercules, CA). Data are given as means ± SEM. Statistical analysis was performed using analysis of variance followed by Fisher's test for multiple comparisons. All P values were derived from at least three independent experiments. Statistical significance was defined by P < 0.05.
Results
MAT2B and GIT1 Interact with c-Raf and B-Raf to Enhance Recruitment to MEK1/2 in Liver and Colon Cancer Cells
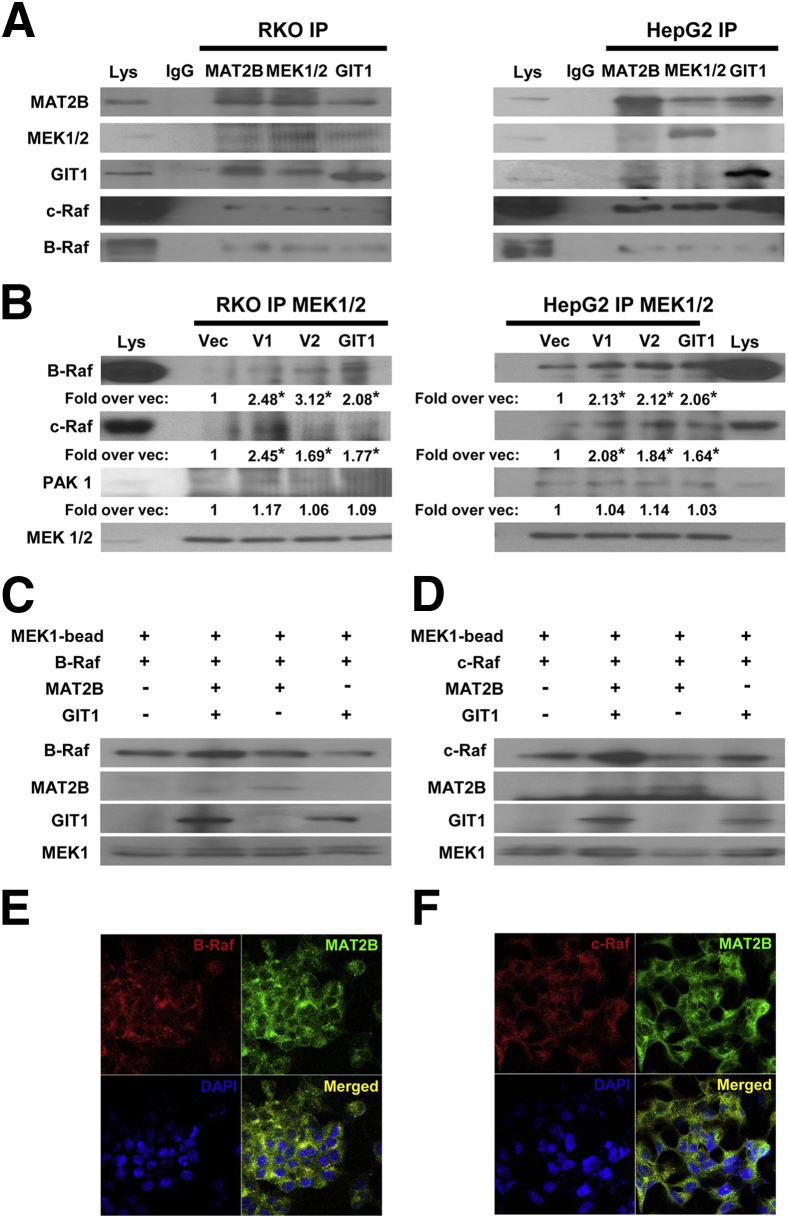
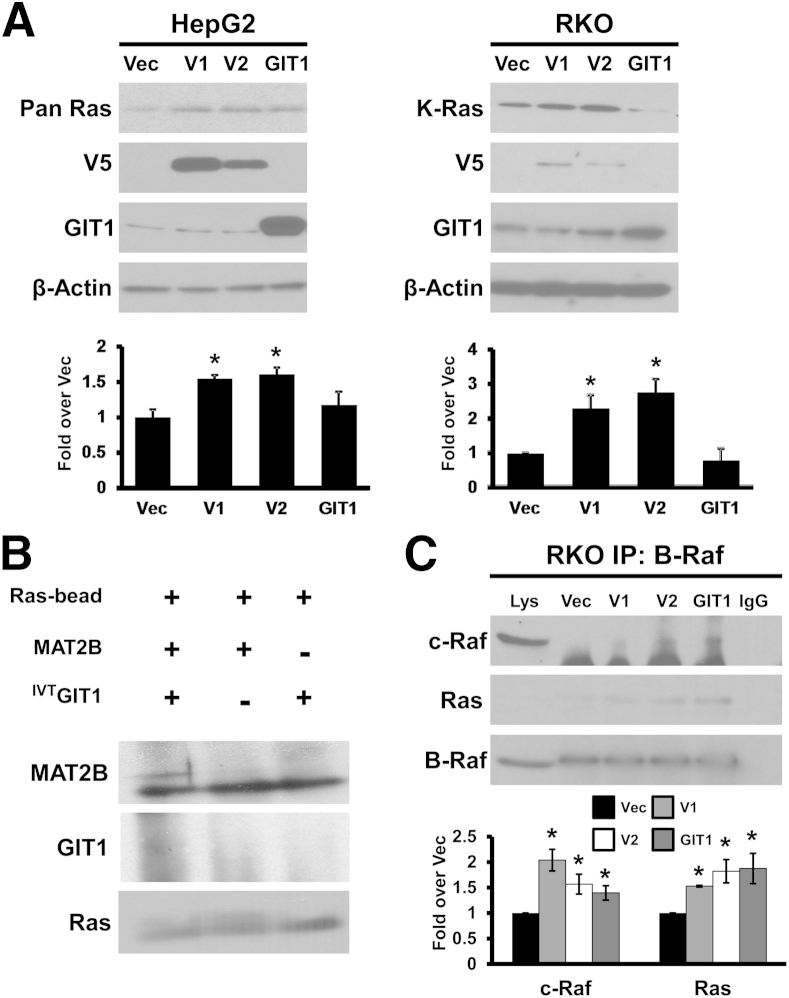
Raf kinases, particularly c-Raf (same as Raf-1) and B-Raf, are well known to activate MEK.5 We first examined whether GIT1 and MAT2B interact with c-Raf and B-Raf. In both RKO and HepG2 cells, co-immunoprecipitation experiments indicate that MAT2B and GIT1 interact with c-Raf and B-Raf (Figure 1A). Furthermore, overexpression of V1, V2, or GIT1 in both cell types led to increased recruitment of both B-Raf and c-Raf to MEK1/2 (Figure 1B). Using an in vitro system, enhanced recruitment of B-Raf and c-Raf to MEK1 occurred only when both MAT2B and GIT1 were present (Figure 1, C and D). Finally, immunofluorescence and confocal microscopy confirmed that endogenous MAT2B interact with B-Raf and c-Raf in HepG2 cells (Figure 1, E and F).
Figure 1.
Methionine adenosyltransferase 2B (MAT2B) and GIT1 interact with c-Raf and B-Raf and enhance their recruitment to MEK when overexpressed. A: Immunoprecipitation (IP) of cell lysates (Lys) from RKO and HepG2 cells for MEK, MAT2B, and GIT1 were immunoblotted with antibodies to MAT2B, MEK1/2, GIT1, c-Raf, and B-Raf. Normal IgG was used as a negative control, whereas 1% of cell lysates used in IP were loaded as input. B: RKO or HepG2 cells were transfected with overexpression vectors for V1, V2, GIT1, or empty vector (Vec) as described in Materials and Methods followed by IP for MEK1/2 and immunoblotting for B-Raf, c-Raf, PAK1, and MEK1/2. Numbers below the blots represent mean densitometric values normalized to MEK1/2 from at least three independent experiments expressed as fold over vector control. C:In vitro pull-down assay using immobilized MEK1 plus B-Raf, with MAT2B or GIT1 alone or together. Note that MAT2B and GIT1 must both be present to enhance recruitment of B-Raf to MEK1. D:In vitro pull-down assay with immobilized MEK1 plus c-Raf, with MAT2B or GIT1 alone or together. Both MAT2B and GIT1 must be present to enhance recruitment of c-Raf to MEK1. E and F: Immunofluorescence detection of MAT2B (green), B-Raf or c-Raf (red), and nuclei (blue) in HepG2 cells. Merged images are shown in the bottom right of each panel, in which yellow color shows cytoplasmic co-localization of MAT2B and B-Raf (E) or c-Raf (F). Data are representative of at least three independent experiments (A–D). ∗P < 0.05 versus Vec.
We also examined whether the same interactions occur in NCM460 cells, which are normal human colon epithelial cells. Interestingly, although both MAT2B and GIT1 interact with each other as well as MEK1/2, c-Raf, and B-Raf (Supplemental Figure S1A), overexpressing V1, V2, or GIT1 failed to activate B-Raf, c-Raf, or MEK1/2 (Supplemental Figure S1B) or enhance recruitment of B-Raf and c-Raf to MEK1/2 (Supplemental Figure S1C). In terms of relative expression of MAT2B and GIT1, NCM460 cells have 40% lower GIT1 mRNA levels compared with two malignant colon cancer cells (RKO and SW480, which have comparable expression). MAT2BV1 mRNA levels are highest in RKO cells, followed by SW480, and lowest in NCM460 cells at ratios of 3.8:2.6:1. However, MAT2BV2 mRNA levels are similar between NCM460 cells and RKO cells and both are approximately twofold higher than in SW480 cells.
Effect of MAT2B-GIT1 on MEK1/2 Activation Is PAK1 and Src Independent
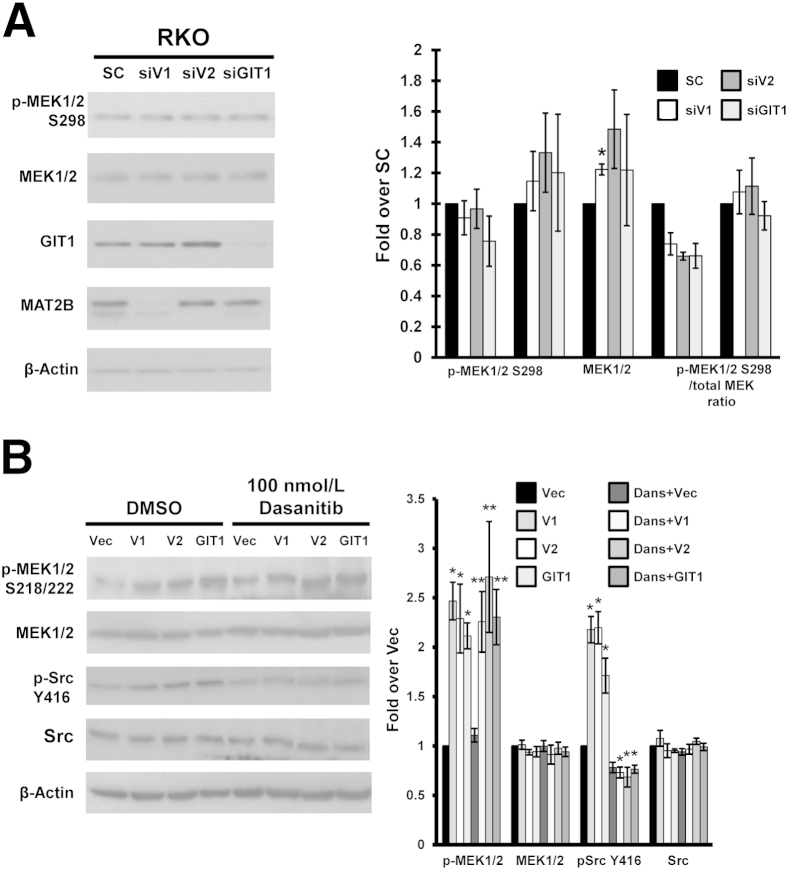
Besides Raf kinases, MEK1/2 activation can occur by PAK1-mediated phosphorylation of MEK1 on S298, which primes activation by c-Raf.6 However, overexpression of V1, V2, or GIT1 did not increase interaction between MEK1/2 and PAK1 (Figure 1B). Whereas knockdown of V1, V2, or GIT1 lowered c-Raf–mediated phosphorylation of MEK1 on S218/S222 that is required for MEK1/2 activity,4,7 they had no effect on PAK1-mediated phosphorylation of MEK1/2 at S298 (Figure 2A). Another potential mediator for increased MEK1/2 activation is Src because it is able to stimulate c-Raf via phosphorylation of tyrosine 341.8,9 We next investigated whether Src family members may be involved using dasatinib, a potent inhibitor of Src kinase activity.10 Overexpression of V1, V2, or GIT1 increased p-MEK1 on S218/S222 and p-Src on Y416 (required for Src activity).11 Treatment with dasanitib completely blocked Src activation as indicated by level of p-Src Y416 but had no influence on MEK1/2 activation (Figure 2B).
Figure 2.
The effect of methionine adenosyltransferase 2B (MAT2B) and GIT1 on MEK is PAK1 and Src independent. A: RKO cells were treated with siRNA against V1 (siV1), V2 (siV2), GIT1 (siGIT1), or scramble control (SC). Protein levels of phospho (p)-MEK1/2 at S298 (site of phosphorylation by PAK1), total MEK1/2, GIT1, and MAT2B were measured by Western blotting after normalizing to β-actin housekeeping control. Graph summarizes the densitometric values expressed as fold over SC from three independent experiments. B: RKO cells were transfected with overexpression vectors of V1, V2, GIT1, or empty vector (Vec) for 24 hours before 100 nmol/L dasanitib (Dans) was added for an additional 24 hours. Protein levels of p-MEK1/2 (S218/222), total MEK1/2, pSrc (Y416), and total Src were measured by Western blotting after normalizing to β-actin housekeeping control. Graph summarizes the densitometric values expressed as fold over Vec. Results are means ± SEM from three to four independent experiments (B). ∗P < 0.05, ∗∗P < 0.01 versus respective controls. DMSO, dimethyl sulfoxide.
MAT2B and GIT1 Up-Regulate c-Raf and B-Raf Activity and Their Heterodimerization
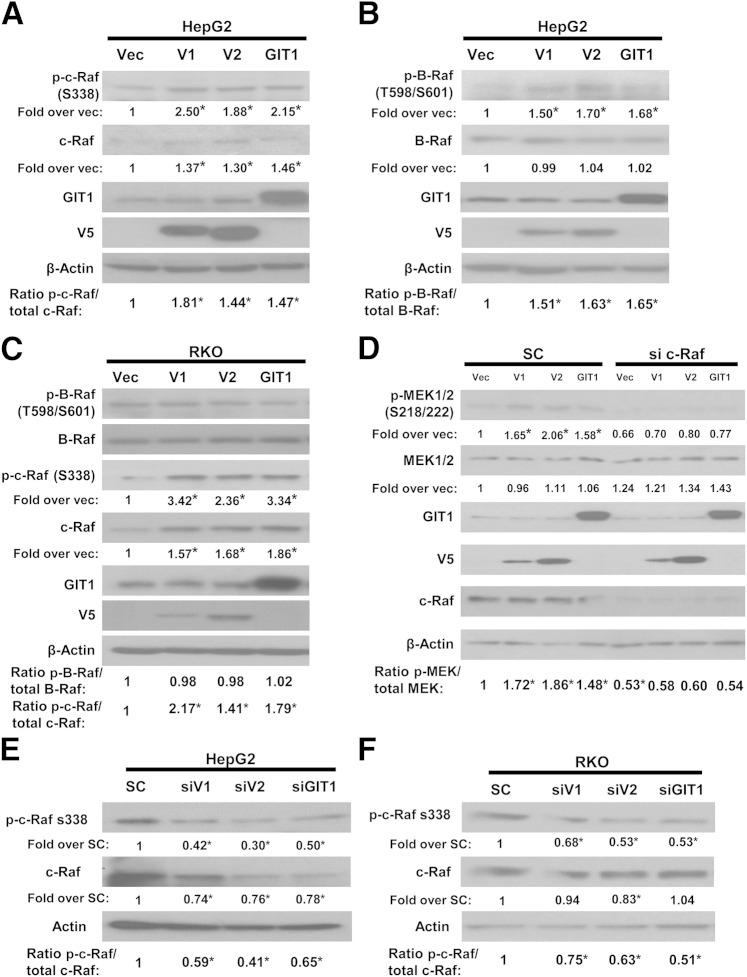
We next investigated whether MAT2B and GIT1 influenced the activity of c-Raf and B-Raf. In HepG2 cells, which express wild-type c-Raf and B-Raf,12 overexpression of V1, V2, or GIT1 activated both Rafs as indicated by p-c-Raf on S338 and p-B-Raf on T598 and S601 (Figure 3, A and B).8 In RKO cells, which express constitutively active B-Raf mutant,13 overexpression of V1, V2, or GIT1 activated c-Raf but had no influence on B-Raf activity (Figure 3C). In both cell types, overexpression of V1, V2, or GIT1 also raised total c-Raf (but not B-Raf) protein levels (Figure 3, A–C). This effect was not mediated at the mRNA level because c-Raf mRNA levels were unchanged (Supplemental Figure S2). To determine whether c-Raf is responsible for V1-, V2-, and GIT1-mediated MEK1/2 activation, RKO cells were treated with scramble or siRNA targeting c-Raf in conjunction to overexpressing V1, V2, or GIT1. Overexpression of V1, V2, or GIT1 activated MEK1/2 in the scramble siRNA treated cells but not in those treated with c-Raf siRNA (Figure 3D). In contrast, knockdown of V1, V2, or GIT1 in both HepG2 (Figure 3E) and RKO (Figure 3F) cells markedly reduced levels of active c-Raf, with less effect on total c-Raf.
Figure 3.
Methionine adenosyltransferase 2B (MAT2B) and GIT1 expression regulate c-Raf and B-Raf activity. A and B: HepG2 cells (expressing wild-type c-Raf and B-Raf) were transfected with V1, V2, GIT1 overexpression vector, or empty vector (Vec) and processed for Western blotting for phospho (p)-c-Raf (S338, active c-Raf) and total c-Raf (A) and p-B-Raf (T598/S601, active B-Raf) and total B-Raf (B). Numbers below the blots are densitometric values expressed as fold over Vec after normalizing to β-actin housekeeping control. Ratios of p-c-Raf to total c-Raf and p-B-Raf to total B-Raf are also shown. C: RKO cells (expressing constitutively active B-Raf mutant but wild-type c-Raf) were transfected with V1, V2, GIT1, or Vec as above and processed for Western blotting for active and total B-Raf and c-Raf. Numbers below the blots are densitometric values expressed as fold over Vec. Ratios of p-c-Raf to total c-Raf and p-B-Raf to total B-Raf are also shown. D: RKO cells were transfected with V1, V2, or GIT1 overexpression vectors concurrent with knockdown of c-Raf by siRNA versus scramble control (SC). Protein levels of p-MEK1/2 (S218/222), total MEK1/2, GIT1, V5 tag, and c-Raf were measured by Western blotting and normalized to β-actin. Numbers below the blots are densitometric values expressed as fold over Vec. Ratios of p-MEK to total MEK are also shown. E and F: HepG2 and RKO cells were treated with SC, V1, V2, or GIT1 siRNA and processed for Western blotting for p-c-Raf (S338, active c-Raf) and total c-Raf. Numbers below the blots are densitometric values expressed as fold over SC after normalizing to β-actin housekeeping control. Ratios of p-c-Raf to total c-Raf are also shown. Results are representative of three independent experiments (C and D). ∗P < 0.05 versus Vec (A–D) or versus SC (E and F).
To make sure the effects of overexpressing V1, V2, or GIT1 on activating B-Raf/c-Raf/MEK or recruiting B-Raf/c-Raf to MEK1/2 were not unique to HepG2 and RKO cells, we also examined this in SW480 and Hep3B cells, which are human colon adenocarcinoma and hepatoma cell lines, respectively. In both SW480 and Hep3B cells, overexpressing V1, V2, or GIT1 activated B-Raf, c-Raf, and MEK1/2 and enhanced recruitment of B-Raf and c-Raf to MEK1/2 (Supplemental Figure S3).
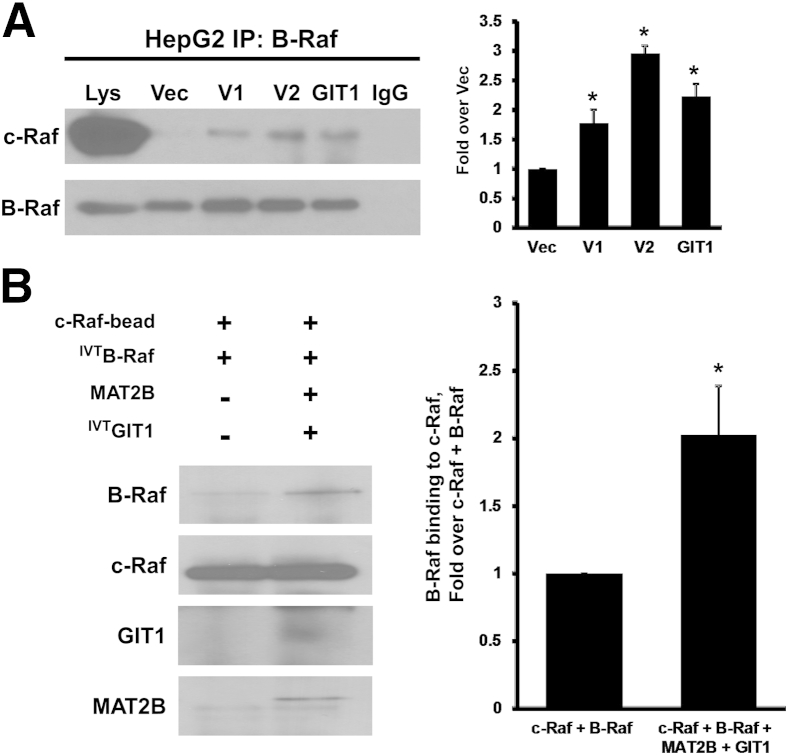
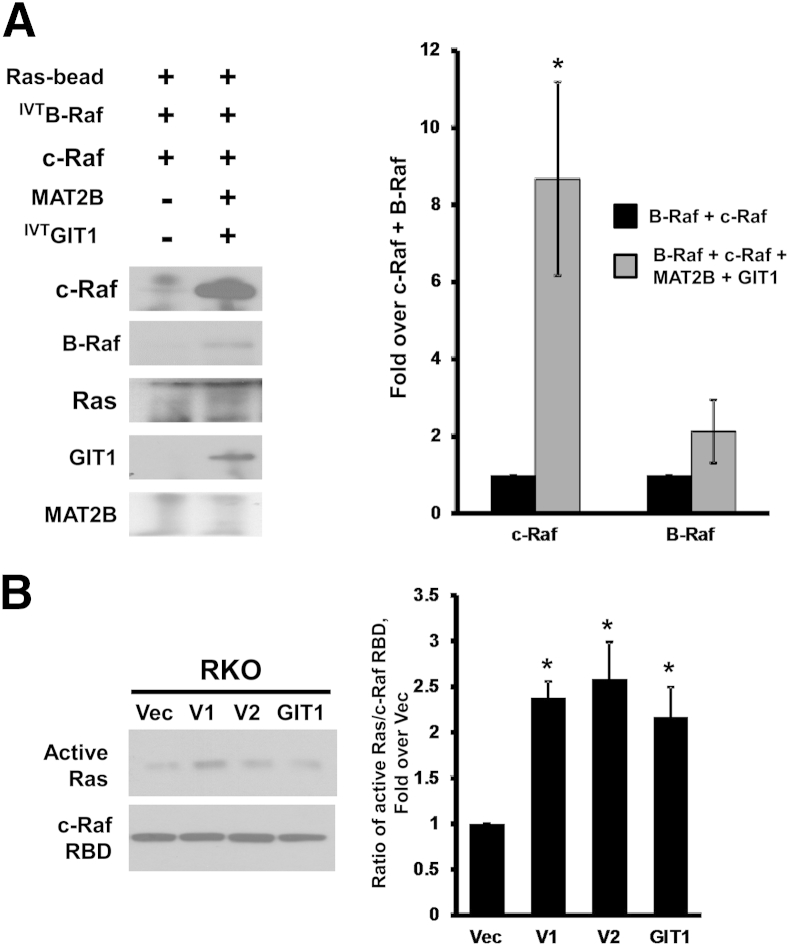
Heterodimerization of c-Raf and B-Raf is known to greatly enhance kinase activity compared with the respective homodimers or monomers.14 Overexpression of V1, V2, or GIT1 in HepG2 cells enhanced interaction between B-Raf and c-Raf (Figure 4A), and in the in vitro system, presence of MAT2B and GIT1 enhanced binding of B-Raf to immobilized c-Raf (Figure 4B).
Figure 4.
Overexpression of methionine adenosyltransferase 2B (MAT2B) and GIT1 enhances heterodimerization of B-Raf and c-Raf. A: HepG2 cells (expressing wild-type c-Raf and B-Raf) overexpressing V1, V2, GIT1, or empty vector (Vec) were subjected to immunoprecipitation (IP) for B-Raf and immunoblotted for c-Raf and B-Raf as described in Materials and Methods. One percentage of cell lysates (Lys) used in IP were loaded as input. Graph summarizes the densitometric values expressed as fold over Vec of c-Raf recruited to B-Raf. B:In vitro pull-down assay with immobilized c-Raf plus in vitro transcription-translation B-raf (IVTB-Raf), with or without MAT2B and GIT1. Graph summarizes the densitometric values expressed as fold over c-Raf+B-Raf alone. Data are expressed as means ± SEM from three independent experiments (A and B). ∗P < 0.05 versus Vec (A) or versus c-Raf+B-Raf (B).
MAT2B and GIT1 Activate Ras and Enhance Recruitment of c-Raf to Ras
Ras proteins lie upstream of c-Raf and B-Raf in the signal-transduction cascade. HepG2 cells express mutant K-Ras and oncogenic dominant-acting N-Ras,12,15 whereas RKO cells express mainly wild-type K-Ras.16 Interestingly, overexpression of V1 or V2, but not GIT1, raised total Ras protein levels in HepG2 cells and K-Ras in RKO cells (Figure 5A). This effect lies at the protein level because the mRNA levels of H-Ras and K-Ras were unaffected (Supplemental Figure S4). Using purified proteins in an in vitro assay, direct interaction occurred between Ras and MAT2B but not between Ras and GIT1 (Figure 5B). In RKO cells, overexpression of V1, V2, or GIT1 enhanced recruitment of not only c-Raf to B-Raf but also Ras to the complex (Figure 5C). The presence of MAT2B and GIT1 also markedly enhanced recruitment of c-Raf and to a smaller extent B-Raf to immobilized Ras (Figure 6A). Finally, overexpression of V1, V2, or GIT1 in RKO cells resulted in higher Ras activity even after normalizing for the effect they have on total Ras expression (Figure 6B).
Figure 5.
Methionine adenosyltransferase 2B (MAT2B) interacts with Ras and enhances Ras expression and recruitment to B-Raf. A: HepG2 (expressing mutant K-Ras and oncogenic dominant-acting N-Ras) and RKO (expressing wild-type K-Ras) cells were transfected with V1, V2, GIT1 overexpression vectors, or empty vector (Vec), and protein levels of pan Ras, K-Ras, V5 tag, and GIT1 were measured by Western blotting after normalizing to β-actin. Graphs summarize the densitometric values expressed as fold over Vec. B:In vitro pull-down assay with immobilized Ras plus MAT2B and/or in vitro transcription-translation GIT1 (IVTGIT1) shows that Ras directly interacts with MAT2B but not GIT1. C: RKO cells overexpressing V1, V2, and GIT1 were subjected to immunoprecipitation (IP) for B-Raf and immunoblotted for c-Raf, pan Ras, and B-Raf. Normal IgG was used as a negative control, whereas 1% of cell lysates (Lys) used in IP were loaded as input. Data are expressed means ± SEM from three independent experiments (A), blots from three independent experiments (B), and fold over Vec from three independent experiments (C). ∗P < 0.05 versus Vec.
Figure 6.
Methionine adenosyltransferase 2B (MAT2B) and GIT1 promote recruitment of c-Raf to Ras and activate Ras. A:In vitro pull-down assay with immobilized Ras and in vitro transcription-translation B-Raf (IVTB-Raf) and c-Raf, with or without MAT2B and IVTGIT1, reveals that the presence of MAT2B and GIT1 enhances recruitment of c-Raf and perhaps also B-Raf to Ras. B: Ras activity assay was measured as described in Materials and Methods in RKO cells (expressing wild-type K-Ras) overexpressing V1, V2, GIT1, or empty vector (Vec) for 48 hours. Results are representative of three independent experiments. ∗P < 0.05 versus Vec.
HCC that Overexpresses V1/V2 Plus GIT1 Exhibits Increased Interaction and Activation of Ras-Raf-MEK Signaling
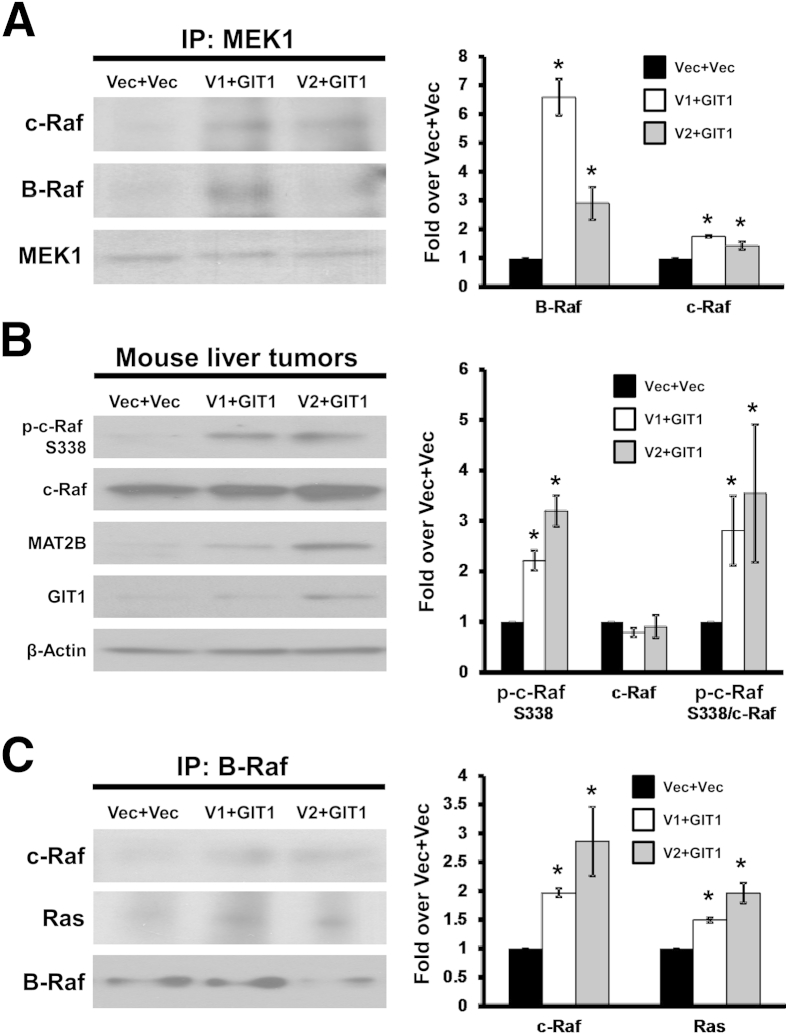
We previously found that use of an in vivo orthotopic HCC model that increased V1, V2, or GIT1 expression individually enhanced tumor growth, but when V1 or V2 was combined with GIT1, tumor growth was further enhanced and distant metastasis occurred.4 Increased expression of V1 or V2 with GIT1 resulted in increased c-Raf and B-Raf recruitment to MEK1 (Figure 7A), higher activation of c-Raf (Figure 7B), and enhanced interaction between B-Raf and c-Raf and with Ras (Figure 7C).
Figure 7.
Increased methionine adenosyltransferase 2B (MAT2B) and GIT1 expression in hepatocellular carcinoma (HCC) enhances recruitment of c-Raf and B-Raf to MEK1, c-Raf activation, and interaction of B-Raf with c-Raf and Ras. Liver tumors were derived from an orthotopic HCC model, where Huh7 cells overexpressing MAT2BV1, V2, or empty vector (Vec) were injected into the livers of nude mice and then subjected to a total of two tail vein injections of a lentiviral vector overexpressing GIT1 or Vec during a 3-week period were used. A: Recruitment of c-Raf and B-Raf to MEK1 using immunoprecipitation for MEK1 followed by immunoblotting for c-Raf and B-Raf. B: Representative Western blots showing the expression level of phospho (p)-c-Raf (S338), total c-Raf, MAT2B, and GIT1 in the tumors of each treatment group. C: Recruitment of c-Raf and Ras to B-Raf using immunoprecipitation (IP) for B-Raf followed by immunoblotting for c-Raf and Ras. Graph summarizes the densitometric values normalized to MEK1 (A), β-actin (B), and B-Raf (C) expressed as fold over Vec+Vec from three tumors per condition. ∗P < 0.05 versus Vec+Vec.
Discussion
Until recently, MAT2B was only known in the regulation of MATII enzymatic activity.1 Both MAT2B variants lower the Michaelis constant of MATII for its substrates and V1 (same as the β subunit) also lowers the inhibitory constant for SAMe.1,3 The first indication that MAT2B is involved in growth regulation of liver cancer cells was reported in 2003 by Martínez-Chantar et al,17 who found increased MAT2B expression in human HCC and cirrhotic livers and correlated increased liver cancer cell growth to lowering of the SAMe level, consistent with its known effect on MATII activity. Because a high SAMe level is growth inhibitory and proapoptotic in liver cancer cells,1 the ability of MAT2B to modulate liver cancer cell growth was thought to be related to a change in the SAMe level.17 In 2008, we first reported the existence of multiple MAT2B splicing variants, with V1 and V2 being dominant.2 The two variants differ only by 20 amino acids at the 5′ end but exhibit differential tissue expression pattern and regulation by tumor necrosis factor-α (positively regulates V1 at the promoter level but not V2).2 In addition, although both variants are overexpressed in HCC and positively regulate growth, only V1 is involved in apoptosis regulation.2 These findings suggest that although they share many common features, they also have distinct functions that may be independent of SAMe regulation. Indeed, we later reported that both variants are predominantly localized in the nuclear compartment and interact with multiple proteins, including HuR, an mRNA-binding protein known to stabilize the mRNA levels of many genes, including cyclins, and GIT1.4,18 Overexpression of V1 or V2 resulted in increased cytoplasmic HuR content and higher mRNA levels of several cyclins, which is one of the key mechanisms of MAT2B's growth inductive effect.18
Another key mechanism we identified is the ability of MAT2B variants to form a scaffold complex with GIT1 in the activation of MEK and ERK in both liver and colon cancer cell lines.4 Overexpression of V1, V2, or GIT1 increased MEK and ERK activity, whereas the opposite occurred when their endogenous expression was reduced by siRNA.4 Interestingly, V1, V2, and GIT1 are all indispensable because ERK activation and growth induction could not occur if any of these were knocked down by siRNA.4 The relevance of MAT2B-GIT1 interaction was confirmed in human HCC and colon cancer specimens, where their expression was increased in most and their interaction documented by co-immunoprecipitation.4 Enhanced expression of V1 or V2 in conjunction with GIT1 was particularly growth inductive in an orthotopic HCC model and resulted in distant metastasis.4 Taken together, we identified MAT2B-GIT1 as a key component required for MEK-ERK signaling, but the mechanism(s) responsible for MEK activation was unknown. The goal of our current work was to elucidate how MAT2B-GIT1 activates MEK, and during this work, we found that MAT2B-GIT1 is a scaffold complex that regulates multiple steps in the Ras-Raf-MEK-ERK signaling cascade.
MEK is an integral part of the Ras-Raf-MEK-ERK MAPK signaling pathway, where activation of Ras followed by Raf kinases phosphorylate two serine residues (S218 and S222) in the activation loop of MEK, which in turn activates ERK1/2.19 The MAPK cascade is not a linear pathway because each member consists of a number of components. In humans, three Ras genes encode four distinct but highly homologous Ras proteins: H-Ras, N-Ras, K-RasA, and K-RasB (the latter two are alternative splice variants of the KRAS gene).20,21 Ras proteins are activated when GTP bound and inactivated when GDP bound.20 There are three Raf kinases (A-Raf, B-Raf, and c-Raf) that share MEK1/2 as substrates, although A-Raf has very low kinase activity toward MEK.5 HepG2 cells express oncogenic dominant-acting N-Ras, and RKO cells express predominantly wild-type K-Ras.15,16 Although both cell types express B-Raf and c-Raf, HepG2 cells express wild-type Rafs and RKO cells express mutant B-Raf that is constitutively active.13 In addition to Ras-mediated signaling, PAK1 and Src can also help activate MEK1/2. PAK1 phosphorylates MEK1 on S298, which primes activation by c-Raf.6 Src activates c-Raf by phosphorylating c-Raf on Y341.8,9 Given these considerations, we began our studies focusing on the key players, namely, c-Raf, B-Raf, PAK1, and Src, which can lead to MEK activation in HepG2 and RKO cells.
We found that MAT2B and GIT1 interact with both c-Raf and B-Raf endogenously in HepG2 and RKO cells. Moreover, overexpression of V1, V2, or GIT1 enhanced recruitment of these Raf kinases to MEK1/2. Importantly, with the use of in vitro assays in which MEK1 was immobilized, MAT2B and GIT1 enhanced recruitment of B-Raf and c-Raf to MEK1 only if they were both present. This result is consistent with our previous observation that the effect on MEK/ERK activation and growth requires the presence of both MAT2B and GIT1.4 We excluded PAK1 being involved in activating MEK for several reasons. Overexpression of V1, V2, or GIT1 did not enhance interaction between MEK1/2 and PAK1. Knockdown of V1, V2, or GIT1 lowered MEK activity4 but did not affect PAK1-mediated phosphorylation of MEK1/2 on S298. We also excluded Src being involved because inhibition of Src activation by dasanitib had no influence on MEK activation induced by overexpressing V1, V2, or GIT1. However, overexpression of MAT2B variants and GIT1 activated Src, the implication of which will require further study.
MAT2B and GIT1 overexpression not only enhanced interaction between the Raf kinases and MEK1/2 but also activated both Raf kinases in HepG2 cells and c-Raf in RKO cells. Because RKO cells express a mutant B-Raf that is constitutively active, overexpression of MAT2B or GIT1 had no effect on B-Raf activation but they still increased MEK1/2 activity. This finding suggests that in RKO cells the mechanism is independent of B-Raf activation. Consistently, knockdown of c-Raf in RKO cells completely blocked MEK1/2 activation by MAT2B and GIT1 overexpression. In addition, knockdown of V1, V2, or GIT1 reduced c-Raf activation in both HepG2 and RKO cells.
B-Raf and c-Raf form homodimers and heterodimers, with heterodimers exhibiting much higher kinase activity.5 Overexpression of MAT2B and GIT1 in both HepG2 and RKO cells enhanced interaction between B-Raf and c-Raf. Overexpressing V1, V2, or GIT1 also activated B-Raf, c-Raf, and MEK1/2 and enhanced recruitment of B-Raf and c-Raf to MEK1/2 in SW480 and Hep3B cells. Several proteins have been reported to enhance B-Raf and c-Raf dimerization, including Ras, 14-3-3, KSR1, and MLK3.5 The in vitro assay using immobilized c-Raf revealed clearly that the presence of the MAT2B-GIT1 complex facilitated recruitment of B-Raf in the absence of any additional proteins. This finding also suggests that MAT2B and GIT1 may activate MEK in RKO cells via facilitating c-Raf/B-Raf heterodimerization. MAT2B-GIT1 complex likely can also stabilize c-Raf because the c-Raf protein but not mRNA levels increased when they were overexpressed and decreased when they were reduced. This can then further contribute to activating Raf and downstream events.
Interestingly, although MAT2B and GIT1 interacted with MEK1/2, c-Raf, and B-Raf in NCM460 cells, which are normal human colonic epithelial cells, overexpressing V1, V2, or GIT1 failed to enhance recruitment of c-Raf and B-Raf to MEK1/2 or activate these kinases. The reasons for the different response to V1, V2, and GIT1 overexpression between malignant and nonmalignant cells are not clear. One possibility is the lower expression level of GIT1 and the dominant MAT2B variant V1 so that even when overexpressed, the scaffold complex may not form efficiently to affect recruitment of the Raf kinases or to activate them. In addition, we and others have reported that NCM460 cells respond differently to agents that induce apoptosis in colon cancer cells, and others have also reported that NCM460 cells have different MAPK and c-Myc signaling responses compared with colon cancer cells.22,23
Because MAT2B and GIT1 overexpression increased the level of active Raf at Ras target sites (phosphorylation of c-Raf on S338 and B-Raf on T598/S601) (Figure 3), the next question was whether Ras was affected. Interestingly, overexpression of MAT2B but not GIT1 increased Ras protein levels in both HepG2 and RKO cells. This finding is likely due to protein stabilization because mRNA levels were unchanged. In terms of Ras protein regulation, it is generally agreed that on activation by growth factors, such as insulin-like growth factor 1, Ras translocates to the plasma membrane and is farnesylated at the COOH terminus, which is required for activation and membrane anchorage.24 When Ras farnesylation is inhibited, Ras is dislodged from the membrane and degraded.25 The insulin-like growth factor 1 receptor stabilizes Ras by enhancing farnesylation.24 More recently, H-Ras was found to be polyubiquitylated and degraded via proteasome by Wnt/β-catenin signaling.26 How MAT2B affects Ras protein stability will require further examination.
In vitro, MAT2B but not GIT1 can directly bind to immobilized Ras, but in cells, overexpression of V1, V2, or GIT1 can enhance recruitment of c-Raf and Ras to B-Raf. This finding can be explained by the fact that overexpression of GIT1 will also recruit more MAT2B to interact with Ras. Overexpression of V1, V2, or GIT1 also activated Ras in RKO cells that express wild-type Ras. Ras activity is regulated by several mechanisms, including guanine nucleotide exchange and lipid-based modifications, such as farnesylation, which are required for membrane translocation and functionality.24 SOS is a well-known Ras activator that translocates to the plasma membrane on growth factor activation, interacts with Ras, and facilitates Ras activation.27 However, we did not detect increased interaction between Ras with SOS in cells overexpressing V1, V2, or GIT1 (data not shown). Thus, the mechanism for MAT2B/GIT1 overexpression to activate Ras is not due to their ability to help Ras also recruit SOS. How MAT2B/GIT1 activates Ras will be the subject of future investigation.
We previously found that increased V1 or V2 expression in conjunction with GIT1 resulted in a much more aggressive liver cancer phenotype as indicated by enhanced growth and distant metastasis.4 We also found that these tumors exhibited increased MEK activity.4 Our current work confirmed the upstream signaling because increased V1, V2, and GIT1 expression in cell lines activated c-Raf and enhanced interaction of Ras with Raf kinases and recruitment of Raf kinases to MEK in these tumors. Mutations in the Ras-Raf-MEK-ERK signaling pathway are well known in many cancers, and targeting this pathway has been the subject of intense research.28 However, clinical success has been disappointing,28 and paradoxical activation in Raf kinase can occur with B-Raf–specific inhibitor (thought to be due to enhanced heterodimerization of B-Raf and c-Raf).8 Our finding that MAT2B and GIT1, which are overexpressed in human liver and colon cancer,4 form a scaffold complex that positively regulates Ras signaling at multiple steps suggests that targeting this complex may be a novel and effective approach in cancer therapy. Although multiple scaffold proteins, such as KSR1/2, IQGAP1, MP1, and β-Arrestin1/2, have been described to regulate Ras-Raf-MEK-ERK signaling, to the best of our knowledge, none of them interact with all the components of this signaling pathway.29
Acknowledgment
Human GIT1 expression plasmid Xpress-GIT1was a gift from Dr. Bradford C. Berk (University of Rochester, Rochester, NY). HepG2, SW480, Hep3B, and RKO cells were provided by the Cell Separation and Culture Core facility. Confocal microscopy was provided by the Imaging Core of the University of Southern California Research Center for Liver Diseases.
Footnotes
Supported by NIH grant R01DK51719 (S.C.L. and J.M.M.); Plan Nacional of I+D SAF 2011-29851, Departamento de Educación del Gobierno Vasco (J.M.M.); and grant P30DK48522 from the Imaging Core of the USC Research Center for Liver Diseases.
Disclosures: M.P.M. is the founder, a co-owner, and a full-time employee of INCELL Corporation, which provided the NCM460 cells.
Current address of S.C.L., Cedar-Sinai Medical Center, Los Angeles, CA.
Supplemental Data
Methionine adenosyltransferase 2B (MAT2B) and GIT1 interact with MEK1/2, c-Raf, and B-Raf but do not regulate their activity in NCM460 cells. A: Immunoprecipitation (IP) of cell lysates from NCM460 cells for MAT2B, MEK1/2, and GIT1 were immunoblotted with antibodies to MAT2B, MEK1/2, GIT1, c-Raf, and B-Raf. Normal IgG was used as a negative control, whereas 1% of cell lysates used in IP were loaded as input. B: NCM460 cells were transfected with overexpression vectors for V1, V2, GIT1, or empty vector (Vec) as described in Materials and Methods and processed for Western blotting for phospho (p)-B-Raf (T598/S601, active B-Raf) and total B-Raf, p-c-Raf (S338, active c-Raf), total c-Raf, p-MEK1/2 (S218/222, active form), and total MEK1/2. Numbers below the blots are densitometric values expressed as fold over Vec after normalizing to β-actin housekeeping control. Ratios of p-B-Raf to total B-Raf, p-c-Raf to total c-Raf, and p-MEK1/2 to total MEK are also shown. C: NCM460 cells were transfected as in B followed by IP for MEK1/2 and immunoblotting for B-Raf, c-Raf, and MEK1/2. Numbers below the blots represent mean densitometric values normalized to MEK1/2 from at least three independent experiments expressed as fold over vector control.
Effect of V1, V2, or GIT1 overexpression on V1, V2, GIT1, and c-Raf mRNA levels. HepG2 cells were transfected with V1, V2, and GIT1 overexpression vector or empty vector (Vec) for 48 hours as described in Materials and Methods, and mRNA levels of V1, V2, GIT1 (A), and c-Raf (B) were measured using real-time quantitative PCR. Results are from duplicate samples from one experiment expressed as means ± SEM. ∗P < 0.05 versus Vec.
Methionine adenosyltransferase 2B (MAT2B) and GIT1 overexpression regulates c-Raf and B-Raf activity in SW480 and Hep3B cells. A: SW480 and Hep3B cells were transfected with V1, V2, GIT1 overexpression vector, or empty vector (Vec) and processed for Western blotting for phospho (p)-B-Raf (T598/S601, active B-Raf), total B-Raf, p-c-Raf (S338, active c-Raf), total c-Raf, p-MEK1/2 (S218/222, active MEK), and total MEK. Ratios of activated forms over total are shown below after normalizing to the β-actin housekeeping control and expressing as fold over Vec. B: SW480 and Hep3B cells were transfected with overexpression vectors for V1, V2, GIT1, or empty vector (Vec) as described in Materials and Methods followed by immunoprecipitation for MEK1/2 and immunoblotting for B-Raf, c-Raf, and MEK1/2. Numbers below the blots represent mean densitometric values normalized to MEK1/2 from at least three independent experiments expressed as fold over vector control. ∗P < 0.05 versus Vec.
Effect of V1, V2, or GIT1 overexpression on H-Ras and K-Ras mRNA levels. HepG2 cells were transfected with V1, V2, GIT1 overexpression vector, or empty vector (Vec) for 48 hours as described in Materials and Methods, and mRNA levels of H-RAS and K-RAS were measured using real-time quantitative PCR. Results are from duplicate samples from one experiment expressed as means ± SEM.
References
- 1.Lu S.C., Mato J.M. S-adenosylmethionine in liver health, injury and cancer. Physiol Rev. 2012;92:1515–1542. doi: 10.1152/physrev.00047.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang H.P., Iglesias Ara A., Magilnick N., Xia M., Ramani K., Chen H., Lee T.D., Mato J.M., Lu S.C. Expression pattern, regulation, and functions of methionine adenosyltransferase 2beta splicing variants in hepatoma cells. Gastroenterology. 2008;134:281–291. doi: 10.1053/j.gastro.2007.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nordgren K.K.S., Peng Y., Pelleymounter L.L., Moon I., Abo R., Feng Q., Eckloff B., Yee V.C., Wieben E., Weinshilboum R.M. Methionine adenosyltransferase 2A/2B and methylation: gene sequence variation and functional genomics. Drug Metab Dispos. 2011;39:2135–2147. doi: 10.1124/dmd.111.040857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peng H., Dara L., Li T.W.H., Zheng Y., Yang H.P., Tomasi M.L., Tomasi I., Giordano P., Mato J.M., Lu S.C. Methionine adenosyltransferase 2B-GIT1 interplay activates MEK1-ERK1/2 to induce growth in human liver and colon cancer. Hepatology. 2013;57:2299–2313. doi: 10.1002/hep.26258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Matallanas D., Birtwistle M., Romano D., Zebisch A., Rauch J., von Kriegsheim A., Kolch W. Raf family kinases: old dogs have learned new tricks. Genes Cancer. 2011;2:232–260. doi: 10.1177/1947601911407323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coles L.C., Shaw P.E. PAK1 primes MEK1 for phosphorylation by Raf-1 kinase during cross-cascade activation of the ERK pathway. Oncogene. 2002;21:2236–2244. doi: 10.1038/sj.onc.1205302. [DOI] [PubMed] [Google Scholar]
- 7.Roskoski R., Jr. MEK1/2 dual-specificity protein kinases: structure and regulation. Biochem Biophys Res Commun. 2012;417:5–10. doi: 10.1016/j.bbrc.2011.11.145. [DOI] [PubMed] [Google Scholar]
- 8.Roskoski R., Jr. RAF protein-serine/threonine kinases: structure and regulation. Biochem Biophys Res Commun. 2010;399:313–317. doi: 10.1016/j.bbrc.2010.07.092. [DOI] [PubMed] [Google Scholar]
- 9.Stokoe D., McCormick F. Activation of c-Raf-1 by Ras and Src through different mechanisms: activation in vivo and in vitro. EMBO J. 1997;16:2384–2396. doi: 10.1093/emboj/16.9.2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buettner R., Mesa T., Vultur A., Lee F., Jove R. Inhibition of Src family kinases with dasatinib blocks migration and invasion of human melanoma cells. Mol Cancer Res. 2008;6:1766–1774. doi: 10.1158/1541-7786.MCR-08-0169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roskoski R., Jr. Src kinase regulation by phosphorylation and dephosphorylation. Biochem Biophys Res Commun. 2005;331:1–14. doi: 10.1016/j.bbrc.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 12.Liu L., Cao Y., Chen C., Zhang X., McNabola A., Wilkie D., Wilhelm S., Lynch M., Carter C. Sorafenib blocks the RAF/MEK/ERK pathway, inhibits tumor angiogenesis, and induces tumor cell apoptosis in hepatocellular carcinoma model PLC/PRF/5. Cancer Res. 2006;66:11851–11858. doi: 10.1158/0008-5472.CAN-06-1377. [DOI] [PubMed] [Google Scholar]
- 13.Deng G., Bell I., Crawley S., Gum J., Terdiman J.P., Allen B.A., Truta B., Sleisenger M.H., Kim Y.S. BRAF mutation is frequently present in sporadic colorectal cancer with methylated hMLH1, but not in hereditary nonpolyposis colorectal cancer. Clin Cancer Res. 2004;10:191–195. doi: 10.1158/1078-0432.ccr-1118-3. [DOI] [PubMed] [Google Scholar]
- 14.Rushworth L.K., Hindley A.D., O'Neill E., Kolch W. Regulation and role of Raf-1/B-Raf heterodimerization. Mol Cell Biol. 2006;26:2262–2272. doi: 10.1128/MCB.26.6.2262-2272.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Richards C.A., Short S.A., Thorgeirsson S.S., Huber B.E. Characterization of a transforming N-ras gene in the human hepatoma cell line HepG2: additional evidence for the importance of c-myc and ras cooperation in hepatocarcinogenesis. Cancer Res. 1990;50:1521–1527. [PubMed] [Google Scholar]
- 16.Suter C.M., Norrie M., Ku S.L., Cheong K.F., Tomlinson I., Ward R.L. CpG island methylation is a common finding in colorectal cancer cell lines. Br J Cancer. 2003;88:413–419. doi: 10.1038/sj.bjc.6600699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martínez-Chantar M.L., Garcia-Trevijano E.R., Latasa M.U., Martin-Duce A., Fortes P., Caballeria J., Avila M.A., Mato J.M. Methionine adenosyltransferase II beta subunit gene expression provides a proliferative advantage in human hepatoma. Gastroenterology. 2003;124:940–948. doi: 10.1053/gast.2003.50151. [DOI] [PubMed] [Google Scholar]
- 18.Xia M., Chen Y., Wang L.C., Zandi E., Yang H.P., Bemanian S., Martínez-Chantar M.L., Mato J.M., Lu S.C. Novel function and intracellular localization of methionine adenosyltransferase 2beta splicing variants. J Biol Chem. 2010;285:20015–20021. doi: 10.1074/jbc.M109.094821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pouyssegur J., Volmat V., Lenormand P. Fidelity and spatio-temporal control in MAP kinase (ERKs) signalling. Biochem Pharmacol. 2002;64:755–763. doi: 10.1016/s0006-2952(02)01135-8. [DOI] [PubMed] [Google Scholar]
- 20.Pylayeva-Gupta Y., Grabocka E., Bar-Sagi D. RAS oncogenes: weaving a tumorigenic web. Nat Rev Cancer. 2011;11:761–774. doi: 10.1038/nrc3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fernández-Medarde A., Santos E. Ras in cancer and developmental diseases. Genes Cancer. 2011;2:344–358. doi: 10.1177/1947601911411084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li T., Zhang Q., Oh P., Xia M., Chen H., Bemanian S., Circ M., Moyer M.P., Mato J.M., Aw T.Y., Lu S.C. S-Adenosylmethionine and methylthioadenosine inhibit cellular FLICE inhibitory protein expression and induce apoptosis in colon cancer cells. Mol Pharmacol. 2009;76:192–200. doi: 10.1124/mol.108.054411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zeng H., Briske-Anderson M., Wu M., Moyer M.P. Methylselenol, a selenium metabolite, plays common and different roles in cancerous colon HCT116 cell and noncancerous NCM460 colon cell proliferation. Nutr Cancer. 2012;64:128–135. doi: 10.1080/01635581.2012.630555. [DOI] [PubMed] [Google Scholar]
- 24.Gatzka M., Prisco M., Baserga R. Stabilization of the Ras oncoprotein by the Insulin-like Growth Factor 1 Receptor during anchorage-independent growth. Cancer Res. 2000;60:4222–4230. [PubMed] [Google Scholar]
- 25.Haklai R., Weisz M.G., Elad G., Paz A., Marciano D., Egozi Y., Ben-Baruch G., Kloog Y. Dislodgment and accelerated degradation of Ras. Biochemistry. 1998;37:1306–1314. doi: 10.1021/bi972032d. [DOI] [PubMed] [Google Scholar]
- 26.Kim S.E., Yoon J.Y., Jeong W.J., Jeon S.H., Park Y., Yoon J.B., Park Y.N., Kim H., Choi K.Y. H-Ras is degraded by Wnt/β-catenin signaling via β-TrCP-mediated polyubiquitylation. J Cell Sci. 2009;122:842–848. doi: 10.1242/jcs.040493. [DOI] [PubMed] [Google Scholar]
- 27.Groves J.T., Kuriyan J. Molecular mechanisms in signal transduction at the membrane. Nat Struct Mol Biol. 2010;17:659–665. doi: 10.1038/nsmb.1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sacco E., Spinelli M., Vanoni M. Approaches to Ras signaling modulation and treatment of Ras-dependent disorders: a patent review (2007-present) Expert Opin Ther Pat. 2012;22:1263–1287. doi: 10.1517/13543776.2012.728586. [DOI] [PubMed] [Google Scholar]
- 29.Roskoski R., Jr. ERK1/2 MAP kinases: structure, function, and regulation. Pharmacol Res. 2012;66:105–143. doi: 10.1016/j.phrs.2012.04.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Methionine adenosyltransferase 2B (MAT2B) and GIT1 interact with MEK1/2, c-Raf, and B-Raf but do not regulate their activity in NCM460 cells. A: Immunoprecipitation (IP) of cell lysates from NCM460 cells for MAT2B, MEK1/2, and GIT1 were immunoblotted with antibodies to MAT2B, MEK1/2, GIT1, c-Raf, and B-Raf. Normal IgG was used as a negative control, whereas 1% of cell lysates used in IP were loaded as input. B: NCM460 cells were transfected with overexpression vectors for V1, V2, GIT1, or empty vector (Vec) as described in Materials and Methods and processed for Western blotting for phospho (p)-B-Raf (T598/S601, active B-Raf) and total B-Raf, p-c-Raf (S338, active c-Raf), total c-Raf, p-MEK1/2 (S218/222, active form), and total MEK1/2. Numbers below the blots are densitometric values expressed as fold over Vec after normalizing to β-actin housekeeping control. Ratios of p-B-Raf to total B-Raf, p-c-Raf to total c-Raf, and p-MEK1/2 to total MEK are also shown. C: NCM460 cells were transfected as in B followed by IP for MEK1/2 and immunoblotting for B-Raf, c-Raf, and MEK1/2. Numbers below the blots represent mean densitometric values normalized to MEK1/2 from at least three independent experiments expressed as fold over vector control.
Effect of V1, V2, or GIT1 overexpression on V1, V2, GIT1, and c-Raf mRNA levels. HepG2 cells were transfected with V1, V2, and GIT1 overexpression vector or empty vector (Vec) for 48 hours as described in Materials and Methods, and mRNA levels of V1, V2, GIT1 (A), and c-Raf (B) were measured using real-time quantitative PCR. Results are from duplicate samples from one experiment expressed as means ± SEM. ∗P < 0.05 versus Vec.
Methionine adenosyltransferase 2B (MAT2B) and GIT1 overexpression regulates c-Raf and B-Raf activity in SW480 and Hep3B cells. A: SW480 and Hep3B cells were transfected with V1, V2, GIT1 overexpression vector, or empty vector (Vec) and processed for Western blotting for phospho (p)-B-Raf (T598/S601, active B-Raf), total B-Raf, p-c-Raf (S338, active c-Raf), total c-Raf, p-MEK1/2 (S218/222, active MEK), and total MEK. Ratios of activated forms over total are shown below after normalizing to the β-actin housekeeping control and expressing as fold over Vec. B: SW480 and Hep3B cells were transfected with overexpression vectors for V1, V2, GIT1, or empty vector (Vec) as described in Materials and Methods followed by immunoprecipitation for MEK1/2 and immunoblotting for B-Raf, c-Raf, and MEK1/2. Numbers below the blots represent mean densitometric values normalized to MEK1/2 from at least three independent experiments expressed as fold over vector control. ∗P < 0.05 versus Vec.
Effect of V1, V2, or GIT1 overexpression on H-Ras and K-Ras mRNA levels. HepG2 cells were transfected with V1, V2, GIT1 overexpression vector, or empty vector (Vec) for 48 hours as described in Materials and Methods, and mRNA levels of H-RAS and K-RAS were measured using real-time quantitative PCR. Results are from duplicate samples from one experiment expressed as means ± SEM.