Abstract
PTEN and PIK3CA mutations occur with high frequency in uterine endometrioid carcinoma (UEC). Although PTEN mutations are present in complex atypical hyperplasia and carcinoma, PIK3CA mutations are restricted to carcinoma. We generated mouse models harboring Pten loss and/or activated Pik3ca in the endometrial epithelium to investigate their respective roles in the pathogenesis of UEC. Presence of an activated mutant Pik3ca on the background of Pten loss led to aggressive disease, with 100% of mice exhibiting carcinoma. Expression of Pik3ca with E545K mutation alone was unable to cause hyperplasia or cancer in the uterus and did not activate Akt as effectively as Pten deletion in short-term cultures of mouse endometrial epithelium, likely explaining the lack of phenotype in vivo. We also report that nuclear localization of FOXO1 correlated with PTEN mutational status irrespective of the PIK3CA status in endometrial cancer cell lines. Furthermore, gene expression profiles resulting from Pten loss or activation of Pik3ca in primary mouse endometrial epithelial cells exhibit minimal overlap. Thus, Pten and Pik3ca have distinct consequences on the activation of the phosphatidylinositol 3-kinase pathway in endometrial epithelium and are likely to affect other nonoverlapping cellular mechanisms involved in the development and progression of the most common type of uterine cancer.
Endometrial carcinoma is a significant cause of morbidity and mortality worldwide, and in the United States, it is the eighth most common cause of cancer-related deaths in women.1 Endometrial carcinoma is most often of endometrioid morphology, and is usually preceded by complex atypical hyperplasia (CAH). The development of both endometrial hyperplasia and uterine endometrioid carcinoma (UEC) is associated with unopposed estrogen stimulation and/or specific genetic alterations.1 Approximately 30% to 80% of primary UECs harbor PTEN mutations,2–4 with a similar frequency detected in CAH. In contrast, PIK3CA mutations occur in 20% to 40% of UECs and are rarely, if ever, found in CAH.
Phosphatase and tensin homolog (PTEN) and PIK3CA are main components of the phosphatidylinositol 3-kinase (PI3K)/AKT/PTEN pathway with opposing actions. PIK3CA encodes the p110α catalytic subunit of the PI3K complex and is considered an oncogene. As a component of PI3K, it is activated downstream of growth factor signaling, resulting in phosphorylation of phosphatidylinositol-4,5-phosphate (PIP2) to generate phosphatidlyinositol-3,4,5-phosphate (PIP3). PIP3 activates AKT, a protein kinase that regulates several downstream pathways that impinge on cell proliferation, cell growth, and apoptosis. In contrast, PTEN is a tumor-suppressor gene encoding a dual-specificity phosphatase, capable of dephosphorylating both proteins and lipids. Its most well-described activity is the conversion of PIP3 to PIP2, indirectly inhibiting the action of PI3K. Thus, by preventing AKT activation, PTEN inhibits cell proliferation.5,6
Most PTEN mutations in CAH and UEC are localized to exons encoding the lipid phosphatase domain, resulting in loss of its ability to dephosphorylate PIP3.2,7,8 PIK3CA mutations, E542K E545K (both in exon 9) and H1047R (in exon 20), are hotspots, found in endometrial carcinoma, as well as several other cancers and lead to constitutive activation of p110α.5 Because loss of PTEN or activation of PIK3CA lead to activation of the pathway, it is reasonable to assume that they have similar downstream effects. However, as noted above, PTEN mutations are found in both CAH and UEC, whereas PIK3CA mutations are almost exclusively found in UEC, suggesting unique and nonoverlapping functions in endometrial carcinoma pathogenesis.
Pten+/− female mice develop CAH by 32 weeks of age, and at approximately 52 weeks of age, 25% of the Pten+/− female mice develop UEC. Both CAH and UEC exhibit complete loss of Pten expression, as a result of biallelic Pten inactivation by either loss of heterozygosity or intragenic mutations in the wild-type allele.9,10 In this mouse model, the frequency of biallelic inactivation is similar in CAH and UEC, and we have previously shown that mismatch repair deficiency hastens inactivation of the remaining Pten allele and promotes the progression of hyperplastic lesions to UEC.9 These observations suggest that, although biallelic inactivation of Pten is an early event in endometrial tumorigenesis, it may not be sufficient for progression to invasive carcinoma. Given the genetic analyses of primary human tumors, it seems likely that mutations in PIK3CA may promote progression to carcinoma.
To determine the effect of loss of PTEN and/or activated PIK3CA in the development of UEC, we have developed several genetic mouse models. The results reported herein demonstrate distinct roles of these two genes in the pathogenesis of endometrial carcinoma.
Materials and Methods
Animals
PtenloxP/loxP mice on a Balbc/129SvJ background and Ksp1.3-Cre+/− mice were obtained from The Jackson Laboratory (Bar Harbor, ME). To investigate the role of Pik3ca, a mouse line with a Cre-inducible mutant form of Pik3ca (designated as Pik3caE545K/+) was constructed in the laboratory of Dr. Suzanne Baker (St. Jude Children's Research Hospital, Memphis, TN). The Pik3caE545K/+ mouse contains a lox-STOP-lox cassette upstream of the Pik3ca gene containing the E545K mutation. In the presence of Cre, the stop codon is excised such that the mutant allele is expressed from its endogenous promoter. These mice were bred with PtenloxP/loxP strain to generate PtenloxP/loxP;Pik3caE545K/+ (Ptenf/f;Pik3caE545K), PtenloxP/loxP;Pik3ca+/+ (Ptenf/f), Pten+/+; Pik3caE545K/+ (Pik3caE545K), and PtenloxP/+;Pik3caE545K/+ (Ptenf/+;Pik3caE545K) mice. Mice expressing Cre under the Wnt7a promoter (Wnt7a-Cre+/−) were obtained from Dr. Kenneth S. Korach (National Institute of Environmental Health Science, Research Triangle Park, NC). Ptenf/f;Pik3caE545K mice were crossed with the Wnt7a-Cre and Ksp1.3-Cre mice to generate the described genotypes. All animal experiments were performed in accordance with Institutional Animal Care and Use Committee guidelines.
IHC Staining
The uteri harvested from all mice at indicated time points were fixed in formalin and embedded in paraffin. Sections (5 μm thick) were processed for hematoxylin and eosin staining and immunohistochemical (IHC) analysis. For IHC, the slides were deparaffinized, rehydrated, and subjected to antigen retrieval by boiling in the microwave in 10 mmol/L sodium citrate. The Vectastain ABC reagent (Vector Labs, Burlingame, CA) was then used according to the manufacturer's protocol. Antibodies used were Pten (1:100), phospho-Akt S473 (1:100), β-catenin (1:800), p-Gsk3β (1:50), p-Pras40 (1:800), Stathmin (1:50), and FoxO1 (1:100), all from Cell Signaling Technologies (Beverly, MA). Two human tissue microarrays (TMAs), one containing 100 cases of normal cycling endometrium and the second with 96 cases of UEC (24 International Federation of Gynecology and Obstetrics grade 1, 37 International Federation of Gynecology and Obstetrics grade 2, and 35 International Federation of Gynecology and Obstetrics grade 3 cases) generated at Weill Cornell Medical College (New York, NY) under Institutional Review Board approval were also stained with FOXO1 antibody, as per the protocol described above.
Microdissection
Areas corresponding to CAH, UEC, or normal epithelium were microdissected from deparaffinized, rehydrated, hematoxylin-stained sections (5 μm thick) under light microscope visualization with a 26-gauge needle. The DNA was extracted using previously published protocols.11,12
Primary Uterine Epithelial Cell Cultures
Epithelial cells from uteri of Ptenf/f;Pik3caE545K, Ptenf/f, and Pik3caE545K mice were prepared as described previously.13 Briefly, the horns were slit lengthwise and digested in a solution containing 0.25% trypsin in Hanks’ balanced salt solution for 1 hour at 4°C, followed by another incubation for 1 hour at 22°C. The tissue pieces were transferred to ice-cold Hanks’ balanced salt solution and vortex mixed to release sheets of epithelial cells. The cell suspension was filtered through a 20 μm nylon mesh, and the epithelial sheets (retained on the filter) were collected and resuspended in Dulbecco’s modified Eagle’s medium/F12 (1:1), 10% charcoal-stripped fetal bovine serum, 20 mmol/L HEPES, 100 μg/mL streptomycin, 100 U/mL penicillin, and 2 mmol/L l-glutamine and insulin-transferrin-selenium supplement (1×). Cells were plated in 6-well dishes coated with 1:10 diluted Matrigel (Life Technologies, Grand Island, NY) and cultured at 37°C in an incubator with 5% CO2. After reaching 80% to 90% confluence, the cells were infected with adenoviruses expressing Cre or green fluorescent protein (GFP; control), as described previously,13 for 48 hours. Cells were harvested for RNA or protein extraction. The RNA was used for RNA sequencing analysis and for subsequent validation of differentially expressed genes (DEGs) by real-time quantitative PCR (qPCR). Whole cell protein lysates were used for immunoblot analysis using Pten, phospho-Akt, and Akt antibodies (Cell Signaling Technologies), all at 1:1000 dilution. Actin antibody (Sigma, St. Louis, MO) was used for normalization.
Nuclear and Cytoplasmic Extract Preparation
Endometrial cancer cell lines Hec1A parental clone (Hec1A WT) and PTEN-deleted Hec1A (Hec1A clone 16) have been described previously.14 KLE and RL95 cells were purchased from ATCC (Manassas, VA) and were cultured as per the ATCC protocol. Nuclear and cytoplasmic extracts were prepared using the NE-PER Nuclear Protein Extraction Kit from Thermo Fisher Scientific (Waltham, MA), as per the manufacturer's protocol. The extracts were resolved by 4% to 20% SDS-PAGE and transferred to a polyvinylidene difluoride membrane, and immunoblot analysis was performed using p-FOXO1 and FOXO1 antibodies. Lamin A/C and glyceraldehyde-3-phosphate dehydrogenase antibodies were used to ensure purity of the nuclear and cytoplasmic extracts, respectively. All antibodies were purchased from Cell Signaling Technologies and used at a dilution of 1:1000.
RNA Sequencing and Analysis
Total RNA was extracted from the cells using the RNeasy Plus Universal Micro kit (Qiagen, Valencia, CA), as per the manufacturer's instructions. After isolation, total RNA integrity was checked using a 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA), and samples with RNA integrity number value greater than eight were used for library preparation and sequencing. Library construction, template hybridization, and cluster amplification were performed using the TruSeq RNA Sample Preparation Kit version 2 (Illumina Inc., San Diego, CA). After cluster generation, sequencing by synthesis was performed on the HiSeq2000/1000 machine (Illumina Inc.).
The RNA sequencing data were analyzed using Genesifter software (Geospiza, Seattle, WA). Raw reads from an Illumina HiSeq2000 in fastq format were filtered to remove any reads that contained primarily adaptor/primer sequences. Reads were then aligned using Burrows-Wheeler Aligner against the genomic reference sequence for Mus musculus (Build 37.2). The Mus musculus (Build 37.2) reference sequence and annotation were pulled from RefSeq database (http://www.ncbi.nlm.nih.gov/refseq; last accessed November 12, 2014), and reads mapping to the genome were characterized as exon, intron, or intergenic (outside any annotated gene). These aligned data were then used to calculate gene expression by taking the total of exon and known splice reads for each annotated gene to generate a count value per gene. For each gene, a normalized expression value was generated in two ways: reads per mapped million (RPMM; which is calculated by taking the count value and dividing it by the number of million mapped reads) and RPMM per kilobase (RPMM value divided by the kilobase length of the longest transcript for each gene). The RPMM values were used for comparing gene expression across samples to remove the bias of different numbers of reads mapped per sample. RPMM per kilobase values were used for comparing relative expression of genes to one another to remove the bias of different numbers of mapped reads and different transcript lengths. Samples were initially analyzed in a pairwise fashion with normalization by the number of million mapped reads (not including rRNA, tRNA, snRNA, or mitochondrial RNA), followed by likelihood ratio test, and Benjamini and Hochberg correction testing for significance via Bioconductor. After the statistical qualification of expression values, the data were filtered for genes that were threefold differentially expressed. A multiple sample analysis was also performed wherein GFP-treated (control) cells were compared to Cre-treated cells from Ptenf/f;Pik3caE545K, Ptenf/f, and Pik3caE545K genotypes. For this analysis, the data were filtered for genes that were also threefold differentially regulated, and a Kyoto Encyclopedia of Genes and Genomes pathway analysis was performed. We initially focused on genes in the endometrial cancer and Wnt signaling pathways.
qPCR Validation of DEGs
Validation of DEGs was done by qPCR. We first used cDNA prepared from the same RNA used for RNA sequencing as technical validation. For biological validation, RNA was extracted from adenovirus-Cre–treated primary cultures of epithelial cells from at least two independent experiments, as described in Materials and Methods. RNA (1.0 μg) was reverse transcribed using the High Capacity RNA-to-cDNA Kit (Life Technologies), as per the manufacturer's instructions. Volume of reverse transcriptase reaction corresponding to 25 ng cDNA was used to set up qPCRs using TaqMan primer-probe sets from Life Technologies in the StepOnePlus PCR machine, and fold differences in gene expression were calculated using the −ΔΔCT method.
Results
Pik3caE545K/+ Mutation Causes Carcinoma in the Setting of Biallelic Pten Deletion
The epithelium-restricted expression levels of the Wnt7a15 and Ksp1.316–18 promoters in the endometrium have been described previously. Therefore, Ptenf/f;Pik3caE545K mice were crossed with two mouse strains expressing Cre under Wnt7a (Wnt-Cre) or Ksp1.3 (Ksp-Cre) promoters to delete Pten or express mutant activated Pik3ca in the uterine epithelium.
Uteri of Wnt-Cre+/−;Ptenf/+;Pik3caE545K, Wnt-Cre+/−;Ptenf/+, and Wnt-Cre+/−;Pik3caE545K mice were analyzed at 5 months of age. This time point was determined due to the development of lymphoma and breast cancers in many of the Wnt-Cre+/−;Ptenf/+;Pik3caE545K mice at this age. The Wnt-Cre and Ptenf/f;Pik3caE545K crosses never produced Wnt-Cre+/−;Ptenf/f;Pik3caE545K or Wnt-Cre+/−;Ptenf/f mice, likely due to embryonic expression of Wnt7a with subsequent lethality. Ten of 15 Wnt7a-Cre+/−;Ptenf/+;Pik3caE545K and 12 of 17 Wnt-Cre+/−;Ptenf/+ female mice developed CAH (Figure 1A and Table 1). Only two Wnt-Cre+/−;Ptenf/+;Pik3caE545K mice exhibited in situ carcinoma and/or myoinvasive carcinoma. We did not observe significant differences between the number of CAH foci and severity of disease between Wnt-Cre+/−;Ptenf/+ and Wnt7a-Cre+/−;Ptenf/+;Pik3caE545K genotypes. Interestingly, uteri of Wnt-Cre+/−;Ptenf/+;Pik3caE545K mice exhibited lymphatic dilation and hemorrhagic cysts (Figure 1B). Of 15 Wnt7a-Cre+/−;Ptenf/+;Pik3caE545K mice, 9 also developed lymphoma and breast lesions, likely due to expression of Wnt-Cre in these tissues. None of the 17 Wnt-Cre+/−;Ptenf/+ mice developed hemorrhagic cysts, lymphomas, or breast lesions. All Wnt-Cre+/−;Pik3caE545K mice (n = 7) exhibited normal uterine histological features at 5 months of age and also lacked hemorrhagic cysts, lymphoma, and breast lesions.
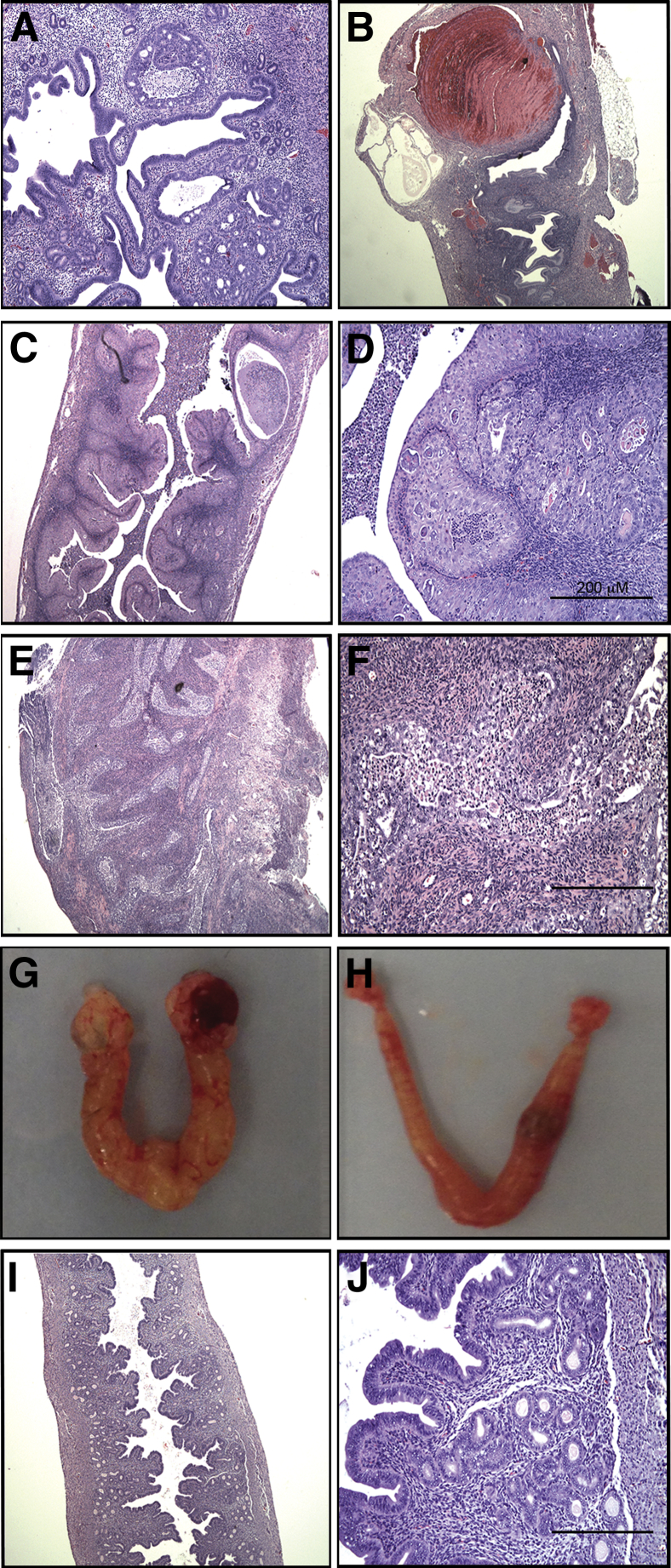
Figure 1.
Photomicrographs of hematoxylin and eosin (H&E)–stained mouse uteri. A and B: Complex atypical hyperplasia (CAH; A) and hemorrhagic cysts (B) in Wnt-Cre+/−;Ptenf/+;Pik3caE545K uteri at 5 months of age. C: Low-magnification H&E image of Ksp-Cre+/−;Ptenf/f mice with CAH involving entire luminal and glandular epithelium. D: High-magnification image of CAH with areas of squamous metaplasia. E and F: Low-magnification (E) and high-magnification (F) image of Ksp-Cre+/−;Ptenf/f;Pik3caE545K uteri with carcinoma and myometrial invasion. G: Gross morphological features of Ksp-Cre+/−;Ptenf/f;Pik3caE545K uteri with both ovaries engulfed in cysts. H: Gross morphological features of Ksp-Cre+/−;Ptenf/f mice exhibiting uterine disease but normal ovaries. I: Low-magnification H&E image of Ksp-Cre+/−;Pik3caE545K uterus with normal histological features. J: High-magnification image of I. Original magnifications: ×200 (A, D, F, and J); ×40 (B, C, E, and I).
Table 1.
Endometrial Lesions in Wnt-Cre+/−;Ptenf/+;Pik3caE545K Mice and Littermates
| Genotype | No. of mice | Mice with endometrial pathological features | Mice with CAH | Average No. of CAH foci | Mice with CA/Inv | Breast/lymph node pathological features |
|---|---|---|---|---|---|---|
| Wnt-Cre+/−;Ptenf/+;Pik3caE545K | 15 | 10 | 8 | 6–7 | 2 | 9 |
| Wnt-Cre+/−;Ptenf/+ | 17 | 12 | 12 | 7–8 | 0 | 0 |
| Wnt-Cre+/−;Pik3caE545K | 7 | 0 | 0 | 0 | 0 | 0 |
CA, carcinoma; CAH, complex atypical hyperplasia; Inv, invasion.
Mating of Ptenf/f;Pik3caE545K and Ksp1.3-Cre mouse strains generated Ksp-Cre+/−;Ptenf/+;Pik3caE545K, Ksp-Cre+/−;Ptenf/+, Ksp-Cre+/−;Ptenf/f, Ksp-Cre+/−;Ptenf/f;Pik3caE545K, and Ksp-Cre+/−;Pik3caE545K/+ genotypes. Previously published work from our laboratory demonstrated that Ptenf/f mice injected with adeno-Cre virus developed carcinoma between 2 and 4 months after injection.13 We, therefore, chose to sacrifice mice of the five genotypes mentioned above at 4 months of age (Table 2). Eleven of 12 Ksp-Cre+/−;Ptenf/+;Pik3caE545K and 12 of 16 Ksp-Cre+/−;Ptenf/+ mice exhibited CAH only. The number of foci of CAH was comparable in Ksp-Cre+/−;Ptenf/+;Pik3caE545K and Ksp-Cre+/−;Ptenf/+ genotypes, to those observed in the Wnt7a-Cre+/−;Ptenf/+;Pik3caE545K mice and the Wnt-Cre+/−;Ptenf/+ mice. However, in the Wnt7a-Cre+/−;Ptenf/+;Pik3caE545K genotype, two mice developed invasive carcinomas that were not observed in the Ksp-Cre+/−;Ptenf/+;Pik3caE545K mice. All Ksp-Cre+/−;Ptenf/f mice (n = 8) developed extensive endometrial disease, with CAH involving the entire luminal and glandular epithelium (Figure 1C). In five of eight mice, CAH exhibited squamous metaplasia (Figure 1D). Only one mouse (12%) showed carcinoma with myometrial invasion. Hematoxylin and eosin analysis of Ksp-Cre+/−;Ptenf/f;Pik3caE545K uteri (n = 6) showed CAH with squamous metaplasia and carcinoma with myometrial invasion (Figure 1, E and F) in all of the mice. In five mice, the gross examination of the uterus demonstrated enlarged uteri with ovaries engulfed by cysts that could not be distinguished from the oviduct (Figure 1G) as compared to Ksp-Cre+/−;Ptenf/f mice (Figure 1H). The carcinoma was well to moderately differentiated and extended into and completely surrounded the ovaries (Figure 1G). Thus, the presence of a mutant Pik3ca on the background of biallelic Pten deletion resulted in extensive invasive carcinoma with 100% penetrance. None of the Ksp-Cre+/−;Pik3caE545K mice developed endometrial disease (Figure 1, I and J), similar to Wnt-Cre+/−;Pik3caE545K mice.
Table 2.
Endometrial Lesions in Ksp-Cre+/-;Ptenf/f;Pik3caE545K Mice and Littermates
| Genotype | No. of mice | Mice with endometrial pathological features | Mice with CAH | Average No. of CAH foci | Mice with CA/Inv |
|---|---|---|---|---|---|
| Ksp-Cre+/−;Ptenf/+;Pik3caE545K | 12 | 11 | 11 | 8 | 0 |
| Ksp-Cre+/−;Ptenf/+ | 16 | 12 | 12 | 9–10 | 0 |
| Ksp-Cre+/−;Pik3caE545K | 6 | 0 | 0 | 0 | 0 |
| Ksp-Cre+/−;Ptenf/f | 8 | 8 | 8 | Extensive | 1 |
| Ksp-Cre+/−;Ptenf/f;Pik3caE545K | 6 | 6 | 6 | Extensive | 6 |
CA, carcinoma; CAH, complex atypical hyperplasia; Inv, invasion.
Expression of Pik3caE545K Does Not Cause Endometrial Carcinoma and Is Less Efficient at Activating Akt in Epithelial Cells
Carcinoma and/or CAH did not develop in mice regardless of the promoter used to activate Pik3ca. This lack of phenotype could be due to inefficient DNA recombination of the mutant Pik3ca allele or lack of clonal selection in endometrial epithelial cells expressing mutant Pik3ca. To determine the reason, we used PCR analysis to ascertain excision of the STOP codon upstream of exon 1 in the Pik3ca mutant allele by Cre recombinase. Presence of a recombined allele leads to amplification of a 735-bp band, whereas the nonrecombined allele amplifies a 637-bp band. Areas of CAH and carcinoma were microdissected from tissue sections of Ksp-Cre+/−;Ptenf/f;Pik3caE545K, Ksp-Cre+/−;Ptenf/f, and Ksp-Cre+/−;Ptenf/+;Pik3caE545K uteri. DNA was extracted and analyzed by PCR. DNA from Ksp-Cre+/−;Ptenf/f;Pik3caE545K and Ksp-Cre+/−;Ptenf/+;Pik3caE545K uteri amplified both 637- and 735-bp bands (Figure 2A), proving that the mutant allele had undergone recombination in the lesions from these mice. DNA extracted from Ksp-Cre+/−;Ptenf/f uteri amplified only a 637-bp band, as expected (Figure 2A). DNA was also extracted from normal epithelium of Ksp-Cre+/−;Pik3caE545K mice and PCR amplified a 637-bp nonrecombined band, but a faint 735-bp band from the recombined allele was detected (Figure 2A). This observation suggested that even though recombination occurred, epithelial cells with the recombined allele do not have a significant growth advantage in this setting.
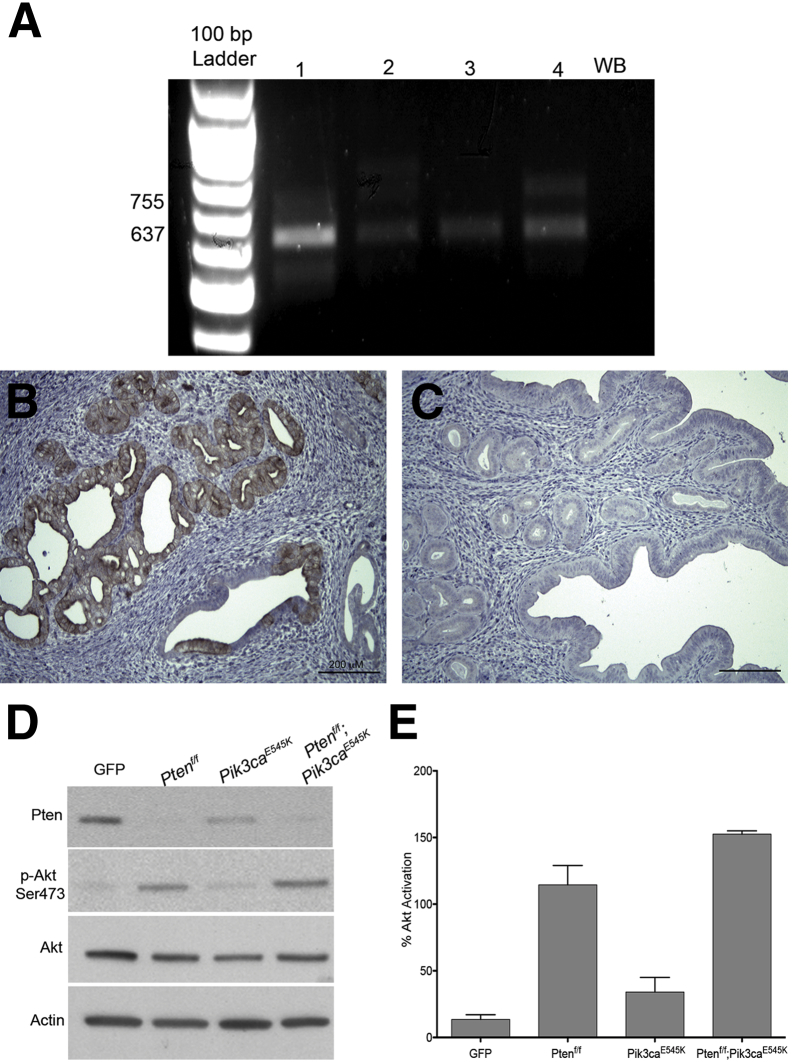
Figure 2.
Recombination of mutant Pik3ca allele and activation of Akt in primary cultures. A: Agarose gel electrophoresis of PCR analysis of DNA extracted from microdissected, formalin-fixed, paraffin-embedded tissue from Ksp-Cre+/−;Pik3caE545K (lane 1), Ksp-Cre+/−;Ptenf/+;Pik3caE545K (lane 2), Ksp-Cre+/−;Ptenf/f (lane 3), and Ksp-Cre+/−;Ptenf/f;Pik3caE545K (lane 4) mice. B and C: p-Akt immunohistochemical analysis of uterine sections of Ksp-Cre+/−;Ptenf/+;Pik3caE545K (B) and Ksp-Cre+/−;Pik3caE545K (C) mice. D: Immunoblot analysis of whole cell extracts of Adeno-GFP or Adeno-Cre–treated primary uterine epithelial cells isolated from Ptenf/f, Pik3caE545K, and Ptenf/f;Pik3caE545K mice using Pten, p-Akt, Akt, and actin antibodies. E: Quantitation of p-Akt in D. Graph represents p-Akt/Akt ratios from three independent experiments. Data represent means ± SD. GFP, green fluorescent protein; WB, water blank.
We also performed IHC analysis on the tissue sections using p-Akt antibody. Although areas of CAH and carcinoma in all of the genotypes with endometrial pathological features (Figure 2B) exhibited characteristic membrane staining for p-Akt, the epithelium or glands in Ksp-Cre+/−;Pik3caE545K mice (Figure 2C) were completely negative for p-Akt. The uteri of Wnt-Cre+/−;Pik3caE545K mice were also negative for p-Akt staining (data not shown).
Pik3ca was activated in vitro by adeno-Cre treatment of primary epithelial cells isolated from Ptenf/f and Ptenf/f;Pik3caE545K mice. Adeno-GFP–treated cells were used as control cells (GFP). We have previously demonstrated that in vitro adeno-Cre–mediated Pten deletion in epithelial cells results in activation of Akt.13 In cells prepared from Pik3caE545K mice, Akt was activated but was severalfold lower than that observed in Ptenf/f or Ptenf/f;Pik3caE545K cells (Figure 2D). Thus, mutant Pik3ca alone results in a much less robust activation of Akt, compared to loss of Pten, in endometrial epithelial cells. Quantitation of the immunoblot analysis is depicted in Figure 2E.
Expression of Downstream Signaling Molecules in the PI3K/Pten/Akt Pathway
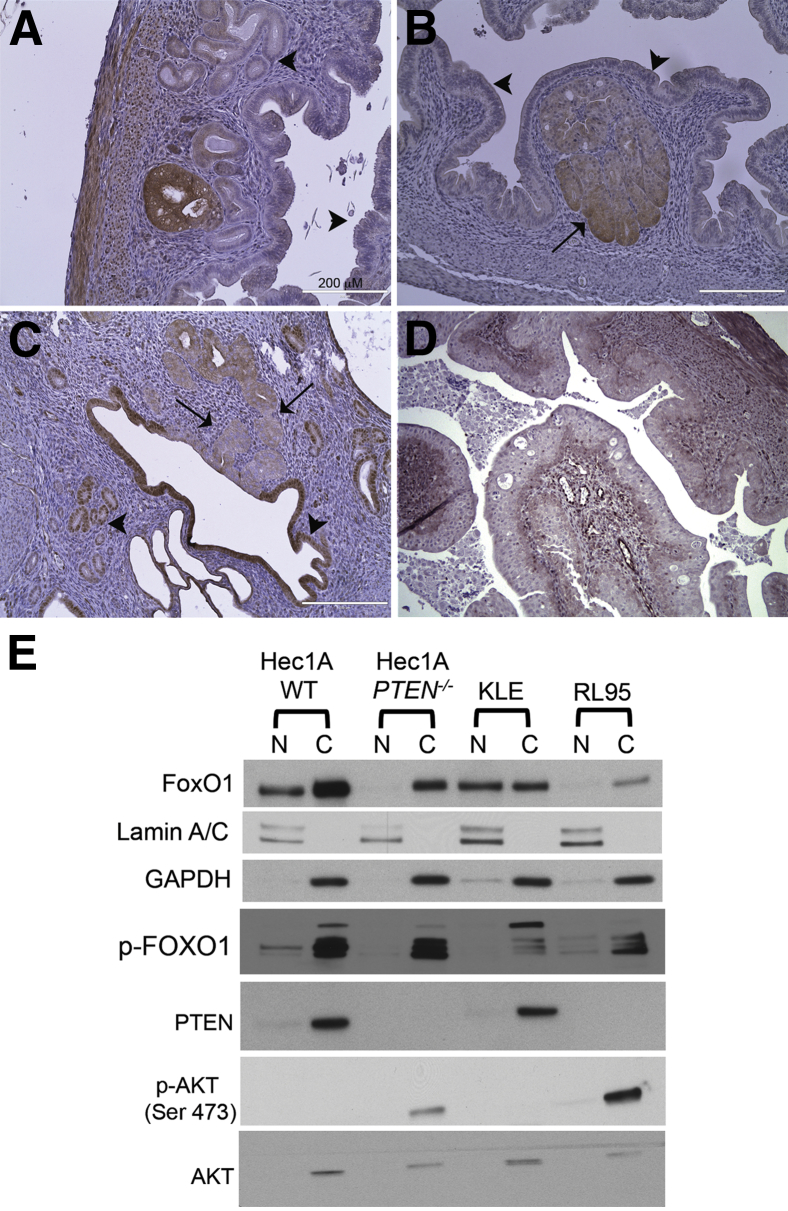
Next, we performed IHC analysis on uterine sections from Ksp-Cre+/−;Ptenf/+, Ksp-Cre+/−;Ptenf/+;Pik3caE545K, Ksp-Cre+/−;Ptenf/f, and Ksp-Cre+/−;Ptenf/f;Pik3caE545K mice with antibodies against p-Gsk3β, p-Pras40, Stathmin, and FoxO1, which are known substrates of p-Akt. IHC analysis was also performed using antibodies against β-catenin because cytoplasmic and nuclear accumulation of β-catenin has been associated with squamous metaplasia. In the genotypes with CAH and carcinoma, the lesions expressed p-Gsk3β and p-Pras40 (Figure 3, A and B), whereas normal epithelium was negative (Figure 3, A and B), correlating with activation of the pathway. Stathmin and β-catenin expression was not altered, and β-catenin was localized to cell membranes in normal and neoplastic cells, even in areas of squamous metaplasia (Supplemental Figure S1). FoxO1 expression was also significantly altered. FoxO1 localization was nuclear in normal epithelium and stromal cells (Figure 3C), but areas of CAH (Figure 3C) and carcinoma (Figure 3D) exhibited complete lack of nuclear FoxO1 with reduced cytoplasmic expression, irrespective of the genotypes. Pras40, Gsk3β, and FoxO1, therefore, are likely downstream targets of Pten inactivation in the uterine epithelium. IHC analysis with FOXO1 antibody was also performed on TMAs of human endometrial carcinomas and normal cycling (proliferative and secretory) endometria. Intense nuclear expression was observed only in the normal secretory epithelium but was absent in normal proliferative epithelium, as reported previously.19 In contrast, 72% of carcinomas (69 of 96 cases) lacked nuclear FOXO1 expression. More important, the results with the human TMA (Supplemental Table S1) corroborate the results found in our genetic mouse model.
Figure 3.
Immunohistochemical (IHC) analysis of downstream signaling molecules. A and B: Positive IHC staining with p-Gsk3β (A) and p-Pras40 (B) antibodies in uteri with complex atypical hyperplasia (CAH; arrow) as compared to normal epithelium (arrowheads). C: Loss of nuclear FoxO1 staining in CAH (arrows) compared to the intense nuclear staining of normal epithelium (arrowheads). D: Loss of nuclear FoxO1 in carcinoma. E: Immunoblot analysis depicting subcellular localization of FOXO1 in endometrial cancer cell lines. Lamin A/C and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) served as controls to determine relative purity of nuclear (N) and cytoplasmic (C) extracts. WT, wild type.
To further investigate lack of nuclear FOXO1, we analyzed several endometrial cancer cell lines for subcellular distribution and phosphorylation status of FOXO1 by immunoblot analysis. Nuclear and cytoplasmic extracts were prepared from KLE, RL95, Hec1A WT, and Hec1A clone16 (isogenic clone with somatic deletion of PTEN) cell lines with unique combinations of mutations (Supplemental Table S2) in PTEN and PIK3CA. FOXO1 was detected in both the cytoplasm and the nucleus in Hec1A WT and KLE cells (Figure 3E). In contrast, the Hec1A clone 16 and RL95 (mutated for PTEN) cell lines lacked nuclear FOXO1 expression and demonstrated reduced cytoplasmic expression. This observation correlates with the mouse studies described above and provides evidence that nuclear localization of FOXO1 is dependent on PTEN. In all of the cell lines, p-FOXO1 was detected maximally in the cytoplasmic compartment. The Hec1A cell lines (parent and clone) have a G1049R mutation in PIK3CA yet fail to activate AKT (Hec1A WT) in the absence of PTEN deletion (Hec1A clone).
Pten and Pik3ca Regulate Nonoverlapping Gene Sets
To delineate downstream changes in gene expression due to deletion of Pten, activation of Pik3ca, or both, we performed RNA sequencing analysis with RNA extracted from adeno-Cre–treated primary epithelial cell cultures prepared from Ptenf/f, Pik3caE545K, and Ptenf/f;Pik3caE545K mice. Deletion of Pten and expression of Pik3caE545K allele were confirmed at the RNA level by qPCR analysis (Supplemental Figure S2) and were submitted for RNA sequencing analysis.
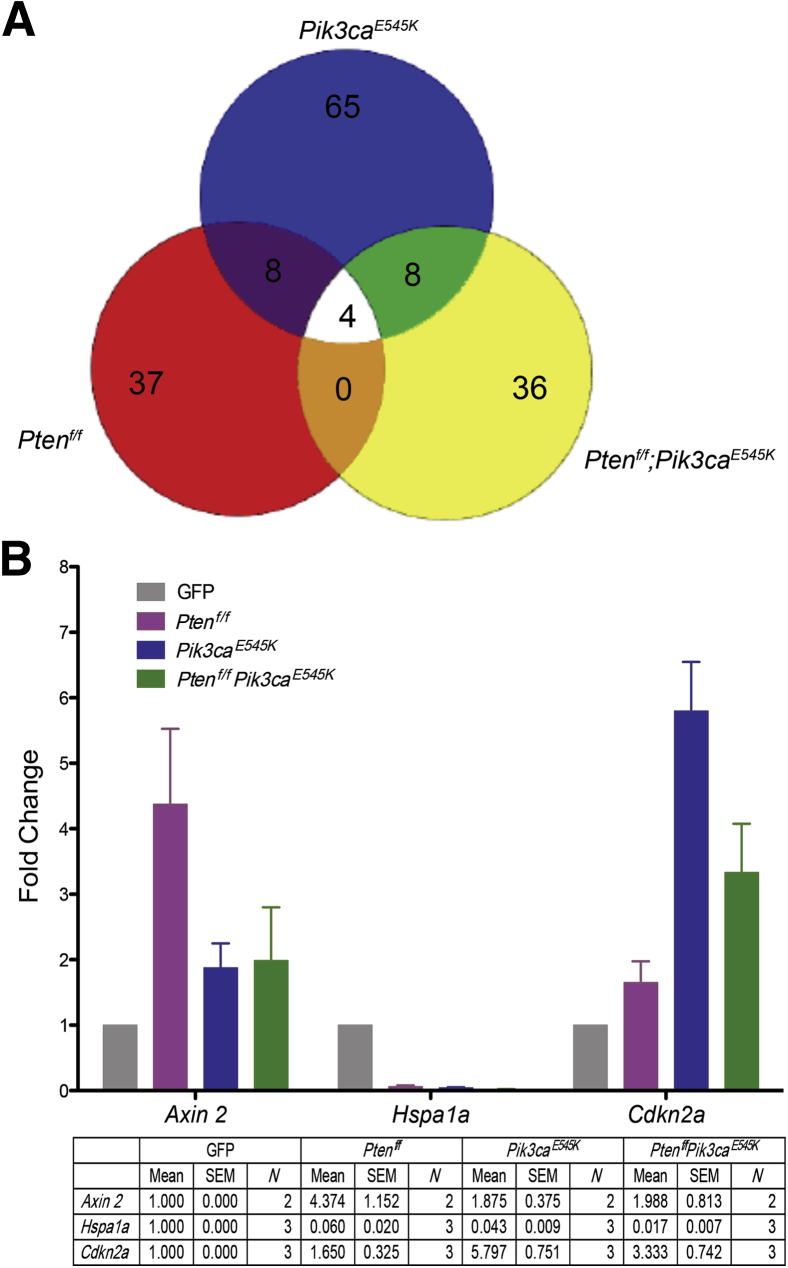
A pairwise analysis was performed to individually compare control (GFP) with Ptenf/f, Pik3caE545K, and Ptenf/f;Pik3caE545K cells by setting a P cutoff at 0.05 and threefold differential expression. Only four genes were altered in all three conditions (Figure 4A). There was also minimal overlap between Ptenf/f versus Pik3caE545K and Pik3caE545K versus Ptenf/f;Pik3caE545K cells. Most of the DEGs were unique to each condition (Supplemental Tables S3–S5). This finding suggests that deletion of Pten and activation of Pik3ca regulate distinct, nonoverlapping genes. For validation, we chose genes from the following groups: i) genes regulated in all of the groups (Hspa1a), ii) genes regulated in Pik3caE545K and Ptenf/f;Pik3caE545K cells (Cdkn2a), and iii) genes regulated maximally in Ptenf/f cells (Axin 2). Validation was performed on cDNA from total RNA extracted from three independent cell preparations. Regulation of the three genes was similar to that observed by RNA sequencing (Figure 4B).
Figure 4.
RNA sequencing analysis. A: Venn diagram depicting the differentially expressed genes (DEGs) common and/or unique to Ptenf/f, Pik3caE545K, and Ptenf/f;Pik3caE545K. B: Real-time quantitative PCR analysis to validate expression of DEGs identified by RNA sequencing. Graph depicts fold change in expression of Axin2, Hspa1a, and Cdkn2a in Ptenf/f, Pik3caE545K, and Ptenf/f;Pik3caE545K cells as compared to green fluorescent protein (GFP)–treated cells. Means ± SEM are summarized. N = number of independent experiments performed for each gene.
Discussion
Although PTEN and PIK3CA mutations occur at high frequencies in endometrial carcinoma, their respective roles in the neoplastic process remain unclear. We have been specifically interested in defining the consequences of these genetic alterations in the development of endometrial carcinoma. With the use of genetic mouse models reported herein, along with human tissue samples and cell lines, we have begun to dissect the impact of these two common genetic alterations on endometrial carcinogenesis.
We have shown that mouse strains with conditional deletion of Pten, as has been previously shown,13,20 predictably develop endometrial hyperplasia that can progress to carcinoma. In contrast, the isolated activation of Pik3ca in the endometrial epithelium does not cause any phenotypic light microscopic changes. However, the activation of Pik3ca in the presence of Pten loss not only accelerates the progression to carcinoma, but leads to more extensive disease with extension into the ovaries. Interestingly, Pik3ca activation does not lead to the development of more extensive hyperplasia or carcinoma in the setting of Pten haploinsufficiency, and the frequency of CAH in Pten-haploinsufficient mice with or without Pik3ca activation was similar to germline Pten+/− mice9 of the same age. This suggests that the rate-limiting step in the development of hyperplasia is the biallelic loss of Pten in the epithelium. The current findings corroborate the mutational analyses of primary human tumors and provide definitive evidence for a role of Pten in the development of hyperplasia, with Pik3ca driving the transition to invasion in the endometrial epithelium.
We present evidence from primary mouse cultures, as well as human cancer cell lines, that the extent of Akt phosphorylation differs between loss of Pten and activation of Pik3ca. Exon 9 and 20 mutations in p110α are hotspots common across all cancer types,21,22 and these mutations are equally effective in phosphorylating AKT in breast23 and colorectal cancer24 cell lines as well as primary urothelial cells.25 However, in NIH3T3 cells, the kinase domain H1047R mutation was severalfold more potent than the helical domain E545K mutation at phosphorylating Akt, suggesting that the extent of pathway activation by these mutations (as seen by Akt phosphorylation) is cell context dependent.25 In our studies, the E545K mutation resulted in a much less robust activation of Akt when compared to Pten in primary mouse endometrial epithelial cells. In human endometrial cancer cell lines as well, phosphorylated AKT was detected only in the cell lines with PTEN mutations (Figure 3E and Supplemental Table S2), despite the presence of a G1049R mutation in PIK3CA, as has been demonstrated previously.26 In endometrial epithelial cells, therefore, both helical or kinase domain mutations were unable to overcome the inhibitory effects of PTEN, which likely explains, at least in part, the lack of phenotype in mice expressing only the activated Pik3ca allele. To our knowledge, this is the first report to provide mechanistic insights into the respective roles of Pten and Pik3ca in the endometrial epithelium in vivo. Recently, studies using mouse models of prostate and breast cancer highlighted the importance of p110β over p110α isoform, particularly in the context of PTEN deficiency.27,28 It has also been suggested that the PI3K isoform dependence is affected by mutations in other genes.29 However, p110β inhibition alone in endometrial cancer cell lines was ineffective at inducing apoptosis, irrespective of the PTEN status.26
FoxO1, a tumor-suppressor protein, is a well-characterized downstream target of Akt. Nuclear FoxO1 is a transcription factor activating expression of cell cycle inhibitors like p27 and p21, while repressing cyclins D1 and D2.30 Activated Akt (as a result of Pten loss) phosphorylates FoxO1, leading to nuclear export, inhibiting its transcriptional activity and thus promoting cell proliferation and blocking apoptosis.30 FOXO1 in humans and mice has been demonstrated to play a role in decidualization of stromal cells in the endometrium,31–33 and its expression in the endometrial epithelium varies with the stage of the menstrual/estrus cycle.34 Although UEC cases have been shown to express reduced FOXO1 as compared to the normal cycling endometrium,19,35 there is a lack of evidence suggesting a direct link between PTEN mutation/inactivation and reduced expression. In our mouse models with CAH and UEC, FoxO1 expression was strikingly absent in the nucleus as compared to the normal epithelium, in agreement with the human data. As mentioned previously in Results, a TMA consisting of primary human tumors also showed lack of nuclear FOXO1 in 72% of cases. However, because activation of Pik3ca alone in the mouse uterus did not lead to endometrial disease, we were unable to dissect out the role of Pten and Pik3ca in regulating FoxO1 using mouse models. The human endometrial cancer cell lines proved ideal to investigate the link between FOXO1 and PTEN. Nuclear localization of FOXO1 correlated with presence of wild-type PTEN, irrespective of PIK3CA status in endometrial cancer cell lines. This was particularly evident in Hec1A WT and Hec1A clone 16 cells, which harbor a mutation in PIK3CA but differ from each other only with respect to the lack of PTEN in clone 16. Furthermore, KLE and RL95 (wild-type and mutant for PTEN, respectively) also exhibited similar localization of FOXO1. These observations provide evidence for a direct link between PTEN and FOXO1 in UEC. The lack of FOXO1 expression in most human UEC cases and its loss in hyperplasia and carcinoma in the mouse model suggest a central role for this transcription factor in the pathogenesis of endometrial carcinoma. Interestingly, FOXO1 expression was up-regulated as a result of progestin treatment of immortalized and transformed endometrial epithelial cells19,35,36 and is thought to mediate the progestin inhibition of epithelial proliferation. Regulating FOXO1 expression may be central to the mechanism by which PTEN promotes endometrial hyperplasia, most likely by inhibiting cell death rather than increasing proliferation. This is consistent with previous studies from our laboratory showing that CAH and UEC arising in the mouse model have lower proliferative indices than proliferative endometrium.14 miRNA-mediated repression has also been implicated in the regulation of FOXO1 levels in the endometrial epithelium,37,38 but its relationship to PTEN is currently unknown.
In addition to being less potent than Pten in activating Akt, Pik3ca also regulates transcription of a distinct set of genes in vitro, as demonstrated by RNA sequencing analysis of adeno-Cre–treated primary endometrial epithelial cell cultures. Furthermore, combined Pten deletion and Pik3ca activation altered expression of an entirely different set of genes as compared to Pten deletion or Pik3ca activation alone. Along with their roles as key players in the PI3K/AKT/PTEN pathway, Pten and Pik3ca may also individually participate in other cellular processes in the endometrial epithelium, providing a possible explanation for the frequent co-occurrence of mutations in primary human tumors.11,39 Although these cell culture experiments are short-term and they may not recapitulate the complex genetic alterations observed in human cancers, they provide unique insights into immediate downstream effects of Pten deletion and/or Pik3ca activation. RNA sequencing on cells isolated from the lesions arising in different genotypes might more closely reflect the situation in human carcinogenesis.
In summary, the data presented herein provide unique insights into the respective roles of PTEN and PIK3CA in endometrial carcinoma. The PI3K/AKT pathway can be activated by many signals like estrogen or growth factors, secreted in a cyclic fashion in the endometrial epithelium, inducing proliferation and differentiation. The results presented above strongly support PTEN as the prominent suppressor of growth signals and show that the loss of PTEN removes the inhibition leading to hyperplasia and that PIK3CA mutations promote a transition to carcinoma. The mechanism underlying this transition may be related to further activation of AKT and likely regulation of other nonoverlapping downstream molecules. Currently, we are pursuing the identification and characterization of molecules and pathways critical to the development of carcinoma.
Acknowledgment
Mice expressing Cre under the Wnt7a promoter (Wnt7a-Cre+/−) were obtained from Dr. Kenneth S. Korach (National Institute of Environmental Health Science, Research Triangle Park, NC).
Footnotes
Supported by NIH/National Cancer Institute grants RO1 CA188516 (S.J.B.) and R01 CA095427 (L.H.E).
Disclosures: None declared.
Supplemental Data
Immunohistochemical analysis with Stathmin and β-catenin antibody. There is no difference between the staining between the normal epithelium and complex atypical hyperplasia (CAH) for both antibodies in all of the genotypes. Original magnification, x40.
Real-time quantitative PCR (qPCR) analysis for RNA extracted from primary epithelial cell cultures treated with adeno-Cre. A:Pten qPCR indicating loss of Pten expression only in Ptenf/f and Ptenf/f;Pik3caE545K cells. B: Expression of the mutant E545K Pik3ca allele. The expression of the mutant allele was detected only in Pik3caE545K cells. GFP, green fluorescent protein; WT, wild type.
References
- 1.Di Cristofano A., Ellenson L.H. Endometrial carcinoma. Annu Rev Pathol. 2007;2:57–85. doi: 10.1146/annurev.pathol.2.010506.091905. [DOI] [PubMed] [Google Scholar]
- 2.Tashiro H., Blazes M.S., Wu R., Cho K.R., Bose S., Wang S.I., Li J., Parsons R., Ellenson L.H. Mutations in PTEN are frequent in endometrial carcinoma but rare in other common gynecological malignancies. Cancer Res. 1997;57:3935–3940. [PubMed] [Google Scholar]
- 3.Salvesen H.B., Stefansson I., Kretzschmar E.I., Gruber P., MacDonald N.D., Ryan A., Jacobs I.J., Akslen L.A., Das S. Significance of PTEN alterations in endometrial carcinoma: a population-based study of mutations, promoter methylation and PTEN protein expression. Int J Oncol. 2004;25:1615–1623. [PubMed] [Google Scholar]
- 4.Risinger J.I., Hayes K., Maxwell G.L., Carney M.E., Dodge R.K., Barrett J.C., Berchuck A. PTEN mutation in endometrial cancers is associated with favorable clinical and pathologic characteristics. Clin Cancer Res. 1998;4:3005–3010. [PubMed] [Google Scholar]
- 5.Cantley L.C. The phosphoinositide 3-kinase pathway. Science. 2002;296:1655–1657. doi: 10.1126/science.296.5573.1655. [DOI] [PubMed] [Google Scholar]
- 6.Song M.S., Salmena L., Pandolfi P.P. The functions and regulation of the PTEN tumour suppressor. Nat Rev Mol Cell Biol. 2012;13:283–296. doi: 10.1038/nrm3330. [DOI] [PubMed] [Google Scholar]
- 7.Risinger J.I., Hayes A.K., Berchuck A., Barrett J.C. PTEN/MMAC1 mutations in endometrial cancers. Cancer Res. 1997;57:4736–4738. [PubMed] [Google Scholar]
- 8.Kanamori Y., Kigawa J., Itamochi H., Shimada M., Takahashi M., Kamazawa S., Sato S., Akeshima R., Terakawa N. Correlation between loss of PTEN expression and Akt phosphorylation in endometrial carcinoma. Clin Cancer Res. 2001;7:892–895. [PubMed] [Google Scholar]
- 9.Wang H., Douglas W., Lia M., Edelmann W., Kucherlapati R., Podsypanina K., Parsons R., Ellenson L.H. DNA mismatch repair deficiency accelerates endometrial tumorigenesis in Pten heterozygous mice. Am J Pathol. 2002;160:1481–1486. doi: 10.1016/S0002-9440(10)62573-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang H., Joshi A., Iaconis L., Solomon G.J., Xiang Z., Verhage H.G., Douglas W., Ronnett B.M., Ellenson L.H. Oviduct-specific glycoprotein is a molecular marker for invasion in endometrial tumorigenesis identified using a relevant mouse model. Int J Cancer. 2009;124:1349–1357. doi: 10.1002/ijc.24022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hayes M.P., Wang H., Espinal-Witter R., Douglas W., Solomon G.J., Baker S.J., Ellenson L.H. PIK3CA and PTEN mutations in uterine endometrioid carcinoma and complex atypical hyperplasia. Clin Cancer Res. 2006;12:5932–5935. doi: 10.1158/1078-0432.CCR-06-1375. [DOI] [PubMed] [Google Scholar]
- 12.Hayes M.P., Ellenson L.H. Molecular alterations in uterine serous carcinoma. Gynecol Oncol. 2010;116:286–289. doi: 10.1016/j.ygyno.2009.11.012. [DOI] [PubMed] [Google Scholar]
- 13.Joshi A., Ellenson L.H. Adenovirus mediated homozygous endometrial epithelial Pten deletion results in aggressive endometrial carcinoma. Exp Cell Res. 2011;317:1580–1589. doi: 10.1016/j.yexcr.2011.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Joshi A., Wang H., Jiang G., Douglas W., Chan J.S., Korach K.S., Ellenson L.H. Endometrial tumorigenesis in Pten(+/-) mice is independent of coexistence of estrogen and estrogen receptor alpha. Am J Pathol. 2012;180:2536–2547. doi: 10.1016/j.ajpath.2012.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Winuthayanon W., Hewitt S.C., Orvis G.D., Behringer R.R., Korach K.S. Uterine epithelial estrogen receptor alpha is dispensable for proliferation but essential for complete biological and biochemical responses. Proc Natl Acad Sci U S A. 2010;107:19272–19277. doi: 10.1073/pnas.1013226107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wild P.J., Ikenberg K., Fuchs T.J., Rechsteiner M., Georgiev S., Fankhauser N., Noske A., Roessle M., Caduff R., Dellas A., Fink D., Moch H., Krek W., Frew I.J. p53 Suppresses type II endometrial carcinomas in mice and governs endometrial tumour aggressiveness in humans. EMBO Mol Med. 2012;4:808–824. doi: 10.1002/emmm.201101063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frew I.J., Minola A., Georgiev S., Hitz M., Moch H., Richard S., Vortmeyer A.O., Krek W. Combined VHLH and PTEN mutation causes genital tract cystadenoma and squamous metaplasia. Mol Cell Biol. 2008;28:4536–4548. doi: 10.1128/MCB.02132-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shao X., Somlo S., Igarashi P. Epithelial-specific Cre/lox recombination in the developing kidney and genitourinary tract. J Am Soc Nephrol. 2002;13:1837–1846. doi: 10.1097/01.asn.0000016444.90348.50. [DOI] [PubMed] [Google Scholar]
- 19.Ward E.C., Hoekstra A.V., Blok L.J., Hanifi-Moghaddam P., Lurain J.R., Singh D.K., Buttin B.M., Schink J.C., Kim J.J. The regulation and function of the forkhead transcription factor, Forkhead box O1, is dependent on the progesterone receptor in endometrial carcinoma. Endocrinology. 2008;149:1942–1950. doi: 10.1210/en.2007-0756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Daikoku T., Hirota Y., Tranguch S., Joshi A.R., DeMayo F.J., Lydon J.P., Ellenson L.H., Dey S.K. Conditional loss of uterine Pten unfailingly and rapidly induces endometrial cancer in mice. Cancer Res. 2008;68:5619–5627. doi: 10.1158/0008-5472.CAN-08-1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Campbell I.G., Russell S.E., Choong D.Y., Montgomery K.G., Ciavarella M.L., Hooi C.S., Cristiano B.E., Pearson R.B., Phillips W.A. Mutation of the PIK3CA gene in ovarian and breast cancer. Cancer Res. 2004;64:7678–7681. doi: 10.1158/0008-5472.CAN-04-2933. [DOI] [PubMed] [Google Scholar]
- 22.Samuels Y., Wang Z., Bardelli A., Silliman N., Ptak J., Szabo S., Yan H., Gazdar A., Powell S.M., Riggins G.J., Willson J.K., Markowitz S., Kinzler K.W., Vogelstein B., Velculescu V.E. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304:554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- 23.Zhao J.J., Liu Z., Wang L., Shin E., Loda M.F., Roberts T.M. The oncogenic properties of mutant p110alpha and p110beta phosphatidylinositol 3-kinases in human mammary epithelial cells. Proc Natl Acad Sci U S A. 2005;102:18443–18448. doi: 10.1073/pnas.0508988102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Samuels Y., Diaz L.A., Jr., Schmidt-Kittler O., Cummins J.M., Delong L., Cheong I., Rago C., Huso D.L., Lengauer C., Kinzler K.W., Vogelstein B., Velculescu V.E. Mutant PIK3CA promotes cell growth and invasion of human cancer cells. Cancer Cell. 2005;7:561–573. doi: 10.1016/j.ccr.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 25.Ross R.L., Askham J.M., Knowles M.A. PIK3CA mutation spectrum in urothelial carcinoma reflects cell context-dependent signaling and phenotypic outputs. Oncogene. 2013;32:768–776. doi: 10.1038/onc.2012.87. [DOI] [PubMed] [Google Scholar]
- 26.Weigelt B., Warne P.H., Lambros M.B., Reis-Filho J.S., Downward J. PI3K pathway dependencies in endometrioid endometrial cancer cell lines. Clin Cancer Res. 2013;19:3533–3544. doi: 10.1158/1078-0432.CCR-12-3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wee S., Wiederschain D., Maira S.M., Loo A., Miller C., deBeaumont R., Stegmeier F., Yao Y.M., Lengauer C. PTEN-deficient cancers depend on PIK3CB. Proc Natl Acad Sci U S A. 2008;105:13057–13062. doi: 10.1073/pnas.0802655105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jia S., Liu Z., Zhang S., Liu P., Zhang L., Lee S.H., Zhang J., Signoretti S., Loda M., Roberts T.M., Zhao J.J. Essential roles of PI(3)K-p110beta in cell growth, metabolism and tumorigenesis. Nature. 2008;454:776–779. doi: 10.1038/nature07091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schmit F., Utermark T., Zhang S., Wang Q., Von T., Roberts T.M., Zhao J.J. PI3K isoform dependence of PTEN-deficient tumors can be altered by the genetic context. Proc Natl Acad Sci U S A. 2014;111:6395–6400. doi: 10.1073/pnas.1323004111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lam E.W., Brosens J.J., Gomes A.R., Koo C.Y. Forkhead box proteins: tuning forks for transcriptional harmony. Nat Rev Cancer. 2013;13:482–495. doi: 10.1038/nrc3539. [DOI] [PubMed] [Google Scholar]
- 31.Buzzio O.L., Lu Z., Miller C.D., Unterman T.G., Kim J.J. FOXO1A differentially regulates genes of decidualization. Endocrinology. 2006;147:3870–3876. doi: 10.1210/en.2006-0167. [DOI] [PubMed] [Google Scholar]
- 32.Kajihara T., Jones M., Fusi L., Takano M., Feroze-Zaidi F., Pirianov G., Mehmet H., Ishihara O., Higham J.M., Lam E.W., Brosens J.J. Differential expression of FOXO1 and FOXO3a confers resistance to oxidative cell death upon endometrial decidualization. Mol Endocrinol. 2006;20:2444–2455. doi: 10.1210/me.2006-0118. [DOI] [PubMed] [Google Scholar]
- 33.Labied S., Kajihara T., Madureira P.A., Fusi L., Jones M.C., Higham J.M., Varshochi R., Francis J.M., Zoumpoulidou G., Essafi A., Fernandez de Mattos S., Lam E.W., Brosens J.J. Progestins regulate the expression and activity of the forkhead transcription factor FOXO1 in differentiating human endometrium. Mol Endocrinol. 2006;20:35–44. doi: 10.1210/me.2005-0275. [DOI] [PubMed] [Google Scholar]
- 34.Fan W., Li S.W., Li W.H., Wang Y., Gong Y., Ma Q.H., Luo S. FOXO1 expression and regulation in endometrial tissue during the menstrual cycle and in early pregnancy decidua. Gynecol Obstet Invest. 2012;74:56–63. doi: 10.1159/000336633. [DOI] [PubMed] [Google Scholar]
- 35.Goto T., Takano M., Albergaria A., Briese J., Pomeranz K.M., Cloke B., Fusi L., Feroze-Zaidi F., Maywald N., Sajin M., Dina R.E., Ishihara O., Takeda S., Lam E.W., Bamberger A.M., Ghaem-Maghami S., Brosens J.J. Mechanism and functional consequences of loss of FOXO1 expression in endometrioid endometrial cancer cells. Oncogene. 2008;27:9–19. doi: 10.1038/sj.onc.1210626. [DOI] [PubMed] [Google Scholar]
- 36.Kyo S., Sakaguchi J., Kiyono T., Shimizu Y., Maida Y., Mizumoto Y., Mori N., Nakamura M., Takakura M., Miyake K., Sakamoto M., Inoue M. Forkhead transcription factor FOXO1 is a direct target of progestin to inhibit endometrial epithelial cell growth. Clin Cancer Res. 2011;17:525–537. doi: 10.1158/1078-0432.CCR-10-1287. [DOI] [PubMed] [Google Scholar]
- 37.Myatt S.S., Wang J., Monteiro L.J., Christian M., Ho K.K., Fusi L., Dina R.E., Brosens J.J., Ghaem-Maghami S., Lam E.W. Definition of microRNAs that repress expression of the tumor suppressor gene FOXO1 in endometrial cancer. Cancer Res. 2010;70:367–377. doi: 10.1158/0008-5472.CAN-09-1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mozos A., Catasus L., D'Angelo E., Serrano E., Espinosa I., Ferrer I., Pons C., Prat J. The FOXO1-miR27 tandem regulates myometrial invasion in endometrioid endometrial adenocarcinoma. Hum Pathol. 2014;45:942–951. doi: 10.1016/j.humpath.2013.12.007. [DOI] [PubMed] [Google Scholar]
- 39.Oda K., Stokoe D., Taketani Y., McCormick F. High frequency of coexistent mutations of PIK3CA and PTEN genes in endometrial carcinoma. Cancer Res. 2005;65:10669–10673. doi: 10.1158/0008-5472.CAN-05-2620. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Immunohistochemical analysis with Stathmin and β-catenin antibody. There is no difference between the staining between the normal epithelium and complex atypical hyperplasia (CAH) for both antibodies in all of the genotypes. Original magnification, x40.
Real-time quantitative PCR (qPCR) analysis for RNA extracted from primary epithelial cell cultures treated with adeno-Cre. A:Pten qPCR indicating loss of Pten expression only in Ptenf/f and Ptenf/f;Pik3caE545K cells. B: Expression of the mutant E545K Pik3ca allele. The expression of the mutant allele was detected only in Pik3caE545K cells. GFP, green fluorescent protein; WT, wild type.